ABSTRACT
Individuals chronically infected with hepatitis C virus (HCV) commonly exhibit hepatic intracellular lipid accumulation, termed steatosis. HCV infection perturbs host lipid metabolism through both cellular and virus-induced mechanisms, with the viral core protein playing an important role in steatosis development. We have recently identified a liver protein, the cell death-inducing DFFA-like effector B (CIDEB), as an HCV entry host dependence factor that is downregulated by HCV infection in a cell culture model. In this study, we investigated the biological significance and molecular mechanism of this downregulation. HCV infection in a mouse model downregulated CIDEB in the liver tissue, and knockout of the CIDEB gene in a hepatoma cell line results in multiple aspects of lipid dysregulation that can contribute to hepatic steatosis, including reduced triglyceride secretion, lower lipidation of very-low-density lipoproteins, and increased lipid droplet (LD) stability. The potential link between CIDEB downregulation and steatosis is further supported by the requirement of the HCV core and its LD localization for CIDEB downregulation, which utilize a proteolytic cleavage event that is independent of the cellular proteasomal degradation of CIDEB.
IMPORTANCE Our data demonstrate that HCV infection of human hepatocytes in vitro and in vivo results in CIDEB downregulation via a proteolytic cleavage event. Reduction of CIDEB protein levels by HCV or gene editing, in turn, leads to multiple aspects of lipid dysregulation, including LD stabilization. Consequently, CIDEB downregulation may contribute to HCV-induced hepatic steatosis.
INTRODUCTION
Hepatitis C virus (HCV) is a positive-strand RNA virus and a significant human pathogen. Chronic HCV infection causes liver complications, such as steatosis, cirrhosis, and hepatocellular carcinoma. The arrival of new directly acting antivirals (DAAs) has resulted in markedly improved virologic response in patients with access to these new drugs, but the high cost of the new therapy and the low diagnosis rate of HCV-infected individuals present new challenges for hepatitis C management (1). Furthermore, chronic liver damage can persist even after the infection has been cleared, so HCV pathogenesis remains an area of research highly significant for human health.
The HCV life cycle and pathogenesis are intimately linked to host lipid metabolism (2). On one hand, lipids are involved in multiple stages of the infection cycle. HCV virions are assembled on lipid droplets (LDs) (3) and associated with host lipoproteins to form lipoviral particles (LVP) for infection (4). The productive entry of HCV is aided by several molecules involved in lipid uptake (5–7); replication of HCV genome critically depends on a lipid kinase (8, 9) and is regulated by lipid peroxidation (10). On the other hand, HCV infection profoundly disturbs lipid metabolism pathways (11). HCV patients exhibit enhanced lipogenesis (12), consistent with in vitro results showing that HCV infection upregulates genes encoding sterol regulatory element binding protein 1c (SREBP-1c) and fatty acid synthase (FASN), both important for the intracellular lipid synthesis pathway (13–16). More recently, the 3′ untranslated region (UTR) of HCV was shown to, upon binding of DDX3, activate IκB kinase α and trigger biogenesis of LDs (17). Consequently, liver steatosis, the intracellular accumulation of lipids, is a common histological feature of patients with chronic hepatitis C, especially in those with genotype 3 (GT3) infection (18, 19). The mechanisms of virus-induced steatosis may involve both increased lipogenesis and reduced lipolysis and secretion (20, 21). The expression of HCV core protein was shown to recapitulate HCV-induced steatosis in a transgenic mouse model (22, 23), and the localization of core protein to LDs may be important for intracellular LD accumulation and steatosis induction (24–26).
The cell death-inducing DFFA-like effector (CIDE) family proteins, CIDEA, CIDEB, and CIDEC/fat-specific protein 27 (Fsp27), were originally identified using a bioinformatics approach based on their homology to the N-terminal domain of DNA fragmentation factors (27). While CIDEA and CIDEC are more widely expressed, CIDEB is mostly expressed in liver cells (27) and induced during hepatic differentiation of stem cells (28, 29). Although these proteins can induce cell death when overexpressed (27, 30, 31), gene knockout (KO) experiments with mice indicate that their function relates mostly to lipid metabolism in vivo (32–34). A role for CIDEB in very-low-density lipoprotein (VLDL) lipidation, VLDL transport, and cholesterol metabolism in nonprimate cell culture models has been reported (34–36). We previously characterized a role for CIDEB in a late step of HCV entry into hepatocytes (29). In this study, we investigated the molecular mechanism and biological consequence of HCV-induced downregulation of CIDEB. We demonstrate that CIDEB protein is normally regulated through the ubiquitin-mediated proteasome pathway and that HCV infection further downregulates CIDEB by inducing CIDEB protein degradation, most likely through proteolytic cleavage. This HCV-mediated degradation of CIDEB requires the expression of the HCV core, and downregulation of CIDEB protein was observed in an HCV-infected humanized mouse model. In addition, we demonstrate that gene knockout of CIDEB in a human hepatoma cell line reduces the secretion of triglycerides (TGs) and stabilizes cytoplasmic LDs in a manner similar to HCV infection. Core-dependent CIDEB downregulation in vivo may contribute to hepatic steatosis in the setting of HCV infection.
MATERIALS AND METHODS
Antibodies, compounds, and inhibitors.
The following antibodies and chemicals were used in this study: anti-JFH-1 core and anti-NS3 (BioFront Technologies Inc., FL); anti-CIDEB, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and Ku80 (Santa Cruz Biotechnology, TX); anti-dengue virus (DENV)-NS3 (Genetex, CA); fluorescein isothiocyanate (FITC)- and tetramethyl rhodamine isothiocyanate (TRITC)-conjugated anti-rabbit and anti-mouse IgGs (Sigma-Aldrich, MO); oleic acid (OA), oil red O (ORO), cycloheximide (CHX), Triacsin C (TriC), and 4-([diethylamino]methyl)-N-[2-(2-methoxyphenyl)ethyl]-N-(3R)-3-pyrrolidinylbenzamide (PF-429242) (Sigma-Aldrich, MO); and MG132 (EMD-Millipore, MA).
Cell culture and HCVcc infection.
Huh-7.5 cells were provided by Charles Rice (Rockefeller University) and Apath LLC (NY). Generation of the CIDEB knockdown and knockout cell lines has been previously described (29). FLAG-CIDEB stable cells were generated using a human immunodeficiency virus (HIV)-based lentiviral vector expressing FLAG-CIDEB cDNA. Briefly, we transduced Huh-7.5 CIDEB-KO cells with pHIV-7-IRES-FLAG-CIDEB, selected stable populations with 1.2 μg/ml of puromycin for 3 weeks, and then obtained single cell clones. For infection with genotype 2 cell culture-derived HCV (HCVcc), cells were inoculated with either wild-type JFH-1 or several high-titer variants—JFH-1/Ad16 (provided by Guangxiang Luo, University of Alabama at Birmingham), Mut4-6, and JLSO-III (37, 38)—for 6 to 8 h at 37°C, followed by three phosphate-buffered saline (PBS) washes before being changed to fresh media. The cell lines harboring infection by genotype 3 viruses have been described previously (39).
Infection with other viruses.
Huh-7.5 cells were seeded onto glass coverslips in 12-well plates and infected for 16 h with green fluorescent protein (GFP)-tagged vesicular stomatitis virus (VSV-GFP) (kindly provided by Fanxiu Zhu, Florida State University) or DENV for 48 h.
Plasmid construction and mutagenesis.
The hemagglutinin (HA)-ubiquitin plasmid was a gift from Fanxiu Zhu (Florida State University). For generation of the K173A CIDEB construct, mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Agilent Technologies, CA) according to the manufacturer's instructions.
Immunofluorescence analysis.
Cells seeded on slides were fixed in 4% paraformaldehyde for 10 min, followed by three 10-min washes in PBS at room temperature and blocking in PBTG (PBS containing 0.1% Triton X-100, 10% normal goat serum, and 1% bovine serum albumin [BSA]) at room temperature for 2 h. Slides were then incubated with primary antibody either at room temperature for 1 h or at 4°C overnight. Following primary antibody incubation, slides were washed with PBS for three subsequent 15-min washes and then incubated with secondary antibody for 1 h at room temperature, followed by three subsequent 15-min washes with PBS. Slides were mounted and nuclei were stained using VECTASHIELD (Vector Laboratories, CA). ORO staining was carried out according to the supplier's instructions.
Western blotting.
Cells were harvested by trypsinization, pelleting, and subsequent lysis in 1× Laemmli buffer and boiled or directly lysed in 1× Laemmli buffer and boiled.
Electroporation of viral RNA.
In vitro transcription and electroporation of HCV RNAs were performed as previously described (29). Viral J6/JFH-1/Gluc and deletion mutant (ΔE1/E2, Δcore, and GNN) RNAs were generated in vitro using a MEGAscript T7 kit (Ambion, TX) and purified by phenol-chloroform extraction. For electroporation, 10 μg of RNA was used for 4 × 106 cells in a volume of 400 μl of low-serum medium in a 4-mm cuvette (VWR) using the Gene Pulser Xcell electroporation system (Bio-Rad, CA). Medium was changed 4 h postelectroporation. Samples were collected at 4 h postelectroporation to control for electroporation efficiency. Samples were collected either by direct lysis with 1× Laemmli buffer and boiling or by incubation with trypsin, followed by pelleting and lysis by boiling in 1× Laemmli buffer for Western blot analysis. The Jc1/GLuc constructs were provided by Brett Lindenbach (Yale University).
Cell viability assay.
Huh-7.5 cells were seeded into 96-well plates before treatment with the desired compound for 24 h. Cells were then assayed in biological triplicate using the CellTiter-Glo luminescent cell viability kit according to the manufacturer's instructions (Promega, WI).
Coimmunoprecipitation.
Huh-7.5 cells were cotransfected with FLAG-CIDEB or HA-ubiquitin constructs. Eighteen to 24 h after transfection, cells were treated with 10 μM MG132, then washed three times with ice-cold PBS, solubilized with lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1.0% NP-40, 5 mM EDTA, 5 mM EGTA, 15 mM MgCl2, 60 mM β-glycerophosphate, 0.1 mM sodium orthovanadate, 0.1 mM NaF, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1× proteinase inhibitor cocktail) for 5 min on ice, and then rotated for 1 h at 4°C. The cell lysate was centrifuged at 12,000 × g for 10 min at 4°C to remove any insoluble material and then subjected to immunoprecipitation (IP) with EZview anti-Flag M2 or anti-HA affinity beads (Sigma-Aldrich, MO) according to the manufacturer's instructions. The beads were collected by centrifugation and then washed gently three times with lysis buffer supplemented with protease inhibitors. Beads and bound protein were boiled in 2× Laemmli buffer and analyzed by Western blotting.
CIDEB stability assays.
To analyze stability of endogenous CIDEB, Huh-7.5 cells were treated with 2.5 μg/ml of CHX for various amounts of time before being directly boiled in 1× Laemmli buffer and analyzed by Western blotting. For CIDEB mutants, cells were transfected with the mutant constructs for 18 h, treated with 2.5 μg/ml of CHX for 4 h, and analyzed similarly. For proteasome inhibition, cells were treated with 10 μM MG132 for 18 h. For lipid loading, cells were electroporated with JFH-1 or infected with JFH-1/AD16 for 24 h and then incubated in media containing 0 μM, 100 μM, or 400 μM OA for 20 h before Western blot analysis.
Lipid droplet purification.
Huh-7.5 cells were infected with JFH-1/AD16 for 42 h and then treated with 375 μM OA for 14 h. Cells were harvested in ice-cold PBS and stored at −80°C. Cell pellets were fractionated by following a modified previously published protocol (40). Briefly, cell pellets were Dounce homogenized in buffer A (20 mM Tricine, 250 mM sucrose, 1 mM PMSF [pH 7.8]), overlaid with buffer B (20 mM HEPES, 100 mM KCl, 2 mM MgCl2, [pH 7.4]), and ultracentrifuged at 270,000 × g for 1 h. The floating crude LD fraction was collected and then centrifuged at 14,000 × g for 10 min, and residual underlying liquid and pellet were removed. Total LD was then washed with buffer B four times. Lipids were dissolved with equal amounts of chloroform and acetone, and the resulting protein pellet was resuspended in 1× Laemmli buffer. The total protein amount was measured using the Bio-Rad DC protein assay. For Western blot analysis, 15 μg of whole-cell lysate and 7.5 μg of purified LD proteins were used.
Lipid droplet stability analysis.
Naive Huh-7.5 cells, Huh-7.5 cells 60 h post-JFH-1/AD16 infection, or naive Huh-7.5 CIDEB-KO cells (single-cell clones 3 and 11) were seeded at low confluence (10%) onto coverslips in 12-well plates and treated with 375 μM OA for 14 to 20 h. The cells were then washed three times with PBS and further incubated in fresh, OA-free medium containing 5.5 μM TriC or 40 μM PF-429242 for 24 h before LDs were visualized using ORO staining.
VLDL density analysis.
Naive Huh-7.5 cells, Huh-7.5 cells 60 h post-JFH-1/AD16 infection, or naive Huh-7.5 CIDEB-KO clone 11 cells were incubated in a lipid-rich medium (Dulbecco modified Eagle medium [DMEM] containing 375 μM OA, 10% fetal bovine serum [FBS], and nonessential amino acids) for 14 h, washed three times with PBS, and then changed to DMEM for 8 h. Collected supernatant was fractionated into 11 fractions using a continuous 10 to 40% iodixanol gradient (77,160 × g for 17 h at 4°C). The fractions were analyzed for ApoB content using an ApoB100 detection kit (Mabtech, OH).
Triglyceride storage and secretion.
Huh-7.5 or Huh-7.5 CIDEB-KO clone 11 cells were incubated in the lipid-rich medium for 20 h, washed three times with PBS, incubated in DMEM, collected by scraping in ice-cold PBS, and lysed via sonication. The cell lysate and supernatant were analyzed for TG content using a TG detection kit (Sigma-Aldrich, MO).
Humanized mouse model sample collection.
Methods for collection of humanized mouse model samples have been described previously (41).
RESULTS
HCV infection downregulates CIDEB in vitro and in vivo.
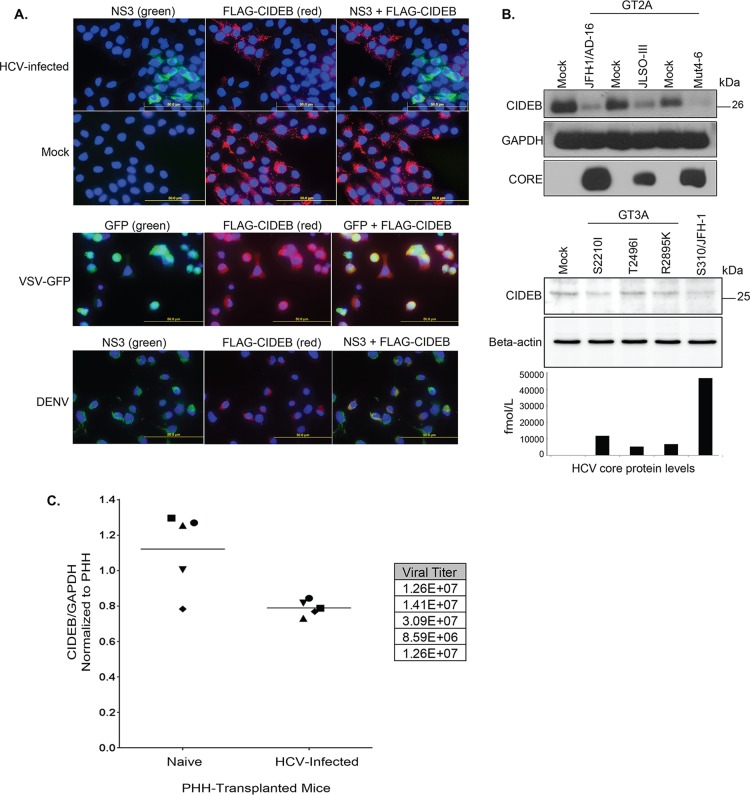
We have previously observed that JFH-1/HCVcc infection resulted in a lower CIDEB protein level on Western blots without reducing CIDEB mRNA in cultured cells (29). To facilitate single-cell, immunofluorescence-based detection of CIDEB, we generated a stable FLAG-CIDEB-expressing Huh-7.5-based cell line (FLAG-CIDEB CBKO#3) that lacked endogenous CIDEB expression. Indeed, infection of this cell line and subsequent dual staining of HCV-infected cells for HCV NS3 and CIDEB revealed a pattern of mutual exclusivity between HCV expression and CIDEB protein (Fig. 1A, top row), consistent with HCV-mediated CIDEB downregulation. The uninfected cells exhibited a more uniform expression pattern of CIDEB (Fig. 1A, second row), and VSV or dengue virus infection of the FLAG-CIDEB CBKO#3 cells did not result in this pattern of exclusion between infection and CIDEB protein (Fig. 1A, third and fourth rows). Western blotting confirmed the downregulation of CIDEB protein by JFH-1, a genotype 2 virus, and its derivatives (Fig. 1B).We also tested a genotype 3 virus (39) for CIDEB downregulation. Infection of Huh-7.5.1 cells, a derivative of the Huh-7.5 cell line, with three different clones of a GT3a virus resulted in virus production with different efficiencies. Higher levels of core protein (as measured by enzyme-linked immunosorbent assay [ELISA]) were correlated with a reduction of the endogenous intracellular CIDEB protein level, similar to what was observed in the JFH-1-infected cells (Fig. 1B). The less pronounced downregulation of CIDEB protein by these GT3 viruses is likely a result of lower infection efficiencies of these isolates, rather than a genotype-specific effect on CIDEB degradation (Fig. 1B). Finally, we investigated if endogenous human CIDEB is downregulated by HCV infection in vivo. We analyzed liver samples of human-liver SCID/uPA mice with or without infection by genotype 1a HCV derived from human serum (41). As shown in Fig. 1C, liver tissues from HCV-infected mice expressed lower overall CIDEB proteins than those from similarly transplanted but uninfected animals. Together, these results suggest that CIDEB downregulation by HCV infection occurs with multiple genotypes and within the liver environment.
FIG 1.
HCV infection downregulates CIDEB in vitro and in vivo. (A) Immunofluorescence staining of viral proteins and CIDEB. Huh-7.5-based CIDEB-KO cells (clone 3) stably expressing FLAG-CIDEB were infected with a high-titer JFH-1 (HCV) variant, JFH-1/AD16 (genotype 2), for 3 days before costaining for FLAG and HCV NS3. FLAG-CIDEB CBKO#3 cells were infected with VSV-GFP for 16 h, stained for FLAG-CIDEB, and analyzed for coexpression of FLAG-CIDEB and GFP. FLAG-CIDEB CBKO#3 cells were infected with DENV for 48 h and costained for FLAG-CIDEB and DENV NS3. (B) Immunoblot of CIDEB in genotype 2-infected (top) or genotype 3-infected (bottom) cells. Cells were infected for 3 days with various genotype 2a viruses before analysis by Western blotting or for 8 days with various genotype 3a-based (S310) clones or the S310/JFH-1 chimera. On day 8 postinfection, core protein was measured from the supernatants by HCV core-specific ELISA, and cell lysates were analyzed by Western blotting for CIDEB. Beta-actin was included as a loading control. (C) CIDEB protein levels in the HCV-infected uPA/SCID humanized mouse model. Liver tissue lysates from primary human hepatocytes (PHH)-transplanted uPA/SCID mice, either uninfected (n = 5) or HCV infected (n = 5), were subjected to Western blotting to measure CIDEB protein levels. Data from individual mice are plotted as CIDEB protein normalized to GAPDH protein (control). Quantification of band intensity was performed using ImageJ software (National Institutes of Health). HCV-infected versus uninfected humanized mice showed statistically significant levels of CIDEB protein (P = 0.01). Measured HCV titers (in RNA copies per milliliter) are listed in the adjacent table.
CIDEB is normally regulated at the cellular level by ubiquitin-mediated proteasomal degradation.
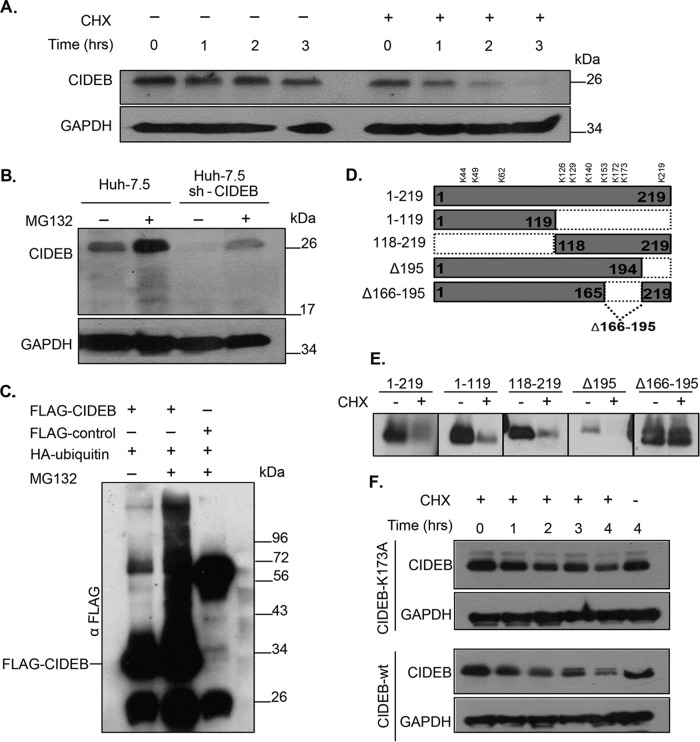
In addition to HCV infection, the expression level of CIDEB in liver cells is also regulated by a variety of stimuli (29, 42). In order to understand whether HCV enhances normal cellular degradation of CIDEB or utilizes a separate, distinct mechanism, we investigated the cellular mechanism of CIDEB regulation in uninfected hepatocytes. We performed CHX inhibition experiments to determine the half-life of the CIDEB protein in uninfected cells. As shown in Fig. 2A, CIDEB is a short-lived protein, with an estimated half-life of <60 min, which is near the low end of the spectrum of mammalian protein half-lives and well below the average protein turnover rate (10 to 20 h) in human cells reported previously (43). The GAPDH protein was used as a loading control because it has been reported to have a long half-life, 40 to 50 h, and was thus unlikely to change for the duration of the experiments performed in this study (44). To determine whether the rapid turnover of CIDEB is mediated by proteasomes, we treated the cells with MG132, a proteasome inhibitor, and determined the CIDEB protein level with immunoblotting. An increase of CIDEB level was observed with MG132 treatment in both Huh-7.5 cells and a derivative cell line that harbored a small hairpin RNA (shRNA) against the CIDEB mRNA (Fig. 2B), suggesting that proteasomes play a role in regulating CIDEB protein stability. Because canonical proteasome targeting is mediated by polyubiquitylation of the target protein, we tested whether inhibition of proteasome-mediated protein degradation results in accumulation of polyubiquitylated CIDEB. Treatment of cells cotransfected with FLAG-CIDEB and HA-ubiquitin with MG132 resulted in accumulation of polyubiquitylated CIDEB (Fig. 2C), indicating that the CIDEB protein level is regulated by ubiquitylation and the proteasome. Ubiquitins are commonly linked to lysine residues on target proteins. To identify the lysine residue(s) responsible for CIDEB ubiquitylation, we generated truncation mutants of CIDEB cDNA in which regions containing specific lysine residues were removed (Fig. 2D). The deletion of amino acids 166 through 195, which contained two lysine residues, K172 and K173, significantly stabilized the protein (Fig. 2E). Site-directed mutagenesis further confirmed that lysine 173 is important for CIDEB degradation, because its replacement by an alanine residue stabilized the protein (Fig. 2F).
FIG 2.
Endogenous CIDEB is a short-lived protein that is posttranslationally regulated via the ubiquitin-proteasome pathway. (A) Cells were treated with the protein synthesis inhibitor CHX for 0, 1, 2, or 3 h, then lysed, and analyzed by Western blotting for CIDEB protein levels. GAPDH was included as a loading control. (B) Treatment with the proteasome inhibitor MG132 blocked CIDEB degradation in both wild-type cells and cells harboring a lentivirus encoding shRNA targeting CIDEB mRNA. (C) Analysis of CIDEB ubiquitination. Cells were cotransfected with FLAG-CIDEB or a FLAG-control protein (Prp31c) and HA-ubiquitin constructs, followed by MG132 treatment, coimmunoprecipitation with anti-FLAG beads, and Western blotting. (D) Schematic of FLAG-CIDEB deletion constructs used for stability analysis. (E) Stability of CIDEB deletion mutants. Cells were transfected with various FLAG-CIDEB-encoding constructs (represented in panel D) and subsequently treated with CHX, 24 h posttransfection, for 4 h, before immunoblot analysis using FLAG-specific antibody. (F) The point mutation K173A enhanced CIDEB stability. Cells transfected with wild-type or K173A CIDEB mutant cDNA were treated with CHX, collected at the indicated times, and analyzed similarly to the cells for panel A.
HCV infection triggers a proteolytic cleavage of CIDEB in a core-dependent manner.
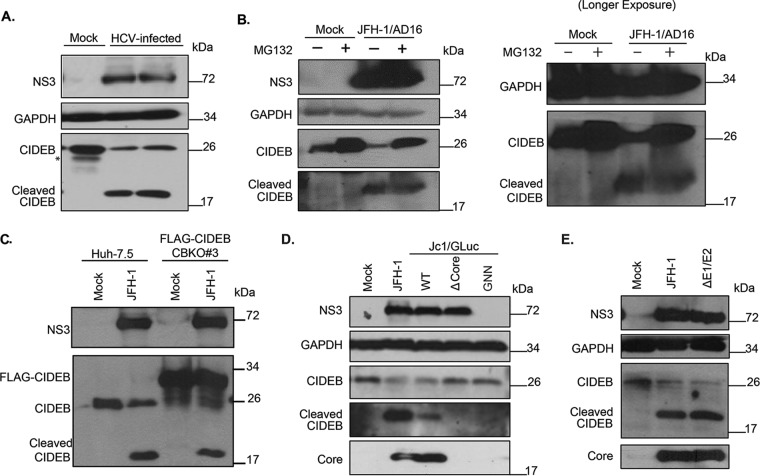
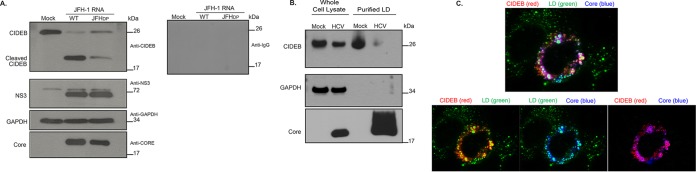
Consistent with our previous report (29), immunoblot analysis of HCVcc-infected Huh-7.5 cells showed reduced CIDEB levels compared to those of uninfected Huh-7.5 cells. Interestingly, in experiments where the samples were directly lysed in protein loading buffer and where protein degradation was minimized after cell lysis, the reduction of CIDEB full-length protein coincided with the appearance of a faster-migrating species that is recognized by the anti-CIDEB antibody (Fig. 3A). This species was not observed when immunoblot analysis was performed with an IgG control instead of anti-CIDEB (data not shown), indicating that this faster-migrating species is a CIDEB cleavage product and that HCV infection may trigger proteolytic cleavage of CIDEB. MG132 treatment did not reduce the HCV-activated proteolytic cleavage of CIDEB (Fig. 3B), indicating that the mechanisms of cellular and virus-induced CIDEB downregulation are distinct. A similar band was also observed in the aforementioned FLAG-CIDEB CBKO#3 cells infected with HCVcc that was detectable by anti-CIDEB antibody (Fig. 3C) but not by anti-FLAG antibody. Note that the FLAG epitope was fused to the N terminus of CIDEB, which is unstable upon cleavage and cannot be detected with the anti-FLAG antibody either in immunofluorescence assays (IFA) (Fig. 1A) or on Western blots (data not shown). To identify viral determinants involved in CIDEB downregulation and cleavage, immunoblot analysis of CIDEB was performed following electroporation of cells with one of three different mutant HCV genomes: one with an internal in-frame deletion in the HCV core gene (Δcore), one with mutations in the active site of the viral polymerase (GNN), and one with deletion of the E1E2 genes (ΔE1/E2). While the wild-type genome efficiently downregulated CIDEB full-length protein and resulted in the appearance of the cleavage product (Fig. 3D), the Δcore mutant failed to reduce the CIDEB level or trigger cleavage despite expressing similar amounts of the HCV NS3 protease (Fig. 3D). The polymerase-deficient GNN mutant failed to replicate and did not express detectable amounts of HCV proteins. It also failed to downregulate CIDEB as expected (Fig. 3D). On the other hand, the ΔE1/E2 mutant was still able to downregulate CIDEB and to activate cleavage (Fig. 3E). These results demonstrate that the HCV core, but not the glycoproteins, is required for CIDEB downregulation via a mechanism independent of the HCV protease.
FIG 3.
HCV-infected cells exhibit lower levels of CIDEB protein than do uninfected cells, likely through a proteolytic cleavage event. (A) Effect of HCV infection on endogenous CIDEB protein. Western blot analysis of Huh-7.5 cells infected with JFH-1/AD16 for 3 days revealed a faster-migrating band recognized by the CIDEB antibody, in addition to the full-length product. The asterisk indicates a degradation product occasionally seen from uninfected (mock) Huh-7.5 cells. (B) Effect of proteasome inhibition on CIDEB protein levels in HCV-infected cells. Huh-7.5 cells were infected with JFH-1/AD16 for 48 h and then treated with MG132 for 18 h before analysis by Western blotting. Results obtained with a longer exposure of GAPDH, CIDEB, and the CIDEB cleavage product are shown to the right. (C) Effect of HCV infection on N-terminally FLAG-tagged CIDEB. Shown are the results of Western blot analysis of CIDEB-KO cells (clone number 3) stably expressing FLAG-CIDEB and electroporated with JFH-1 for 3 days. FLAG-CIDEB was detected by anti-CIDEB antibody. (D) Western blot analysis of endogenous CIDEB and NS3 levels in Huh-7.5 cells with various HCV RNAs. Cells were electroporated with 10 μg of RNA generated from either wild-type or mutant genomes and then analyzed 48 h later for the ability to downregulate CIDEB. The Δcore mutant is a Jc1/GLuc variant lacking the intact HCV capsid protein; the GNN mutant is a Jc1/GLuc variant harboring a replication-deficient replicase. (E) Similar analysis of endogenous CIDEB and NS3 levels in Huh-7.5 cells 48 h after cells were electroporated with 10 μg of RNA generated from either wild-type JFH-1 or the ΔE1/E2-JFH-1 mutant, a viral construct lacking the two HCV envelope proteins.
Role of LDs and the core's LD localization in CIDEB downregulation.
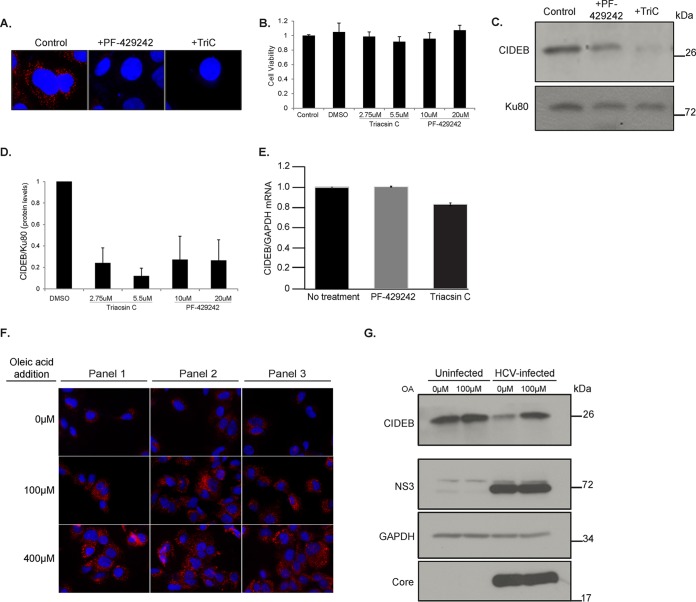
The HCV core localizes to the surface of LDs in HCV infected cells, which may result in a competition with other LD-localized proteins, such as CIDEB. To address this possibility, we investigated whether the amounts of intracellular lipids and LDs affect CIDEB protein stability. First, we inhibited LD formation in Huh-7.5 cells using lipid inhibitors and then determined whether the CIDEB level was reduced. Two structurally distinct inhibitors were used. 4-([Diethylamino]methyl)-N-[2-(2-methoxyphenyl)ethyl]-N-(3R)-3-pyrrolidinylbenzamide (PF-429242) is a reversible inhibitor of cholesterol and fatty acid synthesis, acting by blocking SREBP cleavage; Triacsin C (TriC) is an inhibitor of long-chain-fatty-acid acyl coenzyme A (acyl-CoA) synthetase. Both compounds inhibited LD formation as expected, without significantly affecting cell viability (Fig. 4A and B), and more importantly, they also effectively reduced CIDEB protein in Huh-7.5 cells (Fig. 4C and D). The reduction of CIDEB protein was not due to a decrease in CIDEB mRNA level upon treatment (Fig. 4E), and we did not detect the putative cleavage product (data not shown) in these experiments. In the second line of experiments, we augmented intracellular LD formation by feeding the cells with excess oleic acid (OA) and then determined if this lipid loading, which significantly increased the number of LDs in Huh-7.5 cells (Fig. 4F), could modulate the CIDEB level in HCV-infected cells. Loading the cells with OA restored the CIDEB protein level in HCV-infected cells to a level similar to that in uninfected, unloaded cells (Fig. 4G), consistent with the hypothesis that increased LD availability can counteract the effect of HCV infection on the CIDEB level. To directly address if the localization of HCV core protein to LDs is required for CIDEB downregulation, we took advantage of a core double proline mutant that reduced its LD association without significantly affecting its expression level (45). Electroporation of the full-length HCV genome containing these core mutations (JFHDP) was still capable of activating the pathway that leads to CIDEB cleavage, but the extent of downregulation was reduced compared to that of the wild type (Fig. 5A). The NS3 expression levels were comparable, suggesting that core protein and its association with LDs, but not overall viral protein expression, is correlated with CIDEB downregulation. In addition, the reduction of CIDEB protein upon HCV infection was even more pronounced in the LD fractions (Fig. 5B). Localization of both CIDEB and core onto the surface of the same LDs could also be detected when we coexpressed these proteins to a high level in uninfected cells (Fig. 5C).
FIG 4.
Intracellular lipid abundance affects CIDEB protein stability. (A) Effect of two structurally distinct lipid droplet inhibitors, PF-429242, a reversible inhibitor of cholesterol synthesis, and Triacsin C, an inhibitor of long-chain-fatty-acid acyl-CoA synthetase, on LDs in Huh-7.5 cells. Cells were treated with 40 μM PF-429242 or 5.5 μM TriC for 24 h before staining with ORO. (B) Cell viability was measured after 24 h of treatment with the indicated compounds. (C and D) Western blot analysis of CIDEB protein levels in Huh-7.5 cells treated with PF-429242 or TriC for 24 h. Shown is quantification of CIDEB protein levels on a Western blot by ImageJ analysis (analyzed from biological triplicates). Ku80 was used as a loading control. (E) A 24-h treatment of Huh-7.5 cells with PF-429242 or TriC does not significantly alter CIDEB mRNA levels. (F and G) Effect of exogenous lipid loading on CIDEB protein in infected cells. Uninfected or (24-h) JFH-1/AD16-infected Huh-7.5 cells were treated with 100 μM OA for 20 h before being fixed for ORO staining or lysed for Western blot analysis.
FIG 5.
(A) Effect of a JFH-1 core mutant deficient in LD-targeting on CIDEB cleavage. Western blot analysis of Huh-7.5 cells was performed 48 h after electroporation of cells with 10 μg of wild-type JFH-1 (WT) or JFHDP RNA. (B) Localization of endogenous CIDEB and HCV core after HCV infection assayed by LD fractionation. Fifteen micrograms of whole-cell lysate and 7.5 μg of purified LD-associated proteins were analyzed by Western blotting. (C) Colocalization of exogenously overexpressed FLAG-tagged CIDEB and HCV core protein on lipid droplets.
CIDEB knockout stabilizes LDs in the presence of excess lipids.
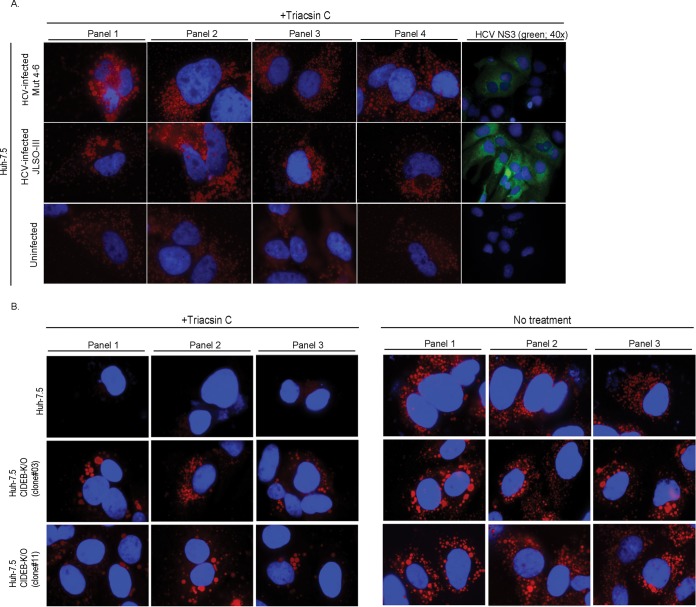
A mechanism by which HCV core may contribute to hepatic steatosis is through the stabilization of cytoplasmic LDs (25, 26). Consistent with previous reports, we also observed increased LD stability in hepatoma cells infected with HCV compared to that in uninfected cells (Fig. 6A). We determined if suppression of CIDEB could similarly affect LD stability. We recently generated Huh-7.5 cell clones that have the CIDEB gene knocked out using transcription activator-like effector nuclease (TALEN)-based genome editing (29). In the absence of lipid loading, both the CIDEB shRNA and the CIDEB KO reduced the number of LDs (29). When the cells were fed with lipid-rich medium that contained oleic acid, however, both parental and KO cells were able to form LDs (Fig. 6B). To determine if CIDEB KO affects LD stability, we used TriC to block the synthesis of fatty acids after lipid loading. Under these conditions, the KO cells contained many more large LDs than the wild-type cells did (Fig. 6C), likely due to the reduced secretion of the VLDLs or LDLs, which can slow down the depletion of their intracellular reservoir (the cytoplasmic LDs) in the absence of new lipid synthesis.
FIG 6.
CIDEB modulates LD stability. (A) LD stability in naive or JFH-1/AD16-infected Huh-7.5 cells. Cells were incubated in lipid-rich medium for 14 h, washed with PBS, and cultured in DMEM with 5.5 μM TriC for 24 h before staining with ORO (red, 100×) and 4′,6-diamidino-2-phenylindole (DAPI) (blue) or with anti-NS3 (green, 40×) and DAPI. (B) Formation of LDs in Huh-7.5 or Huh-7.5 CIDEB-KO cells after lipid loading. Cells were incubated in lipid-rich medium for 20 h, washed with PBS, and stained for LDs using ORO (red, 100×). (C) LD stability in wild-type and CIDEB KO cells. After lipid loading as described above, cells were cultured in DMEM with or without 5.5 μM TriC for 24 h and then stained for LDs using ORO (red, 100×). Panels 1, 2, and 3 are separate frames. DAPI (blue) was used as a counterstain.
CIDEB knockout in vitro phenocopies the alteration of lipid profiles caused by HCV infection.
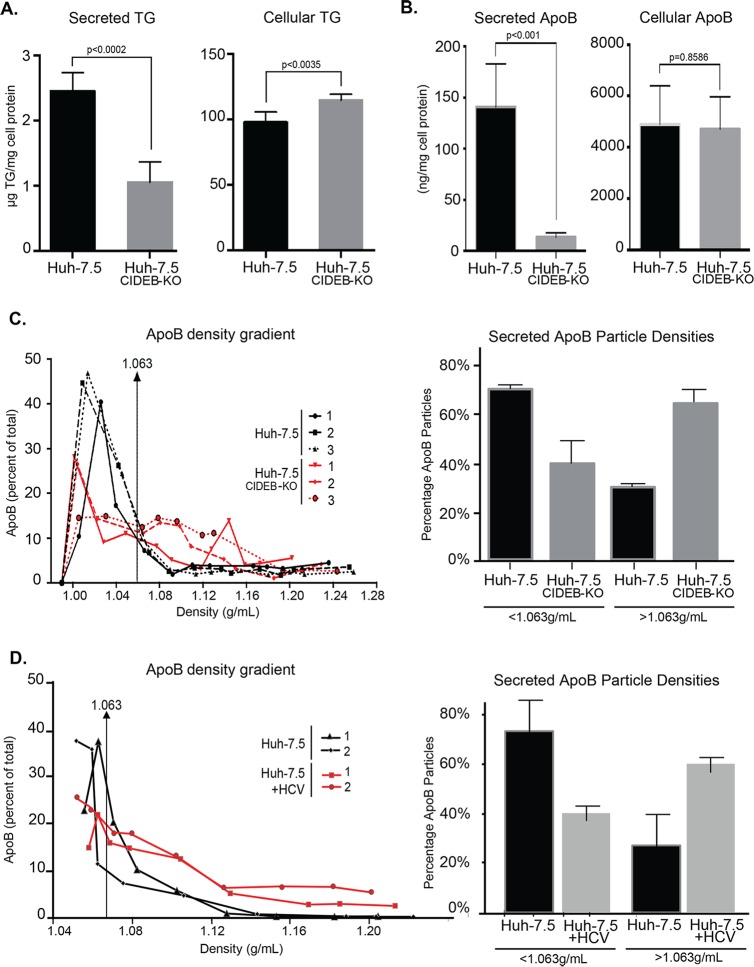
Given the involvement of CIDE family proteins in lipid metabolism, we determined the effect of CIDEB deficiency on the VLDL secretion and intracellular TG levels in a human hepatoma cell line. It has previously been shown that Huh-7.5 cells secrete very small amounts of TG; however, these cells can be used for VLDL studies if supplemented with exogenous OA in order to first expand the intracellular TG pool (21). Therefore, we supplied cells with exogenous OA prior to measuring TG secretion. Compared with wild-type Huh-7.5 cells, the CIDEB KO cells had a reduced level of TG secretion accompanied by a slight increase of intracellular TG level after lipid loading (Fig. 7A). In addition, the CIDEB KO cells secreted less ApoB-containing VLDLs (Fig. 7B). Analysis of secreted ApoB particles revealed an overall increase of density of the ApoB particles secreted from the CIDEB KO cells (Fig. 7C), suggesting a reduction in intracellular lipidation of ApoB. Overall, the VLDL density profile of ApoB particles secreted from CIDEB KO cells is similar to the density profiles for ApoB secreted from HCV-infected cells (Fig. 7D) and differs from that of wild-type, uninfected cells.
FIG 7.
Knockout of CIDEB in human hepatoma cells results in altered VLDL secretion and TG storage. (A) Secreted and intracellular TG in Huh-7.5 or Huh-7.5 CIDEB-KO (clone 11) cells. Cells were cultured in a lipid-rich medium (supplemented with 375 μM OA) for 20 h, washed with PBS, and then either collected for lysate or further incubated in fresh DMEM for 2 h for supernatant collection. Both the lysate and supernatant were analyzed for TG contents. (B) Analysis of secreted and intracellular ApoB levels. Huh-7.5 cells or or Huh-7.5 CIDEB-KO (clone 11) cells were cultured in a lipid-rich medium (supplemented with 375 μM OA) for 14 h, washed with PBS, and then further incubated in fresh DMEM for 8 h for supernatant collection. (C) Lipid density profile of secreted ApoB-associated VLDL particles. Fractions of the supernatant collected as described above were analyzed for the amount of ApoB protein in each fraction. Distribution of ApoB-containing particles of different densities are quantified and plotted. Data from three replicate experiments are shown. (D) Lipid density profile of secreted ApoB-associated VLDL particles in HCV-infected cells. Naive Huh-7.5 cells and and Huh-7.5 cells 60 h post-JFH-1/AD16 infection were cultured in lipid-rich medium (supplemented with 375 μM OA) for 14 h, washed with PBS, and then further incubated in fresh DMEM for 8 h for supernatant collection. Fractions of the supernatant collected as described above were analyzed for the amount of ApoB protein in each fraction. Distributions of ApoB-containing particles of different densities are quantified and plotted. Data from two biological replicate experiments are shown.
DISCUSSION
In this study, we demonstrated that HCV infection reduces full-length CIDEB protein by activating a proteolytic cleavage event. This downregulation is independent of HCV glycoproteins but requires the HCV core and may play a role in HCV-induced lipid accumulation in HCV pathogenesis.
HCV-induced modulation of CIDEB protein is distinct from the normal ubiquitin-proteasomal regulation of the protein, which has also been shown to modulate CIDEA and CIDEC proteins in other cell types (46, 47). The fine-tuning of CIDE family proteins by proteasomes may be critical for maintaining normal cell physiology. On one hand, the CIDE proteins readily induced cell death in a variety of cell types when overexpressed (27, 30, 31); on the other hand, suboptimal levels of CIDEB can lead to an imbalance of lipid metabolism. Notably, apoptosis induction likely accounts for a reported antiviral effect of CIDEB overexpression in cell culture (48); Singaravelu et al. concluded that CIDEB's ability to reduce HCV RNA levels in replicon cells was correlated with its proapoptotic function.
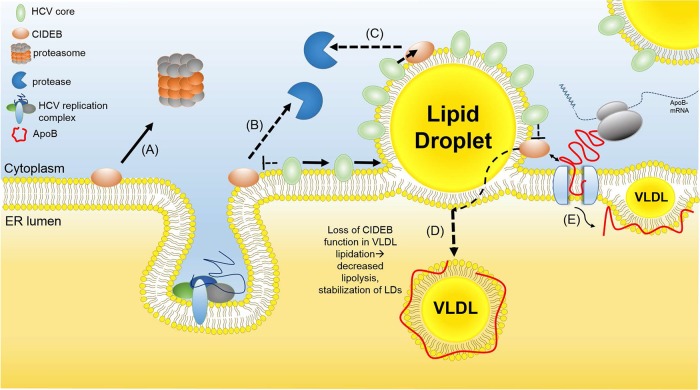
A specific cleavage in response to a stimulus such as viral infection has not been reported for any CIDE proteins but appears to be the mechanism of HCV-mediated downregulation of CIDEB. Previous studies have shown CIDEB KO mice to exhibit increased resistance to diet-induced steatosis and reduced lipogenesis (34). Similarly, we have observed LD formation being impaired in uninfected human hepatoma cells when CIDEB expression is reduced by shRNA or eliminated by gene knockout (29). Upon HCV infection, which increases lipogenesis (12, 14, 23), however, CIDEB depletion causes altered VLDL profiles and leads to intracellular lipid accumulation. Similarly, when cells were fed with excess lipids, suppression of CIDEB stabilized cytoplasmic LDs. The observation that genetic ablation led to perturbations of the lipid secretion pathway similar to those caused by HCV infection suggests a connection between CIDEB modulation and lipid regulation in infected cells. Consistent with this hypothesis, HCV-induced CIDEB downregulation was observed in cultured cells infected with a genotype 3 virus as well as in infected liver tissues of an HCV mouse model. The in vivo data are consistent with a previous report showing that transgenic expression of HCV proteins in mice led to a reduction of CIDEB protein (31), even though the cleavage product was not detectable from the in vivo samples, which could be due to the labile nature of the protein, especially considering the fixing and processing necessary for the live tissues. Combined with a growing body of evidence supporting the function of the CIDE proteins in lipid regulation and the reported role of the HCV core in virus-induced steatosis, our results support a model (Fig. 8) in which chronic HCV infection of human hepatocytes results in sustained reduction of CIDEB protein, which, in turn, contributes to hepatic lipid dysregulation and steatosis.
FIG 8.
Potential mechanism for HCV-mediated CIDEB downregulation. CIDEB protein levels are normally regulated by the ubiquitin-proteasome pathway (A). In the infected cells, HCV core is trafficked onto LDs (B), which may result in competitive displacement of CIDEB off the LD surface (C), exposing CIDEB to a protease. A reduced level of CIDEB results in an altered VLDL lipid profile (D), through a mechanism involving the reported CIDEB-ApoB interaction (E) (34). ER, endoplasmic reticulum.
Localization of HCV core to the surface of LDs is important for HCV assembly (3), and colocalization of an LD-targeted cellular factor, TIP47, with NS5A and the core on LDs may be required for HCV replication and release (49). On the other hand, the extensive coating of the LD surface by HCV core protein in the infected hepatocytes can lead to displacement of native LD proteins, such as CIDEB and perilipins. Such a competition has been shown to play a role in the downregulation of perilipin 2 (PLIN2) protein, also known as adipocyte differentiation-related protein (ADRP). The dissociation of ADRP from LDs, as a result of core competition, led to reduced stability of ADRP (50), presumably due to increased exposure to protease(s). Our results suggest a role for HCV core's LD localization in CIDEB downregulation, consistent with such a competition-displacement model that is distinct from HCV NS3/4A-mediated cleavage of critical antiviral signaling proteins (51, 52). Of note, no specific interaction between CIDEB and the HCV core was detected in co-IP experiments (data not shown).
The displacement of CIDEB from the LD surface may expose the protein to a protease that normally does not have access to CIDEB. Alternatively, the presence of the HCV core during infection may activate a protease that can cleave CIDEB. Preliminary experiments testing different classes of protease inhibitors, including calpain and caspase inhibitors, have yielded inconclusive results. The Δcore mutant genome expresses both NS2 and the NS3/4A proteases encoded by HCV but failed to downregulate CIDEB protein or trigger the proteolytic cleavage, suggesting the involvement of cellular, rather than viral, proteases. The identification of the specific protease(s) that is responsible for cleaving CIDEB in an HCV-dependent manner can provide promising candidates for targeted interventions of lipid dysregulation.
ACKNOWLEDGMENTS
We thank the following colleagues for providing reagents: Guangxiang G. Luo, Charles M. Rice, Fanxiu Zhu, and Brett Lindenbach. We thank Stephen Frausto, Christopher Pu, and Jaime Lewis for technical assistance.
Funding Statement
This work was supported by NIH/NIAID grants R56 AI107763 (H.T.) and R21 AI111250 (H.T.) and CIHR grant 52973 (N.M.K., D.N.D.). E.M.L. is supported by a predoctoral fellowship from the American Heart Association. N.M.K. is also supported by the Capital Health Chair in Transplantation Research from Alberta Health Services, Canada.
REFERENCES
- 1.Pawlotsky JM. 2013. Treatment of chronic hepatitis C: current and future. Curr Top Microbiol Immunol 369:321–342. [DOI] [PubMed] [Google Scholar]
- 2.Schaefer EA, Chung RT. 2013. HCV and host lipids: an intimate connection. Semin Liver Dis 33:358–368. doi: 10.1055/s-0033-1358524. [DOI] [PubMed] [Google Scholar]
- 3.Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol 9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 4.Lindenbach BD, Rice CM. 2013. The ins and outs of hepatitis C virus entry and assembly. Nat Rev Microbiol 11:688–700. doi: 10.1038/nrmicro3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monazahian M, Bohme I, Bonk S, Koch A, Scholz C, Grethe S, Thomssen R. 1999. Low density lipoprotein receptor as a candidate receptor for hepatitis C virus. J Med Virol 57:223–229. doi:. [DOI] [PubMed] [Google Scholar]
- 6.Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J 21:5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sainz B Jr, Barretto N, Martin DN, Hiraga N, Imamura M, Hussain S, Marsh KA, Yu X, Chayama K, Alrefai WA, Uprichard SL. 2012. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat Med 18:281–285. doi: 10.1038/nm.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, Grakoui A, Randall G. 2009. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc Natl Acad Sci U S A 106:7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. 2009. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe 5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamane D, McGivern DR, Wauthier E, Yi M, Madden VJ, Welsch C, Antes I, Wen Y, Chugh PE, McGee CE, Widman DG, Misumi I, Bandyopadhyay S, Kim S, Shimakami T, Oikawa T, Whitmire JK, Heise MT, Dittmer DP, Kao CC, Pitson SM, Merrill AH Jr, Reid LM, Lemon SM. 2014. Regulation of the hepatitis C virus RNA replicase by endogenous lipid peroxidation. Nat Med 20:927–935. doi: 10.1038/nm.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diamond DL, Syder AJ, Jacobs JM, Sorensen CM, Walters KA, Proll SC, McDermott JE, Gritsenko MA, Zhang Q, Zhao R, Metz TO, Camp DG II, Waters KM, Smith RD, Rice CM, Katze MG. 2010. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog 6:e1000719. doi: 10.1371/journal.ppat.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lambert JE, Bain VG, Ryan EA, Thomson AB, Clandinin MT. 2013. Elevated lipogenesis and diminished cholesterol synthesis in patients with hepatitis C viral infection compared to healthy humans. Hepatology 57:1697–1704. doi: 10.1002/hep.25990. [DOI] [PubMed] [Google Scholar]
- 13.Waris G, Felmlee DJ, Negro F, Siddiqui A. 2007. Hepatitis C virus induces proteolytic cleavage of sterol regulatory element binding proteins and stimulates their phosphorylation via oxidative stress. J Virol 81:8122–8130. doi: 10.1128/JVI.00125-07. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Yang W, Hood BL, Chadwick SL, Liu S, Watkins SC, Luo G, Conrads TP, Wang T. 2008. Fatty acid synthase is up-regulated during hepatitis C virus infection and regulates hepatitis C virus entry and production. Hepatology 48:1396–1403. doi: 10.1002/hep.22508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujino T, Nakamuta M, Yada R, Aoyagi Y, Yasutake K, Kohjima M, Fukuizumi K, Yoshimoto T, Harada N, Yada M, Kato M, Kotoh K, Taketomi A, Maehara Y, Nakashima M, Enjoji M. 2010. Expression profile of lipid metabolism-associated genes in hepatitis C virus-infected human liver. Hepatol Res 40:923–929. doi: 10.1111/j.1872-034X.2010.00700.x. [DOI] [PubMed] [Google Scholar]
- 16.Lerat H, Kammoun HL, Hainault I, Merour E, Higgs MR, Callens C, Lemon SM, Foufelle F, Pawlotsky JM. 2009. Hepatitis C virus proteins induce lipogenesis and defective triglyceride secretion in transgenic mice. J Biol Chem 284:33466–33474. doi: 10.1074/jbc.M109.019810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Q, Pene V, Krishnamurthy S, Cha H, Liang TJ. 2013. Hepatitis C virus infection activates an innate pathway involving IKK-alpha in lipogenesis and viral assembly. Nat Med 19:722–729. doi: 10.1038/nm.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leandro G, Mangia A, Hui J, Fabris P, Rubbia-Brandt L, Colloredo G, Adinolfi LE, Asselah T, Jonsson JR, Smedile A, Terrault N, Pazienza V, Giordani MT, Giostra E, Sonzogni A, Ruggiero G, Marcellin P, Powell EE, George J, Negro F, HCV Meta-Analysis (on) Individual Patients' Data Study Group. 2006. Relationship between steatosis, inflammation, and fibrosis in chronic hepatitis C: a meta-analysis of individual patient data. Gastroenterology 130:1636–1642. doi: 10.1053/j.gastro.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 19.Castera L, Chouteau P, Hezode C, Zafrani ES, Dhumeaux D, Pawlotsky JM. 2005. Hepatitis C virus-induced hepatocellular steatosis. Am J Gastroenterol 100:711–715. doi: 10.1111/j.1572-0241.2005.40898.x. [DOI] [PubMed] [Google Scholar]
- 20.Bugianesi E, Salamone F, Negro F. 2012. The interaction of metabolic factors with HCV infection: does it matter? J Hepatol 56(Suppl 1):S56–S65. [DOI] [PubMed] [Google Scholar]
- 21.Nourbakhsh M, Douglas DN, Pu CH, Lewis JT, Kawahara T, Lisboa LF, Wei E, Asthana S, Quiroga AD, Law LM, Chen C, Addison WR, Nelson R, Houghton M, Lehner R, Kneteman NM. 2013. Arylacetamide deacetylase: a novel host factor with important roles in the lipolysis of cellular triacylglycerol stores, VLDL assembly and HCV production. J Hepatol 59:336–343. doi: 10.1016/j.jhep.2013.03.022. [DOI] [PubMed] [Google Scholar]
- 22.Moriya K, Yotsuyanagi H, Shintani Y, Fujie H, Ishibashi K, Matsuura Y, Miyamura T, Koike K. 1997. Hepatitis C virus core protein induces hepatic steatosis in transgenic mice. J Gen Virol 78(Part 7):1527–1531. [DOI] [PubMed] [Google Scholar]
- 23.Lerat H, Honda M, Beard MR, Loesch K, Sun J, Yang Y, Okuda M, Gosert R, Xiao SY, Weinman SA, Lemon SM. 2002. Steatosis and liver cancer in transgenic mice expressing the structural and nonstructural proteins of hepatitis C virus. Gastroenterology 122:352–365. doi: 10.1053/gast.2002.31001. [DOI] [PubMed] [Google Scholar]
- 24.Piodi A, Chouteau P, Lerat H, Hezode C, Pawlotsky JM. 2008. Morphological changes in intracellular lipid droplets induced by different hepatitis C virus genotype core sequences and relationship with steatosis. Hepatology 48:16–27. doi: 10.1002/hep.22288. [DOI] [PubMed] [Google Scholar]
- 25.Harris C, Herker E, Farese RV Jr, Ott M. 2011. Hepatitis C virus core protein decreases lipid droplet turnover: a mechanism for core-induced steatosis. J Biol Chem 286:42615–42625. doi: 10.1074/jbc.M111.285148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camus G, Schweiger M, Herker E, Harris C, Kondratowicz AS, Tsou CL, Farese RV Jr, Herath K, Previs SF, Roddy TP, Pinto S, Zechner R, Ott M. 2014. The hepatitis C virus core protein inhibits adipose triglyceride lipase (ATGL)-mediated lipid mobilization and enhances the ATGL interaction with comparative gene identification 58 (CGI-58) and lipid droplets. J Biol Chem 289:35770–35780. doi: 10.1074/jbc.M114.587816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inohara N, Koseki T, Chen S, Wu X, Nunez G. 1998. CIDE, a novel family of cell death activators with homology to the 45 kDa subunit of the DNA fragmentation factor. EMBO J 17:2526–2533. doi: 10.1093/emboj/17.9.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu X, Robotham JM, Lee E, Dalton S, Kneteman NM, Gilbert DM, Tang H. 2012. Productive hepatitis C virus infection of stem cell-derived hepatocytes reveals a critical transition to viral permissiveness during differentiation. PLoS Pathog 8:e1002617. doi: 10.1371/journal.ppat.1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu X, Lee EM, Hammack C, Robotham JM, Basu M, Lang J, Brinton MA, Tang H. 2014. Cell death-inducing DFFA-like effector b is required for hepatitis C virus entry into hepatocytes. J Virol 88:8433–8444. doi: 10.1128/JVI.00081-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu K, Zhou S, Kim JY, Tillison K, Majors D, Rearick D, Lee JH, Fernandez-Boyanapalli RF, Barricklow K, Houston MS, Smas CM. 2009. Functional analysis of FSP27 protein regions for lipid droplet localization, caspase-dependent apoptosis, and dimerization with CIDEA. Am J Physiol Endocrinol Metab 297:E1395–E1413. doi: 10.1152/ajpendo.00188.2009. [DOI] [PubMed] [Google Scholar]
- 31.Erdtmann L, Franck N, Lerat H, Le Seyec J, Gilot D, Cannie I, Gripon P, Hibner U, Guguen-Guillouzo C. 2003. The hepatitis C virus NS2 protein is an inhibitor of CIDE-B-induced apoptosis. J Biol Chem 278:18256–18264. doi: 10.1074/jbc.M209732200. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Z, Yon Toh S, Chen Z, Guo K, Ng CP, Ponniah S, Lin SC, Hong W, Li P. 2003. Cidea-deficient mice have lean phenotype and are resistant to obesity. Nat Genet 35:49–56. doi: 10.1038/ng1225. [DOI] [PubMed] [Google Scholar]
- 33.Nishino N, Tamori Y, Tateya S, Kawaguchi T, Shibakusa T, Mizunoya W, Inoue K, Kitazawa R, Kitazawa S, Matsuki Y, Hiramatsu R, Masubuchi S, Omachi A, Kimura K, Saito M, Amo T, Ohta S, Yamaguchi T, Osumi T, Cheng J, Fujimoto T, Nakao H, Nakao K, Aiba A, Okamura H, Fushiki T, Kasuga M. 2008. FSP27 contributes to efficient energy storage in murine white adipocytes by promoting the formation of unilocular lipid droplets. J Clin Invest 118:2808–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye J, Li JZ, Liu Y, Li X, Yang T, Ma X, Li Q, Yao Z, Li P. 2009. Cideb, an ER- and lipid droplet-associated protein, mediates VLDL lipidation and maturation by interacting with apolipoprotein B. Cell Metab 9:177–190. doi: 10.1016/j.cmet.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 35.Tiwari S, Siddiqi S, Siddiqi SA. 2013. CideB protein is required for the biogenesis of very low density lipoprotein (VLDL) transport vesicle. J Biol Chem 288:5157–5165. doi: 10.1074/jbc.M112.434258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li JZ, Lei Y, Wang Y, Zhang Y, Ye J, Xia X, Pan X, Li P. 2010. Control of cholesterol biosynthesis, uptake and storage in hepatocytes by Cideb. Biochim Biophys Acta 1801:577–586. doi: 10.1016/j.bbalip.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 37.Jiang J, Luo G. 2012. Cell culture-adaptive mutations promote viral protein-protein interactions and morphogenesis of infectious hepatitis C virus. J Virol 86:8987–8997. doi: 10.1128/JVI.00004-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaul A, Woerz I, Meuleman P, Leroux-Roels G, Bartenschlager R. 2007. Cell culture adaptation of hepatitis C virus and in vivo viability of an adapted variant. J Virol 81:13168–13179. doi: 10.1128/JVI.01362-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim S, Date T, Yokokawa H, Kono T, Aizaki H, Maurel P, Gondeau C, Wakita T. 2014. Development of hepatitis C virus genotype 3a cell culture system. Hepatology 60:1838–1850. doi: 10.1002/hep.27197. [DOI] [PubMed] [Google Scholar]
- 40.Ding Y, Zhang S, Yang L, Na H, Zhang P, Zhang H, Wang Y, Chen Y, Yu J, Huo C, Xu S, Garaiova M, Cong Y, Liu P. 2013. Isolating lipid droplets from multiple species. Nat Protoc 8:43–51. [DOI] [PubMed] [Google Scholar]
- 41.Mercer DF, Schiller DE, Elliott JF, Douglas DN, Hao C, Rinfret A, Addison WR, Fischer KP, Churchill TA, Lakey JR, Tyrrell DL, Kneteman NM. 2001. Hepatitis C virus replication in mice with chimeric human livers. Nat Med 7:927–933. doi: 10.1038/90968. [DOI] [PubMed] [Google Scholar]
- 42.Singaravelu R, Lyn RK, Srinivasan P, Delcorde J, Steenbergen RH, Tyrrell DL, Pezacki JP. 2013. Human serum activates CIDEB-mediated lipid droplet enlargement in hepatoma cells. Biochem Biophys Res Commun 441:447–452. doi: 10.1016/j.bbrc.2013.10.080. [DOI] [PubMed] [Google Scholar]
- 43.Eden E, Geva-Zatorsky N, Issaeva I, Cohen A, Dekel E, Danon T, Cohen L, Mayo A, Alon U. 2011. Proteome half-life dynamics in living human cells. Science 331:764–768. doi: 10.1126/science.1199784. [DOI] [PubMed] [Google Scholar]
- 44.Franch HA, Sooparb S, Du J, Brown NS. 2001. A mechanism regulating proteolysis of specific proteins during renal tubular cell growth. J Biol Chem 276:19126–19131. doi: 10.1074/jbc.M101777200. [DOI] [PubMed] [Google Scholar]
- 45.Boulant S, Targett-Adams P, McLauchlan J. 2007. Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J Gen Virol 88:2204–2213. doi: 10.1099/vir.0.82898-0. [DOI] [PubMed] [Google Scholar]
- 46.Chan SC, Lin SC, Li P. 2007. Regulation of Cidea protein stability by the ubiquitin-mediated proteasomal degradation pathway. Biochem J 408:259–266. doi: 10.1042/BJ20070690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nian Z, Sun Z, Yu L, Toh SY, Sang J, Li P. 2010. Fat-specific protein 27 undergoes ubiquitin-dependent degradation regulated by triacylglycerol synthesis and lipid droplet formation. J Biol Chem 285:9604–9615. doi: 10.1074/jbc.M109.043786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singaravelu R, Delcorde J, Lyn RK, Steenbergen RH, Jones DM, Tyrrell DL, Russell RS, Pezacki JP. 2014. Investigating the antiviral role of cell death-inducing DFF45-like effector B in HCV replication. FEBS J 281:3751–3765. doi: 10.1111/febs.12901. [DOI] [PubMed] [Google Scholar]
- 49.Vogt DA, Camus G, Herker E, Webster BR, Tsou CL, Greene WC, Yen TS, Ott M. 2013. Lipid droplet-binding protein TIP47 regulates hepatitis C virus RNA replication through interaction with the viral NS5A protein. PLoS Pathog 9:e1003302. doi: 10.1371/journal.ppat.1003302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boulant S, Douglas MW, Moody L, Budkowska A, Targett-Adams P, McLauchlan J. 2008. Hepatitis C virus core protein induces lipid droplet redistribution in a microtubule- and dynein-dependent manner. Traffic 9:1268–1282. doi: 10.1111/j.1600-0854.2008.00767.x. [DOI] [PubMed] [Google Scholar]
- 51.Foy E, Li K, Wang C, Sumpter R Jr, Ikeda M, Lemon SM, Gale M Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145–1148. doi: 10.1126/science.1082604. [DOI] [PubMed] [Google Scholar]
- 52.Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. 2005. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A 102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]