Abstract
Disulfide bonds are important for the stability and function of many secreted proteins. In Gram-negative bacteria, these linkages are catalyzed by thiol-disulfide oxidoreductases (Dsb) in the periplasm. Protein oxidation has been well studied in these organisms, but it has not fully been explored in Gram-positive bacteria, which lack traditional periplasmic compartments. Recent bioinformatics analyses have suggested that the high-GC-content bacteria (i.e., actinobacteria) rely on disulfide-bond-forming pathways. In support of this, Dsb-like proteins have been identified in Mycobacterium tuberculosis, but their functions are not known. Actinomyces oris and Corynebacterium diphtheriae have recently emerged as models to study disulfide bond formation in actinobacteria. In both organisms, disulfide bonds are catalyzed by the membrane-bound oxidoreductase MdbA. Remarkably, unlike known Dsb proteins, MdbA is important for pathogenesis and growth, which makes it a potential target for new antibacterial drugs. This review will discuss disulfide-bond-forming pathways in bacteria, with a special focus on Gram-positive bacteria.
INTRODUCTION
Before the discovery of chaperones, it was generally accepted that protein folding was dependent upon primary amino acid sequences and the laws of thermodynamics. This notion was first challenged by Anfinsen's classical RNase A folding experiments in the 1960s. Following its denaturation, Anfinsen and colleagues (1) observed that RNase A could spontaneously refold in vitro, but the process was slow and prone to error. It was discovered that the formation of four disulfide bonds within RNase A was a limiting factor for its folding. In vivo, protein maturation is accelerated by protein disulfide isomerase (PDI) (2, 3). PDI is a multidomain thioredoxin-like enzyme that catalyzes disulfide bonds and reduces nonnative linkages in proteins secreted into the glutathione-based oxidizing environment of the endoplasmic reticulum (ER) (4).
Anfinsen's pioneering work revealed a major protein folding mechanism used by both eukaryotic and prokaryotic organisms. The formation of native disulfide bonds, often referred to as oxidative protein folding, is essential for the stability of many secreted polypeptides (5, 6). In Gram-negative bacteria, protein oxidation occurs in the extracytoplasmic periplasm and is regulated by a family of thioredoxin-like enzymes, DsbA to DsbG (7). DsbA and DsbB work in concert to catalyze disulfide bond formation in nascent proteins secreted into the periplasm by SecYEG (8). In contrast, the reduction or rearrangement of incorrectly formed disulfide bonds in this compartment is controlled by DsbC, DsbG, and DsbD (9–11).
Disulfide bond formation is not fully understood in Gram-positive bacteria, which lack traditional periplasmic spaces. Gram-positive cell envelopes are composed of a single membrane that is surrounded by thick layers of peptidoglycan. Although a space between these regions has been observed in a number of organisms by electron microscopy, it is not considered to be equivalent to those spaces found in Gram-negative bacteria (12, 13). Therefore, it is possible that proteins secreted by Gram-positive bacteria are exposed to oxidative stresses within the extracellular milieu, which can cause misfolding. It was proposed that Gram-positive bacteria avoid this potential stress by simply not catalyzing disulfide bonds in the exoplasm (14). An analysis of bacterial secretomes partially supported this hypothesis (14, 15). Low-GC bacteria, i.e., Firmicutes, were found to secrete few, if any, proteins with multiple Cys residues, which suggests that they lack disulfide bonds. Dsb-like proteins have been identified in some Firmicutes, like Bacillus, but substrates are unknown or arranged with their putative oxidoreductases in gene clusters (16–18). These current data suggest that Firmicutes do not rely on disulfide bond formation as a general tool to fold secreted proteins.
In contrast to Firmicutes, actinobacteria secrete an abundance of proteins with multiple Cys residues, which suggests that they possess oxidative folding pathways (14). Current efforts to elucidate disulfide bond formation in these organisms have focused heavily on Mycobacterium tuberculosis. Four Dsb-like factors, vitamin K epoxide reductase (VKOR), M. tuberculosis DsbA (Mt-DsbA), DsbE, and DsbF, have been identified in this bacterium (19–22). In vitro analyses of these enzymes have been extensive, but their biological functions are not clear. The ability to study these factors in vivo is probably hindered by the slow-growth phenotype of M. tuberculosis and a lack of facile genetic tools.
Recently, the oral pathogens Corynebacterium diphtheriae and Actinomyces oris were introduced as alternative models to study disulfide bond formation in actinobacteria. Using adhesive pili and diphtheria toxin as model substrates, these bacteria were revealed to possess membrane-localized disulfide-bond-forming systems led by the oxidoreductase MdbA (23, 24). Remarkably, mdbA mutants exhibit severe morphological defects, indicating that unlike known Gram-negative Dsb proteins, MdbA is important for growth. In addition, the corynebacterial mdbA mutant is attenuated in virulence due to defective toxin production and pilus assembly. Thus, MdbA may serve as a powerful target for new bactericidal drugs. This review will discuss disulfide bond formation in bacteria, with a special focus on recent efforts to elucidate oxidative folding pathways in actinobacteria. For detailed mechanisms on disulfide bond formation in Gram-negative bacteria, excellent reviews can be found in references 25 and 26.
OXIDATIVE PROTEIN FOLDING IN THE GRAM-NEGATIVE PERIPLASM
DsbA/DsbB disulfide-forming pathway.
The discovery of E. coli DsbA by Bardwell and colleagues (27) was serendipitous, since their experiments were originally designed to identify factors involved with membrane protein insertion. In this screen, cytoplasmic β-galactosidase (β-Gal) was fused to the N terminus of MalF, a known transmembrane protein. Under normal conditions, MalF–β-Gal is transported to the periplasm, but β-Gal attempts to reenter the cytoplasm and becomes embedded in the membrane (28). Blue/white screening was used to identify mutations preventing the initial translocation of this fusion protein. Surprisingly, a functional β-Gal was identified in a strain harboring a mutation in a secreted thioredoxin-like protein named DsbA. When functional, DsbA catalyzed nonnative disulfide bonds in β-Gal within the periplasm, which prevented its reentry into the cytoplasm. The loss of DsbA prevented these linkages from forming, which permitted the translocation of β-Gal.
E. coli DsbA is a 21-kDa monomeric protein that catalyzes the formation of disulfide bonds in unfolded proteins as they are secreted into the periplasm. DsbA harbors a canonical thioredoxin-like fold that is characterized by an N-terminal βαβ motif and a C-terminal ββα motif (29). However, unlike thioredoxin, these motifs are separated by an extended α-helical domain. The DsbA active site is composed of a reactive disulfide bond found in a CXXC (CPHC) consensus sequence that is located in the N terminus and abutted by a cis-proline (29). To catalyze new disulfide bonds, the CXXC linkage within DsbA is broken by a substrate Cys, resulting in the formation of a mixed intermediate (30, 31). In this state, DsbA may serve as a placeholder for substrate folding, as in the case of PDI (32). When folding is near completion, another substrate Cys is positioned to resolve the intermediate, which results in the formation of a new disulfide bond and release of the substrate. In turn, the DsbA CXXC motif is reduced (Fig. 1).
FIG 1.
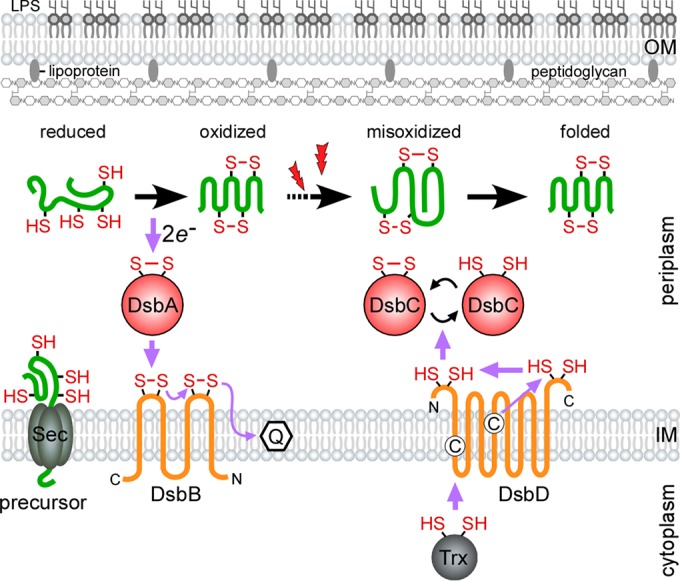
Oxidative protein folding in the Gram-negative periplasm. To catalyze oxidative protein folding, DsbA donates a reactive disulfide bond to reduced protein precursors as they are secreted into the oxidizing periplasm by SecYEG. Following catalysis, the DsbA active site is reoxidized by the transmembrane protein DsbB, which shuttles the gained electrons to the electron transport chain via a conjugated quinone. Extracellular oxidative stress (denoted by lightning bolts) or the lack of DsbA proofreading activity can cause substrates to become misoxidized. Aberrant disulfide bonds are reshuffled by the reductase DsbC. The reducing power of DsbC is maintained by the transmembrane DsbD, which receives electrons from cytoplasmic thioredoxin (adapted from reference 26). The purple arrows denote the direction of the electron flow, and the cysteine residues in the membrane domains of DsbD are shown as circled C's.
The ability of DsbA to form disulfide bonds is dependent on its high redox potential, or tendency to accept electrons (33). This intrinsic property is dependent on the pKa of the solvent-exposed Cys in the CXXC motif and a positively charged His residue in the CXXC consensus sequence (CPHC). The pKa of the solvent-exposed Cys is only 3.5, so it is negatively charged at physiological pH. This charge is stabilized by an electrostatic interaction with His (34, 35). His residues are strong indicators of the redox potential of CXXC motifs of thioredoxin-like proteins. Grauschopf et al. (36) found that substituting a nonpolar or negatively charged amino acid for His reduces the ability of DsbA to accept electrons in vitro. His residues are common features of other known disulfide-bond-forming enzymes. This amino acid is found in the CXXC motifs of eukaryotic PDI and DsbA equivalents in Salmonella, Shigella, Pseudomonas, and Neisseria species (37, 38). Although the strong oxidizing power of DsbA and DsbA-like proteins makes them effective disulfide-bond-forming enzymes, it presents a biological problem. These enzymes are more stable in their reduced forms, so it is unlikely that they spontaneously recycle their disulfide-bond-forming activity (39).
In E. coli, DsbA is regenerated by DsbB (Fig. 1), the discovery of which was simultaneously reported by Missiakas et al. (40) and Bardwell et al. (41). DsbB is a 20-kDa transmembrane protein that harbors two periplasmic loops with redox-active disulfide bonds. Using a series of disulfide exchanges, DsbB reoxidizes DsbA by shuttling electrons to a conjugated quinone, a component of the electron transport chain (42). Although the transfer of electrons between DsbA and DsbB is logical, their comparative redox potentials initially challenged this model. The redox potential of DsbB is lower than that of DsbA, which makes the flow of electrons between these enzymes unfavorable (43). An elegant structural study conducted by Inaba et al. (44) revealed that DsbB compensates for the difference in redox potential by a conformational change. Upon the transfer of DsbA's electrons to the C-terminal redox center, this region shifts toward the N-terminal redox site. This sterically hinders the flow of electrons back to DsbA, thus allowing the reaction to move forward.
Disulfide-reducing pathways.
E. coli DsbA favors the formation of consecutive disulfide bonds in substrates, i.e., oxidation of Cys residues in the order they emerge from the SecYEG (8). This strategy is flawed, since not all secreted proteins possess consecutive disulfide bonds. In these cases, DsbA will catalyze the formation of nonnative disulfide bonds, leading to misfolding. Due to its high redox potential, DsbA cannot reduce nonnative disulfide bonds to correct its errors (45).
Disulfide bond formation in the periplasm is monitored by DsbC (Fig. 1), a 26-kDa isomerase that harbors an N-terminal CXXC motif and C-terminal dimerization domain. This thioredoxin-like enzyme is required for the proper folding of secreted proteins with nonconsecutive disulfide bonds and proteins damaged by oxidative stress (9, 11). In its active form, the DsbC CXXC motif is reduced, which allows the transfer of electrons to misoxidized substrates (46). In turn, DsbC becomes oxidized and is reduced by the transmembrane protein DsbD (Fig. 1). DsbD harbors three domains, the N-terminal IgG-like periplasmic α-loop, the hydrophobic core-β, and the C-terminal thioredoxin-like periplasmic γ-loop, which participate in the transfer of electrons (47–49). The active site with Cys103 and Cys109 is located at the IgG-like domain (50). Electrons derived from cytoplasmic thioredoxin are transported through these domains and then delivered to the CXXC site of DsbC (51) (Fig. 1).
DsbC dimerization is essential for function. Mutational analysis revealed that this higher-order structure prevents cross talk between oxidative and reductive pathways in the periplasm. Bader et al. (52) found that the disruption of DsbC dimerization enables it to rescue an E. coli dsbA-null strain. This suggests that dimerization is required to prevent oxidization by DsbB. In support of this, attempts to model an interaction between DsbB and DsbC revealed a steric clash between one DsbC protomer and the cytoplasmic membrane (53).
In addition to maintaining DsbC in a reduced state, DsbD and DsbD homologs also shuttle electrons to DsbG, DsbE, and peroxiredoxins in the periplasm (54–56). Although DsbG has no known substrates, in vitro data suggest that it is a disulfide isomerase (54). Unlike other Dsb proteins, DsbE is not involved in general secreted protein folding. DsbE, also known as CcmG, is required for the synthesis of cytochrome c, a component of the electron transport chain (57). The reduction of peroxiredoxin is important, as this redox protein scavenges peroxides in the periplasm to combat oxidative stress (56).
Oxidative folding pathways in Gram-negative bacteria are nonessential but required for virulence.
Disulfide-bond-forming pathways in Gram-negative bacteria do not appear to be essential (27). E. coli dsbA mutants are slow growing in minimal medium, but this is attributed to a defect in glucose uptake, because glucose transporters require disulfide bonds (58). Disulfide isomerase pathways are also nonessential, since dsbC-null mutants grow normally under nonstress conditions (59). This is not due to redundancy between oxidative folding factors, since a dsbA dsbC dsbG triple-deletion mutant is also viable (59). dsbD-null mutants are temperature sensitive, but this phenotype is caused by a disruption in DsbE-led cytochrome c synthesis (60).
Although Dsb proteins are not generally important for growth, they are essential for pathogenesis. In addition to E. coli, other important Gram-negative pathogens, including Salmonella enterica, Shigella flexneri, Yersinia pestis, Bordetella pertussis, and Pseudomonas aeruginosa, secrete an arsenal of disulfide-bond-containing virulence factors (61). DsbA- and DsbA-like proteins expressed by these organisms are required for the proper folding of virulence factors, like adhesive pili, secretion systems, flagella, and toxins. dsbA mutations are often associated with decreased virulence, because disulfide-bond-containing secreted proteins are misfolded and degraded. For an in-depth review of this topic, see reference 61.
GRAM-POSITIVE DISULFIDE-BOND-FORMING PATHWAYS
The Firmicutes.
Although a space between the Gram-positive cytoplasmic membrane and cell wall has been observed by cryoelectron microscopy, it is not considered to be equivalent to that in the Gram-negative periplasm (12, 13). Due to the diffusive nature of peptidoglycan, it is possible that this space is exposed to the extracellular milieu. Therefore, environmental stress might cause aberrant oxidation in unfolded proteins with multiple Cys residues.
A survey of Gram-positive secreted proteomes revealed that many of these bacteria avoid oxidative folding stress by simply not utilizing disulfide bond formation (14, 15). The low-GC Firmicutes, including Staphylococcus, Lactobacillus, and Streptococcus spp., were found to secrete few, if any, proteins with two or more Cys residues, suggesting that these bacteria do not fold secreted proteins with disulfide bonds. In support of this, some Firmicutes, including Streptococcus pneumoniae, do not express any dsb-like genes (14, 15). One apparent exception to this trend is Bacillus (for a review, see reference 62). Bacillus brevis BdbA was the first DsbA-like protein discovered in this genus (16). Although BdbA has no known substrates, its overexpression was shown to rescue E. coli ΔdsbA phenotypes. While it is tempting to speculate that this protein is equivalent to DsbA, the overexpression of thioredoxin, DsbC, and DsbG, which are known to reduce proteins, in the periplasm rescues dsbA-null phenotypes (63–65). Therefore, one cannot conclude how B. brevis BdbA functions in vivo by expressing the protein in E. coli.
Bacillus subtilis harbors two gene clusters with the putative oxidoreductase-encoding genes bdbA to bdbD. bdbA and bdbB belong to an operon that encodes sublancin 168, an antibiotic with disulfide bonds (17). BdbA and BdbB, which are proposed to be analogous to DsbA and DsbB, respectively, catalyze disulfide bond formation in sublancin 168. bdbC and bdbD are contained within a competence gene cluster along with the disulfide-bond-containing ComCG pseudopilus (18). BdbC and BdbD, which are also proposed to be DsbB and DsbA equivalents, respectively, are required for production of the ComCG pseudopilus. Since B. subtilis bdb genes are genetically linked with their only known substrates, it is unlikely that they target other secreted proteins. In support of this, Bdb proteins in B. subtilis are not fully interchangeable (17). Furthermore, the transcription of bdbCD was found to be dependent on comX; although expression profiles of bdbCD have not been conducted, this observation suggests that the genes are transcribed in stationary phase. Therefore, BdbC and BdbD are probably not constitutively present to help fold the Bacillus secretome. It is more likely that the disulfide-bond-forming factors in Bacillus are exceptions that were acquired by horizontal gene transfer.
DsbA-like genes have also been identified in Staphylococcus aureus and Streptococcus gordonii (66, 67). S. aureus DsbA (SaDsbA) is a membrane-bound lipoprotein that was shown in vitro to have thiol-disulfide oxidoreductase activity (68). The deletion of S. aureus dsbA is associated with decreased levels of only the ComCG pseudopilus, indicating it may be functionally similar to Bacillus BdbD (69). Interestingly, biochemical analysis has revealed that the SaDsbA CXXC active site is equally stable in its oxidized and reduced forms, which indicates that it does not appear to require a DsbB-like partner (66). This characteristic is highly unusual, since known disulfide-bond-forming factors derive their oxidizing power from an unstable disulfide bond (39). If ComCG is one of few proteins secreted by S. aureus that require disulfide bond formation, it may not be necessary for SaSdbA to be a strong oxidizer. Further analysis is needed to elucidate the role of SaDsbA in vivo. Finally, a single DsbA-like factor has also been identified in S. gordonii (67). Similar to B. subtilis and S. aureus, this putative oxidoreductase has been implicated in competence development. Based upon the current bioinformatics data and limited in vivo evidence, while some Firmicutes appear to encode a few oxidoreductases for specific substrates, their secretomes generally do not rely on disulfide bonds for folding.
The Actinobacteria.
In contrast to the Firmicutes, Gram-positive actinobacteria, like Corynebacterium, Streptomyces, and Mycobacterium spp., secrete an abundance of Cys-containing proteins and encode redox proteins (14, 15). The exploration of disulfide bond formation within these organisms is a relatively recent endeavor and has heavily focused on M. tuberculosis. Disulfide bond formation is hypothesized to be a major folding pathway for this bacterium, as 60% of its secreted proteins contain two or more Cys residues (70). The first novel secreted oxidoreductase discovered in this bacterium was VKOR (15). VKOR is a quinone-conjugated transmembrane protein with two periplasmic loops containing redox-active disulfide bonds (22). Although expression of this enzyme can restore disulfide bond formation in an E. coli dsbB mutant, it is not a DsbB homologue (71). Rather, VKOR is related to mammalian VKOR, an enzyme involved in vitamin K recycling. Due to its functional similarity to E. coli DsbB, VKOR is predicted to reoxidize a DsbA-like enzyme, but its protein folding role in vivo has not been demonstrated.
In addition to M. tuberculosis VKOR, three secreted oxidoreductases, Mt-DsbA, Mt-DsbE, and Mt-DsbF, have also been identified. The structural and biochemical analyses of these proteins have been extensive. Although they are not identical to E. coli DsbA, all three factors display a canonical thioredoxin-like fold, N-terminal CXXC motif, and extended α-helical domain (19–21). Mt-DsbA, a membrane-anchored oxidoreductase, is encoded in an operon with the VKOR gene. Mt-DsbA has been shown to interact with VKOR-derived peptides in vitro, suggesting that the two factors form a redox pair (72). Since VKOR is a DsbB analogue, it is logical that Mt-DsbA would be equivalent to E. coli DsbA. In support of this, the redox potential of Mt-DsbA (−99 mV) is quite high, and the protein might oxidize a substrate in vitro, although it failed to reshuffle disulfide bonds in scrambled RNase A (72). All three traits are reminiscent of E. coli DsbA. However, a conflicting study reported that recombinant Mt-DsbA did not oxidize hirudin but successfully unscrambled RNase A (19). These discrepancies underscore the importance of studying Mt-DsbA in vivo. The identification of Mt-DsbA substrates would further enhance our understanding of its role in M. tuberculosis.
Efforts to study Mt-DsbA thus far have depended on the expression of the protein in E. coli. Interestingly, this factor cannot rescue a flagellar defect associated with the deletion of E. coli dsbA (72). The Mt-DsbA crystal structure revealed that the enzyme has a less flexible and hydrophobic catalytic binding site than that of E. coli DsbA (72). A restrictive binding cleft might have interfered with Mt-DsbA binding to E. coli DsbB for reoxidation. However, this possibility was not explored by testing if the expression of Mt-dsbA and the VKOR gene could restore oxidative protein folding in an E. coli dsbA dsbB double mutant. It was alternatively proposed that the Mt-DsbA binding cleft is optimized for specific substrates in M. tuberculosis, so it could not recognize the E. coli secretome (72). It is noteworthy that since the majority of proteins secreted by M. tuberculosis are predicted to contain disulfide bonds, a DsbA equivalent in this organism should be able to target an array of substrates.
If Mt-DsbA is limited in substrate recognition, M. tuberculosis expresses two additional oxidoreductases (Mt-DsbE and Mt-DsbF) that might participate in oxidative folding. Unlike Mt-DsbA, these factors are predicted to be secreted into the bacterial exoplasm (19). Mt-DsbE is hypothesized to oxidize proteins due to its relatively high redox potential (−128 mV) (20). The observation that Mt-DsbE can oxidize hirudin in vitro but exhibits no isomerase activity corroborates this conjecture. However, it should be noted that the redox potential of Mt-DsbE is similar to that of E. coli DsbC, which functions as a reductase (−130 mV) (73). The identification of an Mt-DsbE mutant phenotype is vital for determining its role in M. tuberculosis. Finally, Mt-DsbF has a higher calculated redox potential (−87 mV) than both Mt-DsbA and Mt-DsbE (21). This suggests that Mt-DsbF is the best suited for oxidizing secreted proteins. However, the gene encoding this factor is adjacent to a putative peroxiredoxin gene. Microarray data showed that these genes have similar expression profiles, and an in vitro pulldown experiment demonstrated that these proteins interact (21). It is possible that Mt-DsbA possesses a novel mechanism for disulfide bond formation or that it belongs to a different redox pathway in the cell. In summary, given the unknown biological functions of Mt-DsbA, Mt-DsbE, and Mt-DsbF, oxidative folding pathways in M. tuberculosis remain to be investigated.
ADHESIVE PILUS PROTEINS REVEAL OXIDATIVE PROTEIN-FOLDING PATHWAYS IN THE ACTINOBACTERIA A. ORIS AND C. DIPHTHERIAE
Recent investigations of pilus assembly in the oral pathogens A. oris and C. diphtheriae have advanced our understanding of disulfide bond formation in actinobacteria. Adhesive pili expressed by Gram-positive bacteria are host colonization factors composed of individual subunits that are covalently linked together and anchored to the cell surface by sortase enzymes (74). Prior to their assembly, pilus precursors are translocated to the exoplasm in unfolded states. Due to the lack of a recognizable periplasm, it was not known how these proteins attained their native conformations outside the cell. Structural studies of the major pilus proteins FimA and FimP expressed by A. oris and SpaA and SpaD of C. diphtheriae provided a clue (75–78). The presence of disulfide bonds in the crystal structures of all three pilins suggested that they required oxidative protein folding.
In vivo, these linkages are essential for pilus assembly. The failure to form disulfide bonds is associated with the absence of pili and the secretion of degradation products into the culture medium (23, 24). This suggests that the covalent linkages are important for the proper folding and/or stability of pilus precursors in the exoplasm. The discovery of disulfide bonds in FimA, FimP, and SpaA conferred a great advantage for studying protein oxidation in actinobacteria. Using A. oris and C. diphtheriae pilus proteins as model substrates, disulfide-bond-forming pathways vital for both virulence and growth were identified.
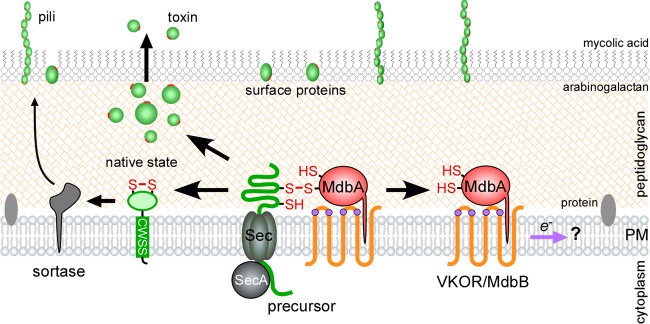
Using a combination of genetics, X-ray crystallization, and biochemical methods, the thiol-disulfide oxidoreductase enzyme called MdbA was identified in A. oris and C. diphtheriae (23, 24). In these organisms, it was shown that pilin precursors are oxidized by the membrane-bound oxidoreductase. In A. oris, MdbA activity is recycled by a VKOR homologue (23), while C. diphtheriae MdbA may be reoxidized by an unknown factor (24) (Fig. 2). MdbA shares low sequence homology with E. coli DsbA but is structurally similar. Crystal structures have revealed that both MdbA factors display thioredoxin-like folds and extended α-helical domains that most closely resemble B. subtilis BdbD and M. tuberculosis Mt-DsbA (23, 24). A. oris and C. diphtheriae MdbA enzymes also contain His residues in their CXXC motifs and possess neutral surface potentials near the predicted sites for substrate binding (23, 24). These observations suggest that MdbA exhibits strong redox potential and broad substrate specificity. Although these findings are supported by in vivo evidence, they have yet to be biochemically tested.
FIG 2.
Oxidative protein folding in the actinobacterial exoplasm. Using pilus proteins and diphtheria toxin as model substrates, oxidative protein-folding pathways have been proposed in the actinobacterial pathogens A. oris and C. diphtheriae. Unfolded pilin precursors are oxidized by the membrane-tethered thiol-disulfide oxidoreductase MdbA. Proper folding is a prerequisite for sortase-mediated assembly of pili on the cell surface. A. oris MdbA is reoxidized by the transmembrane VKOR, while C. diphtheriae MdbA is hypothesized to be recycled by an unidentified factor called MdbB. It is not clear how VKOR or MdbB is reoxidized. The catalytic cysteine residues of VKOR are shown as purple circles, disulfide bonds formed in mature proteins are shown as red lines, and the purple arrow indicates the presumed direction of the electron flow (modified after references 23 and 24).
Since >60% of proteins secreted by A. oris and C. diphtheriae are predicted to contain at least one disulfide linkage (38, 79), it was hypothesized that MdbA proteins target multiple substrates. The disulfide-bond-containing diphtheria toxin (DT) was chosen as an additional model substrate. The deletion of mdbA was associated with the release of reduced and degraded DT. This phenotype, along with the lack of adhesive pili, had profound consequences on C. diphtheriae pathogenesis, as the mdbA mutant was attenuated in a guinea pig model of infection (24). The combined data indicated that MdbA is important for the general folding of secreted virulence factors with disulfide bonds.
Unlike E. coli DsbA and known Gram-negative DsbA-like enzymes, MdbA is also required for proper growth and division. This remarkable feature was discovered when multiple attempts to generate an A. oris mdbA deletion mutant failed. A conditional mdbA deletion mutant was created as an alternative by placing mdbA under the control of an inducible promoter. Decreased mdbA expression was associated with a cell-chaining and altered morphology (23). This surprising phenotype suggested that MdbA has a much broader role in protein folding (i.e., it targets secreted proteins involved with cell division). In contrast, a C. diphtheriae mdbA deletion mutant was successfully generated but resulted in a severe temperature-sensitive defect. The bacteria were observed to grow normally at 30°C but became coccoid, chained, and eventually stopped growing when the temperature was shifted to 37°C (24).
The misfolding of secreted penicillin-binding proteins (PBPs), which contain multiple Cys residues, is one potential basis for the mdbA growth phenotypes. PBPs synthesize peptidoglycan, a determinant of cell shape (80). The inhibition of cell wall synthesis and removal of existing peptidoglycan are known to transform rod-shaped bacteria, like E. coli, Corynebacterium glutamicum, and Bacillus, into cocci (81–84). It is proposed that C. diphtheriae and A. oris PBPs misfold in the absence of MdbA, which prevents normal growth. In support of this, the C. diphtheriae ΔmdbA mutant is more susceptible to antibiotics that target PBP function than the wild-type strain and exhibits abnormal vancomycin BODIPY-FL (Van-FL) staining (24). The reason why A. oris mdbA is essential while C. diphtheriae mdbA is important only for growth at 37°C is not known. It is possible that C. diphtheriae possesses some capability for background protein oxidation that is sufficient for growth at lower temperatures.
Finally, the housekeeping role of disulfide-bond-forming enzymes in actinobacteria is not limited to A. oris and C. diphtheriae. An M. tuberculosis transposon library generated by Sassetti et al. (85) revealed a low insertion frequency within the Mt-DsbA and VKOR genes, suggesting that the genes are important for growth. Consistent with this, slow-growth phenotypes of strains with VKOR gene deletions were observed in M. tuberculosis and Mycobacterium smegmatis (15, 71). These findings suggest that the bacterial thiol-disulfide oxidoreductases would provide excellent targets for the development of antimicrobials. Since dsbA mutants are attenuated in rodent models of infection (86, 87), efforts have recently been made to identify inhibitors of DsbA/DsbB as antivirulence agents. By screening a library of 1,123 fragments for compounds that bind to oxidized DsbA, Adams and colleagues (88) identified a small set of molecules that exhibit high binding affinity to DsbA and inhibit its activity in vitro. One of the compounds was shown to reduce bacterial motility, but it did not affect cell growth (88). Using a virtual screening approach, Duprez et al. (89) identified a set of noncovalent inhibitors of DsbA that exhibit inhibitory activity at millimolar concentrations (89). While the first two approaches are aimed at DsbA, the Beckwith group (90) employed a cell-based screening method to find compounds that target DsbB and revealed several inhibitors with a pyridazinone core. With further modification using a medicinal chemistry approach, they selected more potent inhibitors that have a broad spectrum for inhibition of DsbB in many other Gram-negative bacteria. Finally, by interfering with the DsbB-catalyzed recycling of DsbA, Halili and coworkers (91) synthesized analogues of ubiquinone and found a couple of dimedone derivatives with high inhibitory activity (50% inhibitory concentration [IC50], ∼1 μM) to E. coli DsbA, but none of these compounds inhibited human thioredoxin (91). While the identification of disulfide-bond-forming inhibitors is encouraging, it remains to be seen if these compounds have any antivirulence activity in vivo.
PERSPECTIVES
It was previously proposed that Gram-positive bacteria do not possess periplasmic compartments to regulate the folding of secreted proteins. While this may be true for Firmicutes, actinobacteria may be exceptions. Corynebacteria and mycobacteria exhibit unique cell envelope architectures in which peptidoglycan is cross-linked to arabinogalactan, which is esterifed by mycolic acid (92). Mycolic acid, a type of long-chain fatty acid, forms a hydrophobic surface layer that is visible by thin-section electron microscopy (EM) (93). This layer contributes to the high impermeability of corynebacteria and is known to form liposomes when cells are treated with detergent (93, 94). This layer of fatty acids is proposed to form a so-called mycomembrane, which may be analogous to the Gram-negative outer membrane. It is possible that some Gram-positives, like corynebacteria, contain enclosed compartments to protect thiol-containing secreted proteins from aberrant oxidation.
A. oris is not known to produce mycolic acid. It is possible that this organism possesses an outer lipid layer, but its ultrastructure has not been examined. Alternatively, it is also likely that its environment contributes to its oxidation of proteins in the exoplasm. A. oris, a pioneer colonizer of the oral cavity, inhabits the anaerobic layers of mature biofilm (95, 96). The absence of oxygen in this niche may allow the bacterium to avoid random oxidation of secreted proteins.
Finally, actinobacteria might avoid random Cys oxidation by coordinating translocation and folding events. In C. diphtheriae, the secretion and assembly of pilus subunits is thought to be a tightly coupled process. This is supported by thin-section microscopic data showing that Sec and sortase machinery colocalize (97). In addition, pilin precursors missing their C-terminal membrane anchors are still incorporated into pilus structures, suggesting that the subunits were processed by sortase during or immediately after translocation (98, 99). If MdbA must fold pilins, such as FimA, FimP, SpaA, and SpaD, before sortase-catalyzed assembly, it must also colocalize with Sec machinery. The ability of MdbA to oxidize substrates as they emerge from the cytoplasm might serve as an adaptation to the secretion of proteins into unfavorable environments. Coupling translocation with protein folding may increase the likelihood that disulfide bonds are catalyzed by cellular machinery rather than the extracellular milieu (Fig. 2).
In summary, disulfide bond formation is an important mechanism to fold many secreted proteins. Oxidative protein-folding pathways have been widely studied in Gram-negative bacteria. However, due to the lack of a periplasmic space, disulfide bond formation largely has been unexplored in Gram-positive bacteria. Observations that disulfide-bond-containing proteins are rare and often found in operons with putative oxidoreductases suggest that these bacteria do not rely on disulfide-bond-forming pathways. Bioinformatics analyses proposed that while many Gram-positive bacteria do not use protein oxidation, actinobacteria may be exceptions (14, 15). However, elucidation of the disulfide-bond-forming pathways in these organisms was impeded by a lack of in vivo work. Recent investigations of virulence factors secreted by the oral pathogens A. oris and C. diphtheriae have advanced our understanding of protein oxidation in actinobacteria. Not only are disulfide-bond-forming factors required for virulence in these organisms, but they are also required for proper growth and division. This unexpected discovery might pave the way for new antimicrobial drugs targeting important actinobacterial pathogens, like M. tuberculosis. Furthermore, oxidative protein folding appears to be an Achilles heel of antibiotic resistance in these pathogens, as a mutant defective in disulfide bond formation is highly sensitive to β-lactam antibiotics (24). Perhaps a combination of disulfide-bond-forming inhibitors and known antibiotics would enhance the efficacy of antibiotics and curtail antibiotic resistance.
ACKNOWLEDGMENTS
We thank our laboratory members for critical review of the manuscript and three reviewers for their excellent comments and suggestions.
REFERENCES
- 1.Anfinsen CB, Haber E, Sela M, White FH Jr. 1961. The kinetics of formation of native ribonuclease during oxidation of the reduced polypeptide chain. Proc Natl Acad Sci U S A 47:1309–1314. doi: 10.1073/pnas.47.9.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldberger RF, Epstein CJ, Anfinsen CB. 1963. Acceleration of reactivation of reduced bovine pancreatic ribonuclease by a microsomal system from rat liver. J Biol Chem 238:628–635. [PubMed] [Google Scholar]
- 3.Givol D, Goldberger RF, Anfinsen CB. 1964. Oxidation and disulfide interchange in the reactivation of reduced ribonuclease. J Biol Chem 239:PC3114–PC3116. [PubMed] [Google Scholar]
- 4.Tian G, Xiang S, Noiva R, Lennarz WJ, Schindelin H. 2006. The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites. Cell 124:61–73. doi: 10.1016/j.cell.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 5.Creighton TE, Zapun A, Darby NJ. 1995. Mechanisms and catalysts of disulfide bond formation in proteins. Trends Biotechnol 13:18–23. doi: 10.1016/S0167-7799(00)88896-4. [DOI] [PubMed] [Google Scholar]
- 6.Mamathambika BS, Bardwell JC. 2008. Disulfide-linked protein folding pathways. Annu Rev Cell Dev Biol 24:211–235. doi: 10.1146/annurev.cellbio.24.110707.175333. [DOI] [PubMed] [Google Scholar]
- 7.Denoncin K, Collet JF. 2013. Disulfide bond formation in the bacterial periplasm: major achievements and challenges ahead. Antioxid Redox Signal 19:63–71. doi: 10.1089/ars.2012.4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kadokura H, Beckwith J. 2009. Detecting folding intermediates of a protein as it passes through the bacterial translocation channel. Cell 138:1164–1173. doi: 10.1016/j.cell.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiniker A, Collet JF, Bardwell JC. 2005. Copper stress causes an in vivo requirement for the Escherichia coli disulfide isomerase DsbC. J Biol Chem 280:33785–33791. doi: 10.1074/jbc.M505742200. [DOI] [PubMed] [Google Scholar]
- 10.Shao F, Bader MW, Jakob U, Bardwell JC. 2000. DsbG, a protein disulfide isomerase with chaperone activity. J Biol Chem 275:13349–13352. doi: 10.1074/jbc.275.18.13349. [DOI] [PubMed] [Google Scholar]
- 11.Hiniker A, Bardwell JC. 2004. In vivo substrate specificity of periplasmic disulfide oxidoreductases. J Biol Chem 279:12967–12973. doi: 10.1074/jbc.M311391200. [DOI] [PubMed] [Google Scholar]
- 12.Matias VR, Beveridge TJ. 2006. Native cell wall organization shown by cryo-electron microscopy confirms the existence of a periplasmic space in Staphylococcus aureus. J Bacteriol 188:1011–1021. doi: 10.1128/JB.188.3.1011-1021.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matias VR, Beveridge TJ. 2005. Cryo-electron microscopy reveals native polymeric cell wall structure in Bacillus subtilis 168 and the existence of a periplasmic space. Mol Microbiol 56:240–251. doi: 10.1111/j.1365-2958.2005.04535.x. [DOI] [PubMed] [Google Scholar]
- 14.Daniels R, Mellroth P, Bernsel A, Neiers F, Normark S, von Heijne G, Henriques-Normark B. 2010. Disulfide bond formation and cysteine exclusion in Gram-positive bacteria. J Biol Chem 285:3300–3309. doi: 10.1074/jbc.M109.081398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutton RJ, Boyd D, Berkmen M, Beckwith J. 2008. Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc Natl Acad Sci U S A 105:11933–11938. doi: 10.1073/pnas.0804621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishihara T, Tomita H, Hasegawa Y, Tsukagoshi N, Yamagata H, Udaka S. 1995. Cloning and characterization of the gene for a protein thiol-disulfide oxidoreductase in Bacillus brevis. J Bacteriol 177:745–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorenbos R, Stein T, Kabel J, Bruand C, Bolhuis A, Bron S, Quax WJ, Van Dijl JM. 2002. Thiol-disulfide oxidoreductases are essential for the production of the lantibiotic sublancin 168. J Biol Chem 277:16682–16688. doi: 10.1074/jbc.M201158200. [DOI] [PubMed] [Google Scholar]
- 18.Meima R, Eschevins C, Fillinger S, Bolhuis A, Hamoen LW, Dorenbos R, Quax WJ, van Dijl JM, Provvedi R, Chen I, Dubnau D, Bron S. 2002. The bdbDC operon of Bacillus subtilis encodes thiol-disulfide oxidoreductases required for competence development. J Biol Chem 277:6994–7001. doi: 10.1074/jbc.M111380200. [DOI] [PubMed] [Google Scholar]
- 19.Chim N, Harmston CA, Guzman DJ, Goulding CW. 2013. Structural and biochemical characterization of the essential DsbA-like disulfide bond forming protein from Mycobacterium tuberculosis. BMC Struct Biol 13:23. doi: 10.1186/1472-6807-13-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goulding CW, Apostol MI, Gleiter S, Parseghian A, Bardwell J, Gennaro M, Eisenberg D. 2004. Gram-positive DsbE proteins function differently from Gram-negative DsbE homologs. A structure to function analysis of DsbE from Mycobacterium tuberculosis. J Biol Chem 279:3516–3524. [DOI] [PubMed] [Google Scholar]
- 21.Chim N, Riley R, The J, Im S, Segelke B, Lekin T, Yu M, Hung LW, Terwilliger T, Whitelegge JP, Goulding CW. 2010. An extracellular disulfide bond forming protein (DsbF) from Mycobacterium tuberculosis: structural, biochemical, and gene expression analysis. J Mol Biol 396:1211–1226. doi: 10.1016/j.jmb.2009.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, Schulman S, Dutton RJ, Boyd D, Beckwith J, Rapoport TA. 2010. Structure of a bacterial homologue of vitamin K epoxide reductase. Nature 463:507–512. doi: 10.1038/nature08720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reardon-Robinson ME, Osipiuk J, Chang C, Wu C, Jooya N, Joachimiak A, Das A, Ton-That H. 2015. A disulfide bond-forming machine is linked to the sortase-mediated pilus assembly pathway in the Gram-positive bacterium Actinomyces oris. J Biol Chem 290:21393–21405. doi: 10.1074/jbc.M115.672253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reardon-Robinson ME, Osipiuk J, Jooya N, Chang C, Joachimiak A, Das A, Ton-That H. 21 August 2015. A thiol-disulfide oxidoreductase of the Gram-positive pathogen Corynebacterium diphtheriae is essential for viability, pilus assembly, toxin production and virulence. Mol Microbiol doi: 10.1111/mmi.13172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goemans C, Denoncin K, Collet JF. 2014. Folding mechanisms of periplasmic proteins. Biochim Biophys Acta 1843:1517–1528. doi: 10.1016/j.bbamcr.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 26.Kadokura H, Beckwith J. 2010. Mechanisms of oxidative protein folding in the bacterial cell envelope. Antioxid Redox Signal 13:1231–1246. doi: 10.1089/ars.2010.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bardwell JC, McGovern K, Beckwith J. 1991. Identification of a protein required for disulfide bond formation in vivo. Cell 67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 28.Froshauer S, Green GN, Boyd D, McGovern K, Beckwith J. 1988. Genetic analysis of the membrane insertion and topology of MalF, a cytoplasmic membrane protein of Escherichia coli. J Mol Biol 200:501–511. doi: 10.1016/0022-2836(88)90539-6. [DOI] [PubMed] [Google Scholar]
- 29.Martin JL, Bardwell JC, Kuriyan J. 1993. Crystal structure of the DsbA protein required for disulphide bond formation in vivo. Nature 365:464–468. doi: 10.1038/365464a0. [DOI] [PubMed] [Google Scholar]
- 30.Kadokura H, Tian H, Zander T, Bardwell JC, Beckwith J. 2004. Snapshots of DsbA in action: detection of proteins in the process of oxidative folding. Science 303:534–537. doi: 10.1126/science.1091724. [DOI] [PubMed] [Google Scholar]
- 31.Darby NJ, Creighton TE. 1995. Catalytic mechanism of DsbA and its comparison with that of protein disulfide isomerase. Biochemistry 34:3576–3587. doi: 10.1021/bi00011a012. [DOI] [PubMed] [Google Scholar]
- 32.Kosuri P, Alegre-Cebollada J, Feng J, Kaplan A, Ingles-Prieto A, Badilla CL, Stockwell BR, Sanchez-Ruiz JM, Holmgren A, Fernandez JM. 2012. Protein folding drives disulfide formation. Cell 151:794–806. doi: 10.1016/j.cell.2012.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zapun A, Bardwell JC, Creighton TE. 1993. The reactive and destabilizing disulfide bond of DsbA, a protein required for protein disulfide bond formation in vivo. Biochemistry 32:5083–5092. doi: 10.1021/bi00070a016. [DOI] [PubMed] [Google Scholar]
- 34.Guddat LW, Bardwell JC, Glockshuber R, Huber-Wunderlich M, Zander T, Martin JL. 1997. Structural analysis of three His32 mutants of DsbA: support for an electrostatic role of His32 in DsbA stability. Protein Sci 6:1893–1900. doi: 10.1002/pro.5560060910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson JW, Creighton TE. 1994. Reactivity and ionization of the active site cysteine residues of DsbA, a protein required for disulfide bond formation in vivo. Biochemistry 33:5974–5983. doi: 10.1021/bi00185a039. [DOI] [PubMed] [Google Scholar]
- 36.Grauschopf U, Winther JR, Korber P, Zander T, Dallinger P, Bardwell JC. 1995. Why is DsbA such an oxidizing disulfide catalyst? Cell 83:947–955. doi: 10.1016/0092-8674(95)90210-4. [DOI] [PubMed] [Google Scholar]
- 37.Lu X, Gilbert HF, Harper JW. 1992. Conserved residues flanking the thiol/disulfide centers of protein disulfide isomerase are not essential for catalysis of thiol/disulfide exchange. Biochemistry 31:4205–4210. doi: 10.1021/bi00132a008. [DOI] [PubMed] [Google Scholar]
- 38.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wunderlich M, Jaenicke R, Glockshuber R. 1993. The redox properties of protein disulfide isomerase (DsbA) of Escherichia coli result from a tense conformation of its oxidized form. J Mol Biol 233:559–566. doi: 10.1006/jmbi.1993.1535. [DOI] [PubMed] [Google Scholar]
- 40.Missiakas D, Georgopoulos C, Raina S. 1993. Identification and characterization of the Escherichia coli gene dsbB, whose product is involved in the formation of disulfide bonds in vivo. Proc Natl Acad Sci U S A 90:7084–7088. doi: 10.1073/pnas.90.15.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bardwell JC, Lee JO, Jander G, Martin N, Belin D, Beckwith J. 1993. A pathway for disulfide bond formation in vivo. Proc Natl Acad Sci U S A 90:1038–1042. doi: 10.1073/pnas.90.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kobayashi T, Kishigami S, Sone M, Inokuchi H, Mogi T, Ito K. 1997. Respiratory chain is required to maintain oxidized states of the DsbA-DsbB disulfide bond formation system in aerobically growing Escherichia coli cells. Proc Natl Acad Sci U S A 94:11857–11862. doi: 10.1073/pnas.94.22.11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Inaba K, Ito K. 2002. Paradoxical redox properties of DsbB and DsbA in the protein disulfide-introducing reaction cascade. EMBO J 21:2646–2654. doi: 10.1093/emboj/21.11.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inaba K, Murakami S, Suzuki M, Nakagawa A, Yamashita E, Okada K, Ito K. 2006. Crystal structure of the DsbB-DsbA complex reveals a mechanism of disulfide bond generation. Cell 127:789–801. doi: 10.1016/j.cell.2006.10.034. [DOI] [PubMed] [Google Scholar]
- 45.Zapun A, Creighton TE. 1994. Effects of DsbA on the disulfide folding of bovine pancreatic trypsin inhibitor and alpha-lactalbumin. Biochemistry 33:5202–5211. doi: 10.1021/bi00183a025. [DOI] [PubMed] [Google Scholar]
- 46.Rietsch A, Bessette P, Georgiou G, Beckwith J. 1997. Reduction of the periplasmic disulfide bond isomerase, DsbC, occurs by passage of electrons from cytoplasmic thioredoxin. J Bacteriol 179:6602–6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stewart EJ, Katzen F, Beckwith J. 1999. Six conserved cysteines of the membrane protein DsbD are required for the transfer of electrons from the cytoplasm to the periplasm of Escherichia coli. EMBO J 18:5963–5971. doi: 10.1093/emboj/18.21.5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katzen F, Beckwith J. 2000. Transmembrane electron transfer by the membrane protein DsbD occurs via a disulfide bond cascade. Cell 103:769–779. doi: 10.1016/S0092-8674(00)00180-X. [DOI] [PubMed] [Google Scholar]
- 49.Krupp R, Chan C, Missiakas D. 2001. DsbD-catalyzed transport of electrons across the membrane of Escherichia coli. J Biol Chem 276:3696–3701. doi: 10.1074/jbc.M009500200. [DOI] [PubMed] [Google Scholar]
- 50.Goulding CW, Sawaya MR, Parseghian A, Lim V, Eisenberg D, Missiakas D. 2002. Thiol-disulfide exchange in an immunoglobulin-like fold: structure of the N-terminal domain of DsbD. Biochemistry 41:6920–6927. doi: 10.1021/bi016038l. [DOI] [PubMed] [Google Scholar]
- 51.Rietsch A, Belin D, Martin N, Beckwith J. 1996. An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc Natl Acad Sci U S A 93:13048–13053. doi: 10.1073/pnas.93.23.13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bader MW, Hiniker A, Regeimbal J, Goldstone D, Haebel PW, Riemer J, Metcalf P, Bardwell JC. 2001. Turning a disulfide isomerase into an oxidase: DsbC mutants that imitate DsbA. EMBO J 20:1555–1562. doi: 10.1093/emboj/20.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pan JL, Sliskovic I, Bardwell JC. 2008. Mutants in DsbB that appear to redirect oxidation through the disulfide isomerization pathway. J Mol Biol 377:1433–1442. doi: 10.1016/j.jmb.2008.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bessette PH, Cotto JJ, Gilbert HF, Georgiou G. 1999. In vivo and in vitro function of the Escherichia coli periplasmic cysteine oxidoreductase DsbG. J Biol Chem 274:7784–7792. doi: 10.1074/jbc.274.12.7784. [DOI] [PubMed] [Google Scholar]
- 55.Reid E, Cole J, Eaves DJ. 2001. The Escherichia coli CcmG protein fulfils a specific role in cytochrome c assembly. Biochem J 355:51–58. doi: 10.1042/0264-6021:3550051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cho SH, Parsonage D, Thurston C, Dutton RJ, Poole LB, Collet JF, Beckwith J. 2012. A new family of membrane electron transporters and its substrates, including a new cell envelope peroxiredoxin, reveal a broadened reductive capacity of the oxidative bacterial cell envelope. mBio 3(2):e00291-11. doi: 10.1128/mBio.00291-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fabianek RA, Hennecke H, Thöny-Meyer L. 1998. The active-site cysteines of the periplasmic thioredoxin-like protein CcmG of Escherichia coli are important but not essential for cytochrome c maturation in vivo. J Bacteriol 180:1947–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bessette PH, Qiu J, Bardwell JC, Swartz JR, Georgiou G. 2001. Effect of sequences of the active-site dipeptides of DsbA and DsbC on in vivo folding of multidisulfide proteins in Escherichia coli. J Bacteriol 183:980–988. doi: 10.1128/JB.183.3.980-988.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vertommen D, Depuydt M, Pan J, Leverrier P, Knoops L, Szikora JP, Messens J, Bardwell JC, Collet JF. 2008. The disulphide isomerase DsbC cooperates with the oxidase DsbA in a DsbD-independent manner. Mol Microbiol 67:336–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Missiakas D, Schwager F, Raina S. 1995. Identification and characterization of a new disulfide isomerase-like protein (DsbD) in Escherichia coli. EMBO J 14:3415–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heras B, Shouldice SR, Totsika M, Scanlon MJ, Schembri MA, Martin JL. 2009. DSB proteins and bacterial pathogenicity. Nat Rev Microbiol 7:215–225. doi: 10.1038/nrmicro2087. [DOI] [PubMed] [Google Scholar]
- 62.Sarvas M, Harwood CR, Bron S, van Dijl JM. 2004. Post-translocational folding of secretory proteins in Gram-positive bacteria. Biochim Biophys Acta 1694:311–327. [DOI] [PubMed] [Google Scholar]
- 63.Debarbieux L, Beckwith J. 1998. The reductive enzyme thioredoxin 1 acts as an oxidant when it is exported to the Escherichia coli periplasm. Proc Natl Acad Sci U S A 95:10751–10756. doi: 10.1073/pnas.95.18.10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Missiakas D, Georgopoulos C, Raina S. 1994. The Escherichia coli dsbC (xprA) gene encodes a periplasmic protein involved in disulfide bond formation. EMBO J 13:2013–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andersen CL, Matthey-Dupraz A, Missiakas D, Raina S. 1997. A new Escherichia coli gene, dsbG, encodes a periplasmic protein involved in disulphide bond formation, required for recycling DsbA/DsbB and DsbC redox proteins. Mol Microbiol 26:121–132. doi: 10.1046/j.1365-2958.1997.5581925.x. [DOI] [PubMed] [Google Scholar]
- 66.Heras B, Kurz M, Jarrott R, Shouldice SR, Frei P, Robin G, Cemazar M, Thony-Meyer L, Glockshuber R, Martin JL. 2008. Staphylococcus aureus DsbA does not have a destabilizing disulfide. A new paradigm for bacterial oxidative folding. J Biol Chem 283:4261–4271. [DOI] [PubMed] [Google Scholar]
- 67.Davey L, Ng CK, Halperin SA, Lee SF. 2013. Functional analysis of paralogous thiol-disulfide oxidoreductases in Streptococcus gordonii. J Biol Chem 288:16416–16429. doi: 10.1074/jbc.M113.464578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dumoulin A, Grauschopf U, Bischoff M, Thony-Meyer L, Berger-Bachi B. 2005. Staphylococcus aureus DsbA is a membrane-bound lipoprotein with thiol-disulfide oxidoreductase activity. Arch Microbiol 184:117–128. doi: 10.1007/s00203-005-0024-1. [DOI] [PubMed] [Google Scholar]
- 69.van der Kooi-Pol MM, Reilman E, Sibbald MJ, Veenstra-Kyuchukova YK, Kouwen TR, Buist G, van Dijl JM. 2012. Requirement of signal peptidase ComC and thiol-disulfide oxidoreductase DsbA for optimal cell surface display of pseudopilin ComGC in Staphylococcus aureus. Appl Environ Microbiol 78:7124–7127. doi: 10.1128/AEM.01565-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schroder E, Ponting CP. 1998. Evidence that peroxiredoxins are novel members of the thioredoxin fold superfamily. Protein Sci 7:2465–2468. doi: 10.1002/pro.5560071125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang X, Dutton RJ, Beckwith J, Boyd D. 2011. Membrane topology and mutational analysis of Mycobacterium tuberculosis VKOR, a protein involved in disulfide bond formation and a homologue of human vitamin K epoxide reductase. Antioxid Redox Signal 14:1413–1420. doi: 10.1089/ars.2010.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Premkumar L, Heras B, Duprez W, Walden P, Halili M, Kurth F, Fairlie DP, Martin JL. 2013. Rv2969c, essential for optimal growth in Mycobacterium tuberculosis, is a DsbA-like enzyme that interacts with VKOR-derived peptides and has atypical features of DsbA-like disulfide oxidases. Acta Crystallogr D Biol Crystallogr 69:1981–1994. doi: 10.1107/S0907444913017800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zapun A, Missiakas D, Raina S, Creighton TE. 1995. Structural and functional characterization of DsbC, a protein involved in disulfide bond formation in Escherichia coli. Biochemistry 34:5075–5089. doi: 10.1021/bi00015a019. [DOI] [PubMed] [Google Scholar]
- 74.Hendrickx AP, Budzik JM, Oh SY, Schneewind O. 2011. Architects at the bacterial surface—sortases and the assembly of pili with isopeptide bonds. Nat Rev Microbiol 9:166–176. doi: 10.1038/nrmicro2520. [DOI] [PubMed] [Google Scholar]
- 75.Kang HJ, Paterson NG, Gaspar AH, Ton-That H, Baker EN. 2009. The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds. Proc Natl Acad Sci U S A 106:16967–16971. doi: 10.1073/pnas.0906826106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mishra A, Devarajan B, Reardon ME, Dwivedi P, Krishnan V, Cisar JO, Das A, Narayana SV, Ton-That H. 2011. Two autonomous structural modules in the fimbrial shaft adhesin FimA mediate Actinomyces interactions with streptococci and host cells during oral biofilm development. Mol Microbiol 81:1205–1220. doi: 10.1111/j.1365-2958.2011.07745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Persson K, Esberg A, Claesson R, Stromberg N. 2012. The pilin protein FimP from Actinomyces oris: crystal structure and sequence analyses. PLoS One 7:e48364. doi: 10.1371/journal.pone.0048364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kang HJ, Paterson NG, Kim CU, Middleditch M, Chang C, Ton-That H, Baker EN. 2014. A slow-forming isopeptide bond in the structure of the major pilin SpaD from Corynebacterium diphtheriae has implications for pilus assembly. Acta Crystallogr D Biol Crystallogr 70:1190–1201. doi: 10.1107/S1399004714001400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen T, Abbey K, Deng WJ, Cheng MC. 2005. The bioinformatics resource for oral pathogens. Nucleic Acids Res 33:W734–W740. doi: 10.1093/nar/gki361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cava F, de Pedro MA. 2014. Peptidoglycan plasticity in bacteria: emerging variability of the murein sacculus and their associated biological functions. Curr Opin Microbiol 18:46–53. doi: 10.1016/j.mib.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 81.Valbuena N, Letek M, Ordonez E, Ayala J, Daniel RA, Gil JA, Mateos LM. 2007. Characterization of HMW-PBPs from the rod-shaped actinomycete Corynebacterium glutamicum: peptidoglycan synthesis in cells lacking actin-like cytoskeletal structures. Mol Microbiol 66:643–657. doi: 10.1111/j.1365-2958.2007.05943.x. [DOI] [PubMed] [Google Scholar]
- 82.Botta GA, Buffa D. 1981. Murein synthesis and beta-lactam antibiotic susceptibility during rod-to-sphere transition in a pbpA(Ts) mutant of Escherichia coli. Antimicrob Agents Chemother 19:891–900. doi: 10.1128/AAC.19.5.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tomasz A. 1979. The mechanism of the irreversible antimicrobial effects of penicillins: how the beta-lactam antibiotics kill and lyse bacteria. Annu Rev Microbiol 33:113–137. doi: 10.1146/annurev.mi.33.100179.000553. [DOI] [PubMed] [Google Scholar]
- 84.Carballido-López R, Formstone A. 2007. Shape determination in Bacillus subtilis. Curr Opin Microbiol 10:611–616. doi: 10.1016/j.mib.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 85.Sassetti CM, Boyd DH, Rubin EJ. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol 48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 86.Ireland PM, McMahon RM, Marshall LE, Halili M, Furlong E, Tay S, Martin JL, Sarkar-Tyson M. 2014. Disarming Burkholderia pseudomallei: structural and functional characterization of a disulfide oxidoreductase (DsbA) required for virulence in vivo. Antioxid Redox Signal 20:606–617. doi: 10.1089/ars.2013.5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rosadini CV, Wong SM, Akerley BJ. 2008. The periplasmic disulfide oxidoreductase DsbA contributes to Haemophilus influenzae pathogenesis. Infect Immun 76:1498–1508. doi: 10.1128/IAI.01378-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Adams LA, Sharma P, Mohanty B, Ilyichova OV, Mulcair MD, Williams ML, Gleeson EC, Totsika M, Doak BC, Caria S, Rimmer K, Horne J, Shouldice SR, Vazirani M, Headey SJ, Plumb BR, Martin JL, Heras B, Simpson JS, Scanlon MJ. 2015. Application of fragment-based screening to the design of inhibitors of Escherichia coli DsbA. Angew Chem Int Ed Engl 54:2179–2184. doi: 10.1002/anie.201410341. [DOI] [PubMed] [Google Scholar]
- 89.Duprez W, Premkumar L, Halili MA, Lindahl F, Reid RC, Fairlie DP, Martin JL. 2015. Peptide inhibitors of the Escherichia coli DsbA oxidative machinery essential for bacterial virulence. J Med Chem 58:577–587. doi: 10.1021/jm500955s. [DOI] [PubMed] [Google Scholar]
- 90.Landeta C, Blazyk JL, Hatahet F, Meehan BM, Eser M, Myrick A, Bronstain L, Minami S, Arnold H, Ke N, Rubin EJ, Furie BC, Furie B, Beckwith J, Dutton R, Boyd D. 2015. Compounds targeting disulfide bond forming enzyme DsbB of Gram-negative bacteria. Nat Chem Biol 11:292–298. doi: 10.1038/nchembio.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Halili MA, Bachu P, Lindahl F, Bechara C, Mohanty B, Reid RC, Scanlon MJ, Robinson CV, Fairlie DP, Martin JL. 2015. Small molecule inhibitors of disulfide bond formation by the bacterial DsbA-DsbB dual enzyme system. ACS Chem Biol 10:957–964. doi: 10.1021/cb500988r. [DOI] [PubMed] [Google Scholar]
- 92.Marrakchi H, Laneelle MA, Daffe M. 2014. Mycolic acids: structures, biosynthesis, and beyond. Chem Biol 21:67–85. doi: 10.1016/j.chembiol.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 93.Bayan N, Houssin C, Chami M, Leblon G. 2003. Mycomembrane and S-layer: two important structures of Corynebacterium glutamicum cell envelope with promising biotechnology applications. J Biotechnol 104:55–67. doi: 10.1016/S0168-1656(03)00163-9. [DOI] [PubMed] [Google Scholar]
- 94.Puech V, Chami M, Lemassu A, Laneelle MA, Schiffler B, Gounon P, Bayan N, Benz R, Daffe M. 2001. Structure of the cell envelope of corynebacteria: importance of the non-covalently bound lipids in the formation of the cell wall permeability barrier and fracture plane. Microbiology 147:1365–1382. doi: 10.1099/00221287-147-5-1365. [DOI] [PubMed] [Google Scholar]
- 95.Kolenbrander PE. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu Rev Microbiol 54:413–437. doi: 10.1146/annurev.micro.54.1.413. [DOI] [PubMed] [Google Scholar]
- 96.Kolenbrander PE, Palmer RJ Jr, Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI. 2006. Bacterial interactions and successions during plaque development. Periodontol 2000 42:47–79. doi: 10.1111/j.1600-0757.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- 97.Guttilla IK, Gaspar AH, Swierczynski A, Swaminathan A, Dwivedi P, Das A, Ton-That H. 2009. Acyl enzyme intermediates in sortase-catalyzed pilus morphogenesis in Gram-positive bacteria. J Bacteriol 191:5603–5612. doi: 10.1128/JB.00627-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chang C, Mandlik A, Das A, Ton-That H. 2011. Cell surface display of minor pilin adhesins in the form of a simple heterodimeric assembly in Corynebacterium diphtheriae. Mol Microbiol 79:1236–1247. doi: 10.1111/j.1365-2958.2010.07515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mandlik A, Das A, Ton-That H. 2008. The molecular switch that activates the cell wall anchoring step of pilus assembly in Gram-positive bacteria. Proc Natl Acad Sci U S A 105:14147–14152. doi: 10.1073/pnas.0806350105. [DOI] [PMC free article] [PubMed] [Google Scholar]