ABSTRACT
Understanding how some HIV-infected cells resist the cytotoxicity of HIV replication is crucial to enabling HIV cure efforts. HIV killing of CD4 T cells that replicate HIV can involve HIV protease-mediated cleavage of procaspase 8 to generate a fragment (Casp8p41) that directly binds and activates the mitochondrial proapoptotic protein BAK. Here, we demonstrate that Casp8p41 also binds with nanomolar affinity to the antiapoptotic protein Bcl-2, which sequesters Casp8p41 and prevents apoptosis. Further, we show that central memory CD4 T cells (TCM) from HIV-infected individuals have heightened expression of BCL-2 relative to procaspase 8, possibly explaining the persistence of HIV-infected TCM despite generation of Casp8p41. Consistent with this hypothesis, the selective BCL-2 antagonist venetoclax induced minimal killing of uninfected CD4 T cells but markedly increased the death of CD4 T cells and diminished cell-associated HIV DNA when CD4 T cells from antiretroviral therapy (ART)-suppressed HIV patients were induced with αCD3/αCD28 to reactivate HIV ex vivo. Thus, priming CD4 T cells from ART suppressed HIV patients with a BCL-2 antagonist, followed by HIV reactivation, achieves reductions in cell-associated HIV DNA, whereas HIV reactivation alone does not.
IMPORTANCE HIV infection is incurable due to a long-lived reservoir of HIV+ memory CD4 T cells, and no clinically relevant interventions have been identified that reduce the number of these HIV DNA-containing cells. Since postintegration HIV replication can result in HIV protease generation of Casp8p41, which activates BAK, causing infected CD4 T cell death, we sought to determine whether this occurs in memory CD4 T cells. Here, we demonstrate that memory CD4 T cells can generate Casp8p41 and yet are intrinsically resistant to death induced by diverse stimuli, including Casp8p41. Furthermore, BCL-2 expression is relatively increased in these cells and directly binds and inhibits Casp8p41's proapoptotic effects. Antagonizing BCL-2 with venetoclax derepresses this antagonism, resulting in death, preferentially in HIV DNA containing cells, since only these cells generate Casp8p41. Thus, BCL-2 antagonism is a clinically relevant intervention with the potential to reduce HIV reservoir size in patients.
INTRODUCTION
Intense activity is focused on identifying a clinical intervention that results in a long term, drug free remission of HIV-1 infection. Latently infected CD4 T cells harbor transcriptionally silent, replication-competent HIV. Because these cells persist long term and are unaffected by current therapies, the cells represent an HIV reservoir that remains undiminished by current approaches. Pilot clinical trials have tested whether reactivation of HIV-1 from latency will decrease the number of cells containing HIV DNA due to viral cytopathic effect or immune-mediated clearance. The latency reversal agents vorinostat, panobinostat, and romidepsin result in HIV reactivation, as measured by increases in cell-associated HIV RNA, but no change in cell-associated HIV DNA, indicating that the reactivating cells do not die (1–4). Multiple ongoing studies are testing augmentation of the anti-HIV immune response in combination with viral reactivation as a strategy for HIV eradication.
In an effort to inform these attempts to eradicate the HIV reservoir, we and others have been examining the mechanistic basis for HIV-induced killing of CD4 T cells under different circumstances of activation and HIV replication. Although numerous pathways may contribute to the decline of uninfected CD4 T cells during uncontrolled HIV infection (5), fewer pathways have been implicated in the demise of cells directly infected by HIV. After HIV attachment, at least three distinct pathways can initiate the death of infected cells: (i) RIG-I-mediated sensing of HIV RNA (6, 7), (ii) IFI-16 sensing of unintegrated HIV DNA (8, 9), and (iii) DNA-PK-sensing of HIV integrase nicking of host DNA (10). Once integrated into host DNA, HIV can remain in a latent state for years, or it can reactivate and replicate. When HIV reactivation and replication occur, HIV protease is active within the cytosol (11), where it cleaves both viral and host substrates, leading to apoptotic cell death (12). HIV protease-induced death is dependent upon cleavage of the host protein Procaspase 8 to generate the fragment Casp8p41 (13–15), which contains an α-helical BH3-like domain that binds and activates the proapoptotic BCL-2 family member BAK, triggering mitochondrial outer membrane permeabilization (MOMP) and apoptosis (16, 17). Paradoxically, when latently HIV-infected CD4 T cells reactivate HIV, very few cells die (18). In the present study, we sought to determine whether cells that reactivate HIV can generate Casp8p41 and, if so, how they are able to resist its proapoptotic effects.
MATERIALS AND METHODS
Study design.
The objectives of our studies were (i) to determine whether BCL-2 expression impacted Casp8p41-induced cell death in the context of HIV infection and (ii) to determine whether antagonism of BCL-2, along with viral reactivation, decreased the HIV burden. To accomplish this, we used combinations of in vitro and ex vivo studies involving immortalized T cell lines, primary uninfected CD4 T cells, and primary CD4 T cells from antiretroviral therapy (ART)-suppressed HIV-positive patients (see details below). The number of replicates for each experiment is detailed in the text or figure legends. All human studies were performed with the approval of the Mayo Clinic Institutional Review Board (IRB protocol 1039-03) in accordance with all applicable federal, state, and local regulations. Informed written consent was obtained from all participants prior to inclusion.
Cell culture.
Jurkat cells and HEK 293T cells were obtained from the American Type Culture Collection (Manassas, VA). Jurkat cells stably overexpressing BCL-2 were created by transfecting Jurkat cells with pCDNA3/BCL-2 (kindly provided by Stan Korsmeyer), selecting in Geneticin for 30 days, and confirming overexpression via Western blotting. Jurkat cells stably expressing enhanced green fluorescent protein (eGFP) were constructed by stable transfection with eGFP-N1, followed by selection in G418, and then two rounds of sterile flow sorting for eGFP-positive cells. HIV-uninfected primary peripheral blood mononuclear cells (PBMCs) were harvested by Ficoll-Hypaque gradient centrifugation from leukocyte reduction system apheresis chambers from healthy volunteer blood donors in accordance with Mayo Clinic IRB protocol 1039-03 (19). Primary bulk CD4 T cells were isolated by using a RosetteSep human CD4+ T cell enrichment cocktail (Stem Cell Technologies), activated for 24 h with 1 μg/ml phytohemagglutinin, washed in medium, and incubated for 48 h with 50 U/ml interleukin-2 (IL-2) prior to HIV infection. Central memory CD4 T cells (TCM) and effector memory CD4 T cells (TEM) were treated with CH11 (anti-Fas; 1 μg/ml), cycloheximide (CHX; 10 μg/ml), etoposide (20 μM), camptothecin (20 μM), CCCP (carbonyl cyanide m-chlorophenylhydrazone; 1 μM), or hydrogen peroxide (0.35 mM) for 16 h to induce cell death.
Plasmid and peptide construction.
Casp8p41 in pEGFP-C1, pcDNA3, and glutathione S-transferase (GST) were previously described (15, 20). Plasmids encoding BCL-2, BCL-XL, and MCL-1 were previously described (21). The indicated Casp8p41 and BCL-2 mutations were introduced using site-directed mutagenesis (Agilent Technologies, Santa Clara, CA) and confirmed by sequencing.
Transfection.
HEK 293T cells were transfected using Lipofectamine (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Jurkat cells were transfected using a square-wave electroporator (BXT, San Diego, CA) delivering 320 V for 10 ms.
Immunoprecipitations and Western blots.
293T cells transfected with empty vector, HA-Casp8p41, or HA-Casp8p41 V150E/L157K were collected after 24 h, washed with PBS, lysed (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 2 μg/ml aprotinin, 10 μg/ml leupeptin, 2 μg/ml pepstatin, and 1 mM phenylmethylsulfonyl fluoride [PMSF]) for 10 min on ice, and centrifuged at 15,000 × g for 5 min at 4°C. Aliquots containing 500 μg of protein were precleared with 25 μl of protein A/G-agarose (Santa Cruz Biotechnology, Santa Cruz, CA) and incubated with 5 μg of anti-BCL-2 (C21; Santa Cruz Biotechnology) overnight at 4°C. Samples were supplemented with 10 μl of protein-A/G agarose, followed by incubation for an additional 2 h before sedimentation. Beads were washed three times with 10 volumes of lysis buffer. Bound protein was eluted and subjected to SDS-PAGE, followed by immunoblotting as previously described (16). The primary antibodies used were anti-HA peroxidase high-affinity 3F10 (Roche, St. Louis, MO) and the antibodies listed above.
Protein expression and purification.
Plasmids for GST-tagged proteins were transformed into Escherichia coli BL21 or DH5α by heat shock, grown to an optical density of 0.8, and induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h at 37°C. Bacteria were freeze-thawed in calcium- and magnesium-free Dulbecco phosphate-buffered saline containing 0.1% Triton X-100, 2 μg/ml aprotinin, 10 μg/ml leupeptin, 2 μg/ml pepstatin, and 1 mM PMSF and then sonicated three times for 15 s/min on ice. GST-tagged proteins were purified with glutathione-agarose (Thermo Fisher Scientific, Rockford, IL).
SPR.
Proteins used for surface plasmon resonance (SPR) analyses were further purified by fast-performance liquid chromatography on Superdex S200, concentrated in a centrifugal concentrator (Centricon; Millipore), dialyzed against Biacore buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, 0.05 mM EDTA, 0.005% [wt/vol] Polysorbate 20), and stored at 4°C for <48 h before use. Binding assays were performed at 25°C on a Biacore 3000 biosensor (Biacore, Uppsala, Sweden) using the specified proteins immobilized on a CM5 chip (GE Healthcare). Ligands were injected at 30 μl/min for 1 min in Biacore buffer. Bound protein was allowed to dissociate in Biacore buffer at 30 μl/min for 10 min and then desorbed with 2 M MgCl2. Binding kinetics were derived using BIA evaluation software (Biacore).
Flow cytometry.
Immunophenotyping of T cell subsets was performed using multicolor flow cytometry with monoclonal antibodies to human CD3 (Alexa 700; BD Pharmingen), CD4 (FITC; BD Pharmingen), CD8 (Pacific Blue; BD Pharmingen), CD27 (PE; BD Pharmingen), and CD45RO (ECD; Beckman Coulter). TCM cells were defined as CD3+ CD4+ CD27+ CD45RO+; TEM cells were defined as CD3+ CD4+ CD27− CD45RO+/− (22). Intracellular expression of Casp8p41 was assessed as previously described (23). Cell death was measured using Live/Dead Fixable Aqua dead cell stain (Invitrogen) or TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling; Roche) according to the manufacturer's protocol. Gating for TUNEL staining was based on unstained, untransfected controls. Intracellular staining for active BAK (MAb clone TC-100; Enzo Life Sciences) or active caspase 3 was performed and assessed via flow cytometry as previously described (17). Cell proliferation was measured using CellTrace CFSE cell proliferation kit (Life Technologies) according to the manufacturer's protocol. Fluorescence-activated cell sorting (FACS) analysis was performed on either a FACScan or LSRII flow cytometer (BD Biosciences) based on multiparameter needs. FACS data were analyzed using FlowJo software (Tree Star, Inc.).
HIV infections.
Jurkat and Jurkat-BCL-2 cells were infected overnight with HIV-1IIIb (NIH AIDS Reagent Program). Aliquots of the same infectious supernatant were used for all experiments. Cells were then washed three times and incubated in fresh medium. HIV-1 p24 in the cell culture supernatant was measured by Retrotek ELISA kits (Zeptometrix Corporation) according to the manufacturer's protocol. Cell-associated HIV-1 DNA was measured using a previously validated assay as follows (24). Total DNA was extracted using a Qiagen DNeasy blood and tissue kit (Hilden, Germany) and analyzed by a real-time PCR assay specific for HIV-LTR and β-globin primers. A standard curve of pNL4-3 plasmid from 106 through to 10 copies was used as an internal control. Briefly, a 300 nM concentration of the sense primer RU5-F (5′-TTAAGCCTCAATAAAGCTTGCC-3′) and the antisense primer RU5-R (5′-GTTCGGGCGCCACTGCTAGA-3′) were used in conjunction with a 300 nM concentration of a dually labeled fluorogenic TaqMan probe (5′-FAM-CCAGAGTCACACAACAGACGGGCACA-TAMRA-3′). For a 20-μl reaction, 10 μl of gene expression master mix (Applied Biosystems) was used with 5 μl of genomic DNA. PCR conditions consisted of one cycle of 95°C for 3 min, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. Total HIV-1 DNA was compared and normalized with genomic DNA, determined by β-globin detection using the Applied Biosystems (Carlsbad, CA). HIV-1 proviral DNA levels are expressed as HIV-1 copies/β-globin genomic equivalent of 106 cells. HIV RNA in cell culture supernatant was measured by using a COBAS AmpliPrep/COBAS TaqMan HIV-1 test.
Next-generation sequencing.
Primary TCM and TEM cells were isolated using CD4+ central memory T cell isolation and CD4+ effector memory T cell isolation kits (MACS Miltenyi Biotec). After RNA was extracted (Qiagen RNeasy kit), the quality of total RNA samples was assessed by using an Agilent bioanalyzer (Santa Clara, CA). RNA libraries were prepared according to the manufacturer's instructions for the TruSeq RNA sample preparation kit (version 2; Illumina, San Diego, CA). Poly(A) mRNA was purified from the total RNA using oligo(dT) magnetic beads. The purified mRNA was fragmented at 95°C for 8 min, eluted from the beads, and primed for first-strand cDNA synthesis. The RNA fragments were then copied into first-strand cDNA using SuperScript III reverse transcriptase and random primers (Invitrogen). Second-strand cDNA synthesis was performed using DNA polymerase I and RNase H. The double-stranded cDNA was purified using a single AMPure XP bead (Agencourt, Danvers, MA) cleanup step. cDNA ends were repaired and phosphorylated using Klenow, T4 polymerase, and T4 polynucleotide kinase, followed by a single AMPure XP bead cleanup. The blunt-ended cDNAs were modified to include a single 3′ adenylate (A) residue using Klenow Exo-Minus (3′ to 5′ Exo Minus). Paired-end DNA adaptors (Illumina) with a single “T” base overhang at the 3′ end were ligated to an “A-tailed” cDNA population. Unique indices, included in the standard TruSeq kits (12-Set A and 12-Set B) were incorporated at the adaptor ligation step for multiplex sample loading on the flow cells. The resulting constructs were purified by two consecutive AMPure XP bead cleanup steps. The adapter-modified DNA fragments were then enriched by 12 cycles of PCR using primers included in the Illumina sample preparation kit. Libraries were loaded onto paired end flow cells at concentrations of 8 to 10 pM to generate cluster densities of 700,000/mm2 according to Illumina's standard protocol using the Illumina cBot and cBot paired-end cluster kit (version 3). The flow cells were sequenced as 51×2 paired-end reads on an Illumina HiSeq 2000 using the TruSeq SBS sequencing kit version 3 and HCS v2.0.12 data collection software. Base-calling was performed using Illumina's RTA version 1.17.21.3. Data were analyzed according to a Mayo Clinic-developed protocol for analyzing RNA sequencing data, (http://www.biomedcentral.com/content/pdf/1471-2105-15-224.pdf). Once the gene counts were provided by MAP-RSeq, a differential expression and gene set enrichment analysis was performed. DESeq (25), an r package, was used to normalize and quantify the log fold change, the P values, and the false discovery rate between the groups. A hypergeometric test was then performed on all targets with an absolute log2-fold change of >1 to investigate the enrichment of genes associated with cell death and cell proliferation. Cell death and cell proliferation genes were gathered from DeathBase (26), Gene Ontology (27), and KEGG (28) pathways.
Statistics.
Mean values of experimental results were compared by t tests (nonpaired for transformed cell line experiments and paired for primary cell experiments) or by analysis of variance (ANOVA) or Friedman tests as appropriate. The results of time course experiments were compared by area-under-the-curve (AUC) analyses, with mean AUC values compared by t tests. A P value of <0.05 was considered statistically significant.
Molecular modeling.
The initial structure of the Casp8p41·BCL-2 complex was generated by manually docking the BH3-like domain of Casp8p41 (residues 142 to 162) in the α-helical conformation into the vacated BH3-binding groove of BCL-2 (residues 50–203) that was taken from the crystal structure of human BCL-2 (Protein Data Bank ID 4AQ3; residue 9 of the 4AQ3 structure corresponds to residue 50 of the human BCL-2 sequence of NCBI accession no. P10415). This manual docking placed V150Casp8p41 in the proximity of L137BCL-2 and L157Casp8p41 close to V148BCL-2. All Glu, Asp, Arg, and Lys residues were treated as Glu, Asp, Arg, and Lys, respectively. His94BCL-2 was treated as histidine with hydrogen on the epsilon nitrogen (HIE), and all other His residues were treated as histidine with hydrogens on both nitrogens (HIP). The topology and coordinate files of the complex were generated by using LEAP of AmberTools 1.5 (University of California, San Francisco). The complex was refined by energy minimization using SANDER of AMBER 11 (University of California, San Francisco) with a dielectric constant of 1.0 and 200 cycles of steepest-descent minimization, followed by 300 cycles of conjugate-gradient minimization using Amber Forcefield FF12MC. Developed by Yuan-Ping Pang, FF12MC is based on Amber Forcefield FF99 with changes of (i) reducing all atomic masses by 10-fold to improve configurational sampling (29, 30), (ii) shortening C–H bonds by 10 to 14% (1.09 to 0.98 Å for the aliphatic; 1.08 to 0.93 Å for the aromatic) (31), and (iii) zeroing torsion potentials involving a nonperipheral sp3 atom with reduction of scaling factors of protein backbone torsions ϕ and ψ (from 2.00 to 1.00 for the van der Waals interaction; from 1.20 to 1.18 for the electrostatic interaction) (32). The energy minimized complex was then solvated by using the LEAP module with 5946 TIP3P water molecules with the distance parameter of 8.2 Å for the solvatebox command. The solvated complex system was energy minimized for 100 cycles of steepest-descent minimization, followed by 900 cycles of conjugate-gradient minimization to remove close van der Waals contacts in the system, then heated from 0 to 300 K at a rate of 10 K/ps under constant temperature and constant volume, and finally simulated in 10 unique, independent 10-ns molecular dynamics simulations using PMEMD of AMBER 11 with FF12MC. The 10 unique seed numbers for initial velocities of simulations 1 to 10 are 1804289383, 846930886, 1681692777, 1714636915, 1957747793, 424238335, 719885386, 1649760492, 596516649, and 1189641421, respectively. These all-atom, isothermal-isobaric molecular dynamics simulations used (i) a dielectric constant of 1.0, (ii) the Berendsen coupling algorithm (33), (iii) the particle mesh Ewald method to calculate long-range electrostatic interactions (34), (iv) a time step of 1.0 fs, (v) SHAKE-bond-length constraints applied to all the bonds involving the H atom, (vi) a protocol to save the image closest to the middle of the “primary box” to the restart and trajectory files, (vii) a formatted restart file, (viii) a nonbonded cutoff of 8.0 Å, and (ix) default values of all other inputs of PMEMD. All simulations were performed on 10 12-core Apple Mac Pros with Intel Westmere (2.40 GHz) processors. A cluster analysis of all conformations saved at 100-ps intervals from the 10 10-ns molecular dynamics simulations was performed using PTRAJ of AmberTools 1.5 with the average-linkage algorithm (35) (epsilon of 2.0 Å and root mean square coordinate deviation on all Cα atoms of the BH3-like domain). The occurrences of the three most populated clusters were 67, 6, and 5%, respectively. The structure displayed in Fig. 4D is the representative conformation of the most populated cluster identified by PTRAJ.
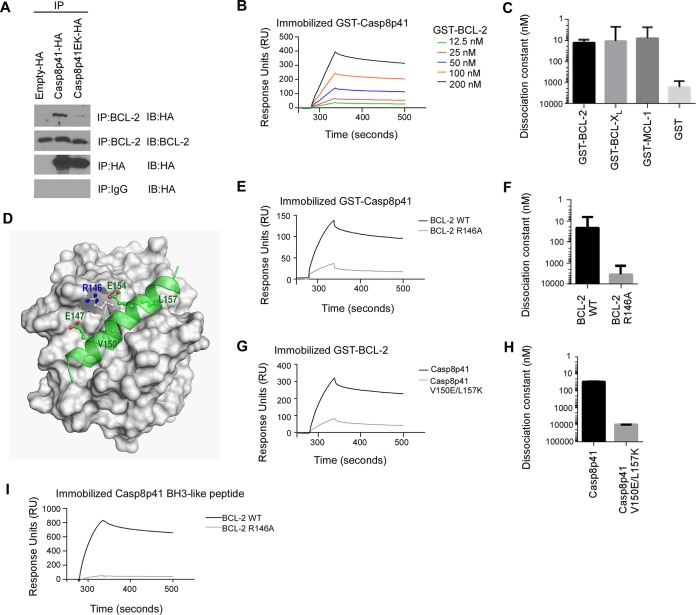
FIG 4.
Casp8p41 binds to the BH3 binding groove of antiapoptotic BCL-2 family members. (A) 293T cells transfected with HA-empty vector, HA-Casp8p41, or HA-Casp8p41 EK were immunoprecipitated with HA-matrix or agarose-conjugated antibody (IgG or BCL-2) and immunoblotted as indicated. (B) Binding of 12.5 to 200 nM GST-BCL-2 to immobilized GST-Casp8p41, as assessed by SPR. (C) Kds for GST-BCL-2, GST-BCL-XL, and GST-MCL-1 binding to GST-Casp8p41. (D) The multiple low-mass molecular dynamics simulation-refined model of the BH3-like domain of Casp8p41 (green) bound at the human BCL-2 BH3-binding groove. (E) Binding of 800 nM GST-BCL-2 or GST-BCL-2 R146A to immobilized GST-Casp8p41. (F) Kds determined as in panel E. (G) Binding of 200 nM GST-Casp8p41 or GST-Casp8p41 Val150Glu/Leu157Glu to immobilized GST-BCL-2. (H) Kds determined as in panel G. Bars in panels C, F, and H: means ± the SD from three independent experiments. (I) Binding of 800 nM GST-BCL-2 or GST-BCL-2 Arg146Ala to immobilized Casp8p41 activator BH3-like peptide. GST, glutathione S-transferase.
RESULTS
Apoptosis resistance of CD4 central memory T cells (TCM).
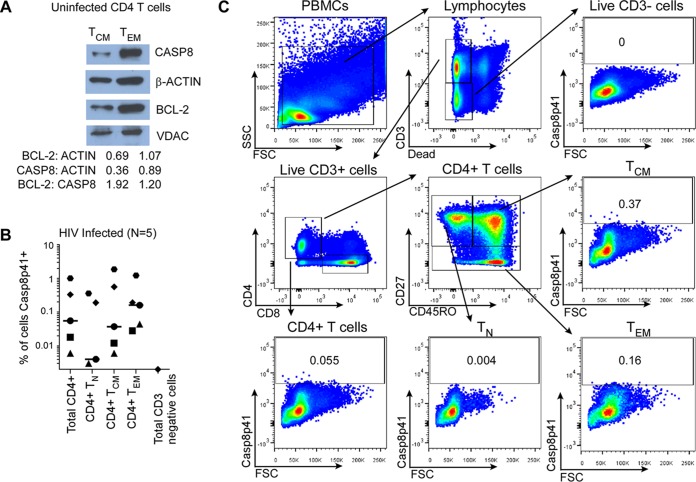
HIV is currently incurable due to the presence of HIV integrated into the genome of resting cells, predominantly TCM, which constitute the major reservoir for HIV in patients on suppressive ART (36). In the setting of HIV reactivation from latency, the “shock and kill hypothesis” (37) predicts that HIV reactivation should cause reactivating cells to die. This is not, however, the case (18). In the context of recent advances in understanding of the biology of Casp8p41, there are two possible reasons why reactivating cells might not die: either Casp8p41 is not generated, or the proapoptotic effects of Casp8p41 are antagonized by intrinsic apoptosis resistance mechanisms. Since TCM express detectable levels of caspase 8 (Fig. 1A), we analyzed whether TCM from five chronically HIV-infected persons generate Casp8p41 in vivo (Fig. 1B and C). Flow cytometry readily detected Casp8p41 in 0.006 to 1.86% of TCM, as well as 0 to 0.36% of TN and 0.006 to 1.22% of TEM, but not in CD3-negative cells. This frequency of Casp8p41 positivity in resting memory CD4 T cells is similar to the frequency of Gag sequences in memory CD4 T cells found in another study which detected 100 to 10,000 Gag copies per 105 highly purified sorted memory CD4 T cells from HIV-infected patients (38). Moreover, our current findings are consistent with our previous analyses where we determined the phenotype of Casp8p41-positive cells and found that Casp8p41 was not detected in cells from HIV-1-negative patients, nor was it detected in CD8 cells, CD14 cells, or CD16 cells from HIV-1-infected patients, and yet both memory and naive CD4 T cells from HIV-infected patients were Casp8p41 positive, with the majority belonging to the memory CD4+ T-cell subset (14, 23, 39). Together, these data suggest that the failure of TCM to die when HIV reactivates is not due to a failure to generate Casp8p41, but is instead perhaps due to antagonism of Casp8p41-mediated killing.
FIG 1.
Central memory CD4 T cells express CASP8 and Casp8p41 in HIV infection. (A) Cytosolic extracts from TCM and TEM were assessed for expression of Casp8 and BCL-2 by Western blotting and densitometry. (B) PBMCs from five HIV-infected patients were assessed for intracellular Casp8p41 expression in CD4+ T cell subsets and in CD3 negative cells. (C) Depicted is representative flow cytometry data showing the gating strategy to determine intracellular Casp8p41 expression in CD4+ T cell subsets: naive (TN), central memory (TCM), effector memory (TEM), and total CD4 T cells. CD3− cells, which are not infected by HIV and thus do not express HIV protease, were used as negative gating controls.
TCM cells are long-lived and proliferate in response to antigenic restimulation. In contrast, TEM, which are a lesser HIV reservoir (40), are found in areas of inflammation and have a shorter half-life than TCM (41). The longer life span of TCM prompted us to speculate that these cells might be intrinsically resistant to death stimuli. To assess this possibility, we treated primary PBMCs from three uninfected donors with a variety of proapoptotic stimuli and analyzed cell death in TCM and TEM (Fig. 2A and B). Not only did TCM die less in response to the Fas receptor activation (agonistic antibody CH11) compared to TEM (absolute difference = −19.0% ± 3.7%, P = 0.006), which is consistent with an earlier report (42), but TCM also died less in response to chemotherapy agents such as etoposide and camptothecin (CPT), suggesting a global death resistance.
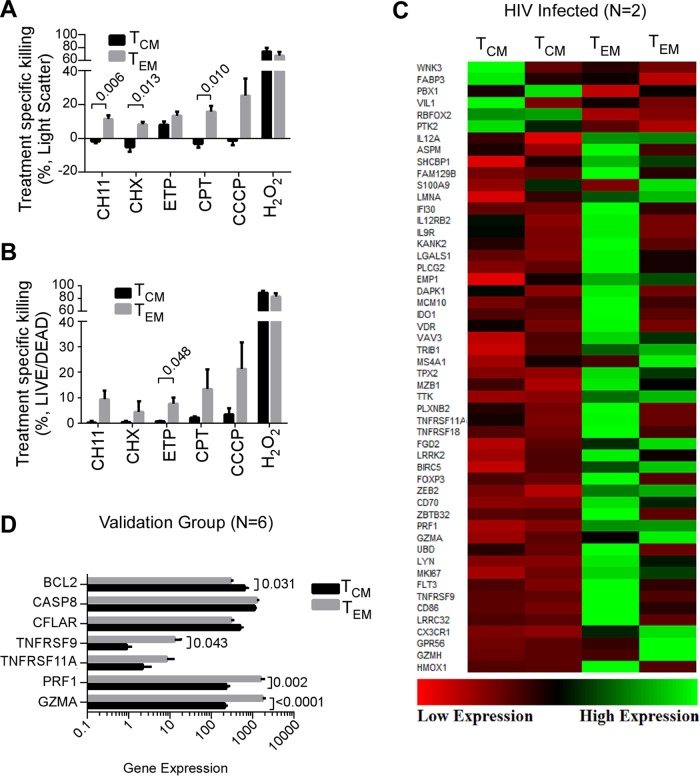
FIG 2.
Central memory CD4 T cells are more resistant to apoptosis than effector memory CD4 T cells. (A and B) PBMCs from three donors were treated overnight with CH11, cycloheximide (CHX), etoposide (ETP), camptothecin (CPT), carbonyl cyanide 3-chlorophenylhydrazone (CCCP), or hydrogen peroxide (H2O2). Cell death was assessed by flow cytometry using light scatter (A) and Live/Dead stains (B); mean (SEM) results of treatment specific killing (treatment-control) from three independent donors. (C) TCM and TEM were isolated from two HIV-infected donors by magnetic bead separation and gene expression measured by RNA sequencing. Depicted are differentially expressed genes associated with cell proliferation and cell death. The genes are sorted by log2-fold change. At the top are the cell proliferation genes. Below are the cell death genes. (D) Gene expression differences in TCM versus TEM were validated in a separate publicly available data set of six uninfected donors.
To understand this apoptosis resistance of TCM, we performed RNA-seq using cells from two HIV-infected donors with long-term suppressed HIV infection. Compared to TEM, TCM upregulated 6 proliferation genes and downregulated 46 cell death genes by at least 2-fold (P = 6.5 × 10−6 for enrichment within the death and proliferation gene sets, Fig. 2C).
Recently, we demonstrated that Casp8p41-induced cell death depends on the interaction between a BH3-like α-helical domain of Casp8p41 and BAK (17). Since BCL-2 antagonizes Bak activation in lymphoid cells (43) by binding and neutralizing proapoptotic proteins containing BH3 domains, we assessed the relative expression of BCL-2 and procaspase 8 protein in TCM versus TEM. This analysis demonstrated that the BCL-2/procaspase 8 ratio is 60% higher in TCM than in TEM (Fig. 1A), suggesting that TCM might resist the prodeath effects of Casp8p41 by virtue of elevated BCL-2 expression. These results are consistent with previously published (44) gene expression data (GSE61697, Gene Expression Omnibus) showing a higher expression of BCL-2 and a lower expression of four other genes that impact T cell survival (TNFRSF9, TNFRSF11A, PRF1, and GZMA) in TCM than in TEM (Fig. 2D). Of note, BCL-2 RNA expression was not significantly different in the two patients analyzed in Fig. 2C.
BCL-2 inhibits Casp8p41-induced cell death.
To assess whether BCL-2 alters the ability of Casp8p41 to kill cells, we compared the responses of parental Jurkat T cells versus Jurkat cells stably overexpressing BCL-2 (Jurkat-BCL-2) to transient GFP-Casp8p41 expression. After transfection, Casp8p41-induced apoptosis, as measured by TUNEL positivity (Fig. 3A and B), was greater in parental Jurkat cells than Jurkat-BCL-2 cells (mean AUC of 1,500 versus 870 [95% confidence interval of difference = −790 to −480; P = 0.004]) despite similar Casp8p41 expression (Fig. 3C). Thus, cells overexpressing BCL-2 die less in response to Casp8p41, indicating that BCL-2 antagonizes Casp8p41-induced killing.
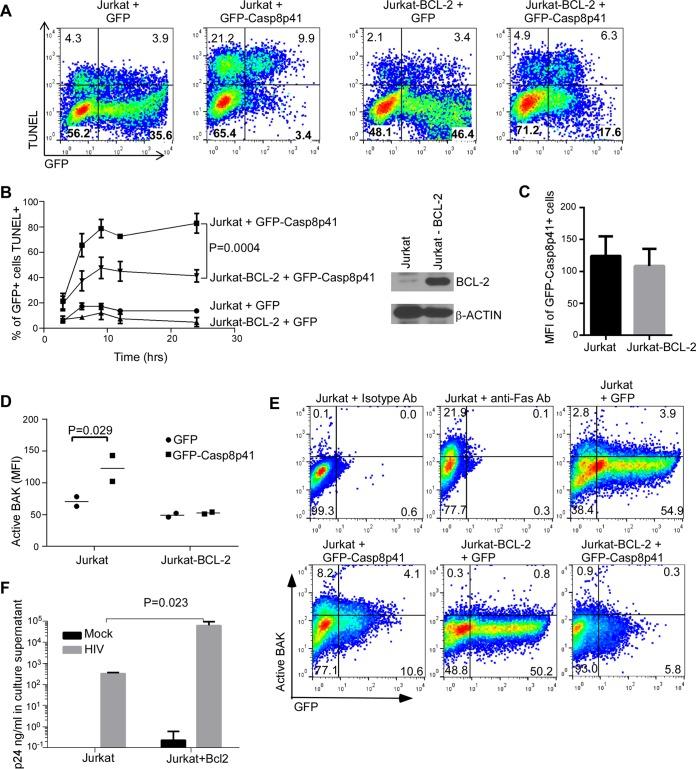
FIG 3.
BCL-2 overexpression decreases cell death induced by Casp8p41 and increases viral replication. (A) Parental Jurkat T cells or Jurkat T cells stably overexpressing BCL-2 (Jurkat-BCL-2) were transfected with GFP-Casp8p41 or GFP alone and assessed for cell death via TUNEL. (B) Mean (standard deviation [SD]) results from three independent experiments as in panel A. (C) The relative GFP-Casp8p41 expression in GFP-positive cells was compared between Jurkat and Jurkat-BCL-2 cells. (D and E) Parental Jurkat cells or Jurkat-BCL-2 cells were transfected with GFP-Casp8p41 or GFP alone and assessed for active BAK expression in GFP-positive cells, using a conformational specific antibody which detects only active BAK. Depicted are the individual mean fluorescence intensities (MFI) of active BAK (D) and representative dot plots (E) from two independent experiments. (F) Parental Jurkat cells or Jurkat-BCL-2 cells were infected with HIVIIIb or mock infected and then assessed for HIV p24 production in culture supernatant at day 9 postinfection. Depicted are the mean (SD) values of three independent experiments.
To determine whether BCL-2 overexpression impacts Casp8p41-mediated BAK activation, Jurkat or Jurkat-BCL-2 cells were transfected with GFP or GFP-Casp8p41 and stained with conformational specific antibodies that recognize activated BAK, but not inactive BAK. As expected, GFP-Casp8p41+ expressing parental Jurkat cells (identified as GFP+ by flow cytometry) contained more activated BAK than parental cells expressing GFP alone (MFI difference = 52 ± 16, P = 0.029; Fig. 3D and E). In contrast, GFP-Casp8p41+ Jurkat-BCL-2 cells did not exhibit increased BAK activation compared to GFP alone (MFI difference 3.5 ± 16, P = 0.834), indicating that BCL-2 prevents BAK activation in response to Casp8p41 expression. Accordingly, the overall number of activated BAK+ cells after GFP-Casp8p41 transfection was lower in Jurkat-BCL-2 cells than in Jurkat cells (P = 0.045). Thus, BCL-2 inhibits Casp8p41-induced BAK activation.
To assess whether BCL-2 overexpression also alters the outcome of acute HIV infection in vitro, Jurkat and Jurkat-BCL-2 cells were infected with HIVIIIb and analyzed for HIV p24 production. BCL-2 overexpression was accompanied by increased HIV p24 production in culture supernatants (2.5-log increase compared to infected parental cell cultures, P = 0.023, Fig. 3F). This suggests that BCL-2 reduces HIV-induced cell death and increases the number of HIV-infected cells, thereby allowing more cells to produce progeny virions (reflected by increased p24).
BCL-2 directly binds Casp8p41.
We next investigated how BCL-2 antagonizes Casp8p41 killing. BCL-2 family members are classified into three groups based on their structure and function (45): the proapoptotic proteins BAX and BAK, which permeabilize the outer mitochondrial membrane when activated; the antiapoptotic proteins, e.g., BCL-2, BCL-XL, and MCL-1, which bind and neutralize activated BAX and BAK, as well as other proteins that contain exposed BH3 domains, and the proapoptotic BH3-only proteins, e.g., BIM, BID, and BAD, which contain only a BH3 domain, serve as sensors of various cellular stresses and trigger apoptosis by (i) directly activating BAX and BAK or (ii) displacing BAX and BAK from antiapoptotic neutralizers.
Because the α-helical BH3-like domain of Casp8p41 directly activates BAK by binding to the BAK BH3 binding groove (17), we investigated whether Casp8p41 also binds antiapoptotic BCL-2, Bcl-xL, and Mcl-1 through their BH3 binding grooves (45, 46). In initial experiments, lysates from 293T cells transfected with empty vector, HA-Casp8p41, or HA-Casp8p41 V150E/L157K (Casp8p41-EK, a variant with decreased affinity for BAK due to disruption of critical binding interactions [17]) were immunoprecipitated for BCL-2 and probed for associated hemagglutinin (HA)-tagged proteins. Immunoprecipitation of BCL-2 demonstrated interaction with HA Casp8p41 that was reduced by the V150E/L157K substitutions (Fig. 4A).
SPR, a widely used technique to assess the affinity of protein-protein interactions, was used for more detailed study of Casp8p41 interactions with BCL-2, BCL-XL, and MCL-1, the antiapoptotic BCL-2 family members expressed in T cells. These experiments indicated equilibrium dissociation constants (Kds) of 13 ± 4 nM for Casp8p41 binding to BCL-2, 11 ± 8 nM for Casp8p41 binding to BCL-XL, and 8 ± 6 nM for Casp8p41 binding to MCL-1 (Fig. 4B and C), which are similar to the affinities of the same proteins for BH3-only proteins such as BIM and PUMA (45, 47). We also confirmed that the peptide corresponding to the α-helical BH3-like domain of Casp8p41 (17) also bound BCL-2, providing evidence that the same Casp8p41 domain is responsible for binding both BAK and BCL-2 (Fig. 4I). Using multiple low-mass molecular dynamic simulations (29, 30), we constructed a three-dimensional model of Casp8p41 bound in the BH3 binding groove of BCL-2 (Fig. 4D). This model predicts that Arg146 of BCL-2 is critical for binding electrostatically to Glu147 and Glu154 of Casp8p41. Mutation of BCL-2 Arg146 to Ala (21) decreased the affinity of BCL-2 for the Casp8p41 activator peptide (Fig. 4I) or full-length Casp8p41 250-fold (Fig. 4E and F), confirming that Casp8p41 is binding to the BH3 binding groove of BCL-2. The model also suggests that Val150 and Leu157 of Casp8p41 bind to two hydrophobic regions of the BCL-2 BH3 binding groove that are similar to the two hydrophobic holes of the BAK BH3 binding groove (48). Consistent with this prediction, the affinity of Casp8p41 for BCL-2 was reduced >300-fold by the introduction of the Val150Glu and Leu157Lys mutations into Casp8p41 (Fig. 4G and H). These observations, combined with the observations in Fig. 3, suggest that BCL-2 inhibits cell death induced by Casp8p41 or HIV by binding and neutralizing the BH3-like domain of Casp8p41, thereby preventing Casp8p41-induced BAK activation.
Venetoclax is a clinically relevant BCL-2 antagonist that impacts Casp8p41-mediated killing.
Because overexpressed antiapoptotic BCL-2 family members have been implicated widely in the pathogenesis of cancer (49), there has been substantial interest in developing therapeutic BCL-2 antagonists. One such molecule, venetoclax (formerly known as ABT-199), potently and selectively inhibits BCL-2 binding to BH3 domains (50), is well tolerated in early phase clinical trials (51), and achieves peak plasma levels of up to 5 μM (52). Accordingly, we assessed the impact of venetoclax on T cells expressing Casp8p41 or reactivating HIV.
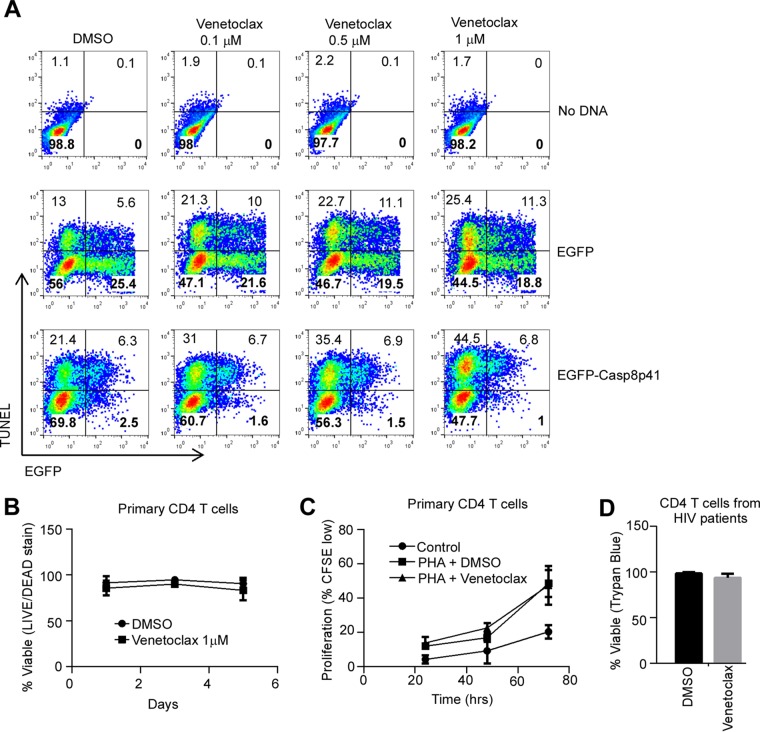
In initial experiments, treatment with increasing doses of venetoclax (up to 1 μM) did not result in a statistically significant increase in apoptosis of either parental Jurkat cells transfected with no DNA or Jurkat cells expressing eGFP compared to diluent control (Fig. 5A). In contrast, venetoclax (1 μM) significantly increased cell death in Jurkat cells expressing eGFP-Casp8p41 (mean difference 25.7% ± 10.3%, P = 0.016, Fig. 5A).
FIG 5.
Venetoclax increases Casp8p41-induced apoptosis but spares cells that do not express Casp8p41. (A) Jurkat T cells pretreated with increasing concentrations of venetoclax or diluent dimethyl sulfoxide (DMSO) were transfected with EGFP-Casp8p41, vector control, or no DNA and assayed for cell death by TUNEL. Data are representative of three independent experiments. (B) Primary CD4 T cells from seven uninfected donors were treated with venetoclax (1 μM) or diluent and assayed for cell viability by flow cytometry over 5 days. (C) Primary CD4 T cells from four uninfected donors were stimulated with phytohemagglutinin (2 μg/ml) in the presence of venetoclax (1 μM) or diluent and assayed for proliferation by CFSE staining. (D) Primary CD4 T cells from seven ART-suppressed HIV-positive patients were treated with venetoclax (1 μM) or DMSO control for 24 h and assessed for viability by trypan blue exclusion.
This selective enhancement of killing induced by Casp8p41 but not generalized toxicity was further evaluated by treating primary uninfected CD4 T cells with venetoclax. Importantly, uninfected CD4 T cells (n = 7 patients) treated with venetoclax (1 μM) for up to 5 days did not exhibit reduced viability compared to control treated cells (mean difference −7.2% ± 4.8% at day 5, P = 0.16, Fig. 5B). Furthermore, uninfected CD4 T cells treated with venetoclax did not exhibit altered mitogen-induced proliferation as measured by CFSE (carboxyfluorescein succinimidyl ester) dilution (mean difference in the percentage of CFSElow cells at day 3 = −1.1% ± 7.9%, P = 0.90, Fig. 5C). The lack of toxicity of venetoclax toward CD4 T cells from HIV uninfected donors was also seen using cells from HIV-infected, but ART-suppressed, patients; venetoclax (1 μM) for 24 h did not adversely affect viability of bulk primary CD4 T cells (mean difference = −4.7% ± 1.6% viable, Fig. 5D). These favorable safety findings in vitro have also been seen in vivo: in 40 non-Hodgkin lymphoma patients receiving venetoclax monotherapy, only two cases of dose-limiting neutropenia were observed (53). These results raise the possibility that venetoclax might potentially be used safely in HIV-infected patients.
Simultaneous detection of markers of HIV infection and apoptosis.
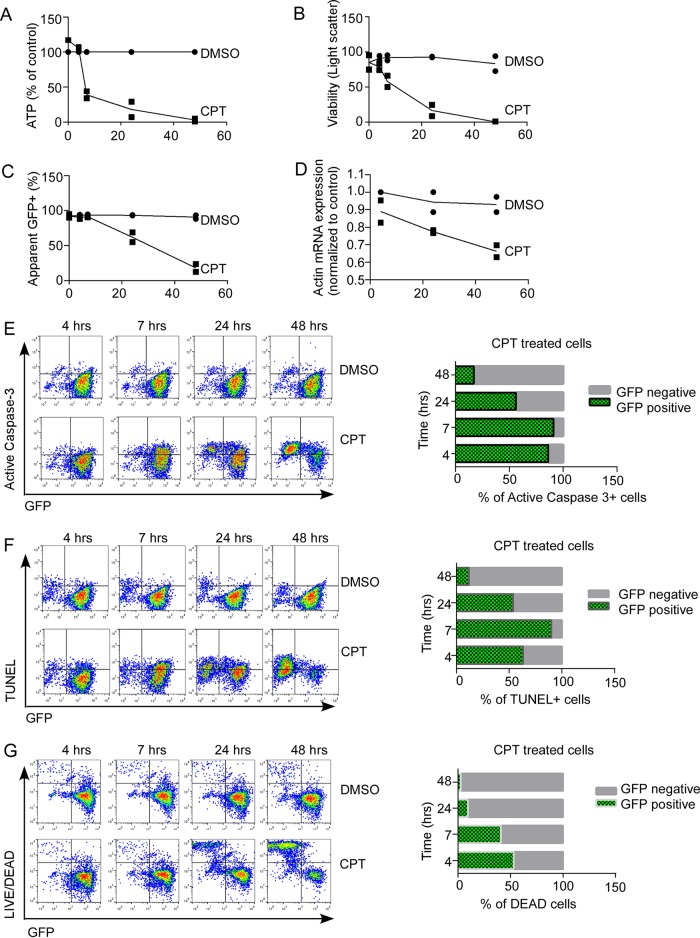
If BCL-2 neutralizes Casp8p41, then antagonizing BCL-2 should augment Casp8p41-induced killing, including when Casp8p41 is generated by latently HIV-infected cells that reactivate the virus. Before testing this possibility, we first assessed whether select markers (protein or nucleic acid based) remain detectable when the cell of interest is undergoing death. Jurkat T cells stably expressing eGFP were treated with vehicle control or the cytotoxic quinolone alkaloid CPT to induce apoptosis and then examined for expression of eGFP (as a prototypic cytoplasmic protein) and actin mRNA (as a prototypic mRNA) over time. Treatment of GFP+ Jurkat cells with CPT decreased cell viability as measured by cellular ATP content (Fig. 6A) and light scatter (Fig. 6B) compared to diluent-treated cells. Coincident with the loss in viability was a decrease in detectable eGFP (17% eGFP+ with CPT versus 91% eGFP+ with diluent, Fig. 6C) and detectable actin message (34% reduction compared to diluent control) as assessed by quantitative reverse transcription-PCR (qRT-PCR) (Fig. 6D). As the proportion of active caspase-3+ cells (Fig. 6E), TUNEL+ cells (Fig. 6F), or membrane-permeable cells (Fig. 6G) increased over time, there was a reciprocal decrease in detectable eGFP (Fig. 6C) despite the fact that virtually all cells were eGFP positive at baseline. For example, by 48 h 97% of CPT-treated cells were eGFP negative, despite having been uniformly eGFP positive. Accordingly, estimating the proportion of dead or dying cells that express a degradable marker of interest (e.g., HIV p24 or HIV RNA/DNA) will substantially underestimate the proportion of cells that expressed the marker before encountering the death stimulus. Therefore, the ability to detect protein- or nucleic acid-based phenotypic markers is impaired as cell death proceeds, rendering this approach insensitive and prone to incorrect phenotyping of the dying cell. Accordingly, to approach the question of whether HIV reactivation in concert with BCL-2 antagonism reduces cell-associated HIV DNA, we compared HIV DNA content in treated versus control samples rather than simultaneously assessing markers of HIV and cell death in the same cells.
FIG 6.
Apoptotic cell death is associated with loss of protein and mRNA markers. Jurkat T cells stably expressing eGFP were treated with DMSO or CPT and assessed for cell viability by ATP content (A) and light scatter (B) over time. (C) Expression of eGFP was measured by flow cytometry. (D) Actin mRNA expression was assessed by qRT-PCR and expressed as mean CT values normalized to baseline DMSO control. (E to G) Cell death was assessed by active-caspase 3 expression (E), TUNEL (F), and Live/Dead viability stain (G) and compared between eGFP-positive and eGFP-negative cells. Values in panels A to D represent the mean (range) of two independent experiments. The results the panels E to G are representative of two independent experiments.
Effect of simultaneous HIV reactivation and venetoclax.
It has previously been established that when cells from HIV-infected patients on suppressive ART are treated with latency reversal agents (LRA), reactivation alone does not alter the amount of cell-associated HIV DNA, nor does it induce markers of cell death in the HIV-containing reactivating cells (18). Moreover, when different LRA have been tested in patients, despite successful reactivation, no studies have achieved a decrease in HIV DNA levels (1–3, 54).
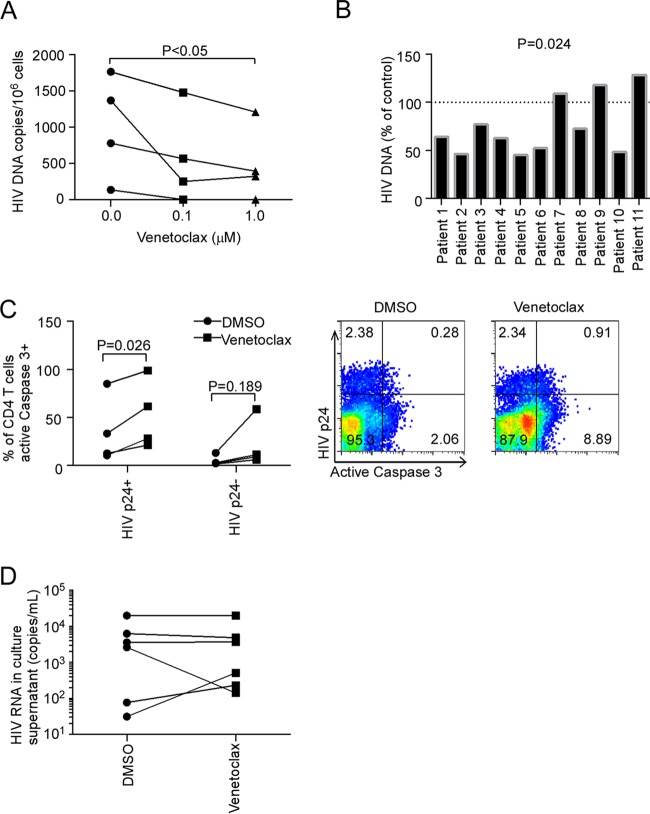
In this context, we assessed whether venetoclax would cause the death of the reactivating cells and decrease the number of HIV DNA-containing cells. For these experiments, we reactivated HIV in primary CD4 T cells from combination antiretroviral therapy (cART)-suppressed patients using αCD3/αCD28, which has previously been observed to induce maximal HIV reactivation in PBLs from HIV-infected cART-suppressed patients (55), which contain some latently infected CD4 T cells. These patient-derived primary CD4 T cells were treated with αCD3/αCD28, along with tenofovir and raltegravir, to prevent spreading infection and in the presence of venetoclax or diluent to assess the impact of BCL-2 inhibition. After 72 h, the cells were harvested and assayed for total cell-associated HIV DNA (56). In experiments using reactivated CD4 T cells from cryopreserved PBMCs, venetoclax caused a dose-dependent reduction in cell-associated HIV DNA, with a mean decrease of 64.4% at 1 μM venetoclax compared to control (Fig. 7A, P < 0.05). To further validate this important result, freshly isolated CD4 T cells from an additional 16 HIV-infected patients were analyzed. Of these, 11 (69%) had measurable cell-associated HIV DNA levels in control samples, and venetoclax caused a reduction in HIV DNA in 8 of the 11 (73%) (median residual DNA = 64.1% of control [interquartile range = 48.6 to 108.9%], P = 0.024, Fig. 7B).
FIG 7.
Selective BCL-2 inhibition reduces cell associated DNA during HIV reactivation ex vivo. (A) Primary CD4 T cells isolated by negative selection from cryopreserved PBMCs of long-term virologically suppressed HIV-infected patients (n = 4) were treated for 16 h with venetoclax or DMSO vehicle in the presence of tenofovir and raltegravir to prevent spreading infection and then exposed to plate-bound αCD3 and soluble αCD28 antibody to induce HIV reactivation. After 72 h, cell-associated HIV DNA was measured. (B) Freshly obtained peripheral CD4 T cells from an additional 16 suppressed HIV-infected patients were treated with venetoclax (1 μM) or DMSO in the presence of tenofovir/raltegravir, followed by αCD3/αCD28 for 72 h, and cell associated HIV DNA was measured. Depicted is the ratio of HIV DNA in venetoclax-treated versus diluent-treated samples for each patient, with measurable HIV DNA in the diluent sample (n = 11). (C) Primary CD4 T cells from four HIV-infected subjects were treated with venetoclax or diluent and induced to reactivate HIV using CD3/CD28, and cell death was measured in HIV P24-positive or -negative cells using activated caspase 3 staining. (D) HIV RNA was measured in cell culture supernatant from 6 of the 11 patient experiments from panel B.
In Fig. 6 we demonstrated that protein and nucleic acid markers can be lost during apoptosis, causing underestimation of the number of cells with those markers that are dying. However, it is still possible to detect p24+ and p24− cells in a population that contains some dying cells. Of these, the p24+ cells will be HIV infected, but p24− cells might be infected (with degraded p24) or uninfected. With this in mind, active caspase 3 (a marker of apoptotic cell death) was assessed in p24+ (HIV infected) and p24− cells that were primed with venetoclax and reactivated with αCD3/αCD28. Median cell death, as measured by active caspase 3 staining in the p24− cells, was 2.5% in the control treated αCD3/αCD28 sample and was not changed significantly by venetoclax (10.3%, P = 0.189, Fig. 7C). In contrast, venetoclax increased the proportion of HIV p24+ CD4+ cells that are active caspase 3+ compared to control treated cells (from 23% active caspase 3+ in control sample to 47% active caspase 3+ in the venetoclax sample, P = 0.026, Fig. 7C). Of note, the frequency of p24-positive cells in reactivated CD4 T cells was similar to other previous reports (57–59). HIV RNA in the cell culture supernatant was not altered by venetoclax treatment compared to control (Fig. 7D), indicating that venetoclax does not impair the ability of cells to reactivate HIV.
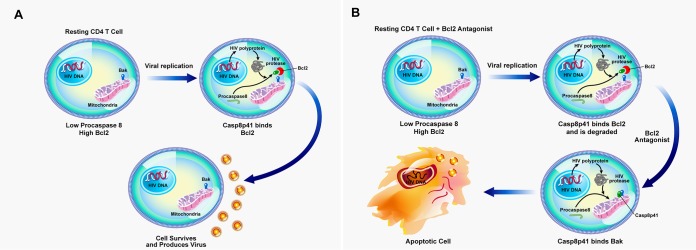
In total, therefore, we demonstrate that venetoclax-mediated inhibition of BCL-2 in all cells has minimal effect on cell viability in uninfected cells (Fig. 5) yet promotes the preferential killing of p24+ (HIV replicating) cells. Moreover, we have successfully identified a clinically relevant treatment that converts HIV reactivation from latency without killing of the reactivating cell (Fig. 8A) into an event that kills the reactivating cell by priming all cells toward an apoptosis-sensitive phenotype and then allows HIV-infected cells, which replicate virus and generate Casp8p41, to undergo apoptosis when BCL-2 is inhibited (Fig. 8B).
FIG 8.
BCL-2 antagonism as a model of “prime, shock, and kill” for decreasing the HIV reservoir. (A) Reactivation of latent HIV does not lead to death of the reactivated cell due to inhibition of Casp8p41 by direct binding to BCL-2. Therefore, the reactivated cell survives and produces progeny virus. (B) Antagonism of BCL-2 activity (as with venetoclax treatment) allows Casp8p41 to bind proapoptotic BAK, leading to apoptotic death of the reactivated cell and to decreased total HIV DNA.
DISCUSSION
Viral infection is often detected by intrinsic host defense mechanisms that activate cell suicide programs in order to limit the production of progeny virions. In response, viruses have evolved mechanisms to inhibit cell death pathways. Prototypic examples include the human papillomavirus E6 and E7 proteins (60), baculovirus inhibitor of apoptosis (IAP) and p35 proteins (61), and adenovirus E1B-55K (62). The role of these viral death inhibitors in viral persistence is well established. In cervical epithelial cells with integrated HPV, for example, chromosomal disruption of either E6 or E7 with CRISPR/Cas9 results in the activation of p53 or retinoblastoma (Rb), respectively, thereby activating cell death cascades (60) and disrupting the persistence of the virally infected cell. In baculovirus-infected cells, p35 and IAP inhibit apoptosis, and baculoviruses deficient in either of these proteins will induce premature death after infection, resulting in fewer progeny viruses (63). Similarly, deletion of antiapoptotic EBV BALF1, BHRF1, or both results in premature cell death during EBV infection of primary B cells (64), and deletion of the adenoviral BCL2 homolog E1B-19K results in premature cell death and loss of persistence of adenoviral infection (65). Thus, virally encoded apoptosis inhibitors are often necessary for infected cells to persist and enable the persisting cells to produce progeny viruses.
Although HIV does not encode an apoptosis inhibitory protein per se, HIV-infected cells are nonetheless resistant to apoptosis (reviewed in references 66 and 67). This apoptosis resistance reflects the ability of various HIV-encoded proteins to upregulate endogenous apoptosis inhibitory proteins and downregulate proapoptotic proteins. For example, Tat upregulates antiapoptotic BCL-2, c-FLIP, XIAP, and C-IAP2 (68), whereas Nef causes phosphorylation and inactivation of proapoptotic BAD (69), thereby conferring apoptosis resistance (70). This shift in apoptotic balance affords an HIV-infected cell the ability to survive and produce more virions. Experimentally, addition of the pan-caspase inhibitor z-VAD-fmk to primary T cells infected with HIV reduces cell death and augments HIV production because fewer HIV-infected cells die (71). Similarly, when primary CD4 T cells are transduced and selected for BCL-2 expression and then infected with GFP/HIV, some cells revert to a quiescent (GFPlow) state, which recapitulates many features of latency, including lack of basal HIV transcription and induction of GFP/HIV by latency-reversing agents (72). Thus, there is an established association between apoptosis resistance, BCL-2 expression, and latency during in vitro HIV infection.
The present work provides a mechanistic basis for such BCL-2 effects. HIV-infected T cell death occurs through multiple mechanisms, including the pathway involving HIV protease-mediated cleavage of the cellular protein procaspase 8 to inactivate the enzyme and unmask a novel α-helical BH3-like domain that interacts with the BH3 binding groove in BAK, thereby inducing BAK oligomerization and MOMP (17). Here, we show for the first time that the BH3-like domain of Casp8p41 also interacts with antiapoptotic BCL-2 family members.
There has previously been considerable debate over whether CD4 T cell loss during HIV infection is due to killing of HIV-infected cells, loss of uninfected (bystander) cells, or both (reviewed in reference 5). The approach used to discriminate infected cells from uninfected cells has been to assess intracellular HIV p24, HIV-specific RNA/DNA sequences, or GFP expression encoded by reporter viruses, in conjunction with phenotypic markers cell death (e.g., TUNEL staining or activated caspase 3). In the years since many of these experiments were first performed, much has been learned about the processes by which cells die and the molecular mechanisms activated within cells to result in the phenotypic changes of cell death (73, 74). For instance, after mitochondrial outer membrane permeabilization, which occurs in CD4 T cells acutely infected with HIV (75), effector caspases that cleave numerous host proteins (76, 77) and effector nucleases that degrade host and viral nucleic acids (78–80) are activated. Indeed, assays used to detect cell death depend upon these processes (e.g., the presence of active caspase 3 indicates that proteases capable of degrading cellular proteins are active, whereas positive TUNEL staining indicates that nucleases degrading host and viral nucleic acids are active). These processes might explain why a seminal paper evaluating apoptosis and HIV/SIV infection in lymph nodes (81) using >2-kb riboprobes in in situ hybridization to identify infected cells failed to identify infected cells that were also TUNEL positive. Interpreted in the context of current understanding of apoptotic processes, it is possible that when TUNEL (or active caspase 3 staining) signals are positive, RNA/DNA and protein have been degraded, rendering riboprobe ISH, PCR detection (or protein detection) assays insensitive for the detection of infected cells.
Previously, there has been no clinically relevant pharmacologic intervention identified that can convert latency reactivation alone (which does not decrease cell-associated HIV DNA levels) to an intervention that reduces cell-associated HIV DNA in the majority of cases. Data presented in here indicate that venetoclax, which has been granted breakthrough therapy designation by the U.S. Food and Drug Administration (82) in recognition of its favorable safety profile and its promising efficacy for patients with hematologic malignancies, is such an agent. It remains to be seen whether unexpected toxicities occur with the use of venetoclax in an HIV-infected population and whether venetoclax would be acceptable to HIV-infected patients. However, the willingness of HIV patients to participate in other cure focused clinical trials involving gene therapy (NCT01769911 and NCT01787994) or chimeric antigen receptors (83), among other unproven technologies, suggest that at least a subset of patients might be willing to participate in trials involving venetoclax, which has shown acceptable toxicity profiles in much sicker patient populations, including those with relapsed malignancies (84). When considering the possible use of venetoclax for HIV, this agent would need be applied in the setting of maximally suppressive ART to prevent reseeding of the reservoir, as has been observed for example with IL-7 therapy (85). Also, in light of observations that HIV reactivating stimuli do not reactivate all replication-competent viruses (86), therapy over a prolonged period, with multiple rounds of reactivation stimuli, might need to be evaluated.
Considering the potential in vivo evaluation of venetoclax for HIV, mouse models of HIV infection appear to be inappropriate for long-term study of latency interventions because of the frequent occurrence of graft-versus-host disease (GVHD) (87). It also remains unknown whether SIV protease generates Casp8p41 in macaques and whether simian Casp8p41, if present, interacts with BAK and BCL-2 similarly to the human counterparts. Thus, it might be of interest to study venetoclax in HIV-infected patients with long-term-suppressed HIV, alone or in conjunction with additional latency-reversing agents, such as the recently described combination of panobinostat and bryostatin (88).
For unknown reasons, not all HIV patient samples treated with venetoclax and αCD3/αCD28 decreased HIV DNA (Fig. 7B). One intriguing possibility is that BCL-2 single-nucleotide variants might result in resistance to the venetoclax. Indeed, BCL-2 somatic mutations that result in enhanced antiapoptotic activity have been observed in follicular lymphoma (89). Importantly, the existence of these mutations correlates with increased activation-induced cytidine deaminase (AID) expression, suggesting that AID-induced somatic hypermutation may have generated the BCL2 mutations. The well-known ability of HIV infection to induce the cytidine deaminase APOBEC3G (90), resulting in increased somatic hypermutation during HIV infection (91), suggests that similar BCL2 mutations might arise during HIV infection. Alternatively, the inability of venetoclax and αCD3/αCD28 to diminish HIV DNA levels in CD4 T cells from a small subset of patient cells might reflect earlier observations that some mutant HIV protease isolates have an impaired ability to cleave procaspase 8 to generate Casp8p41 (92). Both of these possibilities are worthy of further study.
Observations in the Berlin patient (93) and at least two other HIV patients who received allogeneic hematopoietic stem cell transplantation (HSCT) indicate that HSCT alone can induce significant but at times incomplete reductions in HIV reservoir size (94), suggesting that inducing apoptosis in the majority of lymphoid cells by chemotherapy can reduce HIV burden (95). The present study demonstrates that more targeted induction of apoptosis, achieved by combining the intrinsic proapoptotic effect of the HIV-specific death stimulus Casp8p41 (which is only produced in HIV-infected cells) with antagonism of antiapoptotic signals using venetoclax, is effective at reducing HIV burden ex vivo. These observations provide proof of concept that apoptosis sensitization (BCL-2 inhibition), along with HIV reactivation, reduces HIV burden, an observation consistent with our proposed model of “prime, shock, and kill” (96). Further evaluation of apoptosis sensitization strategies in concert with HIV reactivation strategies therefore appear to be warranted.
ACKNOWLEDGMENTS
The contents of this study are solely the responsibility of the authors and do not necessarily represent the official view of NIH. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We thank the patient volunteers from the Mayo HIV Clinic who participated in the study.
N.W.C., A.M.S., S.H.K., and A.D.B. conceived of the project and designed the experiments. N.W.C., A.M.S., H.D., S.N., G.D.B., C.M.D.A.C., R.S., and D.O. performed experiments and analyzed data. Y.-P.P. designed, conducted, and analyzed the computational study. R.S. and S.A.R. recruited human subject participants. N.W.C., A.M.S., and A.D.B. wrote the initial draft of the manuscript. All authors contributed to editing and rewriting the manuscript and agree with the content in the final version. We declare no conflicts of interest.
Funding Statement
The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official view of NIH. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, Richman DD, Hudgens MG, Bosch RJ, Coffin JM, Eron JJ, Hazuda DJ, Margolis DM. 2012. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487:482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elliott JH, Wightman F, Solomon A, Ghneim K, Ahlers J, Cameron MJ, Smith MZ, Spelman T, McMahon J, Velayudham P, Brown G, Roney J, Watson J, Prince MH, Hoy JF, Chomont N, Fromentin R, Procopio FA, Zeidan J, Palmer S, Odevall L, Johnstone RW, Martin BP, Sinclair E, Deeks SG, Hazuda DJ, Cameron PU, Sekaly RP, Lewin SR. 2014. Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog 10:e1004473. doi: 10.1371/journal.ppat.1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rasmussen TA, Tolstrup M, Brinkmann CR, Olesen R, Erikstrup C, Solomon A, Winckelmann A, Palmer S, Dinarello C, Buzon M, Lichterfeld M, Lewin SR, Løstergaard-Søgaard OS. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV 1:e13–e21. doi: 10.1016/S2352-3018(14)70014-1. [DOI] [PubMed] [Google Scholar]
- 4.Sogaard OS, Graversen ME, Leth S, Olesen R, Brinkmann CR, Nissen SK, Kjaer AS, Schleimann MH, Denton PW, Hey-Cunningham WJ, Koelsch KK, Pantaleo G, Krogsgaard K, Sommerfelt M, Fromentin R, Chomont N, Rasmussen TA, Ostergaard L, Tolstrup M. 2015. The depsipeptide romidepsin reverses HIV-1 latency in vivo. PLoS Pathog 11:e1005142. doi: 10.1371/journal.ppat.1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cummins NW, Badley AD. 2010. Mechanisms of HIV-associated lymphocyte apoptosis: 2010. Cell Death Dis 1:e99. doi: 10.1038/cddis.2010.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solis M, Nakhaei P, Jalalirad M, Lacoste J, Douville R, Arguello M, Zhao T, Laughrea M, Wainberg MA, Hiscott J. 2011. RIG-I-mediated antiviral signaling is inhibited in HIV-1 infection by a protease-mediated sequestration of RIG-I. J Virol 85:1224–1236. doi: 10.1128/JVI.01635-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M Jr, Patel SS, Marcotrigiano J. 2011. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature 479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doitsh G, Galloway NL, Geng X, Yang Z, Monroe KM, Zepeda O, Hunt PW, Hatano H, Sowinski S, Munoz-Arias I, Greene WC. 2014. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature 505:509–514. doi: 10.1038/nature12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monroe KM, Yang Z, Johnson JR, Geng X, Doitsh G, Krogan NJ, Greene WC. 2014. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science 343:428–432. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper A, Garcia M, Petrovas C, Yamamoto T, Koup RA, Nabel GJ. 2013. HIV-1 causes CD4 cell death through DNA-dependent protein kinase during viral integration. Nature 498:376–379. doi: 10.1038/nature12274. [DOI] [PubMed] [Google Scholar]
- 11.Kaplan AH, Swanstrom R. 1991. The HIV-1 gag precursor is processed via two pathways: implications for cytotoxicity. Biomed Biochim Acta 50:647–653. [PubMed] [Google Scholar]
- 12.Kaplan AH, Manchester M, Swanstrom R. 1994. The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J Virol 68:6782–6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nie Z, Phenix BN, Lum JJ, Alam A, Lynch DH, Beckett B, Krammer PH, Sekaly RP, Badley AD. 2002. HIV-1 protease processes procaspase 8 to cause mitochondrial release of cytochrome c, caspase cleavage and nuclear fragmentation. Cell Death Differ 9:1172–1184. doi: 10.1038/sj.cdd.4401094. [DOI] [PubMed] [Google Scholar]
- 14.Nie Z, Bren GD, Vlahakis SR, Schimnich AA, Brenchley JM, Trushin SA, Warren S, Schnepple DJ, Kovacs CM, Loutfy MR, Douek DC, Badley AD. 2007. Human immunodeficiency virus type 1 protease cleaves procaspase 8 in vivo. J Virol 81:6947–6956. doi: 10.1128/JVI.02798-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nie Z, Bren GD, Rizza SA, Badley AD. 2008. HIV protease cleavage of procaspase 8 is necessary for death of HIV-infected cells. Open Virol J 2:1–7. doi: 10.2174/1874357900802010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sainski AM, Natesampillai S, Cummins NW, Bren GD, Taylor J, Saenz DT, Poeschla EM, Badley AD. 2011. The HIV-1-specific protein Casp8p41 induces death of infected cells through Bax/Bak. J Virol 85:7965–7975. doi: 10.1128/JVI.02515-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sainski AM, Dai H, Natesampillai S, Pang YP, Bren GD, Cummins NW, Correia C, Meng XW, Tarara JE, Ramirez-Alvarado M, Katzmann DJ, Ochsenbauer C, Kappes JC, Kaufmann SH, Badley AD. 2014. Casp8p41 generated by HIV protease kills CD4 T cells through direct Bak activation. J Cell Biol 207:159. doi: 10.1083/jcb.20140505109252014c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shan L, Deng K, Shroff NS, Durand CM, Rabi SA, Yang HC, Zhang H, Margolick JB, Blankson JN, Siliciano RF. 2012. Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity 36:491–501. doi: 10.1016/j.immuni.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dietz AB, Bulur PA, Emery RL, Winters JL, Epps DE, Zubair AC, Vuk-Pavlovic S. 2006. A novel source of viable peripheral blood mononuclear cells from leukoreduction system chambers. Transfusion 46:2083–2089. doi: 10.1111/j.1537-2995.2006.01033.x. [DOI] [PubMed] [Google Scholar]
- 20.Bren GD, Whitman J, Cummins N, Shepard B, Rizza SA, Trushin SA, Badley AD. 2008. Infected cell killing by HIV-1 protease promotes NF-κB dependent HIV-1 replication. PLoS One 3:e2112. doi: 10.1371/journal.pone.0002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith AJ, Dai H, Correia C, Takahashi R, Lee SH, Schmitz I, Kaufmann SH. 2011. Noxa/Bcl-2 protein interactions contribute to bortezomib resistance in human lymphoid cells. J Biol Chem 286:17682–17692. doi: 10.1074/jbc.M110.189092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, Nguyen PL, Khoruts A, Larson M, Haase AT, Douek DC. 2004. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med 200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cummins NW, Neuhaus J, Sainski AM, Strausbauch MA, Wettstein PJ, Lewin SR, Plana M, Rizza SA, Temesgen Z, Touloumi G, Freiberg M, Neaton J, Badley AD. 2014. Short communication: CD4 T cell declines occurring during suppressive antiretroviral therapy reflect continued production of Casp8p41. AIDS Res Hum Retroviruses 30:476–479. doi: 10.1089/aid.2013.0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liszewski MK, Yu JJ, O'Doherty U. 2009. Detecting HIV-1 integration by repetitive-sampling Alu-gag PCR. Methods 47:254–260. doi: 10.1016/j.ymeth.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diez J, Walter D, Munoz-Pinedo C, Gabaldon T. 2010. DeathBase: a database on structure, evolution and function of proteins involved in apoptosis and other forms of cell death. Cell Death Differ 17:735–736. doi: 10.1038/cdd.2009.215. [DOI] [PubMed] [Google Scholar]
- 27.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M, Goto S. 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pang YP. 2014. Low-mass molecular dynamics simulation: a simple and generic technique to enhance configurational sampling. Biochem Biophys Res Commun 452:588–592. doi: 10.1016/j.bbrc.2014.08.119. [DOI] [PubMed] [Google Scholar]
- 30.Pang YP. 2015. Low-mass molecular dynamics simulation for configurational sampling enhancement: more evidence and theoretical explanation. Biochem Biophys Rep 4:126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pang YP. 2015. At least 10% shorter C-H bonds in cryogenic protein crystal structures than in current AMBER forcefields. Biochem Biophys Res Commun 458:352–355. doi: 10.1016/j.bbrc.2015.01.115. [DOI] [PubMed] [Google Scholar]
- 32.Pang YP. 2015. Use of 1-4 interaction scaling factors to control the conformational equilibrium between alpha-helix and beta-strand. Biochem Biophys Res Commun 457:183–186. doi: 10.1016/j.bbrc.2014.12.084. [DOI] [PubMed] [Google Scholar]
- 33.Berendsen HJC, van Gunsteren PJWF, Di Nola A, Haak JR. 1984. Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690. doi: 10.1063/1.448118. [DOI] [Google Scholar]
- 34.Darden TA, YD& Pedersen LG. 1993. Particle mesh Ewald: An N log(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 35.Shao J, Thompson TSN, Cheatham TE III. 2007. Clustering molecular dynamics trajectories: 1. Characterizing the performance of different clustering algorithms. J Chem Theory Comput 3:2312–2334. [DOI] [PubMed] [Google Scholar]
- 36.Jaafoura S, de Goer de Herve MG, Hernandez-Vargas EA, Hendel-Chavez H, Abdoh M, Mateo MC, Krzysiek R, Merad M, Seng R, Tardieu M, Delfraissy JF, Goujard C, Taoufik Y. 2014. Progressive contraction of the latent HIV reservoir around a core of less-differentiated CD4+ memory T cells. Nat Commun 5:5407. doi: 10.1038/ncomms6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deeks SG. 2012. HIV: Shock and kill. Nature 487:439–440. doi: 10.1038/487439a. [DOI] [PubMed] [Google Scholar]
- 38.Brenchley JM, Hill BJ, Ambrozak DR, Price DA, Guenaga FJ, Casazza JP, Kuruppu J, Yazdani J, Migueles SA, Connors M, Roederer M, Douek DC, Koup RA. 2004. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis. J Virol 78:1160–1168. doi: 10.1128/JVI.78.3.1160-1168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cummins NW, Jiang W, McGinty J, Bren GD, Bosch RJ, Landay A, Deeks SG, Martin JN, Douek D, Lederman MM, Brenchley J, Badley AD. 2010. Intracellular Casp8p41 content is inversely associated with CD4 T cell count. J Infect Dis 202:386–391. doi: 10.1086/653705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sekaly RP. 2009. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macallan DC, Wallace D, Zhang Y, De Lara C, Worth AT, Ghattas H, Griffin GE, Beverley PC, Tough DF. 2004. Rapid turnover of effector-memory CD4+ T cells in healthy humans. J Exp Med 200:255–260. doi: 10.1084/jem.20040341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Grevenynghe J, Procopio FA, He Z, Chomont N, Riou C, Zhang Y, Gimmig S, Boucher G, Wilkinson P, Shi Y, Yassine-Diab B, Said EA, Trautmann L, El Far M, Balderas RS, Boulassel MR, Routy JP, Haddad EK, Sekaly RP. 2008. Transcription factor FOXO3a controls the persistence of memory CD4+ T cells during HIV infection. Nat Med 14:266–274. doi: 10.1038/nm1728. [DOI] [PubMed] [Google Scholar]
- 43.Dai H, Meng XW, Lee SH, Schneider PA, Kaufmann SH. 2009. Context-dependent Bcl-2/Bak interactions regulate lymphoid cell apoptosis. J Biol Chem 284:18311–18322. doi: 10.1074/jbc.M109.004770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeshita M, Suzuki K, Kassai Y, Takiguchi M, Nakayama Y, Otomo Y, Morita R, Miyazaki T, Yoshimura A, Takeuchi T. 2015. Polarization diversity of human CD4+ stem cell memory T cells. Clin Immunol 159:107–117. doi: 10.1016/j.clim.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 45.Czabotar PE, Lessene G, Strasser A, Adams JM. 2014. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol 15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 46.Martinou JC, Youle RJ. 2011. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev Cell 21:92–101. doi: 10.1016/j.devcel.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, Colman PM, Day CL, Adams JM, Huang DC. 2005. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell 17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 48.Dai H, Smith A, Meng XW, Schneider PA, Pang YP, Kaufmann SH. 2011. Transient binding of an activator BH3 domain to the Bak BH3-binding groove initiates Bak oligomerization. J Cell Biol 194:39–48. doi: 10.1083/jcb.201102027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yip KW, Reed JC. 2008. Bcl-2 family proteins and cancer. Oncogene 27:6398–6406. doi: 10.1038/onc.2008.307. [DOI] [PubMed] [Google Scholar]
- 50.Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, Dayton BD, Ding H, Enschede SH, Fairbrother WJ, Huang DC, Hymowitz SG, Jin S, Khaw SL, Kovar PJ, Lam LT, Lee J, Maecker HL, Marsh KC, Mason KD, Mitten MJ, Nimmer PM, Oleksijew A, Park CH, Park CM, Phillips DC, Roberts AW, Sampath D, Seymour JF, Smith ML, Sullivan GM, Tahir SK, Tse C, Wendt MD, Xiao Y, Xue JC, Zhang H, Humerickhouse RA, Rosenberg SH, Elmore SW. 2013. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med 19:202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- 51.Correia C, Lee SH, Meng XW, Vincelette ND, Knorr KL, Ding H, Nowakowski GS, Dai H, Kaufmann SH. 2015. Emerging understanding of Bcl-2 biology: Implications for neoplastic progression and treatment. Biochim Biophys Acta 1853:1658–1671. doi: 10.1016/j.bbamcr.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baccarani M, Deininger MW, Rosti G, Hochhaus A, Soverini S, Apperley JF, Cervantes F, Clark RE, Cortes JE, Guilhot F, Hjorth-Hansen H, Hughes TP, Kantarjian HM, Kim DW, Larson RA, Lipton JH, Mahon FX, Martinelli G, Mayer J, Müller MC, Niederwieser D, Pane F, Radich JP, Rousselot P, Saglio G, Saussele S, Schiffer C, Silver R, Simonsson B, Steegmann JL, Goldman JM, Hehlmann R. 2013. European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood 122:872–874. doi: 10.1182/blood-2013-05-501569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davids MS, Seymour JF, Gerecitano JF, Kahl BS, Pagel JM, Wierda WG, Anderson MA, Rudersdorf N, Gressick LA, Montalvo NP, Yang J, Zhu M, Dunbar M, Cerri E, Enschede SH, Humerickhouse R, Roberts AW. 2014. Phase I study of ABT-199 (GDC-0199) in patients with relapsed/refractory (R/R) non-Hodgkin lymphoma (NHL): responses observed in diffuse large B-cell (DLBCL) and follicular lymphoma (FL) at higher cohort doses. Abstr 8522 J Clin Oncol 32:5S. [PubMed] [Google Scholar]
- 54.Archin NM, Bateson R, Tripathy MK, Crooks AM, Yang KH, Dahl NP, Kearney MF, Anderson EM, Coffin JM, Strain MC, Richman DD, Robertson KR, Kashuba AD, Bosch RJ, Hazuda DJ, Kuruc JD, Eron JJ, Margolis DM. 2014. HIV-1 expression within resting CD4+ T cells after multiple doses of vorinostat. J Infect Dis 210:728–735. doi: 10.1093/infdis/jiu155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spina CA, Anderson J, Archin NM, Bosque A, Chan J, Famiglietti M, Greene WC, Kashuba A, Lewin SR, Margolis DM, Mau M, Ruelas D, Saleh S, Shirakawa K, Siliciano RF, Singhania A, Soto PC, Terry VH, Verdin E, Woelk C, Wooden S, Xing S, Planelles V. 2013. An in-depth comparison of latent HIV-1 reactivation in multiple cell model systems and resting CD4+ T cells from aviremic patients. PLoS Pathog 9:e1003834. doi: 10.1371/journal.ppat.1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eriksson S, Graf EH, Dahl V, Strain MC, Yukl SA, Lysenko ES, Bosch RJ, Lai J, Chioma S, Emad F, Abdel-Mohsen M, Hoh R, Hecht F, Hunt P, Somsouk M, Wong J, Johnston R, Siliciano RF, Richman DD, O'Doherty U, Palmer S, Deeks SG, Siliciano JD. 2013. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog 9:e1003174. doi: 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deng K, Pertea M, Rongvaux A, Wang L, Durand CM, Ghiaur G, Lai J, McHugh HL, Hao H, Zhang H, Margolick JB, Gurer C, Murphy AJ, Valenzuela DM, Yancopoulos GD, Deeks SG, Strowig T, Kumar P, Siliciano JD, Salzberg SL, Flavell RA, Shan L, Siliciano RF. 2015. Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature 517:381–385. doi: 10.1038/nature14053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pegu A, Asokan M, Wu L, Wang K, Hataye J, Casazza JP, Guo X, Shi W, Georgiev I, Zhou T, Chen X, O'Dell S, Todd JP, Kwong PD, Rao SS, Yang ZY, Koup RA, Mascola JR, Nabel GJ. 2015. Activation and lysis of human CD4 cells latently infected with HIV-1. Nat Commun 6:8447. doi: 10.1038/ncomms9447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pallikkuth S, Sharkey M, Babic DZ, Gupta S, Stone GW, Fischl MA, Stevenson M, Pahwa S. 2015. Peripheral T follicular helper cells are the major HIV reservoir within central memory CD4 T cells in peripheral blood from chronic HIV-infected individuals on cART. J Virol [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kennedy EM, Kornepati AV, Goldstein M, Bogerd HP, Poling BC, Whisnant AW, Kastan MB, Cullen BR. 2014. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. J Virol 88:11965–11972. doi: 10.1128/JVI.01879-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clem RJ, Miller LK. 1994. Control of programmed cell death by the baculovirus genes p35 and iap. Mol Cell Biol 14:5212–5222. doi: 10.1128/MCB.14.8.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rubenwolf S, Schutt H, Nevels M, Wolf H, Dobner T. 1997. Structural analysis of the adenovirus type 5 E1B 55-kilodalton-E4orf6 protein complex. J Virol 71:1115–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clem RJ, Fechheimer M, Miller LK. 1991. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science 254:1388–1390. doi: 10.1126/science.1962198. [DOI] [PubMed] [Google Scholar]
- 64.Altmann M, Hammerschmidt W. 2005. Epstein-Barr virus provides a new paradigm: a requirement for the immediate inhibition of apoptosis. PLoS Biol 3:e404. doi: 10.1371/journal.pbio.0030404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pilder S, Logan J, Shenk T. 1984. Deletion of the gene encoding the adenovirus 5 early region 1b 21,000-molecular-weight polypeptide leads to degradation of viral and host cell DNA. J Virol 52:664–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lum JJ, Badley AD. 2003. Resistance to apoptosis: mechanism for the development of HIV reservoirs. Curr HIV Res 1:261–274. doi: 10.2174/1570162033485203. [DOI] [PubMed] [Google Scholar]
- 67.Cummins NW, Badley AD. 2013. Antiapoptotic mechanisms of HIV: lessons and novel approaches to curing HIV. Cell Mol Life Sci 70:3355–3363. doi: 10.1007/s00018-012-1239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lopez-Huertas MR, Mateos E, Sanchez Del Cojo M, Gomez-Esquer F, Diaz-Gil G, Rodriguez-Mora S, Lopez JA, Calvo E, Lopez-Campos G, Alcami J, Coiras M. 2013. The presence of HIV-1 Tat protein second exon delays fas protein-mediated apoptosis in CD4+ T lymphocytes: a potential mechanism for persistent viral production. J Biol Chem 288:7626–7644. doi: 10.1074/jbc.M112.408294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wolf D, Witte V, Laffert B, Blume K, Stromer E, Trapp S, d'Aloja P, Schurmann A, Baur AS. 2001. HIV-1 Nef associated PAK and PI3-kinases stimulate Akt-independent Bad-phosphorylation to induce antiapoptotic signals. Nat Med 7:1217–1224. doi: 10.1038/nm1101-1217. [DOI] [PubMed] [Google Scholar]
- 70.Mahlknecht U, Deng C, Lu MC, Greenough TC, Sullivan JL, O'Brien WA, Herbein G. 2000. Resistance to apoptosis in HIV-infected CD4+ T lymphocytes is mediated by macrophages: role for Nef and immune activation in viral persistence. J Immunol 165:6437–6446. doi: 10.4049/jimmunol.165.11.6437. [DOI] [PubMed] [Google Scholar]
- 71.Chinnaiyan AM, Woffendin C, Dixit VM, Nabel GJ. 1997. The inhibition of proapoptotic ICE-like proteases enhances HIV replication. Nat Med 3:333–337. doi: 10.1038/nm0397-333. [DOI] [PubMed] [Google Scholar]
- 72.Kim M, Hosmane NN, Bullen CK, Capoferri A, Yang HC, Siliciano JD, Siliciano RF. 2014. A primary CD4+ T cell model of HIV-1 latency established after activation through the T cell receptor and subsequent return to quiescence. Nat Protoc 9:2755–2770. doi: 10.1038/nprot.2014.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Widlak P, Garrard WT. 2005. Discovery, regulation, and action of the major apoptotic nucleases DFF40/CAD and endonuclease G. J Cell Biochem 94:1078–1087. doi: 10.1002/jcb.20409. [DOI] [PubMed] [Google Scholar]
- 74.Taylor RC, Cullen SP, Martin SJ. 2008. Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol 9:231–241. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- 75.Laforge M, Limou S, Harper F, Casartelli N, Rodrigues V, Silvestre R, Haloui H, Zagury JF, Senik A, Estaquier J. 2013. DRAM triggers lysosomal membrane permeabilization and cell death in CD4+ T cells infected with HIV. PLoS Pathog 9:e1003328. doi: 10.1371/journal.ppat.1003328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Waterhouse N, Kumar S, Song Q, Strike P, Sparrow L, Dreyfuss G, Alnemri ES, Litwack G, Lavin M, Watters D. 1996. Heteronuclear ribonucleoproteins C1 and C2, components of the spliceosome, are specific targets of interleukin 1beta-converting enzyme-like proteases in apoptosis. J Biol Chem 271:29335–29341. doi: 10.1074/jbc.271.46.29335. [DOI] [PubMed] [Google Scholar]
- 77.Luthi AU, Martin SJ. 2007. The CASBAH: a searchable database of caspase substrates. Cell Death Differ 14:641–650. doi: 10.1038/sj.cdd.4402103. [DOI] [PubMed] [Google Scholar]
- 78.Kelve M, Truve E, Aaspollu A, Kuusksalu A, Dapper J, Perovic S, Muller WE, Schroder HC. 1994. Rapid reduction of mRNA coding for 2′-5′-oligoadenylate synthetase in rat pheochromocytoma PC12 cells during apoptosis. Cell Mol Biol 40:165–173. [PubMed] [Google Scholar]
- 79.Houge G, Robaye B, Eikhom TS, Golstein J, Mellgren G, Gjertsen BT, Lanotte M, Doskeland SO. 1995. Fine mapping of 28S rRNA sites specifically cleaved in cells undergoing apoptosis. Mol Cell Biol 15:2051–2062. doi: 10.1128/MCB.15.4.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Samejima K, Earnshaw WC. 2005. Trashing the genome: the role of nucleases during apoptosis. Nat Rev Mol Cell Biol 6:677–688. doi: 10.1038/nrm1715. [DOI] [PubMed] [Google Scholar]
- 81.Finkel TH, Tudor-Williams G, Banda NK, Cotton MF, Curiel T, Monks C, Baba TW, Ruprecht RM, Kupfer A. 1995. Apoptosis occurs predominantly in bystander cells and not in productively infected cells of HIV- and SIV-infected lymph nodes. Nat Med 1:129–134. doi: 10.1038/nm0295-129. [DOI] [PubMed] [Google Scholar]
- 82.Roche. 2015. U.S. FDA grants breakthrough therapy designation for investigational Bcl-2 inhibitor venetoclax in 17p deletion relapsed-refractory chronic lymphocytic leukemia. Roche Molecular Systems, Branchburg, NJ: http://www.roche.com/investors/updates/inv-update-2015-05-07.htm. [Google Scholar]
- 83.Scholler J, Brady TL, Binder-Scholl G, Hwang WT, Plesa G, Hege KM, Vogel AN, Kalos M, Riley JL, Deeks SG, Mitsuyasu RT, Bernstein WB, Aronson NE, Levine BL, Bushman FD, June CH. 2012. Decade-long safety and function of retroviral-modified chimeric antigen receptor T cells. Sci Transl Med 4:132ra153. doi: 10.1126/scitranslmed.3003761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kumar S, Vij R, Kaufman JL, Mikhael J, Facon T, Moreau P, Amiot M, Alzate S, Morris LJ, Ross JA, Dunbar M, Zhu M, Agarwal SK, Leverson J, Enschede SH, Humerickhouse R, Touzeau C. 2015. Phase I interim safety and efficacy of venetoclax (ABT-199/GDC-0199) monotherapy for relapsed/refractory (R/R) multiple myeloma (MM), abstr. J Clin Oncol. 33:8576. [Google Scholar]
- 85.Vandergeeten C, Fromentin R, DaFonseca S, Lawani MB, Sereti I, Lederman MM, Ramgopal M, Routy JP, Sekaly RP, Chomont N. 2013. Interleukin-7 promotes HIV persistence during antiretroviral therapy. Blood 121:4321–4329. doi: 10.1182/blood-2012-11-465625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, Lai J, Blankson JN, Siliciano JD, Siliciano RF. 2013. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Karpel ME, Boutwell CL, Allen TM. 2015. BLT humanized mice as a small animal model of HIV infection. Curr Opin Virol 13:75–80. doi: 10.1016/j.coviro.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Laird GM, Bullen CK, Rosenbloom DI, Martin AR, Hill AL, Durand CM, Siliciano JD, Siliciano RF. 2015. Ex vivo analysis identifies effective HIV-1 latency-reversing drug combinations. J Clin Invest 125:1901–1912. doi: 10.1172/JCI80142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Correia C, Schneider PA, Dai H, Dogan A, Maurer MJ, Church AK, Novak AJ, Feldman AL, Wu X, Ding H, Meng XW, Cerhan JR, Slager SL, Macon WR, Habermann TM, Karp JE, Gore SD, Kay NE, Jelinek DF, Witzig TE, Nowakowski GS, Kaufmann SH. 2015. BCL2 mutations are associated with increased risk of transformation and shortened survival in follicular lymphoma. Blood 125:658–667. doi: 10.1182/blood-2014-04-571786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 91.Sok D, Laserson U, Laserson J, Liu Y, Vigneault F, Julien JP, Briney B, Ramos A, Saye KF, Le K, Mahan A, Wang S, Kardar M, Yaari G, Walker LM, Simen BB, St John EP, Chan-Hui PY, Swiderek K, Kleinstein SH, Alter G, Seaman MS, Chakraborty AK, Koller D, Wilson IA, Church GM, Burton DR, Poignard P. 2013. The effects of somatic hypermutation on neutralization and binding in the PGT121 family of broadly neutralizing HIV antibodies. PLoS Pathog 9:e1003754. doi: 10.1371/journal.ppat.1003754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Natesampillai S, Nie Z, Cummins NW, Jochmans D, Bren GD, Angel JB, Badley AD. 2010. Patients with discordant responses to antiretroviral therapy have impaired killing of HIV-infected T cells. PLoS Pathog 6:e1001213. doi: 10.1371/journal.ppat.1001213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hutter G, Nowak D, Mossner M, Ganepola S, Mussig A, Allers K, Schneider T, Hofmann J, Kucherer C, Blau O, Blau IW, Hofmann WK, Thiel E. 2009. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med 360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 94.Henrich TJ, Hanhauser E, Marty FM, Sirignano MN, Keating S, Lee TH, Robles YP, Davis BT, Li J Z, Heisey A, Hill AL, Busch MP, Armand P, Soiffer RJ, Altfeld M, Kuritzkes DR. 2014. Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: report of 2 cases. Ann Intern Med 161:319–327. doi: 10.7326/M14-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Henrich TJ, Hanhauser E, Li JZ, Heisey A, Abramson JS, Hamdan A, Armand P, LaCasce AS, Kuritzkes DR. 2014. Impact of systemic cytotoxic chemotherapy for malignancies on HIV-1 reservoir persistence, abstr 418. CROI Conference, Boston, MA http://www.croiconference.org/sessions/impact-systemic-cytotoxic-chemotherapy-malignancies-hiv-1-reservoir-persistence-0. [Google Scholar]
- 96.Badley AD, Sainski A, Wightman F, Lewin SR. 2013. Altering cell death pathways as an approach to cure HIV infection. Cell Death Dis 4:e718. doi: 10.1038/cddis.2013.248. [DOI] [PMC free article] [PubMed] [Google Scholar]