ABSTRACT
Mouse leukemia viruses (MLVs) are found in the common inbred strains of laboratory mice and in the house mouse subspecies of Mus musculus. Receptor usage and envelope (env) sequence variation define three MLV host range subgroups in laboratory mice: ecotropic, polytropic, and xenotropic MLVs (E-, P-, and X-MLVs, respectively). These exogenous MLVs derive from endogenous retroviruses (ERVs) that were acquired by the wild mouse progenitors of laboratory mice about 1 million years ago. We analyzed the genomes of seven MLVs isolated from Eurasian and American wild mice and three previously sequenced MLVs to describe their relationships and identify their possible ERV progenitors. The phylogenetic tree based on the receptor-determining regions of env produced expected host range clusters, but these clusters are not maintained in trees generated from other virus regions. Colinear alignments of the viral genomes identified segmental homologies to ERVs of different host range subgroups. Six MLVs show close relationships to a small xenotropic ERV subgroup largely confined to the inbred mouse Y chromosome. env variations define three E-MLV subtypes, one of which carries duplications of various sizes, sequences, and locations in the proline-rich region of env. Outside the env region, all E-MLVs are related to different nonecotropic MLVs. These results document the diversity in gammaretroviruses isolated from globally distributed Mus subspecies, provide insight into their origins and relationships, and indicate that recombination has had an important role in the evolution of these mutagenic and pathogenic agents.
IMPORTANCE Laboratory mice carry mouse leukemia viruses (MLVs) of three host range groups which were acquired from their wild mouse progenitors. We sequenced the complete genomes of seven infectious MLVs isolated from geographically separated Eurasian and American wild mice and compared them with endogenous germ line retroviruses (ERVs) acquired early in house mouse evolution. We did this because the laboratory mouse viruses derive directly from specific ERVs or arise by recombination between different ERVs. The six distinctively different wild mouse viruses appear to be recombinants, often involving different host range subgroups, and most are related to a distinctive, largely Y-chromosome-linked MLV ERV subtype. MLVs with ecotropic host ranges show the greatest variability with extensive inter- and intrasubtype envelope differences and with homologies to other host range subgroups outside the envelope. The sequence diversity among these wild mouse isolates helps define their relationships and origins and emphasizes the importance of recombination in their evolution.
INTRODUCTION
Mouse leukemia viruses (MLVs) with ecotropic, xenotropic, and polytropic host ranges (E-MLVs, X-MLVs, and P-MLVs, respectively) are readily isolated from the classical strains of the laboratory mouse (reviewed in reference 1). E-MLVs can infect murine cells but not cells of heterologous species; these viruses use the amino acid transporter, CAT-1, as their receptor (2). The X-MLVs and P-MLVs both use the XPR1 receptor but differ in their abilities to infect cells of murine and other mammalian species (3–5); we use the term X/P-MLV to refer to the full set of XPR1-dependent viruses. MLV host range variation is determined by the receptor binding domains (RBDs) of the MLV envelope (env) (6) and by receptor polymorphisms (7, 8).
The classical strains of inbred laboratory mice that harbor MLVs are intersubspecific hybrids of the house mouse subspecies of Mus musculus (9), and MLVs have also been isolated from some wild mouse M. musculus populations (10, 11). MLVs related to the laboratory mouse X-MLVs and the AKV subtype E-MLVs are found in Mus musculus molossinus Japanese house mice (12). California wild mice carry a second subtype of E-MLVs, termed Cas E-MLVs (11), and a novel MLV host range subgroup not found in laboratory mice, amphotropic MLV (A-MLV) (13, 14), which uses the phosphate transporter PiT-2 for entry (15). A virus representing a third E-MLV subtype, HoMuLV, was isolated from the Eastern European mouse Mus spicilegus (16).
MLVs are also found as endogenous retroviruses (ERVs) in laboratory and house mouse subspecies. These ERVs represent relics of past infections and are generated when viral DNA copies insert into germ line cells, becoming a part of the genome. Inbred strains carry E- and X-ERVs, termed Emv and Xmv ERVs, and two subtypes of P-MLV ERVs, termed Pmv and Mpmv ERVs (17, 18). Individual ERVs are identified by their sites of insertion and classified by env sequence homology. Most Emv and Xmv sequences are full-length proviruses with coding region open reading frames, and many are able to produce infectious virus (reviewed in reference 1). In contrast, none of the P-ERVs are demonstrably capable of producing infectious virus, and all MLV isolates with polytropic host ranges are recombinants, with an E-MLV backbone and env substitutions acquired from P-ERVs (19–21).
All M. musculus subspecies carry germ line copies of X/P-MLV env genes (22). Unlike the classical strains of laboratory mice, most of which carry multiple copies of both X- and P-ERVs, the distribution of wild mouse X- and P-ERVs is largely subspecies specific. Thus, X-ERVs are restricted to the Eurasian subspecies M. musculus castaneus, M. m. musculus, and M. m. molossinus, whereas the western European Mus musculus domesticus carries only P-ERVs (22). The classical strains of laboratory mice acquired their MLV ERVs from their wild mouse progenitors, and, in fact, the various Xmv insertions found in the sequenced laboratory mouse genome are also found in Asian wild mice (23). Some of these wild mouse Xmv proviruses are functional (24), and wild mouse P-ERVs can contribute to the production of recombinant P-MLVs that resemble those from laboratory mice (25).
The E- and A-MLVs are more recent acquisitions of Mus and show a more limited distribution in wild mice than the X/P-MLVs. The laboratory mouse AKV E-ERV subtype is found in Japan and northwest Asia, and Cas subtype E-ERVs are found in wild mice found in Japan through Southeast Asia and China to Iran as well as California (22, 26). The HoMuLV E-MLV and A-MLV are found only in small localized mouse subpopulations in Eastern Europe and southern California, respectively, and have not endogenized in their hosts (27, 28). House mice are not native to the Americas but largely derive from animals introduced from Western Europe, M. m. domesticus. These mice carry only P-ERVs, but some populations in California are the natural hybrids of M. m. domesticus and M. m. castaneus brought by Chinese laborers and trade (22, 29), and these hybrid mice carry ERVs of P- and X-MLVs and Cas subtype E-MLVs, and infectious viruses of A- and E-MLV host ranges.
Characterization of wild mouse infectious and endogenous MLVs has been limited. Early studies analyzed some wild mouse MLVs using comparative restriction mapping and partial sequencing (10, 30), and two wild mouse MLVs, Cas-Br-E and A-MLV, were completely sequenced (31, 32). Here, we sequenced the complete genomes of seven MLV isolates obtained from wild mice from Europe, Asia, and California. We sought to define the relationships of these viruses to one another and to known MLVs. Because laboratory mouse MLVs derive from ERVs found in laboratory and wild mice, we also looked to identify possible ERV progenitors of these wild mouse MLVs. We identified segmental intersubgroup relationships between these wild mouse MLVs and various ERVs, indicating that intersubgroup recombination likely contributes to sequence diversity among these MLVs. We found that most of the MLVs are related to a small, distinctive subset of the Xmv X-ERVs, that the non-env segments of the three E-MLV subtypes are related to different nonecotropic MLVs, that E-MLVs of only one subtype have unusual env duplications, and that some MLVs carry shared sequences that are host taxon specific rather than virus host range related.
MATERIALS AND METHODS
Viruses.
We sequenced the full-length genomes of seven MLVs isolated from mice originating from five geographically separated locations (Table 1). CAST-X is an X-MLV isolated from the spleen of a CAST/EiJ mouse (33). Cz524 was isolated from the spleen of a CZECHII/EiJ mouse 2 months after inoculation with Moloney E-MLV (MoMLV) (34). The HoMuLV E-MLV was isolated from cultured tail biopsy specimens of a randomly bred M. spicilegus mouse (formerly Mus hortulanus) (16). CasE#1 was originally isolated from cultured embryo cells from a mouse from Lake Casitas, CA (35). Kyushu X-MLV was induced by 5-iododeoxyuridine (36) from cultured kidney cells of a wild mouse trapped in Kyushu, Japan. Cell lines designated castaneus 17 and 18 were established from individual 22-day-old M. m. castaneus female and male mice from a strain designated CTH, trapped in the Pathumthani region of Thailand, and interbred by F. Bonhomme (Universite de Montpellier, Montpellier, France). E-MLVs initially designated Cast17 and Cast18 were induced from the two CTH lines by treatment with 5-iododeoxyuridine followed by cocultivation with SC-1 cells (37). Other viral genomes analyzed here include Cas-Br-E E-MLV (GenBank accession number X57540), AKV E-MLV (GenBank accession number J01998), and NZB-9-1 X-MLV (GenBank accession numbers K02730 and EU035300).
TABLE 1.
Origins of sequenced viruses isolated from wild mice
| Receptor | Host range | Virusa | Wild mouse origin | Trapping site | Reference or source |
|---|---|---|---|---|---|
| CAT-1 | Ecotropic | Cast17 | M. m. castaneus | Pathumthani, Thailand | This paper |
| Cast18 | M. m. castaneus | Pathumthani, Thailand | This paper | ||
| HoMuLV | M. spicilegus | Halbturn, Austria | 16 | ||
| XPR1 | Xenotropic | CAST-X | M. m. castaneus | Thailand | 33 |
| Kyushu | M. m. molossinus | Kyushu, Japan | This paper | ||
| X/P-MLVb | CasE#1 | California wild mouse | Lake Casitas, CA, USA | 35 | |
| Cz524 | M. m. musculus | Bratislava, Slovakia | 34 |
Cast17 and Cast18 were determined to be nearly identical Cas subtype E-MLVs, and Cz524 X/P-MLV is a recombinant generated in MoMLV-inoculated mice.
CasE#1 and Cz524 do not show a prototypical X- or P-MLV host range.
Mouse genomic DNAs.
Sources of inbred mouse strains, wild-caught and wild-derived mice, and mouse DNAs were described previously (23). Briefly, DNAs were isolated or obtained from mice maintained in our laboratory or from the randomly bred colonies of M. Potter (National Cancer Institute, Bethesda, MD), The Jackson Laboratory (Bar Harbor, ME), S. Rasheed (University of Southern California, Los Angeles, CA), R. Abe (Naval Medical Research Institute, Bethesda, MD), S. Chattopadhyay and H. Morse III (NIAID, Bethesda, MD), and RIKEN BioResource Center (Ibaraki, Japan), which participates in the National Bio-Resources Project of the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan. For this analysis we used DNAs from 28 classical inbred strains and 51 DNAs from wild-trapped or wild-derived colonies and inbred strains of M. musculus.
MLV sequencing.
The four X/P-MLVs were sequenced from genomic DNAs from newly infected mink lung cells (ATCC CCL64). The three E-MLVs were sequenced from DNAs from newly infected Mus dunni cells (38) by reverse transcription-PCR from pelleted virions or, for HoMuLV, from the pHo2.1 molecular clone (28). The primers in Table S1.1 in the supplemental material generated overlapping PCR products that were cloned into pCR2.1-TOPO and sequenced.
We amplified internal MLV ERV segments from DNAs of CZECHII/EiJ; M. m. castaneus strains CAST/EiJ, HMI/Ms, MYS/Mz, and CAST/Ncr; and from three mice trapped in Lake Casitas using primers listed in Table S1 in the supplemental material. Products were cloned into pCR2.1-TOPO and sequenced.
MLV sequences were compared with one another and with previously described or newly sequenced ERVs and MLVs. Segments of high sequence identity (>90%) were identified by BLAST searches of short overlapping genomic segments and in Hypermut plots (http://www.hiv.lanl.gov/content/sequence/HYPERMUT/background.html) (39) constructed with representative ERVs and MLVs of all host range subgroups.
Inbred strain and subspecies origin of the XmvIV1 ERV.
Multiple MLVs contained segments with close homology to a small and distinctive subset of five X-ERVs (Xmv proviruses) that we term XmvIV based on their unique env variable region A (VRA) (40) and other sequence differences; only one copy, XmvIV1, is autosomal (Table 2). DNAs from laboratory strains and M. musculus subspecies were screened for the XmvIV1 X-ERV by PCR using primers designed from the provirus and from cellular flanking sequences. The respective 3′ and 5′ flanking primers were 5′-CGAAACCACGAGAAACACCG and 5′-CTAGTCCTCCCAGAAGAGTGAG. Provirus-specific junction fragments were generated using these flanking primers and forward and reverse primers from the MLV long terminal repeat (LTR) sequence (5′-CAGCTCGCTTCTCGCTTCTG).
TABLE 2.
Location and coding potential of six laboratory mouse X-MLV ERVs
| ERVa | Chromosome locationc | Position (bp) | Strand | ORFs |
|---|---|---|---|---|
| XmvIV1 (Xmv45) | 5:23700580–23709245 | 8667 | − | gag, pol, env |
| XmvIV2 | Y:20663634–20672302 | 8670 | + | gag, pol, stop in env |
| XmvIV3b | Y:30770451–30779129 | 8680 | + | gag, env, stop in pol |
| XmvIV4b | Y:31666648–31675325 | 8679 | + | gag, env, stop in pol |
| XmvIV5 | Y:4795459–4804128 | 8670 | − | gag, pol, env |
| Xmv11 | Y:58846340–58855054 | 8715 | − | gag, pol, env |
Two of the four Y chromosome XmvIV sequences are likely to be previously identified Xmv7 and Xmv40 (56).
Identical ERVs that carry the same pol defect.
According to the GRCm38/mm10 assembly.
We used the Mouse Phylogeny Viewer (MPV) at the University of North Carolina (http://msub.csbio.unc.edu) (41) to determine the subspecies origin of the chromosome 5 (Chr 5) integration site containing XmvIV1. We defined the chromosome coordinates for XmvIV1 by a BLAT search (42) of the NCBI37/mm9 reference assembly used to create the MPV database using the University of California Santa Cruz (UCSC) Genome browser (http://genome.ucsc.edu). We typed 28 of the inbred strains included in the MPV database for XmvIV1 by PCR.
Identification of the NZB-9-1 unique pol sequence in the NZB/BINJ mouse.
Aligned sequence reads for the NZB/BINJ strain (release REL-1502; BAM format, aligned to GRCm38_68 with BWA-MEM, version 0.7.5a-r406 [43]) were provided by the Mouse Genomes group at the Wellcome Trust Sanger Institute and can be obtained from their FTP (file transfer protocol) site (ftp://ftp-mouse.sanger.ac.uk/current_bams) (44). Reads were converted to FASTQ format using SamToFastq, version 1.75 (http://broadinstitute.github.io/picard), and aligned to a reference made from bases 4272 to 5244 of the NZB virus sequence (GenBank accession number EU035300.1) with BWA-MEM, version 0.7.12-r1039 (43), using the default settings. Read pairs where at least one read in the pair aligned to the NZB reference were isolated and converted to FASTQ format with bam2fastx (from the TopHat package [45]) and trimmed with Trimmomatic, version 0.33 (46), using the paired-end (PE) algorithm and the following settings: sliding window, 5:30; leading, 20; trailing, 20; minlen, 50. A new reference fasta file was constructed, including the partial NZB sequence (bases 4272 to 5244, as used in the previous alignment) as well as the top five BLAT (42) matches using the partial NZB sequence as a query (BLAT search performed against GRCm38/mm10 genome at the UCSC Genome Browser [http://genome.ucsc.edu/]) (47); matches were found on Y chromosome positions 20.7, 31.7, 30.8, and 47.98 and on Chr 5:23.7, which we term XmvIV1 to XmvIV5 and which together represent a related, distinctive subgroup of Xmv ERVs (Table 2). Reads were mapped using BWA-MEM, version 0.7.5a-r405 (43), to the new reference; read pairs mapping to the partial NZB sequence were retained, and those mapping to mm10 sequences were discarded. Reads were again converted to FASTQ format with bam2fastx and assembled with SOAPdenovo2 (48) (version LINUX-generic-r240) with a k-mer setting of 49 and from 59 to 81 (odd values), and the largest scaffolds or contigs from each assembly (all over 700 bp) were assembled using Lasergene SeqMan (DNASTAR, Inc., Madison, WI), producing a consensus of 1,255 bp.
Phylogenetic trees.
Four phylogenetic trees were constructed in MEGA, version 6 (49), using the neighbor-joining method (50). The four trees were based on the 5′ end of the MLV env (receptor binding domain/proline-rich region [RBD/PRR] env), the transmembrane segment of env (TM env), the reverse transcriptase domain of pol (RT pol), and the gag gene. The RBD/PRR env segment encodes the surface subunit of env through the end of the PRR. RT pol corresponds to positions 2613 to 4154 in AKV E-MLV. The evolutionary distances were computed using the JTT matrix-based method (51). The rate variation among sites was modeled with a gamma distribution (shape parameter of 1). Trees included MLV ERVs in the sequenced C57BL genome classed as Xmv, Pmv, and Mpmv ERVs (52, 53). Other viral genomes included in the trees are Cas-Br-E (GenBank accession number X57540), AKV (GenBank accession number J01998), Emv2 (Chr 8:123425507 to 123434150; GRCm38/mm10), PreXMRV-1 (GenBank accession number FR871849), PreXMRV-2 (GenBank accession number FR871850), Fv4 (GenBank accession numbers AH001894 and M11052), Frg1 (GenBank accession number AB050720), Frg3 (GenBank accession number AB050721), and NZB-9-1 (GenBank accession numbers K02730 and EU035300). All positions with less than 95% site coverage were eliminated. That is, alignment gaps, missing data, and ambiguous bases greater than 5% were not allowed at any position.
Nucleotide sequence accession numbers.
Sequences of the six distinctive wild mouse MLVs were deposited in GenBank as follows: CasE#1, KU324802; CAST-X, KU324803; Cz524, KU324804; HoMuLV, KU324805; Kyushu, KU324806; Cast17/18, KU324807. Sequences of two ERVs from CZECHII/EiJ, Mmm1 and Mmm2, were deposited under numbers KU324808 and KU324809, and sequences of 20 env-containing ERVs from various wild mice were deposited under accession numbers KU324810 to KU324829.
RESULTS AND DISCUSSION
Genome sequences of seven wild mouse MLV isolates.
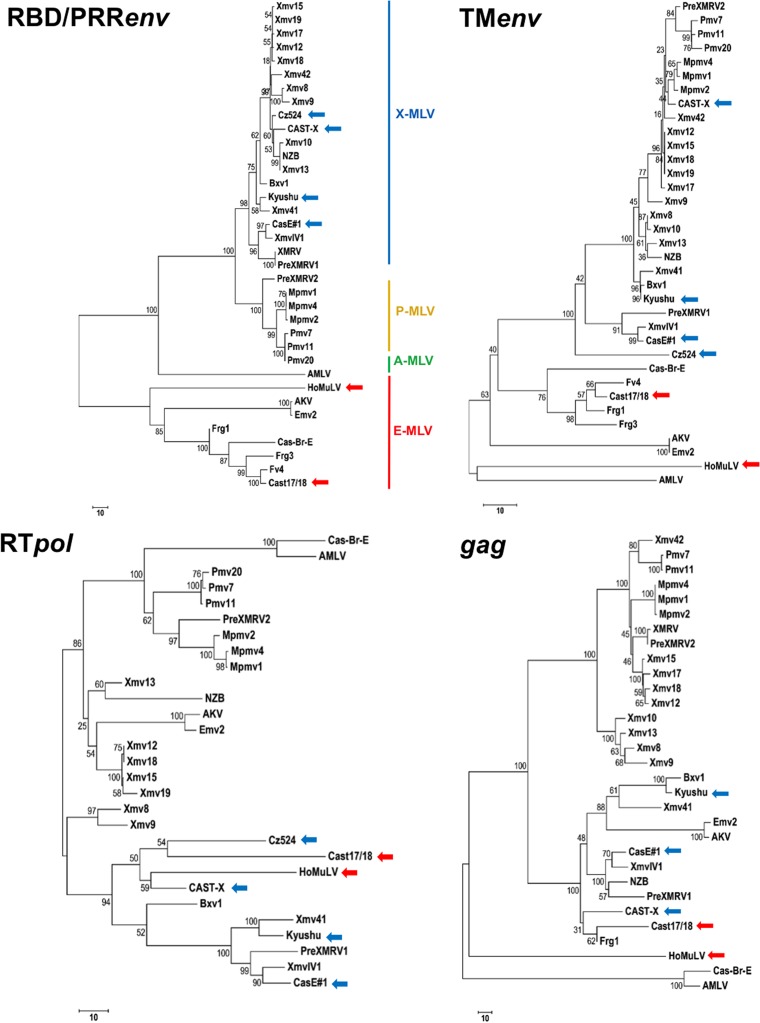
We sequenced the complete genomes of seven wild mouse virus isolates. Six of these MLVs were isolated from house mouse M. musculus subspecies, and one, HoMuLV E-MLV, was isolated from M. spicilegus (Table 1). Three of the seven viruses are E-MLVs, and four are X/P-MLVs capable of infecting cells from nonrodent species; two have prototypical xenotropic host ranges. All of the sequenced genomes show a typical gammaretrovirus genome organization and contain all key functional motifs. Sequences were compared with those of laboratory mouse MLVs, with the ERVs found in the sequenced C57BL genome, and with previously described and newly sequenced ERVs. Phylogenetic trees were constructed from four segments of the viral genome: the 5′ end of env containing the RBD and PRR, the transmembrane subunit of env (TM env), the reverse transcriptase domain of pol (RT pol), and the gag gene (Fig. 1). RBD defines receptor choice (6), and in the RBD/PRR env tree (Fig. 1A), MLVs with known X-, P-, A-, or E-MLV tropism form well-defined clusters with ERVs defined by their sequence homologies to E-, X-, or P-MLVs. The newly sequenced wild mouse E-MLVs group with previously described E-MLVs. The four new wild mouse-derived nonecotropic MLVs all group with X-MLVs although two, Cz524 and CasE#1, have atypical host range phenotypes that are neither classically xenotropic nor polytropic, as reported previously and summarized below; these two MLVs are therefore designated X/P-MLVs (33, 34).
FIG 1.
Phylogenetic trees of four domains of the MLV genome. The optimal tree is shown, and the percentages of replicate trees in which associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches (83). The trees are drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Each analysis included at least 30 MLV DNA sequences. Colored arrows identify the newly sequenced wild mouse E-MLVs (red) and X/P-MLVs (blue). Cz524 X/P-MLV is not included in the gag tree because it contains MoMLV segments in this region.
These phylogenetic relationships are not strictly maintained in trees based on the other segments of the viral genome (Fig. 1). In particular, the E-MLVs show closer relationships with various nonecotropic MLVs than with one another in trees based on RT pol and gag (Fig. 1C and D).
This phylogenetic analysis suggests that some of these wild mouse MLVs may be derived from intersubgroup recombinations, so we examined these sequences in colinear alignments with each other and with other ERVs and MLVs to identify sequence relationships that might elucidate their evolutionary origins as well as describe possible recombinational breakpoints and patterns. For this analysis we also included three previously sequenced MLVs: the AKV and Cas-Br-E E-MLVs and the NZB-9-1 X-MLV. Because few wild mouse MLVs have been sequenced and because sequenced wild mouse genomes are incomplete, these sequence comparisons were largely limited to laboratory mouse MLVs and ERVs, supplemented with newly sequenced ERV segments from selected wild mice.
X/P-MLVs.
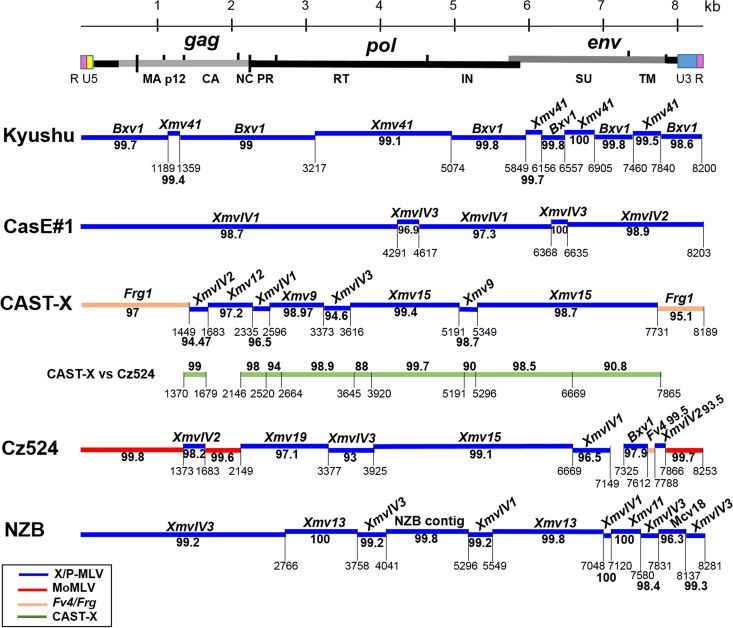
Of the five analyzed X/P-MLVs, only one shows significant homology to the active X-ERV Bxv1 found in laboratory strains and some Asian house mice (54–56). The other four have segments of significant identity to a small, largely uncharacterized subgroup of X-ERVs, here designated XmvIV, and one of these viruses, CasE#1 X/P-MLV, is entirely XmvIV derived.
(i) Kyushu X-MLV.
The Japanese mouse-derived Kyushu X-MLV shows nearly identical (>99%) segmental relationships to two full-length X-ERVs, Bxv1 (also termed Xmv43) (54–56) and Xmv41 (Fig. 2). Kyushu X-MLV has a Bxv1 backbone with five Xmv41-derived substitutions in gag, pol, and env that avoid the two stop codons in the Xmv41 pol. Bxv1 is a nondefective X-ERV capable of producing infectious virus following chemical or immunological stimulation (54). These two ERVs are both found in the sequenced C57BL genome, and both are also found in the Japanese wild mouse M. m. molossinus (23, 55). Bxv1 is the only active X-ERV in most inbred strains of mice (1) and is also thought to be active in Japanese mice (24). Sequence comparisons of Bxv1 and Xmv41 show them to be related X-ERVs that together constitute a distinct clade of Xmv ERVs (53), and these two ERVs are closely linked on distal mouse chromosome 1 (Bxv1, Chr 1:170.9; Xmv41, Chr 1:171.5). The Kyushu X-MLV thus represents an unusual recombinant of two related and closely linked X-ERVs found in inbred and Japanese mice.
FIG 2.
Sequence relationships of four X/P-MLVs from wild mice and the laboratory mouse NZB X-MLV to various ERVs. At the top is a schematic representation of the viral genome. For each of the five genomes, staggered horizontal lines represent regions of greatest identity (over 90%) with ERVs or other MLVs, and these percentages are bolded. Numbering begins with the first nucleotide of the R region of the U5 LTR. Sequence positions are given for the last base pair in each segment of homology. A separate green line is used to show segmental homologies between CAST-X and Cz524.
(ii) CasE#1 X/P-MLV and the XmvIV ERVs.
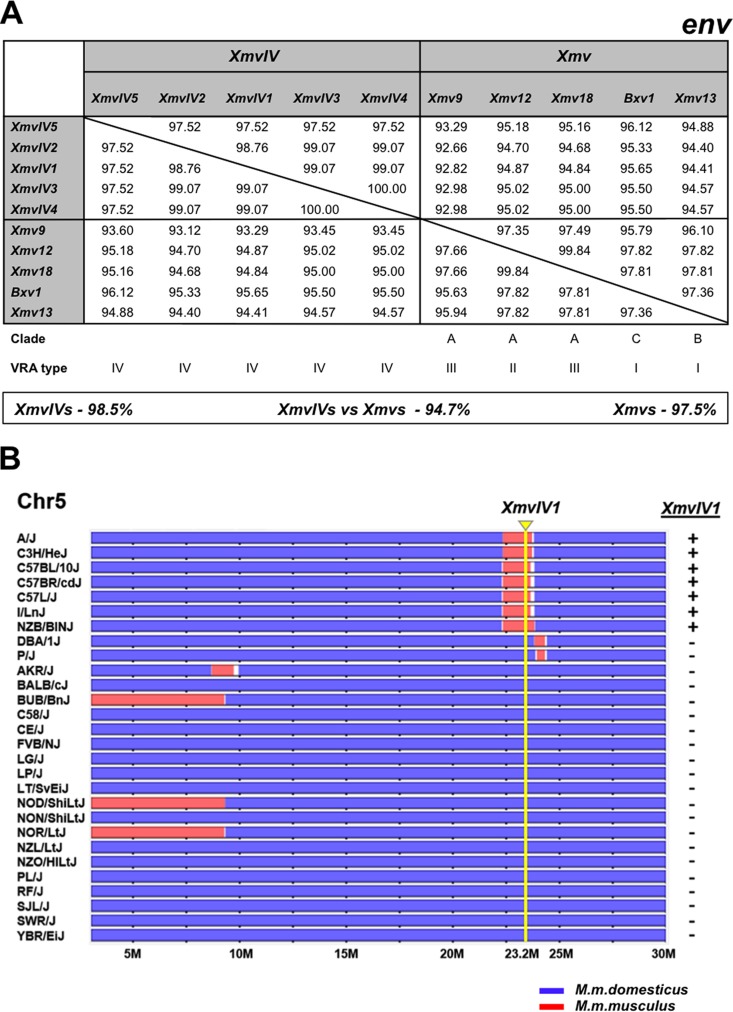
The CasE#1 X/P-MLV is a California wild mouse isolate that shares phenotypic properties with laboratory mouse X- and P-MLVs (35). Like P-MLVs, CasE#1 X/P-MLV induces foci on mink cells and shows a nonreciprocal interference pattern with X-MLVs. While CasE#1 X/P-MLV resembles X-MLVs in its inability to infect laboratory mouse cells, it infects a different subset of mammalian cells than prototypical X- or P-MLVs (35, 57). The CasE#1 genome shows closest sequence identity to a small set of X-ERVs with a distinctive env subtype, previously termed Xeno-IV (40) (Fig. 2). We identified five full-length Xeno-IV copies in the sequenced laboratory mouse genome; there is one autosomal copy, XmvIV1, on Chr 5 and four copies, XmvIV2 to XmvIV5, on the Y chromosome (Table 2). Three of the four Y chromosome ERVs have stop codons in pol or env; the Chr 5 copy, XmvIV1, appears to be nondefective and was earlier identified as the source of gag and pol substitutions in the melanoma-associated recombinant MelARV E-MLV (58). The five XmvIV genomes are 96.9% identical to one another but show 92.2% identity to the Xmv sequences identified in the C57BL genome (53). In env, these five ERVs are 98.5% identical but show 94.7% identity to the various Xmv sequences (Fig. 3A). In comparison, env genes of the P-ERV subgroups designated Mpmv and Pmv are 97 to 98% identical. Most of the CasE#1 genome shows >97% identity to XmvIV1, XmvIV2, and XmvIV3 (Fig. 2).
FIG 3.
Characterization of the five full-length XmvIV ERVs and the Asian mouse origins of XmvIV1. (A) Protein identity matrix for env genes of five XmvIV ERVs and selected Xmv proviruses. At the bottom are average identities between and within groups. Clade and VRA type were taken from Lamont et al. and Jern et al. (40, 53). (B) XmvIV1 (Xmv45) originated in Asian mice. The horizontal tracks represent a 30-Mb segment of Chr 5 for 28 inbred strains of laboratory mice. The mice were typed by PCR for XmvIV1 as indicated to the right. The map location of this ERV, Chr 5:23.7 (NCBI37/mm9 assembly), is marked by a yellow arrow and line. Chromosomal regions originating from M. m. domesticus are in blue; segments from M. m. musculus are in red.
Previous studies identified and mapped over 60 unique laboratory mouse Xmv loci on the basis of their reactivity to an X-MLV-derived oligonucleotide env probe and identified Y-chromosome-linked MLV ERVs in some inbred strains and Mus taxa (56, 59). It is likely that XmvIV1 is the previously identified Xmv45, based on map location and strain distribution, and that two of the four Y-linked XmvIV ERVs correspond to the Y-linked Xmv7 and Xmv40. Xmv7, Xmv40, and Xmv45 all showed poor reactivity to the X-MLV env probe used to identify them (56), reflecting the sequence divergence of the XmvIV subset of Xmv ERVs.
We previously identified the wild mouse origins of laboratory mouse Xmv ERVs (23), and here we used the same two approaches to identify the wild mouse origins of XmvIV1. First, we designed primers that generate diagnostic cell-XmvIV1 junction fragments and screened 51 DNAs from M. musculus subspecies. XmvIV1 was identified in half of the M. m. molossinus DNAs and in five other M. musculus mice trapped in South Korea or mainland China but not in any other house mice (Table 3). Second, we used the same primers to type 28 of the inbred strains included in the Mouse Phylogeny Viewer database. We found that the position of this ERV (Chr 5:23.7; NCBI37/mm9 assembly) maps to a segment of the laboratory mouse genome that is derived from Asian mice (Fig. 3B). Thus, XmvIV1 originated in the same wild mice as the Xmv X-ERV proviruses.
TABLE 3.
Distribution of XmvIV1 in wild-trapped and wild-derived mice
| XmvIV1 (Xmv45) status | M. musculus subspecies | Mouse strain(s)b |
|---|---|---|
| Present | molossinus | JF1, Mol/Li, MOLC, MOLG, MOM, STM1 |
| Unassigneda | BJN3, CHD, IAS-2, KJR, SHH1 | |
| Absent | molossinus | AIZ, KOR5, KOR7, MAE, MOLD, MOLF, STM2 |
| castaneus | BGR1, Cas/Li, Cas18, CASA, CAST/EiJ, CAST/Rp, MYS | |
| musculus | AKT, AST, BLG2, CZECHI, CZECHII, MBT, PWD, PWK, Viborg | |
| domesticus | ABUR, BFM, BQC, HAF, LC107, LC117, LC120, LEWES, PERA, PERC, PWD, PWK, RBF, SKIVE, SOD, TIRANO, ZALENDE |
Mice trapped in South Korea or mainland China and not assigned to any M. musculus subspecies.
LC mice are from Lake Casitas, CA.
The origins and subspecies distribution of the four Y-linked XmvIV ERVs could not be determined. The Phylogeny Viewer does not include the Y chromosome, and these ERVs are embedded in Y-specific repetitive sequences precluding the unambiguous identification of specific ERV insertions by PCR. It is likely, however, that the Y chromosome XmvIV copies are also Asian derived as it has been shown that the Y chromosome of C57BL is of Asian origin (60).
(iii) CAST-X X-MLV.
The CAST-X X-MLV from M. m. castaneus shows a typical X-MLV host range but is not identical to any sequenced X-MLV or X-ERV. Five segments of CAST-X, totaling 5,544 bp, show >97% identity to various Xmv sequences (Fig. 2). The LTR, the gag leader, and the 5′ end of gag, however, most closely resemble Frg1, a deleted ERV also found in M. m. castaneus that has an ecotropic env (30). Three other segments of CAST-X show closest identity (94.5 to 96.5%) to the XmvIV ERVs. This virus also shows significant identity to the genome of Cz524 X/P-MLV, as described below and shown in Fig. 2 (green band).
(iv) Cz524 X/P-MLV.
E-MLV-infected laboratory mice carrying P-ERVs can readily produce recombinant infectious P-MLVs (19), so we attempted to isolate such recombinants from wild mice carrying X/P-ERVs. We inoculated M. m. musculus (CZECHII/EiJ) neonates with Moloney E-MLV (MoMLV) and screened thymus and spleen from 2- to 4-month-old mice for viruses that, unlike MoMLV, can replicate efficiently in M. dunni and mink cells (34). The Cz524 isolate was initially classed as an X-MLV because of its reactivity with X-MLV env probes on Southern blots (data not shown) and because it failed to infect NIH 3T3 cells. However, further testing showed that Cz524 is not a classical X-MLV but has a distinctive host range (34). Like P-MLVs, Cz524 is restricted in many nonrodent species like dog and buffalo, but, unlike P-MLVs, Cz524 can infect cells of bat and guinea pig (61). Cz524 has the cytopathic properties of P-MLVs (5, 38) in that it can induce foci in mink lung cells and is cytolytic in M. dunni cells.
Analysis of the complete Cz524 X/P-MLV sequence shows it to be an MoMLV recombinant (Fig. 2). The Cz524 LTR is nearly identical to that of MoMLV with two copies of the MoMLV enhancer direct repeat. Cz524 also has MoMLV sequences in the gag leader and in the 5′ end of gag, which is interrupted by an ERV sequence that replaces the target site for the host Fv1 restriction factor at position 1509 to 1616. The rest of gag and the entire env and pol genes are completely ERV derived, with close sequence identities to various Xmv or XmvIV ERVs.
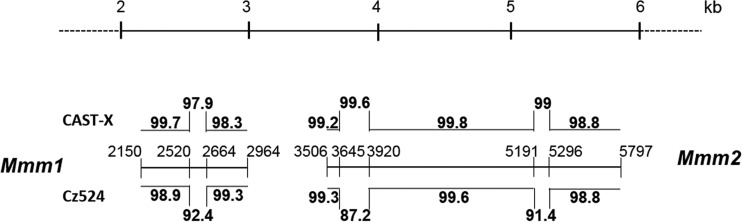
Comparison of Eurasian isolates CAST-X X-MLV and Cz524 X/P-MLV (Fig. 2, green band) shows that these two viruses have regions covering 4,271 bp that show 98 to 99% identity interspersed with four regions that are more divergent (88 to 94%) (Fig. 2 and 4). To determine if ERVs related to either or both of these viruses are found in CAST/EiJ and CZECHII/EiJ mice, we PCR amplified and sequenced pol segments from these mice. All CAST/EiJ-derived ERV segments were equally related to these MLVs (data not shown), but two clones from CZECHII mice, Mmm1 and Mmm2, more closely resembled CAST-X (97.9 to 99.6%) than Cz524 (87.2 to 92.4%) in the three segments that most distinguish these viruses (Fig. 4). This suggests that the different populations of Eurasian mice that produced CAST-X and Cz524 carry highly related X/P-ERVs.
FIG 4.
Relationships of two ERVs from CZECHII/EiJ mice with CAST-X X-MLV and Cz524 X/P-MLV. Mmm1 and Mmm2 ERV pol sequences amplified from CZECHII/EiJ mice are indicated by the central horizontal line and delimited by their positions in Cz524 X/P-MLV. Sequence homologies between these ERVs and the two MLVs are given as percentages (above for CAST-X and below for Cz524). These ERVs include three of the four regions of greatest divergence between the two viruses.
(v) Laboratory mouse-derived X-MLVs.
X-MLVs were first isolated from cells and tissues of various inbred laboratory mice (3), but these viruses have also been found as contaminants in various human cell lines in searches that were prompted by the identification of the xenotropic murine leukemia virus-related virus (XMRV), an X-MLV contaminant in a human prostate xenograft (62, 63). Many of these X-MLVs, such as N417/EKVX, a contaminant of the human SCLC tumor cell line, are virtually identical to Bxv1 (64, 65), whereas XMRV is a novel recombinant of two MLV ERV precursors, PreXMRV-1 and PreXMRV-2 (62, 66). Other X-MLV contaminants, like DG75, found in a human B-lymphoblastoid cell line (67), are related to but distinct from mouse-derived X-MLVs likely due to their origins from unidentified variant ERVs or because of adaptations to their human host cells.
The first X-MLV was isolated from NZB/BINJ mice (3), which produce high levels of X-MLV throughout life (68). NZB mice have at least two active X-ERVs, Nzv1 and Nzv2, one of which is constitutively active and neither of which is Bxv1 (55, 69). While neither of these active ERVs has been sequenced, we have the complete sequence of the infectious NZB-derived NZB-9-1 X-MLV. This virus differs from Bxv1 (94.1% identity) but has segments of substantial homology to XmvIV3, to Xmv13, and to a Y-chromosome-linked Xmv that is likely to be Xmv11 (Fig. 2 and Table 2) (56). Two segments of NZB-9-1 were <95% identical to any laboratory mouse MLVs. One, a 305-bp segment near the 3′ end, shows closest identity to a previously reported ERV sequence found in M. m. molossinus, Mcv18 (70) (GenBank accession number AF070726) (Fig. 2). The second variant segment of 975 bp was used to screen the partially sequenced genome of NZB/BINJ, and this screen identified a contig of 1,254 bp with >99% identity to NZB-9-1 (Fig. 2). Thus, NZB mice carry ERV variants capable of contributing to MLVs that are not found in the sequenced C57BL mouse genome.
E-MLVs.
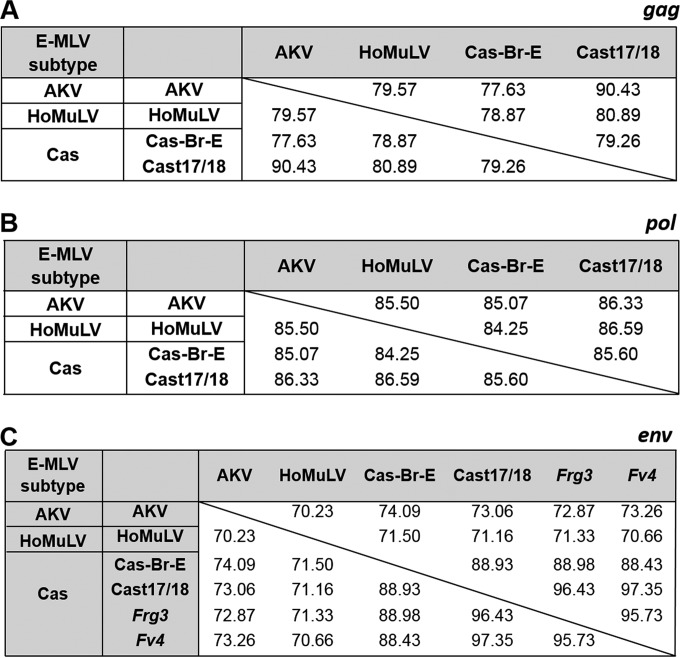
Of the three env sequence subtypes of E-MLVs in wild mice (AKV, Cas, and HoMuLV), only one, AKV, is also found in the classical strains of laboratory mice. In wild mice, the three E-MLV subtypes each have more limited geographic ranges than X/P-MLVs and do not have taxon-wide distributions (22, 26). All three E-MLV subtypes use the CAT-1 receptor (2), but their env genes are quite divergent (∼72% identity compared to ∼89% among X/P-MLVs), while their genomes outside the env gene all show generally closer identity to different nonecotropic MLVs than to one another (Fig. 5 and 6).
FIG 5.
Similarities in the env, gag, and pol genes among members of the three E-MLV subtypes. Similarities are given as percent nucleotide identities. Cas-Br-E, Cast17/18, Frg3, and Fv4 are Cas subtype E-MLVs and are compared with representatives of the other subtypes, AKV and HoMuLV.
(i) HoMuLV E-MLV.
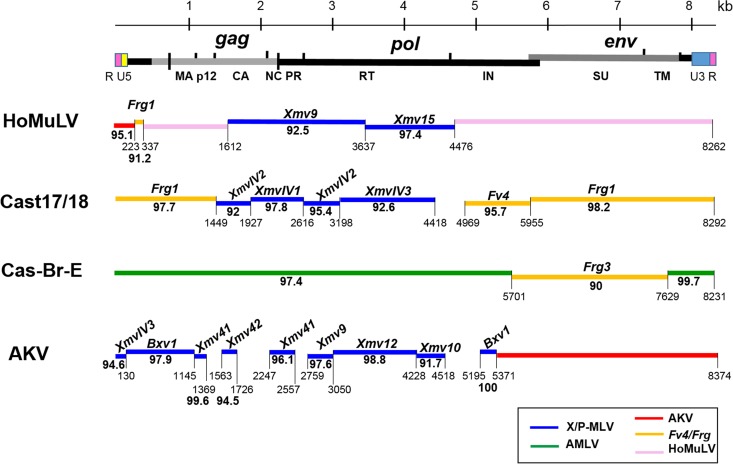
HoMuLV is a pathogenic E-MLV that was isolated from a laboratory mouse colony of M. spicilegus (16) of Eastern Europe. The mice carrying HoMuLV were derived from animals trapped in Halbturn, Austria, but HoMuLV was not detected in M. spicilegus trapped in Pancevo, Serbia, or in any other wild mice. HoMLV is not endogenous in M. spicilegus, and this MLV is no longer carried by the Halbturn-derived colony currently maintained in our laboratory. We previously sequenced the HoMuLV LTR, gag, and env (28), and here we sequenced the entire virus genome. The new sequence is virtually identical (>99%) to our previously reported partial sequences (GenBank accession numbers M26526.1 to M26528.1), but the present sequence corrects frameshifts in the earlier p12gag and env sequences.
Segments of HoMuLV show >92% identity to Xmv15 and Xmv9 (Fig. 6), but other coding region segments of the genome show <90% identity to other MLVs. At the 5′ end of the virus, a segment of the gag leader is related to the Frg1 E-ERV of M. m. castaneus (30), and the R and U5 domains of LTR most closely resemble AKV E-MLV.
FIG 6.
Sequence relationships of three wild mouse E-MLVs and the laboratory mouse AKV E-MLV to ERVs and MLVs. At the top is a schematic representation of the viral genome. For each of the four genomes, horizontal lines represent regions of greatest identities (>90%) with ERVs or other MLVs, annotated as described for Fig. 2. The env genes and some other viral regions are E-MLV subtype specific.
That HoMuLV is the most divergent of the E-MLVs examined here is not surprising, given its isolation from a mouse outside the MLV-carrying house mouse subspecies. M. spicilegus is sympatric with M. m. musculus, but these mice are not interfertile and are unlikely to have much contact because of the different ecologies of the mound-building M. spicilegus and the human-dependent house mice. In any case, no other MLVs of ecotropic host range or E-ERV env genes have been found in European mice although a previous analysis of M. spicilegus found sequences related to MLV LTRs and to a novel gammaretrovirus specific to M. spicilegus, termed hortulanus endogenous MLV (HEMV), that shows an ancestral relationship to the MLVs (22, 70).
(ii) Cast17/18 and Cas-Br-E E-MLVs.
Two E-MLVs were isolated from cultured tail biopsy specimens from two individual mice from the CTH line of M. m. castaneus mice. These isolates, Cast17 and Cast18, are 99.9% identical and are therefore termed Cast17/18 E-MLV. The Cast17/18 env shows significant identity to the four other previously described M. m. castaneus-derived ERVs and MLVs (Fig. 1): the two Asian mouse E-ERVs, Frg1 and Frg3 (30), Cas-Br-E E-MLV, and Fv4, an E-ERV that encodes an Env that acts as a host restriction factor by blocking infection by E-MLVs (71). The Cast17/18 U3 LTR region is related to that of the Frg1 E-ERV (Fig. 5). Cast17/18 shows considerable identity to the XmvIV sequences in gag-pol.
Cas-Br-E and Cast17/18 carry related env genes but were isolated, respectively, from mice from Thailand and California (Fig. 1). It is not surprising to find these viruses in such far-flung locales as mouse populations in California are natural hybrids of M. m. domesticus and M. m. castaneus. Outside the env gene, however, the rest of the Cas-Br-E genome closely resembles that of A-MLV (31, 32), a mouse gammaretrovirus with a novel host range found only in California wild mice (13, 14).
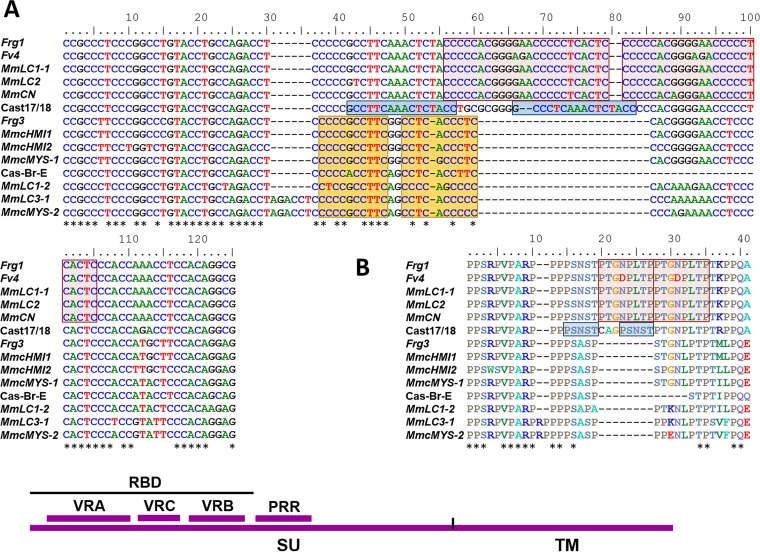
The related env genes of Cast17/18, Cas-Br-E, and the three deleted Cas subtype E-ERVs, Fv4, Frg1, and Frg3, show greatest sequence variation in the PRR of env (Fig. 7). This PRR variation is due largely to indels resulting from sequence duplications of C-rich segments of 10, 16, or 24 bp near the carboxyl (C)-terminal end of the PRR, as previously described for Fv4 and Frg1 (30) (Fig. 7A). The same segment is not duplicated in these five different MLVs, and there are some base pair differences between duplicates. The duplication in Cast17/18 and the duplication shared by Fv4 and Frg1 are all in frame and produce amino acid duplications of 5 and 8 residues, respectively, and the Cast17/18 duplication introduces an additional N-linked glycosylation site (Fig. 7B). A third and different duplication found in the Cas-Br-E PRR is also present in Frg3, but these are not in frame. Although MLVs generally show substantial sequence variation in the 3′ end of the PRR (72, 73), similar PRR duplications are not found in any other MLVs.
FIG 7.
Segmental duplications in the PRR regions of Cas subtype E-MLVs. Three different nucleotide duplications are marked by pink, blue, and yellow (A), and two amino acid duplications are marked by pink and blue (B). The nine Mm entries represent unique ERV PRR sequences amplified from M. m. castaneus or Lake Casitas (LC) mice. Asterisks mark sequence identities. At the bottom is a diagram of env showing the surface (SU) and TM domains, the RBD, the three variable domains within RBD (variable region A to variable region C [VRA-VRC]), and PRR.
To further characterize the range of PRR duplications in Cas subtype E-MLVs, we amplified env genes from wild mice previously shown to carry Cas subtype env genes by Southern blotting. We sequenced 23 env genes representing 20 sequence variants from three mice from Lake Casitas and from four M. m. castaneus mice. Fifteen PRR sequences resembled Fv4, and eight were Frg3-like although four of these eight share a novel 6-bp insertion. The nine unique PRR sequences in this 20-sequence set are shown in Fig. 7.
The PRR is thought to provide a flexible hinge between the RBD and the C-terminal domain of env that stabilizes the viral envelope protein conformation and influences cell fusion (74, 75). Structural studies indicate that the PRRs of feline leukemia virus (FeLV) and A-MLV form a highly ordered polyproline β-turn helix (76, 77) that may function to transmit conformational changes in env after receptor binding. The functional significance of the PRR hypervariability in gammaretroviruses is unknown, but only the largest truncations of the A-MLV PRR adversely affect viral infectivity or env processing (78). On the other hand, residues in PRR have been implicated in the human cell tropism of some porcine ERVs (79), suggesting a role in receptor interactions that could result from the structural flexibility and surface exposure of PRR. That all E-MLVs of the Cas subtype have PRR duplications raises the possibility that this E-MLV subtype-specific change may represent an adaptation to taxon-specific host factors that influence entry.
(iii) AKV E-MLVs.
Over 30 different AKV-type E-ERVs are found in the classical inbred strains of laboratory mice; some of these insertions are shared by strains having common ancestry (18). Several Emv proviruses have been fully sequenced, including the single E-ERV in the sequenced C57BL genome, Emv2, and the E-ERV in NOD mice, Emv30 (80), both of which are virtually identical to the infectious AKV E-MLV from AKR strain mice (GenBank accession number J01998). Partial sequencing, DNA hybridization, and restriction mapping indicate that the other E-ERVs in the laboratory strains are AKV-like (18) and that this virus type is also found in the Japanese mouse M. m. molossinus (12). These ERVs are recent acquisitions in Mus, but they are also closely related to the older X/P-MLVs. Examination of the AKV genome shows that ∼3.8 kb of its genome outside the env region shows significant identity (>95%) to the Xmv and XmvIV proviruses (Fig. 6). This suggests that the AKV E-MLVs are derived from the older and more geographically widespread X-ERVs and MLVs.
Thus, members of the three subtypes of the naturally occurring E-MLVs (AKV, Cas, and HoMLV) carry distinctly different env genes. This env sequence variation may represent separate examples of envelope capture as seen with other mammalian and invertebrate retroviruses (81, 82), or, alternatively, it may be the consequence of a rapidly evolving env glycoprotein in E-MLVs descended from a progenitor ERV in a common ancestor. The discontinuous geographic and taxonomic distribution of these E-MLVs makes it hard to establish phylogeographical links and to account for the present-day distribution of these viruses. Mice carrying these different E-MLVs do not share an environment but are native to widely separated geographic regions (California, eastern Asia, and eastern Europe), and E-MLVs are not necessarily found in mice native to the regions between these virus-infected populations. While this suggests that these retroviruses may have been transmitted through an intermediate vector, E-MLVs have not been found outside Mus.
The E-MLVs are also different from each other outside the env gene (Fig. 7). The A-MLV-like Cas-Br-E is clearly different from the others, and gag-pol sequences of AKV, HoMuLV, and the other Cas subtype E-MLVs are related to different X/P-ERVs, suggesting that they either arose from different progenitors or, more likely, engaged in multiple ongoing rounds of recombination with the other MLVs in their host taxa.
Conclusions.
The nine distinctive virus sequences examined here show significant homologies with laboratory mouse MLV ERVs and with newly described inbred and wild mouse ERVs. Most of these homologies are inexact, rarely approaching 100%, reflecting the fact that the various progenitor M. musculus subspecies carrying these MLVs diverged 0.5 to 1.0 million years ago. The laboratory strains were established only ∼100 years ago from fancy mice that were intersubspecies mosaics, and the sequenced C57BL genome clearly did not capture a fully representative set of wild mouse ERVs.
All of the examined viruses show apparent recombinant genomic structures relative to the known set of MLV ERVs, and in many cases these recombinations cross conventional viral host range subgroups. It is, of course, not clear which of these full-length ERVs and MLVs are the recombinants and which are the progenitors.
The E-MLVs are clearly more recent acquisitions in Mus than the X/P-MLVs but for unknown reasons show substantially more sequence divergence in the subtype-determining env and outside env, where all three E-MLV subtypes were more closely related to nonecotropes than to each other. We also found some taxon-specific sequence commonalities that are unconnected to the virus host range subgroup: CAST X-MLV and Frg1 E-MLV from M. m. castaneus have shared 5′ ends, and the AKV E-MLVS and the Xmv ERVs of Japanese mice show sequence commonalities in non-env regions.
The viruses examined here do not represent the full range of MLV variants found in inbred and wild mice. In addition to this set of naturally occurring viruses, the laboratory mouse has also yielded a number of viruses widely studied for their pathogenic properties and usefulness in constructing retroviral vectors. These viruses, which most famously include Moloney and Friend E-MLVs, were not included here because they are essentially laboratory-generated artifacts. These viruses do not exist in Mus but were isolated after multiple serial passages in mice and cultured cells and therefore offer no insight into the evolution of viruses in natural populations.
Supplementary Material
ACKNOWLEDGMENTS
We thank Herbert Morse III for helpful discussions.
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, Bethesda, MD.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03186-15.
REFERENCES
- 1.Kozak CA. 2014. Origins of the endogenous and infectious laboratory mouse gammaretroviruses. Viruses 7:1–26. doi: 10.3390/v7010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albritton LM, Tseng L, Scadden D, Cunningham JM. 1989. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 57:659–666. doi: 10.1016/0092-8674(89)90134-7. [DOI] [PubMed] [Google Scholar]
- 3.Levy JA. 1973. Xenotropic viruses: murine leukemia viruses associated with NIH Swiss, NZB, and other mouse strains. Science 182:1151–1153. doi: 10.1126/science.182.4117.1151. [DOI] [PubMed] [Google Scholar]
- 4.Fischinger PJ, Nomura S, Bolognesi DP. 1975. A novel murine oncornavirus with dual eco- and xenotropic properties. Proc Natl Acad Sci U S A 72:5150–5155. doi: 10.1073/pnas.72.12.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartley JW, Wolford NK, Old LJ, Rowe WP. 1977. A new class of murine leukemia virus associated with development of spontaneous lymphomas. Proc Natl Acad Sci U S A 74:789–792. doi: 10.1073/pnas.74.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Battini JL, Heard JM, Danos O. 1992. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J Virol 66:1468–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eiden MV, Farrell K, Warsowe J, Mahan LC, Wilson CA. 1993. Characterization of a naturally occurring ecotropic receptor that does not facilitate entry of all ecotropic murine retroviruses. J Virol 67:4056–4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marin M, Tailor CS, Nouri A, Kozak SL, Kabat D. 1999. Polymorphisms of the cell surface receptor control mouse susceptibilities to xenotropic and polytropic leukemia viruses. J Virol 73:9362–9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang H, Wang JR, Didion JP, Buus RJ, Bell TA, Welsh CE, Bonhomme F, Yu AH, Nachman MW, Pialek J, Tucker P, Boursot P, McMillan L, Churchill GA, de Villena FP. 2011. Subspecific origin and haplotype diversity in the laboratory mouse. Nat Genet 43:648–655. doi: 10.1038/ng.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chattopadhyay SK, Oliff AI, Linemeyer DL, Lander MR, Lowy DR. 1981. Genomes of murine leukemia viruses isolated from wild mice. J Virol 39:777–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardner MB. 1978. Type C viruses of wild mice: characterization and natural history of amphotropic, ecotropic, and xenotropic MuLV. Curr Top Microbiol Immunol 79:215–259. doi: 10.1007/978-3-642-66853-1_5. [DOI] [PubMed] [Google Scholar]
- 12.Chattopadhyay SK, Lander MR, Rowe WP. 1980. Close similarity between endogenous ecotropic virus of Mus musculus molossinus and AKR virus. J Virol 36:499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartley JW, Rowe WP. 1976. Naturally occurring murine leukemia viruses in wild mice: characterization of a new “amphotropic” class. J Virol 19:19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rasheed S, Gardner MB, Chan E. 1976. Amphotropic host range of naturally occurring wild mouse leukemia viruses. J Virol 19:13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller DG, Edwards RH, Miller AD. 1994. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc Natl Acad Sci U S A 91:78–82. doi: 10.1073/pnas.91.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voytek P, Kozak C. 1988. HoMuLV: a novel pathogenic ecotropic virus isolated from the European mouse, Mus hortulanus. Virology 165:469–475. doi: 10.1016/0042-6822(88)90590-9. [DOI] [PubMed] [Google Scholar]
- 17.Stoye JP, Coffin JM. 1987. The four classes of endogenous murine leukemia virus: structural relationships and potential for recombination. J Virol 61:2659–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenkins NA, Copeland NG, Taylor BA, Lee BK. 1982. Organization, distribution, and stability of endogenous ecotropic murine leukemia virus DNA sequences in chromosomes of Mus musculus. J Virol 43:26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoye JP, Moroni C, Coffin JM. 1991. Virological events leading to spontaneous AKR thymomas. J Virol 65:1273–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chattopadhyay SK, Cloyd MW, Linemeyer DL, Lander MR, Rands E, Lowy DR. 1982. Cellular origin and role of mink cell focus-forming viruses in murine thymic lymphomas. Nature 295:25–31. doi: 10.1038/295025a0. [DOI] [PubMed] [Google Scholar]
- 21.Evans LH, Cloyd MW. 1985. Friend and Moloney murine leukemia viruses specifically recombine with different endogenous retroviral sequences to generate mink cell focus-forming viruses. Proc Natl Acad Sci U S A 82:459–463. doi: 10.1073/pnas.82.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozak CA, O'Neill RR. 1987. Diverse wild mouse origins of xenotropic, mink cell focus-forming, and two types of ecotropic proviral genes. J Virol 61:3082–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bamunusinghe D, Liu Q, Lu X, Oler A, Kozak CA. 2013. Endogenous gammaretrovirus acquisition in Mus musculus subspecies carrying functional variants of the XPR1 virus receptor. J Virol 87:9845–9855. doi: 10.1128/JVI.01264-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozak CA, Hartley JW, Morse HC 3rd. 1984. Laboratory and wild-derived mice with multiple loci for production of xenotropic murine leukemia virus. J Virol 51:77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jung YT, Wu T, Kozak CA. 2003. Characterization of recombinant nonecotropic murine leukemia viruses from the wild mouse species Mus spretus. J Virol 77:12773–12781. doi: 10.1128/JVI.77.23.12773-12781.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inaguma Y, Miyashita N, Moriwaki K, Huai WC, Jin ML, He XQ, Ikeda H. 1991. Acquisition of two endogenous ecotropic murine leukemia viruses in distinct Asian wild mouse populations. J Virol 65:1796–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Neill RR, Hartley JW, Repaske R, Kozak CA. 1987. Amphotropic proviral envelope sequences are absent from the Mus germ line. J Virol 61:2225–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voytek P, Kozak CA. 1989. Nucleotide sequence and mode of transmission of the wild mouse ecotropic virus, HoMuLV. Virology 173:58–67. doi: 10.1016/0042-6822(89)90221-3. [DOI] [PubMed] [Google Scholar]
- 29.Orth A, Adama T, Din W, Bonhomme F. 1998. Natural hybridization between two subspecies of the house mouse, Mus musculus domesticus and Mus musculus castaneus, near Lake Casitas, California. Genome 41:104–110. (In French.) doi: 10.1139/g97-109. [DOI] [PubMed] [Google Scholar]
- 30.Ikeda H, Kato K, Kitani H, Suzuki T, Yoshida T, Inaguma Y, Yamamoto N, Suh JG, Hyun BH, Yamagata T, Namikawa T, Tomita T. 2001. Virological properties and nucleotide sequences of Cas-E-type endogenous ecotropic murine leukemia viruses in south Asian wild mice, Mus musculus castaneus. J Virol 75:5049–5058. doi: 10.1128/JVI.75.11.5049-5058.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Howard TM, Sheng Z, Wang M, Wu Y, Rasheed S. 2006. Molecular and phylogenetic analyses of a new amphotropic murine leukemia virus (MuLV-1313). Virol J 3:101. doi: 10.1186/1743-422X-3-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perryman SM, McAtee FJ, Portis JL. 1991. Complete nucleotide sequence of the neurotropic murine retrovirus CAS-BR-E. Nucleic Acids Res 19:1707. doi: 10.1093/nar/19.7.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan Y, Knoper RC, Kozak CA. 2007. Wild mouse variants of envelope genes of xenotropic/polytropic mouse gammaretroviruses and their XPR1 receptors elucidate receptor determinants of virus entry. J Virol 81:10550–10557. doi: 10.1128/JVI.00933-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan Y, Liu Q, Kozak CA. 2009. Six host range variants of the xenotropic/polytropic gammaretroviruses define determinants for entry in the XPR1 cell surface receptor. Retrovirology 6:87. doi: 10.1186/1742-4690-6-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cloyd MW, Thompson MM, Hartley JW. 1985. Host range of mink cell focus-inducing viruses. Virology 140:239–248. doi: 10.1016/0042-6822(85)90362-9. [DOI] [PubMed] [Google Scholar]
- 36.Lowy DR, Rowe WP, Teich N, Hartley JW. 1971. Murine leukemia virus: high-frequency activation in vitro by 5-iododeoxyuridine and 5-bromodeoxyuridine. Science 174:155–156. doi: 10.1126/science.174.4005.155. [DOI] [PubMed] [Google Scholar]
- 37.Hartley JW, Rowe WP. 1975. Clonal cell lines from a feral mouse embryo which lack host-range restrictions for murine leukemia viruses. Virology 65:128–134. doi: 10.1016/0042-6822(75)90013-6. [DOI] [PubMed] [Google Scholar]
- 38.Lander MR, Chattopadhyay SK. 1984. A Mus dunni cell line that lacks sequences closely related to endogenous murine leukemia viruses and can be infected by ecotropic, amphotropic, xenotropic, and mink cell focus-forming viruses. J Virol 52:695–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rose PP, Korber BT. 2000. Detecting hypermutations in viral sequences with an emphasis on G → A hypermutation. Bioinformatics 16:400–401. doi: 10.1093/bioinformatics/16.4.400. [DOI] [PubMed] [Google Scholar]
- 40.Lamont C, Culp P, Talbott RL, Phillips TR, Trauger RJ, Frankel WN, Wilson MC, Coffin JM, Elder JH. 1991. Characterization of endogenous and recombinant proviral elements of a highly tumorigenic AKR cell line. J Virol 65:4619–4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang JR, de Villena FP, McMillan L. 2012. Comparative analysis and visualization of multiple collinear genomes. BMC Bioinformatics 13(Suppl 3):S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res 12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H. 2014. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics 30:2843–2851. doi: 10.1093/bioinformatics/btu356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, Furlotte NA, Eskin E, Nellåker C, Whitley H, Cleak J, Janowitz D, Hernandez-Pliego P, Edwards A, Belgard TG, Oliver PL, McIntyre RE, Bhomra A, Nicod J, Gan X, Yuan W, van der Weyden L, Steward CA, Bala S, Stalker J, Mott R, Durbin R, Jackson IJ, Czechanski A, Guerra-Assunção JA, Donahue LR, Reinholdt LG, Payseur BA, Ponting CP, Birney E, Flint J, Adams DJ. 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res 12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu SM, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam TW, Wang J. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 51.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. [DOI] [PubMed] [Google Scholar]
- 52.Frankel WN, Stoye JP, Taylor BA, Coffin JM. 1990. A linkage map of endogenous murine leukemia proviruses. Genetics 124:221–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jern P, Stoye JP, Coffin JM. 2007. Role of APOBEC3 in genetic diversity among endogenous murine leukemia viruses. PLoS Genet 3:e183. doi: 10.1371/journal.pgen.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kozak C, Rowe WP. 1978. Genetic mapping of xenotropic leukemia virus-inducing loci in two mouse strains. Science 199:1448–1449. doi: 10.1126/science.204014. [DOI] [PubMed] [Google Scholar]
- 55.Baliji S, Liu Q, Kozak CA. 2010. Common inbred strains of the laboratory mouse that are susceptible to infection by mouse xenotropic gammaretroviruses and the human-derived retrovirus XMRV. J Virol 84:12841–12849. doi: 10.1128/JVI.01863-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frankel WN, Stoye JP, Taylor BA, Coffin JM. 1989. Genetic analysis of endogenous xenotropic murine leukemia viruses: association with two common mouse mutations and the viral restriction locus Fv-1. J Virol 63:1763–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yan Y, Liu Q, Wollenberg K, Martin C, Buckler-White A, Kozak CA. 2010. Evolution of functional and sequence variants of the mammalian XPR1 receptor for mouse xenotropic gammaretroviruses and the human-derived XMRV. J Virol 84:11970–11980. doi: 10.1128/JVI.01549-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pothlichet J, Mangeney M, Heidmann T. 2006. Mobility and integration sites of a murine C57BL/6 melanoma endogenous retrovirus involved in tumor progression in vivo. Int J Cancer 119:1869–1877. doi: 10.1002/ijc.22066. [DOI] [PubMed] [Google Scholar]
- 59.Eicher EM, Hutchison KW, Phillips SJ, Tucker PK, Lee BK. 1989. A repeated segment on the mouse Y chromosome is composed of retroviral-related, Y-enriched and Y-specific sequences. Genetics 122:181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang H, Bell TA, Churchill GA, Pardo-Manuel de Villena F. 2007. On the subspecific origin of the laboratory mouse. Nat Genet 39:1100–1107. doi: 10.1038/ng2087. [DOI] [PubMed] [Google Scholar]
- 61.Kozak CA. 2010. The mouse “xenotropic” gammaretroviruses and their XPR1 receptor. Retrovirology 7:101. doi: 10.1186/1742-4690-7-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paprotka T, Delviks-Frankenberry KA, Cingoz O, Martinez A, Kung HJ, Tepper CG, Hu WS, Fivash MJ Jr, Coffin JM, Pathak VK. 2011. Recombinant origin of the retrovirus XMRV. Science 333:97–101. doi: 10.1126/science.1205292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dong B, Kim S, Hong S, Das Gupta J, Malathi K, Klein EA, Ganem D, DeRisi JL, Chow SA, Silverman RH. 2007. An infectious retrovirus susceptible to an IFN antiviral pathway from human prostate tumors. Proc Natl Acad Sci U S A 104:1655–1660. doi: 10.1073/pnas.0610291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang YA, Maitra A, Hsieh JT, Rudin CM, Peacock CD, Karikari C, Brekken RA, Stastny V, Gao B, Girard L, Wistuba I, Frenkel E, Minna JD, Gazdar AF. 2011. Frequent detection of infectious xenotropic murine leukemia virus (XMLV) in human cultures established from mouse xenografts. Cancer Biol Ther 12:617–628. doi: 10.4161/cbt.12.7.15955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sfanos KS, Aloia AL, Hicks JL, Esopi DM, Steranka JP, Shao W, Sanchez-Martinez S, Yegnasubramanian S, Burns KH, Rein A, De Marzo AM. 2011. Identification of replication competent murine gammaretroviruses in commonly used prostate cancer cell lines. PLoS One 6:e20874. doi: 10.1371/journal.pone.0020874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Courgnaud V, Battini JL, Sitbon M, Mason AL. 2010. Mouse retroviruses and chronic fatigue syndrome: does X (or P) mark the spot? Proc Natl Acad Sci U S A 107:15666–15667. doi: 10.1073/pnas.1007944107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Raisch KP, Pizzato M, Sun HY, Takeuchi Y, Cashdollar LW, Grossberg SE. 2003. Molecular cloning, complete sequence, and biological characterization of a xenotropic murine leukemia virus constitutively released from the human B-lymphoblastoid cell line DG-75. Virology 308:83–91. doi: 10.1016/S0042-6822(02)00074-0. [DOI] [PubMed] [Google Scholar]
- 68.Datta SK, Schwartz RS. 1977. Mendelian segregation of loci controlling xenotropic virus production in NZB crosses. Virology 83:449–452. doi: 10.1016/0042-6822(77)90193-3. [DOI] [PubMed] [Google Scholar]
- 69.Elder JH, Gautsch JW, Jensen FC, Lerner RA, Chused TM, Morse HC, Hartley JW, Rowe WP. 1980. Differential expression of two distinct xenotropic viruses in NZB mice. Clin Immunol Immunopathol 15:493–501. doi: 10.1016/0090-1229(80)90061-6. [DOI] [PubMed] [Google Scholar]
- 70.Tomonaga K, Coffin JM. 1999. Structures of endogenous nonecotropic murine leukemia virus (MLV) long terminal repeats in wild mice: implication for evolution of MLVs. J Virol 73:4327–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ikeda H, Laigret F, Martin MA, Repaske R. 1985. Characterization of a molecularly cloned retroviral sequence associated with Fv-4 resistance. J Virol 55:768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koch W, Hunsmann G, Friedrich R. 1983. Nucleotide sequence of the envelope gene of Friend murine leukemia virus. J Virol 45:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ott D, Friedrich R, Rein A. 1990. Sequence analysis of amphotropic and 10A1 murine leukemia viruses: close relationship to mink cell focus-inducing viruses. J Virol 64:757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gray KD, Roth MJ. 1993. Mutational analysis of the envelope gene of Moloney murine leukemia virus. J Virol 67:3489–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lavillette D, Maurice M, Roche C, Russell SJ, Sitbon M, Cosset FL. 1998. A proline-rich motif downstream of the receptor binding domain modulates conformation and fusogenicity of murine retroviral envelopes. J Virol 72:9955–9965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fontenot JD, Tjandra N, Ho C, Andrews PC, Montelaro RC. 1994. Structure and self assembly of a retrovirus (FeLV) proline rich neutralization domain. J Biomol Struct Dyn 11:821–836. doi: 10.1080/07391102.1994.10508035. [DOI] [PubMed] [Google Scholar]
- 77.Valsesia-Wittmann S, Morling FJ, Hatziioannou T, Russell SJ, Cosset FL. 1997. Receptor co-operation in retrovirus entry: recruitment of an auxiliary entry mechanism after retargeted binding. EMBO J 16:1214–1223. doi: 10.1093/emboj/16.6.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weimin Wu B, Cannon PM, Gordon EM, Hall FL, Anderson WF. 1998. Characterization of the proline-rich region of murine leukemia virus envelope protein. J Virol 72:5383–5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Argaw T, Wilson CA. 2012. Detailed mapping of determinants within the porcine endogenous retrovirus envelope surface unit identifies critical residues for human cell infection within the proline-rich region. J Virol 86:9096–9104. doi: 10.1128/JVI.00738-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Triviai I, Ziegler M, Bergholz U, Oler AJ, Stubig T, Prassolov V, Fehse B, Kozak CA, Kroger N, Stocking C. 2014. Endogenous retrovirus induces leukemia in a xenograft mouse model for primary myelofibrosis. Proc Natl Acad Sci U S A 111:8595–8600. doi: 10.1073/pnas.1401215111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim FJ, Battini JL, Manel N, Sitbon M. 2004. Emergence of vertebrate retroviruses and envelope capture. Virology 318:183–191. doi: 10.1016/j.virol.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 82.Malik HS, Henikoff S, Eickbush TH. 2000. Poised for contagion: evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res 10:1307–1318. doi: 10.1101/gr.145000. [DOI] [PubMed] [Google Scholar]
- 83.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.