ABSTRACT
The hepatitis E virus (HEV) sheds into feces as nonenveloped virions but circulates in the blood in a membrane-associated, quasi-enveloped form (eHEV). Since the eHEV virions lack viral proteins on the surface, we investigated the entry mechanism for eHEV. We found that compared to nonenveloped HEV virions, eHEV attachment to the cell was much less efficient, requiring a longer inoculation time to reach its maximal infectivity. A survey of cellular internalization pathways identified clathrin-mediated endocytosis as the main route for eHEV entry. Unlike nonenveloped HEV virions, eHEV entry requires Rab5 and Rab7, small GTPases involved in endosomal trafficking, and blocking endosomal acidification abrogated eHEV infectivity. However, low pH alone was not sufficient for eHEV uncoating, suggesting that additional steps are required for entry. Supporting this concept, eHEV infectivity was substantially reduced in cells depleted of Niemann-Pick disease type C1, a lysosomal protein required for cholesterol extraction from lipid, or in cells treated with an inhibitor of lysosomal acid lipase. These data support a model in which the quasi-envelope is degraded within the lysosome prior to virus uncoating, a potentially novel mechanism for virus entry.
IMPORTANCE The recent discovery of quasi-enveloped viruses has shifted the paradigm of virus-host interactions. The impact of quasi-envelopment in the virus life cycle and pathogenesis is largely unknown. HEV is a highly relevant model to study these questions. HEV circulates as quasi-enveloped virions in the blood that are hidden from neutralizing antibodies. eHEV particles most likely are responsible for the cell-to-cell spread of the virus. Given the increasing concerns about persistent HEV infection and its potential for transmission via the blood supply, understanding how eHEV infects cells is important for understanding its pathogenesis and developing therapies. Our data provide evidence that eHEV uses a potentially novel mechanism for cellular entry. Several steps critical to eHEV entry were identified and may provide a basis for developing treatments for hepatitis E. Because quasi-enveloped viruses resemble exosomes, these data also may provide insights into the exosome-mediated intercellular communications.
INTRODUCTION
Hepatitis E virus (HEV) infection is a major cause of liver disease worldwide (1). HEV belongs to the Hepeviridae, a growing virus family consisting of members that infect a wide range of hosts, including mammals, chicken, and trout (2). Four genotypes have been associated with serious liver disease in humans. Infection with HEV genotypes (gt) 1 and 2 occurs mostly in developing countries where fecal-oral transmission results in epidemic spread via contaminated water (1, 3). HEV gt 3 and 4 are most common in developed countries and are transmitted to humans who ingest contaminated meat from domestic and wild animals (4, 5). Seroprevalence studies suggest these zoonotic infections are more common in the United States and parts of Europe than previously appreciated, although they appear to be self-limited and asymptomatic in most healthy adults (6). There is, however, an increasing concern with the persistence of HEV, particularly gt 3 viruses, in individuals with weakened immune systems, resulting in accelerated progression to cirrhosis (7, 8). In addition, extrahepatic manifestations have been reported, raising the possibility that infection involves multiple cell types (9). Because HEV gt 3 infection appears to be more common than expected and sometimes can persist, the possibility of transmission through the blood supply has been recognized recently (10).
The 7.2-kb positive-stranded RNA genome of HEV contains three open reading frames (ORFs) (11). ORF1 encodes a polyprotein (pORF1) that contains several putative functional domains and is involved in viral RNA replication. ORF2 encodes a 72-kDa protein that forms the capsid. ORF3 encodes a small multifunctional protein (12–15). Although originally recognized as a nonenveloped virus, recent studies show that HEV circulating in blood is cloaked in host membranes (16). The biogenesis of the membrane-associated, quasi-enveloped HEV particles (eHEV) involves a specific interaction between pORF3 and Tsg101, a cellular component of the endosomal sorting complex required for transport (ESCRT) that is commonly involved in the budding of many classic enveloped viruses (17–19). As a result, pORF3 is incorporated into the eHEV virion but is not found in virions in the feces (20). Neither polyclonal anti-pORF2 nor anti-pORF3 antibodies capture eHEV, except following detergent treatment, indicating that viral proteins are completely hidden within the eHEV membrane (16, 21). Despite lacking viral proteins on the surface, eHEV remains infectious and most likely is responsible for cell-to-cell spread within the host (21, 22). This dual life style is similar to that of hepatitis A virus (HAV), a phylogenetically unrelated hepatotropic virus that we recently discovered usurps the ESCRT components during its release (23). While in both cases cloaking of the virus capsids in host-derived membranes protects against antibody-mediated neutralization, how these virions mediate infection is unclear.
The entry process of HEV is poorly understood. HEV has been difficult to grow in culture, and replication usually takes weeks (24). The cellular receptor is not known. Previous studies using HEV-like particles (HEV-LPs) have shown that HEV-LPs attach to cells via heparan sulfate proteoglycans (HSPGs), after which HEV-LPs are internalized via a dynamin-2-, clathrin-, and membrane cholesterol-dependent pathway (25, 26). However, the entry mechanism of infectious HEV virions, particularly eHEV, is unclear.
In this study, we used a recently developed gt 3 HEV culture system to investigate the entry mechanisms of HEV. This cell culture-adapted HEV strain (Kernow C1/p6) replicates much faster than the parental wild-type virus (27), enabling dissection of the entry process of HEV. Moreover, a direct comparison between the two virion forms facilitated studies of the specific role of the quasi-envelope in virus entry. Our results indicate that nonenveloped HEV and quasi-enveloped HEV utilize different mechanisms for cell entry. Notably, the entry of eHEV appears to rely on the degradation of its membrane in the lysosome, suggesting a potentially novel mechanism for virus entry.
MATERIALS AND METHODS
Reagents and antibodies.
Chemical reagents were purchased from Sigma unless otherwise noted. The gt 3 Kernow-C1/p6 strain of HEV infectious clones was described previously (27). Lalistat 2 (an inhibitor of the lysosomal acid lipase) was a gift from Paul Helquist (University of Notre Dame). Rabbit anti-pORF2 antibody was a gift from X. J. Meng (Virginia Tech). Other antibodies obtained include the following: Rab5A and Rab7A (376801 and 6706-1; Epitomics), clathrin-1 heavy chain (ab21679; Abcam), NPC1 (ab134113; Abcam), dynamin-2 (A303-513A; Bethyl Laboratories), caveolin-1 (ab2910; Abcam), GFP (632375; Clontech), and β-actin (A2228; Sigma). Alexa Fluor-labeled reagents, including transferrin (T-13342) and cholera toxin B subunit (C-34777), were purchased from Invitrogen.
Cells and viruses.
Huh-7 cells and HepG2 cells (CRL-10741; ATCC) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin. To generate HepG2 cells stably expressing caveolin-1 (CAV1), the caveolin-1 fragment was amplified from pGex-Cav1 (33364; Addgene) and subcloned into pEGFP-N1 vector using HindIII and PstI restriction sites. The construct was transfected into HepG2 cells using a TransIT-X2 dynamic delivery system (Mirus Bio, LLC) and selected with 700 μg/ml G418. HEV stocks were generated by transfecting Huh-7 cells with in vitro-transcribed HEV Kernow C1/p6 RNA (27). The transfected cells were incubated at 34.5°C in DMEM with 10% FBS. Virus in pooled culture supernatant fluids collected between day 5 and 20 after transfection was concentrated by differential centrifugation (23). Transfected cells were lysed by freeze-thawing 3 times in distilled H2O, and a 1/10 volume of 10× phosphate-buffered saline (PBS) was added. Cell debris was removed by centrifugation at 10,000 × g for 30 min. Concentrated culture supernatant fluids and cell lysates were subjected to isopycnic iodixanol gradient centrifugation for virus purification. Vaccinia virus (MVA-T7 strain) stock was kindly provided by Bernard Moss and was grown in DF-1 cells.
Vaccinia virus infection.
HepG2 cells grown in 24-well plates were pretreated with inhibitors for 1 h prior to inoculation with vaccinia virus (MVA strain) for 2 h at 37°C in the presence of inhibitors. A 1:200 dilution of the stock was chosen because it resulted in replication with minimal cytopathic effects. Following inoculation, cells were washed three times with PBS and refed with DMEM supplemented with 2% FBS. Infection was allowed to proceed for 2 days at 37°C before the determination of viral RNA by quantitative reverse transcription-PCR (qRT-PCR).
Isopycnic gradient centrifugation.
Virus in culture supernatant fluids and cell lysates was concentrated by ultracentrifugation at 100,000 × g for 2 h at 4°C. Pellets were resuspended in PBS, loaded onto an 8 to 40% iodixanol (Opti-Prep) step gradient, and concentrated by ultracentrifugation at 120,000 × g in an SW55i rotor for 18 h at 4°C. Approximately 20 fractions were collected from the top of the gradient. The density of each fraction was determined with a Bausch & Lomb Abbé refractometer.
Real-time qRT-PCR.
Total RNA was isolated from cell lysates with the RNeasy kit (Qiagen) in accordance with the manufacturer's instructions. Viral RNA from culture supernatants and gradient fractions were extracted with the QIAamp viral RNA isolation kit (Qiagen). Real-time qRT-PCR was performed to quantify the HEV RNA using the iTaq universal probes one-step kit (Bio-Rad) using forward primer HEV-F (5′-GGTGGTTTCTGGGGTGAC-3′), reverse primer HEV-R (5′-AGGGGTTGGTTGGATGAA-3′), and probe HEV-P (5′-6-carboxyfluorescein-TGATTCTCAGCCCTTCGC-6-carboxytetramethylrhodamine-3′). A synthetic full-length HEV Kernow C1/p6 RNA was used as the standard. The expression levels of caveolin-1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and replication of vaccinia virus were determined by real-time qRT-PCR with an iTaq Universal SYBR green one-step kit (Bio-Rad) using caveolin-1-specific primers (HP205553; OriGene Technologies) and, for vaccinia virus, forward primer Vaccinia-F (5′-GCCAATGAGGGTTCGAGTTC-3′) and reverse primer Vaccinia-R (5′-CAACATCCCGTCGTTCATCA-3′). The primer sequences for vaccinia virus RNA measurement were adopted from a previous report (28). The mRNA levels of GAPDH were determined in the same samples for normalization using primers GAPDH-F (5′-CATGAGAAGTATGACAACAGCCT-3′) and GAPDH-R (5′-AGTCCTTCCACGATACCAAAGT-3′).
Virus infection.
HepG2 cells (4 × 104) were seeded onto eight-well Lab-Tek II CC2 slides (Nunc) a day before infection. Cells were inoculated with HEV or eHEV for 6 h at 34.5°C, washed three times with PBS, and refed with DMEM supplemented with 10% FBS and 2% dimethyl sulfoxide (DMSO). Cells were returned to 34.5°C and incubated for 5 days. For experiments involving inhibitors, cells were pretreated with inhibitors for 1 h at 34.5°C, followed by inoculation with HEV or eHEV for 6 h in the presence of the inhibitors unless otherwise stated. The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was used to measure the cytotoxicity of these inhibitors in HepG2 cells. Cells were treated with inhibitors at the indicated concentrations for 20 h and subjected to MTT assays by following the manufacturer's recommendations.
Immunoblotting.
Cells were lysed in 100 mM Tris-HCl (pH 7.5), 50 mM NaCl, 5 mM EDTA, and 1% Triton X-100 in the presence of protease inhibitor cocktail (Roche). SDS-polyacrylamide gel electrophoresis and immunoblotting were performed as described previously (23). Protein bands were detected with an Odyssey infrared imaging system (LI-COR Biosciences).
IFA and confocal microscopy.
For indirect immunofluorescence assay (IFA) and confocal microscopy, cells were washed once with PBS, fixed with 4% paraformaldehyde for 20 min, and permeabilized with 0.2% Triton X-100 for 15 min. After blocking in 10% goat serum diluted in 1% BSA solution for 1 h, cells were stained with rabbit anti-pORF2 antibody for 1 h and treated with goat-anti-rabbit IgG conjugated with Alexa Fluor 488 or 594 (Invitrogen) for 1 h. After adding antifade-4,6-diamidino-2-phenylindole (DAPI) mounting solution (Sigma), slides were viewed with a Zeiss Axiovert 25 compact fluorescent light (CFL) fluorescence microscope. In the confocal microscopy experiment, HepG2 cells were transfected with plasmids carrying mRFP-Rab5 (14437; Addgene) or DsRed-Rab7 WT (12661; Addgene). One day after transfection, cells were inoculated with HEV or eHEV (3 × 108 genome equivalents [GE]) for 1 h, processed as described above, and viewed with a Zeiss laser scanning microscope (LSM) 510 confocal microscope with a 63× (numeric aperture, 1.2) apochromatic water objective. Images were acquired using ZEN 2009 software.
Gene knockdown.
Cells were transfected with gene-specific SmartPool short interfering RNAs (siRNAs) (Dharmacon) with Lipofectamine RNAiMAX (Invitrogen). Knockdown efficiency was determined at 48 h by real-time qRT-PCR with a SYBR green one-step qRT-PCR kit (Bio-Rad) or by immunoblotting. Two days posttransfection, the cells were inoculated with HEV and the number of infected cells was determined by indirect IFA.
Statistical analysis.
Statistical analyses were performed using GraphPad Prism (GraphPad, San Diego, CA). Unpaired Student t tests were used to obtain P values between groups.
RESULTS
HEV released from infected cells is predominantly quasi-enveloped, but the virus within the cells is predominantly nonenveloped.
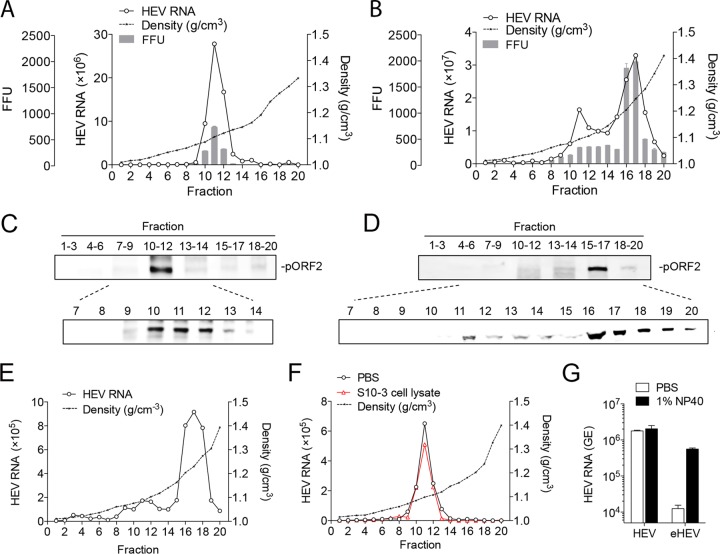
To generate stocks for quasi-enveloped and nonenveloped HEV, we characterized the virions released into the culture supernatant fluids as well as those within the cells. Virus was produced by transfecting Huh-7 cells (a human hepatoma cell line) with an in vitro-transcribed gt 3 HEV (Kernow-C1/p6) RNA (27). Supernatant fluids collected between day 3 and day 20 after transfection were pooled and concentrated by ultracentrifugation. Intracellular virions were released by freeze-thawing the cell pellets in a hypotonic buffer (27). Both extracellular and intracellular viruses were subjected to isopycnic iodixanol gradient centrifugation. Fecal material from a macaque experimentally infected with the Kernow C1 virus was included as a positive control for nonenveloped virions. HEV RNA and pORF2 (capsid protein) in gradient fractions were determined by quantitative RT-PCR and Western blot analysis, respectively.
The majority of the virus particles released into the culture supernatant were quasi-enveloped virions, with a peak density of 1.10 g/cm3 (Fig. 1A and C), as reported by other groups (21). In contrast, most of the virus particles in cell lysates banded at fractions 16 and 17, with a peak density of 1.25 g/cm3 (Fig. 1B and D), similar to virions in the feces (Fig. 1E). A minor portion of the intracellular virions banded at fractions 10 to 12, with a density similar to that found in the culture supernatant. The gradient profiles of virus RNA and protein correlated well with virus infectivity, confirming these measurements represent authentic virions (Fig. 1A and B, bars). To ensure that nonenveloped HEV was not a by-product of the freeze-thaw process, gradient-purified eHEV was incubated with uninfected Huh-7 cell lysates, followed by the freeze-thaw treatment using the same conditions. The freeze-thaw process did not alter the density of eHEV (Fig. 1F). To confirm eHEV was coated with lipid membrane, gradient-purified virions were subjected to immunoprecipitation with a polyclonal antibody against pORF2, either in the presence or absence of detergent. Bound virus particles were quantified by qRT-PCR. Precipitation of nonenveloped HEV did not change, but there was a substantial increase (>40-fold) in the binding of eHEV after 1% NP-40 treatment (Fig. 1G). Collectively, these results suggest that two populations of HEV particles are present in the Huh-7 cells, and only quasi-enveloped virions are released into the supernatant. Similar findings were described for PLC/PRF/5 cells (a human hepatoma cell line expressing hepatitis B virus surface antigen) in a recent report (19). In subsequent experiments, gradient-purified eHEV and nonenveloped HEV were used to study the HEV entry process. For simplicity, nonenveloped HEV is referred to as HEV.
FIG 1.
Characterization of extracellular and intracellular HEV virions. (A and B) Buoyant density of HEV virions released into supernatant fluids (A) or in cell lysates (B) of Huh-7 cells transfected with HEV RNA. HEV RNA in fractions was determined by qRT-PCR. HEV infectivity was determined by IFA in HepG2 cells. (C and D) Distribution of the HEV capsid protein pORF2 in gradient fractions of panels A and B. (E) Buoyant density of HEV in a fecal sample of a rhesus macaque experimentally infected with HEV. (F) Buoyant density of eHEV after incubation with either PBS or uninfected Huh-7 cell lysates. (G) HEV and eHEV were either treated with 1% NP-40 or left untreated and immunoprecipitated with rabbit anti-pORF2 antibody. Bound virus was quantified by HEV-specific qRT-PCR. The results show the means ± SEM from 2 independent experiments. FFU, focus-forming units.
eHEV infects cells at a lower rate than HEV due to inefficient cell attachment.
To assess the infectivity of HEV and eHEV, we used HepG2 cells, which are more permissive for HEV infection (27). In the first experiment, HepG2 cells grown on 8-well chamber slides were inoculated with equal genome equivalents (GE) of HEV and eHEV (∼3 × 107 GE/well) for 6 h. The number of infected cells was determined 5 days later by IFA using a polyclonal antibody against pORF2. HEV spread poorly in cell culture, and most foci consisted of only one or a few cells (Fig. 2A). About 2,000 foci were detected in wells inoculated with HEV. However, only ∼750 foci were detected in wells inoculated with eHEV (Fig. 2B). Therefore, the specific infectivity (GE/FFU) of HEV is 2.7-fold higher than that of eHEV. Consistent with this, culture supernatant fluids from HEV-inoculated wells also contained 2-fold more virus genomes than wells inoculated with eHEV (Fig. 2C). This difference was observed in multiple independent experiments. We reasoned that the relatively low infectivity of eHEV is due to the lack of viral proteins on the surface, resulting in inefficient cell attachment. To test this hypothesis, prechilled HepG2 cells were inoculated with equal amounts of HEV and eHEV (3 × 107 GE/well) at 4°C for 1 h. After washing cells with cold PBS to remove the unbound virus, total RNA was extracted from the cells. The number of virus particles bound to the cell surface was determined by qRT-PCR. The binding efficiency of eHEV was 10-fold lower than that of HEV (Fig. 2D), suggesting a lack of a specific interaction between the virus and the cell. Heparan sulfate proteoglycan (HSPG) has been shown to be involved in cell attachment of HEV virus-like particles (25). In agreement with this, preincubation of HEV with increasing amounts of heparan sulfate resulted in a dose-dependent reduction in virus infectivity. However, eHEV infectivity was not significantly inhibited (Fig. 2E), indicating that in contrast to the HEV virion, eHEV attachment to the cells does not require HSPG.
FIG 2.
Cell attachment and entry kinetics of HEV and eHEV. (A) Immunofluorescence images showing the expression of the HEV pORF2 protein in HepG2 cells 5 days after inoculation with equal amounts of HEV and eHEV (3 × 107 HEV GE). (B) Virus yields in wells inoculated with HEV and eHEV for 5 days. Shown are the means ± SEM from 2 independent experiments each performed in duplicate. *, P < 0.05; **, P < 0.01. (C) HEV RNA in supernatants of HEV-infected HepG2 culture. HEV RNA was detected by qRT-PCR. The results show the means ± SEM from 2 independent experiments performed in duplicate. *, P < 0.05; **, P < 0.01. (D) HepG2 cells were prechilled on ice for 15 min prior to inoculation with equal amounts of HEV and eHEV (3 × 107 GE) at 4°C for 1 h. Cells were washed with cold PBS three times. Total RNA was extracted and subjected to HEV-specific qRT-PCR analysis. The results show means ± SEM from 2 independent experiments performed in duplicate. *, P < 0.05; **, P < 0.01. (E) HEV and eHEV (3 × 107 GE) were incubated with heparan sulfate (HS) at the indicated concentrations for 1 h prior to inoculation. Virus yields were determined by IFA after 5 days. The results show the means ± SEM from 2 independent experiments performed in duplicate. *, P < 0.05; **, P < 0.01. (F) HepG2 cells were inoculated with HEV and eHEV (3 × 107 GE). The inoculum was removed at different times. Cells were washed extensively with PBS, and fresh medium was added. Virus yields were determined by IFA after 5 days. Shown are representative results from two independent experiments.
The >10-fold lower cell-binding efficiency of eHEV at 4°C prompted us to compare the kinetics of HEV and eHEV entry at a physiological temperature. To this end, HepG2 cells were inoculated with equal amounts of HEV or eHEV (∼1,000 GE/cell) at 34.5°C, a standard temperature to grow HEV in culture (27, 29). The inoculum was removed at various times, and the cells were washed extensively before adding fresh medium. Virus yields were determined 5 days after inoculation by IFA. HEV and eHEV exhibited different binding kinetics (Fig. 2F). While the binding of HEV exhibited a rapid increase within 1 h, reaching 90% of its maximum by 3 h after inoculation, the binding of eHEV exhibited a slow and more linear increase, reaching 90% of its maximum by 6 h. Finally, the maximum infectivity of eHEV was half that of HEV, even with an extended inoculation period (up to 12 h). In conclusion, these data indicate that the envelopment of HEV reduces its ability to attach to the cell, resulting in a slower entry kinetic.
Entry of eHEV is dependent on clathrin-mediated endocytosis but not on caveola-dependent endocytosis or macropinocytosis.
Previous studies indicated that the internalization of Escherichia coli-expressed HEV-like particles (HEV-LPs) is dependent on dynamin-2 and the clathrin-mediated pathway (26). To identify the pathways involved in the entry of infectious HEV and eHEV, we used small-molecule inhibitors that are commonly used to target dynamin-2 (dynasore), clathrin-mediated endocytosis (chlorpromazine or CPZ), caveola-mediated endocytosis (filipin), or macropinocytosis (LY294002 and EIPA) (30). The effects of these entry inhibitors were confirmed with respective positive controls (transferrin for chlorpromazine and dynasore, cholera toxin B for filipin, and vaccinia virus for EIPA and LY294002 [Fig. 3D to F]).
FIG 3.
Role of clathrin-mediated endocytosis, caveola-mediated endocytosis, and macropinocytosis in HEV and eHEV entry. (A and B) HepG2 cells were pretreated with the indicated inhibitors for 1 h prior to inoculation with HEV or eHEV. Inoculum was removed after 6 h, and culture was continued for 5 days. Virus yields were determined by IFA after 5 days. Data are represented as a percentage of the DMSO-treated control. The results show the means ± SEM from at least 2 independent experiments in duplicate. *, P < 0.05; **, P < 0.01. (C) HepG2 cells were treated with inhibitors at the indicated concentrations for 20 h, and cell viability was measured by an MTT assay. The results show the mean ± SEM results from triplicate wells. (D) HepG2 cells were treated with CPZ (10 μg/ml) or Dynasore (80 μM) for 1 h prior to incubation with Alexa-488-labeled transferrin (25 μg/ml). After 15 min of incubation, cells were fixed and analyzed by confocal microscopy. Scale bar, 10 μm. (E) HepG2 cells transfected with plasmids carrying GFP or CAV1-GFP for 48 h were treated with filipin (3 μg/ml) for 1 h and incubated with Alexa-594-labeled cholera toxin subunit B (CTxB; 20 μg/ml). After 1 h of incubation, cells were fixed and analyzed by confocal microscopy. Scale bar, 10 μm. (F) HepG2 cells were pretreated with EIPA or LY294002 for 1 h prior to inoculation with vaccinia virus (MVA-T7 strain) for 1 h. mRNA levels of the vaccinia DNA-directed RNA polymerase were determined 12 h after inoculation by qRT-PCR. Data are represented as a percentage of the DMSO-treated control. The data represent the means ± SEM from 2 independent experiments, each in duplicate. *, P < 0.05; **, P < 0.01. (G and H) HepG2 cells were transfected with nontargeting siRNA (siCtrl) or siRNA targeting the clathrin-1 heavy chain (CLTC) (G) or dynamin-2 (DNM2) (H). Thirty-six hours posttransfection, cells were inoculated with HEV or eHEV. (Upper) The knockdown efficiency was determined by immunoblotting. Virus yields were determined by IFA 5 days after inoculation. (Lower) Relative infection (percentage of control siRNA-transfected culture) was calculated based on FFU. Shown are the means ± SEM from 3 independent experiments, each in duplicate. *, P < 0.05; **, P < 0.01. (I, upper) Western blot showing the expression of GFP and CAV1-GFP in HepG2 cells transfected with the corresponding plasmids. Unmodified HepG2 cells were included as a control. Cells stably expressing GFP or CAV1-GFP were inoculated with HEV or eHEV in the presence or absence of filipin (in the case of cells expressing CAV1-GFP). (Lower) Virus yields were determined by IFA 5 days after inoculation. Shown are representative results from two independent experiments.
HepG2 cells were pretreated with these inhibitors for 1 h, followed by virus inoculation in the presence of inhibitors for 6 h. Virus yields were determined at 5 days after inoculation. Pretreatment with chlorpromazine or dynasore inhibited the entry of both HEV and eHEV (Fig. 3A and B). However, the extent of inhibition was greater on eHEV. MTT assays did not reveal obvious cytotoxicity at the concentrations tested (Fig. 3C). Consistent with these results, depletion of the clathrin-1 heavy chain (CTLC) or dynamin-2 (DNM2) by siRNA also had a greater inhibitory effect on eHEV infectivity (Fig. 3G and H). To test the role of caveola-mediated endocytosis on the entry of HEV and eHEV, filipin and siRNA targeting caveolin-1 (CAV1) were used. We were unable to detect caveolin-1 protein expression in HepG2 cells, consistent with a previous report showing that hepatoma cells, including HepG2 cells, express low levels of caveolin-1 (26). Transfection of caveolin-1-specific siRNA significantly reduced the caveolin-1 mRNA level. However, the infectivities of HEV and eHEV were not reduced (data not shown). To corroborate this result, we created HepG2 cell lines stably expressing GFP or CAV1-GFP. The expression and function of ectopically expressed caveolin-1 was confirmed by Western blotting and confocal microscopy, demonstrating the colocalization of CAV1-GFP and Alexa-594-labbled cholera toxin subunit B (CTxB) (a positive control for caveola-dependent endocytosis). Moreover, CTxB uptake by the HepG2/CAV1-GFP cells was effectively blocked by filipin (Fig. 3E). However, the infectivity of HEV or eHEV was not enhanced in the HepG2/CAV1-GFP cells compared to that of the control HepG2/GFP cells (Fig. 3I). Importantly, filipin did not inhibit virus infectivity in the HepG2/CAV1-GFP cells (Fig. 3I). These results indicate that caveola-mediated endocytosis does not play a significant role in HEV and eHEV entry. Finally, EIPA (a Na+/H+ exchanger inhibitor) and LY294002 (a phosphatidylinositol 3-kinase inhibitor) were used to investigate the role of macropinocytosis in HEV and eHEV entry. The treatment of cells with these inhibitors effectively reduced vaccinia virus entry (Fig. 3F). However, neither reduced HEV or eHEV infectivity (Fig. 3A and B), suggesting an insignificant role for macropinocytosis in HEV and eHEV entry.
eHEV entry is dependent on Rab5 and Rab7.
The above-described analyses suggested that both HEV and eHEV are internalized after initial cell attachment. Since the small GTPases Rab5 and Rab7 are commonly involved in virus trafficking through the early endosome (Rab5) and the late endosome (Rab7), we investigated whether HEV and eHEV traffic via these compartments. While no significant colocalization of HEV with Rab5 and Rab7 was detected, colocalization of eHEV with Rab5 and Rab7 was readily detectable 1 h after virus inoculation (Fig. 4A). The virions detected at 1 h reflected the input virus, since the newly synthesized HEV antigens usually are not detectable until 48 h, and similar localization patterns were observed with UV-inactivated virus (data not shown).
FIG 4.
Role of Rab5 and Rab7 during HEV and eHEV entry. (A) Confocal images showing the localization of the HEV capsid protein pORF2 (green) in HepG2 cells previously transfected with mRFP-Rab5 or DsRed-Rab7 (red). Cells were inoculated with HEV or eHEV (1 × 104 GE per cell) for 1 h and processed for confocal imaging analysis using rabbit anti-pORF2 antibody. Cells were counterstained with DAPI (blue). Scale bar, 10 μm. (B and C) HepG2 cells were transfected with control siRNA or siRNA targeting Rab5A or Rab7A. Thirty-six hours posttransfection, cells were inoculated with HEV or eHEV. (B) Immunoblots showing the knockdown efficiency 2 days after transfection. (C) Virus yields determined by IFA 5 days after inoculation. Data are represented as a percentage of the nontargeting siRNA (siCtrl)-treated control. The results represent the means ± SEM from 2 independent experiments, each in duplicate. *, P < 0.05; **, P < 0.01.
To further define the role of Rab5 and Rab7 in HEV and eHEV entry, HepG2 cells were transfected with siRNA targeting these genes. Two days later, cells were inoculated with HEV and eHEV. At this time, siRNA-transfected cells had significantly reduced levels of the targeted proteins (Fig. 4B). On day 5 postinoculation, virus yields were determined by IFA. Depletion of Rab5 or Rab7 resulted in a 20% or 70% reduction in eHEV infectivity, respectively. In contrast, HEV infectivity was not significantly reduced in cells depleted of Rab5 or Rab7 (Fig. 4C).
eHEV entry requires endosomal acidification.
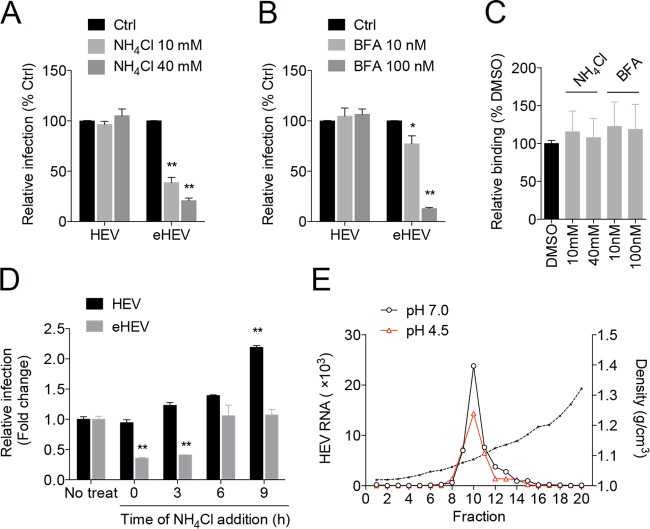
One of the major changes along the endocytic pathway is the gradual reduction in pH, an essential requirement for the entry of certain viruses (e.g., influenza virus) (30). To test whether low pH is required for eHEV entry, cells were pretreated with NH4Cl (which neutralizes endosomal pH) and bafilomycin A1 (which inhibits endosomal H+ ATPases) followed by inoculation with HEV or eHEV. While NH4Cl and bafilomycin A1 did not reduce HEV infectivity, these treatments dramatically reduced eHEV infectivity (Fig. 5A and B). Similar results were obtained with chloroquine, a weak base that also neutralizes pH in the lysosomes (not shown). Treatment with NH4Cl or bafilomycin A1 did not affect eHEV cell binding (Fig. 5C), suggesting that eHEV entry requires endosomal acidification.
FIG 5.
Effect of lysosomotropic agents on HEV and eHEV entry. (A and B) HepG2 cells were pretreated with NH4Cl (A) or bafilomycin A1 (BFA) (B) for 1 h prior to inoculation with HEV or eHEV. Virus yields were determined 5 days after inoculation. Data are represented as a percentage relative to the nontreated control (Ctrl). The results represent the means ± SEM from 3 independent experiments, each in duplicate. *, P < 0.05; **, P < 0.01. (C) HepG2 cells were inoculated with eHEV (3 × 107 GE) in the presence or absence of NH4Cl or bafilomycin A1 (BFA) at 4°C for 1 h. Cells were washed with cold PBS three times. Total RNA was extracted and subjected to HEV-specific qRT-PCR analysis. Data represent the means ± SEM from 2 independent experiments, each in duplicate. (D) HepG2 cells were inoculated with HEV or eHEV. At different times postinoculation, NH4Cl was added and infection was continued in the presence of the drug. Virus yields were determined as indicated above. Data are represented as fold changes relative to the nontreated control. Data represent the means ± SEM from 2 independent experiments, each in duplicate. (E) Buoyant density of eHEV after incubation with solution of low (pH 4.5) or neutral (pH 7.0) pH for 30 min. HEV RNA in gradient fractions was measured by qRT-PCR.
We reasoned that lysosomotropic agents might be used to probe the timing of eHEV uncoating. Since uncoated eHEV should not be affected by these agents, NH4Cl was added to the culture medium at different times after virus inoculation. As expected, the addition of NH4Cl at 0 h (same time as virus inoculation) resulted in a significant reduction in eHEV infectivity. However, when NH4Cl was added 6 h or later after inoculation, eHEV infectivity was no longer impaired. This result suggests that eHEV uncoating is completed by 6 h after inoculation. However, treatment with NH4Cl did not reduce HEV infectivity (Fig. 5D). The slight increase at later times possibly is due to the ability of NH4Cl to enhance HEV release (31).
To determine whether the low pH affects eHEV membrane integrity, eHEV was incubated in an acid solution (pH 4.5) or a solution with neutral pH (pH 7.0) for 30 min and subjected to isopycnic gradient centrifugation. The density of eHEV between the two treatments did not differ (Fig. 5E), indicating that the lipid membrane of eHEV was not removed by the lower pH. Furthermore, the acidic treatment had no impact on the infectivity of eHEV (not shown). These data suggest that low pH in the endosome alone is insufficient for removing the eHEV membrane.
eHEV entry is dependent on lysosomal lipid degradation.
The above-described results suggest that a low-pH-dependent event in the endosome is required for eHEV entry. We postulate that this event is the degradation of the eHEV membrane, a step that would allow the viral capsid to interact with its receptor on the inner leaflet of the endolysosome and subsequently uncoat. Lipid degradation in lysosomes is a complex process (32). Among the initial steps, Niemann-Pick disease, type C1 (NPC1)-mediated extraction of cholesterol is a critical step for membrane degradation (32). To determine whether NPC1 has a role in eHEV entry, cells were transfected with siRNA targeting NPC1 and inoculated with HEV or eHEV. The depletion of NPC1 reduced infection by eHEV by 50% without significantly affecting HEV infectivity (Fig. 6A and B).
FIG 6.
Role of lysosomal lipid degradation in HEV and eHEV entry. (A and B) HepG2 cells were transfected with control siRNA or siRNA targeting NPC1. Thirty-six hours after transfection, cells were inoculated with HEV or eHEV. (A) Immunoblots showing the knockdown efficiency of NPC1. (B) Virus yields (FFU) 5 days after inoculation. Data are represented as a percentage relative to a nontargeting siRNA-treated control (siCtrl). Shown are the means ± SEM from 2 independent experiments, each in duplicate. *, P < 0.05; **, P < 0.01. (C) HepG2 cells were pretreated with Lalistat 2 at the indicated concentrations for 1 h prior to virus inoculation. Data are represented as a percentage of the DMSO-treated control. The result represents the means ± SEM from 2 independent experiments, each in duplicate. *, P < 0.05; **, P < 0.01.
The lysosomal acid lipase (LAL) has a key role in lipid metabolism through the hydrolysis of cholesteryl esters and triglycerides in lysosomes (33). We reasoned that the LAL might be important for degradation of the eHEV membrane within the lysosome. To test this hypothesis, we pretreated HepG2 cells with Lalistat 2, a specific inhibitor of LAL (34), prior to virus inoculation. Treatment with Lalistat 2 resulted in a dose-dependent reduction in eHEV infectivity but not in HEV infectivity.
DISCUSSION
There is a gap in our knowledge of how quasi-enveloped virions differ from their nonenveloped counterparts in cell entry. By analyzing sequential steps in the entry process of nonenveloped and quasi-enveloped HEV in parallel, our data have revealed several important distinctions between the entry mechanisms for the two forms of the virion. Compared with HEV virions, eHEV attachment to cells was much less efficient, resulting in a slow and more linear entry kinetic. Unlike HEV, eHEV entry required Rab5- and Rab7-dependent endosomal trafficking and was critically dependent on endosomal acidification. Moreover, entry of eHEV was substantially reduced when lysosomal lipid degradation was blocked. These results support a model in which the quasi-envelope is degraded in the endolysosome prior to virus uncoating, a potentially novel mechanism for virus entry.
A key difference between the entry of HEV and eHEV is that the initial cell attachment of eHEV is much less efficient (∼10-fold less) than that of the nonenveloped virions. The low efficiency of eHEV in cell attachment likely is due to the lack of viral glycoproteins on the eHEV surface. Kinetic analysis revealed that cell attachment of eHEV reached a plateau by 6 h, correlating with the time when the majority of virus particles likely uncoated (Fig. 5C). These results suggest that the initial cell attachment is a major limiting factor during eHEV entry. This difference in cell attachment between eHEV and HEV may provide an explanation for the lower infectivity of patient's serum-derived virus than virus derived from patient's feces reported in a recent study (35). While soluble heparan sulfate blocked HEV entry, we observed no effect on eHEV entry. The molecules on the eHEV surface that mediate its cell attachment remain to be determined. The receptor for HEV is unknown but is unlikely to have a role in eHEV cell attachment. The biogenesis of eHEV is similar to that of exosomes, cell-derived membrane vesicles that mediate intercellular communication (36, 37), suggesting commonalities exist in cell attachment and uptake mechanisms. Previous work has implicated the roles of phosphatidylserine, tetraspanins, and integrins in cellular uptake of exosomes (38–41). Therefore, the roles of these molecules in eHEV entry are of further interest.
Despite inefficient cell attachment, the infectivity of eHEV reached half of the maximum infectivity of HEV given sufficient time (≥6 h) (Fig. 2F). In addition, copackaging of multiple capsids into single vesicles may lower the numbers of foci associated with eHEV, as seen in the case of poliovirus (42). Hence, the quasi-envelopment of HEV does not appear to substantially hamper the virus infectivity under culture conditions. However, serum-derived HEV was much less infectious than fecal HEV when tested in vivo (35). The reason for the difference between the in vitro and the in vivo findings is unclear, but it is possible that the movement of the blood reduces the contact time and therefore the efficiency of cell attachment of eHEV.
Our study provides insights into how quasi-enveloped virus particles penetrate the cell membrane. The entry of eHEV requires clathrin-mediated internalization and endosomal acidification, indicating that eHEV does not penetrate at the plasma membrane. An important question is whether eHEV entry requires membrane fusion within internal vesicles. Unlike classic enveloped viruses, eHEV does not encode surface glycoproteins to mediate membrane fusion. Therefore, the eHEV membrane either carries host-derived fusion proteins or is removed during the entry process. Experimental evidence from this work supports the latter. Depletion of the lysosomal cholesterol transporter NPC1 reduced eHEV infectivity, suggesting that eHEV entry involves NPC1-mediated extraction of cholesterol from the eHEV membrane. NPC1 is an unlikely receptor for eHEV, as in the case for ebolavirus (43), since the depletion of NPC1 had no effect on HEV entry. Furthermore, blocking the lysosomal acid lipase also reduced eHEV infectivity, providing additional evidence that the eHEV membrane is degraded in lysosomes. Since genetic defects in NPC1 and LAL often are associated with lysosomal storage disorders (44, 45), patients with these diseases could demonstrate a natural resistance to HEV infection.
Finally, our results have further advanced the elucidation of the entry mechanism for HEV. While eHEV most likely is responsible for cell-to-cell spread within the host and blood-borne transmission between hosts, HEV most likely mediates fecal-oral transmission between hosts (22). We found that inhibiting dynamin-2- and clathrin-dependent endocytosis also reduced HEV entry, but the effect was less dramatic than that of eHEV. Importantly, the entry of HEV was unaffected by treatments that inhibited eHEV entry (Fig. 3 to 6). Since HEV enters cells independently of Rab5/7 and endosomal acidification, the mechanism of cell penetration may occur at or near the plasma membrane, as reported for poliovirus (46). Although HEV virions were found in the perinuclear region after inoculation (Fig. 4), these virions may not represent infectious virions, given that the particle-to-FFU ratio for HEV is very high (∼15,000:1) (27). The observation that HEV virions did not colocalize with Rab5 or Rab7 suggests that these virions do not follow a classic endocytic pathway during entry.
In summary, we have shown that nonenveloped and quasi-enveloped HEV use distinct mechanisms for cell entry. eHEV efficiently infects cells without virally encoded glycoproteins. Our data demonstrate that eHEV entry requires virus internalization and trafficking to an acidic compartment within the lysosome to degrade the viral quasi-envelope prior to penetration, a potentially novel mechanism for virus entry. Overall, our data provide a framework for future studies of the HEV entry process. Understanding the entry mechanism of eHEV is important to interpret the pathogenesis and to develop therapies for HEV-associated diseases. This knowledge also may be applicable to other quasi-enveloped viruses, such as quasi-enveloped hepatitis A virus (eHAV) and certain members of enteroviruses (23, 42).
ACKNOWLEDGMENTS
This work was supported by the Pinnacle Research Award in Liver Diseases from the American Association for the Study of Liver Diseases Foundation, the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R21AI122228), and internal startup funds from the Nationwide Children's Hospital (Z.F.).
We are grateful to Suzanne Emerson for providing the HEV Kernow C1/p6 infectious clone, X. J. Meng for providing the rabbit anti-ORF2 antibody, Christopher Walker for providing monkey fecal samples, Paul Helquist for providing the LAL inhibitor Lalistat 2, Bernard Moss for providing the vaccinia virus stock, and Jacqueline Corry for helping with the preparation of the vaccinia virus stock. We thank Mark Peeples, Christopher Walker, and Kevin Cassady for helpful discussions and readings of the manuscript.
Funding Statement
Z.F. is a recipient of the 2015 Pinnacle Research Award in Liver Diseases.
REFERENCES
- 1.Purcell RH, Emerson SU. 2008. Hepatitis E: an emerging awareness of an old disease. J Hepatol 48:494–503. doi: 10.1016/j.jhep.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 2.Smith DB, Simmonds P, Jameel S, Emerson SU, Harrison TJ, Meng XJ, Okamoto H, Van der Poel WH, Purdy MA. 2014. Consensus proposals for classification of the family Hepeviridae. J Gen Virol 95:2223–2232. doi: 10.1099/vir.0.068429-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalton HR, Bendall R, Ijaz S, Banks M. 2008. Hepatitis E: an emerging infection in developed countries. Lancet Infect Dis 8:698–709. doi: 10.1016/S1473-3099(08)70255-X. [DOI] [PubMed] [Google Scholar]
- 4.Pavio N, Meng XJ, Doceul V. 2015. Zoonotic origin of hepatitis E. Curr Opin Virol 10:34–41. doi: 10.1016/j.coviro.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 5.Meng XJ. 2013. Zoonotic and foodborne transmission of hepatitis E virus. Semin Liver Dis 33:41–49. doi: 10.1055/s-0033-1338113. [DOI] [PubMed] [Google Scholar]
- 6.Kuniholm MH, Engle RE, Purcell RH, Nelson KE. 2014. Hepatitis E virus seroprevalence in the United States: no easy answers. Hepatology 61:1441–1442. doi: 10.1016/j.jhep.2014.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalton HR, Bendall RP, Keane FE, Tedder RS, Ijaz S. 2009. Persistent carriage of hepatitis E virus in patients with HIV infection. N Engl J Med 361:1025–1027. doi: 10.1056/NEJMc0903778. [DOI] [PubMed] [Google Scholar]
- 8.Kamar N, Selves J, Mansuy JM, Ouezzani L, Peron JM, Guitard J, Cointault O, Esposito L, Abravanel F, Danjoux M, Durand D, Vinel JP, Izopet J, Rostaing L. 2008. Hepatitis E virus and chronic hepatitis in organ-transplant recipients. N Engl J Med 358:811–817. doi: 10.1056/NEJMoa0706992. [DOI] [PubMed] [Google Scholar]
- 9.Woolson KL, Forbes A, Vine L, Beynon L, McElhinney L, Panayi V, Hunter JG, Madden RG, Glasgow T, Kotecha A, Dalton HC, Mihailescu L, Warshow U, Hussaini HS, Palmer J, McLean BN, Haywood B, Bendall RP, Dalton HR. 2014. Extra-hepatic manifestations of autochthonous hepatitis E infection. Aliment Pharmacol Ther 40:1282–1291. doi: 10.1111/apt.12986. [DOI] [PubMed] [Google Scholar]
- 10.Gallian P, Lhomme S, Piquet Y, Sauné K, Abravanel F, Assal A, Tiberghien P, Izopet J. 2014. Hepatitis E virus infections in blood donors, France. Emerg Infect Dis 20:1914–1917. doi: 10.3201/eid2011.140516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emerson SU, Purcell RH. 2003. Hepatitis E virus. Rev Med Virol 13:145–154. doi: 10.1002/rmv.384. [DOI] [PubMed] [Google Scholar]
- 12.Xu J, Wu F, Tian D, Wang J, Zheng Z, Xia N. 2014. Open reading frame 3 of genotype 1 hepatitis E virus inhibits nuclear factor-κappa B signaling induced by tumor necrosis factor-α in human A549 lung epithelial cells. PLoS One 9:e100787. doi: 10.1371/journal.pone.0100787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geng Y, Yang J, Huang W, Harrison TJ, Zhou Y, Wen Z, Wang Y. 2013. Virus host protein interaction network analysis reveals that the HEV ORF3 protein may interrupt the blood coagulation process. PLoS One 8:e56320. doi: 10.1371/journal.pone.0056320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandra V, Holla P, Ghosh D, Chakrabarti D, Padigaru M, Jameel S. 2011. The hepatitis E virus ORF3 protein regulates the expression of liver-specific genes by modulating localization of hepatocyte nuclear factor 4. PLoS One 6:e22412. doi: 10.1371/journal.pone.0022412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagashima S, Takahashi M, Jirintai Tanaka T, Yamada K, Nishizawa T, Okamoto H. 2011. A PSAP motif in the ORF3 protein of hepatitis E virus is necessary for virion release from infected cells. J Gen Virol 92:269–278. doi: 10.1099/vir.0.025791-0. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi M, Yamada K, Hoshino Y, Takahashi H, Ichiyama K, Tanaka T, Okamoto H. 2008. Monoclonal antibodies raised against the ORF3 protein of hepatitis E virus (HEV) can capture HEV particles in culture supernatant and serum but not those in feces. Arch Virol 153:1703–1713. doi: 10.1007/s00705-008-0179-6. [DOI] [PubMed] [Google Scholar]
- 17.Hurley JH. 2010. The ESCRT complexes. Crit Rev Biochem Mol Biol 45:463–487. doi: 10.3109/10409238.2010.502516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kenney SP, Wentworth JL, Heffron CL, Meng XJ. 2015. Replacement of the hepatitis E virus ORF3 protein PxxP motif with heterologous late domain motifs affects virus release via interaction with TSG101. Virology 486:198–208. doi: 10.1016/j.virol.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagashima S, Takahashi M, Jirintai S, Tanggis Kobayashi T, Nishizawa T, Okamoto H. 2014. The membrane on the surface of hepatitis E virus particles is derived from the intracellular membrane and contains trans-Golgi network protein 2. Arch Virol 159:979–991. doi: 10.1007/s00705-013-1912-3. [DOI] [PubMed] [Google Scholar]
- 20.Nagashima S, Takahashi M, Jirintai S, Tanaka T, Nishizawa T, Yasuda J, Okamoto H. 2011. Tumour susceptibility gene 101 and the vacuolar protein sorting pathway are required for the release of hepatitis E virions. J Gen Virol 92:2838–2848. doi: 10.1099/vir.0.035378-0. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi M, Tanaka T, Takahashi H, Hoshino Y, Nagashima S, Jirintai Mizuo H, Yazaki Y, Takagi T, Azuma M, Kusano E, Isoda N, Sugano K, Okamoto H. 2010. Hepatitis E virus (HEV) strains in serum samples can replicate efficiently in cultured cells despite the coexistence of HEV antibodies: characterization of HEV virions in blood circulation. J Clin Microbiol 48:1112–1125. doi: 10.1128/JCM.02002-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng Z, Lemon SM. 2014. Peek-a-boo: membrane hijacking and the pathogenesis of viral hepatitis. Trends Microbiol 22:59–64. doi: 10.1016/j.tim.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong SH, Walker C, Lanford RE, Lemon SM. 2013. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496:367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okamoto H. 2013. Culture systems for hepatitis E virus. J Gastroenterol 48:147–158. doi: 10.1007/s00535-012-0682-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalia M, Chandra V, Rahman SA, Sehgal D, Jameel S. 2009. Heparan sulfate proteoglycans are required for cellular binding of the hepatitis E virus ORF2 capsid protein and for viral infection. J Virol 83:12714–12724. doi: 10.1128/JVI.00717-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holla P, Ahmad I, Ahmed Z, Jameel S. 2015. Hepatitis E virus enters liver cells through a dynamin-2, clathrin and membrane cholesterol-dependent pathway. Traffic 16:398–416. doi: 10.1111/tra.12260. [DOI] [PubMed] [Google Scholar]
- 27.Shukla P, Nguyen HT, Torian U, Engle RE, Faulk K, Dalton HR, Bendall RP, Keane FE, Purcell RH, Emerson SU. 2011. Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus-host recombinant. Proc Natl Acad Sci U S A 108:2438–2443. doi: 10.1073/pnas.1018878108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howell MD, Jones JF, Kisich KO, Streib JE, Gallo RL, Leung DY. 2004. Selective killing of vaccinia virus by LL-37: implications for eczema vaccinatum. J Immunol (Baltimore) 172:1763–1767. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka T, Takahashi M, Kusano E, Okamoto H. 2007. Development and evaluation of an efficient cell-culture system for Hepatitis E virus. J Gen Virol 88:903–911. doi: 10.1099/vir.0.82535-0. [DOI] [PubMed] [Google Scholar]
- 30.Helenius A, Moss B. 2013. Virus entry–an unwilling collaboration by the cell. Curr Opin Virol 3:1–2. doi: 10.1016/j.coviro.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagashima S, Jirintai S, Takahashi M, Kobayashi T, Tanggis Nishizawa T, Kouki T, Yashiro T, Okamoto H. 2014. Hepatitis E virus egress depends on the exosomal pathway, with secretory exosomes derived from multivesicular bodies. J Gen Virol 95:2166–2175. doi: 10.1099/vir.0.066910-0. [DOI] [PubMed] [Google Scholar]
- 32.Kolter T, Sandhoff K. 2010. Lysosomal degradation of membrane lipids. FEBS Lett 584:1700–1712. doi: 10.1016/j.febslet.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 33.Dubland JA, Francis GA. 2015. Lysosomal acid lipase: at the crossroads of normal and atherogenic cholesterol metabolism. Front Cell Dev Biol 3:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenbaum AI, Cosner CC, Mariani CJ, Maxfield FR, Wiest O, Helquist P. 2010. Thiadiazole carbamates: potent inhibitors of lysosomal acid lipase and potential niemann-pick type C disease therapeutics. J Med Chem 53:5281–5289. doi: 10.1021/jm100499s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allweiss L, Gass S, Giersch K, Groth A, Kah J, Volz T, Rapp G, Schöbel A, Lohse AW, Polywka S, Pischke S, Herker E, Dandri M, Lütgehetmann M. 21 January 2016. Human liver chimeric mice as a new model of chronic hepatitis E virus infection and preclinical drug evaluation. J Hepatol doi: 10.1016/j.jhep.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 36.Mathivanan S, Ji H, Simpson RJ. 2010. Exosomes: extracellular organelles important in intercellular communication. J Proteomics 73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 37.Colombo M, Raposo G, Théry C. 2014. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 38.Hoshino A, Costa-Silva B, Shen T-L, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, Singh S, Williams C, Soplop N, Uryu K, Pharmer L, King T, Bojmar L, Davies AE, Ararso Y, Zhang T, Zhang H, Hernandez J, Weiss JM, Dumont-Cole VD, Kramer K, Wexler LH, Narendran A, Schwartz GK, Healey JH, Sandstrom P, Jørgen Labori K, Kure EH, Grandgenett PM, Hollingsworth MA, de Sousa M, Kaur S, Jain M, Mallya K, Batra SK, Jarnagin WR, Brady MS, Fodstad O, Muller V, Pantel K, Minn AJ, Bissell MJ, Garcia BA, Kang Y, Rajasekhar VK, Ghajar CM, Matei I, Peinado H, Bromberg J, Lyden D. 2015. Tumour exosome integrins determine organotropic metastasis. Nature 527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rana S, Zöller M. 2011. Exosome target cell selection and the importance of exosomal tetraspanins: a hypothesis. Biochem Soc Trans 39:559–562. doi: 10.1042/BST0390559. [DOI] [PubMed] [Google Scholar]
- 40.Martínez ZA, Yáñez-Mó M. 2014. Tetraspanins in extracellular vesicle formation and function. Front Immunol 5:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keller S, König A-K, Marmé F, Runz S, Wolterink S, Koensgen D, Mustea A, Sehouli J, Altevogt P. 2009. Systemic presence and tumor-growth promoting effect of ovarian carcinoma released exosomes. Cancer Lett 278:73–81. doi: 10.1016/j.canlet.2008.12.028. [DOI] [PubMed] [Google Scholar]
- 42.Chen Y-H, Du W, Hagemeijer Marne C, Takvorian Peter M, Pau C, Cali A, Brantner Christine A, Stempinski Erin S, Connelly Patricia S, Ma H-C, Jiang P, Wimmer E, Altan-Bonnet G, Altan-Bonnet N. 2015. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell 160:619–630. doi: 10.1016/j.cell.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cote M, Misasi J, Ren T, Bruchez A, Lee K, Filone CM, Hensley L, Li Q, Ory D, Chandran K, Cunningham J. 2011. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schultz ML, Tecedor L, Chang M, Davidson BL. 2011. Clarifying lysosomal storage diseases. Trends Neurosci 34:401–410. doi: 10.1016/j.tins.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fouchier SW, Defesche JC. 2013. Lysosomal acid lipase A and the hypercholesterolaemic phenotype. Curr Opin Lipidol 24:332–338. doi: 10.1097/MOL.0b013e328361f6c6. [DOI] [PubMed] [Google Scholar]
- 46.Brandenburg B, Lee LY, Lakadamyali M, Rust MJ, Zhuang X, Hogle JM. 2007. Imaging poliovirus entry in live cells. PLoS Biol 5:e183. doi: 10.1371/journal.pbio.0050183. [DOI] [PMC free article] [PubMed] [Google Scholar]