ABSTRACT
The papillomavirus (PV) E1 helicase contains a conserved C-terminal domain (CTD), located next to its ATP-binding site, whose function in vivo is still poorly understood. The CTD is comprised of an alpha helix followed by an acidic region (AR) and a C-terminal extension termed the C-tail. Recent biochemical studies on bovine papillomavirus 1 (BPV1) E1 showed that the AR and C-tail regulate the oligomerization of the protein into a double hexamer at the origin. In this study, we assessed the importance of the CTD of human papillomavirus 11 (HPV11) E1 in vivo, using a cell-based DNA replication assay. Our results indicate that combined deletion of the AR and C-tail drastically reduces DNA replication, by 85%, and that further truncation into the alpha-helical region compromises the structural integrity of the E1 helicase domain and its interaction with E2. Surprisingly, removal of the C-tail alone or mutation of highly conserved residues within the domain still allows significant levels of DNA replication (55%). This is in contrast to the absolute requirement for the C-tail reported for BPV1 E1 in vitro and confirmed here in vivo. Characterization of chimeric proteins in which the AR and C-tail from HPV11 E1 were replaced by those of BPV1 indicated that while the function of the AR is transferable, that of the C-tail is not. Collectively, these findings define the contribution of the three CTD subdomains to the DNA replication activity of E1 in vivo and suggest that the function of the C-tail has evolved in a PV type-specific manner.
IMPORTANCE While much is known about hexameric DNA helicases from superfamily 3, the papillomavirus E1 helicase contains a unique C-terminal domain (CTD) adjacent to its ATP-binding site. We show here that this CTD is important for the DNA replication activity of HPV11 E1 in vivo and that it can be divided into three functional subdomains that roughly correspond to the three conserved regions of the CTD: an alpha helix, needed for the structural integrity of the helicase domain, followed by an acidic region (AR) and a C-terminal tail (C-tail) that have been shown to regulate the oligomerization of BPV1 E1 in vitro. Characterization of E1 chimeras revealed that, while the function of the AR could be transferred from BPV1 E1 to HPV11 E1, that of the C-tail could not. These results suggest that the E1 CTD performs multiple functions in DNA replication, some of them in a virus type-specific manner.
INTRODUCTION
Human papillomaviruses (HPVs) are small viruses with double-stranded DNA (dsDNA) genomes that infect both outer and mucosal stratified epithelium (for a recent review see reference 1). Of the more than 180 HPV genotypes described to date, about 20% infect the anogenital tract, causing either benign or precancerous lesions (2). In addition to their well-established role in inducing cervical and other anogenital cancers, HPVs have more recently been shown to be the causative agents for the majority of oropharyngeal cancers (3, 4).
Only two HPV proteins, E1 and E2, are required for replication of the viral circular dsDNA genome, with the remaining functions being provided by host cell proteins that are recruited to the viral episomes through the actions of E1 and E2 (5–9). DNA replication is initiated by E2 binding both to specific sequences within the viral origin of replication and to E1, the DNA helicase motor protein that drives the progression of the HPV DNA replication fork (9–11). At the origin, E2 assists E1 to assemble into a double-hexameric complex that unwinds the dsDNA ahead of the DNA replication forks (9–11). The E1 helicase also recruits and coordinates the actions of several cellular DNA replication factors required for HPV DNA synthesis, including topoisomerase I, the single-stranded-DNA (ssDNA)-binding complex (RPA), and the polymerase α-primase complex (7, 9, 10).
The E1 helicase is a member of helicase superfamily 3 (SF3), which contains replication initiator proteins from other small DNA and RNA genome viruses, and is also related to the cellular hexameric DNA helicases involved in genome replication (MCMs) (12, 13). The many biochemical studies on the enzymatic functions of various E1 proteins and the availability of high-resolution crystal structures and functional assays to probe E1's DNA replication activity both in vitro and in vivo have made E1 one of the best paradigms for the study of hexameric replicative DNA helicases (9, 10).
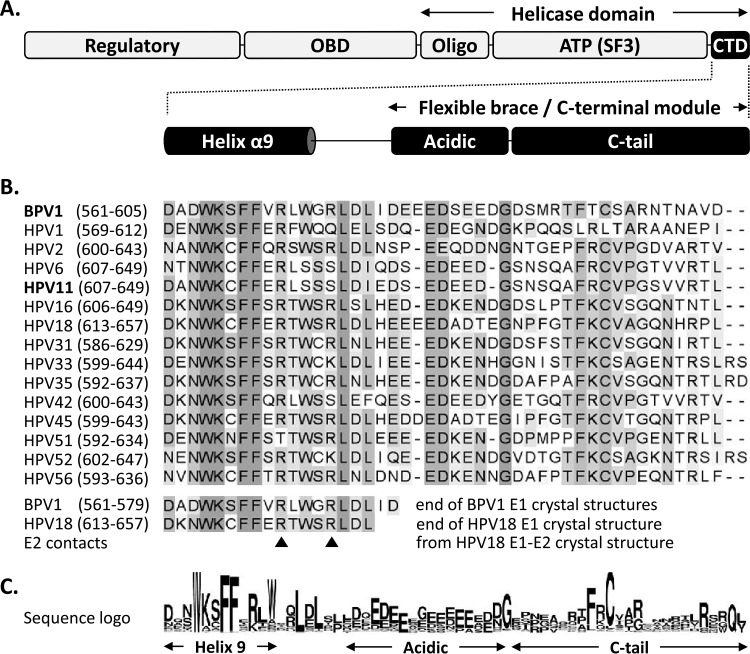
E1 can be divided into three major functional regions: an N-terminal regulatory region, a central origin-binding domain (OBD), and a C-terminal helicase domain (HD) (Fig. 1A) (10). At the core of the E1 HD lies the nucleoside triphosphate (NTP)-binding site characteristic of superfamily 3 DNA helicases (amino acids [aa] 407 to 557 for bovine papillomavirus 1 [BPV1] E1 and aa 452 to 603 for HPV11 E1; PROSITE domain profile PS51206 [13, 14]), flanked on the N-terminal side by the oligomerization domain (Oligo) (15) and on the other side by a C-terminal domain (CTD) comprised of an alpha helix followed by an approximately 45-amino-acid-long region that is predicted to be unstructured and that has been called either the “flexible brace” (FB) (16) or the “C-terminal module” (CTM) (17), depending on the study. The E1 FB/CTM has been shown to be sensitive to protease digestion, consistent with its predicted disordered nature (18). As a result, the FB/CTM was either missing from the E1 HD fragment used for crystallography (19, 20) or not resolved when included (18), further attesting to its flexibility. The FB/CTM has been posited to play a role in the assembly and stabilization of the E1 hexamer (16). This suggestion was based, in part, on the finding that deletion of the region impairs, but does not completely prevent, the assembly of E1 hexamers in vitro and that the resulting oligomers are less stable (15). Analysis of the solution structure of BPV1 E1 hexamers by small-angle X-ray scattering (SAXS) was used to create a model of the FB/CTM in the context of the assembled helicase; in this model, a negatively charged region of the FB/CTM from each monomer was proposed to contact a positively charged cleft on the adjacent monomer so that each CTD would act as an intermonomer brace to help stabilize the E1 hexamer (16). It was also suggested that the FB/CTM might play a role in helping maintain the oligomeric state of E1 during conformational changes induced by ATP binding and hydrolysis. This may account for the reduced unwinding activity of E1 proteins lacking the FB/CTM, particularly on longer DNA substrates (15). A recent study has proposed that the FB/CTM may actually consist of two subdomains, an acidic region (AR) and a C-terminal tail (C-tail), which play independent and antagonistic roles in the assembly of E1 into either hexamers around ssDNA or double hexamers at the origin (17). In this model, supported by substantial biochemical data, the AR functions primarily as an inhibitor of E1 assembly by interacting directly with the E1 oligomerization domain, while the C-tail is needed to counteract this negative effect of the AR to allow hexamer and double-hexamer formation (17). Exactly how these interactions are involved in regulating the assembly and activity of E1 at the replication fork remains unclear. As the E1 proteins from all papillomavirus types contain a CTD similar to that of BPV E1, it will be important to determine whether the proposed mechanisms of CTD function are generally conserved.
FIG 1.
Schematic representation of the papillomavirus E1 helicase highlighting the conservation of the C-terminal domain. (A) Diagram of the E1 protein showing the locations of the N-terminal regulatory region, OBD, Oligo, and ATPases associated with diverse cellular activities (AAA+) SF3 helicase/ATPase domain [ATP(SF3)] and of the ∼45-amino-acid-long CTD, which encompasses both the last alpha helix resolved in the available E1 crystal structures (helix α9) and the region previously called the flexible brace or C-terminal module, itself comprised of an acidic region (Acidic) and a C-tail subdomain separated by a linker sequence (10, 16, 17). (B) Amino acid sequence alignment of the E1 C termini from the indicated papillomavirus types (amino acid boundaries are indicated in parentheses). A large alignment of all of the E1 reference sequences deposited in the PAVE database was generated, but only those from BPV1 and 15 selected HPV types are presented for brevity. Residues are highlighted with increasingly darker shades of gray according to their degrees of conservation. The bottom of the alignment indicates the last region of the E1 C terminus that is resolved in the crystal structures of the E1 helicase domain from BPV1 (up to aa 577 and 579 in PBD structures 2V9P and 2GXA, respectively) and from HPV18 in complex with the E2 transactivation domain (PDB 1TUE) (18–20). While the major contacts between HPV18 E1 and E2 involve residues located in the ATPase domain, two potential minor contact points were identified within the C terminus (arrowheads) (20). (C) Amino acid sequence logo of the E1 C terminus generated from the alignment of all E1 reference sequences in the PAVE database (33).
While the models for the FB/CTM of BPV1 E1 are attractive and are supported by substantial biochemical results, there are few data available to date on the role of the E1 CTD in vivo, especially for E1 proteins from HPV types. In this study, we analyzed the requirement for the CTD of HPV11 E1 in DNA replication, using a series of truncations and selected amino acid substitutions at highly conserved residues of the CTD, in a cell-based HPV11 DNA replication assay. While our results certainly highlight the importance of the E1 CTD in HPV DNA replication in vivo, they also raise the possibility that the contribution of the C-tail subdomain may differ between papillomavirus (PV) types. As such, these results add important information to the growing understanding of the role of the C terminus of the E1 DNA helicase and may provide further insights into the regulation of hexameric DNA helicases in general.
MATERIALS AND METHODS
Plasmid construction and mutagenesis.
Plasmid p11E1, which expresses HPV11 E1 fused at its N terminus to green fluorescent protein (GFP), was constructed by inserting an XhoI-BamHI fragment including the E1 open reading frame (ORF) between the XhoI and BamHI sites of pEGFP-C2 (Clontech). The XhoI-BamHI E1 fragment used for this construction contains the previously reported mutations inactivating the major splicing donor site (21), as well as a NotI restriction site between the stop codon of E1 and the BamHI site. The nucleotide sequence of this fragment can be summarized as follows, with the XhoI, NotI, and BamHI sites underlined and the translation initiation and termination codons of the E1 ORF in brackets and italicized: 5′-CTCGAGCCACC[ATG-E1ORF-TAG]TGAGCGGCCGCTAGTAACATATGTAGTAAGGATCC-3′. Derivatives of p11E1 encoding C-terminally truncated HPV11 E1 proteins were created by PCR amplification of the desired portion of the E1 ORF from p11E1 with a primer hybridizing upstream of a naturally occurring PstI restriction site in the E1 ORF (5′-CCTGACACTGGGAAGTCGTGCTTTT-3′) and a second, variable primer containing a synthetic NotI restriction site (underlined) and two termination codons (5′-CTAGCGGCCGCTCACTA[variable sequence]-3′); digestion of the PCR product with PstI and NcoI; and ligation of the digested DNA between the PstI and NotI sites of p11E1 to replace the C-terminal portion of the wild-type E1 ORF with the truncated version. Plasmid p11E2, which encodes a codon-optimized version of HPV11 E2 fused at its N terminus with a triple Flag epitope, was constructed in two steps. First, a version of the E2 ORF that is codon optimized for expression in mammalian cells was commercially synthesized (Genscript). Second, this optimized sequence, starting at the second codon of E2, was amplified by PCR with oligonucleotides incorporating BamHI and HindIII sites (underlined) (5′-CCCGGATCCGAGGCAATCGCCAAGCG-3′ and 5′-GGGAAGCTTTCACAGCAGGTGCAGGCTC-3′) and inserted between the BamHI and HindIII sites of pCMV-3Tag-1a (Stratagene). Plasmid pFLORI11, which encodes the minimal HPV11 origin of DNA replication, was constructed by PCR amplification of the origin (nucleotides 7876 to 97 of the reference HPV11 genome obtained from ATCC; GenBank accession no. M14119) with a pair of primers containing synthetic NgoMIV restriction sites (underlined) (5′-CCAGCCGGCGTCACACACCTGCAAC-3′ and 5′-CCCGCCGGCCCTCGTCTGCTAATTTTTTGG-3′), digestion of the amplified product with NgoMIV, and ligation of the digested DNA into the NgoMIV site of plasmid pCI-FLuc (22). The orientation of the origin fragment in pFLORI11 is such that nucleotide 7876 of the origin is closest to the simian virus 40 (SV40) polyadenylation site of pCI-FLuc. HPV11/BPV1 E1 chimeras Ch617 and Ch620 were constructed by PCR in two steps. First, the regions spanning amino acids 1 to 617 and 1 to 620 of HPV11 E1 were each amplified from p11E1 with a common primer spanning the initiation codon of HPV11 E1 (boldface), 5′-GCTTCTCGAGCCACCATGGCGGACGATTCT-3′, and a second, chimeric primer encoding the desired HPV11/BPV1 E1 junction sequence, either 5′-GTCTAAACGCCCCCACAGTCTTTCAAAGAAACATTTCCAGTTTGCATC-3′ for the amplification of E1(1–617) (the codon for leucine 617 is in boldface; the BPV1 E1 sequence is italicized) or 5′-GTCAATCAGGTCTAAGCTGGACGACAGTCTTTCAAAGAAACATT-3′ for E1(1–620) (the codon for serine 620 is in boldface; the BPV1 E1 sequence is italicized). Similarly, the regions encoding residues 572 to 605 and 575 to 605 of the BPV1 E1 C terminus were amplified from pCG E1Eag 1235− (23) with a chimeric primer, either 5′-TTCTTTGAAAGACTGTGGGGGCGTTTAGACCTGATTGACG-3′ for E1(572–605) (the codon for tryptophan 572 is in boldface; the HPV11 E1 sequence is italicized) or 5′-AGACTGTCGTCCAGCTTAGACCTGATTGACGAGGAGGAGG-3′ for E1(575–605) (the codon for leucine 575 is in boldface; the HPV11 E1 sequence is italicized), and with a second, common primer overlapping the BPV1 E1 translation termination codon, 5′-CCAGCGGCCGCTCACTAATCAACTGCATTTGTGTTTCTTGCGCTACA-3′ (the codon for translation termination is in boldface; the NotI site is underlined). In the second step, the resulting PCR fragments encoding HPV11 E1(1–617) and BPV1(572–605), or those encoding HPV11 E1(1–620) and BPV1(575–605), were mixed in a 1:1 ratio and used as the template for a second round of PCR using the following pair of primers: 5′-GCTTCTCGAGCCACCATGGCGGACGATTCT-3′ and 5′-CCAGCGGCCGCTCACTAATCAACTGCATTTGTGTTTCTTGCGCTACA-3′ (the codons for translation initiation and termination are in boldface; the XhoI and NotI sites are underlined). The resulting PCR fragments were digested with XhoI and NotI and subcloned into the XhoI and NotI sites of dephosphorylated p11E1 to replace the wild-type HPV11 E1 ORF with that of each chimera. Mutant versions of p11E1 and chimeric derivatives were created using overlapping oligonucleotides encoding the desired mutations either by extension of these oligonucleotides using the KOD Hot Start DNA polymerase (Novagen) and overnight digestion with DpnI prior to transformation or using a site-directed mutagenesis kit (Agilent Technologies) following the instructions provided by the manufacturer. All DNA constructs were confirmed by sequencing. More details on the construction of the above-mentioned plasmids will be provided upon request.
Cell culture and transfections.
C33A human cervical carcinoma cells were grown at 37°C with 5% CO2 in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 50 IU/ml penicillin, and 50 μg/ml streptomycin. Transfection of C33A cells was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
Luciferase-based HPV11 and BPV1 DNA replication assays.
The HPV11 DNA replication assay was developed and performed essentially as described previously for HPV31 and BPV1 (22, 24, 25). Briefly, 2.5 × 104 C33A cells/well were seeded in white flat-bottom 96-well plates (Corning) and transfected, 24 h later, with the following four plasmids: for HPV11, p11E1 (10 ng), p11E2 (10 ng), pFLORI11 (2.5 ng), and the Renilla luciferase (RLuc) control plasmid pRL (0.5 ng) (22); for BPV1, pCG E1Eag 1235− (10 ng; BPV1 E1 expression plasmid [23]), pBPV1E2 (10 ng [25]), pFLORI-BPV1short (2.5 ng [25]), and pRL (0.5 ng). In all experiments, the total amount of transfected DNA was adjusted to 100 ng with pCI (Promega) as the carrier DNA. Firefly (FLuc) and Renilla luciferase activities were measured using the Dual-Glo luciferase assay system (Promega) 72 h posttransfection unless otherwise indicated. The results are reported as the mean of at least three independent experiments, each performed in triplicate. For most experiments presented in this study, RLuc/FLuc ratios were determined for increasing quantities of E1 expression vector (as indicated in the figure legends) so that the area under the curve (AUC) could be used as an overall measure of DNA replication activity when assessing statistical significance.
Statistical analysis.
The AUC value derived from each dose-response curve was used as an integrated measure of DNA replication activity. Statistical significance was assessed by comparing mean AUC values using one-way analysis of variance (ANOVA) followed either by Dunnett's post hoc analysis for comparison to a reference E1 protein or by Bonferroni's post hoc analysis when no single E1 protein was used as a reference, as indicated in the figure legends. Statistics and AUC were calculated using GraphPad Prism version 6.00.
Inhibition of BPV1 DNA replication by dominant-negative E1 proteins.
When testing for the dominant-negative activity of BPV1 E1 mutant proteins, the BPV1 DNA replication assay was performed as described above but using, in addition to the four standard plasmids, increasing amounts of mutant E1 expression vector (0, 3.13, 6.26, 12.5, 25, 50, 100, and 200 ng) and adjusting the final amount of transfected DNA to 200 ng with pCI as the carrier DNA. DNA replication levels (FLuc/RLuc values) were measured 48 h posttransfection and fitted with GraphPad Prism 6 software (GraphPad Software) to a sigmoidal dose-response curve described by the following equation: y = bottom + (top − bottom)/[1 × 10(logEC50 − x) × hill slope], where y is the DNA replication value, top is the DNA replication value obtained in the absence of the dominant-negative inhibitory E1, bottom is the maximal level of inhibition, x is the amount of dominant-negative E1 expression plasmid (in nanograms), and EC50 is the 50% effective concentration.
LUMIER E1-E2 coimmunoprecipitation assay.
The LUMIER (luminescence-based mammalian interactome mapping) E1-E2 coimmunoprecipitation assay was adapted from Blasche and Koegl (26) using plasmid p11E1 and its derivatives, together with a previously described plasmid encoding HPV11 E2 fused to Renilla luciferase (RLuc-E2) (24). Briefly, 8.5 × 105 C33A cells were transfected with 2 μg of p11E1 expression plasmid, along with 2 μg of RLuc-E2 expression plasmid, in a well of a 6-well plate. Cells were harvested 24 h posttransfection in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 10 μg/ml antipain, 2 μg/ml leupeptin, 2 μg/ml aprotinin, 1 μg/ml pepstatin A, 1 mM phenylmethylsulfonyl fluoride, 10 μM proteasome inhibitor MG132). GFP-E1 and truncated derivatives were immunoprecipitated from cleared cellular extracts for 3 h with 40 μl of protein G-Sepharose (GE Healthcare) conjugated to 1 μg of anti-GFP antibody (Roche). The beads were then washed three times with Tris-buffered saline (TBS) buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl) and resuspended in 50 μl of lysis buffer. For each E1 protein, the levels of Renilla luciferase activity present in the input cellular extract (50 μl) and in the corresponding immunoprecipitate (50 μl of bead-bound material) were determined using the Renilla luciferase assay system (Promega). The fraction of RLuc-E2 coimmunoprecipitated with GFP-E1 was then determined by calculating the ratio of Renilla luciferase activity bound to the beads over the total amount of Renilla luciferase activity measured in the input cellular extract; for wild-type GFP-E1, this ratio was assigned a value of 100% and was used as the reference to which all other truncated E1 proteins were compared.
Antibodies and immunoblotting.
3×Flag-tagged E2 protein and β-tubulin were detected using mouse monoclonal antibodies from Sigma-Aldrich (catalog no. F1804 and T0426, respectively). The anti-GFP antibody used to detect GFP-tagged E1 was purchased from Roche (catalog no. 11814460001). For immunoblot analysis, cell extracts were separated on SDS-10% or 12% PAGE gels, as indicated in the figure legends, and the proteins were transferred onto polyvinylidene difluoride membranes prior to incubation with the above-mentioned primary antibodies. Sheep anti-mouse (catalog no. NA931) or donkey anti-rabbit (NA934V) IgGs conjugated to horseradish peroxidase (GE Healthcare) were used as secondary antibodies and detected with an enhanced-chemiluminescence detection kit (GE Healthcare).
Confocal fluorescence microscopy.
C33A cells (5 × 105) were seeded on coverslips and then transfected using Lipofectamine 2000 (Invitrogen) with 1 μg of p11E1 in the absence or presence of p11E2. Twenty-four hours later, the cells were fixed with 4% formaldehyde, permeabilized with 0.1% Triton X-100, and mounted using Vectashield mounting medium, which contains DAPI (4′,6-diamidino-2-phenylindole) to stain nuclei (Vector Laboratories). Images were acquired using a Zeiss LSM700 laser scanning confocal microscope and analyzed using Zen 2009 LE software.
Bioinformatics tools.
Amino acid sequence alignments were generated with Clustal Omega (27). Sequence logos were generated with WebLogo (28). The structural model of the HPV11 E1 helicase domain (aa 353 to 623) was generated using the I-TASSER suite (29) and refined with ModRefiner (30). The resulting model was submitted to the GalaxyWeb server (31) to predict the structure of the HPV11 E1 HD in its hexameric form using the GalaxyGemini program or, alternatively, to predict its structure in complex with the HPV11 E2 transactivation domain (TAD) (Protein Data Bank [PDB] 1R6K) using the Galaxy Refine Complex algorithm (29). A representation of the HPV11 E1 AR and C-tail in their extended conformation was obtained using the IntFOLD Server (32). Structure representations were generated with the PyMOL Molecular Graphics System (Schrödinger, LLC).
RESULTS
Deletion of the C terminus of E1 is detrimental to HPV DNA replication in vivo.
A schematic linear representation of the PV E1 protein is shown in Fig. 1A. To identify conserved subdomains within the E1 C-terminal domain (CTD), an alignment of the last ∼45 amino acids from all of the reference E1 proteins present in the Papillomavirus Episteme (PaVE) database (33; http://pave.niaid.nih.gov) was performed; a subset of this alignment and a sequence logo representation of all E1 proteins are presented in Fig. 1B and C, respectively. Four subdomains were identified: an alpha helix (termed helix α9 in the crystal structure of the HPV18 E1 HD in complex with the E2 TAD [20]; this helix is also present in the crystal structures of the BPV1 E1 HD [18, 19]), followed by a short (∼6-aa) linker, an ∼10-aa acidic region, and an ∼15-aa region designated the C-tail (17). Together, the AR and C-tail constitute the previously described FB/CTM (16, 17). While others have evaluated the effects of truncations and selected amino acid substitutions within the FB/CTM on the function of BPV1 E1 in vitro (23), there has not yet been any mutational study addressing the functions of these domains in the ability of HPV E1 to support DNA replication in live cells (in vivo).
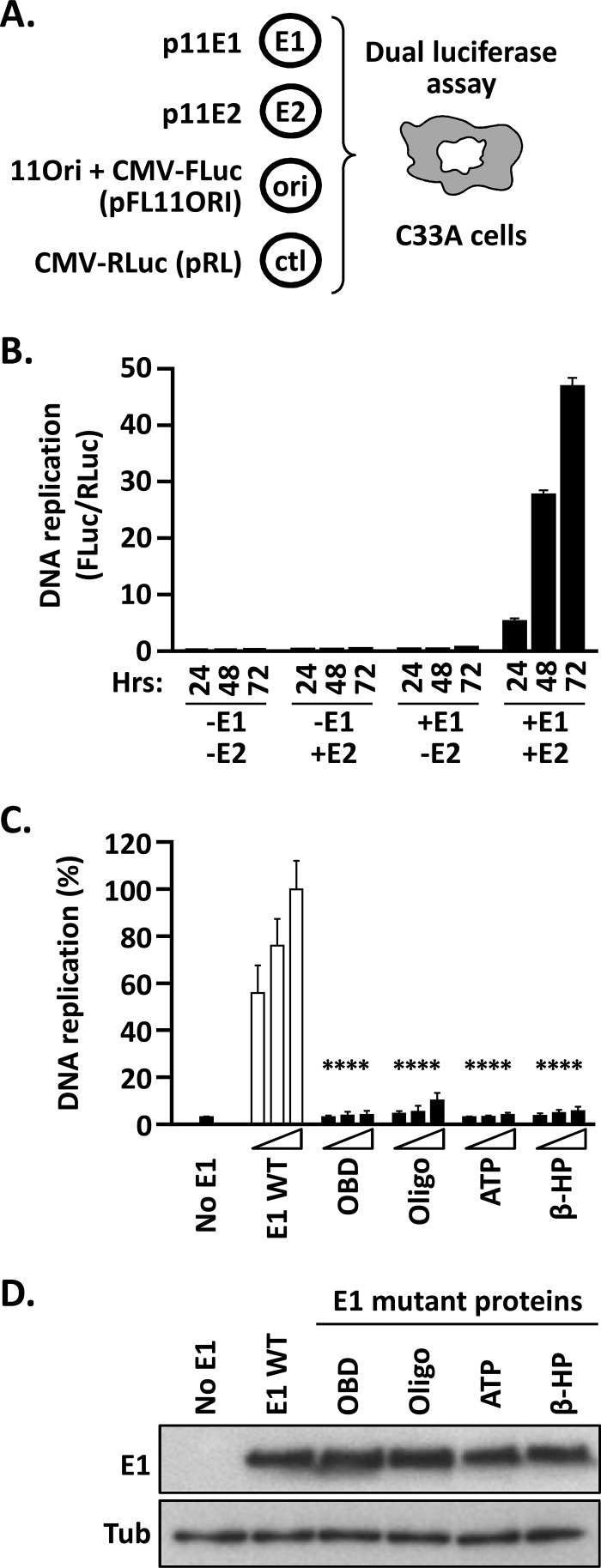
To fill this gap, we first implemented a transient HPV11 DNA replication assay using the same luciferase reporter gene methodology previously described for HPV31 and BPV1 (Fig. 2) (22, 24, 25). We chose the HPV11 system to evaluate the in vivo activity of the E1 CTD because it has proven to be more amenable to subsequent biochemical studies. The HPV11 cell-based assay relies on the transfection of four different plasmids into C33A cervical carcinoma cells (HPV negative). Two of these plasmids, p11E1 and p11E2, express E1 and E2, respectively, from the cytomegalovirus (CMV) promoter. The third plasmid, pFL11ORI, directs expression of FLuc from the CMV promoter and also contains the HPV11 minimal origin of DNA replication. The fourth plasmid, pRL, expresses RLuc from the CMV promoter and is used as an origin-minus control to normalize for luciferase expression from a transfected but unreplicated plasmid. Upon introduction of these four plasmids into C33A cells, pFL11ORI is replicated to a higher copy number by E1 and E2, resulting in increased expression of the FLuc reporter gene. Hence, the ratio of the activities of FLuc and RLuc, which is typically measured 24 to 72 h posttransfection, provides a robust surrogate readout of the levels of HPV DNA replication that occurred during the time frame (22).
FIG 2.
Luciferase reporter cell-based assay for HPV11 DNA replication. (A) Principle of the HPV11 DNA replication assay. Expression vectors for GFP-tagged HPV11 E1 (p11E1) and triple-Flag-tagged HPV11 E2 (p11E2) are transfected into C33A cells, together with a plasmid containing the HPV11 minimal origin of replication (ori) linked in cis to an FLuc reporter gene (pFLORI11). A nonreplicating plasmid (pRL) encoding RLuc is used as an internal control (ctl). In this assay, replication of the origin-containing plasmid pFLORI11 is detected as an E1- and E2-dependent increase in FLuc activity relative to the level of RLuc expression from the control plasmid, pRL. DNA replication levels are presented as FLuc/RLuc activity ratios measured using a dual-luciferase assay. (B) Dependence of the HPV11 DNA replication assay on E1 and E2 expression as a function of time. DNA replication levels are presented as FLuc/RLuc ratios, measured in C33A cells that were cotransfected with 2.5 ng of pFLORI11 and 0.5 ng of pRL, together with (+) or without (−) 10 ng of p11E1 (E1) and 10 ng p11E2 (E2), as indicated. DNA replication levels were measured at different time points posttransfection (24, 48, and 72 h). Standard deviations are indicated by the error bars. Control assays performed in the absence of p11E1, p11E2, or both expression vectors showed no viral DNA replication. (C) Validation of the assay with replication-defective E1 mutant proteins. Shown are DNA replication activities supported by the indicated E1 proteins carrying deleterious amino acid substitutions in the OBD (K286A/R288A), Oligo (F393A), Walker A box (ATP) (K484A), and β-hairpin (β-HP) (K551A/H552A). DNA replication levels were measured from cells transfected with three different amounts of E1 expression plasmid (2.5, 5, and 10 ng) or with an empty vector as a negative control (No E1). Replication activity was measured 72 h posttransfection and is reported as a percentage of the FLuc/RLuc ratio obtained with 10 ng of wild-type E1 expression plasmid (WT), which was set to 100%. Standard deviations are indicated. Statistical significance was assessed by comparing the DNA replication activity of each E1 mutant protein to that of wild-type E1 (white bars), using one-way ANOVA followed by Dunnett's post hoc analysis. Significant differences are indicated (****, P ≤ 0.0001). (D) Expression levels of the indicated E1 mutant proteins compared to wild-type E1. Extracts from transfected cells were separated on an SDS-12% PAGE gel prior to immunoblotting with an anti-GFP antibody. Tubulin (Tub) was used as a loading control.
Our validation studies confirmed the well-established dependence of HPV DNA replication on both E1 and E2 (Fig. 2B). To further verify the usefulness of this assay for analysis of HPV11 E1 function, we used site-directed mutagenesis of p11E1 to create vectors that express HPV11 E1 proteins carrying deleterious amino acid substitutions in either the OBD (K286A/R288A), the Oligo (F393A), the nucleotide-binding site (ATP; K484A in the Walker A box) or the β-hairpin required for helicase tracking along ssDNA (β-HP) (K551A/H552A). The results clearly show that E1 proteins with substitutions known to compromise PV DNA synthesis are unable to support HPV11 DNA replication in vivo, in contrast to the wild-type protein (P < 0.0001) (Fig. 2C), even though they are expressed at levels similar to those of wild-type E1 (Fig. 2D).
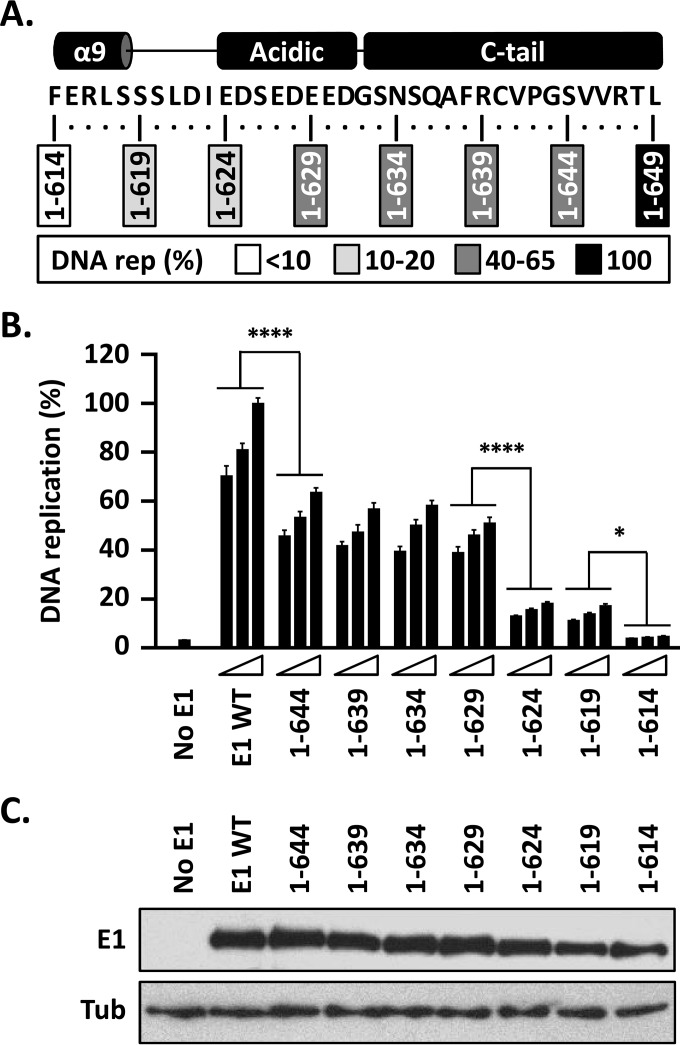
To address the function of the E1 CTD, seven deletions that removed increasingly larger portions of the domain, in 5-amino-acid increments, were created and tested for their effects on the ability of E1 to support HPV DNA replication (Fig. 3A). Even a minimal deletion of only 5 amino acids from the CTD had a substantial negative effect, resulting in a >35% decrease in DNA replication activity (P < 0.0001) (Fig. 3B). Similar levels of DNA replication (50 to 65%) were obtained with E1 proteins with deletions further into the CTD, and even into the C-terminal part of the AR up to amino acid 629. An additional 10 to 20% reduction in DNA replication levels was seen when the deletions completely removed the AR [compare E1(1–624) to E1(1–629); P < 0.0001] (Fig. 3B). Once the deletions encroached into helix 9, the replication capacity of E1 became negligible [compare E1(1–614) to E1(1–619); P < 0.05] (Fig. 3B) and virtually indistinguishable from the background seen in the absence of E1 expression plasmid. Immunoblotting for the N-terminal GFP tag on E1 showed minimal variation in the expression levels of these truncated E1 proteins (we have noted that slight variations in E1 expression have little effect on DNA replication levels, consistent with the <25% decrease in replication activity observed when the amounts of transfected GFP-E1 expression vector are decreased 2-fold). Collectively, the results of this deletion analysis defined three functional regions within the E1 CTD that span, respectively, the C-tail and the last quarter of the AR (aa 630 to 649), the N-terminal three-quarters of the AR and the linker region (aa 620 to 629), and the C-terminal portion of helix 9 (aa 615 to 619). Because these three functional regions roughly correspond to the different conserved subdomains of the E1 CTD, we simply refer to them as the C-tail, AR, and helix 9, respectively, here. Below, we examine in greater detail the contribution of each of these functional regions to HPV DNA replication.
FIG 3.
The C terminus of E1 contains three subdomains that play roles in HPV11 DNA replication. (A) Schematic representation and amino acid sequence of the C terminus of HPV11 E1. The boxes indicate the amino acid residues retained in the C-terminally truncated E1 proteins; 1 to 649 refers to the wild-type protein. The boxes are shaded according to the levels of DNA replication (rep) supported by each E1 protein (as shown in panel B), with the darkest shade indicating the highest level of DNA replication and an open box indicating background levels of activity (<10%). (B) The DNA replication activities of the indicated E1 proteins were evaluated as described in the legend to Fig. 2, using three amounts of E1 expression vector (2.5, 5, and 10 ng). DNA replication levels were measured 72 h posttransfection and are reported as percentages of the levels obtained with 10 ng of wild-type E1 expression plasmid (WT). Statistical significance was assessed by comparing the DNA replication activity of each truncated E1 protein to that of the preceding (i.e., 5-amino-acid-longer) deletion using one-way ANOVA followed by Bonferroni's post hoc analysis. Significant differences are indicated (*, P ≤ 0.05; ****, P ≤ 0.0001). (C) Expression of GFP-tagged wild-type E1 and truncated derivatives. Extracts from transfected cells were separated on an SDS-10% PAGE gel prior to immunoblotting with an anti-GFP antibody. Tubulin was used as a loading control.
Deletion into the AR and helix 9 reduces HPV DNA replication.
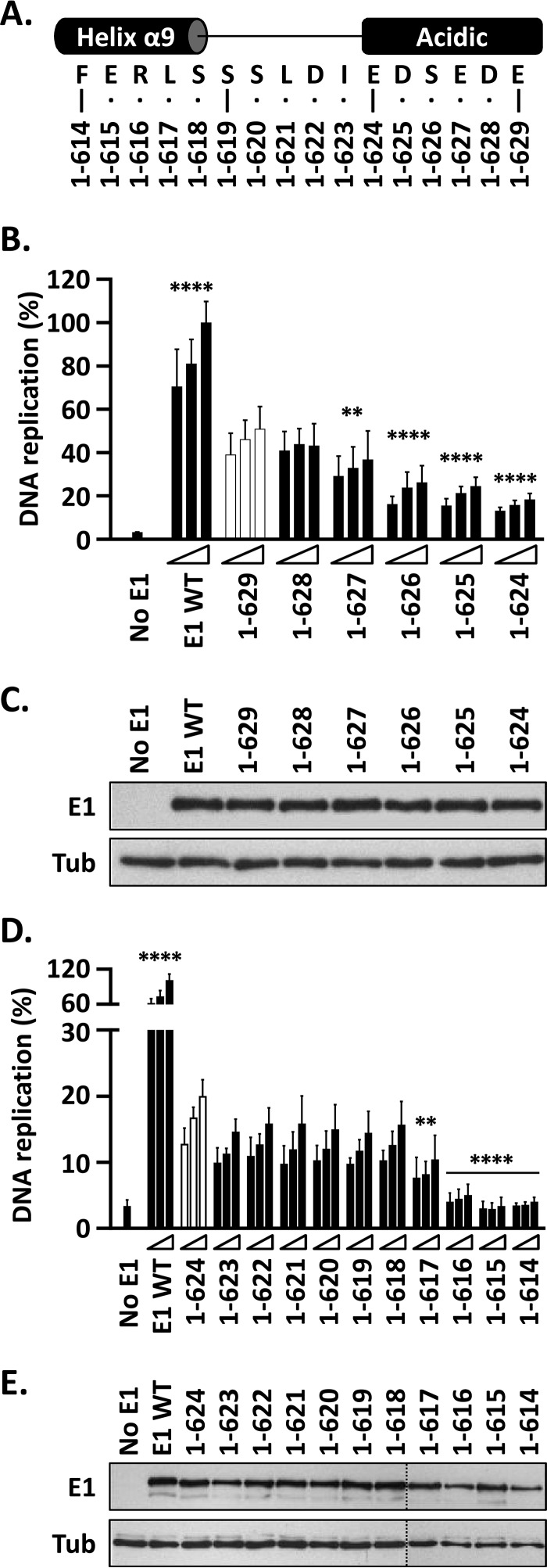
The results discussed above indicated that E1(1–624) and E1(1–629) support levels of DNA replication that are approximately 20% and 55%, respectively, of those obtained with wild-type E1. Thus, deletion of the C-terminal portion of the AR (aa 624 to 629) results in a 35% drop in DNA replication activity, highlighting the importance of the region for this process. To further investigate the function of the AR and to map more precisely the boundary between the region and the C-tail, we created four additional E1 mutant proteins ending at amino acids 625, 626, 627, and 628 (Fig. 4A). The DNA replication levels supported by these E1 truncations decreased incrementally as the protein became shorter, with E1(1–627) being the first deletion showing a statistically significant reduction in activity compared to E1(1–629) (P < 0.01) (Fig. 4B). This decrease in activity was not due to lower protein expression, as all the E1 truncations accumulated to comparable levels when analyzed by immunoblotting (Fig. 4C). The lack of an obvious breakpoint in activity is consistent with the net negative charge of the AR, rather than its precise amino acid sequence, being the main determinant of its function. It is noteworthy that complete removal of the 8-amino-acid AR results in a 40 to 45% drop in replication activity [compare E1(1–623) to E1(1–634)], while removal of four acidic residues [E1(1–627) versus E1(1–634)] results in half that decrease.
FIG 4.
The DNA replication activity of E1 is reduced by C-terminal deletions that remove the AR and abolished by those that encroach into helix 9. (A) Representation and amino acid sequence of a portion of the E1 C terminus encompassing the end of helix α9, the linker region, and the acidic region. The C-terminal boundaries of the different truncated E1 proteins are indicated. (B) Deletions into the AR. Shown are the DNA replication activities of E1 proteins ending between residues 629 and 624. In addition to lacking the C-tail, these proteins lack increasingly larger portions, in single-amino-acid increments, of the AR. The activity of each protein was determined using three amounts of E1 expression vector (2.5, 5, and 10 ng) 72 h posttransfection and is reported as a percentage of the activity obtained with 10 ng of wild-type E1 expression plasmid (WT), as described in the legend to Fig. 3B. Statistical significance was assessed by comparing the DNA replication activity of each E1 protein to that of E1(1–629) (white bars) using one-way ANOVA followed by Dunnett's post hoc analysis. Significant differences are indicated (**, P ≤ 0.01; ****, P ≤ 0.0001). (C) Expression of GFP-tagged wild-type E1 and truncated derivatives. Extracts from transfected cells were separated on an SDS-10% PAGE gel prior to immunoblotting with an anti-GFP antibody. Tubulin was used as a loading control. (D) Deletions into the linker region and helix 9. Shown are the DNA replication activities of truncated E1 proteins lacking increasingly larger portions of the linker region and helix 9. DNA replication levels and statistical significance relative to E1(1–624) (white bars) were determined as described for panel A. (E) Expression of the indicated GFP-E1 proteins was determined as described for panel C.
Next, we aimed to delineate more precisely the boundary between helix 9 and the AR, using a series of intermediate deletions, in single-amino-acid increments, spanning residues 614 to 624 (Fig. 4A). DNA replication assays performed with these truncated E1 proteins clearly demonstrated that the drop from 15 to 20% DNA replication levels observed with E1(1–624) to the less than 10% background levels measured for E1(1–614) occurs when the deletions begin to encroach upon helix 9 (Fig. 4D), with E1(1–617) being the largest protein showing a statistically significant reduction in activity relative to E1(1–624) (P < 0.01) (Fig. 4D). This drop in activity was not due to a reduction in protein expression, as all the truncated E1 proteins accumulated to comparable levels when analyzed by immunoblotting (Fig. 4E). Together, these fine-mapping experiments revealed a gradual decrease in DNA replication activity with progressive deletions into the AR, from amino acids 629 to 623, and a second drop in activity when deletions extended into helix 9, beginning at residue 617.
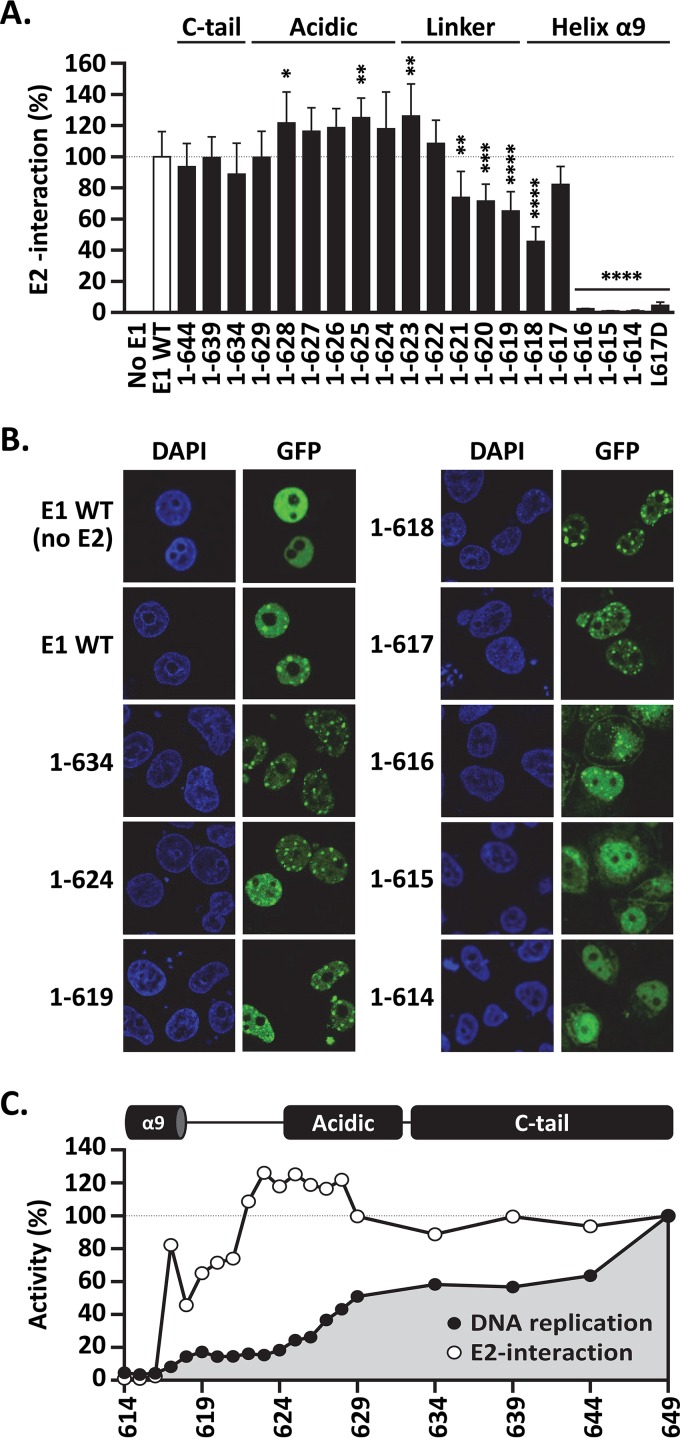
C-terminal deletions that encroach into helix 9 compromise interaction with E2.
An obvious possibility that would account for the low DNA replication activity of some of the truncated E1 proteins would be failure to productively bind to E2. This prompted us to investigate the E2 interaction capacities of GFP-E1 and of the 19 truncated derivatives using a quantitative coimmunoprecipitation assay based on the LUMIER protocol (26). In these assays, GFP-E1 was transiently expressed in C33A cells, together with HPV11 E2 fused to RLuc, so that the levels of RLuc-E2 that were coimmunoprecipitated with GFP-E1, as well as those present in the input cellular extract, could be measured using a standard luciferase assay. Approximately one-third of the RLuc-E2 luciferase activity detected in the input extract was coprecipitated with GFP-E1 under our assay conditions (data not shown). The results presented in Fig. 5A show that E1(1–629) was as proficient as wild-type E1 at interacting with RLuc-E2. This indicated that the ∼40% drop in DNA replication activity that is observed upon deletion of the C-tail and the C-terminal part of the AR is not caused by a defect in binding to E2. Further truncations into the AR and the C-terminal part of the linker region, from amino acids 629 to 622, also did not reduce E2 interaction and even enhanced it slightly, although this ∼25% increase in coprecipitated E2 reached statistical significance only for E1(1–628), E1(1–625), and E1(1–623) (P < 0.05, P < 0.01, and P < 0.01, respectively) (Fig. 5A). Finally, deletions that extended further into the linker region and the C-terminal part of helix 9 progressively reduced E2 binding, with E1(1–618) and E1(1–617) being the two smallest proteins that retained an appreciable degree of interaction with E2. Surprisingly, E1(1–617) consistently showed higher E2 interaction activity than E1(1–618), perhaps because helix 9 is stabilized upon deletion of serine 618. The importance of leucine 617 for E2 interaction was further demonstrated by changing the residue to glutamic acid in the context of the full-length protein. The resulting L617D E1 was severely defective for E2 interaction in the LUMIER coprecipitation assay (Fig. 5A) and, accordingly, was unable to support viral DNA replication in vivo (<5% activity), although it was expressed at levels comparable to those of the wild-type protein (data not shown).
FIG 5.
C-terminal deletions that extend into helix 9 reduce interaction with E2. (A) LUMIER E1-E2 coimmunoprecipitation assay. Expression vectors for GFP-tagged wild-type E1 and truncated derivatives, as well as for the full-length L617D mutant protein, were cotransfected with a vector encoding E2 fused to Renilla luciferase (RLuc-E2). An empty vector encoding GFP alone (No E1) was used as a negative control. For each E1 protein, the amount of RLuc-E2 that was coprecipitated by GFP-E1 was determined by measuring the levels of Renilla luciferase activity and normalized to the amount of RLuc-E2 present in the input cellular extract. The amount of RLuc-E2 coprecipitated by wild-type GFP-E1 was set at 100%. Each bar represents the average of three independent experiments, each performed in duplicate. Standard deviations are indicated by the error bars. Statistical significance was assessed by comparing the DNA replication activity of each E1 protein to that of wild-type GFP-E1 (white bar) using one-way ANOVA followed by Dunnett's post hoc analysis. Significant differences are indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). (B) Intracellular localization of the indicated GFP-E1 proteins in the presence of E2 (unless otherwise indicated). C33A cells transiently expressing the wild-type or truncated E1 proteins were visualized by fluorescence confocal microscopy. DNA was stained with DAPI to visualize the cell nuclei. Note the presence of GFP-E1 in E2-induced nuclear foci and the absence of such foci for E1(1–615) and E1(1–614). (C) Diagram summarizing the DNA replication and E2 interaction activities of the 19 truncated E1 proteins characterized in this study.
Immunofluorescence confocal microscopy was then used to verify that all of the GFP-E1 proteins were properly localized to the nucleus and, in addition, that they could associate with E2 in nuclear foci, which are the precursors of viral DNA replication centers (34) (Fig. 5B and data not shown). Most truncated proteins were nuclear and behaved like wild-type E1 in these assays, with E1(1–617) being the shortest protein that could associate with E2 in nuclear foci (Fig. 5B). Thus, E1 truncations that interact with E2 in coimmunoprecipitation assays also retain the ability to accumulate in E2-induced nuclear foci. In contrast, the two smallest proteins, which did not appreciably coprecipitate with E2, E1(1–615) and E1(1–614), were never found in nuclear foci and showed signs of precipitation in the cytoplasm (Fig. 5B), suggesting that their folding may have been compromised. E1(1–616), which also did not coprecipitate with E2, showed an intermediate phenotype, with some of the protein being localized in foci and some diffusely distributed throughout the nucleus, with the remainder found in brightly fluorescent aggregates (Fig. 5B and data not shown). Thus, it seems that E1(1–616) retains some low level of interaction with E2 that cannot be detected under the harsher conditions of the coimmunoprecipitation assay but that is perhaps stabilized by the formaldehyde fixation step used to prepare cells for microscopy. Collectively, the results from the coimmunoprecipitation experiments and microscopic examination of E1-E2 nuclear focus formation indicated that C-terminal deletions that encroach into helix 9 impair E2 binding and likely the structural integrity of the E1 HD.
A graph summarizing the DNA replication and E2-binding activities of the 19 truncated E1 proteins tested in this study is presented in Fig. 5C. The graph emphasizes the ∼40% reduction in DNA replication activity that is brought about by partial or complete deletion of the C-tail and the fact that it is not caused by a defect in interaction with E2. It also underscores the gradual decrease in DNA replication activity of C-terminal deletions that progressively extend into the AR, resulting in an ∼85% net drop in DNA replication levels when both the AR and C-tail are completely removed. Finally, it highlights the deleterious effect on E2 interaction caused by deletions that extend into the N-terminal half of the linker region and the C-terminal portion of helix 9.
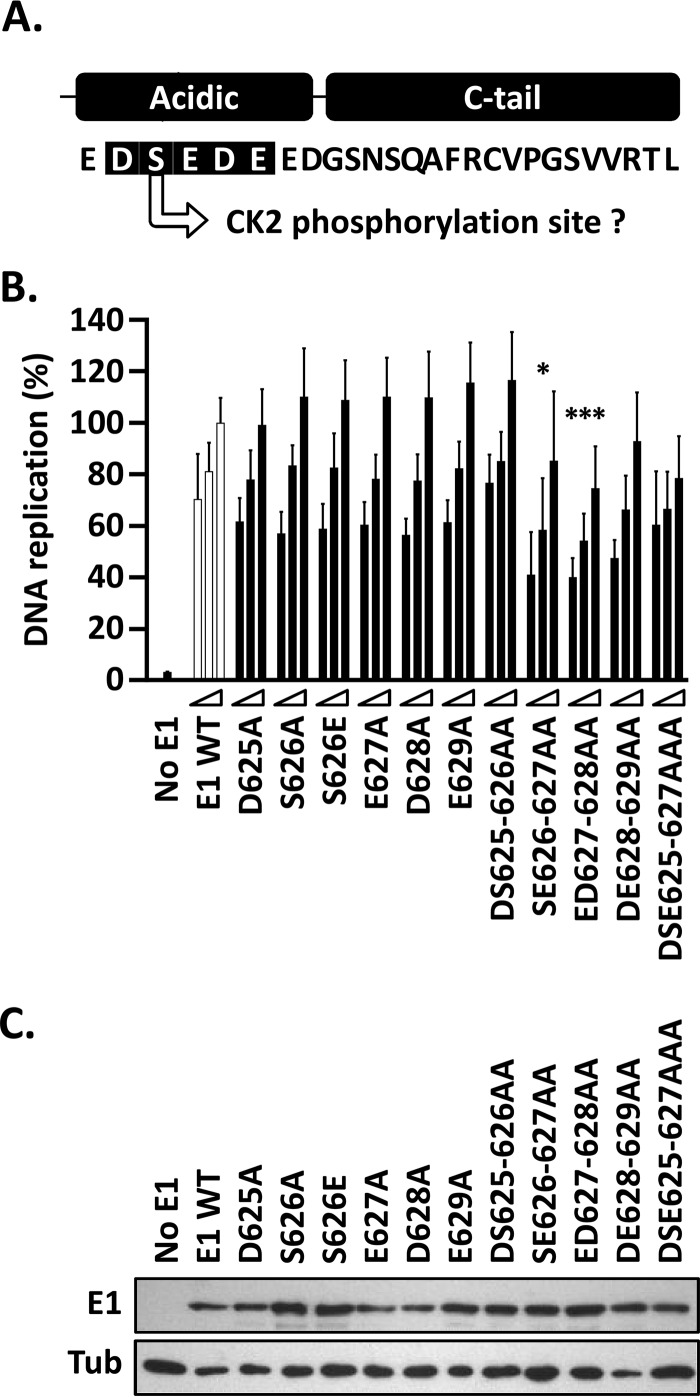
Mutational analysis of the AR.
The deletion analyses presented above clearly indicate the importance of the AR and the C-tail to the overall DNA replication activity of E1 and set the stage for a finer mutational study of the two regions. Amino acid substitutions within the AR were introduced within the context of the full-length protein (aa 1 to 649) to better evaluate the role of the region in HPV DNA replication (Fig. 6A). Individual replacements of amino acid residues 625 to 629 with alanine had no noticeable effect on the ability of E1 to support HPV DNA replication compared to wild-type E1 (Fig. 6B). Serine 626, which lies within a putative CK2 phosphorylation site, was also changed to glutamate as a phosphomimetic residue, but this, too, had little effect on DNA replication. The fact that none of the single-amino-acid substitutions had a significant effect on HPV DNA replication reinforces the concept from the AR deletion analysis that the function of the AR depends primarily on its overall negative charge rather than its specific amino acid sequence per se. Accordingly, only the double-amino-acid substitutions SE626–627AA and ED627–628AA were able to reduce the DNA replication activity of E1 in a statistically significant manner (P < 0.05 and P < 0.001, respectively) (Fig. 6B). These results were not attributable to variation in E1 levels, as all the E1 mutants were expressed similarly (Fig. 6C). The modest decrease in DNA replication activity brought about by the SE626–627AA and ED627–628AA substitutions may be due to the fact that they remove only one or two of the seven negatively charged amino acids (D or E) present in the AR. As such, these results are consistent with those of the AR deletion analysis (Fig. 5), which indicated that E1 proteins lacking two and three negatively charged residues from the AR [E1(1–129) and E1(1–628), respectively] show only a 10 to 15% reduction in DNA replication activity compared to a larger E1 containing the entire AR [E1(1–634)]. Collectively, the results of the deletion and mutational analyses of the AR suggest that the overall negative charge of the region underlies its function and that at least four acidic residues must be changed to appreciably alter its activity.
FIG 6.
Single-amino-acid substitutions in the E1 AR have little effect on HPV11 DNA replication. (A) Representation and amino acid sequence of the C terminus of HPV11 E1. The 5 residues within the AR that were subjected to mutagenesis are shaded in black. Also indicated is the fact that serine 626 is a putative CK2 phosphorylation site. (B) Mutant E1 proteins were evaluated for HPV11 DNA replication activity (as for Fig. 3B). Only the mutant proteins that had two or three amino acid substitutions showed a decrease in activity, albeit moderate (10 to 20%). Statistical significance was assessed by comparing the DNA replication activity of each E1 mutant protein to that of wild-type E1 (white bars) using one-way ANOVA followed by Dunnett's post hoc analysis. Significant differences are indicated (*, P ≤ 0.05; ***, P ≤ 0.001). Standard deviations are indicated by the error bars. (C) Expression of GFP-tagged wild-type E1 and mutant derivatives. Extracts from transfected cells were separated on an SDS-12% PAGE gel prior to immunoblotting with an anti-GFP antibody. Tubulin was used as a loading control.
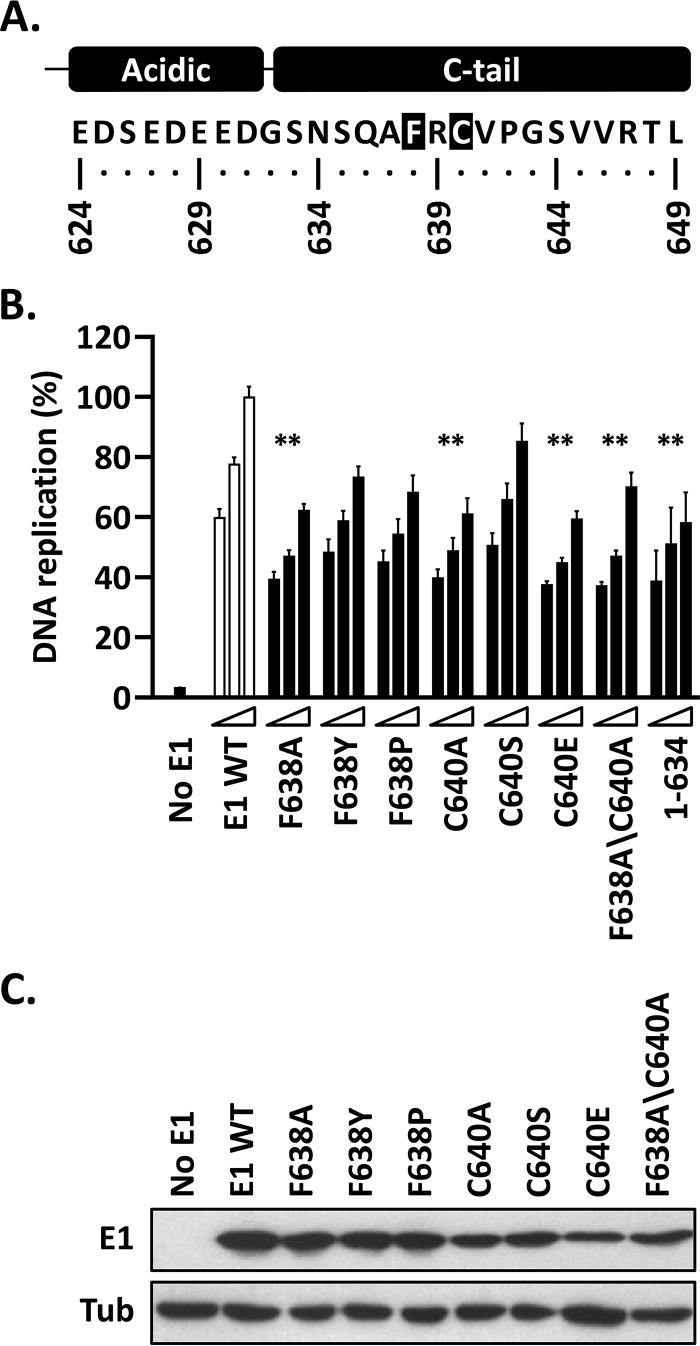
Effect of mutation of conserved residues within the C-tail domain on HPV DNA replication.
Alignment of the C-tail regions of E1 proteins revealed conservation of some amino acid residues, with the most conserved being F638 and C640 (of HPV11 E1) (Fig. 7A). To evaluate the roles of these two residues, we mutated each to an alanine, in addition to making other more or less conservative replacements. The results presented in Fig. 7B show that the most conservative changes (F638Y, F638P, and C640S) had little or no effect on the activity of E1, while the more drastic ones (F638A, C640A, C640E, and F638A/F640A) reduced DNA replication levels significantly (P < 0.01) and to the same extent as deletion of the entire C-tail [E1(1–634)]. All the mutant proteins were expressed at comparable levels (Fig. 7C). The finding that amino acid substitutions such as F368A and C640A and their combination, F368A/C640A, almost completely eliminated the function of the C-tail highlights the importance of the conserved phenylalanine and cysteine residues for the activity and/or structure of the domain.
FIG 7.
Replacement of the highly conserved F638 and C640 residues in HPV11 E1 abrogates the function of the C-tail. (A) Representation and amino acid sequence of the C terminus of HPV11 E1 highlighting the two most highly conserved amino acid residues in the C-tail, F638 and C640. (B) Mutant E1 proteins carrying the indicated amino acid substitutions at F638 and/or C640 were evaluated for HPV11 DNA replication activity (as for Fig. 3B). The activity of E1(1–634), which lacks a functional C-tail, is shown for comparison. Statistical significance was assessed by comparing the DNA replication activity of each E1 protein to that of wild-type E1 (white bars) using one-way ANOVA followed by Dunnett's post hoc analysis. Significant differences are indicated (**, P ≤ 0.01). (C) Expression of GFP-tagged wild-type E1 and mutant derivatives. Extracts from transfected cells were separated on an SDS-12% PAGE gel prior to immunoblotting with an anti-GFP antibody. Tubulin was used as a loading control.
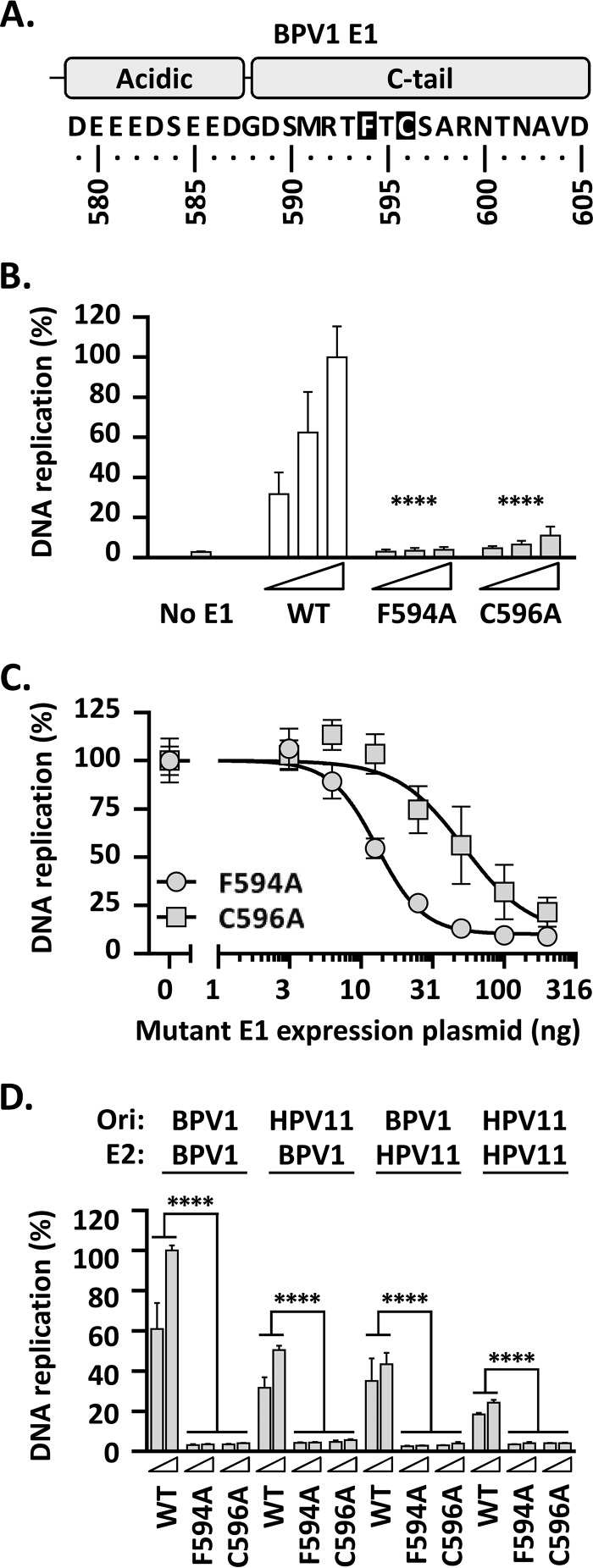
Mutation of the conserved F/C residues in the BPV1 E1 C-tail dramatically inhibits BPV1 DNA replication in vivo.
Our analysis of the HPV11 E1 C-tail has shown that truncation of the domain decreases DNA replication activity by 40%, an effect that could be recapitulated by mutation of the conserved F638 or C640 residue. Shortly after we made these observations, it was reported that mutation of the analogous residues in BPV1 E1, F594 and C596, had much more dramatic effects on BPV1 DNA replication in vitro, inhibiting the process almost completely (17). To evaluate if this could be due to differences between the in vitro and cell-based assays, we created the equivalent F594A and C596A substitutions in BPV1 E1 (Fig. 8A) and analyzed the activities of the resulting mutant proteins in our previously described cell-based BPV1 DNA replication assay (which is based on the same luciferase readout used in the HPV11 assay shown in Fig. 2) (25). Unlike the results obtained with HPV11, replacement of the conserved F594 or C596 with alanine had a dramatic effect on BPV1 DNA replication, decreasing replication levels by more than 90% (P < 0.0001) (Fig. 8B), consistent with what was reported for these substitutions in vitro (25). Unfortunately, we have been unable to assess the expression levels of any BPV1 E1 proteins (wild type or mutant) in these experiments due to the lack of a suitable antibody sensitive enough for immunoblotting. As an alternative, however, we have been able to demonstrate that the two BPV1 mutant proteins, E1 F594A and E1 C596A, act as dominant-negative inhibitors of BPV1 DNA replication (Fig. 8C). The amount of mutant E1 expression plasmid required to inhibit BPV1 DNA replication by 50% was determined, using nonlinear regression analysis of the data, to be 13 ng for E1 F638A and 37 ng for E1 C640A; the fact that these values are close to the 10-ng amount of wild-type E1 expression plasmid used in the assays suggests that both mutant proteins compete efficiently with wild-type E1 and, hence, that they must be expressed at comparable levels.
FIG 8.
Replacement of the highly conserved F594 and C596 residues in BPV1 E1 indicates an essential role for the C-tail in BPV1 DNA replication. (A) Depiction and amino acid sequence of the BPV1 E1 C terminus highlighting the two most highly conserved amino acid residues in the C-tail, F594 and C596, analogous to F638 and C640 in HPV11 E1 (Fig. 7). (B) The DNA replication activities of mutant BPV1 E1 proteins carrying the F594A and C596A substitutions were tested for the ability to support BPV1 DNA replication using a luciferase-based assay (25) similar to the one described in this study for HPV11 and using three amounts of BPV1 E1 expression vector (2.5, 5, and 10 ng) 72 h posttransfection. DNA replication levels are reported as percentages of the activity obtained with 10 ng of wild-type BPV1 E1 expression plasmid (WT). Note the more profound effect of the two C-tail substitutions on the activity of BPV1 E1 compared to HPV11 (Fig. 7B). Statistical significance was assessed by comparing the DNA replication activity of each BPV1 E1 mutant protein to that of wild-type E1 (white bars) using one-way ANOVA followed by Dunnett's post hoc analysis. Significant differences are indicated (****, P ≤ 0.0001). (C) Dominant-negative inhibition of BPV1 DNA replication by E1 F638A and E1 C640A. BPV1 DNA replication was performed using 10 ng of wild-type E1 expression plasmid and increasing amounts of expression vector (0, 3.13, 6.26, 12.5, 25, 50, 100, and 200 ng) for either E1 F638A or E1 C640A, as indicated. (D) Abilities of the WT and mutant BPV1 E1 proteins to support DNA replication using the origin and/or E2 from HPV11. HPV11 or BPV1 DNA replication assays were performed with 5 and 10 ng of BPV1 E1 expression plasmid (wild-type or mutant proteins), together with 2.5 ng of origin plasmid (Ori) and 10 ng of E2 expression plasmid from either BPV1 or HPV11, as indicated at the top. DNA replication levels were determined 72 h posttransfection and are reported as percentages of the activity measured with wild-type E1, E2, and the origin from BPV1. Statistical significance was assessed by comparing the DNA replication activity of each BPV1 E1 mutant protein to that of wild-type E1, using one-way ANOVA followed by Dunnett's post hoc analysis. Significant differences are indicated (****, P ≤ 0.0001). Standard deviations are indicated by the error bars.
The fact that the F638A and C640A substitutions completely abrogate the activity of BPV1 E1 suggests that the C-tail may play a more important role in BPV1 DNA replication than in HPV11 DNA replication, at least under our assay conditions. To address this question more directly, we tested if the two substitutions in the BPV1 E1 C-tail had a similar effect when the BPV1 origin and/or BPV1 E2 were replaced by their counterparts from HPV11 (Fig. 8D). While the levels of DNA replication driven by the BPV1 E1 wild-type protein were lower when using the HPV11 origin (P < 0.0001) (Fig. 8D), and even more so when using the HPV11 E2 protein in place of BPV1 E2 (P < 0.0001) (Fig. 8D), no combination of HPV11 origin or E2 protein alleviated the dramatic inhibition of PV DNA replication imposed by the F594A or C596A substitution in BPV1 E1 (Fig. 8D). This led us to conclude that the drastic effects of these substitutions on BPV1 E1 are inherent in the BPV1 E1 protein and not a function of the way it cooperates with E2 or the origin.
Chimeric HPV11 E1 proteins with BPV1 E1 C termini only partially restore HPV11 DNA replication in vivo.
The finding that the HPV11 and BPV1 E1 proteins show differences in their sensitivities to mutations of the most highly conserved amino acids in the C-tail was surprising, given the evolutionary conservation of the E1 C terminus. This prompted us to investigate if the C terminus of BPV1 E1 could replace that of HPV11 E1 in supporting HPV11 E1-driven DNA replication. Two chimeras (Ch617 and Ch629) were created, in which the BPV1 E1 C terminus was fused to HPV11 E1 at residues 617 and 620, respectively (Fig. 9A). The residue 617 chimera fusion point was chosen because it corresponds to the end of helix 9 and of the region necessary for E2 interaction, in addition to being the last part of E1 that is resolved in the different crystal structures of the protein (18–20). Residue 620 was chosen as a fusion point because it includes the three serines at HPV11 E1 residues 618 to 620, which are conserved in many of the low-risk HPV types but not in BPV1 E1 (Fig. 1). In both chimeras, the AR and C-tail are derived entirely from BPV1 E1. The DNA replication activities of both chimeras were compared to the replication activities of full-length HPV11 E1 and truncations ending at residues 617 and 620. The replication activities of the two truncated E1 proteins were minimal (<20% of that of HPV11 E1), consistent with the results presented in Fig. 4. The DNA replication activities of the E1 Ch617 and Ch620 chimeras (50 to 60% of wild-type levels) were substantially higher than those of the parental truncations (P < 0.001 and P < 0.001, respectively) (Fig. 9B) but were not completely restored compared to that of the full-length HPV11 E1 protein (Fig. 9B). The replication levels supported by Ch617 and Ch620 were comparable to those measured for HPV11 E1 proteins lacking the C-tail but retaining an intact AR (Fig. 5). Consistent with this, critical mutations that abolish the function of the BPV1 E1 C-tail (F594A and C596A) (Fig. 8B) had no effect on this partial rescue by Ch617 and Ch620 (Fig. 9B). All E1 proteins were expressed at comparable levels (Fig. 9C). Together, these results suggest that the partial rescue of activity observed with both E1 chimeras is due to the BPV1 AR functionally replacing the HPV11 E1 AR, with little or no contribution from the BPV1 E1 C-tail.
FIG 9.
The C terminus of HPV11 E1 can be partially replaced by the analogous domain from BPV1 E1. (A) Depiction and amino acid sequence of the C termini of the two HPV11/BPV1 E1 chimeras created in this study. The regions highlighted in black are from HPV11 E1, while those in gray are from BPV1 E1. Chimera 617 (Ch617) is comprised of residues 1 to 617 of HPV11 E1 fused to amino acids 572 to 605 of BPV1 E1. Ch620 contains residues 1 to 120 of HPV11 E1 fused to amino acids 575 to 605 of BPV1 E1. Mutant derivatives of both chimeras were also created by replacing the two conserved C-tail residues F594 and C596 (boxed) with alanines. (B) DNA replication levels supported by the Ch617 and Ch620 chimeras and mutant derivatives were measured using the HPV11 DNA replication assay essentially as described in the legend to Fig. 3B but using 2.5, 1.25, and 0.625 ng of E1 expression vector. DNA replication activities were measured 72 h posttransfection and are reported as percentages of the levels obtained with 2.5 ng of wild-type HPV11 E1 expression plasmid [E1(1–649)]. Also presented are the levels of DNA replication supported by HPV11 truncated E1 proteins ending at residues 617 and 620 to depict the “baseline” DNA replication value for each chimera in the absence of the BPV1 E1 C terminus. Statistical significance was assessed by comparing the DNA replication activity of each chimera to that of its parental truncated protein (white bars) using one-way ANOVA followed by Dunnett's post hoc analysis. Significant differences are indicated (***, P ≤ 0.001; ****, P ≤ 0.0001). (C) Expression of the indicated E1 proteins and chimeras. Extracts from transfected cells were separated on an SDS-10% PAGE gel prior to immunoblotting with an anti-GFP antibody. Tubulin was used as a loading control.
DISCUSSION
Structure-function studies of the papillomavirus E1 helicase have revealed the modular nature of the enzyme, likely arising from the acquisition of functional domains during the course of evolution (10). Amino acid sequence alignment of E1 proteins from different PV types showed that the enzyme has acquired a CTD at the end of the SF3 ATP-binding site, comprised of an alpha helix followed by an approximately 45-amino-acid region termed the FB or CTM (16, 17). Recent biochemical studies have shown that the FB/CTM of BPV1 E1, which is not observed or present in any of the crystal structures available for the protein, plays an important role in the DNA replication function, in particular for the assembly of BPV1 E1 as a double hexamer at the origin (17).
The current study complements previous reports in three ways. First, it assesses the function of the CTD of a second E1 protein, that of HPV11. Second, it evaluates the function of the E1 CTD in vivo in a cell-based viral DNA replication assay. Third, it extends the deletion and mutational analyses of the E1 CTD to helix 9 to assess its role in DNA replication and E2 binding. Figure 10A shows a synopsis of our findings, highlighting the fact that removal of as few as 5 amino acids from the C-terminal end of HPV11 E1 results in a substantial decrease in DNA replication activity (to approximately 60% of wild-type levels), which is maintained upon further deletion throughout the C-tail and even of a few residues into the AR. However, as deletions proceed further into the AR, there is another drop in DNA replication activity, down to approximately 20% of wild-type levels. Eventually, as deletions begin to encroach into helix 9, DNA replication is abrogated to background levels (<10%), and these truncated E1 proteins no longer bind to the HPV E2 protein (Fig. 4 and 5). Thus, our deletion analysis defined three functional regions of the E1 CTD, which roughly correspond to the C-tail, the AR, and helix 9.
FIG 10.
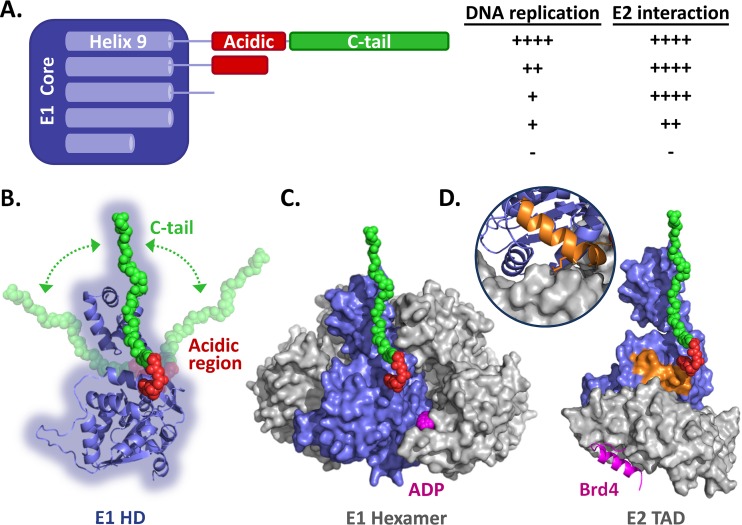
Summary of the data and homology models of the E1 helicase domain. (A) Schematic representation of the HPV11 E1 core domain (aa 1 to 617) fused to the linker sequence, AR, and C-tail domain. Helix 9, which is required for the proper folding of the helicase domain, is diagrammed within the E1 core region. Also shown is a summary of the results presented here for the effects of removing the C-tail, AR, and linker sequence and of deleting part of helix 9 on the ability of HPV11 E1 to support viral DNA replication and to interact with E2 (++++, like wild-type E1; ++, ∼50% reduction; +, ∼75% reduction; −, inactive). (B) Homology model of the HPV11 E1 HD based on the structures of the analogous regions from BPV1 and HPV18 (18–20). A cartoon representation of the AR (red) and C-tail (green), drawn to scale, is shown in an extended conformation to suggest how far in space these two regions could possibly reach. The arrows are meant to represent the flexible nature of the E1 C terminus. (C) Model of the HPV11 E1 HD in its hexameric form. ADP is colored magenta. (D) Model of the HPV11 E1 HD (blue) in complex with the E2 TAD (gray) and Brd4 helix (magenta). The model is based on the structures of the HPV18 E1-E2 complex and the HPV18 E2-Brd4 complex (35). The C-terminal alpha helix of Brd4, which binds on the opposite face of the E2 TAD (gray), is colored magenta. The inset shows the interaction of helix 9 (orange) with the E2 TAD. Also shown in orange is the side chain of arginine 616 that likely contacts the E2 TAD directly, as observed for the analogous residue (arginine 622) in the HPV18 E1-E2 TAD structure (20).
Some of our findings are consistent with the previous biochemical studies of the BPV1 E1 FB/CTM, including the overall importance of the AR and C-tail region for E1's DNA replication function. However, there are also some notable differences. Many of the truncations and point mutations that we analyzed within the E1 FB/CTM had much less of an effect on HPV11 DNA replication in vivo than has been reported for BPV1 in vitro (17). For example, complete removal of the BPV1 E1 C-tail [BPV1 E1(1–584)] or mutation of the two conserved residues F594 and C596 to alanine completely eliminates the ability of E1 to support DNA replication in vitro (17), while the corresponding truncations and point mutations in HPV11 E1 (F638A and C640A) only reduce DNA replication activity by 40 to 50% (Fig. 7). Furthermore, whereas the combined deletion of the AR and C-tail totally abrogates the DNA replication activity of the BPV1 protein [E1(1–577)] in vitro (16), it stills allows approximately 15% replication in the HPV11 cell-based assay [E1(1–623)] (Fig. 4C). According to the model proposed by Schuck and Stenlund (17), the reduction in viral DNA replication resulting from the combined deletion of the AR and C-tail is due to the inability of the truncated E1 to assemble into double trimers at the origin. Likewise, the decrease in PV DNA replication brought about by deletion or mutation of the C-tail is attributable to a failure to convert E1 double trimers into double hexamers at the origin, a suggestion consistent with the dominant-negative phenotype of the BPV1 E1 F594A and C596A mutant proteins (Fig. 8C). However, the fact that HPV11 E1 mutant proteins with a truncated or mutated C-tail can still support DNA replication up to 60% of wild-type levels indicates that double hexamers must be able to form and thus that the conversion of double trimers into double hexamers may not be as stringently regulated by the C-tail in the HPV11 system. Importantly, we have shown that these disparities are not due to differences in assay conditions, since mutation of conserved residues in the BPV1 E1 C-tail results in a more dramatic inhibition of DNA replication in vivo than the corresponding mutations in HPV11 E1 (Fig. 8). This suggests that while the C terminus is clearly important for both HPV11 and BPV1 E1 proteins, there are differences in the precise roles of the E1 C-tail for different E1 molecules. Further evidence that the C-tails of HPV11 and BPV1 E1 proteins are not equivalent came from the observation that a simple 5-amino-acid deletion from the C terminus of HPV11 E1 had a significant effect on DNA replication (a decrease of approximately 35% to 40%), while a 5-amino-acid deletion from BPV1 E1 had virtually no effect (17). The uniqueness of the C-tail also became apparent in our characterization of the chimeric HPV11/BPV1 E1 proteins, which revealed that the FB/CTM of BPV1 E1 can only partially substitute for the analogous region of HPV11 E1 (Fig. 9). We attributed this partial rescue to the function of the AR and not to that of the C-tail, as mutation of the latter domain had no effect on the DNA replication activities of both chimeric enzymes (Fig. 9). Thus, while the function of the HPV11 E1 AR can be replaced by that of BPV1 E1, the activity of the C-tail is not interchangeable between these viral types. Overall, these findings suggest that the FB/CTM does not function as an entirely autonomous module and that the function of the C-tail is not limited to its interaction with the AR but likely also involves other determinants in E1 that differ between the BPV1 and HPV11 enzymes. One such determinant underlying the “type specificity” of the C-tail might be the reported interaction between the AR and the oligomerization domain, which ultimately needs to be counteracted by the C-tail to allow conversion of the E1 double trimer into a double hexamer (17). Slight differences in the sequence, structure, and/or strength of the E1 oligomerization domain may therefore account for the specificity of the C-tail. Additional experiments will be needed to address this possibility. Finally, it is also worth keeping in mind that the portion of the E1 ORF that corresponds to the C-tail overlaps the beginning of the E2 ORF in most PV genomes. This pressure for the C-tail to coevolve with the E2 N terminus could also contribute to its type specificity.
In addition to investigating the function of the FB/CTM in vivo, our study also addressed the requirement for the linker region and for helix 9 in PV DNA replication and E2 interaction. The fact that a truncation removing only a few amino acid residues in helix 9 [such as HPV11 E1(1–615)] resulted in impaired binding to E2 was somewhat surprising, given that it had been shown previously for HPV16 E1 that larger deletions extending upstream of this region (up to HPV16 E1 aa 583, corresponding to HPV11 E1 aa 584) did not significantly alter its interaction with HPV16 E2 in vitro at 20°C (34). However, the same study also indicated that helix 9 was required for E2 interaction when binding assays were performed at 4°C (34). The latter result is more in line with our findings and with the crystal structure of the HPV18 E1-E2 complex, which revealed at least one minor contact between the E2 TAD and a conserved arginine residue located at the end of helix 9 (R622 of HPV18 E1, corresponding to R616 in HPV11 E1 and to R615 in HPV16 E1 [highlighted in Fig. 10D]), although the major contacts with E2 involve R454 and surrounding residues (20). Abbate et al. also suggested that helix 9 might be required for the proper folding of the E1 HD (20), a suggestion supported by our findings that HPV11 E1 proteins with truncations into helix 9 appeared to precipitate in the cytoplasm (Fig. 5B). Overall, our results with HPV11 E1 suggests that helix 9 is required for E2 interaction and for the folding of the E1 HD, with E1(1–617) being the shortest E1 truncation that retained the ability to bind to E2 and to efficiently localize with it into nuclear foci (Fig. 5B). A detailed mutational analysis of helix 9 and flanking regions is currently being performed in our laboratories and has already confirmed that mutations of conserved residues in helix 9 and in the first half of the linker region can impair E2 binding (unpublished data).
To help conceptualize where the E1 FB/CTM might be located in space relative to the other subdomains of E1, we generated a homology model of the HPV11 E1 HD based on the structures of the analogous regions from BPV1 and HPV18 (Fig. 10B) (18–20). Figure 10C and D shows this model of the HPV11 E1 HD monomer (blue) either in the context of an E1 HD hexamer (with the five other E1 HDs colored gray) (Fig. 10C) or in complex with both the HPV11 E2 transactivation domain (gray; also obtained by homology modeling) and the Brd4-interacting helix (purple) (35) and with E1 helix 9 highlighted (orange) (Fig. 10D). The AR (red) and the C-tail (green), which are predicted to be disordered, were added to each structure in an extended conformation to depict the possible “reach” that the FB/CTM could have relative to the other E1 subdomains and/or interacting partners. Clearly, an extended E1 C terminus could contact a neighboring E1 monomer in the context of an E1 hexamer (Fig. 10C). The results of SAXS studies have indicated that the E1 FB/CTM does indeed interact with an adjacent E1 monomer within an E1 hexamer, likely as a means of stabilizing the hexameric helicase conformation (16). Schuck and Stenlund, on the other hand, suggested that the role of the E1 FB/CTM is mediated primarily through interaction of the AR with the E1 oligomerization domain to prevent premature oligomerization of the helicase domain and thus favor the assembly of E1 into a double trimer at the origin (17). In this context, we note that the BPV1 E1 AR is predicted to lie near a positively charged patch on the surface of the oligomerization domain, formed in part by lysines 383 and 387; mutagenesis of these residues could be used to probe their involvement in binding to the AR. Schuck and Stenlund also suggested that the function of the C-tail in counteracting the effect of the AR might be triggered upon binding to single-stranded DNA (17). If so, the general location of the C-tail presented in Fig. 10C would suggest that it is best positioned to bind to the DNA strand that is routed on the surface of the E1 hexamer rather than to the one encircled by the β-hairpins. Finally, we note that the E1 FB/CTM is also well situated to occlude and/or modulate the ATP-binding cleft, an attractive possibility given the stimulatory role of ATP in E1 oligomerization (Fig. 10C). As an extended E1 FB/CTM would be long enough to contact the E2 TAD, even reaching as far as the Brd4-binding surface (Fig. 10D), one should also keep in mind the possibility that the C-tail could modulate the activity of E2, although this should be regarded as highly speculative at this time given the lack of evidence in support of the hypothesis. Ultimately, the less stably structured nature of the E1 FB/CTM certainly makes it possible, if not likely, that the region could be playing multiple roles in E1 function and that this may even vary to some degree between E1 molecules of different PV types, as shown in this study by the comparison of BPV1 and HPV11 E1 proteins.
ACKNOWLEDGMENTS
We thank Caleb Homiski for the construction of the HPV11 E1 F638A expression plasmid and all the members of the Archambault and Melendy laboratories for critical readings of the manuscript.
This work was supported by a grant from the Canadian Institutes of Health Research (CIHR) to J.A. and a grant from the National Institutes of Health (R01 AI095632) to T.M. that supported M.B. and T.M.
REFERENCES
- 1.Egawa N, Egawa K, Griffin H, Doorbar J. 2015. Human papillomaviruses; epithelial tropisms, and the development of neoplasia. Viruses 7:3863–3890. doi: 10.3390/v7072802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bzhalava D, Guan P, Franceschi S, Dillner J, Clifford G. 2013. A systematic review of the prevalence of mucosal and cutaneous human papillomavirus types. Virology 445:224–231. doi: 10.1016/j.virol.2013.07.015. [DOI] [PubMed] [Google Scholar]
- 3.Gillison ML, Alemany L, Snijders PJ, Chaturvedi A, Steinberg BM, Schwartz S, Castellsagué X. 2012. Human papillomavirus and diseases of the upper airway: head and neck cancer and respiratory papillomatosis. Vaccine 30(Suppl 5):F34–F54. doi: 10.1016/j.vaccine.2012.05.070. [DOI] [PubMed] [Google Scholar]
- 4.Cubie HA. 2013. Diseases associated with human papillomavirus infection. Virology 445:21–34. doi: 10.1016/j.virol.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Kadaja M, Silla T, Ustav E, Ustav M. 2009. Papillomavirus DNA replication: from initiation to genomic instability. Virology 384:360–368. doi: 10.1016/j.virol.2008.11.032. [DOI] [PubMed] [Google Scholar]
- 6.Archambault J, Melendy T. 2013. Targeting human papillomavirus genome replication for antiviral drug discovery. Antivir Ther 18:271–283. doi: 10.3851/IMP2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisk JC, Chojnacki MD, Melendy T. 2013. Replicative helicases as the central organizing motor proteins in the molecular machines of the elongating eukaryotic replication fork. INTECH Open Access Publisher, Rijeka, Croatia. [Google Scholar]
- 8.D'Abramo CM, Fradet-Turcotte A, Archambault J. 2012. Human papillomavirus DNA replication: insights into the structure and regulation of a eukaryotic DNA replisome, p 217–239. In Gaston K (ed), Small DNA tumour viruses. Horizon Scientific Press, Norfolk, United Kingdom. [Google Scholar]
- 9.Stenlund A. 2003. Initiation of DNA replication: lessons from viral initiator proteins. Nat Rev Mol Cell Biol 4:777–785. doi: 10.1038/nrm1226. [DOI] [PubMed] [Google Scholar]
- 10.Bergvall M, Melendy T, Archambault J. 2013. The E1 proteins. Virology 445:35–56. doi: 10.1016/j.virol.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McBride AA. 2013. The papillomavirus E2 proteins. Virology 445:57–79. doi: 10.1016/j.virol.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duderstadt KE, Berger JM. 2013. A structural framework for replication origin opening by AAA+ initiation factors. Curr Opin Struct Biol 23:144–153. doi: 10.1016/j.sbi.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorbalenya AE, Koonin EV, Wolf YI. 1990. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett 262:145–148. doi: 10.1016/0014-5793(90)80175-I. [DOI] [PubMed] [Google Scholar]
- 14.Sigrist CJ, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, Bougueleret L, Xenarios I. 2013. New and continuing developments at PROSITE. Nucleic Acids Res 41:D344–D347. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Titolo S, Pelletier A, Pulichino AM, Brault K, Wardrop E, White PW, Cordingley MG, Archambault J. 2000. Identification of domains of the human papillomavirus type 11 E1 helicase involved in oligomerization and binding to the viral origin. J Virol 74:7349–7361. doi: 10.1128/JVI.74.16.7349-7361.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whelan F, Stead JA, Shkumatov AV, Svergun DI, Sanders CM, Antson AA. 2012. A flexible brace maintains the assembly of a hexameric replicative helicase during DNA unwinding. Nucleic Acids Res 40:2271–2283. doi: 10.1093/nar/gkr906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuck S, Stenlund A. 2015. A conserved regulatory module at the C terminus of the papillomavirus E1 helicase domain controls E1 helicase assembly. J Virol 89:1129–1142. doi: 10.1128/JVI.01903-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanders CM, Kovalevskiy OV, Sizov D, Lebedev AA, Isupov MN, Antson AA. 2007. Papillomavirus E1 helicase assembly maintains an asymmetric state in the absence of DNA and nucleotide cofactors. Nucleic Acids Res 35:6451–6457. doi: 10.1093/nar/gkm705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enemark EJ, Joshua-Tor L. 2006. Mechanism of DNA translocation in a replicative hexameric helicase. Nature 442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 20.Abbate EA, Berger JM, Botchan MR. 2004. The X-ray structure of the papillomavirus helicase in complex with its molecular matchmaker E2. Genes Dev 18:1981–1996. doi: 10.1101/gad.1220104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng W, Jin G, Lin BY, Van Tine BA, Broker TR, Chow LT. 2003. mRNA splicing regulates human papillomavirus type 11 E1 protein production and DNA replication. J Virol 77:10213–10226. doi: 10.1128/JVI.77.19.10213-10226.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fradet-Turcotte A, Morin G, Lehoux M, Bullock PA, Archambault J. 2010. Development of quantitative and high-throughput assays of polyomavirus and papillomavirus DNA replication. Virology 399:65–76. doi: 10.1016/j.virol.2009.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ustav M, Stenlund A. 1991. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J 10:449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gagnon D, Fradet-Turcotte A, Archambault J. 2015. A quantitative and high-throughput assay of human papillomavirus DNA replication. Methods Mol Biol 1249:305–316. doi: 10.1007/978-1-4939-2013-6_23. [DOI] [PubMed] [Google Scholar]
- 25.Gagnon D, Senechal H, D'Abramo CM, Alvarez J, McBride AA, Archambault J. 2013. Genetic analysis of the E2 transactivation domain dimerization interface from bovine papillomavirus type 1. Virology 439:132–139. doi: 10.1016/j.virol.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blasche S, Koegl M. 2013. Analysis of protein-protein interactions using LUMIER assays. Methods Mol Biol 1064:17–27. doi: 10.1007/978-1-62703-601-6_2. [DOI] [PubMed] [Google Scholar]
- 27.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J, Zhang Y. 2015. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res 43:W174–W181. doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu D, Zhang Y. 2011. Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys J 101:2525–2534. doi: 10.1016/j.bpj.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ko J, Park H, Heo L, Seok C. 2012. GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res 40:W294–W297. doi: 10.1093/nar/gks493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGuffin LJ, Atkins JD, Salehe BR, Shuid AN, Roche DB. 2015. IntFOLD: an integrated server for modelling protein structures and functions from amino acid sequences. Nucleic Acids Res 43:W169–W173. doi: 10.1093/nar/gkv236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Doorslaer K, Tan Q, Xirasagar S, Bandaru S, Gopalan V, Mohamoud Y, Huyen Y, McBride AA. 2013. The Papillomavirus Episteme: a central resource for papillomavirus sequence data and analysis. Nucleic Acids Res 41:D571–578. doi: 10.1093/nar/gks984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masterson PJ, Stanley MA, Lewis AP, Romanos MA. 1998. A C-terminal helicase domain of the human papillomavirus E1 protein binds E2 and the DNA polymerase alpha-primase p68 subunit. J Virol 72:7407–7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abbate EA, Voitenleitner C, Botchan MR. 2006. Structure of the papillomavirus DNA-tethering complex E2:Brd4 and a peptide that ablates HPV chromosomal association. Mol Cell 24:877–889. doi: 10.1016/j.molcel.2006.11.002. [DOI] [PubMed] [Google Scholar]