ABSTRACT
Baculovirus DNAs are synthesized and inserted into preformed capsids to form nucleocapsids at a site in the infected cell nucleus, termed the virogenic stroma. Nucleocapsid assembly of Autographa californica multiple nucleopolyhedrovirus (AcMNPV) requires the major capsid protein VP39 and nine minor capsid proteins, including VP1054. However, how VP1054 participates in nucleocapsid assembly remains elusive. In this study, the VP1054-encoding gene (ac54) was deleted to generate the ac54-knockout AcMNPV (vAc54KO). In vAc54KO-transfected cells, nucleocapsid assembly was disrupted, leading to the formation of abnormally elongated capsid structures. Interestingly, unlike cells transfected with AcMNPV mutants lacking other minor capsid proteins, in which capsid structures were distributed within the virogenic stroma, ac54 ablation resulted in a distinctive location of capsid structures and VP39 at the periphery of the nucleus. The altered distribution pattern of capsid structures was also observed in cells transfected with AcMNPV lacking BV/ODV-C42 or in cytochalasin d-treated AcMNPV-infected cells. BV/ODV-C42, along with PP78/83, has been shown to promote nuclear filamentous actin (F-actin) formation, which is another requisite for nucleocapsid assembly. Immunofluorescence using phalloidin indicated that the formation and distribution of nuclear F-actin were not affected by ac54 deletion. However, immunoelectron microscopy revealed that BV/ODV-C42, PP78/83, and 38K failed to integrate into capsid structures in the absence of VP1054, and immunoprecipitation further demonstrated that in transient expression assays, VP1054 interacted with BV/ODV-C42 and VP80 but not VP39. Our findings suggest that VP1054 plays an important role in the transport of capsid proteins to the nucleocapsid assembly site prior to the process of nucleocapsid assembly.
IMPORTANCE Baculoviruses are large DNA viruses whose replication occurs within the host nucleus. The localization of capsids into the capsid assembly site requires virus-induced nuclear F-actin; the inhibition of nuclear F-actin formation results in the retention of capsid structures at the periphery of the nucleus. In this paper, we note that the minor capsid protein VP1054 is essential for the localization of capsid structures, the major capsid protein VP39, and the minor capsid protein 38K into the capsid assembly site. Moreover, VP1054 is crucial for correct targeting of the nuclear F-actin factors BV/ODV-C42 and PP78/83 for capsid maturation. However, the formation and distribution of nuclear F-actin are not affected by the lack of VP1054. We further reveal that VP1054 interacts with BV/ODV-C42 and a capsid transport-related protein, VP80. Taken together, our findings suggest that VP1054 plays a unique role in the pathway(s) for transport of capsid proteins.
INTRODUCTION
The family Baculoviridae includes a diverse group of insect-specific viruses that contain circular double-stranded DNA within a rod-shaped protein capsid enclosed by a lipid envelope (1, 2). Two forms of virions (budded virions [BV] and occlusion-derived virions [ODV]) are generated during the baculovirus life cycle; the main difference between them is the origin and constitution of their lipid envelopes (3, 4). During the early phase of infection, nucleocapsids assembled in the host cell nucleus are transported to the cytoplasm and bud into the extracellular environment to form BVs. BVs are required for spread among cells and tissues (5). During the late phase of infection, nucleocapsids are enclosed by virus-induced intranuclear membranes to form ODVs, which are subsequently embedded in the paracrystalline protein matrix to generate occlusion bodies (OBs). Due to the protection of the outer protein shell, OBs are durable and easily disseminated in nature and therefore are responsible for horizontal infections among insect hosts (3, 6, 7).
Baculovirus DNA replication occurs in a specific subnuclear region of the host cells, called the virogenic stroma (VS). The VS grows until it gradually occupies most of the nucleus and marginalizes the host chromatin (8). The VS is composed of a homogenous fibrillar electron-dense mat and electron-lucent intrastromal spaces (9). Capsids grow from the edge of the stromal mat. New viral DNA is synthesized within the electron-dense mat and subsequently packaged into capsids to form nucleocapsids (8–10).
A baculovirus nucleocapsid consists of an apical cap, a cylindrical sheath, and a basal structure (8). The capsid sheath appears to form on the basal structure, and the apical cap structure is thought to mediate the incorporation of nucleoprotein into the preassembled capsid sheath (8). Although the mechanism of nucleocapsid assembly utilized by baculoviruses is poorly understood, an increasing number of components of the nucleocapsid have been identified. Autographa californica multiple nucleopolyhedrovirus (AcMNPV) is the most intensively studied baculovirus. In addition to the major capsid protein VP39 (11, 12), AcMNPV nucleocapsids contain several minor components (6, 13), such as the following: P6.9, a protamine-like protein that is responsible for the condensation of viral DNA to form the nucleocapsid core (14–16); PK-1, a virus-encoded protein kinase that is required for the hyperphosphorylation of P6.9, which facilitates the release of viral DNA from the nucleocapsid (17); and 38K, Ac53, VP91 (encoded by ac83), and VLF-1, which are also classified as minor capsid proteins although their exact functions are poorly understood (18–21). Single deletion of the genes encoding each of the six minor capsid proteins mentioned above results in the failure of nucleocapsid assembly and the distribution of abnormally long electron-lucent tubular structures at the electron-dense edges of the stroma, indicating that the packaging of DNA and nucleoproteins into capsids is interrupted (16, 18–22).
Previous studies have suggested that nuclear filamentous actin (F-actin) provides a scaffold to position capsids for proper assembly and DNA packaging (23). Treatment with cytochalasin D (CD), which is capable of binding specifically to the growing end of the actin filament, was found to prevent proper nucleocapsid assembly in AcMNPV-infected nuclei (24, 25). Long, electron-lucent, tubular capsid precursors were observed to locate always at the periphery of the nucleus (25), suggesting that the transport of VP39 to the VS was blocked by CD treatment; therefore, excess VP39 proteins accumulated to form the tubular structures.
F-actin is primarily distributed in the cytoplasm. Nuclear F-actin formation in AcMNPV-infected cells requires that three key elements (globular actin [G-actin], nucleation promoting factor [NPF], and the actin-related protein 2/3 complex [Arp2/3]) are relocated from the cytoplasm into the nucleus for actin polymerization (26). Both PP78/83 and BV/ODV-C42 (encoded by ac101) are located at the basal end of the nucleocapsid (27). PP78/83 is a WASP (Wiskott-Aldrich syndrome protein family)-like protein (28). When activated by PP78/83 and other related activators of actin nucleation, Arp2/3 initiates G-actin polymerization into F-actin in the nucleus (29, 30). Of note, the entry of PP78/83 into the nucleus is mediated by the nuclear localization signal of BV/ODV-C42 (31). Deletion of the BV/ODV-C42-encoding gene in AcMNPV prevents nucleocapsid assembly and results in the aggregation of tubular capsid precursors at the periphery of the nucleus (30).
VP1054 is a capsid-associated protein that is conserved in all sequenced baculoviruses (4, 32, 33). VP1054 is encoded by the ac54 gene in AcMNPV (34), and one single base pair mutation in ac54 is sufficient to prohibit the production of infectious BV at a nonpermissive temperature, suggesting that ac54 plays a crucial role in the viral life cycle (32). VP1054 is able to interact with GGN-rich nucleic acid sequences, similar to PURα family proteins (33). An AcMNPV strain with a deletion of ac54 lacked the ability to initiate nucleocapsid assembly and infectious BV propagation although viral DNA replication was unaffected (33). Moreover, the interaction between VP1054 and 38K has been identified via coimmunoprecipitation, indicating the interplay between VP1054 and capsid proteins (35). However, the precise mechanism of how VP1054 mediates the nucleocapsid assembly is not clear yet.
In the present study, an ac54-knockout AcMNPV (vAc54KO) mutant was constructed via ET homologous recombination. ET recombination is a DNA engineering method based on homologous recombination mediated by phage proteins RecE/RecT (36) or Redα/β/γ (37). Nucleocapsid assembly was abrogated in mutant-transfected cells, which was in accordance with former reports (32, 33). However, we observed that the abnormally long tubular capsid precursors containing VP39 were localized at the periphery of the nucleus instead of within the VS, a distribution pattern that was similar to that in AcMNPV-infected cells treated with CD (38) as well as in cells transfected with an ac101-null mutant (30). We demonstrated that neither capsid structures nor VP39 was present within the VS, suggesting that VP1054 was required for the transport of VP39 into the VS. Even though F-actin polymerization was not affected, loss of ac54 drastically affected the localization of BV/ODV-C42, PP78/83, and 38K to the capsid structures. Furthermore, immunoprecipitation assay showed that in transient expression assays, VP1054 was able to interact with the capsid proteins BV/ODV-C42 and VP80 but not VP39. Our findings suggest that VP1054 plays an important role in the transport of VP39 to the baculovirus nucleocapsid assembly site and in the integration of some minor capsid proteins into the capsid structures.
MATERIALS AND METHODS
Construction of an ac54-knockout virus.
In this study, the bacmid bMON14272, which contains an AcMNPV genome (39), was used to generate an ac54-knockout AcMNPV (bAc54KO) through ET homologous recombination in Escherichia coli as previously described (18). A 558-bp highly conserved region of the ac54 open reading frame (ORF) (AcMNPV nucleotides [nt] 45552 to 46109) was replaced with a chloramphenicol resistance (Cmr) gene cassette; the 5′-end 330-bp region and the 3′-end 210-bp region of the ac54 ORF were preserved to avoid any effects on the transcription of neighboring genes. Briefly, we designed two long primers (ac54-CmF and ac54-CmR) that contained the left and right flanking sequences of the deletion region, respectively, and the sequences to amplify the Cmr gene cassette (primers used in this study are listed in Table 1). Then, a linear 1,177-bp fragment was generated by PCR using the plasmid pKOV-kanF, which contained the Cmr gene cassette (40) as the template. Homologous recombination between the Cmr gene cassette and the deletion region was performed as previously described (18). The replacement was confirmed by PCR and sequencing, and the resulting bacmid was designated bAc54KO.
TABLE 1.
List of primers used in this paper
| Primer | Sequencea | Restriction enzyme |
|---|---|---|
| ac54-CmF | 5′ -GATCAAGCCGAGGCGAGAAAAACAATTCTCAACGCCGACAGCGTGTACGAGTGCATGTTAATTAGACCAATTCGTACGTAAAATACCTGTGACGGAAGA | |
| ac54-CmR | 5′-ATCCGCCGCCGGTTTCTGATTGGCGCGCGATTAACTCGCTGTATCGTTTGTACTTGTTGGGATTTCGAACCCACTTTGCTAAACCAGCAATAGACATAA | |
| CmUS | 5′-TAAAATACCTGTGACGGAAGA | |
| CmDS | 5′-TAAACCAGCAATAGACATAA | |
| ac54US | 5′-ATGTGTTCGACCAAGAAACC | |
| ac54DS | 5′- CTACACGTTGTGTGCGTGCA | |
| REPac54US | 5′-TCTAGAGAGATCCCTTTAATCGTGT | XbaI |
| REPac54DS | 5′-CTGCAGCTAAGCGTAATCTGGAACATCGTATGGGTAAGCGTAATCTGGAACATCGTATGGGTACACGTTGTGTGCGTGCAGAC | PstI |
| SV40DS | 5′-GAGCTCGATCCAGACATGATAAGATA | SacI |
| ac54US-T.E. | 5′-GGATCCGAGATCCCTTTAATCGTGT | BamHI |
| ac54DS-T.E. | 5′-GATATCCACGTTGTGTGCGTGCAG | EcoRV |
| ac54HAUS-T.E. | 5′-GGATCCGCCACCATGTGTTCGACCAAGAAACCG | BamHI |
| ac54HADS-T.E. | 5′-GATATCTTAGGCGTAGTCGGGCACGTCGTAGGGGTACACGTTGTGTGCGTGCAGACCAA | EcoRV |
| c42US-T.E. | 5′-GGATCCATGGCCACCAGCGCTATCGCGTTGTAT | BamHI |
| c42DS-T.E. | 5′-CTCGAGTCACTTATCGTCGTCATCCTTGTAATC ATATTTTTTACGCTTTGC | XhoI |
| vp39 US-T.E. | 5′-GGATCCATGGCCACCGCGCTAGTGCCCGTGGGT | BamHI |
| vp39 DS-T.E. | 5′-CTCGAGTCACTTATCGTCGTCATCCTTGTAATC GACGGCTATTCCTCCACC | XhoI |
| vp80 US-T.E. | 5′-GGATCCATGGCCACCAACGATTCCAATTCTCTG | BamHI |
| vp80 DS-T.E. | 5′-CTCGAGTCACTTATCGTCGTCATCCTTGTAATC TATAACATTGTAGTTTGC | XhoI |
Underlining indicates sequence complementary to the chloramphenicol resistance gene; boldface indicates restriction enzyme sites.
Because a mini-attTn7 site has been integrated into the AcMNPV polh locus in bMON14272, foreign genes can be easily inserted therein via site-specific transposition (39). Thus, by transforming pFB1-PH-GFP (where GFP is green fluorescent protein) (41) into competent DH10B cells that contained bAc54KO and the helper plasmid pMON7124 as previously described (18), an AcMNPV polyhedrin (polh) gene for restoration and a green fluorescent protein (gfp) gene for observation were inserted into the mini-attTn7 site of bAc54KO. The resulting AcMNPV ac54 deletion mutant was named vAc54KO.
To demonstrate that any defective phenotype for vAc54KO was attributable to the ac54 deletion and not to effects exerted on the transcription of neighboring genes, a repair bacmid, vAc54:2HA (where HA is hemagglutinin) was constructed by ectopically inserting the ac54 ORF into the mini-attTn7 site of vAc54KO as follows. A 1,451-bp fragment containing the promoter and the ORF of ac54 tagged with two HA epitopes at its C terminus was PCR amplified using the primers REPac54US and REPac54DS. The PCR product was cloned into the XbaI/PstI-digested plasmid pFast-Bac1 (Invitrogen Life Technologies) to obtain pFB-pac54HA. A simian virus 40 (SV40) poly(A) signal was added by PCR amplification using pFB-pac54HA as the template and the primers REPac54US and SV40DS. The product was digested with XbaI/SacI and subcloned into pFB1-PH-GFP (41) to generate the donor plasmid pFB1-54REP-PH-GFP. Site-specific transposition was performed as described above. Similarly, a wild-type AcMNPV (vAcWT) was also constructed by inserting the polh and gfp genes into the mini-attTn7 site of bMON14272.
Analysis of viral propagation.
The Spodoptera frugiperda cell strain Sf9 was cultured in TMM-FH medium (Invitrogen Life Technologies) supplemented with 10% fetal bovine serum (Invitrogen Life Technologies), 100 μg/ml penicillin, and 30 μg/ml streptomycin at 27°C. To analyze the effects of the ac54 deletion, Sf9 cells (1 × 106) were transfected with 2 μg of vAc54KO, vAc54:2HA, or vAcWT DNA using Cellfectin II reagent (Invitrogen Life Technologies). After the culture was incubated for 5 h, the transfection buffer was replaced with TMM-FH medium, and the time point was designated 0 h posttransfection (p.t.). The supernatants were collected at different time points, and the BV titers were determined by a 50% tissue culture infective dose (TCID50) endpoint dilution assay as previously described (42). For infection, Sf9 cells were infected with BVs harvested from transfections at a multiplicity of infection of 5 TCID50s/cell.
Analysis of viral DNA synthesis by qPCR.
Quantitative PCR (qPCR) was performed to investigate viral DNA replication. Sf9 cells (1 × 106) were transfected in triplicate with 1 μg of 38K-null recombinant AcMNPV (v38KKO), ie1-null recombinant AcMNPV (vIE1KO), or vAc54KO bacmid DNA and collected at different time points. Total DNA from each sample was extracted using a Universal Genomic DNA Extraction kit (TaKaRa) and resuspended in 50 μl of sterile water. To eliminate input bacmid DNA, 1 μg of DNA was digested with 20 units of the DpnI restriction enzyme (NEB) overnight in a 40-μl reaction volume. qPCR was performed using Hot Start PCR master mix III (Chaoshi-Bio) and primers targeting a 100-bp region of the gp41 gene under conditions described previously (43).
Immunofluorescence.
Sf9 cells (5 × 105) were seeded into 35-mm glass-bottom culture dishes (MatTek) and incubated at 27°C for 12 h, followed by transfection with 2 μg of bacmid DNA. Cells were processed for immunofluorescence microscopy as previously described at different time points (44). Mouse monoclonal antibody targeting actin (1:200; Abmart), antiserum against IE1 (1:100; obtained from L. A. Guarino, Texas A&M University), and rabbit polyclonal antisera (1:100) against VP39 (45), PP78/83 (31), BV/ODV-C42 (30), and 38K (35) were used as primary antibodies. Alexa Fluor 555-conjugated donkey anti-mouse antibody (1:400; Invitrogen/Molecular Probes) and Alexa Fluor 647-conjugated goat anti-rabbit antibody (1:400; Invitrogen/Molecular Probes) were used as secondary antibodies. Prior to observation, the cells were stained with Hoechst 33342 (Invitrogen/Molecular Probes) and/or phalloidin-fluorescein isothiocyanate (FITC) (Beyotime Biotechnology) and washed four times with 1× phosphate-buffered saline (PBS). Cells were visualized with a Zeiss LSM 7 Duo NLO laser scanning confocal microscope (Carl Zeiss, Germany).
Electron microscopy.
For electron microscopy, 1 × 106 Sf9 cells were transfected with 1 μg of bacmid DNA or infected with BVs at a multiplicity of infection of 5 TCID50s/cell. At different time points posttransfection or postinfection (p.i.), the cells were collected with a cell scraper and pelleted at 3,000 × g for 5 min. The cells were fixed, dehydrated, embedded, sectioned, and stained as previously described (18). For immunoelectron microscopy, Sf9 cells were transfected with bacmid DNA and labeled with bromodeoxyuridine (BrdU) (1:100; Invitrogen) at 12 h p.t. Then, the cells were harvested at 72 h p.t. and fixed in 100 mM PBS containing 1% (vol/vol) glutaraldehyde and 3% paraformaldehyde (pH 7.2) for 2 h at 4°C. The cells were subsequently dehydrated and embedded as previously described (18). Ultrathin sections were collected on Formvar-coated nickel grids and immunostained with primary antibodies including anti-BrdU (Abmart), anti-38K (35), anti-P6.9 (16), anti-BV/ODV-C42 (30), anti-PP78/83 (31), and anti-VP39 (45) (1:100). Then, colloidal gold particles (10 nm) coated with goat anti-mouse IgG were introduced as secondary antibodies (1:50; Sigma). The ultrathin sections were observed with a JEM-100CX/II transmission electron microscope at an accelerating voltage of 80 kV.
Generation of transient expression plasmids.
Plasmids transiently expressing BV/ODV-C42, VP80, VP39, or VP1054 were constructed for immunoprecipitation assays. Briefly, the ORFs of ac101, vp80, and vp39 with a Kozak consensus sequence at the 5′ ends and FLAG-coding sequence at the 3′ ends were PCR amplified from bMON14272 and cloned into the transient expression vector pIB/V5-His (Invitrogen). The primers c42US-T.E. and c42DS-T.E. were used to amplify the coding sequence of ac101, the primers vp80US-T.E. and vp80DS-T.E. were used for vp80, and the primers vp39US-T.E. and vp39DS-T.E. were used for vp39. The constructed plasmids were named pIB-BV/ODV-C42:FLAG, pIB-VP80:FLAG, and pIB-VP39:FLAG, respectively. Similarly, primer pairs ac54US-T.E./ac54DS-T.E. and ac54:HAUS-T.E./ac54:HADS-T.E. were used to PCR amplify the ac54 ORF from bMON14272; the PCR products were then cloned into pIB-GFP (referred to as pIB-nGFP in the manuscript) (20) and pIB/V5-His (Invitrogen), respectively. The constructed plasmids were named pIB-nGFP-VP1054 and pIB-VP1054:HA.
Immunoprecipitation.
In the immunoprecipitation assays with anti-FLAG, Sf9 cells (6 × 106) were cotransfected with 18 μg of pIB-BV/ODV-C42:FLAG, pIB-VP80:FLAG, or pIB-VP39:FLAG combined with 6 μg of pIB-nGFP-VP1054. To achieve a higher transfection efficiency, a second cotransfection as described above was performed in the same batch of Sf9 cells at 18 h after the first transfection. Cells were harvested at 36 h after the second transfection. After one wash in PBS, cells were lysed in 1% NP-40 buffer supplemented with 2 μg/ml of complete EDTA-free protease inhibitor cocktail (Roche) and then incubated on a rotary table. After samples were rotated for 1.5 h, the lysates were centrifuged at 13,000 × g for 15 min at 4°C. The extract was harvested and precleared by the addition of 30 μl of a 50% slurry of protein A/G-agarose beads (Abmart), followed by incubation at 4°C for 1.5 h on a rotary table. The supernatant was transferred to a fresh 1.5-ml Eppendorf tube and mixed with 20 μl of protein A/G-agarose beads (50% slurry) conjugated to an anti-FLAG antibody (Abmart). After samples were rotated at 4°C for 12 h, the beads were collected by centrifugation at 1,000 × g for 15 min at 4°C, washed four times with 1 ml of 1% NP-40 buffer, and boiled in 25 μl of 2× PBS for 10 min. The dissolved immunoprecipitates and the input cell lysates were analyzed by Western blotting with mouse monoclonal anti-FLAG (1:2,000; Abmart) or rabbit polyclonal anti-GFP (1:2,000; Abmart) antibody. For negative controls, pIB-nGFP was cotransfected with pIB-BV/ODV-C42:FLAG or pIB-VP80:FLAG into Sf9 cells. In the immunoprecipitation assay of VP1054 and VP39, cells cotransfected with pIB-nGFP-VP1054 and pIB-BV/ODV-C42 were used as positive controls.
In the reciprocal pulldown experiments, Sf9 cells (6 × 106) were cotransfected with 9 μg of pIB-BV/ODV-C42:FLAG, pIB-VP80:FLAG, or pIB-VP39:FLAG combined with 9 μg of pIB-VP1054:HA. The procedures were the same as described above except that protein A/G-agarose beads were conjugated to an anti-HA antibody (Covance). Western blotting was performed with mouse monoclonal anti-FLAG (1:2,000; Abmart) or anti-HA (1:2000; Covance) antibody. For negative controls, pIB/V5:His was cotransfected with pIB-BV/ODV-C42:FLAG, pIB-VP80:FLAG, or pIB-VP39:FLAG into Sf9 cells.
RESULTS
Capsid structures cannot localize to the VS following ac54 deletion.
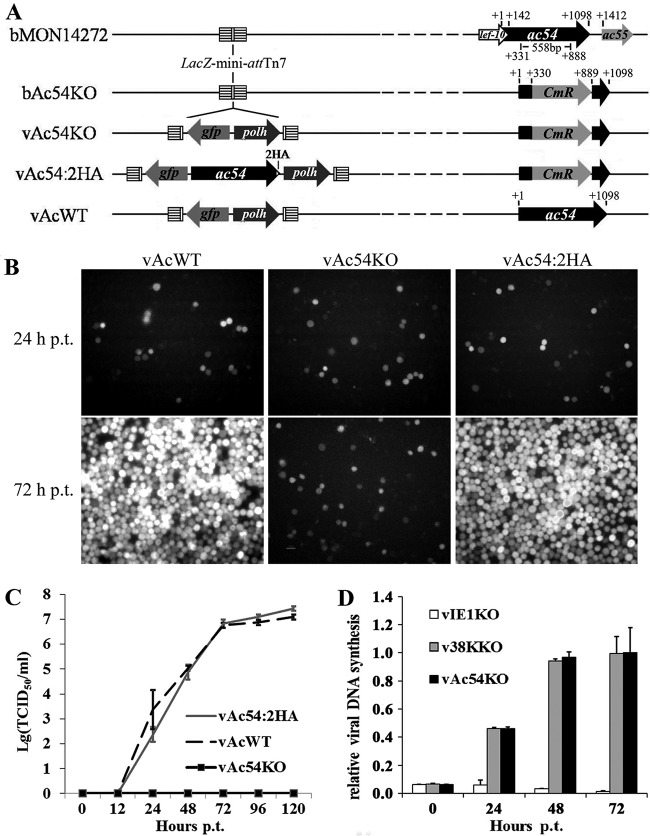
To investigate the role of VP1054 in nucleocapsid assembly, an AcMNPV ac54 deletion mutant (vAc54KO) was generated via ET recombination in E. coli (Fig. 1A). To demonstrate that any defective phenotype resulting from vAc54KO was attributable to the ac54 deletion, an ac54-repaired bacmid (vAc54:2HA) and a wild-type AcMNPV (vAcWT) were constructed as positive controls (Fig. 1A). An AcMNPV polh gene for restoration and a gfp gene for better observation were inserted into the mini-attTn7 site of each of the recombinant viruses described above (Fig. 1A).
FIG 1.
Characterization of the ac54-knockout bacmid. (A) Strategy for the construction of the recombinant viruses vAc54KO, vAc54:2HA, and vAcWT. (B) Sf9 cells were transfected with vAc54KO, vAc54:2HA, and vAcWT bacmid DNA and observed at 72 h p.t. (C) Virus growth curves. The supernatants of transfected cells were harvested at different time points, and viral titers were determined by TCID50 endpoint dilution assays. Each data point represents the average titer of three independent biological repeats; error bars indicate standard deviations. (D) Real-time PCR analysis of viral DNA synthesis in Sf9 cells. Total DNA was extracted from Sf9 cells transfected with vAc54KO, vIE1KO, or v38KKO bacmid DNA at indicated time points, further digested with the restriction enzyme DpnI to eliminate input bacmid DNA, and quantified by real-time PCR. Data are shown as mean values ± standard deviations.
Sf9 cells were transfected with vAc54KO, vAc54:2HA, or vAcWT bacmid DNA. As shown in Fig. 1B, at 24 h p.t., similar amounts of fluorescent cells were observed in the three groups, indicating comparable transfection efficiencies. At 72 h p.t., in vAc54KO-transfected cells no obvious increase in fluorescent cell numbers was observed compared to the level at 24 h p.t., whereas typical, well-spread infections were observed in vAc54:2HA- and vAcWT-transfected cells, suggesting that viral propagation was inhibited by ac54 deletion. Analysis of viral propagation further determined that no infectious BVs were produced in cells transfected with vAc54KO, whereas the vAc54:2HA- and vAcWT-transfected cells showed similar capabilities in the generation of infectious BVs (Fig. 1C). qPCR was then performed to investigate if the deletion of ac54 affects viral DNA synthesis. Loss of the 38K gene of AcMNPV led to a defect in BV production but had no influence on viral DNA synthesis (18); thus a 38K-null recombinant AcMNPV (v38KKO) (18) was used as a positive control. On the other hand, the baculovirus transactivator IE1 was shown to be essential for DNA replication (46, 47); thus, an ie1-null recombinant AcMNPV (vIE1KO) (48) was used as a negative control. As shown in Fig. 1D, the viral DNA replication levels of vAc54KO-transfected cells and v38KKO-transfected cells were comparable; in the negative control, as expected, viral DNA replication was not triggered in vIE1KO-transfected cells. Therefore, the qPCR results demonstrated that deletion of ac54 did not affect the synthesis of viral DNA.
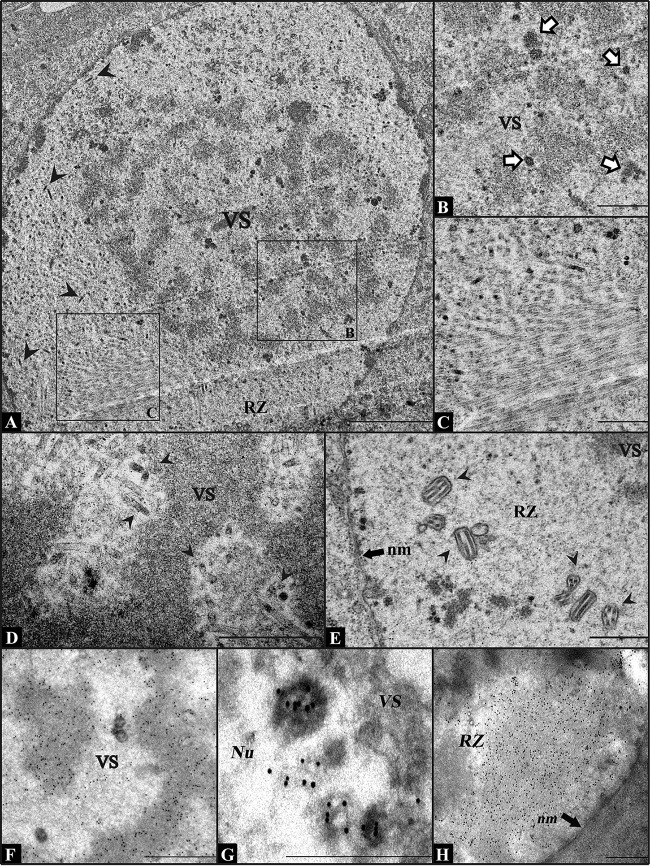
By using electron microscopy, a defect in the generation of nucleocapsids was identified in the nuclei of vAc54KO-transfected cells at 24 h p.t., whereas the formation of VS was not influenced (Fig. 2A). A large quantity of anomalous electron-dense bodies was observed at the edge of the electron-dense mats (Fig. 2B), and abnormally long electron-lucent tubules were found in the ring zone (RZ) (Fig. 2C). The RZ is the peristroma area where nucleocapsids are enveloped to form ODVs (49). On the other hand, in wild-type AcMNPV-infected Sf9 cells, nucleocapsids with DNA content were observed in the electron-lucent space of the VS (Fig. 2D), and formed ODVs were observed in the RZ (Fig. 2E) at 24 h p.i. The composition of the electron-dense bodies and the electron-lucent tubules in vAc54KO-transfected cells was further investigated by immunoelectron microscopy. At 12 h p.t., BrdU was added to the culture medium. Antibodies against BrdU, P6.9, and VP39 were used as the primary antibodies, respectively, and colloidal gold particles coated with protein A were used as the secondary antibody. It was shown that both BrdU and P6.9 were detected in the electron-dense bodies and the electron-dense mat of the VS (Fig. 2F and G, respectively), indicating the colocalization of P6.9 with the newly synthesized viral DNA in vAc54KO-transfected cells. The anti-VP39 antibody showed that VP39 specifically located to the abnormally long electron-lucent tubules that accumulated in the RZ, indicating that the electron-lucent tubules were capsid structures (Fig. 2H). Taking these observations together, the vAc54KO phenotype described above is consistent with previous reports of ac54 mutants (32, 33).
FIG 2.
Electron micrographs of Sf9 cells transfected with vAc54KO or infected with wt AcMNPV. (A) Nucleus of a vAc54KO-transfected Sf9 cell at 24 h p.t. No capsid structures (with or without DNA content) exist in the VS. Black arrowheads indicate normal-appearing nucleocapsids in the RZ. (B) Partial view of the VS magnified from the cell shown in panel A, showing a large quantity of abnormal electron-dense bodies (indicated with white arrows) at the edge of the electron-dense mats. (C) Magnified view from panel A. Abnormally long, electron-lucent tubules can be found in the RZ. (D and E) Partial view of an Sf9 cell infected with wt AcMNPV at 24 h p.i. Nucleocapsids with DNA content are found in the VS (D), and ODVs are found in the RZ (E). Black arrowheads indicate normal nucleocapsids. (F to H) Immunoelectron microscopy analysis of vAc54KO-transfected Sf9 cells at 72 h p.t. (cells were labeled with BrdU at 12 h p.t.). Antiserum against BrdU (F), P6.9 (G), or VP39 (H) was used as the primary antibody. VS, virogenic stroma; RZ, ring zone; Nu, nucleus; nm, nuclear membrane. Scale bars, 2 μm (A) and 500 nm (B to H).
Similarly, the cessation of nucleocapsid assembly can also be induced by individual deletion of other AcMNPV genes, such as 38K, ac53, ac83, p6.9, and pk-1, as indicated by the presence of empty capsid structures. However, the capsid structures resulting from loss of the aforementioned genes were located not only in the RZ but also in the VS (interspace of the electron-dense mats) (16, 18–20, 22), whereas in vAc54KO-transfected cells, no capsid structures were observed in the VS (Fig. 2B). The different distribution patterns of capsid structures suggest that capsid structures cannot localize to the VS following ac54 deletion.
ac54 deletion blocks the entry of VP39 into the VS.
On the basis of the finding that capsid structures containing VP39 are distinctively located in the RZ but not the VS, immunofluorescence assays were subsequently performed to investigate whether the ac54 deletion affected the entry of VP39 monomers into the VS. The ac54-repaired recombinant virus vAc54:2HA was previously demonstrated to be comparable to the wild-type AcMNPV (vAcWT) in analyses of both viral propagation (Fig. 1B and C) and electron microscopy (50); thus, vAc54:2HA was used as a positive control. On the other hand, the 38K deletion did not affect the localization of the capsid structures in the VS (18); thus, a 38K deletion mutant (here, v38KKO) (see reference 18 for details, where this mutant is designated vAc38K-KO-PH-GFP) was also used as a control. Sf9 cells were transfected with vAc54KO, vAc54:2HA, or v38KKO bacmid DNA and labeled with VP39 antiserum at 36 h p.t. The transfected cells were also immunostained with antiserum against IE1 to visualize the VS (51). VP39 localized only in the RZ of the vAc54KO-transfected cells, and no signals were detected within the VS (Fig. 3A). In contrast, VP39 was associated with the VS and RZ in vAc54:2HA-transfected cells (Fig. 3B), indicating the incorporation of VP39 into the maturing nucleocapsids in the VS and the existence of VP39 in the mature nucleocapsids in the RZ. In v38KKO-transfected cells, VP39 was predominantly localized within the RZ, indicating that the long tubules were incorporated with VP39 in the RZ. VP39 was also detected within the VS, appearing as long belts (Fig. 3C), similar to the observed long tubules in electron microscopy results described previously (18). These results clearly indicate that the entry of VP39 into the VS was blocked by the deletion of ac54.
FIG 3.
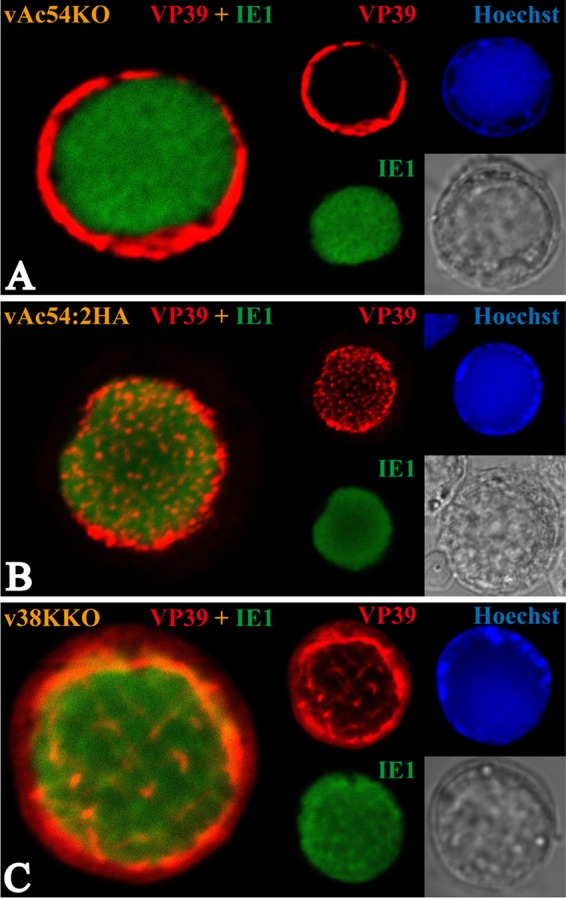
Subcellular localization of the major capsid protein VP39. Sf9 cells were transfected with vAc54KO (A), vAc54:2HA (B), or v38KKO (C) bacmid DNA. Transfected cells were fixed at 36 h p.t. and incubated with antisera against VP39 (rabbit) and IE-1 (mouse) as the primary antibody mixture. The primary antibodies were visualized with Alexa Fluor 555-conjugated goat anti-rabbit (VP39; red) and Alexa Fluor 647-conjugated donkey anti-mouse (IE1; green) antibodies. Hoechst 33342 was used to visualize DNA-rich regions of the nucleus (blue).
ac54 deletion does not affect the formation of nuclear F-actin and its localization.
It has been demonstrated that baculovirus nucleocapsid assembly is an F-actin-dependent process as AcMNPV-infected Sf9 cells treated with CD resulted in a failure of nucleocapsid assembly (38). Therefore, we hypothesized that the compromised transport of VP39 from the RZ to VS might be associated with the dysfunction of actin. To verify this hypothesis, the distribution of actin in vAc54KO-transfected cells was investigated using immunofluorescence assays. As shown in Fig. 4A, actin was observed either throughout the nucleus or with a high concentration in the RZ in vAc54KO-transfected cells. The distribution patterns of actin were similar to those observed in vAc54:2HA- or v38KKO-transfected cells, suggesting that deletion of either ac54 or 38K exerted no effect on the localization of actin.
FIG 4.
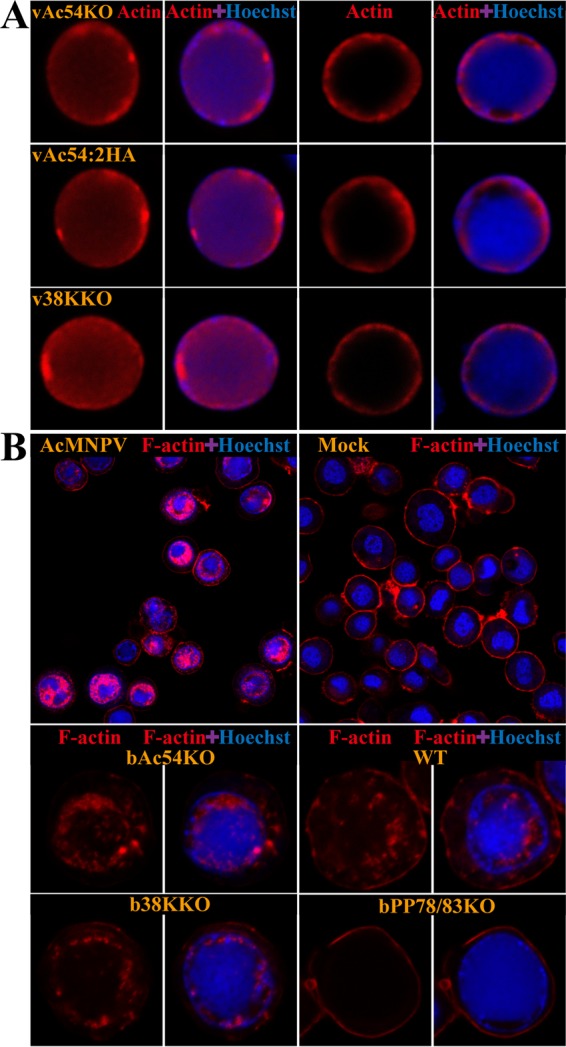
Subcellular localization of total actin and F-actin in Sf9 cells transfected with different recombinant viruses. (A) Total actin localization. Sf9 cells were transfected with vAc54KO, vAc54:2HA, or v38KKO bacmid DNA. At 36 h p.t., the cells were fixed, incubated with an anti-β-actin antibody and visualized with Alexa Fluor 647-conjugated donkey anti-mouse antibody (red). Hoechst 33342 was used to visualize DNA-rich regions of the nucleus (blue). (B) F-actin localization. Sf9 cells were infected with AcMNPV or transfected with bAc54KO, bMON14272 (indicated as WT in the figure), b38KKO, or bPP78/83KO bacmid DNA. At 36 h p.i. or 48 h p.t., cells were fixed and stained with both phalloidin-FITC (red) and Hoechst 33342 (blue).
Phalloidin-FITC was applied as a specific dye for F-actin. Phalloidin is a well-established F-actin probe that binds only to filaments more than seven monomers long (52). As shown in Fig. 4B, phalloidin-stainable nuclear F-actin localized mostly in the RZ and was occasionally distributed throughout the nucleus in AcMNPV-infected Sf9 cells. This result is in agreement with a previous report concerning AcMNPV-induced F-actin (53). Similarly, nuclear F-actin was observed in bMON14272-, bAc54KO-, and b38KKO-transfected cells. In cells transfected with the negative control bPP78/83KO (17), F-actin was distributed mostly near the plasma membrane, indicating the actin-rich cortex; no nuclear F-actin was found in the negative-control or mock-infected cells. These data demonstrated that, in contrast to PP78/83, which is crucial for the genesis of nuclear F-actin, ac54 and 38K are dispensable for the formation and distribution of nuclear F-actin.
ac54 deletion results in the failure of PP78/83, BV/ODV-C42, and 38K to integrate into capsid structures and affects the transport of 38K into the VS.
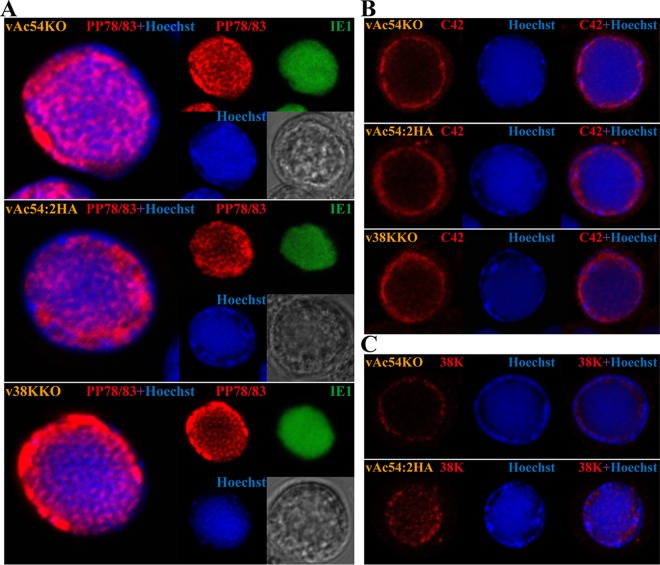
The distributions of minor capsid proteins PP78/83, BV/ODV-C42, and 38K were further investigated using immunofluorescence and immunoelectron microscopy. Sf9 cells were transfected with vAc54KO, vAc54:2HA, or v38KKO bacmid DNA, followed by the labeling of the anti-PP78/83 and -IE1 antisera as well as Hoechst 33342 staining to visualize the DNA. As shown in Fig. 5A, no significant differences were detected in the distributions of PP78/83 among cells transfected with the aforementioned viruses, wherein PP78/83 localized in both the VS and RZ. Within the VS, PP78/83 and the viral DNA generally exhibited complementary distribution patterns, suggesting that PP78/83 was present within the interspace of the electron-dense mats where nucleocapsid matures. Analogously, the distribution of BV/ODV-C42 was similar to that of PP78/83, albeit it appeared to localize mainly within the RZ (Fig. 5B). Notably, substantial differences in the distribution of 38K were observed in vAc54KO- and vAc54:2HA-transfected cells: 38K was present in the VS of the vAc54:2HA-transfected cells, whereas no obvious 38K was found in the VS of the vAc54KO-transfected cells (Fig. 5C).
FIG 5.
Subcellular localization of the minor capsid proteins PP78/83, BV/ODV-C42 (C42), and 38K, as indicated. Sf9 cells were transfected with vAc54KO, vAc54:2HA, or v38KKO bacmid DNA and fixed at 36 h p.t. (A) Transfected cells were incubated with antisera against PP78/83 (rabbit) and IE-1 (mouse) as the primary antibody mixture. The primary antibodies were visualized with Alexa Fluor 555-conjugated goat anti-rabbit (PP78/83; red) and Alexa Fluor 647-conjugated donkey anti-mouse (IE1; green) antibodies. (B) Transfected cells were incubated with antiserum against BV/ODV-C42 and visualized with Alexa Fluor 555-conjugated goat anti-rabbit (red). (C) vAc54KO- or vAc54:2HA-transfected cells were incubated with antiserum against 38K and visualized with Alexa Fluor 555-conjugated goat anti-rabbit (red). Hoechst 33342 was used to visualize DNA-rich regions of the nucleus (blue).
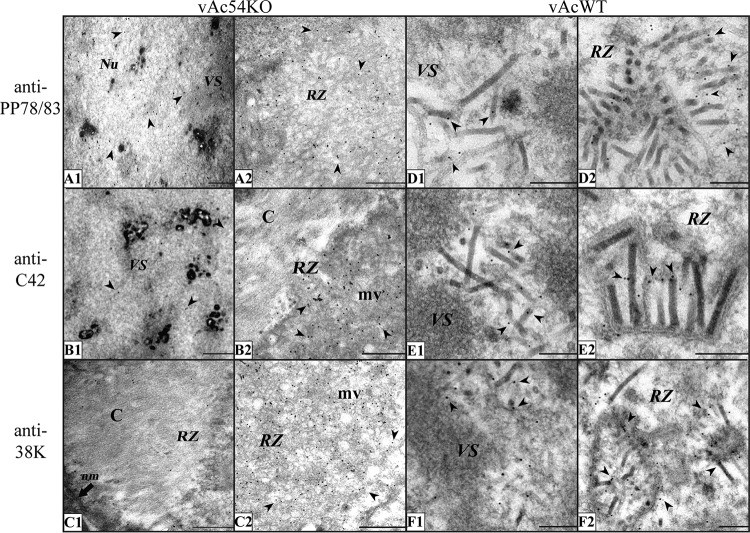
To obtain a higher-resolution view of the distribution of the target proteins, we next carried out immunoelectron microscopy. Sf9 cells were transfected with vAc54KO and vAcWT bacmid DNA, and antisera against PP78/83, BV/ODV-C42, and 38K were used as primary antibodies. In the vAcWT-infected cells, the three proteins localized specifically to the nucleocapsids within the VS and the RZ (Fig. 6 D to F), which was consistent with the results of previous studies (27, 35, 54). However, PP78/83 was found to distribute nonspecifically in the nuclei in vAc54KO-transfected Sf9 cells (Fig. 6A), and BV/ODV-C42 and 38K primarily localized to the microvesicles but not capsid structures within the RZ (Fig. 6B and C). Together these observations indicate that ac54 deletion results in a failure in integrating minor capsid proteins PP78/83, BV/ODV-C42, and 38K into capsid structures and affects the transport of 38K into the VS.
FIG 6.
Immunoelectron microscopy analysis of Sf9 cells transfected with vAc54KO or vAcWT bacmid DNA at 72 h p.t. using antisera against different nucleocapsid structural proteins. For the experiments shown in panels A to C, Sf9 cells were transfected with vAc54KO bacmid DNA. (A) Antiserum against PP78/83 was used as the primary antibody. The entire nucleus was equally stained with gold particles. (B) Antiserum against BV/ODV-C42 was used as the primary antibody. A minimum BV/ODV-C42 signal is visible in the VS; in the RZ, gold particles specifically colocalize with microvesicles but not with capsid structures. (C) Antiserum against 38K was used as the primary antibody. 38K mainly colocalizes with microvesicles; only a background level of gold particles was observed in the VS or on capsid structures. For the experiments shown in panels D to F, Sf9 cells were transfected with vAcWT. Signals for PP78/83 (D), BV/ODV-C42 (E), and 38K (F) specifically localize to the nucleocapsids. Arrowheads, gold particles; Nu, nucleus; nm, nuclear membrane; mv, microvesicles; C, capsid structures. Scale bars, 500 nm (A to C) and 200 nm (D to F).
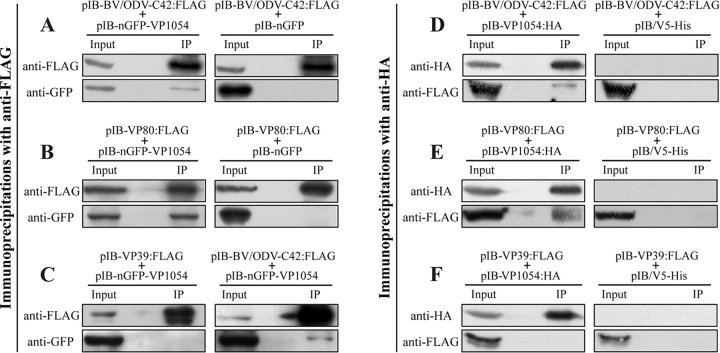
VP1054 interacts with BV/ODV-C42 and VP80.
The changes in the distributions of VP39, PP78/83, BV/ODV-C42, and 38K upon ac54 deletion implied possible protein-protein interactions between VP1054 and these capsid proteins. Since the interaction between 38K and VP1054 has been demonstrated previously (35) and since PP78/83 does not locate in the nucleus without BV/ODV-C42 (31), in the present study we investigated primarily whether VP1054 interacts with BV/ODV-C42 or VP39. In addition, VP80 is a nuclear F-actin-related capsid protein that is required for transport of mature nucleocapsids (55), and both VP80 and VP1054 were proved to interact with 38K (35). So we also investigated whether VP1054 interacts with VP80. The expression vectors pIB-nGFP-VP1054, pIB-VP39:FLAG, pIB-BV/ODV-C42:FLAG, and pIB-VP80:FLAG were generated, and pIB-nGFP-VP1054 was cotransfected with each FLAG-tagged expression plasmid into Sf9 cells. Cells were harvested at 36 h p.t. and subjected to immunoprecipitation with anti-FLAG. For negative controls, pIB-nGFP was cotransfected with pIB-BV/ODV-C42:FLAG or pIB-VP80:FLAG into Sf9 cells. As shown in Fig. 7A, nGFP-VP1054 and BV/ODV-C42:FLAG were detected in the lysates of the cotransfected cells (input lane). Both nGFP-VP1054 and BV/ODV-C42:FLAG were detected in immunocomplexes precipitated by the anti-FLAG beads (Fig. 7A, IP lane), indicating that nGFP-VP1054 was coimmunoprecipitated with BV/ODV-C42:FLAG. In the negative control, although nGFP and BV/ODV-C42:FLAG were both detected in the cell lysates (Fig. 7A, input lane), nGFP was not coimmunoprecipitated with BV/ODV-C42:FLAG (IP lane). These results indicated that in the transient expression assay, VP1054 interacts with BV/ODV-C42. Similarly, as shown in Fig. 7B, nGFP-VP1054, but not nGFP, was coimmunoprecipitated with VP80:FLAG (IP lane), suggesting that VP1054 also interacts with VP80. To determine whether VP1054 interacts with VP39, cells were cotransfected with pIB-nGFP-VP1054 and pIB-nGFP-VP39. Cells cotransfected with pIB-nGFP-VP1054 and pIB-BV/ODV-C42:FLAG were applied as a positive control. As shown in Fig. 7C, nGFP-VP1054 was coimmunoprecipitated with BV/ODV-C42:FLAG but not with VP39:FLAG, suggesting that VP1054 does not interact with VP39 in this transient expression assay. To confirm the immunoprecipitation results, reciprocal pulldown assays were subsequently carried out with anti-HA. A transient expression plasmid, pIB-VP1054:HA, was generated and cotransfected with pIB-VP80:FLAG, pIB-BV/ODV-C42:FLAG, or pIB-VP39:FLAG into Sf9 cells. As shown in Fig. 7D and E, both VP80:FLAG and BV/ODV-C42:FLAG were detected in the immunocomplexes (IP lanes), indicating that they were both coimmunoprecipitated with VP1054:HA. However, the results in Fig. 7F show that VP39 was not coimmunoprecipitated with VP1054:HA. For negative controls, Sf9 cells were cotransfected with pIB/V5-His (Invitrogen) and the corresponding FLAG-tagged plasmid; none of the three FLAG-tagged proteins was detected in the immunocomplexes (Fig. 7D to F, IP lanes), indicating that they were not coimmunoprecipitated with anti-HA. To sum up, the immunoprecipitation results suggested that VP1054 interacts with VP80 and BV/ODV-C42.
FIG 7.
VP1054 is able to interact with BV/ODV-C42 and VP80. (A and B) Sf9 cells were cotransfected with pIB-nGFP-VP1054 and pIB-BV/ODV-C42:FLAG or pIB-VP80:FLAG, as indicated. Cells cotransfected with pIB-nGFP and the corresponding FLAG-tagged expression plasmid were used as negative controls. (C) Sf9 cells were cotransfected with pIB-nGFP-VP1054 and pIB-VP39:FLAG. Cells cotransfected with pIB-nGFP-VP1054 and pIB-BV/ODV-C42 were used as a positive control. (D to F) Sf9 cells were cotransfected with pIB-VP1054:HA and pIB-BV/ODV-C42:FLAG, pIB-VP80:FLAG, or pIB-VP39:FLAG, as indicated. Cells cotransfected with pIB/V5-His and the corresponding FLAG-tagged expression plasmid were used as negative controls. For all cotransfections, cells were collected at 36 h p.t., lysed, immunoprecipitated with anti-FLAG or anti-HA antibody, and subjected to Western blot analysis.
DISCUSSION
VP1054 is a minor capsid protein of AcMNPV (32) (33), and its homologs have been identified in all fully sequenced baculovirus genomes (4). VP1054 is crucial for AcMNPV nucleocapsid assembly because either deletion of ac54 (the VP1054-encoding gene) or a single nucleotide substitution that alters the proline at position 286 to a serine results in a massive decrease in the formation of nucleocapsids and a total loss of infective progeny virions (32, 33). The importance of ac54 in nucleocapsid assembly has been proposed to be related to its DNA binding activity because VP1054 is an acquired PURα that specifically binds to the GGN repeats on single-stranded DNA and RNA (33). PURα has been strongly conserved throughout evolution and has multiple functions, such as the initiation of DNA replication and regulation of transcription and the cell cycle (56). Moreover, in an in vitro analysis, the conserved residues (VP1054 R152, Y154, and D156) of the PURα domain of VP1054 are crucial for the interaction between VP1054 and the GGN-rich region of AcMNPV pp78/83 (ac9) (33).
In this study, the ac54-knockout mutant vAc54KO was generated to explore the role of VP1054 in the AcMNPV life cycle. Consistent with previous reports, the deletion of ac54 resulted in an inability to initiate cell-to-cell infection but did not affect the replication of viral DNA. In addition, abnormal electron-dense bodies were found in the VS, and empty capsid structures were found in the RZ in cells transfected with vAc54KO, indicating that ablation of ac54 disrupted the normal assembly of nucleocapsids. In addition to ac54, some other AcMNPV genes (i.e., 38K, ac53, ac83, pk-1, and p6.9) have also been implicated in nucleocapsid assembly, and their individual deletion results in the cessation of assembly and the appearance of empty capsid structures in both the RZ and VS (16–20, 22). However, in the present study, neither capsid structures nor VP39 was observed inside the VS in Sf9 cells transfected with vAc54KO, implying a special role of VP1054 in the baculovirus life cycle besides nucleocapsid assembly.
Actin polymerization can be induced by diverse pathogens to facilitate the infection of host cells (28). In the AcMNPV life cycle, virus-induced F-actin is essential for nucleocapsid assembly and viral replication (57). Shortly after the entry of a nucleocapsid in the cytoplasm, transient actin cables are formed toward the nucleus and help transport the nucleocapsid through the cytoplasm and into the nucleus. Nucleocapsids nucleate actin polymerization both in vitro and in vivo, possibly through the action of the two actin-binding proteins VP39 and PP78/83 (23). During the later phase of viral infection, G-actin and the Arp2/3 complex are recruited into the nucleus, and a dynamic microfilament network is formed (23) (28). BV/ODV-C42 and PP78/83 have been demonstrated to be essential for this process because deletion of either BV/ODV-C42 or PP78/83 disrupts the formation of microfilaments in the nucleus (28, 30). Deletion of ac101 did not affect viral DNA replication and the entry of VP39 into the nucleus. However, elongated tubular structures composed of VP39 were observed in parallel and adjoined to the nuclear membrane and could not be transported into the VS (30). Treatment with CD or latrunculin A results in a similar phenomenon; thus, the formation of actin filaments is considered to be crucial for the morphogenesis of progeny virions and especially for nucleocapsid assembly (25, 57, 58). Immunofluorescence assays showed that the distributions of nuclear F-actin, BV/ODV-C42, and PP78/83 were not affected by ac54 deletion, whereas the distribution of 38K was changed by the deletion of ac54 in a pattern similar to the change in VP39. However, by using immunoelectron microscopy, we found that the subcellular distributions of BV/ODV-C42 and PP78/83 were also affected by the deletion of ac54. In vAcWT-infected cells, BV/ODV-C42 and PP78/83 located specifically to the basal ends of the assembled nucleocapsids and showed almost no distribution around the vesicles in the RZ. In contrast, in the vAc54KO-transfected cells, neither BV/ODV-C42 nor PP78/83 was specifically attached to the capsid structures. BV/ODV-C42 was localized mainly in the microvesicles in the RZ, similar to 38K, whereas PP78/83 was equally distributed in the nucleus. The alterations in the distribution of these capsid proteins imply that ac54 deletion may affect the integration of BV/ODV-C42, PP78/83, and 38K into the capsid structures. Because BV/ODV-C42 and PP78/83 are essential for nuclear F-actin formation, ac54 deletion may affect the connection between capsid structures and the virus-induced cytoskeleton, thereby preventing the capsid structures and VP39 from entering the VS. Moreover, although nuclear F-actin was observed in vAc54KO-transfected cells, we cannot rule out the possibility that ac54 deletion exerted a minor influence on nuclear F-actin that could not be detected by phalloidin staining.
Taken together, our findings suggest that VP1054 plays an important role in the transport of capsid proteins to the baculovirus nucleocapsid assembly site, which is prior to the nucleocapsid assembly process. VP1054 is also responsible for the integration of BV/ODV-C42, PP78/83, and 38K into the capsid structures, implying that VP1054 is responsible for the maturation of capsids. Studies to further characterize the transport pathway(s) of baculovirus capsid proteins and to elucidate the details of nucleocapsid assembly are under way.
ACKNOWLEDGMENTS
We acknowledge Xinwen Chen (Wuhan Institute of Virology) for providing the BV/ODV-C42 and PP78/83 polyclonal antisera. We thank L. A. Guarino (Texas A&M University) for providing antiserum against IE1.
This research was supported by the National Natural Science Foundation of China (31370188).
REFERENCES
- 1.Hayakawa T, Ko R, Okano K, Seong SI, Goto C, Maeda S. 1999. Sequence analysis of the Xestia c-nigrum granulovirus genome. Virology 262:277–297. doi: 10.1006/viro.1999.9894. [DOI] [PubMed] [Google Scholar]
- 2.Lauzon HA, Lucarotti CJ, Krell PJ, Feng Q, Retnakaran A, Arif BM. 2004. Sequence and organization of the Neodiprion lecontei nucleopolyhedrovirus genome. J Virol 78:7023–7035. doi: 10.1128/JVI.78.13.7023-7035.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slack J, Arif BM. 2007. The baculoviruses occlusion-derived virus: virion structure and function. Adv Virus Res 69:99–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Oers MM, Vlak JM. 2007. Baculovirus genomics. Curr Drug Targets 8:1051–1068. doi: 10.2174/138945007782151333. [DOI] [PubMed] [Google Scholar]
- 5.Blissard GW, Wenz JR. 1992. Baculovirus gp64 envelope glycoprotein is sufficient to mediate pH-dependent membrane fusion. J Virol 66:6829–6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braunagel SC, Russell WK, Rosas-Acosta G, Russell DH, Summers MD. 2003. Determination of the protein composition of the occlusion-derived virus of Autographa californica nucleopolyhedrovirus. Proc Natl Acad Sci U S A 100:9797–9802. doi: 10.1073/pnas.1733972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braunagel SC, Summers MD. 1994. Autographa californica nuclear polyhedrosis virus, PDV, and ECV viral envelopes and nucleocapsids: structural proteins, antigens, lipid and fatty acid profiles. Virology 202:315–328. doi: 10.1006/viro.1994.1348. [DOI] [PubMed] [Google Scholar]
- 8.Fraser M. 1986. Ultrastructural observations of virion maturation in Autographa californica nuclear polyhedrosis virus infected Spodoptera frugiperda cell cultures. J Ultrastruct Mol Struct Res 95:189–195. doi: 10.1016/0889-1605(86)90040-6. [DOI] [Google Scholar]
- 9.Young JC, Mackinnon EA, Faulkner P. 1993. The architecture of the virogenic stroma in isolated nuclei of Spodoptera frugiperda cells in vitro infected by Autographa californica nuclear polyhedrosis virus. J Struct Biol 110:141–153. doi: 10.1006/jsbi.1993.1015. [DOI] [Google Scholar]
- 10.Harrap KA. 1972. The structure of nuclear polyhedrosis viruses. 3. Virus assembly. Virology 50:133–139. [DOI] [PubMed] [Google Scholar]
- 11.Thiem SM, Miller LK. 1989. Identification, sequence, and transcriptional mapping of the major capsid protein gene of the baculovirus Autographa californica nuclear polyhedrosis virus. J Virol 63:2008–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blissard GW, Quant-Russell RL, Rohrmann GF, Beaudreau GS. 1989. Nucleotide sequence, transcriptional mapping, and temporal expression of the gene encoding p39, a major structural protein of the multicapsid nuclear polyhedrosis virus of Orgyia pseudotsugata. Virology 168:354–362. doi: 10.1016/0042-6822(89)90276-6. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Stojiljkovic N, Jehle JA. 2010. Cloning of complete genomes of large dsDNA viruses by in vitro transposition of an F factor containing transposon. J Virol Methods 167:95–99. doi: 10.1016/j.jviromet.2009.11.026. [DOI] [PubMed] [Google Scholar]
- 14.Funk CJ, Consigli RA. 1993. Phosphate cycling on the basic protein of Plodia interpunctella granulosis virus. Virology 193:396–402. doi: 10.1006/viro.1993.1136. [DOI] [PubMed] [Google Scholar]
- 15.Tweeten KA, Bulla LA, Consigli RA. 1980. Characterization of an extremely basic protein derived from granulosis virus nucleocapsids. J Virol 33:866–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X, Zhao H, Fang Z, Yuan M, Yang K, Pang Y. 2012. Distribution and phosphorylation of the basic protein P6.9 of Autographa californica nucleopolyhedrovirus. J Virol 86:12217–12227. doi: 10.1128/JVI.00438-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li A, Zhao H, Lai Q, Huang Z, Yuan M, Yang K. 2015. Posttranslational modifications of baculovirus protamine-like protein P6.9 and the significance of its hyperphosphorylation for viral very late gene hyperexpression. J Virol 89:7649–7659. doi: 10.1128/JVI.00333-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu W, Lin T, Pan L, Yu M, Li Z, Pang Y, Yang K. 2006. Autographa californica multiple nucleopolyhedrovirus nucleocapsid assembly is interrupted upon deletion of the 38K gene. J Virol 80:11475–11485. doi: 10.1128/JVI.01155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu C, Li Z, Wu W, Li L, Yuan M, Pan L, Yang K, Pang Y. 2008. Autographa californica multiple nucleopolyhedrovirus ac53 plays a role in nucleocapsid assembly. Virology 382:59–68. doi: 10.1016/j.virol.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Zhu S, Wang W, Wang Y, Yuan M, Yang K. 2013. The baculovirus core gene ac83 is required for nucleocapsid assembly and per os infectivity of Autographa californica nucleopolyhedrovirus. J Virol 87:10573–10586. doi: 10.1128/JVI.01207-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vanarsdall AL, Okano K, Rohrmann GF. 2006. Characterization of the role of very late expression factor 1 in baculovirus capsid structure and DNA processing. J Virol 80:1724–1733. doi: 10.1128/JVI.80.4.1724-1733.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang C, Li M, Dai X, Zhao S, Hou Y, Zhang Y, Lan D, Wang Y, Chen X. 2013. Autographa californica multiple nucleopolyhedrovirus PK-1 is essential for nucleocapsid assembly. Virology 443:349–357. doi: 10.1016/j.virol.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 23.Charlton CA, Volkman LE. 1991. Sequential rearrangement and nuclear polymerization of actin in baculovirus-infected Spodoptera frugiperda cells. J Virol 65:1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Volkman LE, Goldsmith PA, Hess RT. 1987. Evidence for microfilament involvement in budded Autographa californica nuclear polyhedrosis virus production. Virology 156:32–39. doi: 10.1016/0042-6822(87)90433-8. [DOI] [PubMed] [Google Scholar]
- 25.Volkman LE. 1988. Autographa californica MNPV nucleocapsid assembly: inhibition by cytochalasin D. Virology 163:547–553. doi: 10.1016/0042-6822(88)90295-4. [DOI] [PubMed] [Google Scholar]
- 26.Campellone KG, Welch MD. 2010. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol 11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russell RL, Funk CJ, Rohrmann GF. 1997. Association of a baculovirus-encoded protein with the capsid basal region. Virology 227:142–152. doi: 10.1006/viro.1996.8304. [DOI] [PubMed] [Google Scholar]
- 28.Goley ED, Ohkawa T, Mancuso J, Woodruff JB, D'Alessio JA, Cande WZ, Volkman LE, Welch MD. 2006. Dynamic nuclear actin assembly by Arp2/3 complex and a baculovirus WASP-like protein. Science 314:464–467. doi: 10.1126/science.1133348. [DOI] [PubMed] [Google Scholar]
- 29.Ohkawa T, Volkman LE, Welch MD. 2010. Actin-based motility drives baculovirus transit to the nucleus and cell surface. J Cell Biol 190:187–195. doi: 10.1083/jcb.201001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li K, Wang Y, Bai H, Wang Q, Song J, Zhou Y, Wu C, Chen X. 2010. The putative pocket protein binding site of Autographa californica nucleopolyhedrovirus BV/ODV-C42 is required for virus-induced nuclear actin polymerization. J Virol 84:7857–7868. doi: 10.1128/JVI.00174-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Wang Q, Liang C, Song J, Li N, Shi H, Chen X. 2008. Autographa californica multiple nucleopolyhedrovirus nucleocapsid protein BV/ODV-C42 mediates the nuclear entry of P78/83. J Virol 82:4554–4561. doi: 10.1128/JVI.02510-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olszewski J, Miller LK. 1997. Identification and characterization of a baculovirus structural protein, VP1054, required for nucleocapsid formation. J Virol 71:5040–5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marek M, Romier C, Galibert L, Merten OW, van Oers MM. 2013. Baculovirus VP1054 is an acquired cellular PURα, a nucleic acid-binding protein specific for GGN repeats. J Virol 87:8465–8480. doi: 10.1128/JVI.00068-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ayres MD, Howard SC, Kuzio J, Lopez-Ferber M, Possee RD. 1994. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202:586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- 35.Wu W, Liang H, Kan J, Liu C, Yuan M, Liang C, Yang K, Pang Y. 2008. Autographa californica multiple nucleopolyhedrovirus 38K is a novel nucleocapsid protein that interacts with VP1054, VP39, VP80, and itself. J Virol 82:12356–12364. doi: 10.1128/JVI.00948-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet 20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 37.Muyrers JP, Zhang Y, Testa G, Stewart AF. 1999. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res 27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hess RT, Goldsmith PA, Volkman LE. 1989. Effect of cytochalasin D on cell morphology and AcMNPV replication in a Spodoptera frugiperda cell line. J Invertebr Pathol 53:169–182. doi: 10.1016/0022-2011(89)90005-0. [DOI] [PubMed] [Google Scholar]
- 39.Luckow VA, Lee SC, Barry GF, Olins PO. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J Virol 67:4566–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lalioti M, Heath J. 2001. A new method for generating point mutations in bacterial artificial chromosomes by homologous recombination in Escherichia coli. Nucleic Acids Res 29:E14. doi: 10.1093/nar/29.3.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dai X, Stewart TM, Pathakamuri JA, Li Q, Theilmann DA. 2004. Autographa californica multiple nucleopolyhedrovirus EXON0 (ORF141), which encodes a RING finger protein, is required for efficient production of budded virus. J Virol 78:9633–9644. doi: 10.1128/JVI.78.18.9633-9644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Reilly DR, Miller LK, Luckow VA. 1994. Baculovirus expression vectors: a laboratory manual. Oxford University Press, New York, NY. [Google Scholar]
- 43.Vanarsdall AL, Okano K, Rohrmann GF. 2005. Characterization of the replication of a baculovirus mutant lacking the DNA polymerase gene. Virology 331:175–180. doi: 10.1016/j.virol.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 44.Yuan M, Huang Z, Wei D, Hu Z, Yang K, Pang Y. 2011. Identification of Autographa californica nucleopolyhedrovirus ac93 as a core gene and its requirement for intranuclear microvesicle formation and nuclear egress of nucleocapsids. J Virol 85:11664–11674. doi: 10.1128/JVI.05275-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X, Pang A, Lauzon HA, Sohi SS, Arif BM. 1997. The gene encoding the capsid protein P82 of the Choristoneura fumiferana multicapsid nucleopolyhedrovirus: sequencing, transcription and characterization by immunoblot analysis. J Gen Virol 78:2665–2673. doi: 10.1099/0022-1317-78-10-2665. [DOI] [PubMed] [Google Scholar]
- 46.Kool M, Ahrens CH, Goldbach RW, Rohrmann GF, Vlak JM. 1994. Identification of genes involved in DNA replication of the Autographa californica baculovirus. Proc Natl Acad Sci U S A 91:11212–11216. doi: 10.1073/pnas.91.23.11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stewart TM, Huijskens I, Willis LG, Theilmann DA. 2005. The Autographa californica multiple nucleopolyhedrovirus ie0-ie1 gene complex is essential for wild-type virus replication, but either IE0 or IE1 can support virus growth. J Virol 79:4619–4629. doi: 10.1128/JVI.79.8.4619-4629.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y. 2010. AcMNPV IE-1 plays an important role in the development of virogenic stroma in the baculovirus life cycle. M.A. thesis. Sun Yat-sen University, Guangzhou, China. [Google Scholar]
- 49.Rohrmann GF. 2013. Baculovirus molecular biology, 3rd ed National Center for Biotechnology Information, Bethesda, MD. [PubMed] [Google Scholar]
- 50.Zhong L. 2010. Studies on baculovirus conserved gene AcMNPV ac54. M.A. thesis Sun Yat-sen University, Guangzhou, China. [Google Scholar]
- 51.Nagamine T, Saito T, Osada H, Matsumoto S. 2015. Dissection of two modes of IE1 sub-nuclear localization in baculovirus-infected cells. Virus Res 208:120–128. doi: 10.1016/j.virusres.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 52.Visegrady B, Lorinczy D, Hild G, Somogyi B, Nyitrai M. 2005. A simple model for the cooperative stabilisation of actin filaments by phalloidin and jasplakinolide. FEBS Lett 579:6–10. doi: 10.1016/j.febslet.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 53.Talhouk SN, Volkman LE. 1991. Autographa californica M nuclear polyhedrosis virus and cytochalasin D: antagonists in the regulation of protein synthesis. Virology 182:626–634. doi: 10.1016/0042-6822(91)90603-9. [DOI] [PubMed] [Google Scholar]
- 54.Braunagel SC, Guidry PA, Rosas-Acosta G, Engelking L, Summers MD. 2001. Identification of BV/ODV-C42, an Autographa californica nucleopolyhedrovirus orf101-encoded structural protein detected in infected-cell complexes with ODV-EC27 and p78/83. J Virol 75:12331–12338. doi: 10.1128/JVI.75.24.12331-12338.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marek M, Merten OW, Galibert L, Vlak JM, van Oers MM. 2011. Baculovirus VP80 protein and the F-actin cytoskeleton interact and connect the viral replication factory with the nuclear periphery. J Virol 85:5350–5362. doi: 10.1128/JVI.00035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.White MK, Johnson EM, Khalili K. 2009. Multiple roles for Purα in cellular and viral regulation. Cell Cycle 8:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ohkawa T, Volkman LE. 1999. Nuclear F-actin is required for AcMNPV nucleocapsid morphogenesis. Virology 264:1–4. doi: 10.1006/viro.1999.0008. [DOI] [PubMed] [Google Scholar]
- 58.Kasman LM, Volkman LE. 2000. Filamentous actin is required for lepidopteran nucleopolyhedrovirus progeny production. J Gen Virol 81:1881–1888. doi: 10.1099/0022-1317-81-7-1881. [DOI] [PubMed] [Google Scholar]