Abstract
Mammalian arenaviruses are zoonotic viruses that cause asymptomatic, persistent infections in their rodent hosts but can lead to severe and lethal hemorrhagic fever with bleeding and multiorgan failure in human patients. Lassa virus (LASV), for example, is endemic in several West African countries, where it is responsible for an estimated 500,000 infections and 5,000 deaths annually. There are currently no FDA-licensed therapeutics or vaccines available to combat arenavirus infection. A hallmark of arenavirus infection (e.g., LASV) is general immunosuppression that contributes to high viremia. Here, we discuss the early host immune responses to arenavirus infection and the recently discovered molecular mechanisms that enable pathogenic viruses to suppress host immune recognition and to contribute to the high degree of virulence. We also directly compare the innate immune evasion mechanisms between arenaviruses and other hemorrhagic fever-causing viruses, such as Ebola, Marburg, Dengue, and hantaviruses. A better understanding of the immunosuppression and immune evasion strategies of these deadly viruses may guide the development of novel preventative and therapeutic options.
INTRODUCTION
Arenaviruses are enveloped negative-sense RNA viruses with a bisegmented genome composed of the L (large; ca. 7.3 kb) and S (small; ca. 3.5 kb) segments. The S segment encodes the nucleoprotein (NP) and glycoprotein precursor complex (GPC), and the L segment encodes the L RNA-dependent RNA polymerase (RdRp) and the matrix RING-finger protein (Z) (1, 2).
A newly suggested taxonomy divides the Arenaviridae family into the genera Mammarenavirus and Reptarenavirus, whose reservoirs are mainly rodents and reptiles (i.e., snakes), respectively (3). Mammalian arenaviruses are grouped into New World (NW) and Old World (OW) arenaviruses based on their geographical distributions, as well as their serological and phylogenetic differences (1, 3). While the majority of these viruses do not cause human disease, nine species are associated with neurological and hemorrhagic diseases in humans. Lassa virus (LASV) accounts for the highest number of cases, with an estimated 300,000 to 500,000 infections and ∼5,000 deaths annually (4, 5). There is currently no FDA-licensed vaccine or therapeutic agent to protect against arenavirus infection. The upregulation of type I interferon (IFN-α and IFN-β), followed by the upregulation of a subset of cellular gene products, namely, interferon-stimulated genes (ISGs), is an essential mechanism to control viral infections (reviewed in reference 6). Two of the four arenavirus-encoded proteins, namely, the nucleoprotein NP and the RING-finger Z protein, have recently been described as type I IFN antagonists that contribute to the general immunosuppression observed during the course of arenavirus infection. A better understanding of how the host responds to arenaviral infection and how some of these viruses manage to evade host immune surveillance will offer important insights for the development of effective preventative and treatment options.
INNATE AND CELLULAR IMMUNE RESPONSES DURING VIRAL INFECTIONS
An important line of defense against intracellular pathogens, such as viruses, is the innate immune system, with the IFN induction and IFN signaling pathways at its core (6). The pattern recognition receptors (PRRs), which consist of the three main protein families retinoic acid-inducible gene RIG-I-like receptors (RLRs), Toll-like receptors (TLRs), and NOD-like receptors (NLRs), are responsible for the initial sensing of different pathogens (7). It is thought that these PRRs recognize specific features of a virus, some of which are generated during viral replication, such as double-stranded RNA or 5′-triphosphorylated RNA, collectively referred to as pathogen-associated molecular patterns (PAMPs) (8, 9). After initial sensing of these PAMPs, the PRRs initiate the activation of different signaling pathways, using several essential cellular adaptor proteins, that eventually results in the activation of transcription factors, namely, interferon regulatory factor 3 (IRF3), IRF7, or NF-κB (10, 11). Subsequently, these pathways lead to the expression or upregulation of specific subsets of gene products, including type I IFN, proapoptotic factors, and cytokines. The secreted type I IFN is recognized by target cells in an autocrine or paracrine manner, initiating the IFN signaling pathway (6, 12). The IFN signaling activation leads to the expression or upregulation of hundreds of ISGs, which have specific functions to counteract a given viral infection (6). ISG expression can affect global cellular and/or specific viral transcription or translation, as well as interfere with various steps of viral replication (reviewed in reference 13). Some components of the IFN induction pathway (e.g., IRF7) are also considered ISGs and, thus, are upregulated, which in turn forms an amplifying feedback loop to prolong and strengthen the antiviral effects (14, 15).
In addition to expressing type I IFN and ISGs, infected cells express and secrete a large variety of cytokines, which play crucial roles in the regulation of cellular immune responses (16–18). Cytokines are responsible for the activation and maturation of many professional immune cells, such as macrophages, dendritic cells (DCs), and T cells, which are thought to be crucial in the control and clearance of many viral infections, including arenaviruses (19–23).
IMMUNE RESPONSES DURING ARENAVIRUS INFECTIONS IN MOUSE MODELS
To combat viral infections, different components of the immune response, including innate, cellular, and humoral systems, are activated. Studies in mice have shown that a functional innate immune system with a productive IFN response is essential to control and eventually clear arenavirus infections (24–28). IFN-α/β or IFN-α/βγ receptor knockout mice fail to control arenavirus infections, which result in systemic viral infections with more severe and lethal pathological phenotypes than in wild-type mice (24, 27, 28). Although the activation of IRF7 has been shown to be important for the production of IFN-α (25, 26), which is crucial to control lymphocytic choriomeningitis virus (LCMV) infection, it is not involved in the eventual clearance of the virus by CD8+ T cells. This stands in agreement with the finding that IFN-γ produced early in LCMV infection subsequently activates CD8+ T cells, thus favoring virus clearance from mice (29). Interestingly, LCMV (clone 13) can maintain persistent infection, which depends on a low level of type I IFN signaling that induces the anti-inflammatory interleukin (IL-10), as well as programmed cell death 1 ligand 1 (PD-L1) (30, 31). The balance of these two cytokines is critical for the successful establishment of persistence or the switch to an acute infection. IL-10 can interfere with the NF-κB pathway, downregulate cytokines secreted by helper T cells (Th1), decrease major histocompatibility complex (MHC) class II presentation, and interfere with macrophage stimulation (30). The inhibition of innate immune pathways during arenavirus infections can interfere with the function of plasmacytoid DCs (pDCs), leading to a failure of CD8+ T cell activation due to a lack of cross-priming by pDCs and a reduction in immunopathological damages (32), which can result in an overall reduction of immunopathological damages. A recent study showed that IFN-α and IFN-β have different effects on arenavirus infection: IFN-α may control virus spread in early infection, while IFN-β may be involved in viral clearance later in infection (33).
CELLULAR IMMUNE RESPONSES UPON IFN INDUCTION DURING ARENAVIRUS INFECTIONS OF RODENTS AND HUMANS
During arenavirus infection, the initial innate immune response and the activation of the type I IFN system are essential to prime subsequent cellular and adaptive immune responses to eventually overcome and clear the infection (34–37). In adult mice infected with LCMV, an IFN burst was observed from 6 to 48 h postinfection (38–40). Although the viral titers peak at 3 to 5 days postinfection, long after IFN levels have declined, this initial IFN burst can be enough to restrict viral titers to levels manageable primarily by cellular immune responses, which can clear the virus after 8 days of infection (39, 40). The type I IFN released mainly by pDCs can subsequently affect the activation and maturation of B and T cells (41, 42). Apart from secreting IFN, the activation of pDCs also increases the presentation of MHC class I and II peptides and leads to the production of costimulatory molecules and inflammatory cytokines. Before CD8+ T lymphocytes can become activated against arenavirus infection, the early expression of and downregulation of viral replication by the ISGs is paramount, but the exact subset of ISGs required needs further investigation.
Studies of human survivors of LASV infections indicate a general immunosuppression characterized by low levels of type I IFN and proinflammatory cytokines, such as tumor necrosis factor alpha (TNF-α) or IL-1β (28, 43, 44). Fatal LASV infections are often directly associated with altered levels of IFN-γ-induced protein 10 (IP-10), IL-6, or IL-8 (28, 45–49). The reduction of cytokines goes hand in hand with reduced expression of costimulatory proteins in DCs, such as CD86, which results in the failure to activate CD4+ and CD8+ T lymphocytes (32, 45, 48). Finally, in LASV-infected patients, lymphopenia and lymphoid depletion are often observed in the spleen and lymph nodes as a result of reductions in DCs and CD8+ T lymphocytes (50–52), which may explain the overall poor immune response throughout the course of LASV infection.
The immune response during infection by NW arenaviruses, such as Junin virus (JUNV), appears to differ from immune responses during OW arenavirus infections, observed in LASV patients. The differences in immune responses upon infection by OW or NW arenaviruses have been discussed in detail elsewhere (53). Available data from in vitro and in vivo studies suggest that NW arenavirus infections with JUNV or Machupo virus (MACV) trigger an initial IFN response similar to that of LASV (54). But rather than suppressing the innate immune pathways after the initial IFN induction, as observed for LASV, the response during JUNV infections shifts toward a proinflammatory response mediated mainly by IFN-α and TNF-α (55, 56). Although in vivo results support the importance of IFN-α and IFN-γ, work with JUNV and MACV in cell culture indicates that viral titers were mainly affected by IFN-β (57). Currently, it is unclear whether these NW arenaviruses are unable to inhibit pathways that result in the expression of IFN-α and IFN-γ or whether these viruses might even actively direct the innate immune responses toward an inflammatory response to activate potential target cells at the site of infection.
The differences in immune responses upon infection by OW or NW arenaviruses can partly be explained by the difference in the mechanisms of viral cell entry and/or cellular tropisms. While OW arenaviruses (e.g., LASV) use cellular α-dystroglycan (α-DG) as their receptors, clade B NW arenaviruses (e.g., JUNV and MACV) that cause human disease use human transferrin receptor 1 (hTfR1) (58), which can result in a change of target cells in vivo. This is supported by the lack of cytokine release from macrophages and dendritic cells during JUNV infection (21), results that have previously been observed in Argentine hemorrhagic fever patients (59–61).
MOLECULAR IMMUNOSUPPRESSIVE MECHANISMS OF ARENAVIRUSES
NP as an IFN antagonist.
The arenaviral nucleoprotein is the most abundantly expressed viral protein in infected cells (62). NP binds and encapsidates viral genomic RNA to form the viral nucleocapsid (63). The nucleocapsid forms a complex with the viral polymerase (L protein), called the viral ribonucleoprotein (vRNP) complex, to synthesize viral genomic RNAs during viral replication and to transcribe viral mRNAs (63). Through interactions with other viral and cellular proteins, NP also plays important roles in the proper assembly and budding of progeny virions from infected cells (64, 65). Besides serving essential roles in viral replication, genome encapsidation, and virion assembly, NP has also been shown to be intimately involved in regulating host innate immunity (Fig. 1) (66–68).
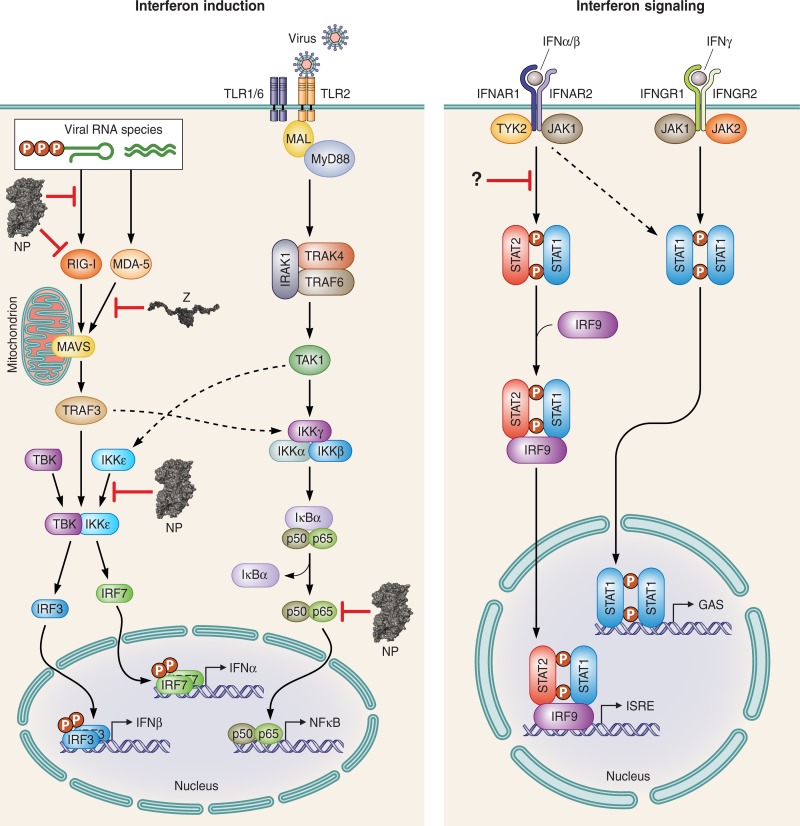
FIG 1.
Inhibition of innate immunity by arenavirus NP and Z. (A) Diagram of the type I interferon (IFN) induction and signaling pathways that play important roles as part of innate immunity against arenavirus infection. The IFN induction pathway triggered by RIG-I and MDA5 (collectively known as the RIG-I-like receptors or RLRs) is an important viral-defense pathway against arenaviruses. This RLR pathway can be blocked by arenavirus Z protein when it binds directly to RIG-I and MDA-5 and prevents the activation of the mitochondrial antiviral signaling (MAVS) protein. The action of NP in this pathway is more complex, as it exerts important effects at multiple steps. It is thought that the exoribonuclease function of NP can destroy pathogen-associated molecular pattern (PAMP) RNAs to blunt RIG-I and MDA-5 activation. At the same time, NP might be able to bind directly to RIG-I to prevent its activation. It can also bind to IKKε and prevent the activation of the IFN-responsive factor IRF3. NP is also capable of blocking NF-κB activation, a pathway that is thought to be activated by arenavirus glycoprotein GP via the Toll-like receptors (TLRs). Mouse experiments have also shown that arenaviruses can interfere with the p65 subunit of NF-κB, as well as STAT2, via an unknown mechanism.
Initial studies looking at IFN antagonism by different arenaviral proteins have identified NP as the major viral protein that can interfere with the IFN induction pathways (66, 67). NP is comprised of two domains separated by a flexible linker region (Fig. 2A). The N-terminal domain seems to function as the RNA-binding domain, contributing to viral genomic-RNA synthesis and encapsidation (68, 69). Structural and biochemical analysis of several arenaviral NPs, as well as bioinformatics analyses, have shown that the C-terminal domain is mainly comprised of a highly conserved 3′-to-5′ exonuclease of the DEDDh family (68, 69). Members of this exonuclease family are often involved in various nucleic acid repair or proof-reading mechanisms (70, 71), but for arenavirus NP, this exoribonuclease activity appears to mediate type I IFN suppression (66, 72, 73). When any of the amino acids of the active site of the exoribonuclease (RNase) is mutated, the immunosuppressive function by NP is abolished, resulting in a virus that can induce strong IFN expression and shows a reduced growth potential in immunocompetent cells and an attenuated phenotype in infected animals (66–68). The important role of the NP RNase function in suppressing immune response in order to favor virus replication is evidenced by the appearance of wild-type virus revertants in many of the sick animals with detectable levels of viremia (72). It is thought that double-stranded RNAs (dsRNAs) are generated during viral replication in the cytoplasm (Fig. 1 and 2C). These dsRNAs serve as PAMPs for ligands such as retinoic acid-inducible gene 1 (RIG-I) or melanoma differentiation-associated protein 5 (MDA5), which in turn would induce an antiviral signaling cascade (74, 75).
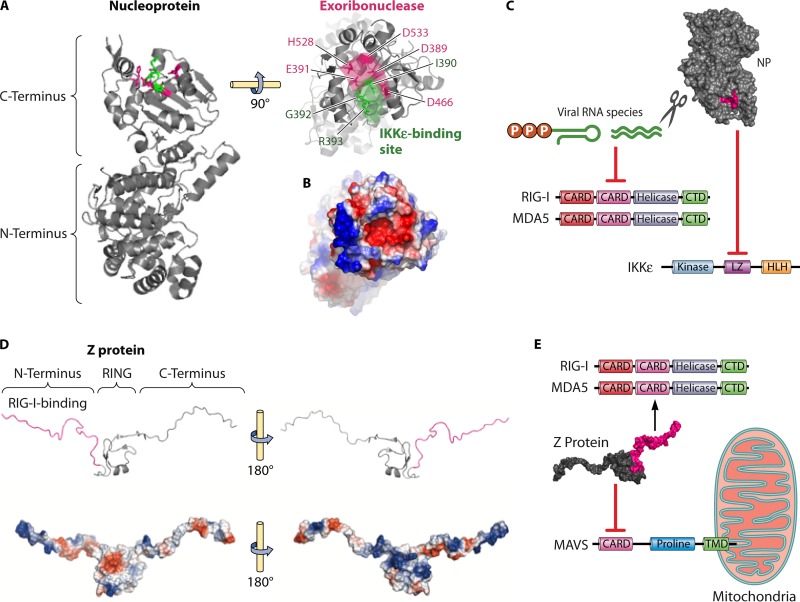
FIG 2.
Molecular basis for arenavirus immune evasion. (A) Crystal structure of LASV NP (PDB code 3MWP). The C terminus of NP comprises a 3′-to-5′ exoribonuclease whose active-site residues (magenta) are involved in mediating type I IFN suppression. The direct binding to IKKε might be facilitated by the highly conserved DIEGR motif (green) of NP. (B) The DIEGR motif involved in the possible IKKε binding and the catalytic site of the exoribonuclease form a single positively charged binding pocket (red), as can be seen in the surface charge representation. (C) The C terminus of NP is involved in two known immune evasion mechanisms, namely, the reduction of PAMP RNA species through its exoribonuclease function to prevent RIG-I and MDA5 activation and the inhibition of IKKε to prevent downstream activation of IRF3. RIG-I and MDA5 consist of two CARD domains, a helicase domain, and a C-terminal domain (CTD), whereas IKKε possesses a kinase domain, a leucine zipper (LZ), and a helix-loop-helix (HLH) domain. (D) Nuclear magnetic resonance structure of LASV Z (PDB code 2M1S). The small matrix Z protein possesses a central RING domain flanked by highly flexible N- and C-terminal extensions. The immunosuppressive function of Z has recently been mapped to the N-terminal domain (magenta), which contains negatively (red) and positively (blue) charged regions that could facilitate different protein-protein interactions. (E) The immunosuppressive function of the Z protein N-terminal domain has been shown to be via direct binding to the CARD domains of RIG-I and MDA5, resulting in a disruption in the interaction and activation of the mitochondrial antiviral signaling (MAVS) protein with its CARD domain, proline-rich domain, and transmembrane domain (TMD).
NP has also been shown to disrupt the IFN induction pathway by directly binding to effector proteins of this pathway (Fig. 2A to C). Biochemical assays have shown that LCMV NP can directly interact with RIG-I and MDA5 to suppress the IFN induction pathway (76). Further downstream in this pathway, LCMV and Pichinde virus (PICV) NPs have also been shown to interact directly with the IκB kinase ε (IKKε) (72, 77). This interaction disrupts the binding of IKKε and the TANK-binding kinase 1 (TBK1) to form a heterodimer, which results in the failure of IRF3 phosphorylation and, therefore, prevents its activation (Fig. 1). Mutations in either the NP exoribonuclease active site (D391, E393, D466, D533, and H528 in LASV NP) or a nearby conserved DIEGR amino acid motif in NP (Fig. 2A) result in a defect in the ability of NP to block nuclear translocation of IRF3 as a potential consequence of the reduced affinity to IKKε (Fig. 2C) (72, 77). In addition to interfering with the IRF3 pathway, NP can also block the NF-κB pathway (Fig. 1), the exact mechanism of which is currently unknown (72). A major caveat is that the majority of experiments performed in these studies used transiently expressed NPs. Therefore, the importance of the difference of the NP-directed inhibition of either the IRF3 or the NF-κB pathways remains to be analyzed in more detail.
Z as an IFN antagonist.
The smallest arenaviral protein is the zinc finger protein (Z), encoded on the L genomic RNA segment. The Z proteins are 90 to 99 amino acids in size, depending on the virus species, and belong to the RING finger protein family (78) (Fig. 2D). RING finger proteins are involved in many protein-protein interactions (79). Correspondingly, the arenavirus Z protein has been shown to interact with cellular proteins, such as the promyelocytic leukemia protein (PML), eukaryotic elongation factor 4E (eIF4E), or components of the endosomal sorting complexes required for transport (ESCRT) (79, 80). In addition to interactions with cellular proteins, Z also interacts with the viral L protein to “lock” the protein onto the genome (81), as well as serving as a main component for viral budding by oligomerizing and forming the viral matrix of progeny virus particles (79).
Z has also recently been shown to inhibit the IFN induction pathway by directly binding to the N-terminal CARD domains of RIG-I and MDA5 (82) (Fig. 2E). Interestingly, Z proteins from all known highly pathogenic arenaviruses (e.g., LASV, Lujo virus, MACV, Sabia virus, Chapare virus, Guanarito virus, Dandenong virus, and JUNV), as well as the relatively low-pathogenicity LCMV, are able to bind to RIG-I and MDA5 and, thus, suppress the production of IFN-β (82). In contrast, Z proteins of nonpathogenic arenaviruses do not bind to RIG-I or MDA5 and fail to inhibit the IFN induction pathway (82). This reported interference with IFN induction can lead to the inhibition of human macrophage activation, which subsequently affects the proper function of human NK cells and T cells (83). Therefore, it is hypothesized that the type I IFN-antagonistic action of the Z protein may actively contribute to a successful infection by increasing viral replication and disease pathogenesis in human patients.
COMPARISON OF INNATE IMMUNE EVASION STRATEGIES OF DIFFERENT HEMORRHAGIC FEVER-CAUSING VIRUSES AND ARENAVIRUSES
Viral IFN antagonists have been described for almost all known virus families (84), so that the inhibition of different IFN pathways seems to be a common theme among all viruses to successfully establish infections. Besides arenaviruses, other hemorrhagic fever-causing viruses, including members of the filoviruses (Marburg virus and Ebola virus [EBOV]) (85), flaviviruses (yellow fever virus, Dengue virus [DENV], Omsk hemorrhagic fever virus, and Kyasanur forest disease virus) (86–88, 104, 105), and bunyaviruses (Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, and severe fever with thrombocytopenia syndrome virus [SFTSV], and hantaviruses) (89–93) are also known to encode factors to suppress host IFN responses. It would appear that viral replication and disease progression that eventually lead to symptoms like vascular leakage, hemorrhages, and multiorgan failure (85, 94, 95) originated with a strong inhibition of IFN induction and/or signaling by these viruses. Thus, for several of these viruses, it has been shown that the inhibition of the innate immune pathways correlates with disease progression and outcome in humans (94, 95).
Interestingly, it appears that immune suppression by these viruses is not perfect, as early IFN responses are usually triggered that result in the reduction of viral titers in patients' sera, which has been linked to improved survival rates (89, 96). However, different viruses encode IFN antagonists that can effectively suppress type I IFN expression (89, 94–96), leading to high viral titers. In order to increase the efficiency of immunosuppression, these hemorrhagic fever-causing viruses appear to utilize several of their proteins to inhibit IFN induction and/or IFN signaling pathways (89, 97, 98). Some of these viral IFN antagonists target the early stages of the IFN induction pathway by reducing PAMP RNAs, as well as reducing the activation of the RLRs (i.e., RIG-I or MDA5) (7). Similarly to the exoribonuclease function of arenaviral NPs and the inhibitory effect of the Z protein on RIG-I and MDA5, other viruses target this important induction step to prevent an innate immune response by other means. For example, the filoviral VP35 has been shown to sequester dsRNAs that could serve as PAMPs for RIG-I and MDA5 activation (97). At the same time, VP35 interferes with the RIG-I cofactor known as the protein activator of the interferon-induced protein kinase (PACT) to further aid the inhibitory effect. Hantaviruses remove the triphosphorylated termini of their genomes to evade RIG-I recognition (99), highlighting an active role for viral enzymatic functions to evade recognition by RLRs.
Besides directly interfering with the upstream stages of the IFN induction pathway, hemorrhagic fever viral IFN antagonists seem to target at least one other step of this pathway, which is further downstream and is involved in the phosphorylation and, thus, the activation of IRF3. Similar to arenavirus NP, other hemorrhagic fever viruses, such as filoviruses, hantaviruses, and SFTSV, also prevent the formation of the TBK1-IKKε heterodimer (97, 99), which phosphorylates IRF3.
While published studies have only indicated that arenaviruses interfere with IFN production and consequently suppress the induction of ISGs (100, 101), other viruses have been shown to also inhibit the IFN signaling pathway. It has been shown that MARV VP40 inhibits the phosphorylation of STAT1 by Jak1, while EBOV VP24 blocks this pathway further downstream from STAT1/Jak1 and prevents the nuclear translocation of the interferon-stimulated gene factor 3 (ISGF3) complex (97). DENV utilizes several of its proteins to antagonize IFN signaling. This virus uses its NS2A, NS4A, NS4B, and NS5 proteins to downregulate the phosphorylation of Jak1, Tyk2, and STAT1, while targeting STAT2 for proteasomal degradation (102) and, thus, preventing any form of STAT-mediated downstream signaling in this important antiviral pathway. The mechanism(s) by which arenaviruses might inhibit the IFN signaling pathway still need to be demonstrated.
SUMMARY
The outcomes of human arenavirus infections are tightly linked to cellular immune responses, which are controlled by the activation of the innate immune pathways. In LASV-infected patients, the immune response is generally suppressed, while an inflammatory response is observed in JUNV-infected patients (44, 56). While having limited coding capacity on a relatively small genome, these viruses remarkably possess two IFN antagonists, namely, NP and Z proteins. Comparing the functions of these arenaviral IFN antagonists with other hemorrhagic fever-causing viruses highlights the fact that this seems to be a common strategy displayed by these viruses. It is conceivable that multiple viral proteins acting as IFN antagonists increase the chances of a complete inhibition of these antiviral pathways in order to optimize viral replication and transmission (84, 94, 103). This emphasizes the importance of regulating host immunity during the course of the infection, especially the innate immune responses early in the infection. The current knowledge about the immune antagonism displayed by arenaviruses is almost exclusively limited to the inhibition of the IFN induction pathway by NP and Z proteins. It was shown that arenaviruses are capable of inhibiting IRF3 to prevent the production of type I IFN (66, 67), whereas the role of and interaction with IRF7 was implicated as important (25) but remains less clear. Future research will require extending the knowledge about potential interactions between arenaviral proteins and the IFN signaling pathway to explore potential mechanisms of inhibition as indicated in mouse studies (101). For other hemorrhagic fever-causing viruses, the inhibition of the IFN signaling pathway has been characterized (84, 89, 97) and highlights potential starting points for the development of potential immunotherapeutic options.
ACKNOWLEDGMENTS
We apologize to colleagues whose works contributed a great deal to the field but could not be cited in this article due to space constraints.
This work was supported in part by NIH grants R01AI093580 and R56AI091805 to H.L.
Biographies

Bjoern Meyer received his B.Sc. (Hons) degree in Microbiology from the University of Liverpool (United Kingdom) in 2010. He was awarded the Society of General Microbiology (now Society for Microbiology) Undergraduate Prize in 2009. He obtained his Ph.D. degree in Molecular Virology from the University of St. Andrews (United Kingdom) in 2014. During this time, he was supported by the Medical Research Council (MRC, United Kingdom). He is currently serving as a postdoctoral research associate in Dr. Ly's laboratory at the University of Minnesota, Twin Cities, where he focuses on investigations of arenavirus immune evasion, virus-host protein-protein interactions, and the development of screening platforms for universal antiarenaviral compounds.

Hinh Ly received his B.S. and M.A. degrees with honors in Microbiology and Molecular Genetics from the University of California, Los Angeles (UCLA), in 1995 and 1998, respectively, and his Ph.D. degree in Microbiology and Immunology from the University of North Carolina at Chapel Hill (UNC) in 2000. He carried out his postdoctoral work at the University of California, San Francisco (UCSF), in the laboratories of Dr. Tristram Parslow (HIV researcher) and Dr. Elizabeth Blackburn (telomere researcher and 2009 Nobel prize winner in Physiology or Medicine). Prior to his current position as an Associate Professor of Virology and Immunology in the Department of Veterinary Biomedical Sciences at the University of Minnesota, Twin Cities, Dr. Ly served as an Assistant Professor in the Department of Pathology and Laboratory Medicine at Emory University in Atlanta. Dr. Ly's research interest has focused on virus-host interactions with an emphasis on host immune suppression by arenaviruses.
REFERENCES
- 1.Buchmeier MJ, de la Torre JC, Peters CJ. 2013. Arenaviridae: the viruses and their replication, p 1791–1828. In Knipe DM, Howley P, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Salvato MS, Clegg JCS, Buchmeier MJ, Charrell RN, Gonzalez JP, Lukasevich IS, Peters CJ, Romanovski V. 2011. In King AM, Lefkowitz E, Adams MJ, Carstens EB (ed), 9th report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 3.Radoshitzky SR, Bao Y, Buchmeier MJ, Charrel RN, Clawson AN, Clegg CS, DeRisi JL, Emonet S, Gonzalez JP, Kuhn JH, Lukashevich IS, Peters CJ, Romanowski V, Salvato MS, Stenglein MD, de la Torre JC. 2015. Past, present, and future of arenavirus taxonomy. Arch Virol 160:1851–1874. doi: 10.1007/s00705-015-2418-y. [DOI] [PubMed] [Google Scholar]
- 4.Charrel RN, de Lamballerie X, Emonet S. 2008. Phylogeny of the genus Arenavirus. Curr Opin Microbiol 11:362–368. doi: 10.1016/j.mib.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Gunther S, Lenz O. 2004. Lassa virus. Crit Rev Clin Lab Sci 41:339–390. doi: 10.1080/10408360490497456. [DOI] [PubMed] [Google Scholar]
- 6.Randall RE, Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol 89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 7.Sparrer KMJ, Gack MU. 2015. Intracellular detection of viral nucleic acids. Curr Opin Microbiol 26:1–9. doi: 10.1016/j.mib.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcus PI, Sekellick MJ. 1977. Defective interfering particles with covalently linked [+/−]RNA induce interferon. Nature 266:815–819. doi: 10.1038/266815a0. [DOI] [PubMed] [Google Scholar]
- 9.Marcus PI, Svitlik C, Sekellick MJ. 1983. Interferon induction by viruses. X. A model for interferon induction by Newcastle Disease virus. J Gen Virol 64(Pt 11):2419–2431. doi: 10.1099/0022-1317-64-11-2419. [DOI] [PubMed] [Google Scholar]
- 10.Ikushima H, Negishi H, Taniguchi T. 2013. The IRF family transcription factors at the interface of innate and adaptive immune responses. Cold Spring Harbor Symp Quant Biol 78:105–116. doi: 10.1101/sqb.2013.78.020321. [DOI] [PubMed] [Google Scholar]
- 11.Oeckinghaus A, Ghosh S. 2009. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol 1:a000034. doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isaacs A, Lindenmann J. 1957. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci 147:258–267. doi: 10.1098/rspb.1957.0048. [DOI] [PubMed] [Google Scholar]
- 13.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haller O, Kochs G, Weber F. 2006. The interferon response circuit: induction and suppression by pathogenic viruses. Virology 344:119–130. doi: 10.1016/j.virol.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marie I, Durbin JE, Levy DE. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J 17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crouse J, Kalinke U, Oxenius A. 2015. Regulation of antiviral T cell responses by type I interferons. Nat Rev Immunol 15:231–242. doi: 10.1038/nri3806. [DOI] [PubMed] [Google Scholar]
- 17.Glass CK, Natoli G. 2015. Molecular control of activation and priming in macrophages. Nat Immunol 17:26–33. doi: 10.1038/ni.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swiecki M, Colonna M. 2015. The multifaceted biology of plasmacytoid dendritic cells. Nat Rev Immunol 15:471–485. doi: 10.1038/nri3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carnec X, Baize S, Reynard S, Diancourt L, Caro V, Tordo N, Bouloy M. 2011. Lassa virus nucleoprotein mutants generated by reverse genetics induce a robust type I interferon response in human dendritic cells and macrophages. J Virol 85:12093–12097. doi: 10.1128/JVI.00429-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flatz L, Rieger T, Merkler D, Bergthaler A, Regen T, Schedensack M, Bestmann L, Verschoor A, Kreutzfeldt M, Bruck W, Hanisch UK, Gunther S, Pinschewer DD. 2010. T cell-dependence of Lassa fever pathogenesis. PLoS Pathog 6:e1000836. doi: 10.1371/journal.ppat.1000836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groseth A, Hoenen T, Weber M, Wolff S, Herwig A, Kaufmann A, Becker S. 2011. Tacaribe virus but not Junin virus infection induces cytokine release from primary human monocytes and macrophages. PLoS Negl Trop Dis 5:e1137. doi: 10.1371/journal.pntd.0001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott EP, Aronson JF. 2008. Cytokine patterns in a comparative model of arenavirus haemorrhagic fever in guinea pigs. J Gen Virol 89:2569–2579. doi: 10.1099/vir.0.2008/002048-0. [DOI] [PubMed] [Google Scholar]
- 23.Wieland SF, Takahashi K, Boyd B, Whitten-Bauer C, Ngo N, de la Torre JC, Chisari FV. 2014. Human plasmacytoid dendritic cells sense lymphocytic choriomeningitis virus-infected cells in vitro. J Virol 88:752–757. doi: 10.1128/JVI.01714-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolokoltsova OA, Yun NE, Poussard AL, Smith JK, Smith JN, Salazar M, Walker A, Tseng CT, Aronson JF, Paessler S. 2010. Mice lacking alpha/beta and gamma interferon receptors are susceptible to Junin virus infection. J Virol 84:13063–13067. doi: 10.1128/JVI.01389-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li W, Hofer MJ, Nocon AL, Manders P, Campbell IL. 2013. Interferon regulatory factor 7 (IRF7) is required for the optimal initial control but not subsequent clearance of lymphocytic choriomeningitis virus infection in mice. Virology 439:152–162. doi: 10.1016/j.virol.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 26.Malmgaard L, Salazar-Mather TP, Lewis CA, Biron CA. 2002. Promotion of alpha/beta interferon induction during in vivo viral infection through alpha/beta interferon receptor/STAT1 system-dependent and -independent pathways. J Virol 76:4520–4525. doi: 10.1128/JVI.76.9.4520-4525.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rieger T, Merkler D, Gunther S. 2013. Infection of type I interferon receptor-deficient mice with various Old World arenaviruses: a model for studying virulence and host species barriers. PLoS One 8:e72290. doi: 10.1371/journal.pone.0072290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yun NE, Poussard AL, Seregin AV, Walker AG, Smith JK, Aronson JF, Smith JN, Soong L, Paessler S. 2012. Functional interferon system is required for clearance of Lassa virus. J Virol 86:3389–3392. doi: 10.1128/JVI.06284-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pien GC, Nguyen KB, Malmgaard L, Satoskar AR, Biron CA. 2002. A unique mechanism for innate cytokine promotion of T cell responses to viral infections. J Immunol 169:5827–5837. doi: 10.4049/jimmunol.169.10.5827. [DOI] [PubMed] [Google Scholar]
- 30.Ng CT, Oldstone MB. 2014. IL-10: achieving balance during persistent viral infection. Curr Top Microbiol Immunol 380:129–144. doi: 10.1007/978-3-662-43492-5_6. [DOI] [PubMed] [Google Scholar]
- 31.Teijaro JR, Ng C, Lee AM, Sullivan BM, Sheehan KC, Welch M, Schreiber RD, de la Torre JC, Oldstone MB. 2013. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science 340:207–211. doi: 10.1126/science.1235214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pannetier D, Reynard S, Russier M, Journeaux A, Tordo N, Deubel V, Baize S. 2011. Human dendritic cells infected with the nonpathogenic Mopeia virus induce stronger T-cell responses than those infected with Lassa virus. J Virol 85:8293–8306. doi: 10.1128/JVI.02120-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng CT, Sullivan BM, Teijaro JR, Lee AM, Welch M, Rice S, Sheehan KCF, Schreiber RD, Oldstone MBA. 2015. Blockade of interferon beta, but not interferon alpha, signaling controls persistent viral infection. Cell Host Microbe 17:653–661. doi: 10.1016/j.chom.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baize S, Marianneau P, Loth P, Reynard S, Journeaux A, Chevallier M, Tordo N, Deubel V, Contamin H. 2009. Early and strong immune responses are associated with control of viral replication and recovery in Lassa virus-infected cynomolgus monkeys. J Virol 83:5890–5903. doi: 10.1128/JVI.01948-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carrion R Jr, Brasky K, Mansfield K, Johnson C, Gonzales M, Ticer A, Lukashevich I, Tardif S, Patterson J. 2007. Lassa virus infection in experimentally infected marmosets: liver pathology and immunophenotypic alterations in target tissues. J Virol 81:6482–6490. doi: 10.1128/JVI.02876-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hensley LE, Smith MA, Geisbert JB, Fritz EA, Daddario-DiCaprio KM, Larsen T, Geisbert TW. 2011. Pathogenesis of Lassa fever in cynomolgus macaques. Virol J 8:205. doi: 10.1186/1743-422X-8-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lukashevich IS. 2013. The search for animal models for Lassa fever vaccine development. Expert Rev Vaccines 12:71–86. doi: 10.1586/erv.12.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biron CA, Nguyen KB, Pien GC. 2002. Innate immune responses to LCMV infections: natural killer cells and cytokines. Curr Top Microbiol Immunol 263:7–27. [DOI] [PubMed] [Google Scholar]
- 39.Lee LN, Burke S, Montoya M, Borrow P. 2009. Multiple mechanisms contribute to impairment of type 1 interferon production during chronic lymphocytic choriomeningitis virus infection of mice. J Immunol 182:7178–7189. doi: 10.4049/jimmunol.0802526. [DOI] [PubMed] [Google Scholar]
- 40.Zuniga EI, Liou LY, Mack L, Mendoza M, Oldstone MB. 2008. Persistent virus infection inhibits type I interferon production by plasmacytoid dendritic cells to facilitate opportunistic infections. Cell Host Microbe 4:374–386. doi: 10.1016/j.chom.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jung A, Kato H, Kumagai Y, Kumar H, Kawai T, Takeuchi O, Akira S. 2008. Lymphocytoid choriomeningitis virus activates plasmacytoid dendritic cells and induces a cytotoxic T-cell response via MyD88. J Virol 82:196–206. doi: 10.1128/JVI.01640-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montoya M, Edwards MJ, Reid DM, Borrow P. 2005. Rapid activation of spleen dendritic cell subsets following lymphocytic choriomeningitis virus infection of mice: analysis of the involvement of type 1 IFN. J Immunol 174:1851–1861. doi: 10.4049/jimmunol.174.4.1851. [DOI] [PubMed] [Google Scholar]
- 43.Baize S, Kaplon J, Faure C, Pannetier D, Georges-Courbot MC, Deubel V. 2004. Lassa virus infection of human dendritic cells and macrophages is productive but fails to activate cells. J Immunol 172:2861–2869. doi: 10.4049/jimmunol.172.5.2861. [DOI] [PubMed] [Google Scholar]
- 44.Geisbert TW, Jahrling PB. 2004. Exotic emerging viral diseases: progress and challenges. Nat Med 10:S110–S121. doi: 10.1038/nm1142. [DOI] [PubMed] [Google Scholar]
- 45.Baize S, Pannetier D, Faure C, Marianneau P, Marendat I, Georges-Courbot MC, Deubel V. 2006. Role of interferons in the control of Lassa virus replication in human dendritic cells and macrophages. Microbes Infect 8:1194–1202. doi: 10.1016/j.micinf.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 46.Lukashevich IS, Maryankova R, Vladyko AS, Nashkevich N, Koleda S, Djavani M, Horejsh D, Voitenok NN, Salvato MS. 1999. Lassa and Mopeia virus replication in human monocytes/macrophages and in endothelial cells: different effects on IL-8 and TNF-α gene expression. J Med Virol 59:552–560. doi:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahanty S, Bausch DG, Thomas RL, Goba A, Bah A, Peters CJ, Rollin PE. 2001. Low levels of interleukin-8 and interferon-inducible protein-10 in serum are associated with fatal infections in acute Lassa fever. J Infect Dis 183:1713–1721. doi: 10.1086/320722. [DOI] [PubMed] [Google Scholar]
- 48.Mahanty S, Hutchinson K, Agarwal S, McRae M, Rollin PE, Pulendran B. 2003. Cutting edge: impairment of dendritic cells and adaptive immunity by Ebola and Lassa viruses. J Immunol 170:2797–2801. doi: 10.4049/jimmunol.170.6.2797. [DOI] [PubMed] [Google Scholar]
- 49.Peters CJ, Liu CT, Anderson GW Jr, Morrill JC, Jahrling PB. 1989. Pathogenesis of viral hemorrhagic fevers: Rift Valley fever and Lassa fever contrasted. Rev Infect Dis 11(Suppl 4):S743–S749. doi: 10.1093/clinids/11.Supplement_4.S743. [DOI] [PubMed] [Google Scholar]
- 50.Edington GM, White HA. 1972. The pathology of Lassa fever. Trans R Soc Trop Med Hyg 66:381–389. doi: 10.1016/0035-9203(72)90268-4. [DOI] [PubMed] [Google Scholar]
- 51.Fisher-Hoch S, McCormick JB, Sasso D, Craven RB. 1988. Hematologic dysfunction in Lassa fever. J Med Virol 26:127–135. doi: 10.1002/jmv.1890260204. [DOI] [PubMed] [Google Scholar]
- 52.Walker DH, McCormick JB, Johnson KM, Webb PA, Komba-Kono G, Elliott LH, Gardner JJ. 1982. Pathologic and virologic study of fatal Lassa fever in man. Am J Pathol 107:349–356. [PMC free article] [PubMed] [Google Scholar]
- 53.McLay L, Liang Y, Ly H. 2014. Comparative analysis of disease pathogenesis and molecular mechanisms of New World and Old World arenavirus infections. J Gen Virol 95:1–15. doi: 10.1099/vir.0.057000-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang C, Kolokoltsova OA, Yun NE, Seregin AV, Ronca S, Koma T, Paessler S. 2015. Highly pathogenic New World and Old World human arenaviruses induce distinct interferon responses in human cells. J Virol 89:7079–7088. doi: 10.1128/JVI.00526-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Levis SC, Saavedra MC, Ceccoli C, Falcoff E, Feuillade MR, Enria DA, Maiztegui JI, Falcoff R. 1984. Endogenous interferon in Argentine hemorrhagic fever. J Infect Dis 149:428–433. doi: 10.1093/infdis/149.3.428. [DOI] [PubMed] [Google Scholar]
- 56.Levis SC, Saavedra MC, Ceccoli C, Feuillade MR, Enria DA, Maiztegui JI, Falcoff R. 1985. Correlation between endogenous interferon and the clinical evolution of patients with Argentine hemorrhagic fever. J Interferon Res 5:383–389. doi: 10.1089/jir.1985.5.383. [DOI] [PubMed] [Google Scholar]
- 57.Huang C, Walker AG, Grant AM, Kolokoltsova OA, Yun NE, Seregin AV, Paessler S. 2014. Potent inhibition of Junin virus infection by interferon in murine cells. PLoS Negl Trop Dis 8:e2933. doi: 10.1371/journal.pntd.0002933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rojek JM, Kunz S. 2008. Cell entry by human pathogenic arenaviruses. Cell Microbiol 10:828–835. doi: 10.1111/j.1462-5822.2007.01113.x. [DOI] [PubMed] [Google Scholar]
- 59.Ambrosio AM, Enria DA, Maiztegui JI. 1986. Junin virus isolation from lympho-mononuclear cells of patients with Argentine hemorrhagic fever. Intervirology 25:97–102. doi: 10.1159/000149662. [DOI] [PubMed] [Google Scholar]
- 60.Gonzalez PH, Cossio PM, Arana R, Maiztegui JI, Laguens RP. 1980. Lymphatic tissue in Argentine hemorrhagic fever. Pathologic features. Arch Pathol Lab Med 104:250–254. [PubMed] [Google Scholar]
- 61.Maiztegui JI, Laguens RP, Cossio PM, Casanova MB, de la Vega MT, Ritacco V, Segal A, Fernandez NJ, Arana RM. 1975. Ultrastructural and immunohistochemical studies in five cases of Argentine hemorrhagic fever. J Infect Dis 132:35–53. doi: 10.1093/infdis/132.1.35. [DOI] [PubMed] [Google Scholar]
- 62.Fuller-Pace FV, Southern PJ. 1988. Temporal analysis of transcription and replication during acute infection with lymphocytic choriomeningitis virus. Virology 162:260–263. doi: 10.1016/0042-6822(88)90419-9. [DOI] [PubMed] [Google Scholar]
- 63.Young PR, Howard CR. 1983. Fine structure analysis of Pichinde virus nucleocapsids. J Gen Virol 64(Pt 4):833–842. doi: 10.1099/0022-1317-64-4-833. [DOI] [PubMed] [Google Scholar]
- 64.Eichler R, Strecker T, Kolesnikova L, ter Meulen J, Weissenhorn W, Becker S, Klenk HD, Garten W, Lenz O. 2004. Characterization of the Lassa virus matrix protein Z: electron microscopic study of virus-like particles and interaction with the nucleoprotein (NP). Virus Res 100:249–255. doi: 10.1016/j.virusres.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 65.Groseth A, Wolff S, Strecker T, Hoenen T, Becker S. 2010. Efficient budding of the Tacaribe virus matrix protein Z requires the nucleoprotein. J Virol 84:3603–3611. doi: 10.1128/JVI.02429-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martinez-Sobrido L, Zuniga EI, Rosario D, Garcia-Sastre A, de la Torre JC. 2006. Inhibition of the type I interferon response by the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol 80:9192–9199. doi: 10.1128/JVI.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martinez-Sobrido L, Giannakas P, Cubitt B, Garcia-Sastre A, de la Torre JC. 2007. Differential inhibition of type I interferon induction by arenavirus nucleoproteins. J Virol 81:12696–12703. doi: 10.1128/JVI.00882-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qi XX, Lan SY, Wang WJ, Schelde LM, Dong HH, Wallat GD, Ly H, Liang YY, Dong CJ. 2010. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature 468:779–765. doi: 10.1038/nature09605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Levingston Macleod JM, D'Antuono A, Loureiro ME, Casabona JC, Gomez GA, Lopez N. 2011. Identification of two functional domains within the arenavirus nucleoprotein. J Virol 85:2012–2023. doi: 10.1128/JVI.01875-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bernad A, Blanco L, Lazaro JM, Martin G, Salas M. 1989. A conserved 3′–5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell 59:219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- 71.Minskaia E, Hertzig T, Gorbalenya AE, Campanacci V, Cambillau C, Canard B, Ziebuhr J. 2006. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci U S A 103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang QF, Shao JJ, Lan SY, Zhou YQ, Xing JJ, Dong CJ, Liang YY, Ly H. 2015. In vitro and in vivo characterizations of Pichinde viral nucleoprotein exoribonuclease functions. J Virol 89:6595–6607. doi: 10.1128/JVI.00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martinez-Sobrido L, Emonet S, Giannakas P, Cubitt B, Garcia-Sastre A, de la Torre JC. 2009. Identification of amino acid residues critical for the anti-interferon activity of the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol 83:11330–11340. doi: 10.1128/JVI.00763-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jensen S, Thomsen AR. 2012. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J Virol 86:2900–2910. doi: 10.1128/JVI.05738-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 76.Zhou S, Cerny AM, Zacharia A, Fitzgerald KA, Kurt-Jones EA, Finberg RW. 2010. Induction and inhibition of type I interferon responses by distinct components of lymphocytic choriomeningitis virus. J Virol 84:9452–9462. doi: 10.1128/JVI.00155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pythoud C, Rodrigo WW, Pasqual G, Rothenberger S, Martinez-Sobrido L, de la Torre JC, Kunz S. 2012. Arenavirus nucleoprotein targets interferon regulatory factor-activating kinase IKKε. J Virol 86:7728–7738. doi: 10.1128/JVI.00187-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Salvato MS, Shimomaye EM. 1989. The completed sequence of lymphocytic choriomeningitis virus reveals a unique RNA structure and a gene for a zinc finger protein. Virology 173:1–10. doi: 10.1016/0042-6822(89)90216-X. [DOI] [PubMed] [Google Scholar]
- 79.Fehling SK, Lennartz F, Strecker T. 2012. Multifunctional nature of the arenavirus RING finger protein Z. Viruses 4:2973–3011. doi: 10.3390/v4112973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Campbell Dwyer EJ, Lai HK, MacDonald RC, Salvato MS, Borden KLB. 2000. The lymphocytic choriomeningitis virus RING protein Z associates with eukaryotic initiation factor 4E and selectively represses translation in a RING-dependent manner. J Virol 74:3293–3300. doi: 10.1128/JVI.74.7.3293-3300.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kranzusch PJ, Whelan SP. 2011. Arenavirus Z protein controls viral RNA synthesis by locking a polymerase-promoter complex. Proc Natl Acad Sci U S A 108:19743–19748. doi: 10.1073/pnas.1112742108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xing JJ, Ly H, Liang YY. 2015. The Z proteins of pathogenic but not nonpathogenic arenaviruses inhibit RIG-I-like receptor-dependent interferon production. J Virol 89:2944–2955. doi: 10.1128/JVI.03349-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xing J, Chai Z, Ly H, Liang Y. 2015. Differential inhibition of macrophage activation by lymphocytic choriomeningitis virus and Pichinde virus is mediated by the Z protein N-terminal domain. J Virol 89:12513–12517. doi: 10.1128/JVI.01674-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Versteeg GA, Garcia-Sastre A. 2010. Viral tricks to grid-lock the type I interferon system. Curr Opin Microbiol 13:508–516. doi: 10.1016/j.mib.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Feldmann H, Geisbert TW. 2011. Ebola haemorrhagic fever. Lancet 377:849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Diamond MS. 2009. Mechanisms of evasion of the type I interferon antiviral response by flaviviruses. J Interferon Cytokine Res 29:521–530. doi: 10.1089/jir.2009.0069. [DOI] [PubMed] [Google Scholar]
- 87.Green AM, Beatty PR, Hadjilaou A, Harris E. 2014. Innate immunity to dengue virus infection and subversion of antiviral responses. J Mol Biol 426:1148–1160. doi: 10.1016/j.jmb.2013.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ye J, Zhu B, Fu ZF, Chen H, Cao S. 2013. Immune evasion strategies of flaviviruses. Vaccine 31:461–471. doi: 10.1016/j.vaccine.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 89.Elliott RM, Weber F. 2009. Bunyaviruses and the type I interferon system. Viruses 1:1003–1021. doi: 10.3390/v1031003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Flick R, Bouloy M. 2005. Rift Valley fever virus. Curr Mol Med 5:827–834. doi: 10.2174/156652405774962263. [DOI] [PubMed] [Google Scholar]
- 91.Liu S, Chai C, Wang C, Amer S, Lv H, He H, Sun J, Lin J. 2014. Systematic review of severe fever with thrombocytopenia syndrome: virology, epidemiology, and clinical characteristics. Rev Med Virol 24:90–102. doi: 10.1002/rmv.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Maes P, Clement J, Gavrilovskaya I, Van Ranst M. 2004. Hantaviruses: immunology, treatment, and prevention. Viral Immunol 17:481–497. doi: 10.1089/vim.2004.17.481. [DOI] [PubMed] [Google Scholar]
- 93.Weber F, Mirazimi A. 2008. Interferon and cytokine responses to Crimean Congo hemorrhagic fever virus; an emerging and neglected viral zonoosis. Cytokine Growth Factor Rev 19:395–404. doi: 10.1016/j.cytogfr.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bray M. 2005. Pathogenesis of viral hemorrhagic fever. Curr Opin Immunol 17:399–403. doi: 10.1016/j.coi.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 95.Paessler S, Walker DH. 2013. Pathogenesis of the viral hemorrhagic fevers. Annu Rev Pathol 8:411–440. doi: 10.1146/annurev-pathol-020712-164041. [DOI] [PubMed] [Google Scholar]
- 96.McCormick JB, Fisher-Hoch SP. 2002. Lassa fever. Curr Top Microbiol Immunol 262:75–109. [DOI] [PubMed] [Google Scholar]
- 97.Messaoudi I, Amarasinghe GK, Basler CF. 2015. Filovirus pathogenesis and immune evasion: insights from Ebola virus and Marburg virus. Nat Rev Microbiol 13:663–676. doi: 10.1038/nrmicro3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zinzula L, Tramontano E. 2013. Strategies of highly pathogenic RNA viruses to block dsRNA detection by RIG-I-like receptors: hide, mask, hit. Antiviral Res 100:615–635. doi: 10.1016/j.antiviral.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Weber M, Weber F. 2014. Segmented negative-strand RNA viruses and RIG-I: divide (your genome) and rule. Curr Opin Microbiol 20:96–102. doi: 10.1016/j.mib.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 100.Bowick GC, Fennewald SM, Scott EP, Zhang L, Elsom BL, Aronson JF, Spratt HM, Luxon BA, Gorenstein DG, Herzog NK. 2007. Identification of differentially activated cell-signaling networks associated with Pichinde virus pathogenesis by using systems kinomics. J Virol 81:1923–1933. doi: 10.1128/JVI.02199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hahm B, Trifilo MJ, Zuniga EI, Oldstone MB. 2005. Viruses evade the immune system through type I interferon-mediated STAT2-dependent, but STAT1-independent, signaling. Immunity 22:247–257. doi: 10.1016/j.immuni.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 102.Morrison J, Aguirre S, Fernandez-Sesma A. 2012. Innate immunity evasion by Dengue virus. Viruses 4:397–413. doi: 10.3390/v4030397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bowie AG, Unterholzner L. 2008. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol 8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Diamond MS, Pierson TC. 2015. Molecular insight into dengue virus pathogenesis and its implications for disease control. Cell 162:488–492. doi: 10.1016/j.cell.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gould EA, Solomon T. 2008. Pathogenic flaviviruses. Lancet 371:500–509. doi: 10.1016/S0140-6736(08)60238-X. [DOI] [PubMed] [Google Scholar]