Abstract
The von Hippel-Lindau tumor suppressor protein (pVHL) negatively regulates hypoxia-inducible mRNAs such as the mRNA encoding vascular endothelial growth factor (VEGF). This activity has been linked to its ability to form multimeric complexes that contain elongin C, elongin B, and Cul2. To understand this process in greater detail, we performed a series of in vitro binding assays using pVHL, elongin B, and elongin C variants as well as synthetic peptide competitors derived from pVHL or elongin C. A subdomain of elongin C (residues 17–50) was necessary and sufficient for detectable binding to elongin B. In contrast, elongin B residues required for binding to elongin C were not confined to a discrete colinear domain. We found that the pVHL (residues 157–171) is necessary and sufficient for binding to elongin C in vitro and is frequently mutated in families with VHL disease. These mutations preferentially involve residues that directly bind to elongin C and/or alter the conformation of pVHL such that binding to elongin C is at least partially diminished. These results are consistent with the view that diminished binding of pVHL to the elongins plays a causal role in VHL disease.
J. Clin. Invest. 104:1583–1591 (1999).
Introduction
An hereditary cancer syndrome, von Hippel-Lindau (VHL) disease, displays an autosomal dominant pattern of inheritance (1). At the molecular level, however, this disorder is autosomal recessive insofar as tumor development in this setting is attributable to biallelic inactivation of the VHL tumor suppressor gene. Specifically, VHL disease is caused by germline mutations of the VHL gene (1). Tumors develop when a susceptible cell somatically mutates or deletes the remaining wild-type VHL allele.
Patients with VHL disease are at risk for retinal and central nervous system hemangioblastomas, renal cell carcinomas of the clear cell type, pheochromocytomas, endolymphatic sac tumors, and pancreatic islet cell tumors (1). Genotype-phenotype patterns are emerging such that VHL families can be categorized based on the absence (type I VHL disease) or presence (type II VHL disease) of pheochromocytoma (2). Type II VHL disease has been further subdivided based on the absence (type IIA VHL disease) or presence (type IIB VHL disease) of renal cell carcinoma. In general, frameshift mutations and gross deletions are associated with type I disease whereas specific missense mutations are associated with type II disease (2).
In keeping with Knudson’s 2-hit model of carcinogenesis, biallelic inactivation of the VHL gene also occurs commonly in sporadic clear cell carcinoma of the kidney and central nervous system hemangioblastoma (1). On the other hand, VHL mutations are rare in sporadic pheochromocytoma (3–5). With respect to renal cell carcinoma, loss of VHL appears to occur as an early event and can be detected in premalignant renal cysts (1). Thus, VHL may play a “gatekeeper” role similar to the purported role of the APC gene in polyp formation and colorectal carcinogenesis (6).
The VHL gene gives rise to 2 protein products of approximately 30 and 19 kD (7, 8). The latter arises as a result of internal translation from an in-frame methionine codon. Both of these VHL gene products can inhibit the accumulation of hypoxia-inducible mRNAs such as the vascular endothelial growth factor (VEGF) mRNA under normoxic conditions and can suppress renal cell carcinoma growth in nude mouse xenograft assays (9). Deregulation of VEGF likely contributes to the vascular nature of VHL-associated neoplasms (9). Conversely, suppression of angiogenesis may contribute to tumor suppression by the VHL suppressor protein (pVHL). For simplicity, we will use the term “pVHL” when referring generically to both VHL gene products.
The pVHL resides primarily, but not exclusively, in the cytosol (9). Recent studies suggest that pVHL can shuttle between the nucleus and the cytosol and have demonstrated that the ability to shuttle is required for pVHL to regulate hypoxia-inducible mRNAs (10, 11).
The control of hypoxia-inducible mRNAs by pVHL may involve both transcriptional and posttranscriptional effects and has been linked to the ability of pVHL to form multimeric complexes that contain elongin C, elongin B, and Cul2 (1). Elongin C and Cul2 are similar to 2 yeast proteins, Skp1 and Cdc53, respectively, that form multimeric complexes targeting specific proteins for ubiquitin-dependent proteolysis (1). Thus, one model, which remains to be confirmed, is that pVHL, through its interaction with elongin C and Cul2, regulates the ubiquitination of proteins involved in hypoxia-inducible mRNA production and turnover (12). In this regard, Maxwell et al. recently showed that cells lacking pVHL fail to degrade 2 hypoxia-inducible transcription factors, HIF1α and HIF2α, under well-oxygenated conditions (13).
Elongin C binds directly to pVHL and recruits Cul2 to the complex (12, 14, 15). Elongin B stabilizes the interaction of pVHL and elongin C (14). The region of pVHL that binds to elongin C is frequently altered by naturally occurring VHL mutations (12, 16–18). We set out to understand in greater detail the structural requirements for the formation of a pVHL/elongin C/elongin B complex by using elongin C and pVHL-derived peptides as competitors in in vitro binding assays. Our results, in conjunction with the recently solved crystal structure of pVHL (19) bound to elongin C and elongin B, identify certain critical contact sites that are formed in this complex. In addition, these studies provide information that may provide further insights into the genotype-phenotype relationships that are emerging in VHL disease.
Methods
Cell culture and metabolic labeling.
First, 786-O cells were obtained from the American Type Culture Collection (ATCC; Rockford, Maryland, USA) and grown in DMEM supplemented with 10% FBS; 786-O subclones producing wild-type pVHL, pVHL Y98H, W117R, and C162F were described previously and were grown in the same media supplemented with 1 mg/mL G418 (12, 20, 21). Generation and maintenance of polyclonal 786-O cell transfectants producing pVHL L158S, pVHL R161G, C162F, Q164R, V166F, and R167W and monoclonal 786-O cell transfectants producing pVHL L188V were carried out essentially in the same manner. Metabolic labeling was performed by growing cells in DMEM supplemented with 5% dialyzed FBS and 0.5 mCi/mL of 35S-translabel following 1 hour of serum starvation. Cell extracts were made as described previously (16).
Plasmids.
The following plasmids were described previously (12, 20): pGEX2K, pGEX2TK-VHL(117–213), pGEX2TK-elongin C, pGEX2TK-elongin C (16–50), pcDNAIII-T7-elongin C, pRc-CMV-VHL, pRc-CMV-VHL(C162F), pRc-CMV-VHL(L158S), and pRc-CMV-VHL(R167W). Plasmids pRc-CMV-VHL(R161G), pRc-CMV-VHL(Q164R), pRc-CMV-VHL(V166F), pRc-CMV(L188V) were made by site-directed mutagenesis of pRc-CMV-VHL using a MutaGene kit (Bio-Rad, Mellville, New York, USA) according to the manufacturer’s instructions and were confirmed by DNA sequencing.
To make pGEX2TK-elongin B, the human elongin B cDNA spanning the complete open reading frame was PCR amplified with a sense primer containing a BamHI site and an antisense primer containing an EcoRI site. The product was digested with BamHI and EcoRI and ligated into pGEX2TK (Amersham Pharmacia Biotech, Piscataway, New Jersey, USA) that had been cut with these 2 enzymes.
To make pSP72-elongin B, pSP72-elongin B (15–118), pSP72-elongin B (1–107), pSP72-elongin B (1–91), and pSP72-elongin B (1–82), the corresponding segments of the human elongin B open reading frame were amplified with sense primers containing BamHI sites and antisense primers containing EcoRI sites. The products were digested with BamHI and EcoRI and ligated into pSP72 (Promega Corp., Madison, Wisconsin, USA) that had been cut with these 2 enzymes.
The plasmids encoding the elongin C internal deletion mutants are described elsewhere (22).
Peptides.
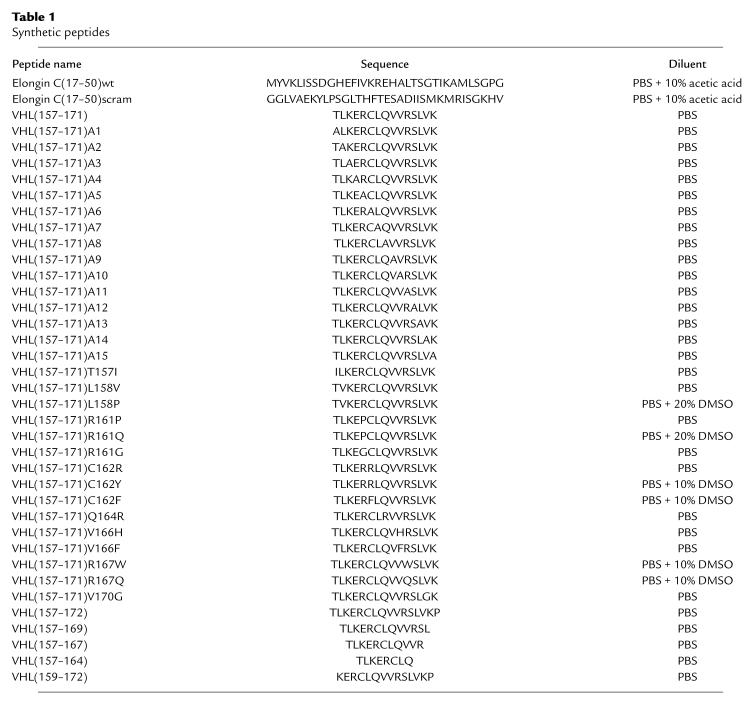
The VHL(157–172), VHL(157–164), VHL(157–167), and VHL(159–169) were a generous gift of Sushi Sharma at the Novartis Research Institute (Summit, New Jersey, USA). The remaining peptides were purchased from Bio-Synthesis (Lewisville, Texas, USA).
Antibodies.
Anti-hemagglutinin (HA) antibody (12CA5) was purchased from Boehringer Mannheim Biochemicals Inc. (Indianapolis, Indiana, USA). The anti-Cul2 (12) and anti–elongin B antibodies (23) were described previously. Anti-GLUT1 affinity-purified rabbit sera was purchased from Alpha Diagnostics (San Antonio, Texas, USA).
Glutathione S-transferase pull-down assays.
Glutathione S-transferase (GST) pulldown assays were performed essentially as described previously (24). To assay binding to GST-VHL, 1 mL of radiolabeled 786-O extract was precleared by incubation with glutathione Sepharose preloaded with GST. Binding reactions contained 100 μL of precleared 786-O cell radiolabeled extract and approximately 1 μg of immobilized GST-VHL(117–213) in a total volume of 500 μL. Where indicated, competitor peptides were added before the addition of the cell extract. Following a 1-hour incubation, the glutathione Sepharose was washed 4 times with NETN containing 900 mM NaCl and once with NETN alone. Bound proteins were eluted by boiling in an SDS-containing protein sample buffer, resolved by a 15% SDS-PAGE, and detected by fluorography.
To assay binding to the elongins, approximately 1 μg of GST-elongin C or GST-elongin B, immobilized on glutathione Sepharose, was incubated with 4 (Figure 1) or 5 μL (Figure 2) of the indicated in vitro translate for 1 hour and then washed 5 times with NETN. Bound proteins were eluted and detected as described above. To assay binding of elongin C internal deletion mutants to GST-VHL(157–177), 3 μg of elongin C or elongin C mutant was mixed with 3 μg of GST or GST-VHL(157–177) in a buffer containing 20 mM Tris-HCl, pH 7.6, 150 mM KCl, 10% glycerol, and 1% Triton X-100. After a 1-hour incubation, the resin was washed twice with the same buffer. Bound proteins were eluted and detected as described above.
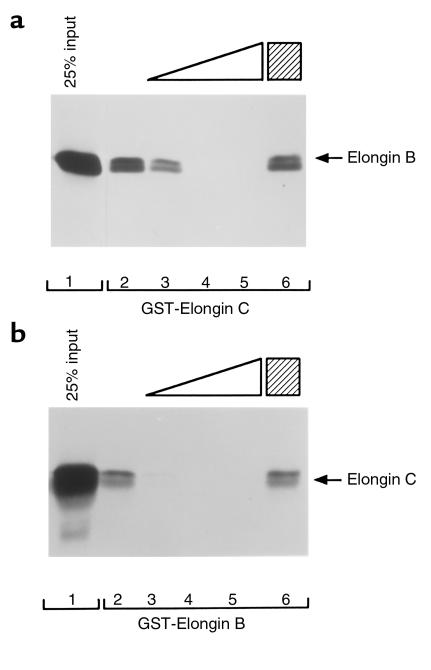
Figure 1.
Disruption of elongin B and elongin C interaction by an elongin C–derived peptide. (a) 35S-labeled elongin B in vitro translate was incubated with glutathione Sepharose preloaded with GST-elongin C in the absence of peptide (lane 2), in the presence of increasing amounts of a synthetic peptide corresponding to elongin C residues 17–50 (lanes 3–5; 0.5, 2.5, 10 μg, respectively), or in the presence of 10 μg of a sequence-scrambled elongin (17–50) peptide. (b) 35S-labeled elongin C in vitro translate was incubated with glutathione Sepharose preloaded with GST-elongin B in the absence (lane 2) or presence (lanes 3–6) of elongin C peptides as in a. Bound proteins were resolved by SDS-PAGE and detected by fluorography. Comparable recovery of the GST fusion proteins in each lane was confirmed by Coomassie blue staining. Twenty-five percent of the in vitro translate used per binding reaction was loaded directly in lane 1.
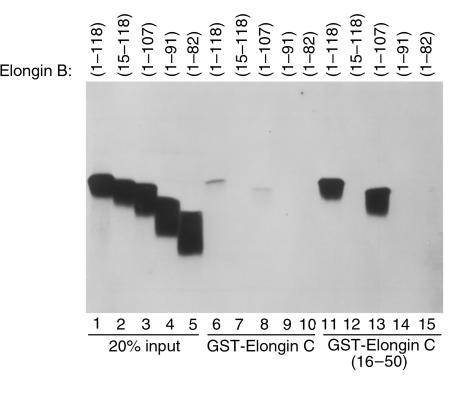
Figure 2.
Binding of elongin B mutants to elongin C. 35S-labeled wild-type elongin B (1–118) or the indicated elongin B mutants incubated with glutathione Sepharose preloaded with GST-elongin C (lanes 6–10) or GST-elongin C (16–50) (lanes 11–15). Comparable recovery of GST-elongin C in each lane was confirmed by Coomassie blue staining and bound proteins were detected by fluorography. Twenty percent of the in vitro translate used per binding reaction was loaded directly in lane 1–5.
Preparation of RNA polymerase II, basal transcription factors, and elongins.
RNA polymerase II and TFIIH from rat liver, recombinant TBP, TFIIB, TFIIF, and TFIIE, purified soluble elongin A, elongin C, and elongin BC complexes were prepared as described (22).
In vitro transcriptional elongation assays.
Preinitiation complexes were assembled as described (14) by preincubation of 50 ng of the EcoRI to NdeI fragment of pDN-AdML and approximately 50 ng of recombinant yeast TBP, 10 ng of recombinant TFIIB, 10 ng of recombinant TFIIF, 7 ng of recombinant TFIIE, 40 ng of TFIIH, and 0.01 unit of RNA polymerase II. Transcription was initiated by addition of 7 mM MgCl2 and the concentrations of ribonucleoside triphosphates indicated in Figure 6. Where indicated, reactions contained approximately 4 nM purified elongin A and approximately 4 nM purified elongin BC complex and various amounts of VHL peptide. After an 8-minute incubation at 28°C, runoff transcripts were analyzed by electrophoresis through 6% polyacrylamide, 7.0 M urea gels. Full-length runoff transcripts were quantitated by PhosphorImager analysis.
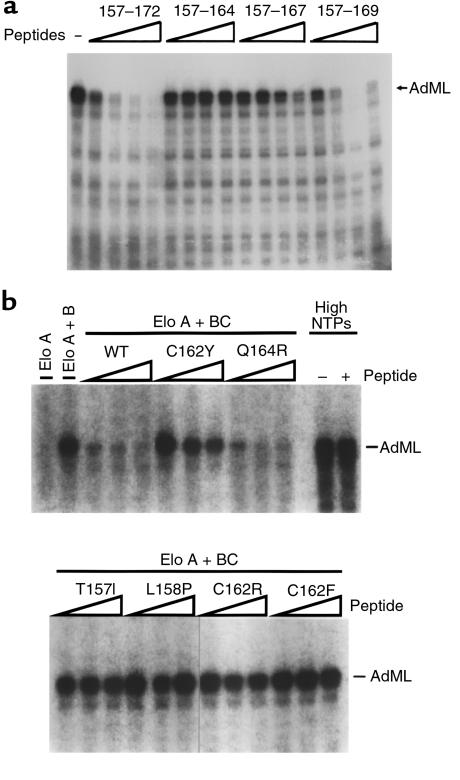
Figure 6.
Inhibition of elongin/SIII activity by pVHL-derived peptides. (a) Runoff transcription reactions containing elongin A and elongin BC were performed as described in Methods. All reactions included 50 μM ATP, 50 μM GTP, 10 μM CTP, 10 μCi [α-32P] CTP, and 2 μM UTP. The reaction in the first lane did not include VHL peptide. Reactions in the remaining lanes included 12 nM, 40 nM, 120 nM, or 400 nM of the indicated peptide. AdML indicates the position of the approximately 250 nucleotides runoff transcript initiated at the AdML promoter. (b) Runoff transcription reactions were performed in the presence of elongin A or elongin A and elongin BC as indicated, with 0, 100 nM, 200 nM, and 400 nM of the indicated peptide, with 50 μM ATP, 50 μM GTP, 10 μM CTP, 10 μCi [α-32P] CTP, and 2 μM UTP. Reactions labeled High NTPs were performed in the absence of elongin A and elongin BC and in the presence of 0 (–) or 600 nM (+) of the wild-type peptide and 50 μM ATP, 50 μM GTP, 10 μM CTP, 10 μCi [α-32P] CTP, and 50 μM UTP. Elo, elongin.
Results
Regulation of hypoxia-inducible genes by pVHL (213 aa) is tightly linked to its ability to form complexes with elongin C (112 aa) and elongin B (118 aa). Elongin C binds directly to pVHL. This interaction is stabilized by binding of elongin B to elongin C. To understand the structural requirements for the formation of a pVHL/elongin C/elongin B complex, we performed a series of biochemical experiments using synthetic peptides derived from pVHL or elongin C.
We showed previously that a GST fusion protein containing elongin C residues 18–50 was sufficient to bind to elongin B (12). To confirm that this region of elongin C mediated binding to elongin B, a peptide corresponding to elongin C residues 17–50 was synthesized along with a sequence scrambled version thereof. The wild-type, but not mutant, elongin C (17–50) peptide blocked the interaction of elongin B with GST-elongin C (Figure 1a) and the interaction of elongin C with GST-elongin B (Figure 1b). Thus, elongin C residues 17–50 constitutes a colinear elongin B binding domain.
To ask which region of elongin B bound to elongin C, a series of NH2-terminal and COOH-terminal elongin B truncation mutants were translated in vitro in the presence of 35S-methionine and assayed for binding to GST-elongin C (16–50) (Figure 2). As expected, wild-type elongin B bound to GST-elongin C (16–50) as did elongin B (1–107). Deletion of 14 residues from the elongin B NH2-terminus or 17 residues from the elongin B COOH-terminus led to a loss of elongin C binding capability. Qualitatively similar results were obtained using GST fused to full-length elongin C (Figure 2). Therefore, elongin B residues necessary for stable binding to elongin C extend throughout much of the molecule. The increased binding of elongin B to elongin C (16–50), compared with wild-type elongin C, might suggest that residues outside elongin (16–50) negatively regulate elongin B binding.
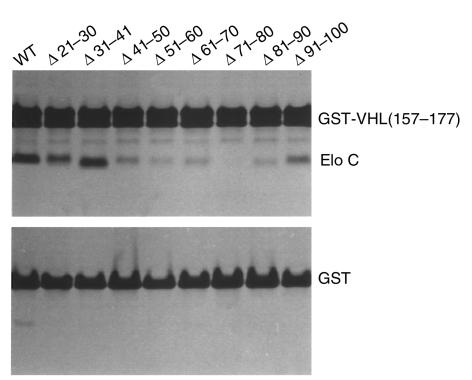
We showed previously that pVHL residues (157–177) bind to elongin C in vitro. To ask which region(s) of elongin C interacted with pVHL residues (157–177), a series of elongin C internal deletion mutants were translated in vitro in the presence of 35S-methionine and assayed for binding to GST-VHL(157–177) (Figure 3). Sequential 10 amino acid deletions affecting elongin C residues 41–90 led to a marked reduction in binding to pVHL(157–177), suggesting that elongin C (41–90) is a major contact region for binding to pVHL or is important for the structural integrity of elongin C.
Figure 3.
Binding of elongin C deletion mutants to pVHL(157–177). Binding assays with GST-VHL(157–177) (upper panel) or GST (lower panel) and wild-type or mutant elongin C were performed as described in Methods. Elo, elongin.
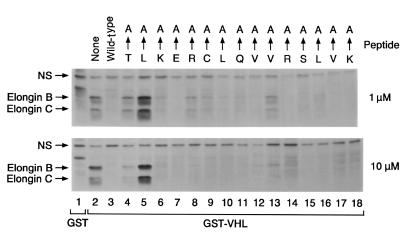
We found that pVHL(157–171) is a hotspot for mutations in VHL disease. In pilot experiments we confirmed that a synthetic peptide spanning residues 157–171 blocked the interaction of pVHL with elongins B and C, in keeping with our earlier results with a peptide encompassing residues 157–172 (16), whereas peptides corresponding to pVHL(159–172) and pVHL(157–164) did not (data not shown). In the next set of experiments, we made a series of pVHL(157–171) peptides in which single amino acids were converted to alanine and tested their ability to block the interaction of elongins B and C with GST-VHL (Figure 4). Alanine substitution of L158 completely inactivated the pVHL peptide (Figure 4, lane 5). Alanine substitutions at T157, K159, R161, C162, and V166 measurably reduced the activity of the corresponding peptides in this assay (Figure 4, lanes 4, 6, 8, 9, 13, respectively).
Figure 4.
Effect of single amino acid alanine substitutions on interaction of VHL(157–171) peptide with elongin B and C. The 786-O renal carcinoma cells were metabolically labeled with 35S-methionine, lysed, and incubated with glutathione Sepharose preloaded with GST (lane 1) or GST-VHL(117–213) (lanes 2–18) in the absence (lanes 1–2) or presence (lanes 3–18) of the indicated VHL(157–171) peptides (final peptide concentration of 1 and 10 μM in upper and lower panels, respectively). The arrows at the top of the figure indicate which amino acid residue was changed to alanine in the mutated VHL(157–171) peptides. Bound proteins were resolved by SDS-PAGE and detected by fluorography. Comparable recovery of the GST fusion proteins in each lane was confirmed by Coomassie blue staining. NS = nonspecific.
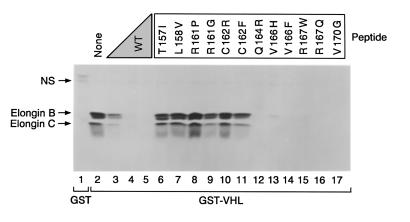
Some of the residues that tolerated alanine substitutions in this assay, such as Q164, R167, and V170, are altered by VHL-associated mutations. We therefore repeated these experiments with a series of pVHL(157–171) peptides corresponding to naturally occurring VHL mutations (Figure 5). As expected from the alanine scanning experiment, mutations affecting residues 157 (lane 6), 158 (lane 7), 161 (lanes 8 and 9), and 162 (lanes 10 and 11) led to a loss of elongin-binding activity. Surprisingly, but in keeping with the alanine scanning data, naturally occurring mutations affecting residues 164, 167, and 170 had little effect on the behavior of the pVHL(157–171) peptide in this assay. In addition, the 2 peptides in which residue 166 was altered (V166H and V166F) retained the ability to interact with elongin B and C in this assay despite the fact that alanine substitution of this residue led to a modest, but reproducible, decrease in elongin binding (Figure 4, lane 13).
Figure 5.
Effect of tumor-derived single amino acid substitutions on interaction of VHL(157–171) peptide with elongin B and C. The 786-O renal carcinoma cells were metabolically labeled with 35S-methionine, lysed, and incubated with glutathione Sepharose preloaded with GST (lane 1) or GST-VHL(117–213) (lanes 2–17) in the absence (lanes 1–2) or presence (lanes 3–17) of the indicated VHL(157–171) peptides. Wild-type VHL (0.1, 1, 10 μg of residues 157–171) was added in lanes 3–5, respectively. Mutant peptides were added to 10 μg. The single amino acid substitutions in the mutated VHL(157–171) peptides are indicated at the top of the figure. Bound proteins were resolved by SDS-PAGE and detected by fluorography. Comparable recovery of the GST fusion proteins in each lane was confirmed by Coomassie blue staining. NS = nonspecific.
To corroborate these data, we developed an additional assay for the interaction of pVHL with elongin C based on the earlier observation that recombinant pVHL can inhibit transcriptional elongation by elongin/SIII in vitro (25). Full activity of elongin/SIII requires that elongin A, the enzymatic subunit, bind to elongin B and C using a motif that is similar to pVHL(157–172) (25). Thus, inhibition of elongin/SIII by pVHL likely is due to its ability to competitively inhibit the binding of elongin B/C to elongin A. This model predicted that the pVHL(157–172) peptide would suffice to inhibit elongin/SIII. To test this, elongin/SIII in vitro transcription assays were carried out in the presence of the pVHL(157–172) or a truncated version thereof (Figure 6a). As expected, addition of the pVHL(157–172) peptide led to a dose-dependent inhibition of elongin/SIII whereas pVHL(157–164) and pVHL(159–172) peptides did not (Figure 6a and data not shown). In keeping with the in vitro binding studies described above, the pVHL(157–169) peptide also inhibited elongin/SIII whereas the pVHL(157–167) was clearly impaired in this assay.
We next tested the tumor-derived pVHL(157–171) peptides in this assay (Figure 6b). As predicted by the in vitro binding assays, these peptides fell into 2 classes. Peptides in which residues T157, L158, or C162 were altered could not inhibit elongin/SIII. In contrast, pVHL(157–171) Q164R, which retained the ability to bind to elongin B and C (Figure 5), was seemingly wild type in this assay (Figure 6b).
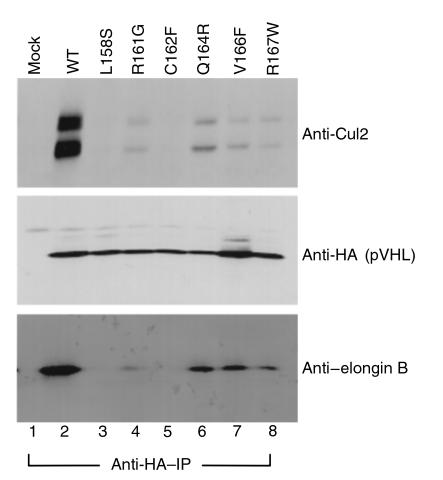
Taken together, these results suggested that either elongin binding was not the target of mutations such as Q164R or R167W, or that these mutations might affect elongin binding in vivo in the context of full-length pVHL. To this end, 786-O renal carcinoma cells were transiently transfected with mammalian expression plasmids encoding HA-tagged wild-type pVHL (Figure 7, lane 2), or missense mutants thereof (Figure 7, lanes 3–8), lysed and immunoprecipitated with anti-HA antibody. Given the available antibodies, the ability to form a productive complex with elongin C was monitored by performing anti-Cul2 and anti-elongin B immunoblot analysis of the anti-HA immunoprecipitates. Both elongin B and Cul2 bind to pVHL through elongin C (12, 14). All of the pVHL mutants displayed a diminished ability to bind elongin/Cul2 in this assay relative to wild-type pVHL. As predicted from our in vitro analysis, mutants such as L158S and C162F were unable to bind to elongin/Cul2. The remaining pVHL displayed detectable but clearly diminished binding to elongin B and Cul2. The results with polyclonal cells producing pVHL C162F, pVHL L158S, and R167W are in keeping with our earlier characterization of clonal cell lines producing these mutants (12, 16). Of note, the intracellular concentration of ectopically produced pVHL proteins in these studies likely exceed the normal concentration of pVHL by approximately 10-fold (7). Thus, it is possible that the residual binding activity of mutants such as R167W might not be measurable under physiological conditions. In sum, these results are consistent with the view that partial or complete loss of elongin/Cul2 binding by pVHL leads to VHL disease.
Figure 7.
Naturally occurring pVHL(157–171) domain mutants exhibit reduced binding to elongin B and Cul2. The 786-O renal carcinoma cells were transfected to produce HA-tagged versions of wild-type pVHL (lane 2) or the indicated pVHL mutants. Anti-HA immunoprecipitates were resolved by SDS-PAGE and immunoblotted with anti-Cul2 (top panel), anti-HA (middle panel), or anti–elongin B (lower panel).
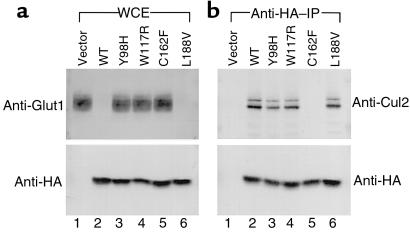
Many naturally occurring VHL mutations map outside pVHL(157–171). For example, surface residues in a region of the protein referred to as the beta domain are also frequently mutated in VHL disease (19). These residues do not appear to play a role in maintaining the conformation of pVHL but instead may be involved in substrate recognition by the pVHL/elongin/Cul2 complex (19). In the last set of experiments, we studied 2 such beta domain mutants, pVHL Y98H and W117R, as well as a third mutant, pVHL L188V. The mutant pVHL L188V has been described in multiple independent kindreds that display a pheochromocytoma only, or type IIC, VHL disease phenotype (26). Wild-type pVHL and pVHL C162F were tested in parallel as positive and negative controls, respectively. The mutants pVHL Y98H, W117R, and L188V all retained the ability to bind productively with elongin C as shown by their ability to coimmunoprecipitate Cul2 (Figure 8). Nonetheless, pVHL Y98H and pVHL W117R, like pVHL C162F, was unable to suppress hypoxia-inducible gene expression as shown by immunoblot analysis for Glut1. In contrast, pVHL L188V retained both the ability to bind to Cul2 and the ability to suppress hypoxia-inducible protein accumulation.
Figure 8.
Characterization of naturally occurring pVHL mutants with an intact pVHL(157–171) domain. The 786-O renal carcinoma cells were transfected to produce HA-tagged versions of wild-type pVHL (lane 2) or the indicated pVHL mutants. (a) Whole-cell extracts were resolved by SDS-PAGE and analyzed for Glut1 (top panel) and pVHL (lower panel) production by anti-Glut1 and anti-HA immunoblot analysis, respectively. (b) In parallel, anti-HA immunoprecipitates were resolved by SDS-PAGE and immunoblotted with anti-Cul2 antibodies (top panel) or anti-HA antibodies (lower panel).
Discussion
VHL mutations can be detected in approximately 100% of patients that carry a clinical diagnosis of VHL disease (27). The nature of the VHL mutation, however, profoundly influences the spectrum of tumors that will develop in a particular VHL family (2). Conceivably, the organ-specific risk of cancer associated with different VHL alleles is due to the biochemical properties that are gained or lost by their protein products. Binding to elongins B and C, and subsequent recruitment of Cul2, has been linked to the regulation of hypoxia-inducible genes and to tumor suppression by pVHL (12). We therefore sought to understand the structural requirements for formation of a pVHL/elongin C/elongin B complex in greater detail using standard biochemical approaches. Our data can now also be interpreted in light of the 3-dimensional structure of this complex based on x-ray crystallography (19).
The elongin C/B interface.
Our studies suggest that elongin C residues 17–50 are sufficient to bind to elongin B. In contrast to elongin C, we were unable to identify a discrete, colinear, subdomain within elongin B that is sufficient to mediate its binding to elongin C. In the crystal structure, the elongin B/C interaction is mediated by edge-to-edge interactions between beta sheets in these 2 molecules (19). Consistent with our peptide studies, elongin C residues Y18, H27, F29, and V31 form contacts with elongin B (19), and deletion of elongin C residues 21–30 leads to a loss of elongin B–binding activity (22). Although residues outside elongin C 18–50 also contact elongin B (for example T78 and Y83), these contacts are apparently not essential (19, 22). In contrast, residues in elongin B that contact elongin C span most of the elongin B primary sequence, including T13, I14, F15, I34, E91, and F93, perhaps accounting for our results with specific elongin B NH2-terminal and COOH-terminal deletion mutants. It should be noted, however, that elongin B (1–84) is sufficient to bind to elongin C under assay conditions that differed from those used in the present study (28). This latter result underscores that the ubiquitin-like domain in elongin B is important for elongin C binding.
The elongin C/pVHL interface.
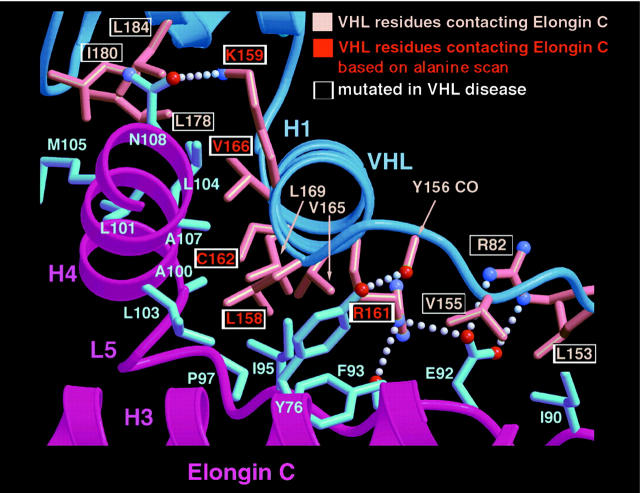
The pVHL residues 157–172 are sufficient to bind to elongin C in vitro (16). In the crystal structure, this region of pVHL forms a helix (H1) that fits into a concave surface present on elongin C (Figure 9; ref. 19). The most extensive contact is made by Leu158 in keeping with our alanine scanning data. Also in approximate agreement with our alanine scanning data, C162 and R161 make critical contacts with elongin C with less significant contributions from K159, V165, V166, and L169. The H1 region of pVHL interacts with elongin C helices H3 and H4 as well as extended loop structure (L5). H3 and L5 are encompassed by elongin C residues 40–90 in approximate agreement with our analysis of elongin C internal deletion mutants. Note that H4, corresponding to elongin C residues 101–108, was present in all of the elongin C internal deletion mutants we assayed.
Figure 9.
Structure of pVHL-elongin C interface. The pVHL and elongin C secondary structure elements are shown in dark blue and fuchsia, respectively. The pVHL side chains contacting elongin C are shown in salmon, and elongin C side chains are shown in light blue. The pVHL residue T157 is not shown because it would obscure the depiction of some of the other contacts formed between pVHL and elongin C in this figure. Note that pVHL T157 makes low-density van der Waals contacts with elongin C residues C112 and Y76.
In vitro versus in vivo binding of pVHL to elongin C.
Although pVHL(157–172) is sufficient to bind to elongin C in vitro, residues COOH-terminal to pVHL residue 172 are required for stable binding to elongin C in vivo (12). Specifically, pVHL(1–197) binds to elongin C in cells whereas pVHL(1–187) and pVHL(1–172) do not (12). Similarly, thioredoxin fusion proteins in which pVHL residues 157–197 were inserted bound to elongin C in cells, whereas fusions containing pVHL residues 157–172 did not (data not shown). Although this discrepancy is not fully understood, the crystal structure reveals that the interaction of pVHL with elongin C is stabilized by 2 additional pVHL helices (H2 and H3) extending to pVHL residue 189 (19). This finding may account for the occurrence of VHL-associated frameshift and missense mutations COOH-terminal to residue 172 (16–18).
A second discrepancy between our in vitro results obtained with peptides and the behavior of full-length pVHL mutants in cells relates to pVHL mutants such as Q164R, V166F, and R167W. The crystal structure predicts that Q164 and R167 participate in an intramolecular interface that stabilizes the pVHL structure without directly contacting elongin C. This presumably accounts for the fact that these mutations did not measurably affect the behavior of the pVHL(157–172) peptides and yet led to diminished (but not absent) binding to elongin C in the context of full-length pVHL ectopically produced in cells. V166 also makes a low-density contact with elongin C. We hypothesize that loss of this minor contact does not affect the behavior of the pVHL(157–172) peptide but that the substitution of the bulky phenylalanine moiety of valine at residue 166 alters the conformation of full-length pVHL and/or sterically clashes with elongin C.
Inhibition of elongin/SIII by a pVHL-derived peptide in vitro.
We confirmed the prediction that a pVHL-derived peptide (157–172) would suffice to inhibit elongin/SIII activity (16, 25). This activity, which likely reflects the fact that this peptide is highly similar to the elongin B/C–binding domain present in elongin A (16, 25), was used to corroborate our results in standard in vitro binding assays. Nonetheless, it seems highly unlikely that inhibition of elongin A contributes to pVHL function in vivo. Specifically, elongin B and C are in vast molar excess of elongin A and pVHL (29). Thus, it is unlikely that pVHL could sequester sufficient elongin B and C to affect elongin A function.
Insights into pathogenesis in VHL disease.
Our peptide experiments showed that alanine substitutions of pVHL residues 157, 158, 159, 161, 162, and 166 affected binding to elongins B and C. Interestingly, multiple mutations affecting residues 158, 161, 162, 167, and 170 have been detected in VHL kindreds whereas mutations affecting residues 160, 163, 165, 168, and 169 have not been reported (2, 18). Furthermore, we confirmed that VHL-associated mutations affecting residues 167 and 170 lead to diminished elongin binding in the context of an otherwise full-length pVHL. Thus, the genetic and biochemical data are consistent with diminished elongin binding playing a causal role in the pathogenesis of VHL disease.
VHL mutations associated with a risk of pheochromocytoma (type II VHL disease) are typically missense mutations, whereas mutations associated with a low risk of pheochromocytoma include frameshifts and gross deletions. This suggests several non–mutually exclusive hypotheses. For example, complete loss of pVHL function may be lethal for those cells that give rise to pheochromocytoma. It is also possible that pheochromocytoma develops as a manifestation of a gain of function by certain pVHL mutants. In this model, this putative gain of function may require that the overall structure of pVHL be at least partially preserved. In this regard, it is of interest that VHL-associated mutations affecting residues 157–172 fall into several classes. Residues L158 and C162 directly contact the elongins. Mutations affecting these residues completely abrogate elongin binding. Residues Q164, V166, and R167 do not directly contact the elongins (Q164, R167) or make minor contacts (V166). Mutations affecting these residues lead to a partial loss of elongin-binding activity, presumably by altering the conformation of pVHL or through steric effects. Finally, R161 contacts the elongin C and also participates in the folding of pVHL. Thus, mutations affecting this residue can have both direct and indirect effects on elongin binding. More “at-risk” patients with these various VHL mutations will need to be studied before it can be determined whether or not this biochemical heterogeneity contributes to the phenotypic heterogeneity observed in VHL disease.
Many VHL missense mutations map outside the core elongin-binding domain. Two such mutants, pVHL Y98H and pVHL W117R, retain the ability to bind to the elongins and Cul2 and yet, as shown here, do not suppress hypoxia-inducible protein accumulation. This result is in keeping with the earlier demonstration that binding to elongins and Cul2 is necessary, but not sufficient, for regulation of hypoxia-inducible mRNAs by pVHL (12) and is at least consistent with the hypothesis that these mutants are defective for substrate recognition (19). Finally, a type IIC mutant, pVHL L188V, retained the ability to bind to elongins and Cul2 and suppressed hypoxia-inducible mRNA accumulation. Together, these results are consistent with the notion that deregulation of hypoxia-inducible mRNAs is necessary for the development of the vascular stigmata of VHL disease. In contrast, deregulation of hypoxia-inducible mRNAs would not appear to be necessary for pheochromocytoma development as manifested by individuals with germ-line pVHL L188V mutations. These clues, in conjunction with the identification of other biochemical activities of pVHL (21, 30–32), should provide a clearer understanding of the biochemical basis of VHL disease.
Table 1.
Synthetic peptides
Acknowledgments
We thank the members of Kaelin and Conaway labs for useful discussions. We also thank Minou Modabber for her assistance in the art work. M. Ohh is a recipient of the Medical Research Council of Canada Fellowship. W.G. Kaelin, Jr. is an Assistant Investigator of the Howard Hughes Medical Institute.
References
- 1.Ohh M, Kaelin WG., Jr The von Hippel-Lindau tumour suppressor protein: new perspectives. Mol Med Today. 1999;5:257–263. doi: 10.1016/s1357-4310(99)01481-1. [DOI] [PubMed] [Google Scholar]
- 2.Crossey PA, et al. Identification of intragenic mutations in the von Hippel-Lindau disease tumor suppressor gene and correlation with disease phenotype. Hum Mol Genet. 1994;3:1303–1308. doi: 10.1093/hmg/3.8.1303. [DOI] [PubMed] [Google Scholar]
- 3.Gnarra JR, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 4.Eng C, et al. Mutations in the RET proto-oncogene and the von Hippel-Lindau disease tumour suppressor gene in sporadic and syndromic phaeochromocytomas. J Med Genet. 1995;32:934–937. doi: 10.1136/jmg.32.12.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hofstra RMW, et al. Extensive mutation scanning of RET in sporadic medullary thyroid carcinoma and of RET and VHL in sporadic pheochromocytoma reveals involvement of these genes in only a minority of cases. J Clin Endocrinol Metab. 1996;81:2881–2884. doi: 10.1210/jcem.81.8.8768845. [DOI] [PubMed] [Google Scholar]
- 6.Kinzler K, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 7.Iliopoulos O, Ohh M, Kaelin W. pVHL19 is a biologically active product of the von Hippel-Lindau gene arising from internal translation initiation. Proc Natl Acad Sci USA. 1998;95:11661–11666. doi: 10.1073/pnas.95.20.11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schoenfeld A, Davidowitz E, Burk R. A second major native von Hippel-Lindau gene product, initiated from an internal translation start site, functions as a tumor suppressor. Proc Natl Acad Sci USA. 1998;95:8817–8822. doi: 10.1073/pnas.95.15.8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maher E, Kaelin WG. von Hippel-Lindau Disease. Medicine (Baltimore) 1997;76:381–391. doi: 10.1097/00005792-199711000-00001. [DOI] [PubMed] [Google Scholar]
- 10.Lee S, et al. Nuclear/cytoplasmic localization of the von Hippel-Lindau tumor suppressor gene product is determined by cell density. Proc Natl Acad Sci USA. 1996;93:1770–1775. doi: 10.1073/pnas.93.5.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S, et al. Transcription-dependent nuclear-cytoplasmic trafficking is required for the function of the von Hippel-Lindau tumor suppressor protein. Mol Cell Biol. 1999;19:1486–1497. doi: 10.1128/mcb.19.2.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lonergan KM, et al. Regulation of hypoxia-inducible mRNAs by the von Hippel-Lindau protein requires binding to complexes containing elongins B/C and Cul2. Mol Cell Biol. 1998;18:732–741. doi: 10.1128/mcb.18.2.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maxwell P, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 14.Aso T, Lane WS, Conaway JW, Conaway RC. Elongin (SIII): a multisubunit regulator of elongation by RNA polymerase II. Science. 1995;269:1439–1443. doi: 10.1126/science.7660129. [DOI] [PubMed] [Google Scholar]
- 15.Pause A, et al. The von Hippel-Lindau tumor-suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc Natl Acad Sci USA. 1997;94:2156–2161. doi: 10.1073/pnas.94.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kibel A, Iliopoulos O, DeCaprio JD, Kaelin WG. Binding of the von Hippel-Lindau tumor suppressor protein to elongin B and C. Science. 1995;269:1444–1446. doi: 10.1126/science.7660130. [DOI] [PubMed] [Google Scholar]
- 17.Kishida T, Stackhouse TM, Chen F, Lerman MI, Zbar B. Cellular proteins that bind the von Hippel-Lindau disease gene product: mapping of binding domains and the effect of missense mutations. Cancer Res. 1995;55:4544–4548. [PubMed] [Google Scholar]
- 18.Zbar B, et al. Germline mutations in the von Hippel-Lindau (VHL) gene in families from North America, Europe, and Japan. Hum Mutat. 1996;8:348–357. doi: 10.1002/(SICI)1098-1004(1996)8:4<348::AID-HUMU8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 19.Stebbins C, Kaelin W, Jr, Pavletich N. Structure of the VHL-Elongin C-Elongin B complex: implications for von Hippel-Lindau tumor suppressor function. Science. 1999;284:455–461. doi: 10.1126/science.284.5413.455. [DOI] [PubMed] [Google Scholar]
- 20.Iliopoulos O, Kibel A, Gray S, Kaelin WG. Tumor suppression by the human von Hippel-Lindau gene product. Nat Med. 1995;1:822–826. doi: 10.1038/nm0895-822. [DOI] [PubMed] [Google Scholar]
- 21.Ohh M, et al. The von Hippel-Lindau tumor suppressor protein is required for proper assembly of an extracellular fibronectin matrix. Mol Cell. 1998;1:959–968. doi: 10.1016/s1097-2765(00)80096-9. [DOI] [PubMed] [Google Scholar]
- 22.Takagi Y, Conaway R, Conaway J. Characterization of elongin C functional domain required for interaction with elongin B and activation of elongin A. J Biol Chem. 1996;271:1–7. doi: 10.1074/jbc.271.41.25562. [DOI] [PubMed] [Google Scholar]
- 23.Garrett K, et al. Positive regulation of general transcription factor SIII by a tailed ubiquitin homolog. Proc Natl Acad Sci USA. 1995;92:7172–7176. doi: 10.1073/pnas.92.16.7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaelin WG, Pallas DC, DeCaprio JA, Kaye FJ, Livingston DM. Identification of cellular proteins that can interact specifically with the T/E1A-binding region of the retinoblastoma gene product. Cell. 1991;64:521–532. doi: 10.1016/0092-8674(91)90236-r. [DOI] [PubMed] [Google Scholar]
- 25.Duan DR, et al. Inhibition of transcriptional elongation by the VHL tumor suppressor protein. Science. 1995;269:1402–1406. doi: 10.1126/science.7660122. [DOI] [PubMed] [Google Scholar]
- 26.Zbar B, et al. Germline mutations in the Von Hippel-Lindau disease (VHL) gene in families from North America, Europe, and Japan. Hum Mutat. 1996;8:348–357. doi: 10.1002/(SICI)1098-1004(1996)8:4<348::AID-HUMU8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 27.Stolle C, et al. Improved detection of germline mutations in the von Hippel-Lindau disease tumor suppressor gene. Hum Mutat. 1998;12:417–423. doi: 10.1002/(SICI)1098-1004(1998)12:6<417::AID-HUMU8>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 28.Brower C, et al. The elongin B ubiquitin homology domain: identification of elongin B sequences important for interaction with elongin C. J Biol Chem. 1999;274:13629–13636. doi: 10.1074/jbc.274.19.13629. [DOI] [PubMed] [Google Scholar]
- 29.Kamura T, et al. The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes Dev. 1998;12:3872–3881. doi: 10.1101/gad.12.24.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsuchiya H, Iseda T, Hino O. Identification of a novel protein (VBP-1) binding to the von Hippel-Lindau (VHL) tumor suppressor gene product. Cancer Res. 1996;56:2881–2885. [PubMed] [Google Scholar]
- 31.Pal S, Claffey K, Cohen H, Mukhopadhyay D. Activation of Sp1-mediated vascular permeability factor/vascular endothelial growth factor transcription requires specific interaction with protein kinase C zeta. J Biol Chem. 1998;273:26277–26280. doi: 10.1074/jbc.273.41.26277. [DOI] [PubMed] [Google Scholar]
- 32.Mukhopadhyay D, Knebelmann B, Cohen H, Ananth S, Sukhatme V. The von Hippel-Lindau tumor suppressor gene product interacts with Sp1 to repress vascular endothelial growth factor promoter activity. Mol Cell Biol. 1997;17:5629–5639. doi: 10.1128/mcb.17.9.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]