ABSTRACT
Clostridium difficile spore germination is essential for colonization and disease. The signals that initiate C. difficile spore germination are a combination of taurocholic acid (a bile acid) and glycine. Interestingly, the chenodeoxycholic acid class (CDCA) bile acids competitively inhibit taurocholic acid-mediated germination, suggesting that compounds that inhibit spore germination could be developed into drugs that prophylactically prevent C. difficile infection or reduce recurring disease. However, a recent report called into question the utility of such a strategy to prevent infection by describing C. difficile strains that germinated in the apparent absence of bile acids or germinated in the presence of the CDCA inhibitor. Because the mechanisms of C. difficile spore germination are beginning to be elucidated, the mechanism of germination in these particular strains could yield important information on how C. difficile spores initiate germination. Therefore, we quantified the interaction of these strains with taurocholic acid and CDCA, the rates of spore germination, the release of DPA from the spore core, and the abundance of the germinant receptor complex (CspC, CspB, and SleC). We found that strains previously observed to germinate in the absence of taurocholic acid correspond to more potent 50% effective concentrations (EC50 values; the concentrations that achieve a half-maximum germination rate) of the germinant and are still inhibited by CDCA, possibly explaining the previous observations. By comparing the germination kinetics and the abundance of proteins in the germinant receptor complex, we revised our original model for CspC-mediated activation of spore germination and propose that CspC may activate spore germination and then inhibit downstream processes.
IMPORTANCE Clostridium difficile forms metabolically dormant spores that persist in the health care environment. In susceptible hosts, C. difficile spores germinate in response to certain bile acids and glycine. Blocking germination by C. difficile spores is an attractive strategy to prevent the initiation of disease or to block recurring infection. However, certain C. difficile strains have been identified whose spores germinate in the absence of bile acids or are not blocked by known inhibitors of C. difficile spore germination (calling into question the utility of such strategies). Here, we further investigate these strains and reestablish that bile acid activators and inhibitors of germination affect these strains and use these data to suggest another role for the C. difficile bile acid germinant receptor.
INTRODUCTION
Spore formation and germination by Peptoclostridium difficile spores (1) (referred to here as Clostridium difficile for simplicity) are significant hurdles for overcoming the transmission of this pathogen within the hospital environment. Due to the strict anaerobic nature of C. difficile vegetative cells, spores are thought to be the main reservoir for transmission within the health care setting (2, 3). Prior antibiotic treatment is the greatest risk factor for C. difficile infection (4). Broad-spectrum antibiotics alter the balance of the colonic microbiota allowing C. difficile an opportunity to colonize the newly generated niche (5–7). Once in a host, C. difficile spores germinate to form the toxin-producing vegetative cells that colonize a host's colonic environment. In susceptible hosts, C. difficile vegetative cells secrete two toxins that damage the colonic epithelium and lead to the primary symptoms of disease (8, 9). C. difficile infections are commonly treated with more antibiotics (e.g., metronidazole, vancomycin, or fidaxomicin) (10). After disease symptoms are alleviated and the antibiotics are discontinued, patients frequently relapse with recurring disease due to germination by spores that remain in the colon or that reinfect the host from the surrounding environment (10). Because germination by the spore form is required for pathogenesis, compounds that prevent spore germination could be an attractive way to prevent the primary or recurring infections (11–13).
Endospores are dormant forms of bacteria and are formed by vegetative cells in response to environmental stress (14, 15). In Bacillus subtilis, sporulation is a tightly regulated process and involves the formation of a forespore within a mother cell (16). The forespore and mother cell communicate through a cascade of sigma factor activation and waves of transcription/translation (16). The result of this developmental program is a dormant spore consisting of a DNA-containing core where much of the water has been replaced by dipicolinic acid as a calcium chelate (CaDPA) (17). Surrounding the spore core is an inner spore membrane, a thin germ cell peptidoglycan layer, a thick cortex peptidoglycan layer, an outer spore membrane, and layers of coat protein (15). The coat proteins, cortex, and the contents of the desiccated core help protect the spore from environmental stressors (e.g., heat, radiation, chemicals, and antibiotics) and keep the spore in a metabolically dormant state (15, 17).
Endospore germination has been extensively studied in B. subtilis (15). In B. subtilis, spore germination can be initiated when l-alanine (a germinant) interacts with the GerAA-AB-AC germinant receptor which is embedded within the inner spore membrane (15). This interaction triggers the release of the large depot of CaDPA, presumably through the SpoVA membrane channel (15). This results in the partial hydration of the spore's core. As CaDPA passes through the cortex layer, CaDPA activates the CwlJ cortex hydrolase and the actions of the CwlJ and SleB hydrolases degrade the spore cortex, resulting in core expansion and full rehydration of the spore core (15).
The receptors with which spore germinants interact are conserved between most of the studied spore-forming bacteria (e.g., B. subtilis, B. anthracis, and C. perfringens), while the germinants which activate spore germination vary between organisms (but germinants are generally amino acids, nucleotides, ions, or sugars) (18). C. difficile spore germination is initiated by certain bile acids (presumably a host signal) and glycine (presumably a nutrient signal) (14, 19–21). Bile acids are small, steroid acids that are released by the gallbladder into the digestive tract to aid in the absorption of fats and cholesterol (22). Two families of bile acids are synthesized by the liver: cholic acid derivatives and chenodeoxycholic acid derivatives (CDCA). Each of these families is conjugated with either a taurine (taurocholic acid [TA] or taurochenodeoxycholic acid) or a glycine (glycocholic acid or glycochenodeoxycholic acid) (22). C. difficile spore germination is stimulated by cholic acid, while CDCA derivatives inhibit cholic acid-mediated germination (11, 19, 23, 24). Based on previous studies, C. difficile spores correspond to 50% effective concentrations (EC50 values; the concentrations that achieve a half-maximum germination rate) in the low-millimolar range for TA and the high-micromolar range for CDCA, suggesting that C. difficile spores may have a tighter interaction with inhibitors of germination than with the activators of germination (11, 23–25).
Although the signals that stimulate spore germination have been studied for some time, only recently was the receptor with which the bile acids interact identified (13). C. difficile does not encode the classical, membrane-embedded, ger-type, germinant receptor (26). Instead, C. difficile spore germination proceeds through a novel germination pathway involving direct stimulation of cortex hydrolysis and subsequent release of CaDPA from the spore core (opposite to what is observed for germinant receptor-mediated germination in B. subtilis or Clostridium perfringens) (13–15, 27, 28). In our working model, C. difficile spore germination is initiated when the germination-specific, pseudoprotease, CspC, interacts with the cholic acid class of bile acids (13). Activated CspC transmits the bile acid activating signal to the CspB protease, which then activates the spore cortex lytic enzyme SleC (deposited in the spore as a zymogen) (13, 29). Activated SleC then begins to degrade the spore cortex, and CaDPA is subsequently released from the spore core (27, 28).
Based on the model described above, all C. difficile isolates should have the requirement for bile acids to stimulate germination. Indeed, spores derived from most clinical isolates require taurocholic acid (TA) to initiate spore germination in rich medium (30, 31). However, a few isolates have been reported not to require TA as a spore germinant or not to be inhibited the CDCA antigerminant (31). If these strains have no requirement for TA or are not inhibited by CDCA, by what mechanism would they be activated for germination (i.e., would the mechanisms for germination in these strains be different from what has been described previously)?
We sought here to reexamine the germination of spores derived from C. difficile strains that were reported previously to germinate in rich medium alone or germinate in the presence of a known inhibitor of C. difficile spore germination, CDCA (31) and compare this germination to that of spores that clearly require TA for germination (11, 25, 27, 32). Upon reinvestigation of these strains, we found that strains previously thought not to be inhibited by CDCA are actually inhibited by CDCA and interact with CDCA with inhibitor constant values similar to those for other strains. However, these strains do exhibit greater EC50 values for TA (EC50,TA values), possibly explaining previous observations (31). We further characterize these strains by determining the abundance of the germinant receptor complex and show that the levels of CspC, CspB, and SleC differ between strains. By comparing the generated kinetic data with the abundance of spore proteins, we revise our model for the mechanism of initiating C. difficile spore germination. Based on our prior genetic data and the data herein, we propose that CspC acts to initiate spore germination in the presence of TA but then to inhibit downstream processes until another signal is received.
MATERIALS AND METHODS
Strains and growth conditions.
C. difficile strains (see Table S1 in the supplemental material) were grown in a Coy Laboratories model B anaerobic chamber (10% H2, 5% CO2, 85% N2) at 37°C in brain heart infusion supplemented with 5 g/liter yeast extract and 0.1% l-cysteine (BHIS), as described previously (13, 27, 32, 33). Escherichia coli routinely was grown at 37°C in Luria-Bertani (LB) medium or in 2×TY medium (5 g of NaCl, 10 g of yeast extract, and 16 g of tryptone/liter) for protein expression (see below). Antibiotics were supplemented as needed (100 μg/ml ampicillin).
Molecular biology.
The C. difficile UK1 sleC gene was amplified with Phusion DNA polymerase (New England BioLabs, Beverly, MA) using the primers 5′pET_SleC (TTTTGTTTAACTTTAAGAAGGAGATATACATATGCAAGATGGTTTCTTAACAGTAAGC) and 3′pET_SleC (ATCTCAGTGGTGGTGGTGGTGGTGCTCGAGAATTAAAGGATTTAAAGAAGCTATT). The resulting PCR product was inserted into pET22b between the XhoI and NdeI restriction sites using Gibson Assembly (34) and transformed into E. coli DH5α to generate pKS08. To construct a strain producing CspB for recombinant protein expression, the cspB gene was amplified with Phusion DNA polymerase from C. difficile UK1 genomic DNA with the primers 5′CO_cspB (GTTTAACTTTAAGAAGGAGATATACATATGATTATCATTAATTACGAACTGATTGTGAAG) and 3′CO_cspB_CPD (TTCCATCCGCGAGCTCGCTTTTATTAATGCTGCG). To aid in solubility and allow for the generation of an untagged CspB protein, the cysteine protease domain from the Vibrio cholerae MARTX protease (CPD) was fused to the 3′ end of cspB (35). cpd was amplified from the pcspC-CO-Δ50CPD plasmid using the primers 5′CPD_cspB (CGCAGCATTAATAAAAGCGAGCTCGCGGATGGAA) and 3′CPD_pET (ATCTCAGTGGTGGTGGTGGTGGTGCTCGAGACCTTGCGCGTCCCA). The resulting cspB and cpd PCR products were inserted into pET22b between restriction sites XhoI and NdeI using Gibson Assembly (34), and the resulting plasmid was named pKS02. The C. difficile cspC gene was codon optimized for expression in E. coli (Eurofins Genomics, Ebersberg, Germany). The resulting gene was subcloned into pET22b-CPD (35). The sequences of all inserts were verified.
Spore preparation.
Spores were generated and purified as described previously (13, 25, 27, 32). Briefly, C. difficile strains were streaked onto 10 to 15 plates of BHIS agar medium. After 4 days, the growth from each plate was scraped into 1 ml of sterile water and incubated overnight at 4°C. The following day, the growth suspension was washed five times in sterile water. During each wash step, the layer of white cell debris was removed. After washing, the spores/cell debris were suspended in 25% (wt/vol) HistoDenz (Sigma-Aldrich, St. Louis, MO) and layered onto a 50% (wt/vol) HistoDenz solution. The resulting gradient was centrifuged at 18,900 × g for 30 min. During centrifugation, spores migrate through the 50% HistoDenz, while the less-dense vegetative cells and cell debris remain at the interface. After centrifugation, the solution is removed, and the pellet, containing the purified spores, was washed five times in sterile water before being resuspended in a final volume of 1 ml. Purified spores were examined microscopically and found to be >99.9% pure and phase bright (dormant).
Germination.
To quantify the interaction between the bile acids and C. difficile spores, purified spores were heat activated at 65°C for 30 min and then placed on ice. Ten microliters of the heat-activated spores were added to a final optical density at 600 nm (OD600) of 0.5 in 990 μl of BHIS medium alone or medium supplemented with the indicated concentrations of bile acids and germination was monitored at 600 nm for 15 min in a Perkin-Elmer (Waltham, MA) Lambda25 UV/Vis spectrophotometer. The data points at OD600 (Tx) were normalized to the starting OD600 value (T0). Germination rates and EC50 values were calculated using the slopes of the linear portions of the germination plots, as described previously (11, 25). The data shown in Fig. 1 and 2 are representative of three independent experiments (the data cannot be averaged between experiments due to differences in time points generated by the Perkin-Elmer spectrophotometer). (For transparency, all germination plots are included Fig. S1 to S3 in the supplemental material.) The rates and EC50 values were individually calculated from each germination experiment and are reported as averages with the standard error of the mean.
FIG 1.
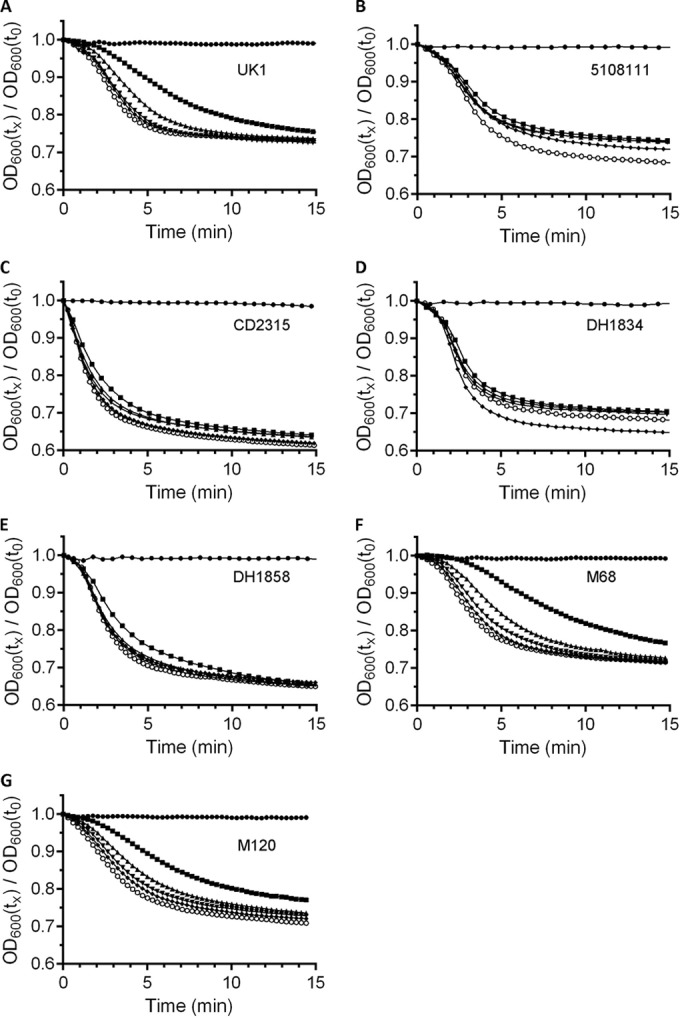
Bile acid-mediated spore germination of several C. difficile strains. Purified C. difficile spores were suspended in BHIS medium alone (●) or medium supplemented with 2 mM (■), 5 mM (▲), 10 mM (▼), 20 mM (◆), or 50 mM (○) TA. Germination was monitored at OD600 as described previously. (A) C. difficile UK1; (B) C. difficile 5108111; (C) C. difficile CD2315; (D) C. difficile DH1834; (E) C. difficile DH1858; (F) C. difficile M68; (G) C. difficile M120. A representative sample from three independent experiments is shown. For transparency, all plots are shown in Fig. S1 in the supplemental material.
FIG 2.
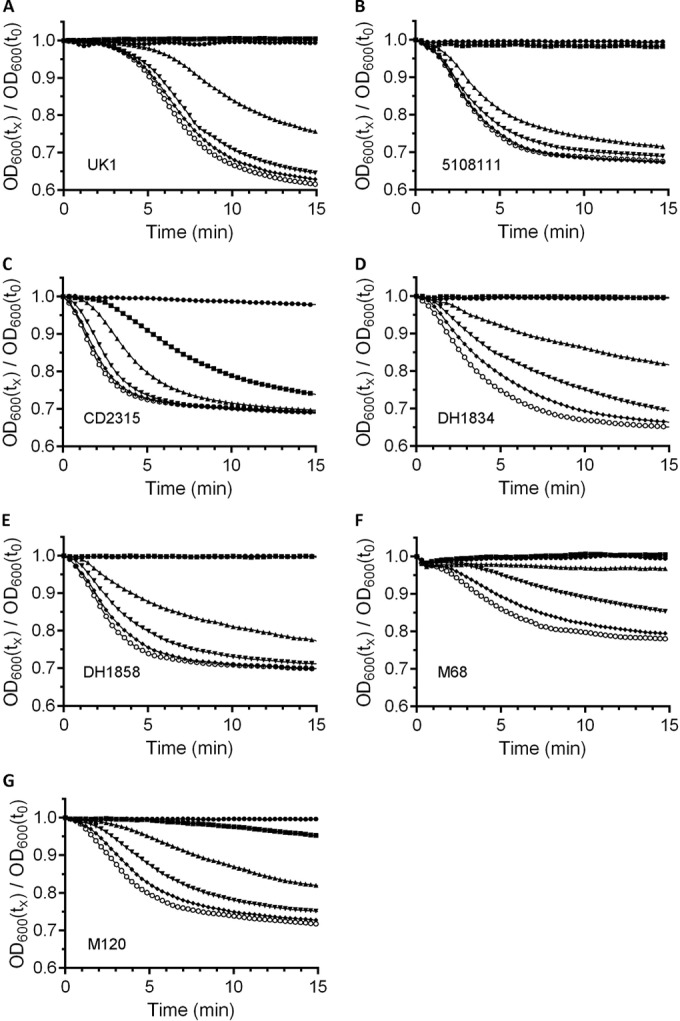
Chenodeoxycholic acid inhibits C. difficile spore germination. Purified C. difficile spores were suspended in BHIS medium supplemented with 1 mM CDCA (●) or medium supplemented with 2 mM (■), 5 mM (▲), 10 mM (▼), 20 mM (◆), or 50 mM (○) TA and 1 mM CDCA. Germination was monitored at OD600 as described previously. (A) C. difficile UK1; (B) C. difficile 5108111; (C) C. difficile CD2315; (D) C. difficile DH1834; (E) C. difficile DH1858; (F) C. difficile M68; (G) C. difficile M120. A representative sample from three independent experiments is shown. For transparency, all plots are shown in Fig. S3 in the supplemental material.
The release of CaDPA from germinating C. difficile spores was monitored in real time using terbium fluorescence (36). An opaque, 96-well plate was prepared with 125 μl of 10 mM Tris (pH 7.5), 150 mM NaCl, 800 μM TbCl3, and 100 mM glycine alone or supplemented with 10 mM TA, as described previously (27). Heat-activated spores were then sedimented for 1 min at 14,000 × g and resuspended in an equal volume of water to remove any CaDPA that may have been released due to autogerminating spores. A 5-μl sample of a spore suspension (OD600 = 60) was added to each well, and the CaDPA release was monitored in a Molecular Devices (Sunnydale, CA) Spectramax M3 fluorescence plate reader. CaDPA release was monitored with the following settings: excitation, 270 nm; emission, 545 nm; and cutoff, 420 nm. The CaDPA release data are reported as the average of three independent experiments, and error bars represent the standard errors of the mean.
The total DPA content of the spores was determined by suspending 105 spores in buffer, followed by incubation at 95°C for 30 min. The spores were cooled and sedimented for 2 min at 14,000 × g. The amount of released DPA was determined as described above using terbium fluorescence.
Protein purification.
Overnight cultures of the appropriate BL21(DE3) strains were diluted 1:200 in 2×TY medium and incubated at 200 rpm and 37°C. When an OD600 of 0.6 to 0.8 was reached, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a 250 μM final concentration, and cultures were grown at 16°C for 12 to 14 h. Subsequently, the cultures were pelleted at 6,000 rpm and 4°C for 30 min and then frozen at −80°C. Cell pellets were resuspended in LIB1 (50 mM Tris [pH 7.5], 500 mM NaCl, 15 mM imidazole, and 10% glycerol) and lysed by sonication. HisPur Ni-NTA resin (Thermo Scientific) was added to the clarified supernatants and washed with LIB2 (50 mM Tris [pH 7.5], 300 mM sodium chloride, 30 mM imidazole, and 10% glycerol). For SleC6His, the resulting protein was eluted from the Ni-resin in LIB1 supplemented with 500 mM imidazole. For CspB-CPD6His and CspC-CPD6His, all proteins were cleaved from the CPD tag in LIB1 supplemented with 75 μM (final concentration) phytic acid at room temperature for 20 min with shaking (four cleavage reactions were sufficient to cleave the respective proteins from the CPD tag). The volume of the phytic acid cleavings was equal to the Ni-bead bed volume. The resulting proteins were analyzed for their purity by SDS-PAGE and Coomassie blue staining and quantified by using a NanoDrop apparatus.
Protein extraction and Western blotting.
NuPAGE soluble proteins (which include CspB, CspC, and SleC) were extracted from 2 × 109/ml purified spores as described previously (37). A separate spore extract from 2 × 109 C. difficile JSC11 (cspBA::ermB) spores or C. difficile CAA5 (sleC::ermB) spores was also generated. Standard amounts of NuPAGE-solubilized recombinant proteins (generated above) were added to a 10% sodium dodecyl sulfate (SDS)–polyacrylamide gel (a standard curve was generated on every gel run). An equal volume of extract from mutant spores was added to the wells containing the recombinant protein standard to allow for equal transfer efficiencies between recombinant protein-containing wells and extract wells. Proteins were separated by SDS-PAGE. After separation, the gels were transferred overnight at 30 V to a low-fluorescence polyvinylidene difluoride membrane (Bio-Rad). Subsequently, the membrane was blocked for 1 h at room temperature with 10% dried, skimmed milk in Tris-buffered saline (TBS). Each membrane was washed thrice for 15 min each with TBS containing 0.1% (vol/vol) Tween 20 (TBST). The membrane was then incubated at room temperature with either rabbit anti-CspC, anti-CspB, or anti-SleC antibodies. After incubation with the primary antibody, membranes were washed as described above in TBST. The membranes were then labeled with Alexa Fluor 555-labeled donkey anti-rabbit IgG (Life Technologies) in the dark for 2 h at room temperature. The membranes were again washed as described above but in the dark. Finally, the fluorescent signal on the membranes were detected by scanning on a GE Typhoon Scanner using the Cy3 setting (appropriate wavelength for Alexa Fluor 555). The resulting scans were quantified using ImageQuant TL 7.0 image analysis software provided by GE.
Statistical analyses.
Statistical analysis between the calculated EC50,TA or inhibitor constant (CDCA) values was accomplished with a one-way analysis of variance with Tukey's test for multiple comparisons. A 99% confidence interval was set for significance (P < 0.01).
RESULTS
Taurocholic acid-induced germination by several C. difficile isolates.
There have been recent reports describing heterogeneity of spore germination between C. difficile strains (30, 31). Some of these strains appeared to germinate in rich medium in the absence of the TA germinant. Other strains were not inhibited by CDCA (31). This calls into question the impact bile acids may have on C. difficile infection and the utility of germination/antigermination-based therapies. Although there are undoubtedly differences in germination responses between C. difficile strains, the lack of inhibition by CDCA and germination in the absence of TA warranted further investigation.
Germination by C. difficile strains UK1 and M68 have been described previously by our lab and spore germination by these strains is activated by TA and inhibited by CDCA (25). We also analyzed germination by C. difficile strains 5108111, DH1834, DH1858, CD2315 (generously provided by Nigel Minton), and M120. Of these, spores derived from C. difficile strains 5108111, DH1834, and CD2315 were reported not to be inhibited by CDCA (spore germination by DH1858 was inhibited by CDCA) (31). To quantitate the differences in germination responses between isolates, we determined the EC50 values for taurocholic acid and spores from different C. difficile strains. These types of assays have been useful to determine the potency of activators and inhibitors of germination (11, 24, 25, 38–40). Although not traditional enzyme kinetics (spore germination is a multienzyme process), these studies can provide a quantitative measure for the interaction of the bile acids and C. difficile spores. C. difficile spores were suspended in BHIS medium alone or in medium supplemented with increasing TA concentrations (2, 5, 10, 20, or 50 mM) and germination was monitored at OD600. These TA concentrations are routinely used by our laboratory to assign EC50 values for bile acids. As shown in Fig. 1 and in Fig. S1 in the supplemental material, BHIS medium alone did not stimulate germination of the seven strains tested, suggesting a requirement for TA to initiate spore germination in rich medium. However, when supplemented with increasing concentrations of TA, C. difficile UK1, M68, and M120 (Fig. 1A, F, and G, respectively) exhibited a concentration-dependent increase in the rate of germination. Interestingly, C. difficile strains 5108111, CD2315, DH1834, and DH1858 achieved near-maximum germination rates at the lowest tested TA concentration (BHIS + 2 mM TA) (Fig. 1B to E; see also Fig. S1B to E in the supplemental material).
Because these strains germinated at near-maximum rates in the lowest tested TA concentration, we modified our germination conditions to determine whether these strains are capable of germinating at even lower TA concentrations (see Fig. S2 in the supplemental material). When germination by C. difficile strains 5108111, CD2315, DH1834, and DH1858 were tested at lower TA concentrations, we observed dose-dependent increases in germination rates. From the newly generated germination plots, we generated rate curves and EC50,TA values (Table 1). Spores from C. difficile UK1 and C. difficile M68 generated EC50,TA values similar to those described previously (25). C. difficile DH1834, DH1858, and M120 had similar EC50,TA values (3.0, 3.03, and 2.37, respectively), and these values are similar to those for UK1 and M68. Interestingly, compared to the other tested strains, C. difficile 5108111 and CD2315 had statistically significant increased EC50 values for TA (Table 1), suggesting that spores derived from these strains require less TA to activate spore germination.
TABLE 1.
Quantifying the interaction of C. difficile spores with bile acids
| Strain | Mean ± SDa |
Inhibition ratiob | |
|---|---|---|---|
| Apparent EC50,TA (mM) | Inhibitor constant (mM) | ||
| UK1 | 2.95 ± 0.23 | 0.21 ± 0.07 | 14.0 |
| 5108111 | 0.76 ± 0.14* | 0.52 ± 0.03* | 1.5 |
| CD2315 | 0.42 ± 0.07* | 0.18 ± 0.03 | 2.3 |
| DH1834 | 3.00 ± 0.45 | 0.21 ± 0.08 | 14.3 |
| DH1858 | 3.03 ± 0.36 | 0.29 ± 0.07 | 10.4 |
| M68 | 3.35 ± 0.39 | 0.12 ± 0.02 | 27.9 |
| M120 | 2.37 ± 0.14 | 0.17 ± 0.02 | 13.9 |
The apparent EC50,TA and inhibitor constant values were calculated from the germination curves, as described previously (11, 25, 32). Inhibitor constant = [inhibitor]/{(EC50,TA with inhibitor)/[(EC50,TA without inhibitor) − 1]}. For statistical comparisons, EC50,TA values were compared to each other, and inhibitor constant values were compared to each other as described in Materials and Methods. *, P < 0.01.
Inhibition ratio = EC50,TA/inhibitor constant.
CDCA-dependent inhibition of C. difficile spore germination.
Because we observed that some C. difficile strains (5108111 and CD2315) exhibited a more potent interaction with TA, we tested whether there were differences in the interaction with CDCA. C. difficile spores were suspended in BHIS medium or BHIS medium supplemented with 1 mM CDCA and increasing concentrations of TA (2, 5, 10, 20, or 50 mM), and germination was monitored at OD600. CDCA completely inhibited germination of the 2 mM TA sample for spores derived from all C. difficile strains except C. difficile strain CD2315. Interestingly, CDCA had less of an effect on germination by C. difficile CD2315 spores (Fig. 2C). However, CDCA still affected its ability to respond to TA as a germinant. Upon comparing Fig. 1C to Fig. 2C, it is evident that germination by C. difficile CD2315 spores clearly is affected by CDCA.
Inhibition constants were generated from the curves in Fig. 2 and Fig. S3 in the supplemental material and the calculated EC50,TA (Table 1). C. difficile UK1 spores and C. difficile M68 spores generated inhibitor constant values similar to those reported previously (Table 1) (25), and all strains generated values near or below 0.5 mM. Spores derived from C. difficile 5108111 had the weakest interaction with CDCA (0.52 mM; P < 0.01) compared to the other strains. These results suggest that CDCA inhibits germination by spores of C. difficile strains previously thought not to be affected by the inhibitor.
Although the germination of all strains was inhibited by CDCA, the effectiveness of the inhibitor varied. When the inhibition ratio (the ratio of the EC50,TA and the inhibitor constant) was calculated, the ratio for most strains was greater than 10, suggesting that germination is strongly inhibited by CDCA in these strains (Table 1). Germination by C. difficile 5108111 and CD2315 spores was not strongly inhibited by CDCA (the same strains with an increased EC50,TA) (Table 1). Taken together, the results suggest that, although there are differences in the interactions of the spores with the bile acids, all tested strains are inhibited by CDCA and are activated by TA.
To determine whether the observed differences in the interaction of the bile acids with the C. difficile spores correlated with specific alterations in the CspB, CspA, or CspC protein sequences, we sequenced the cspBAC locus from each strain (see Fig. S4 to S6 in the supplemental material). All ribotype 027 isolates (UK1, 5108111, DH1834, and DH1858) had identical CspC sequences, whereas ribotypes 078 (CD2315 and M120) and 017 (M68) had 26 substitutions between them (see Fig. S4 in the supplemental material). C. difficile CD2315 had a P317L substitution that was not present in any of the other tested strains (see Fig. S4 in the supplemental material). The CspB sequence only varied in ribotype 078 strains (see Fig. S5 in the supplemental material) with 13 substitutions. The CspA proteins varied most between the three Csp proteins, with a total of 42 substitutions (see Fig. S6 in the supplemental material). With the exception of the CD2315 CspCP317L substitution, all substitutions were consistent between ribotypes. Because substitutions were ribotype specific but the EC50,TA and inhibitor constants were not, these results suggest that the observed differences are not due to specific changes in the CspB, CspA, or CspC sequences.
Taurocholic acid is required for C. difficile spore germination.
Previously, germination by C. difficile strains 5108111, DH1834, DH1858, and CD2315 occurred slowly but in the absence of the TA germinant (31). To further investigate the phenomenon, we utilized an assay which measures the release of CaDPA from the spore core during germination (CaDPA release is a requirement for spore germination). Because this assay is (i) based on the detection of fluorescent CaDPA-Tb3+ complexes and not OD and (ii) each well is mixed prior to analysis, this assay eliminates any chance of spores settling, clumping, and/or localizing to the center of a well during the 4-h incubation. C. difficile spores were suspended in germination buffer supplemented with glycine only (nongerminating conditions) or TA and glycine (germination-inducing conditions) and Tb3+ fluorescence was monitored for 4 h (see Fig. S7 in the supplemental material). Spores from all C. difficile strains tested released CaDPA, which resulted in Tb3+ fluorescence when suspended in both TA and glycine. Spores suspended in glycine alone did not release CaDPA during the 4-h assay. These results suggest that these strains require TA to induce germination in combination with glycine and have a low frequency of autogermination.
The data presented in Fig. S7 in the supplemental material suggest that all tested strains have a requirement for TA to stimulate germination in combination with the glycine cogerminant, but not if these strains require glycine to germinate. Therefore, we tested CaDPA release in the absence of glycine (Fig. 3). C. difficile strains UK1 (Fig. 3A), 5108111 (Fig. 3B), DH1834 (Fig. 3D), DH1858 (Fig. 3E), M68 (Fig. 3F), and M120 (Fig. 3G) all required the presence of both TA and glycine to germinate. Interestingly, C. difficile CD2315 (Fig. 3C) slowly, but significantly, germinated in the presence of TA alone. However, germination by CD2315 spores was enhanced strongly by the presence of glycine (Fig. 3C). These results suggest that glycine is a cogerminant for spores of the tested strains but that C. difficile CD2315 spores can release CaDPA slowly in the absence of glycine.
FIG 3.
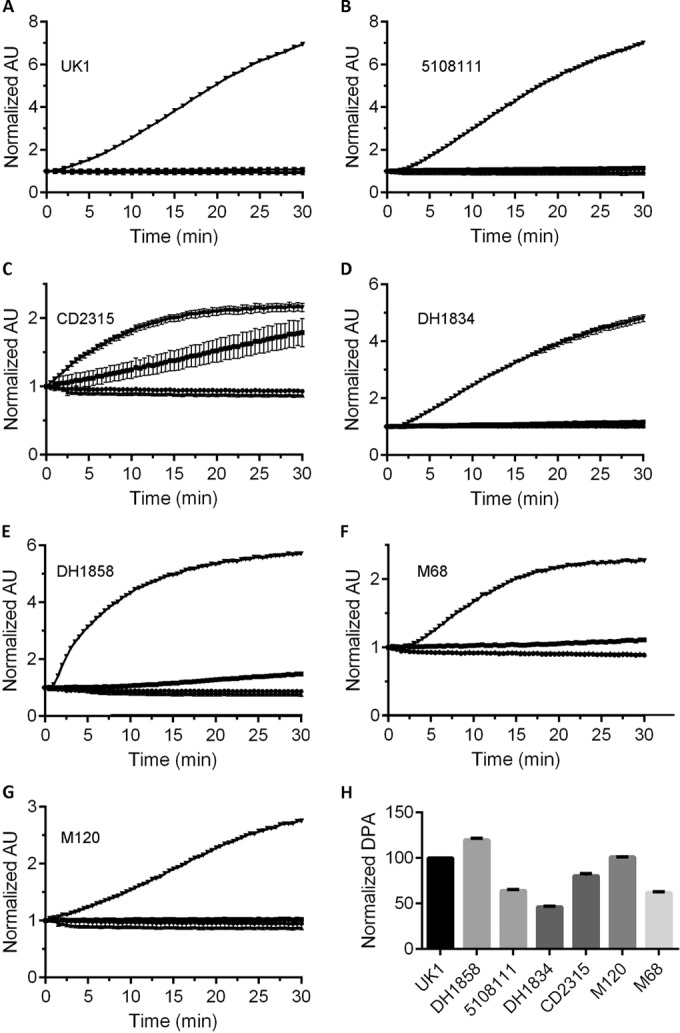
DPA release by several C. difficile strains. Purified C. difficile spores were suspended in HEPES buffer (●) or buffer supplemented with 10 mM TA (■), 100 mM glycine (▲), or 10 mM TA and 100 mM glycine (▼). DPA release during spore germination was monitored using Tb3+ fluorescence, as described previously. (A) C. difficile UK1; (B) C. difficile 5108111; (C) C. difficile CD2315; (D) C. difficile DH1834; (E) C. difficile DH1858; (F) C. difficile M68; (G) C. difficile M120. (H) Purified C. difficile spores were suspended in buffer and incubated at 100°C for 30 min. Total DPA content was normalized to the amount of DPA found in C. difficile UK1. All data represent the average from three independent experiments, and error bars represent the standard deviations from the mean.
To further characterize the differences between these strains, we analyzed the amount of CaDPA present within the core of each strain and normalized this to the amount found in C. difficile UK1 (Fig. 3H). For all strains, 105 spores were boiled in buffer and sedimented, and the amount of CaDPA was determined using Tb3+ fluorescence. As shown in Fig. 3H, the amount of CaDPA present in C. difficile spores varied between strains (although only by 2-fold in the lowest DPA-containing strain, C. difficile DH1834).
Germinant receptor levels vary between C. difficile strains.
To begin to understand how different C. difficile isolates display different EC50 or inhibitor constant values for bile acids, we analyzed the abundance of CspB, CspC, and SleC in purified C. difficile spores. Purified C. difficile spores were extracted with NuPAGE buffer, as described previously (37). We found that by boiling spores in NuPAGE buffer, the amount of remaining CspB, CspC, or SleC in the NuPAGE insoluble fraction (containing unbroken spores) was below the limit of detection. Known amounts of the recombinant CspC, CspB, or SleC protein and spore extracts were separated by SDS-PAGE and detected using rabbit polyclonal antisera, followed by Alexa Fluor 555-conjugated donkey anti-rabbit IgG. The standard curves generated from the signals on each SDS-PAGE were linear (Fig. 4). From the signal intensities of the extracted CspB (Fig. 4A), CspC (Fig. 4B), and SleC (Fig. 4C), the volume loaded onto the SDS-PAGE, the molecular weights of each protein, and the amount of spores extracted, we were able to determine the average abundance of each protein in a C. difficile spore (Table 2).
FIG 4.
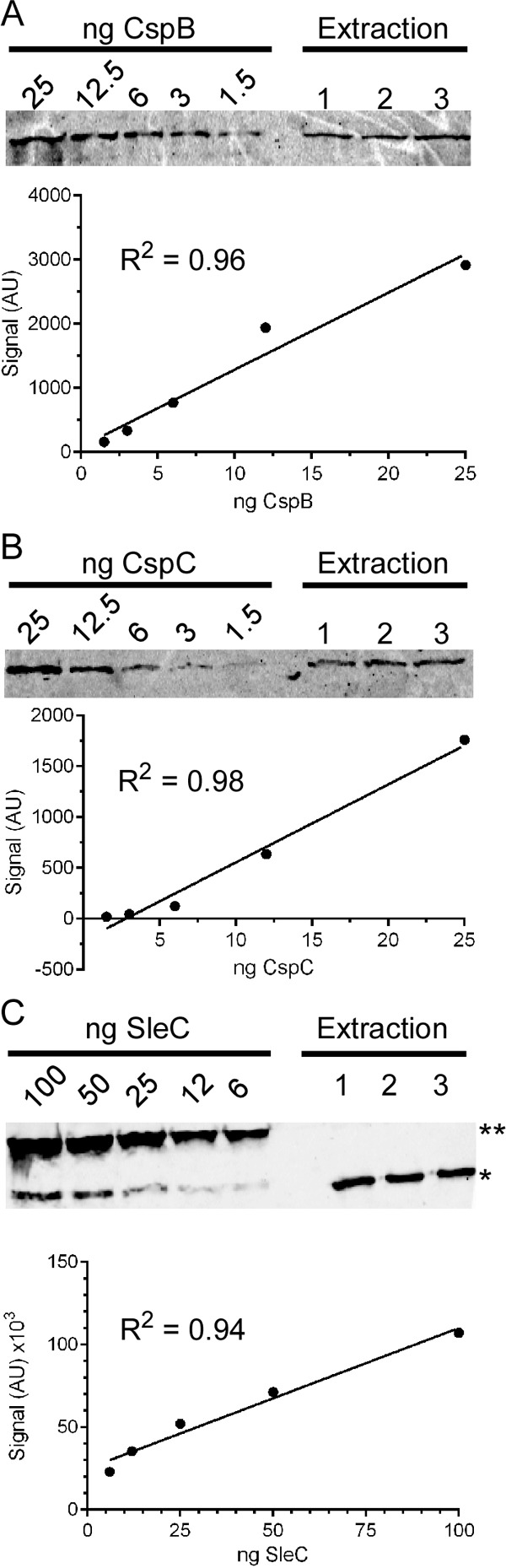
Quantifying the abundance of CspB, CspC, and SleC in C. difficile spores. A total of 109 C. difficile UK1 spores were extracted with NuPAGE buffer, as described previously. NuPAGE-soluble protein from three independent extracts was loaded onto a 10% SDS-polyacrylamide gel, along with recombinantly expressed and purified protein, as a protein standard. Samples were separated and detected as described in Materials and Methods. Signal intensities were quantified and used to generate a standard curve for CspB (A), CspC (B), and SleC (C). Signal intensities in the spore extract samples were quantified, and the generated standard curve was used to quantify the abundance of the specified proteins in the spore extracts. Independent standard curves were generated for each strain, and the results of the quantification can be found in Table 2. **, Full-length pre-pro-SleC; *, pro-SleC.
TABLE 2.
Number of Csp molecules per C. difficile spore
| Strain | Avg ± SDa |
SleC/CspC ratio | ||
|---|---|---|---|---|
| CspB | CspC | SleC | ||
| UK1 | 1,394 ± 426 | 2,141 ± 164 | 46,090 ± 5,089 | 21.5 |
| 5108111 | 3081 ± 154 | 2,304 ± 209 | 31,614 ± 3,497 | 13.7 |
| CD2315 | 435 ± 86 | 1,689 ± 137 | 41,239 ± 1,832 | 24.4 |
| DH1834 | 1,178 ± 120 | 3,155 ± 139 | 37,876 ± 2,027 | 12.0 |
| DH1858 | 1,572 ± 60 | 1,435 ± 154 | 37,557 ± 827 | 26.2 |
| M68 | 1,284 ± 160 | 3,836 ± 541 | 46,685 ± 3,507 | 12.2 |
| M120 | 633 ± 36 | 2,423 ± 203 | 29,473 ± 1,724 | 12.2 |
The average numbers of molecules of CspB, CspC, and SleC were calculated as described in the legend to Fig. 4. The numbers represent three independent extractions.
The abundance of these proteins varied between C. difficile isolates. For example, C. difficile UK1 had 2,000 CspC molecules per spore, while C. difficile M68 had more than 3,800. Interestingly, C. difficile CD2315 and C. difficile M120 had a very low abundance of CspB (435 and 633 molecules per spore, respectively). As described previously for C. perfringens, the average amount of SleC per spore was greater than that of both CspC and CspB but also varied between strains (Table 2) (41).
Prior work has suggested that much of the OD change observed during spore germination is due to the release of CaDPA from the spore core (42). During C. difficile spore germination, this event is dependent on the hydrolysis of the spore cortex by SleC (27). Because SleC is deposited into the spore as a zymogen, SleC must be activated by CspB in order to stimulate germination. In our original model for bile acid mediated spore germination, CspB is activated by the CspC germinant receptor. Because the abundance of CspC varied between isolates, we determined whether the maximum rate of spore germination correlated with the abundance of CspC. Thus, for each strain we calculated the maximum rate of germination from the germination curves used to calculate the EC50,TA (Table 3). Surprisingly, there was a slight trend for an inverse correlation of the abundance of CspC and rate (R2 = 0.32) (Fig. 5A), suggesting that CspC may have an inhibitory effect on spore germination.
TABLE 3.
Rates of C. difficile spore germination
| Strain | Germination rate (mean OD × 10−3/s ± SD)a |
|---|---|
| UK1 | 1.47 ± 0.092 |
| 5108111 | 1.30 ± 0.082 |
| CD2315 | 1.67 ± 0.047 |
| DH1834 | 1.23 ± 0.047 |
| DH1858 | 1.62 ± 0.11 |
| M68 | 1.35 ± 0.054 |
| M120 | 1.04 ± 0.043 |
FIG 5.
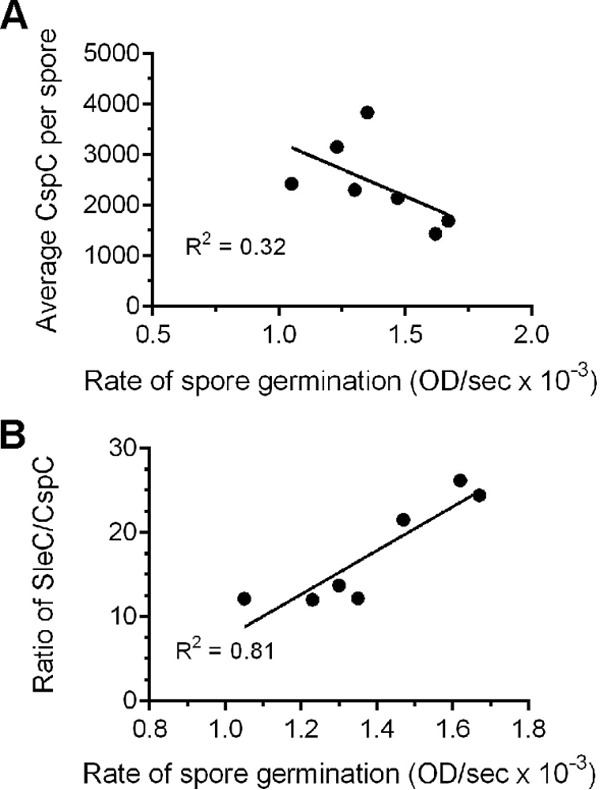
Correlating the abundance of C. difficile proteins with the kinetics of spore germination. (A) The calculated average per spore abundance of CspC was plotted versus the calculated rates of spore germination. (B) The ratio of SleC to CspC was plotted versus the calculated rates of spore germination. For all plots, GraphPad Prism was used to generate the linear fit to the curves, and the listed R2 values represent the fit of the curve to the data.
Because SleC-mediated cortex hydrolysis is required for CaDPA release during C. difficile spore germination, if the above trend is true we should observe a positive correlation between the ratio of SleC to CspC and germination rate (more SleC molecules to CspC molecules should result in an increased rate of germination). As shown in Fig. 5B, we observed a strong correlation (R2 = 0.81) between SleC/CspC and germination rate, again suggesting that CspC has an inhibitory effect on spore germination.
DISCUSSION
Spore germination is an important step in the life cycle of any spore-forming organism. The metabolically dormant spores must sense when conditions again become favorable for growth and quickly respond to the germinants used by that bacterium. Germination in an environment unfavorable for growth could prevent the outgrowth of a vegetative cell from the germinated spore or prevent the vegetative growth of that bacterium. Thus, the signals that stimulate germination (germinants) can vary between organisms depending on their growth niche. For example, C. difficile vegetative cells are not normally found growing outside a host; thus, it would be logical for C. difficile spores to respond to a molecule that is host specific. In fact, C. difficile spore germination is activated by cholic acid-class bile acids (e.g., TA) and is inhibited by chenodeoxycholic acid-class bile acids (e.g., CDCA) (11, 14, 19, 23). Although essential, TA is not sufficient to stimulate C. difficile spore germination. Glycine is also required, presumably as a measure of the nutrient status of the surrounding environment (19, 43, 44).
Until recently, most of the studies on C. difficile spore germination have relied on identifying germinants and their interactions with bile acids, bile acid analogs, amino acids, and amino acid analogs (11, 19, 23–25, 39, 45, 46). Only recently have the mechanisms of C. difficile spore germination been studied, largely due to breakthroughs in genetics (47–51). These breakthroughs allowed us to determine that the germination-specific, pseudoprotease, CspC, is the bile acid germinant receptor.
The germination-specific proteases have largely been studied in C. perfringens (41, 52–54). In C. perfringens the CspA, CspB, and CspC proteins are all catalytically active proteases that can cleave the cortex hydrolase, SleC, to an active form. In C. difficile, the cspB and cspA genes have been translationally fused and yield CspBA upon translation (26, 29). The CspBA protein undergoes interdomain processing by the YabG protease to generate CspB and CspA (55). We proposed a model whereby activated CspC transmits the signal to the CspB protease, which then activates SleC. In support of this model, we found that CaDPA release from the core is dependent on hydrolysis of the spore cortex (27).
If SleC activation occurs prior to CaDPA release from the spore core and if SleC-mediated cortex hydrolysis is essential for germination to begin, all C. difficile strains should have the requirement for bile acids to initiate germination (unless some C. difficile strains germinate using yet another novel mechanism for spore germination). However, some C. difficile strains were reported to have no apparent requirement for TA-mediated germination or whose germination was not inhibited by CDCA (31). If germination in these strains was not influenced by bile acids, by what mechanism(s) were they germinating?
To characterize the germination phenotypes in these strains, we determined the EC50,TA and inhibitor constant values for these strains and strains previously shown to be activated by TA and inhibited by CDCA. As suggested previously, the germination characteristics of these strains varied and did not correlate with ribotype (31). Importantly, though, we did observe that some strains have increased EC50,TA values, while another had a reduced inhibitor constant for CDCA (Table 1). These strains' increased potency toward TA could possibly explain previous observations (31). Spores derived from strains 5108111, DH1834, DH1858, and CD2315 were found previously to lose OD600 in rich medium in the absence of apparent TA (bile acids can be found in blood and, thus, animal products). Moreover, the authors found that germination of spores derived from strains 5108111, DH1834, and CD2315 was not inhibited by the CDCA antigerminant. Importantly, strains whose germination was not inhibited by CDCA had increased EC50,TA values. Because the prior work used only one concentration of TA and CDCA (0.1% [1.8 mM] and 2 mM, respectively), to test the effects of CDCA as an inhibitor, it is likely that these strains' increased EC50,TA overcame the inhibitory effect of CDCA (31). By quantifying the interactions between spores derived from these strains and TA/CDCA, we reestablish the importance of bile acids in promoting and inhibiting germination in these strains.
The differences in EC50,TA and inhibitor constants were not consistent between ribotypes. A growing body of evidence suggests that toxin production, sporulation and germination are not correlated with ribotype (30, 31, 56). In support of this observation, the calculated EC50,TA and inhibitor constant values were not attributed to specific substitutions within the CspB, CspA, or CspC protein sequences, and the substitutions were ribotype specific. Of the three proteins, CspB had the fewest substitutions. Because CspB is required to cleave pro-SleC, CspB is likely under evolutionary pressure to perform this function. Interestingly, CspC and CspA had 26 and 42 substitutions, respectively. The many substitutions in CspA could suggest that of the Csp proteins, it is under the least amount of selective pressure. Although the role of CspA is unknown, the presence of CspA is important for controlling the levels of the germinant receptor, CspC (55). Only one strain, C. difficile CD2315, encoded an extra substitution in the CspC protein sequence that was not present in the other 078 ribotype, P317L. Curiously, C. difficile CD2315 is the only strain that germinated in buffered TA without the need for the glycine cogerminant (although glycine still enhanced germination in this strain, suggesting the spore still responds to glycine). Clearly, more studies are needed to characterize the effects of these substitutions on C. difficile spore germination.
We further characterized these strains by determining the average per spore abundance of CspB, CspC, and SleC (we could not generate a standard curve for CspA—it is insoluble when recombinantly expressed). As expected, the number of molecules per spore varied between isolates and did not correlate with ribotype (Table 2). The abundance of each of these proteins is greater than the amount of germinant receptors found in B. subtilis (57). However, the increased abundance of CspB and SleC found in the C. difficile spore, compared to the B. subtilis germinant receptors, is consistent with a recent publication describing the amount of these proteins in C. perfringens (41). Moreover, as seen in C. perfringens, the amount of SleC was greater than the amount of CspB. The amount of CspC in the C. difficile spore was similar to CspB but varied (Table 2).
By determining the kinetics of spore germination for several C. difficile strains, we were positioned to determine whether the kinetic properties of germination could be attributed to differences in the abundance of germination-specific proteins (CspB, CspC, and SleC). In B. subtilis, overexpression of the GerAA germinant receptor results in an increase in the germination rate (as measured by the decrease in OD600 and by the release of CaDPA) (58). Prior work has shown that much of the OD600 change that occurs during germination is due to the release of CaDPA from the spore core (42). In C. difficile, CaDPA release is dependent on SleC-mediated cortex hydrolysis (27, 28). Thus, we hypothesized that an increased abundance of SleC would correlate with an increased rate of germination. Also, because SleC activity is dependent on activation by CspB, an increased abundance of CspB could result in more active SleC during germination and, thus, an increased rate of germination. However, we did not observe any correlation between the rate of spore germination and the abundance of the SleC hydrolase or CspB protease. One possible explanation for this could be that the abundance of both SleC and CspB in the C. difficile spores are far past saturating levels, and any alteration in the observed levels would have minimal effects on the rate of germination. Surprisingly, we observed a trend for an inversed correlation between the rate of germination and CspC abundance (the more CspC molecules/spore, the slower the rate of germination [Fig. 5A]).
If this correlation is true, we predicted that more SleC molecules would be needed to overcome the potential inhibitory effect CspC would have on C. difficile spore germination. Indeed, we observed a strong correlation between SleC/CspC and the rate of germination (Fig. 5B) (R2 = 0.81). These results suggest that, instead of the activator function we originally hypothesized, CspC may be acting to inhibit C. difficile spore germination.
If true, how would CspC function during germination? Our prior genetic data suggest that CspC transmits the bile acid signal to begin the germination process, though the mechanism is unclear (13). Recently, GerS was identified and recognized to play a role during germination (59). GerS is a protein that is anchored to the inner leaflet of the outer spore membrane (though the lipidation of GerS is not required for it to function during germination). Interestingly, C. difficile ΔgerS strains have a defect in germination but still cleave SleC to the active hydrolase form. This suggested that SleC activity, somehow, is inhibited in this strain (59). Based on the correlation between CspC and SleC abundance and our kinetic data, we hypothesize that CspC activates CspB but inhibits SleC activity in the absence of GerS. Investigating the biochemistry of this germinant receptor complex could lead to a greater understanding of the mechanism of initiating C. difficile spore germination.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nigel Minton (University of Nottingham, United Kingdom) for generously providing the strains used in this study, Aimee Shen (University of Vermont) for sharing reagents and methods, and Matthew Sachs (Texas A&M University) for the use of the Typhoon Scanner.
Funding Statement
The content is solely the responsibility of the authors and does not necessarily reflect the views of the NIAID or the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00908-15.
REFERENCES
- 1.Yutin N, Galperin MY. 2013. A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ Microbiol 15:2631–2641. doi: 10.1111/1462-2920.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jump RLP, Pultz MJ, Donskey CJ. 2007. Vegetative Clostridium difficile survives in room air on moist surfaces and in gastric contents with reduced acidity: a potential mechanism to explain the association between proton pump inhibitors and C. difficile-associated diarrhea? Antimicrob Agents Chemother 51:2883–2887. doi: 10.1128/AAC.01443-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, West MR, Wren BW, Fairweather NF, Dougan G, Lawley TD. 2012. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect Immun 80:2704–2711. doi: 10.1128/IAI.00147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leffler DA, Lamont JT. 2015. Clostridium difficile infection. N Engl J Med 373:287–288. doi: 10.1056/NEJMc1506004. [DOI] [PubMed] [Google Scholar]
- 5.Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. 2009. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun 77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Theriot CM, Koenigsknecht MJ, Carlson PE Jr, Hatton GE, Nelson AM, Li B, Huffnagle GB, Young JB. 2014. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun 5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Theriot CM, Koumpouras CC, Carlson PE, Bergin II, Aronoff DM, Young VB. 2011. Cefoperazone-treated mice as an experimental platform to assess differential virulence of Clostridium difficile strains. Gut Microbes 2:326–334. doi: 10.4161/gmic.19142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voth DE, Ballard JD. 2005. Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev 18:247–263. doi: 10.1128/CMR.18.2.247-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouillaut L, Dubois T, Sonenshein AL, Dupuy B. 2015. Integration of metabolism and virulence in Clostridium difficile. Res Microbiol 166:375–383. doi: 10.1016/j.resmic.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shields K, Araujo-Castillo RV, Theethira TG, Alonso CD, Kelly CP. 2015. Recurrent Clostridium difficile infection: from colonization to cure. Anaerobe 34:59–73. doi: 10.1016/j.anaerobe.2015.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sorg JA, Sonenshein AL. 2010. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J Bacteriol 192:4983–4990. doi: 10.1128/JB.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howerton A, Patra M, Abel-Santos E. 2013. A new strategy for the prevention of Clostridium difficile infection. J Infect Dis 207:1498–1504. doi: 10.1093/infdis/jit068. [DOI] [PubMed] [Google Scholar]
- 13.Francis MB, Allen CA, Shrestha R, Sorg JA. 2013. Bile acid recognition by the Clostridium difficile germinant receptor, CspC, is important for establishing infection. PLoS Pathog 9:e1003356. doi: 10.1371/journal.ppat.1003356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paredes-Sabja D, Shen A, Sorg JA. 2014. Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends Microbiol 22:406–416. doi: 10.1016/j.tim.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Setlow P. 2014. Germination of spores of Bacillus species: what we know and do not know. J Bacteriol 196:1297–1305. doi: 10.1128/JB.01455-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan IS, Ramamurthi KS. 2014. Spore formation in Bacillus subtilis. Environ Microbiol Rep 6:212–225. doi: 10.1111/1758-2229.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Setlow P. 2014. Spore resistance properties. Microbiol Spectr 2014:2. doi: 10.1128/microbiolspec. [DOI] [PubMed] [Google Scholar]
- 18.Paredes-Sabja D, Setlow P, Sarker MR. 2011. Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol 19:85–94. doi: 10.1016/j.tim.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Sorg JA, Sonenshein AL. 2008. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol 190:2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson KH. 1983. Efficiency of various bile salt preparations for stimulation of Clostridium difficile spore germination. J Clin Microbiol 18:1017–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson KH, Kennedy MJ, Fekety FR. 1982. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J Clin Microbiol 15:443–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ridlon JM, Kang D, Hylemon PB. 2006. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47:241–259. [DOI] [PubMed] [Google Scholar]
- 23.Sorg JA, Sonenshein AL. 2009. Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J Bacteriol 191:1115–1117. doi: 10.1128/JB.01260-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howerton A, Ramirez N, Abel-Santos E. 2011. Mapping interactions between germinants and Clostridium difficile spores. J Bacteriol 193:274–282. doi: 10.1128/JB.00980-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Francis MB, Allen CA, Sorg JA. 2013. Muricholic acids inhibit Clostridium difficile spore germination and growth. PLoS One 8:e73653. doi: 10.1371/journal.pone.0073653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeno-Tarraga AM, Wang H, Holden MTG, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E, Sharp S, Simmonds M, Stevens K, Unwin L, Whithead S, Dupuy B, Dougan G, Barrell B, Parkhill J. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet 38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 27.Francis MB, Allen CA, Sorg JA. 2015. Spore cortex hydrolysis precedes dipicolinic acid release during Clostridium difficile spore germination. J Bacteriol 197:2276–2283. doi: 10.1128/JB.02575-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S, Shen A, Setlow P, Li YQ. 2015. Characterization of the dynamic germination of individual Clostridium difficile spores using Raman spectroscopy and differential interference contrast microscopy. J Bacteriol 197:2361–2373. doi: 10.1128/JB.00200-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adams CM, Eckenroth BE, Putnam EE, Doublie S, Shen A. 2013. Structural and functional analysis of the CspB protease required for Clostridium spore germination. PLoS Pathog 9:e1003165. doi: 10.1371/journal.ppat.1003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carlson PE Jr, Kaiser AM, McColm SA, Bauer JM, Young VB, Aronoff DM, Hanna PC. 2015. Variation in germination of Clostridium difficile clinical isolates correlates to disease severity. Anaerobe 33:64–70. doi: 10.1016/j.anaerobe.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heeg D, Burns DA, Cartman ST, Minton NP. 2012. Spores of Clostridium difficile clinical isolates display a diverse germination response to bile salts. PLoS One 7:e32381. doi: 10.1371/journal.pone.0032381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allen CA, Babakhani F, Sears P, Nguyen L, Sorg JA. 2013. Both fidaxomicin and vancomycin inhibit outgrowth of Clostridium difficile spores. Antimicrob Agents Chemother 57:664–667. doi: 10.1128/AAC.01611-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sorg JA, Dineen SS. 2009. Laboratory maintenance of Clostridium difficile. Curr Protoc Microbiol Chapter 9:Unit 9A.1. doi: 10.1002/9780471729259.mc09a01s12. [DOI] [PubMed] [Google Scholar]
- 34.Gibson DG, Young L, Chuang RY, Venter JC, CAHutchison 3rd, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 35.Shen A, Lupardus PJ, Albrow VE, Guzzetta A, Powers JC, Garcia KC, Bogyo M. 2009. Mechanistic and structural insights into the proteolytic activation of Vibrio cholerae MARTX toxin. Nat Chem Biol 5:469–478. doi: 10.1038/nchembio.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hindle AA, Hall EA. 1999. Dipicolinic acid (DPA) assay revisited and appraised for spore detection. Analyst 124:1599–1604. doi: 10.1039/a906846e. [DOI] [PubMed] [Google Scholar]
- 37.Lawley TD, Croucher NJ, Yu L, Clare S, Sebaihia M, Goulding D, Pickard DJ, Parkhill J, Choudhary J, Dougan G. 2009. Proteomic and genomic characterization of highly infectious Clostridium difficile 630 spores. J Bacteriol 191:5377–5386. doi: 10.1128/JB.00597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liggins M, Ramirez N, Magnuson N, Abel-Santos E. 2011. Progesterone analogs influence germination of Clostridium sordellii and Clostridium difficile spores in vitro. J Bacteriol 193:2776–2783. doi: 10.1128/JB.00058-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramirez N, Liggins M, Abel-Santos E. 2010. Kinetic evidence for the presence of putative germination receptors in Clostridium difficile spores. J Bacteriol 192:4215–4222. doi: 10.1128/JB.00488-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akoachere M, Squires RC, Nour AM, Angelov L, Brojatsch J, Abel-Santos E. 2007. Identification of an in vivo inhibitor of Bacillus anthracis spore germination. J Biol Chem 282:12112–12118. doi: 10.1074/jbc.M611432200. [DOI] [PubMed] [Google Scholar]
- 41.Banawas S, Korza G, Paredes-Sabja D, Li Y, Hao B, Setlow P, Sarker MR. 2015. Location and stoichiometry of the protease CspB and the cortex-lytic enzyme SleC in Clostridium perfringens spores. Food Microbiol 50:83–87. doi: 10.1016/j.fm.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 42.Zhang P, Kong L, Wang G, Setlow P, Li YQ. 2010. Combination of Raman tweezers and quantitative differential interference contrast microscopy for measurement of dynamics and heterogeneity during the germination of individual bacterial spores. J Biomed Optics 15:056010. doi: 10.1117/1.3494567. [DOI] [PubMed] [Google Scholar]
- 43.Karasawa T, Ikoma S, Yamakawa K, Nakamura S. 1995. A defined growth medium for Clostridium difficile. Microbiology 141(Pt 2):371–375. [DOI] [PubMed] [Google Scholar]
- 44.Bouillaut L, Self WT, Sonenshein AL. 2013. Proline-dependent regulation of Clostridium difficile Stickland metabolism. J Bacteriol 195:844–854. doi: 10.1128/JB.01492-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wheeldon LJ, Worthington T, Lambert PA. 2011. Histidine acts as a co-germinant with glycine and taurocholate for Clostridium difficile spores. J Appl Microbiol 110:987–994. doi: 10.1111/j.1365-2672.2011.04953.x. [DOI] [PubMed] [Google Scholar]
- 46.Wheeldon LJ, Worthington T, Hilton AC, Elliot TS, Lambert PA. 2008. Physical and chemical factors influencing the germination of Clostridium difficile spores. J Appl Microbiol 105:2223–2230. doi: 10.1111/j.1365-2672.2008.03965.x. [DOI] [PubMed] [Google Scholar]
- 47.Ng YK, Ehsaan M, Philip S, Collery MM, Janoir C, Collignon A, Cartman ST, Minton NP. 2013. Expanding the repertoire of gene tools for precise manipulation of the Clostridium difficile genome: allelic exchange using pyrE alleles. PLoS One 8:e56051. doi: 10.1371/journal.pone.0056051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cartman ST, Kelly ML, Heeg D, Heap JT, Minton NP. 2012. Precise manipulation of the Clostridium difficile chromosome reveals a lack of association between the tcdC genotype and toxin production. Appl Environ Microbiol 78:4683–4690. doi: 10.1128/AEM.00249-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heap JT, Pennington OJ, Cartman ST, Minton NP. 2009. A modular system for Clostridium shuttle plasmids. J Microbiol Methods 78:79–85. doi: 10.1016/j.mimet.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 50.Heap JT, Pennington OJ, Cartmant ST, Carter GP, Minton NP. 2007. The ClosTron: A universal gene knock-out system for the genus Clostridium. J Microbiol Methods 79:452–464. [DOI] [PubMed] [Google Scholar]
- 51.Dembek M, Barquist L, Boinett CJ, Cain AK, Mayho M, Lawley TD, Fairweather NF, Fagan RP. 2015. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio 6:e02383. doi: 10.1128/mBio.02383-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paredes-Sabja D, Setlow P, Sarker MR. 2009. The protease CspB is essential for initiation of cortex hydrolysis and dipicolinic acid (DPA) release during germination of spores of Clostridium perfringens type A food poisoning isolates. Microbiology 155:3464–3472. doi: 10.1099/mic.0.030965-0. [DOI] [PubMed] [Google Scholar]
- 53.Masayama A, Hamasaki K, Urakami K, Shimamoto S, Kato S, Makino S, Yoshimura T, Moriyama M, Moriyama R. 2006. Expression of germination-related enzymes, CspA, CspB, CspC, SleC, and SleM, of Clostridium perfringens S40 in the mother cell compartment of sporulating cells. Genes Genet Syst 81:227–234. doi: 10.1266/ggs.81.227. [DOI] [PubMed] [Google Scholar]
- 54.Shimamoto S, Moriyama R, Sugimoto K, Miyata S, Makino S. 2001. Partial characterization of an enzyme fraction with protease activity which converts the spore peptidoglycan hydrolase (SleC) precursor to an active enzyme during germination of Clostridium perfringens S40 spores and analysis of a gene cluster involved in the activity. J Bacteriol 183:3742–3751. doi: 10.1128/JB.183.12.3742-3751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kevorkian Y, Shirley DJ, Shen A. 2015. Regulation of Clostridium difficile spore germination by the CspA pseudoprotease domain. Biochimie pii:S0300–9084(15)00231-X. doi: 10.1016/j.biochi.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burns DA, Heap JT, Minton NP. 2010. The diverse sporulation characteristics of Clostridium difficile clinical isolates are not associated with type. Anaerobe 16:618–622. doi: 10.1016/j.anaerobe.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 57.Paidhungat M, Setlow P. 2001. Localization of a germinant receptor protein (GerBA) to the inner membrane of Bacillus subtilis spores. J Bacteriol 183:3982–3990. doi: 10.1128/JB.183.13.3982-3990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cabrera-Martinez R-M, Tovar-Rojo F, Vepachedu VR, Setlow P. 2003. Effects of overexpression of nutrient receptors on germination of spores of Bacillus subtilis. J Bacteriol 185:2457–2464. doi: 10.1128/JB.185.8.2457-2464.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fimlaid KA, Jensen O, Donnelly ML, Francis MB, Sorg JA, Shen A. 2015. Identification of a novel lipoprotein regulator of Clostridium difficile spore germination. PLoS Pathog 11:e1005239. doi: 10.1371/journal.ppat.1005239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


