ABSTRACT
Human cytomegalovirus (HCMV) counteracts host defenses that otherwise act to limit viral protein synthesis. One such defense is the antiviral kinase protein kinase R (PKR), which inactivates the eukaryotic initiation factor 2 (eIF2) translation initiation factor upon binding to viral double-stranded RNAs. Previously, the viral TRS1 and IRS1 proteins were found to antagonize the antiviral kinase PKR outside the context of HCMV infection, and the expression of either pTRS1 or pIRS1 was shown to be necessary for HCMV replication. In this study, we found that expression of either pTRS1 or pIRS1 is necessary to prevent PKR activation during HCMV infection and that antagonism of PKR is critical for efficient viral replication. Consistent with a previous study, we observed decreased overall levels of protein synthesis, reduced viral protein expression, and diminished virus replication in the absence of both pTRS1 and pIRS1. In addition, both PKR and eIF2α were phosphorylated during infection when pTRS1 and pIRS1 were absent. We also found that expression of pTRS1 was both necessary and sufficient to prevent stress granule formation in response to eIF2α phosphorylation. Depletion of PKR prevented eIF2α phosphorylation, rescued HCMV replication and protein synthesis, and reversed the accumulation of stress granules in infected cells. Infection with an HCMV mutant lacking the pTRS1 PKR binding domain resulted in PKR activation, suggesting that pTRS1 inhibits PKR through a direct interaction. Together our results show that antagonism of PKR by HCMV pTRS1 and pIRS1 is critical for viral protein expression and efficient HCMV replication.
IMPORTANCE To successfully replicate, viruses must counteract host defenses that limit viral protein synthesis. We have identified inhibition of the antiviral kinase PKR by the viral proteins TRS1 and IRS1 and shown that this is a critical step in HCMV replication. Our results suggest that inhibiting pTRS1 and pIRS1 function or restoring PKR activity during infection may be a successful strategy to limit HCMV disease.
INTRODUCTION
Human cytomegalovirus (HCMV), like all viruses, requires host ribosomes and translation factors for the synthesis of viral proteins. Consequently, upon sensing infection, host antiviral defenses inactivate critical translation factors, leading to reduced viral replication. To circumvent these defenses, HCMV manipulates antiviral signaling pathways to allow for efficient viral protein synthesis. Thus, the interface of HCMV with the host translation machinery lies at the front line of the battle between host and virus for control of the infected cell.
Perhaps the best-studied antiviral defense targeting viral mRNA translation is the RNA-dependent protein kinase R (PKR). PKR binds to double-stranded RNAs (dsRNAs) produced during viral infections, resulting in PKR dimerization and activating autophosphorylation (1–4). Activated PKR in turn inhibits mRNA translation by phosphorylating its substrate the eukaryotic initiation factor 2 alpha (eIF2α) (5–8). eIF2α plays a critical role in translation initiation as a regulatory subunit of the trimeric eIF2 complex, which mediates binding of the ternary complex, consisting of eIF2, GTP, and tRNAMet, to the ribosome (9). eIF2α phosphorylation by PKR prevents recycling of the ternary complex after initiation, resulting in an overall decrease in translation initiation and diminished viral protein synthesis and replication (10).
Phosphorylation of eIF2α further limits protein synthesis by sequestering actively translating mRNAs into cytoplasmic ribonucleoprotein complexes called stress granules (11). During viral infection, stress granules are most often induced by activated PKR; however, additional virus-induced stressors such as the accumulation of unfolded proteins and/or nutrient depletion are also involved (12). Prolonged periods of stress lead to the degradation of stress granule-associated mRNAs, which further inhibits viral protein expression (13). Despite the induction of stress response pathways known to trigger stress granule formation, stress granules do not form in HCMV-infected cells (14, 15). This suggests that HCMV encodes viral proteins that inhibit stress granule formation. However, a role for HCMV proteins in the inhibition of stress granule formation has not been described.
Many viruses generate dsRNA ligands recognized by PKR during infection, and thus viruses commonly encode PKR antagonists. Human cytomegalovirus encodes two PKR antagonists, the TRS1 and IRS1 proteins (pTRS1 and pIRS1, respectively). The amino-terminal 550 amino acids of pTRS1 and pIRS1 are encoded by the short-repeat regions of the viral genome and are therefore identical, while the remainder of pTRS1 and pIRS1 are encoded by the unique short segment of the genome and thus diverge. However, the unique regions of pTRS1 and pIRS1 are highly similar, sharing approximately 50% amino acid conservation. Both proteins limit PKR activation outside the context of HCMV infection (16), and the expression of either pTRS1 or pIRS1 is necessary for HCMV replication (17). Several functional domains have been identified in pTRS1 and pIRS1, including an RNA binding domain between amino acids 86 and 246 (18) and a PKR binding domain in the unique carboxyl terminus (19). While both domains are necessary for PKR antagonism in heterologous systems and in vitro, the contribution of each domain to PKR inhibition during HCMV infection is currently unknown.
Efforts to study pTRS1 functions in the context of HCMV infection have been complicated by the functional redundancy of pIRS1. Expression of either protein is sufficient to support HCMV replication at high multiplicities of infection (MOI) (17, 20), although a TRS1 deletion virus is attenuated after infection at low MOI (20). However, deletion of both TRS1 and IRS1 results in a replication-deficient virus. As a result, the majority of studies defining functional roles for pTRS1 and pIRS1 have been performed in vitro or in heterologous systems outside the context of HCMV infection. Both pTRS1 and pIRS1 complement the growth defect of vaccinia virus mutants lacking the E3L protein (16, 18, 21–24), a known PKR inhibitor, and pTRS1 complements the growth of herpes simplex virus (HSV) mutants lacking the ICP34.5 protein (25), which stimulates eIF2α dephosphorylation through the recruitment of a cellular phosphatase (26, 27). Similarly, pTRS1 complements the replication of murine cytomegalovirus (MCMV) strains lacking the PKR antagonists m142 or m143 (28, 29). While these studies show that pTRS1 and pIRS1 act as PKR antagonists, the role of PKR in suppressing HCMV replication has not been defined. In addition, the requirement for pTRS1 and pIRS1 to prevent PKR activation during HCMV infection has not been described.
A previous study by Marshall et al. (17) generated an HCMV mutant with TRS1 and IRS1 deleted and found that in the absence of both pTRS1 and pIRS1, eIF2α is phosphorylated and HCMV protein synthesis and replication are dramatically inhibited. However, this study did not measure PKR activation in the absence of both pTRS1 and pIRS1, nor was it determined if depleting PKR could prevent eIF2α phosphorylation or restore viral protein synthesis and replication. Fibroblasts express three additional eIF2α kinases, GCN2, HRI, and PERK, which phosphorylate eIF2α in response to amino acid stress, reactive oxygen species (ROS), and activation of the unfolded protein response (UPR), respectively (30, 31). Several studies show that HCMV induces ROS (32–34) and activates PERK as part of the virus-induced UPR (35, 36). In addition, HCMV potentially induces amino acid stress due to high levels of ongoing host and viral protein synthesis. Consistent with this idea, mice lacking GCN2 are more susceptible to infection with the related murine cytomegalovirus (MCMV) (37). The role of these additional eIF2α kinases in HCMV replication is unknown, as is the ability of pTRS1 to regulate their activity. In addition, pTRS1 and pIRS1 have other functions potentially unrelated to PKR antagonism, including suppression of autophagy (38, 39), transactivation of gene expression (40), and packaging of viral DNA into capsids (41). pTRS1 also interacts with the HCMV DNA polymerase accessory subunit and promotes ori-Lyt-dependent DNA replication through an unknown mechanism (42–46). How these functions contribute to the replication defect observed in the absence of pTRS1 and pIRS1 has not been tested.
In this study, we used a combination of short hairpin RNAs (shRNAs) and viral mutants to evaluate the role of antagonism of PKR by pTRS1 and pIRS1 during HCMV infection. Our results confirm previous studies showing that either IRS1 or TRS1 is sufficient for HCMV replication (17). We found that expression of either pTRS1 or pIRS1 is both necessary and sufficient to suppress PKR activation and eIF2α phosphorylation and maintain overall levels of protein synthesis in HCMV-infected cells. In addition, we found that expression of either pTRS1 or pIRS1 is necessary to prevent stress granule formation during HCMV infection and that pTRS1 alone is sufficient to prevent stress granule formation in transfected cells. Using a pTRS1 mutant lacking a portion of the unique carboxyl terminus, we found that the pTRS1 PKR binding domain is necessary to prevent PKR activation during HCMV infection. Depletion of PKR restored virus replication and viral protein synthesis in the absence of pTRS1 and pIRS1 and prevented stress granule formation. Thus, our findings demonstrate that antagonism of PKR by pTRS1 and pIRS1 is critical for efficient HCMV protein expression and replication.
MATERIALS AND METHODS
Viruses, cells, and reagents.
MRC-5 fibroblasts, human foreskin fibroblasts (HFFs), and HeLa cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 U/ml penicillin-streptomycin. MRC-5 fibroblasts and HFFs were used between passages 8 and 20. HCMV strain AD169 lacking the TRS1 open reading frame (ORF) was a generous gift of Adam Geballe (University of Washington) and has been described previously (17). All mutations in the HCMV genome were generated in the AD169BADinGFP bacterial artificial chromosome (BAC) using recombineering in Escherichia coli strain DY380 as previously described (47, 48). To generate the PKR binding domain deletion mutant (HCMVΔPBD), a FLAG-FRT-KAN-FRT cassette was PCR amplified using primers FRT FWD (5′ TTAGATTATAAAGATGATGATGATAAA 3′) and PBDFRT REV (5′ TCCCCGGAGAAGATTCCTGGTACGACTTGGACGAGACTTTCTGGGTTCTTTTAGATTATAAAGATGATGATGATAAA 3′) containing 50 nucleotides of homology flanking the insertion site at nucleotide 228885 (corresponding to amino acid 679; GenBank accession number FJ527563.1). Following recombination, expression of the Flp recombinase was induced with arabinose to excise the kanamycin cassette. The resulting recombinant contains a FLAG epitope tag followed by a translation stop codon in frame with the 3′ end of the TRS1 ORF. The gross integrity of the recombinant viral genomes was confirmed by restriction digest, and the sequences of the entire TRS1 open reading frame and the flanking 500 nucleotides were confirmed by Sanger sequencing. For each mutant, two independent recombinants were isolated, sequenced, and characterized to control for potential spurious mutations introduced during recombination. BAC DNA was purified using the Nucleobond Midiprep kit (Macherey-Nagel) according to the manufacturer's directions. One microgram of BAC DNA together with one microgram of pCGN pp71 (49) was electroporated into MRC-5 fibroblasts to reconstitute infectious virus. Viral stocks were propagated on MRC-5 fibroblasts, and their titers were determined using the tissue culture infective dose (TCID50) method. Unless otherwise noted, all infections were performed at a multiplicity of infection of 1 in 300 μl of medium. The amount of cell-free virus in the supernatant was quantified by the TCID50 method on MRC-5 fibroblasts as previously described (50).
The plasmid pcDNA-pTRS1 expressing the TRS1 protein with a carboxyl-terminal 6× His epitope tag was a kind gift of Adam Geballe (18). Cells were transfected with the indicated plasmids using polyethyleneimine (PEI) and analyzed at 24 h after transfection unless otherwise noted.
siRNA- and shRNA-mediated depletion.
Small interfering RNA (siRNA) targeting the PKR transcript (Dharmacon SMARTpool ON-TARGETplus EIF2AK2 siRNA) was transfected at a final concentration of 20 nM in 100 μl Opti-MEM using 6 μl Mission siRNA transfection reagent (Sigma). Complexes were incubated at room temperature for 15 min prior to dropwise addition to 30 to 50% confluent MRC-5 cells. Cells were used in subsequent experiments at 72 h after transfection. Efficient knockdown was routinely monitored by Western blotting. MRC-5 cells stably expressing scrambled or IRS1-specific shRNAs were generated using the pSUPERretro system (Oligoengine). An shRNA hairpin (5′ GGAGTTCATGTTTCGCGAACATTCAAGAGATGTTCGCGAAACATGAACTCC 3′), which generates the mature shRNA (5′ GGAGTTCATGTTTCGCGAACA 3′) targeting the unique 3′ end of the IRS1 ORF was cloned into the pSUPERretro vector. Retrovirus stocks were generated by transfecting the Phoenix packaging cell line (51) with the pSUPERretro plasmids. Supernatants were collected at 48 h posttransfection, filtered through a 0.45-μm filter, and used to transduce cells in the presence of Polybrene (4 μg/ml). Stable cell lines were obtained by selection in puromycin (1 μg/ml) for at least 1 week prior to use.
Western blot analysis.
Cells were collected by scraping at the indicated times and frozen as dry pellets at −80°C until use. Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 0.25% deoxycholate, 1 mM EDTA) containing protease and phosphatase inhibitors (Roche), and the protein concentration was determined by the Bradford assay (Amresco). Equivalent amounts of protein were resolved on SDS-PAGE gels and transferred to nitrocellulose membranes (Amersham). For monoclonal antibodies, membranes were blocked for 1 h at room temperature in 1% bovine serum albumin (BSA) in TBS-T (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.1% Tween 20) before incubation with primary antibody in TBS-T with 1% BSA for 1 h at room temperature or overnight at 4°C. For polyclonal antibodies, membranes were blocked in 1% BSA followed by incubation with primary antibody overnight at 4°C in 5% BSA in TBS-T. For Western blots using the phospho-eIF2α (Ser51) antibody, membranes were blocked in 5% BSA in TBS-T overnight at 4°C, followed by overnight incubation at 4°C with primary antibody diluted in TBS-T containing 5% BSA. Antibodies to the following proteins were used in this study: IE1 (52) (1:10,000), pUL44 (1:1,000; Virusys), pp28 (53) (1:5,000), pTRS1 (40) (1:100), pIRS1 (40) (1:100), P99 antibody against pTRS1 and pIRS1 (17) (1:10,000), tubulin (1:50,000; Sigma T6199), total PKR (1:1,000; Santa Cruz sc-707), phospho-PKR (Thr446) (1:1,000; Abcam ab32036), total eIF2α (1:1,000; catalog no. 3072; Cell Signaling), and phospho-eIF2α (Ser51) (1:1,000; catalog no. 3398; Cell Signaling).
Indirect immunofluorescence.
Cells were seeded into wells containing glass coverslips. Where noted, cells were infected at a multiplicity of infection of 0.5. At the time of harvest, coverslips were washed three times for 5 min with 37°C phosphate-buffered saline (PBS) and then fixed in 2% paraformaldehyde (PFA) for 15 min at 37°C. The cells were washed three times in room temperature PBS and subsequently permeabilized with 0.1% Triton X-100. The cells were again washed three times with PBS containing 0.2% Tween 20 (PBS-T) at room temperature and then incubated overnight in blocking buffer (2% BSA in TBS-T). The cells were stained for 1 h at room temperature with the following primary antibodies diluted in blocking buffer: IE1 (1:10); pTRS1 (1:10); G3BP1 (Santa Cruz sc-98561; 1:50). The coverslips were washed three times with PBS-T and then incubated with fluorescent secondary antibody (Invitrogen goat anti-mouse or rabbit; 1:500). The coverslips were again washed three times, mounted on glass slides in VectaShield containing 4′,6-diamidino-2-phenylindole (DAPI) to stain nuclei (Vector Laboratories), and sealed with nail polish. Images were captured using a Zeiss 710 confocal microscope with the help of the UNC Microscopy Core.
Quantification of stress granule formation.
Control or PKR-deficient HeLa cells were transfected with either a green fluorescent protein (GFP) or pTRS1 expression vector. Twenty-four hours after transfection, cells were treated with arsenite for 1 h and then processed for immunofluorescence microscopy as above. Transfected cells (i.e., GFP or pTRS1 positive) were identified, and the relative fluorescence intensity of the GFP or pTRS1 signal in each cell was quantified using ImageJ software. The relative pTRS1 or GFP expression level in each cell was scored on a scale of 1 to 100, with 100 representing the highest observed expression. The presence of stress granules in each transfected cell was then determined by measuring the formation of G3BP1 puncta. Transfected cells containing two or more G3BP1 puncta were considered positive for stress granules, while those with fewer than two puncta were considered negative. The range of fluorescence intensity for GFP and pTRS1 was divided into quartiles (1 to 25% of maximum, 26 to 50% of maximum, etc.), and the percentage of cells containing stress granules within each expression quartile was calculated. A minimum of 100 GFP- or pTRS1-positive cells were analyzed. The data are displayed as the percent transfected cells containing stress granules within each quartile of GFP or pTRS1 expression.
Metabolic labeling of nascent proteins.
Nascent proteins were metabolically labeled and quantified as described previously (54). Briefly, cells were incubated in methionine- and cysteine-free medium (Sigma) for 15 min. 35S-labeled methionine and cysteine (125 μCi; PerkinElmer EasyTag Express Labeling Mix) were added and allowed to incorporate for 30 min. Cells were then washed twice in ice-cold PBS, scraped, and collected by centrifugation. Cell pellets were lysed in RIPA medium containing protease inhibitors (Roche), and protein concentrations were determined by the Bradford assay (Amresco). Trichloroacetic acid (TCA) was added to a final concentration of 20%, and precipitated proteins were captured on glass microfiber filters by filtration under vacuum. The filters were washed twice with 20% TCA and once with 100% ethanol and allowed to air dry. The filters were then transferred to vials containing scintillation fluid (EcoScint), and radioactivity was quantified using a scintillation counter. The amount of radioactivity was normalized to the protein concentration for each sample.
RESULTS
HCMV mutant strains lacking either pTRS1 or pIRS1 replicate to levels similar to those of wild-type virus (20, 55), while a mutant lacking both pTRS1 and pIRS1 replicates poorly (17). Previous studies of a pTRS1/pIRS1 double knockout virus were complicated by the inability to efficiently complement virus replication by expression of pTRS1 in trans (17). This likely reflected insufficient expression of the pTRS1 transgene or improper timing of expression. We reasoned that the failure of the pTRS1 transgene to fully complement the replication of a pTRS1/pIRS1 double deletion virus would limit our ability to generate and study recombinant viruses lacking domains of pTRS1 required for virus replication. Thus, we sought to develop a system that allowed us to control pIRS1 expression and express pTRS1 mutants to wild-type levels with wild-type kinetics from the context of the viral genome. We therefore used a combination of viral genetics and shRNA-mediated gene silencing to manipulate pTRS1 and pIRS1 expression during HCMV infection. We first generated primary human fibroblasts expressing either a scrambled shRNA (control cells) or an shRNA specific for the pIRS1 transcript (shIRS1-HFs). Infecting the cells with wild-type virus allowed us to study the contribution of pTRS1 to HCMV infection in the absence of pIRS1. Conversely, infection of control cells with a pTRS1 deletion virus (HCMVΔTRS1, described in reference 17; kindly provided by Adam Geballe) provided a system to study specific roles for pIRS1, while infection of shIRS1-HFs with HCMVΔTRS1 allowed us to determine how the absence of both proteins affected HCMV replication. Importantly, in this system both pTRS1 and pIRS1 were expressed from their native location in the viral genome under the control of their endogenous promoters, allowing wild-type kinetics and expression levels of the pTRS1 and pIRS1 proteins.
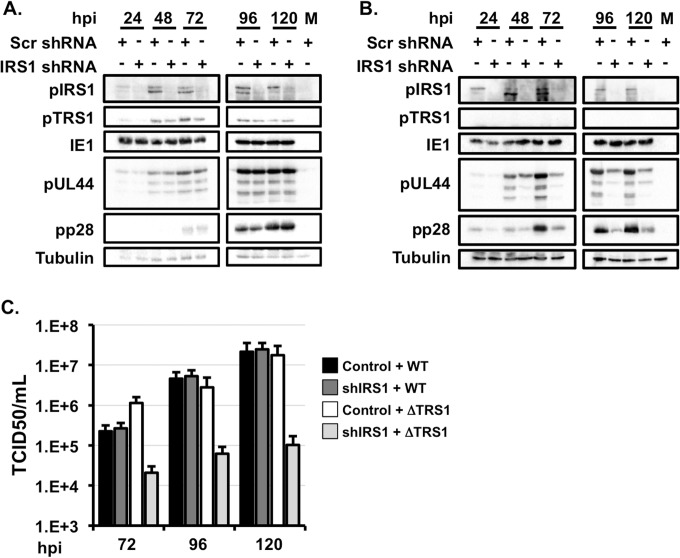
Infection of shIRS1-HFs with wild-type HCMV resulted in a significant reduction in pIRS1 expression at all times after infection (Fig. 1A). Importantly, pTRS1 expression was not affected by the IRS1-specific shRNA. Consistent with previous reports showing that pIRS1 is dispensable for HCMV replication (20), wild-type virus replicated to equivalent titers in the two cell types despite efficient depletion of pIRS1 in shIRS1-HFs (Fig. 1C). These data also demonstrate that the IRS1-specific shRNA did not target additional host or viral proteins necessary for HCMV replication. Both wild-type and HCMVΔTRS1 replicated to similar levels following infection of control cells. However, HCMVΔTRS1 infection on shIRS1-HFs reduced viral replication by >2 orders of magnitude (Fig. 1C). These data demonstrate that our system confirms previous results showing that expression of either pTRS1 or pIRS1 is necessary for efficient HCMV replication.
FIG 1.
Expression of either pTRS1or pIRS1 is necessary for efficient HCMV replication. (A) Control cells (Scr) or shIRS1-HFs were infected with wild-type HCMV at an MOI of 3. Cells were harvested at the indicated times and analyzed by Western blotting (n = 3). (B) Control cells and shIRS1-HFs were infected with HCMV lacking TRS1 (HCMVΔTRS1) at an MOI of 3. Cells were harvested at the indicated times and analyzed by Western blotting (n = 3). (C) Cells were infected as described for panel A, and the amount of virus in cell free supernatants was measured by the TCID50 method (n = 3).
We next examined viral protein expression after infection of control HFs or shIRS1-HFs with wild-type or HCMVΔTRS1 virus. Similar levels of representative immediate early (IE), early, and late proteins were expressed after infection of control cells or shIRS1-HFs with wild-type virus. Viral proteins of each kinetic class were also efficiently expressed after infection of control cells with HCMVΔTRS1 virus (Fig. 1A and B); however, HCMV early and late protein expression was reduced after infection of shIRS1-HFs with the HCMVΔTRS1 virus (Fig. 1B). While the immediate early protein IE1 was expressed equivalently in shIRS1-HFs and control cells, we observed decreased expression of the early protein pUL44 and the late protein pp28 after infection of shIRS1-HFs with HCMVΔTRS1 (Fig. 1B). Consistent with previous results (17), we conclude that the expression of either pTRS1 or pIRS1 is necessary for the efficient expression of HCMV early and late proteins.
pTRS1 or pIRS1 prevents PKR activation during HCMV infection.
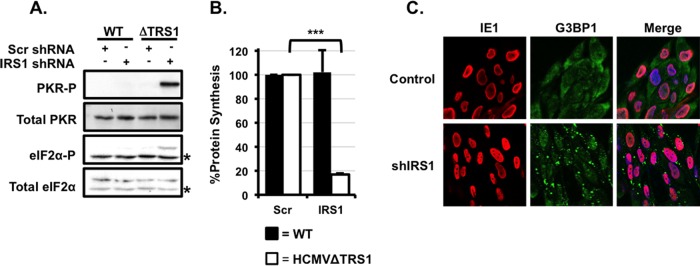
pTRS1 and pIRS1 inhibit the antiviral kinase PKR in the context of vaccinia virus infection and in transfected cells (16, 24, 56). To determine if pTRS1 and pIRS1 similarly limit PKR activation during HCMV infection, we measured PKR autophosphorylation and phosphorylation of the PKR substrate eIF2α after infection of control cells or shIRS1-HFs with wild-type or HCMVΔTRS1 virus. Infection with wild-type virus did not induce PKR or eIF2α phosphorylation in either cell type (Fig. 2A), demonstrating that the expression of pTRS1 is sufficient to limit PKR activation. Similarly, neither PKR nor eIF2α was phosphorylated in control cells infected with HCMVΔTRS1, showing that pIRS1 expression is sufficient to suppress PKR. However, infection of shIRS1-HFs with HCMVΔTRS1 resulted in robust PKR and eIF2α phosphorylation (Fig. 2A). Consistent with the inhibition of protein synthesis induced by eIF2α phosphorylation, we observed an 80% decrease in the overall levels of protein synthesis when shIRS1-HFs were infected with HCMVΔTRS1 (Fig. 2B). These results demonstrate that the expression of either pTRS1 or pIRS1 is necessary to antagonize PKR activation, limit eIF2α phosphorylation, and maintain protein synthesis in HCMV-infected cells.
FIG 2.
HCMV pTRS1 or pIRS1 is necessary to antagonize PKR, maintain infected-cell protein synthesis, and inhibit stress granule formation. (A) Control cells (Scr) or shIRS1-HFs (shIRS1) were infected with either wild-type virus or HCMVΔTRS1 at an MOI of 3. Cells were harvested at 24 h after infection and analyzed by Western blotting (n = 3). Asterisks indicate nonspecific background bands. (B) Cells were infected as described for panel A, and the amount of radiolabeled amino acids incorporated into acid-insoluble protein in 30 min was quantified at 24 h after infection (n = 3; P < 0.05). Filled bars indicate wild-type infection, and open bars indicate HCMVΔTRS1 infection. (C) Control cells or shIRS1-HFs were infected with HCMVΔTRS1, and the formation of G3BP1 puncta was measured by indirect immunofluorescence at 24 h after infection. A representative image from one of three independent experiments is shown.
pTRS1 inhibits stress granule formation.
eIF2α phosphorylation results in the redistribution of actively translating mRNAs into cytoplasmic puncta termed stress granules (11, 57). While stress granules do not form after infection with wild-type HCMV (14), the increase in eIF2α phosphorylation after infection in the absence of both pTRS1 and pIRS1 suggested that pTRS1 and pIRS1 might limit stress granule formation during HCMV infection. In response to cellular stress, the G3BP1 protein localizes to cytoplasmic stress granules (58); therefore, we used the formation of G3BP1-positive cytoplasmic puncta as a measure of stress granule formation. We found that G3BP1 puncta do not accumulate in control cells or shIRS1-HFs infected with wild-type virus (data not shown). Similarly, few if any G3BP1 puncta formed after infection of control cells with the HCMVΔTRS1 virus (Fig. 2C). In contrast, infection of shIRS1-HFs with HCMVΔTRS1 resulted in the accumulation of G3BP1 puncta (Fig. 2C). Therefore, the expression of either pTRS1 or pIRS1 is necessary to prevent stress granule formation during HCMV infection.
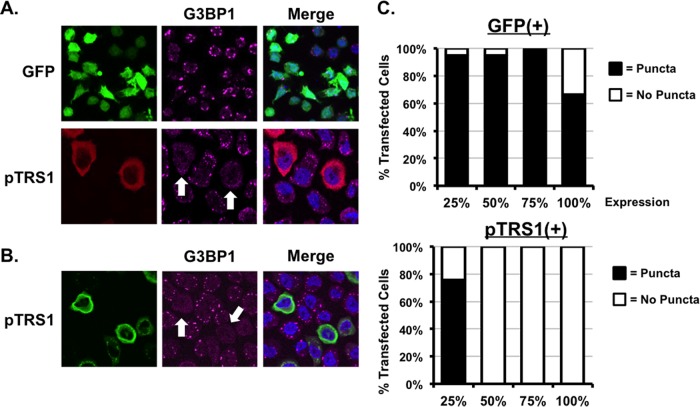
To determine if pTRS1 was sufficient to prevent stress granule formation, we measured the accumulation of stress granules in cells transfected with a pTRS1 expression vector. Cells were transfected with either a pTRS1 expression vector or a control vector expressing GFP and then treated with arsenite to induce stress granule accumulation. G3BP1 puncta readily accumulated in control cells expressing GFP. However, very few G3BP1-positive granules were found in cells expressing pTRS1 (Fig. 3A). Interestingly, arsenite induces stress granule formation through activation of the eIF2α kinase HRI (59), suggesting that pTRS1 could inhibit stress granule formation induced by eIF2α kinases other than PKR. To confirm that PKR was dispensable for arsenite-induced stress granule formation, we measured the formation of G3BP1 puncta after arsenite treatment in cells lacking PKR expression. The PKR gene in these cells was mutated using CRISPR/Cas9-mediated mutagenesis (56). Arsenite readily induced stress granule formation in PKR-deficient cells, and this induction was inhibited by pTRS1 expression (Fig. 3B). In order to measure the effect of pTRS1 expression on stress granule formation, we determined the number of cells containing stress granules among cells expressing pTRS1 or GFP. Stress granules formed in >90% of cells expressing GFP regardless of GFP expression levels (Fig. 3C, top panel). In contrast, a limited number of stress granule-positive cells (<45%) were found in cells expressing low levels of pTRS1. Higher levels of pTRS1 expression were sufficient to completely inhibit stress granule formation (Fig. 3C, bottom panel). Together our results demonstrate that pTRS1 is necessary and sufficient to prevent stress granule formation and suggest that pTRS1 also prevents stress granule formation induced by additional eIF2α kinases.
FIG 3.
pTRS1 is sufficient to inhibit stress granule formation. (A) HeLa cells were transfected with a GFP or pTRS1 expression vector. Twenty-four hours after transfection, cells were treated with sodium arsenite (0.5 mM) for 1 h, and G3BP1 punctum formation was measured by indirect immunofluorescence. (B) PKR-deficient HeLa cells were transfected with a GFP or pTRS1 expression vector. Cells were treated with arsenite as described for panel A and analyzed by indirect immunofluorescence as above. (C) Western blotting was used to measure PKR expression in PKR-deficient HeLa cells. PKR-deficient cells were transfected and treated as described for panel A. The presence of G3BP1-positive puncta in transfected cells was determined by indirect immunofluorescence. The range of transgene expression was divided into quartiles (1 to 25% of maximum, 26 to 50% of maximum, etc.), and the results are shown as the percentage of GFP-expressing (top panel) or pTRS1-expressing (bottom panel) cells containing two or more G3BP1 puncta.
PKR depletion prevents eIF2α phosphorylation and restores HCMV protein expression and replication in the absence of pTRS1 and pIRS1.
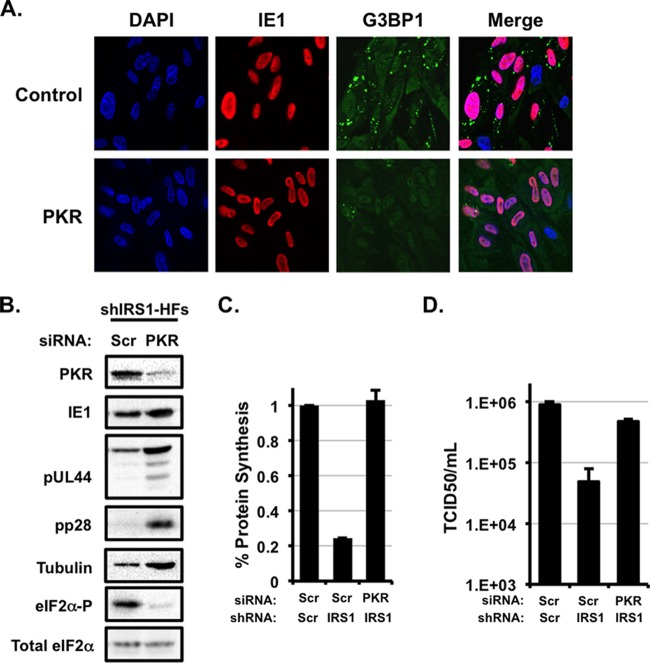
The increase in eIF2α and PKR phosphorylation observed in the absence of pTRS1 and pIRS1 during infection suggested that stress granules might form as a consequence of PKR activation. To determine if PKR was necessary for stress granule formation during infection, we measured G3BP1 punctum formation in PKR-depleted cells in the absence of pTRS1 and pIRS1. We found that PKR depletion reversed stress granule accumulation in shIRS1-HFs infected with HCMVΔTRS1 (Fig. 4A). Thus, pTRS1 and pIRS1 expression counteracted PKR-induced stress granule formation during HCMV infection. In addition, PKR depletion increased the expression of HCMV early and late proteins and prevented eIF2α phosphorylation (Fig. 4B). The increased expression of viral proteins after PKR depletion correlated with an increase in the overall levels of protein synthesis in infected cells (Fig. 4C). Consistent with the increase in protein synthesis, PKR depletion partially rescued HCMVΔTRS1 replication in shIRS1-HFs (Fig. 4D). We conclude that antagonism of PKR by pTRS1 and/or pIRS1 is necessary for efficient viral protein synthesis and replication.
FIG 4.
PKR inhibits viral protein synthesis and induces stress granule formation in the absence of pTRS1 and pIRS1. (A) shIRS1-HFs were transfected with scrambled or PKR-specific siRNAs prior to infection with HCMVΔTRS1. The formation of G3BP1 (+) puncta was monitored by indirect immunofluorescence at 24 h after infection. A representative image from one of three independent experiments is shown. (B) shIRS1-HFs were transfected with scrambled (scr) or PKR-specific (PKR) siRNAs prior to infection with HCMVΔTRS1. Expression of the indicated proteins was measured by Western blotting at 96 h after infection, except for eIF2α-P and total eIF2α, which were measured at 24 h after infection (n = 3). (C) Cells stably expressing scrambled (Scr) or pIRS1-specific (IRS1) shRNAs were transfected with scrambled (scr) or PKR-specific (PKR) siRNAs and then infected with HCMVΔTRS1. The amount of radiolabeled amino acids incorporated into acid-insoluble protein in 30 min was quantified at 24 h after infection (n = 3). (D) Cells were treated and infected as described for panel C. The amount of virus in the culture supernatants at 96 h after infection was determined by the TCID50 method (n = 3).
The pTRS1 PKR binding domain is required during infection to antagonize PKR.
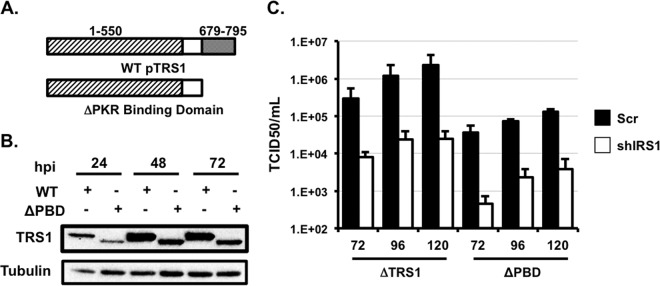
As discussed above, several pTRS1 functional domains have been described outside the context of HCMV infection (18, 21) including a PKR binding domain in the carboxyl-terminal 116 amino acids of pTRS1 (19). As our system allows for the efficient reconstitution and propagation of pTRS1 mutant viruses, we next determined the role of the pTRS1 PKR binding domain in HCMV replication. We first generated a mutant virus lacking the pTRS1 PKR binding domain (HCMVΔPDB) (Fig. 5A). We used BAC-mediated recombineering to delete the final 116 amino acids of pTRS1, which contains the previously described PKR binding domain (19), and fuse a FLAG epitope to the pTRS1 carboxyl terminus to allow us to monitor expression of the mutant pTRS1 protein. This mutant expresses pTRS1 from its native location in the viral genome under the control of its endogenous promoter allowing for wild-type kinetics and levels of pTRS1 expression. Infectious virus was recovered after electroporation of the mutant genome into primary human fibroblasts, indicating that the HCMVΔPDB virus was viable. The truncated pTRS1 was expressed throughout infection, although to slightly lower levels than wild-type pTRS1 (Fig. 5B). In addition, the pTRS1 mutant displayed diffuse cytoplasmic localization, similar to wild-type pTRS1 (data not shown).
FIG 5.
The pTRS1 PKR binding domain is required for efficient virus replication. (A) Cartoon showing the region of pTRS1 deleted in the HCMVΔPBD virus. (B) Western blot showing expression of pTRS1 after infection with wild-type HCMV or HCMVΔPBD virus at 72 h after infection. (C) Control HFs (scr; filled bars) or shIRS1-HFs (open bars) were infected with HCMVΔTRS1 or HCMVΔPBD virus at an MOI of 1, and the amount of cell-free virus in the culture supernatants was quantified by the TCID50 method (n = 3).
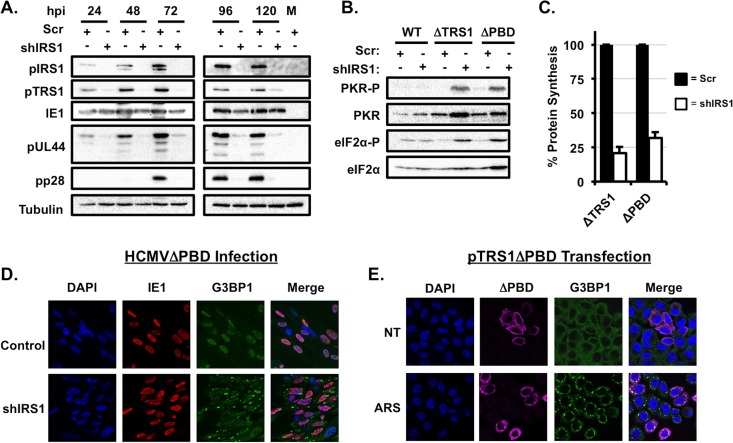
We then measured the replication of the HCMVΔPDB virus over a single round of virus replication. HCMVΔPDB replicated to lower titers than HCMVΔTRS1 on control cells, and virus replication was further reduced 10-fold in the absence of pIRS1 (Fig. 5C). While the HCMV early protein pUL44 and late protein pp28 were expressed in control cells during HCMVΔPDB infection, the expression of both viral proteins was significantly decreased in shIRS1-HFs (Fig. 6A). Interestingly, the expression of the mutant pTRS1 protein itself was also significantly reduced in pIRS1-depleted cells. To determine if PKR activation might account for diminished viral protein expression, we measured PKR and eIF2α phosphorylation after infection of control or shIRS1-HFs with the HCMVΔPDB virus. We found no increase in PKR or eIF2α phosphorylation after infection of control cells with HCMVΔPDB (Fig. 6B), indicating that truncation of the pTRS1 carboxyl terminus did not result in PKR activation in the presence of pIRS1. However, the HCMVΔPDB virus induced robust PKR and eIF2α phosphorylation when pIRS1 was depleted (Fig. 6B), with a concomitant reduction in the overall levels of protein synthesis (Fig. 6C). In addition, HCMVΔPDB infection induced stress granule accumulation in the absence of pIRS1, but not in control cells (Fig. 6D), and expression of the pTRS1ΔPBD protein was not sufficient to prevent arsenite-induced stress granule formation in PKR-deficient cells (Fig. 6E). We conclude that the carboxyl terminus of pTRS1 is required for efficient virus replication, even in the presence of pIRS1. Furthermore, our results suggest that the pTRS1 PKR binding domain is necessary to antagonize PKR during HCMV infection.
FIG 6.
The pTRS1 PKR binding domain is necessary to antagonize PKR in the absence of pIRS1. (A) Control HFs (Scr) or shIRS1-HFs (shIRS1) were infected with the HCMVΔPBD virus at an MOI of 1. Cells were harvested at the indicated times after infection, and the expression of the indicated proteins was measured by Western blotting (n = 3). (B) Control HFs or shIRS1-HFs were infected with the indicated virus as described for panel A. Cells were harvested at 24 h after infection and analyzed by Western blotting (n = 3). (C) Control HFs (filled bars) or shIRS1-HFs (open bars) were infected with HCMVΔTRS1 or HCMVΔPBD virus, and the amount of radiolabeled amino acids incorporated into acid-insoluble protein in 30 min was quantified 24 h after infection (n = 3). (D) Control HFs or shIRS1-HFs were infected with HCMVΔPBD, and the presence of G3BP1 puncta was determined by indirect immunofluorescence 24 h after infection. Representative images from one of three independent experiments are shown. (E) Cells were transfected with a pTRS1ΔPBD expression vector and treated and analyzed as described for Fig. 3. Representative results from one of three independent experiments are shown.
DISCUSSION
In this study, we developed a novel system for the study of pTRS1 and its functional domains in the context of viral infection. Using this system, we confirmed the results of a previous study (17) by showing that expression of either pTRS1 or pIRS1 is necessary for efficient HCMV replication and protein expression. We also found that in the absence of pTRS1 and pIRS1, HCMV infection activates the antiviral kinase PKR. We further show that antagonism of PKR by pTRS1 or pIRS1 is critical for ongoing protein synthesis during HCMV infection, as PKR depletion prevented eIF2α phosphorylation and the shutoff of protein synthesis in the absence of pTRS1 and pIRS1. In addition, depletion of PKR restored the expression of HCMV proteins and rescued virus replication, in the absence of pTRS1 and pIRS1. We also identified a novel role for pTRS1 as an inhibitor of stress granule formation. Expression of either pTRS1 or pIRS1 was sufficient to suppress PKR-dependent stress granule accumulation during HCMV infection, and pTRS1 alone was sufficient to prevent PKR-independent stress granule formation in transfected cells. The ability of pTRS1 to antagonize PKR and prevent stress granule formation during infection required the previously identified PKR binding domain, suggesting that a direct interaction between pTRS1 and PKR is critical for efficient virus replication.
Our data demonstrate that pTRS1 and pIRS1 inhibit PKR activation during HCMV infection and that PKR antagonism is a critical function of pTRS1 and pIRS1, required for efficient viral replication. PKR autophosphorylation and substrate phosphorylation were increased in the absence of both pTRS1 and pIRS1 during infection (Fig. 2A), and overall levels of protein synthesis were greatly reduced (Fig. 2B). As a result, virus replication and HCMV protein synthesis were significantly impaired (Fig. 1B and C). Depleting PKR prior to infection rescued HCMV replication (Fig. 4D) and restored viral protein expression (Fig. 4B) and overall levels of protein synthesis in the absence of pTRS1 and pIRS1 (Fig. 4C). Therefore, our data show that inhibition of PKR is a critical function of pTRS1 and pIRS1 during HCMV infection.
Our data also suggest pTRS1 may have additional roles in HCMV replication. The HCMVΔPDB virus replicated less efficiently than wild-type virus even in the presence of pIRS1 (Fig. 5C). The HCMVΔPDB mutant did not induce PKR or eIF2α phosphorylation or stimulate stress granule formation when pIRS1 was present (Fig. 6B and D), and thus the truncated pTRS1 does not appear to act in a dominant negative manner toward pIRS1, at least in regard to PKR inhibition. These results suggest that pTRS1 may have additional roles in HCMV replication. Previous studies found that replication of an HCMV pTRS1/pIRS1 double mutant virus was rescued by the vaccinia virus E3L protein, a known PKR antagonist (17). However, E3L also suppresses additional aspects of the antiviral response (60–65). Perhaps pTRS1 shares these PKR-independent activities with E3L. The truncation could also impact the ability of pTRS1 to facilitate packaging of viral DNA into nascent nucleocapsids (41) or perhaps limit the ability of pTRS1 to regulate autophagy (38, 39) or transactivate viral gene expression (40). Alternatively, the truncated pTRS1 isoform might partially inhibit pIRS1 function, resulting in low levels of PKR activation that are sufficient to limit viral mRNA translation. Additional studies in primary human cells completely devoid of PKR expression will be needed to determine what, if any, role pTRS1 plays in HCMV replication in addition to inhibiting PKR activation.
We also identified a novel role for pTRS1 in preventing stress granule formation. Previous studies found that stress granules do not form during HCMV infection (14, 56), despite the presence of dsRNAs (17). Our results provide a molecular mechanism for the lack of stress granules during HCMV infection, namely, the antagonism of PKR by pTRS1 and pIRS1. Stress granules did not form in infected cells expressing either pTRS1 or pIRS1; however, the absence of both proteins from infected cells triggered robust stress granule accumulation (Fig. 2C). Thus, expression of either pTRS1 or pIRS1 was necessary to prevent stress granule accumulation during HCMV infection. PKR depletion limited stress granule accumulation in infected cells in the absence of pTRS1 and pIRS1 (Fig. 4A), demonstrating that PKR activation triggers stress granule formation during HCMV infection. In addition, expression of pTRS1 outside the context of infection was sufficient to prevent stress granule accumulation in response to arsenite treatment (Fig. 3A), showing that additional viral proteins are not required for this activity. pTRS1 also blocked stress granule accumulation in response to arsenite treatment in PKR-deficient cells (Fig. 3B and C), suggesting that pTRS1 can prevent stress granule accumulation independent of its ability to antagonize PKR. Stress granules inhibit virus replication by sequestering viral RNAs and preventing their translation (12, 13, 57). While additional studies using depletion of specific stress granule components will be required to understand the impact of stress granule accumulation on HCMV infection, our results suggest that stress granules could similarly regulate HCMV protein synthesis.
Our data also provide new insight into the pTRS1 functional domains necessary for PKR inhibition during HCMV infection. The carboxyl-terminal 116 amino acids of pTRS1 contains a PKR binding domain that is necessary to complement growth in a vaccinia virus mutant lacking its major PKR antagonist, the E3L protein (18). Deletion of the pTRS1 PKR binding domain resulted in PKR activation, decreased viral protein expression, and limited virus replication in the absence of pIRS1 (Fig. 6). It was previously suggested that pTRS1 inhibits PKR in part by competing with PKR for binding to dsRNA (21). Our finding that PKR was autophosphorylated after infection with the HCMVΔPBD virus does not contradict this conclusion but rather suggests that during HCMV infection the ability of pTRS1 to bind dsRNA is not sufficient to antagonize PKR. However, we cannot rule out that these mutations alter pTRS1 folding to abrogate its function. Structural analysis of pTRS1 and PKR will likely provide a better understanding of how these domains contribute to PKR inhibition. Nevertheless, in light of the failure of the HCMVΔPBD pTRS1 mutant to antagonize PKR, it seems likely that pTRS1 binding to PKR is critical for PKR inhibition during HCMV infection. Confirmation of this hypothesis will require careful mapping of pTRS1 point mutants that specifically disrupt pTRS1 binding to PKR and the analysis of recombinant viruses carrying these mutations.
ACKNOWLEDGMENTS
We thank Adam Geballe for providing reagents and for helpful discussions. We also thank the members of the UNC virology community for their continued support, especially the members of the Burnett Womack PKR Club.
This work was supported by NIH grant R01 AI03311 to N.J.M., the North Carolina University Cancer Research Fund, and a UNC Virology Training grant (T32 AI07419) to B.Z.
REFERENCES
- 1.Black TL, Safer B, Hovanessian A, Katze MG. 1989. The cellular 68,000-Mr protein kinase is highly autophosphorylated and activated yet significantly degraded during poliovirus infection: implications for translational regulation. J Virol 63:2244–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katze MG, DeCorato D, Safer B, Galabru J, Hovanessian AG. 1987. Adenovirus VAI RNA complexes with the 68 000 Mr protein kinase to regulate its autophosphorylation and activity. EMBO J 6:689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romano PR, Garcia-Barrio MT, Zhang X, Wang Q, Taylor DR, Zhang F, Herring C, Mathews MB, Qin J, Hinnebusch AG. 1998. Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2alpha kinases PKR and GCN2. Mol Cell Biol 18:2282–2297. doi: 10.1128/MCB.18.4.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor DR, Lee SB, Romano PR, Marshak DR, Hinnebusch AG, Esteban M, Mathews MB. 1996. Autophosphorylation sites participate in the activation of the double-stranded-RNA-activated protein kinase PKR. Mol Cell Biol 16:6295–6302. doi: 10.1128/MCB.16.11.6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samuel CE. 1979. Mechanism of interferon action: phosphorylation of protein synthesis initiation factor eIF-2 in interferon-treated human cells by a ribosome-associated kinase processing site specificity similar to hemin-regulated rabbit reticulocyte kinase. Proc Natl Acad Sci U S A 76:600–604. doi: 10.1073/pnas.76.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lebleu B, Sen GC, Shaila S, Cabrer B, Lengyel P. 1976. Interferon, double-stranded RNA, and protein phosphorylation. Proc Natl Acad Sci U S A 73:3107–3111. doi: 10.1073/pnas.73.9.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kimchi A, Zilberstein A, Schmidt A, Shulman L, Revel M. 1979. The interferon-induced protein kinase PK-i from mouse L cells. J Biol Chem 254:9846–9853. [PubMed] [Google Scholar]
- 8.Zilberstein A, Federman P, Shulman L, Revel M. 1976. Specific phosphorylation in vitro of a protein associated with ribosomes of interferon-treated mouse L cells. FEBS Lett 68:119–124. doi: 10.1016/0014-5793(76)80418-8. [DOI] [PubMed] [Google Scholar]
- 9.Hinnebusch AG. 2014. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem 83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 10.Williams BR. 2001. Signal integration via PKR. Sci STKE 2001:re2. [DOI] [PubMed] [Google Scholar]
- 11.Anderson P, Kedersha N. 2008. Stress granules: the Tao of RNA triage. Trends Biochem Sci 33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Beckham CJ, Parker R. 2008. P bodies, stress granules, and viral life cycles. Cell Host Microbe 3:206–212. doi: 10.1016/j.chom.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White JP, Lloyd RE. 2012. Regulation of stress granules in virus systems. Trends Microbiol 20:175–183. doi: 10.1016/j.tim.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isler JA, Maguire TG, Alwine JC. 2005. Production of infectious human cytomegalovirus virions is inhibited by drugs that disrupt calcium homeostasis in the endoplasmic reticulum. J Virol 79:15388–15397. doi: 10.1128/JVI.79.24.15388-15397.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seto E, Inoue T, Nakatani Y, Yamada M, Isomura H. 2014. Processing bodies accumulate in human cytomegalovirus-infected cells and do not affect viral replication at high multiplicity of infection. Virology 458-459:151–161. doi: 10.1016/j.virol.2014.04.022. [DOI] [PubMed] [Google Scholar]
- 16.Child SJ, Hakki M, De Niro KL, Geballe AP. 2004. Evasion of cellular antiviral responses by human cytomegalovirus TRS1 and IRS1. J Virol 78:197–205. doi: 10.1128/JVI.78.1.197-205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marshall EE, Bierle CJ, Brune W, Geballe AP. 2009. Essential role for either TRS1 or IRS1 in human cytomegalovirus replication. J Virol 83:4112–4120. doi: 10.1128/JVI.02489-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hakki M, Geballe AP. 2005. Double-stranded RNA binding by human cytomegalovirus pTRS1. J Virol 79:7311–7318. doi: 10.1128/JVI.79.12.7311-7318.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hakki M, Marshall EE, De Niro KL, Geballe AP. 2006. Binding and nuclear relocalization of protein kinase R by human cytomegalovirus TRS1. J Virol 80:11817–11826. doi: 10.1128/JVI.00957-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blankenship CA, Shenk T. 2002. Mutant human cytomegalovirus lacking the immediate-early TRS1 coding region exhibits a late defect. J Virol 76:12290–12299. doi: 10.1128/JVI.76.23.12290-12299.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bierle CJ, Semmens KM, Geballe AP. 2013. Double-stranded RNA binding by the human cytomegalovirus PKR antagonist TRS1. Virology 442:28–37. doi: 10.1016/j.virol.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Child SJ, Brennan G, Braggin JE, Geballe AP. 2012. Species specificity of protein kinase R antagonism by cytomegalovirus TRS1 genes. J Virol 86:3880–3889. doi: 10.1128/JVI.06158-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Child SJ, Geballe AP. 2009. Binding and relocalization of protein kinase R by murine cytomegalovirus. J Virol 83:1790–1799. doi: 10.1128/JVI.01484-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Child SJ, Jarrahian S, Harper VM, Geballe AP. 2002. Complementation of vaccinia virus lacking the double-stranded RNA-binding protein gene E3L by human cytomegalovirus. J Virol 76:4912–4918. doi: 10.1128/JVI.76.10.4912-4918.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cassady KA. 2005. Human cytomegalovirus TRS1 and IRS1 gene products block the double-stranded-RNA-activated host protein shutoff response induced by herpes simplex virus type 1 infection. J Virol 79:8707–8715. doi: 10.1128/JVI.79.14.8707-8715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chou J, Roizman B. 1994. Herpes simplex virus 1 gamma(1)34.5 gene function, which blocks the host response to infection, maps in the homologous domain of the genes expressed during growth arrest and DNA damage. Proc Natl Acad Sci U S A 91:5247–5251. doi: 10.1073/pnas.91.12.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He B, Gross M, Roizman B. 1997. The gamma(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci U S A 94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Budt M, Niederstadt L, Valchanova RS, Jonjic S, Brune W. 2009. Specific inhibition of the PKR-mediated antiviral response by the murine cytomegalovirus proteins m142 and m143. J Virol 83:1260–1270. doi: 10.1128/JVI.01558-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valchanova RS, Picard-Maureau M, Budt M, Brune W. 2006. Murine cytomegalovirus m142 and m143 are both required to block protein kinase R-mediated shutdown of protein synthesis. J Virol 80:10181–10190. doi: 10.1128/JVI.00908-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He B. 2006. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ 13:393–403. doi: 10.1038/sj.cdd.4401833. [DOI] [PubMed] [Google Scholar]
- 31.Baird TD, Wek RC. 2012. Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Adv Nutr 3:307–321. doi: 10.3945/an.112.002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Speir E. 2000. Cytomegalovirus gene regulation by reactive oxygen species. Agents in atherosclerosis. Ann N Y Acad Sci 899:363–374. [DOI] [PubMed] [Google Scholar]
- 33.Speir E, Shibutani T, Yu ZX, Ferrans V, Epstein SE. 1996. Role of reactive oxygen intermediates in cytomegalovirus gene expression and in the response of human smooth muscle cells to viral infection. Circ Res 79:1143–1152. doi: 10.1161/01.RES.79.6.1143. [DOI] [PubMed] [Google Scholar]
- 34.Dhaunsi GS, Kaur J, Turner RB. 2003. Role of NADPH oxidase in cytomegalovirus-induced proliferation of human coronary artery smooth muscle cells. J Biomed Sci 10:505–509. doi: 10.1007/BF02256111. [DOI] [PubMed] [Google Scholar]
- 35.Isler JA, Skalet AH, Alwine JC. 2005. Human cytomegalovirus infection activates and regulates the unfolded protein response. J Virol 79:6890–6899. doi: 10.1128/JVI.79.11.6890-6899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu Y, Pierciey FJ Jr, Maguire TG, Alwine JC. 2013. PKR-like endoplasmic reticulum kinase is necessary for lipogenic activation during HCMV infection. PLoS Pathog 9:e1003266. doi: 10.1371/journal.ppat.1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Won S, Eidenschenk C, Arnold CN, Siggs OM, Sun L, Brandl K, Mullen TM, Nemerow GR, Moresco EM, Beutler B. 2012. Increased susceptibility to DNA virus infection in mice with a GCN2 mutation. J Virol 86:1802–1808. doi: 10.1128/JVI.05660-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaumorcel M, Lussignol M, Mouna L, Cavignac Y, Fahie K, Cotte-Laffitte J, Geballe A, Brune W, Beau I, Codogno P, Esclatine A. 2012. The human cytomegalovirus protein TRS1 inhibits autophagy via its interaction with Beclin 1. J Virol 86:2571–2584. doi: 10.1128/JVI.05746-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mouna L, Hernandez E, Bonte D, Brost R, Amazit L, Delgui LR, Brune W, Geballe AP, Beau I, Esclatine A. 2015. Analysis of the role of autophagy inhibition by two complementary human cytomegalovirus BECN1/Beclin 1-binding proteins. Autophagy doi: 10.1080/15548627.2015.1125071:0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Romanowski MJ, Shenk T. 1997. Characterization of the human cytomegalovirus irs1 and trs1 genes: a second immediate-early transcription unit within IRS1 whose product antagonizes transcriptional activation. J Virol 71:1485–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adamo JE, Schroer J, Shenk T. 2004. Human cytomegalovirus TRS1 protein is required for efficient assembly of DNA-containing capsids. J Virol 78:10221–10229. doi: 10.1128/JVI.78.19.10221-10229.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iskenderian AC, Huang L, Reilly A, Stenberg RM, Anders DG. 1996. Four of eleven loci required for transient complementation of human cytomegalovirus DNA replication cooperate to activate expression of replication genes. J Virol 70:383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pari GS, Anders DG. 1993. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J Virol 67:6979–6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pari GS, Kacica MA, Anders DG. 1993. Open reading frames UL44, IRS1/TRS1, and UL36-38 are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA synthesis. J Virol 67:2575–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strang BL, Geballe AP, Coen DM. 2010. Association of human cytomegalovirus proteins IRS1 and TRS1 with the viral DNA polymerase accessory subunit UL44. J Gen Virol 91:2167–2175. doi: 10.1099/vir.0.022640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strang BL, Bender BJ, Sharma M, Pesola JM, Sanders RL, Spector DH, Coen DM. 2012. A mutation deleting sequences encoding the amino terminus of human cytomegalovirus UL84 impairs interaction with UL44 and capsid localization. J Virol 86:11066–11077. doi: 10.1128/JVI.01379-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moorman NJ, Cristea IM, Terhune SS, Rout MP, Chait BT, Shenk T. 2008. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe 3:253–262. doi: 10.1016/j.chom.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moorman NJ, Sharon-Friling R, Shenk T, Cristea IM. 2010. A targeted spatial-temporal proteomics approach implicates multiple cellular trafficking pathways in human cytomegalovirus virion maturation. Mol Cell Proteomics 9:851–860. doi: 10.1074/mcp.M900485-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baldick CJ Jr, Marchini A, Patterson CE, Shenk T. 1997. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J Virol 71:4400–4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lenarcic EM, Ziehr B, De Leon G, Mitchell D, Moorman NJ. 2014. Differential role for host translation factors in host and viral protein synthesis during human cytomegalovirus infection. J Virol 88:1473–1483. doi: 10.1128/JVI.02321-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swift S, Lorens J, Achacoso P, Nolan GP. 2001. Rapid production of retroviruses for efficient gene delivery to mammalian cells using 293T cell-based systems. Curr Protoc Immunol Chapter 10:Unit 10.17C. doi: 10.1002/0471142735.im1017cs31. [DOI] [PubMed] [Google Scholar]
- 52.Zhu H, Shen Y, Shenk T. 1995. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J Virol 69:7960–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silva MC, Yu QC, Enquist L, Shenk T. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J Virol 77:10594–10605. doi: 10.1128/JVI.77.19.10594-10605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lenarcic EM, Ziehr BJ, Moorman NJ. 2015. An unbiased proteomics approach to identify human cytomegalovirus RNA-associated proteins. Virology 481:13–23. doi: 10.1016/j.virol.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu D, Silva MC, Shenk T. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc Natl Acad Sci U S A 100:12396–12401. doi: 10.1073/pnas.1635160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ziehr B, Lenarcic E, Vincent HA, Cecil C, Garcia B, Shenk T, Moorman NJ. 2015. Human cytomegalovirus TRS1 protein associates with the 7-methylguanosine mRNA cap and facilitates translation. Proteomics 15:1983–1994. doi: 10.1002/pmic.201400616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buchan JR, Parker R. 2009. Eukaryotic stress granules: the ins and outs of translation. Mol Cell 36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol 160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.McEwen E, Kedersha N, Song B, Scheuner D, Gilks N, Han A, Chen JJ, Anderson P, Kaufman RJ. 2005. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J Biol Chem 280:16925–16933. doi: 10.1074/jbc.M412882200. [DOI] [PubMed] [Google Scholar]
- 60.Guerra S, Caceres A, Knobeloch KP, Horak I, Esteban M. 2008. Vaccinia virus E3 protein prevents the antiviral action of ISG15. PLoS Pathog 4:e1000096. doi: 10.1371/journal.ppat.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eduardo-Correia B, Martinez-Romero C, Garcia-Sastre A, Guerra S. 2014. ISG15 is counteracted by vaccinia virus E3 protein and controls the proinflammatory response against viral infection. J Virol 88:2312–2318. doi: 10.1128/JVI.03293-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim YG, Muralinath M, Brandt T, Pearcy M, Hauns K, Lowenhaupt K, Jacobs BL, Rich A. 2003. A role for Z-DNA binding in vaccinia virus pathogenesis. Proc Natl Acad Sci U S A 100:6974–6979. doi: 10.1073/pnas.0431131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Langland JO, Kash JC, Carter V, Thomas MJ, Katze MG, Jacobs BL. 2006. Suppression of proinflammatory signal transduction and gene expression by the dual nucleic acid binding domains of the vaccinia virus E3L proteins. J Virol 80:10083–10095. doi: 10.1128/JVI.00607-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Y, Wolff KC, Jacobs BL, Samuel CE. 2001. Vaccinia virus E3L interferon resistance protein inhibits the interferon-induced adenosine deaminase A-to-I editing activity. Virology 289:378–387. doi: 10.1006/viro.2001.1154. [DOI] [PubMed] [Google Scholar]
- 65.Valentine R, Smith GL. 2010. Inhibition of the RNA polymerase III-mediated dsDNA-sensing pathway of innate immunity by vaccinia virus protein E3. J Gen Virol 91:2221–2229. doi: 10.1099/vir.0.021998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]