Abstract
The recently identified arenavirus Lujo virus (LUJV) causes fatal hemorrhagic fever in humans. We analyzed its mechanism of viral release driven by matrix protein Z and the cell surface glycoprotein precursor GPC. The L domains in Z are required for efficient virus-like particle release, but Tsg101, ALIX/AIP1, and Vps4A/B are unnecessary for budding. LUJV GPC is cleaved by site 1 protease (S1P) at the RKLM motif, and treatment with the S1P inhibitor PF-429242 reduced LUJV production.
TEXT
The family Arenaviridae is divided into Old World (OW) and New World (NW) arenaviruses on the basis of the results of geographical, serological, and phylogenetic analyses. The OW arenaviruses include Lassa virus (LASV), the causative agent of Lassa fever, which is endemic to West Africa and has been estimated to infect several hundred thousand individuals annually. The OW arenavirus Lujo virus (LUJV) was associated with a fatal outbreak of hemorrhagic fever in South Africa in 2008. The first patient was transferred from Zambia to South Africa, and four of five patients died (1). LUJV represents a new member of the family Arenaviridae and is the second OW arenavirus reported to cause hemorrhagic fever (1–3). Arenaviruses are enveloped viruses with bisegmented negative-strand RNA genomes consisting of L and S segments. The L segment encodes viral matrix protein Z and RNA-dependent RNA polymerase L, while the S segment encodes nucleoprotein NP and the cell surface glycoprotein precursor GPC in the opposite direction (2). Whole-genome sequence data revealed a unique placement of LUJV among currently known arenaviruses, predicting altered pathogenesis and replication strategies (3). The alignment of LUJV Z is especially unique, as it possesses PT/SAP and YREL sequences, which are known as L-domain motifs critical for virus budding in other viruses and are not conserved among other arenaviruses (4). In addition, although LUJV belongs to the OW arenaviruses, phylogenetic analysis showed that LUJV GPC shows sequence similarity to NW arenaviruses. Interestingly, a recent study showed that the GP-mediated cell entry of LUJV utilizes mechanisms different from those of both OW and NW arenaviruses (5). These observations prompted us to analyze the late stage of LUJV replication, driven by Z and GPC. As the late step of the LUJV life cycle in the cell has not been characterized, we examined the molecular mechanism of LUJV assembly and budding.
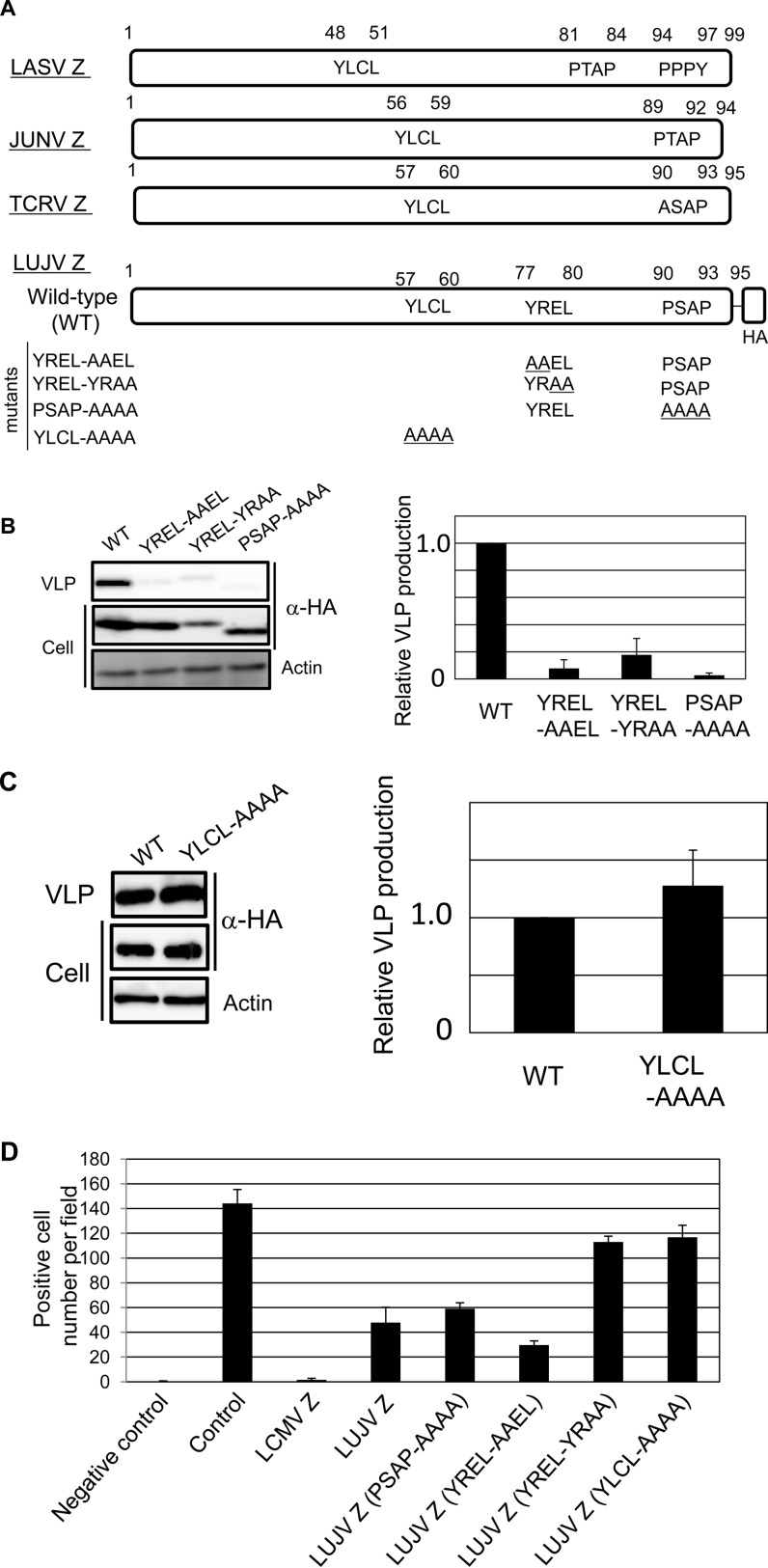
First, we examined whether the putative L domains within LUJV Z have roles in virus-like particle (VLP) production. The L domain is a short amino acid motif that interacts with cellular factors to facilitate the budding process (4, 6, 7). There are three well-characterized L-domain motifs, PT/SAP, PPxY, and YPXnL (or YxxL), that interact with Tsg101 (8), Nedd4-like E3 ligases (9–14), and ALIX/AIP1 (15–19), respectively. The roles of L domains in arenavirus budding have been well characterized (20–22). LUJV Z possesses the YREL motif, which is not conserved in other arenaviruses, and a PSAP motif at the C terminus (Fig. 1A) (4). LUJV Z also possesses the YLCL (YxxL) motif in the RING domain, similar to other arenavirus Z proteins (4). First, we focused on the roles of the YREL and PSAP motifs, which are located at the C terminus of LUJV Z, in VLP production. The LUJV Z-encoding gene, with a hemagglutinin (HA) tag at the C terminus of Z, was inserted into the pCAGGS plasmid (23) to construct pC-LUJV-Z-HA. Three mutant forms with alanine substitutions within the L domains were constructed (YREL-AAEL, YREL-YRAA, and PSAP-AAAA) (Fig. 1A). The wild type (WT) and all of the mutant forms were transfected into 293T cells. At 48 h posttransfection, Z expression in cells and VLP fractions was examined as described previously (24, 25). Samples were separated by SDS-PAGE, followed by Western blotting (WB) to detect Z with an anti-HA antibody (6E2; Cell Signaling Technology, Danvers, MA). All L-domain mutants showed significantly less VLP production than the WT (Fig. 1B), suggesting the importance of the YREL and PSAP motifs within LUJV Z for VLP production. Labeling with an anti-actin antibody (A1978 clone AC-15; Sigma, St. Louis, MO) was used as a loading control. Previously, we and other groups showed that the YLCL motif of Tacaribe virus (TCRV) Z does not play a role in VLP production (24, 26). As LUJV Z also possesses the YLCL motif in the middle of the Z protein, we next examined whether the YLCL motif in LUJV Z plays a role in VLP production. We constructed another plasmid containing AAAA instead of YLCL (YLCL-AAAA) (Fig. 1A), which we transfected into 293T cells as described above to examine the efficiency of VLP production. The normalized results showed that there was no significant reduction in VLP production compared to that of the WT control (Fig. 1C).
FIG 1.
Characterization of L-domain mutant forms of LUJV Z. (A) Schematic representation of LASV Z, JUNV Z, TCRV Z, and WT and mutant forms of LUJV Z. The WT and all mutant forms of LUJV Z possess an HA tag at the C terminus. (B) VLP production driven by the WT and mutant forms was analyzed by WB. Actin (used as a loading control) was also detected (bottom). Normalized VLP production (VLPs per cell) is shown on the right. The data shown are averages and standard deviations of three independent experiments. (C) VLP production driven by the WT or mutant (YLCL-AAAA) protein was analyzed as described above. Actin (used as a loading control) was also detected (bottom). Normalized VLP production (VLPs per cell) is shown on the right. The data shown are averages and standard deviations of three independent experiments. (D) Inhibition of LCMV MG expression by LUJV Z and its mutant forms. BHK-21 cells were transfected with the expression plasmids for LCMV NP and L and with an LCMV MG containing the gene for mCherry together with the expression plasmid for LCMV Z, LUJV Z, or mutant LUJV Z. At 48 h posttransfection, cells were fixed and the numbers of mCherry-positive cells per microscopic field were automatically determined by BZ-X700 (Keyence). The data shown are averages and standard deviations of three independent experiments.
To further examine whether the defects in VLP production induced by either the YREL or the PSAP mutation were due to protein misfolding, we investigated the inhibitory effects of these Z mutations on arenavirus genome replication/transcription. Although it is not known whether LUJV Z protein affects lymphocytic choriomeningitis virus (LCMV) genome replication/transcription, we used the LCMV minigenome (MG) system for these experiments (27). NP and L are known to be sufficient for arenavirus genome replication/transcription (27). BHK-21 cells were cotransfected with pC-LCMV-NP, pC-LCMV-L, and pPol-I-LCMV-SmCherry, which is the expression plasmid for the artificial S segment encoding mCherry together with the expression plasmid for WT or mutant Z. At 48 h posttransfection, the numbers of cells expressing mCherry were determined by BZ-X700 (Keyence, Osaka, Japan). As expected, the expression of LCMV Z significantly reduced the number of mCherry-positive cells, indicating inhibition of MG expression by LCMV Z (Fig. 1D) (28–30). In addition, LUJV Z expression also reduced the number of mCherry-positive cells to approximately one-third of that of the control. Similarly, both the PSAP-AAAA and YREL-AAEL mutations reduced the number of mCherry-positive cells, suggesting that the loss of VLP production capability in these mutants was not due to overall protein misfolding. On the other hand, the inhibitory effects of the YREL-YRAA and YLCL-AAAA mutations were not significant. The YLCL motif is located in the RING finger domain. Therefore, introduction of a mutation into this motif would affect the role of Z in viral gene transcription/replication. The weak inhibitory effect of YREL-YRAA may have been due to the lower expression level in the cells (Fig. 1B).
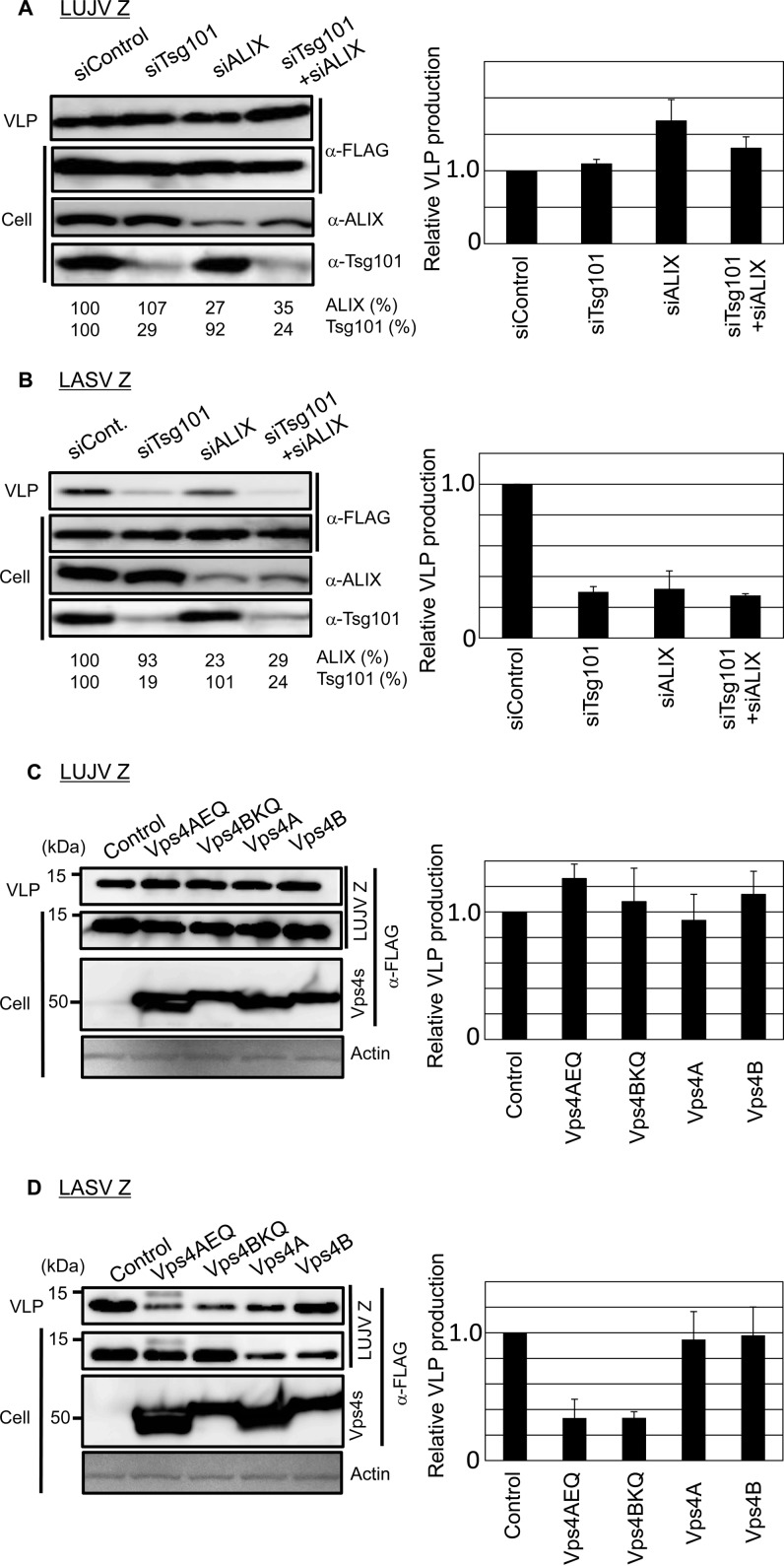
Next, we examined whether Tsg101 (21, 25), ALIX/AIP1 (31), and Vps4A/B (24, 25) are involved in LUJV VLP production. 293T cells were pretreated with small interfering RNA (siRNA) (200 nM) against Tsg101 (8, 25) and/or ALIX/AIP1 (31). At 1 day after siRNA transfection, cells were transfected with the same siRNAs (100 nM) and pC-LUJV-Z-FLAG. At 48 h after the second transfection, VLPs and cell lysates were collected and proteins were detected by WB. Endogenous Tsg101 and ALIX/AIP1 expression was detected with polyclonal antibodies GTX118736 (GeneTex, Irvine, CA) and ab76608 (Abcam, Cambridge, United Kingdom), respectively. The normalized Tsg101 and ALIX/AIP1 protein expression levels based on the bands seen on WB analysis are indicated at the bottom of Fig. 2A and B. To examine the role of Vps4A/B in LUJV Z-mediated VLP production in 293T cells, pC-LUJV-Z-FLAG was transfected with dominant negative (DN) forms of Vps4A/B (Vps4AEQ and Vps4BKQ) or WT Vps4A/B (Vps4A and Vps4B), and at 48 h posttransfection, VLPs and cell lysates were collected and proteins were detected by WB. As both the DN and WT forms of the Vps4A/B plasmids possess the FLAG tag at the N terminus (32), the expression of both the WT and DN forms of Vps4A/B and LUJV Z was detected with an anti-FLAG antibody (M2; Sigma) (Fig. 2C and D). Tsg101 and ALIX/AIP1 knockdown by siRNA and overexpression of DN forms of Vps4A and Vps4B reduced LASV Z VLP production, while overexpression of WT Vps4A/B did not affect LASV Z VLP production (Fig. 2B and D) (25). However, LUJV VLP production was not affected by any of these treatments (Fig. 2A and C), indicating that LUJV Z does not utilize these host factors for budding. These data suggest that LUJV utilizes mechanisms for budding that are different from those of other arenaviruses.
FIG 2.
Requirements of cellular factors for LUJV VLP production. (A, B) Tsg101 and/or ALIX/AIP1 were depleted by siRNA treatment, and their effects on LUJV (A) or LASV (B) Z-mediated VLP production were examined. Endogenous Tsg101 and ALIX/AIP1 were detected with specific antibodies. The LUJV and LASV Z proteins were detected with anti-FLAG antibody. Normalized Tsg101 and ALIX protein levels are shown at the bottom. (C, D) LUJV and LASV VLPs produced by pC-LUJV-Z-FLAG or pC-LASV-Z-FLAG with overexpression of DN (Vps4AEQ and Vps4BKQ) and WT Vps4A/B forms were examined by WB. Overexpressed DN and WT forms of Vps4A/B, LUJV Z, and LASV Z were detected with anti-FLAG antibody. Normalized VLP production (VLPs per cell) is shown on the right. Actin (used as a loading control) was also detected in the experiments whose results are shown in panels C and D. The data shown are averages and standard deviations of three independent experiments.
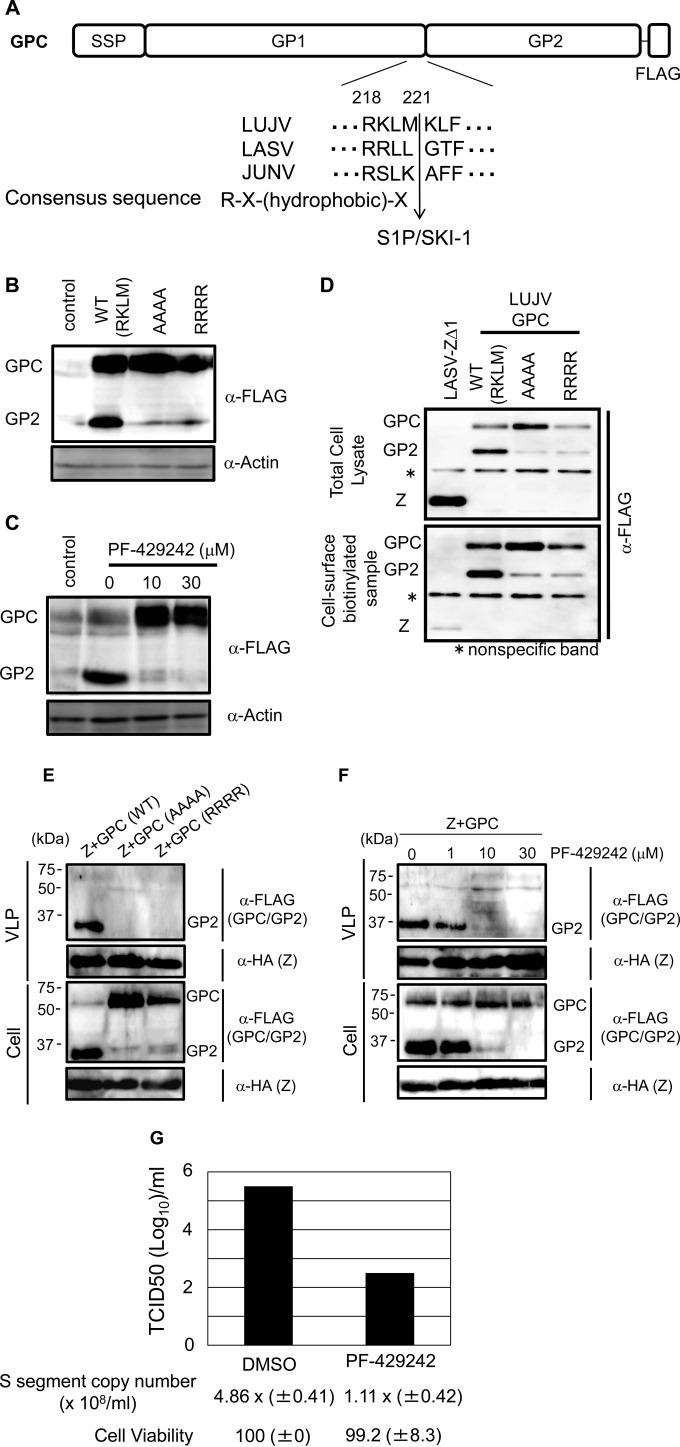
Next, we focused on the molecular mechanisms of action of GPC at the late stage of the virus life cycle. Phylogenetic analysis showed that LUJV GPC is located at similar distances between OW and NW arenaviruses (3). Alignment of LUJV GPC showed a potential S1P/SKI-1 cleavage site at RKLM (Fig. 3A). pC-LUJV-GPC-FLAG, with a FLAG tag at the C terminus of LUJV GPC, was constructed to detect both GPC and GP2 with an anti-FLAG antibody. To examine the role of the RKLM motif in GPC cleavage, two GPC mutant plasmids with RKLM replaced with AAAA or RRRR were constructed. Although GP2 was detected in the WT, no GPC cleavage products were observed in either mutant, suggesting that the RKLM motif is important for GPC cleavage (Fig. 3B). Next, to examine whether LUJV GPC is cleaved by site 1 protease (S1P/SKI-1), similar to other arenavirus GPCs (33–35), PF-429242 (catalog number 3354; Tocris Bioscience, Bristol, United Kingdom), a small chemical compound inhibitor of S1P/SKI-1, was used (36, 37). LUJV GPC cleavage was dose-dependently inhibited by PF-429242 in 293T cells (Fig. 3C). We also examined whether LUJV GPC cleavage affects GP transport to the cell surface. 293T cells were transfected with expression plasmids for WT or mutant GPCs. At 48 h posttransfection, the culture medium was replaced with fresh medium containing 0.25 mg/ml EZ-Link Sulfo-NHS-SS-Biotin (catalog number 89881; Thermo Scientific, Waltham, MA) and incubated for 30 min on ice for cell surface protein biotinylation, followed by isolation of biotinylated proteins according to the manufacturer's protocol. LASV ZΔ1, a deletion mutant form of LASV Z lacking amino acids 3 to 10 preventing it from attaching to the cell membrane because of the defect in its myristoylation (32), was used as a negative control. The levels of WT and mutant GPC expression in cells were similar (Fig. 3D, top). WT and mutant GPCs were efficiently biotinylated, while LASV ZΔ1 was weakly biotinylated (Fig. 3D, bottom), indicating that GPC cleavage is unnecessary for cell surface GPC transport. We examined whether GPC cleavage affected the incorporation of GP into Z-mediated VLPs. When WT or mutant GPC and Z were coexpressed in 293T cells, uncleaved GPCs were not incorporated into VLPs (Fig. 3E) (38). Furthermore, GP incorporation into VLP was markedly reduced by treatment with 10 or 30 μM PF-429242 (Fig. 3F). These results suggest that GPC cleavage is important for incorporation of GP1/GP2 (cleavage products of GPC) into VLPs. We next examined whether PF-429242 inhibits LUJV propagation. Vero cells were infected with LUJV at a multiplicity of infection (MOI) of 0.1 in the presence or absence (Dulbecco's modified Eagle's medium) of PF-429242 (30 μM) in a biosafety level 4 laboratory. After 2 days of incubation, culture media were collected. The 50% tissue culture infective dose was determined by endpoint dilution assay and the Spearman-Karber calculation. Briefly, an initial 1:5 dilution and subsequent 10-fold dilutions of the culture media were used to infect Vero cell monolayers in quadruplicate in the wells of a 96-well plate. At 10 days postinfection, the cells were fixed with 80% ice-cold acetone. The fixed cells were then stained with mouse anti-LUJV IgG for 60 min at 37°C; this was followed by incubation with fluorescein isothiocyanate-conjugated goat anti-mouse IgG (Invitrogen, Carlsbad, CA) for 60 min at 37°C. In addition, the viral genome copy number was determined by real-time reverse transcription (RT)-PCR with a OneStep SYBR PrimeScript RT-PCR kit (TaKaRa, Kyoto, Japan) and a LightCycler 480 (Roche, Penzberg, Germany) for virion RNA extracted from the culture media with specific primers targeting the S segment ( 5′-TCGGGGTGCCCATACCAATC-3′ and 5′-AAGCCAGAGGCCCTGGAGTC-3′). PF-429242 treatment markedly reduced LUJV production, as measured by both LUJV infectious titer and genome copy number, suggesting that S1P/SKI-1 may be a good antiviral target for LUJV, as suggested for LASV and Junin virus (JUNV) (Fig. 3G) (36, 37). Cell viabilities under these experimental conditions were examined by CellTiter-Glo Luminescent Cell Viability Assay (G7570; Promega, Madison, WI). There was no significant cell toxicity under these experimental conditions, indicating that the decrease in LUJV production was not due to cell toxicity of PF-429242 treatment (Fig. 3G).
FIG 3.
Involvement of S1P/SKI-1 in LUJV assembly and production. (A) Schematic representation of the LUJV, LASV, and JUNV GPC cleavage site. The consensus sequence for S1P/SKI-1 cleavage is also shown. (B) 293T cells were transfected with plasmid pC-LUJV-GPC-FLAG or mutant forms thereof with RKLM changed to AAAA or RRRR. GPCs were detected with anti-FLAG antibody. Actin (used as a loading control) was also detected. (C) 293T cells were transfected with pC-LUJV-GPC-FLAG and treated with increasing concentrations of PF-429242 (0, 10, or 30 μM). At 48 h posttransfection, cell lysates were collected and the protein was detected by WB. Actin (used as a loading control) was also detected. (D) WT and mutant forms of LUJV GPC were analyzed for cell surface expression. Total cell lysate (top) and a cell surface biotinylated sample (bottom), pulled down by avidin beads, are shown. LASV ZΔ1 was used as a negative control. (E and F) GPC cleavage is important for its incorporation into VLPs. 293T cells were transfected with pC-LUJV-Z-HA, together with pC-LUJV-GPC-FLAG or its mutant forms, and at 48 h posttransfection, cell lysate and VLP fractions were analyzed by WB (E). 293T cells were transfected with pC-LUJV-Z-HA, together with pC-LUJV-GPC-FLAG, and at 6 h posttransfection, the culture medium was replaced with fresh medium containing increasing amounts of PF-429242 (0, 1, 10, or 30 μM). At 48 h posttransfection, cell lysate and VLP fractions were collected and the prepared samples were analyzed by WB (F). (G) Vero cells, which were seeded at 100% confluence, were infected with infectious LUJV at an MOI of 0.1, and the culture medium was replaced with fresh medium containing 30 μM PF-429242 at 1 h postinfection. After 48 h of incubation, the culture media were collected and infectious titers of LUJV were determined by the procedure described in the text. RNA was also extracted from the culture media for quantitative RT-PCR targeting the NP gene (S segment) to quantify LUJV genome copy numbers. Cell viabilities following the same treatment except virus infection are also shown at the bottom. TCID50, 50% tissue culture infective doses; DMSO, Dulbecco's modified Eagle's medium.
In summary, L domains in LUJV Z are required for efficient VLP release, but the requirements of cellular factors for LUJV Z-mediated budding are different from those for LASV Z-mediated budding. In addition, treatment of LUJV-infected cells with the S1P inhibitor PF-429242 decreased virus production in vitro because of the involvement of S1P in GPC cleavage.
ACKNOWLEDGMENTS
We thank S. Fukushi for providing the LUJV GPC plasmid and T. Ksiazek for providing the mouse anti-LUJV IgG antibody. We are grateful to J. C. de la Torre for providing the LCMV MG system. We also thank all of the members of the Department of Emerging Infectious Diseases, Institute of Tropical Medicine (NEKKEN), Nagasaki University, and the Special Viral Pathogens Laboratory, National Institute for Communicable Diseases (NICD).
This work was supported by grants from the Ministry of Health, Labor, and Welfare of Japan to S.U. and J.Y.
REFERENCES
- 1.Paweska JT, Sewlall NH, Ksiazek TG, Blumberg LH, Hale MJ, Lipkin WI, Weyer J, Nichol ST, Rollin PE, McMullan LK, Paddock CD, Briese T, Mnyaluza J, Dinh TH, Mukonka V, Ching P, Duse A, Richards G, de Jong G, Cohen C, Ikalafeng B, Mugero C, Asomugha C, Malotle MM, Nteo DM, Misiani E, Swanepoel R, Zaki SR, Outbreak Control and Investigation Teams . 2009. Nosocomial outbreak of novel arenavirus infection, southern Africa. Emerg Infect Dis 15:1598–1602. doi: 10.3201/eid1510.090211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchmeier MJ, de la Torre JC, Peters CJ. 2013. Arenaviridae, p 1283–1303. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 3.Briese T, Paweska JT, McMullan LK, Hutchison SK, Street C, Palacios G, Khristova ML, Weyer J, Swanepoel R, Egholm M, Nichol ST, Lipkin WI. 2009. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog 5:e1000455. doi: 10.1371/journal.ppat.1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Urata S, Yasuda J. 2012. Molecular mechanism of arenavirus assembly and budding. Viruses 4:2049–2079. doi: 10.3390/v4102049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tani H, Iha K, Shimojima M, Fukushi S, Taniguchi S, Yoshikawa T, Kawaoka Y, Nakasone N, Ninomiya H, Saijo M, Morikawa S. 2014. Analysis of Lujo virus cell entry using pseudotype vesicular stomatitis virus. J Virol 88:7317–7330. doi: 10.1128/JVI.00512-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolff S, Ebihara H, Groseth A. 2013. Arenavirus budding: a common pathway with mechanistic differences. Viruses 5:528–549. doi: 10.3390/v5020528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fehling SK, Lennartz F, Strecker T. 2012. Multifunctional nature of the arenavirus RING finger protein Z. Viruses 4:2973–3011. doi: 10.3390/v4112973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, Myszka DG, Sundquist WI. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55–65. doi: 10.1016/S0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 9.Martin-Serrano J. 2007. The role of ubiquitin in retroviral egress. Traffic 8:1297–1303. doi: 10.1111/j.1600-0854.2007.00609.x. [DOI] [PubMed] [Google Scholar]
- 10.Urata S, Yasuda J. 2010. Regulation of Marburg virus (MARV) budding by Nedd4.1: a different WW domain of Nedd4.1 is critical for binding to MARV and Ebola virus VP40. J Gen Virol 91:228–234. doi: 10.1099/vir.0.015495-0. [DOI] [PubMed] [Google Scholar]
- 11.Chen BJ, Lamb RA. 2008. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yasuda J, Hunter E. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J Virol 72:4095–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasuda J, Hunter E, Nakao M, Shida H. 2002. Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep 3:636–640. doi: 10.1093/embo-reports/kvf132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yasuda J, Nakao M, Kawaoka Y, Shida H. 2003. Nedd4 regulates egress of Ebola virus-like particles from host cells. J Virol 77:9987–9992. doi: 10.1128/JVI.77.18.9987-9992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689–699. doi: 10.1016/S0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 16.Puffer BA, Parent LJ, Wills JW, Montelaro RC. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J Virol 71:6541–6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci U S A 100:12414–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irie T, Shimazu Y, Yoshida T, Sakaguchi T. 2007. The YLDL sequence within Sendai virus M protein is critical for budding of virus-like particles and interacts with Alix/AIP1 independently of C protein. J Virol 81:2263–2273. doi: 10.1128/JVI.02218-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ciancanelli MJ, Basler CF. 2006. Mutation of YMYL in the Nipah virus matrix protein abrogates budding and alters subcellular localization. J Virol 80:12070–12078. doi: 10.1128/JVI.01743-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strecker T, Eichler R, Meulen J, Weissenhorn W, Dieter Klenk H, Garten W, Lenz O. 2003. Lassa virus Z protein is a matrix protein and sufficient for the release of virus-like particles [corrected]. J Virol 77:10700–10705. doi: 10.1128/JVI.77.19.10700-10705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez M, Craven RC, de la Torre JC. 2003. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc Natl Acad Sci U S A 100:12978–12983. doi: 10.1073/pnas.2133782100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J, Danzy S, Kumar N, Ly H, Liang Y. 2012. Biological roles and functional mechanisms of arenavirus Z protein in viral replication. J Virol 86:9794–9801. doi: 10.1128/JVI.00385-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niwa H, Yamamura K, Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199. doi: 10.1016/0378-1119(91)90434-D. [DOI] [PubMed] [Google Scholar]
- 24.Urata S, Yasuda J, de la Torre JC. 2009. The Z protein of the New World arenavirus Tacaribe virus has bona fide budding activity that does not depend on known late domain motifs. J Virol 83:12651–12655. doi: 10.1128/JVI.01012-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urata S, Noda T, Kawaoka Y, Yokosawa H, Yasuda J. 2006. Cellular factors required for Lassa virus budding. J Virol 80:4191–4195. doi: 10.1128/JVI.80.8.4191-4195.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Groseth A, Wolff S, Strecker T, Hoenen T, Becker S. 2010. Efficient budding of the Tacaribe virus matrix protein Z requires the nucleoprotein. J Virol 84:3603–3611. doi: 10.1128/JVI.02429-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee KJ, Novella IS, Teng MN, Oldstone MB, de La Torre JC. 2000. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J Virol 74:3470–3477. doi: 10.1128/JVI.74.8.3470-3477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cornu TI, de la Torre JC. 2001. RING finger Z protein of lymphocytic choriomeningitis virus (LCMV) inhibits transcription and RNA replication of an LCMV S-segment minigenome. J Virol 75:9415–9426. doi: 10.1128/JVI.75.19.9415-9426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cornu TI, de la Torre JC. 2002. Characterization of the arenavirus RING finger Z protein regions required for Z-mediated inhibition of viral RNA synthesis. J Virol 76:6678–6688. doi: 10.1128/JVI.76.13.6678-6688.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cornu TI, Feldmann H, de la Torre JC. 2004. Cells expressing the RING finger Z protein are resistant to arenavirus infection. J Virol 78:2979–2983. doi: 10.1128/JVI.78.6.2979-2983.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI. 2011. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe 9:235–242. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urata S, Yasuda J. 2015. Cis- and cell type-dependent trans-requirements for Lassa virus-like particle production. J Gen Virol 96:1626–1635. doi: 10.1099/vir.0.000105. [DOI] [PubMed] [Google Scholar]
- 33.Rojek JM, Lee AM, Nguyen N, Spiropoulou CF, Kunz S. 2008. Site 1 protease is required for proteolytic processing of the glycoproteins of the South American hemorrhagic fever viruses Junin, Machupo, and Guanarito. J Virol 82:6045–6051. doi: 10.1128/JVI.02392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beyer WR, Popplau D, Garten W, von Laer D, Lenz O. 2003. Endoproteolytic processing of the lymphocytic choriomeningitis virus glycoprotein by the subtilase SKI-1/S1P. J Virol 77:2866–2872. doi: 10.1128/JVI.77.5.2866-2872.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lenz O, ter Meulen J, Klenk HD, Seidah NG, Garten W. 2001. The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc Natl Acad Sci U S A 98:12701–12705. doi: 10.1073/pnas.221447598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Urata S, Yun N, Pasquato A, Paessler S, Kunz S, de la Torre JC. 2011. Antiviral activity of a small-molecule inhibitor of arenavirus glycoprotein processing by the cellular site 1 protease. J Virol 85:795–803. doi: 10.1128/JVI.02019-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pasquato A, Rochat C, Burri DJ, Pasqual G, de la Torre JC, Kunz S. 2012. Evaluation of the anti-arenaviral activity of the subtilisin kexin isozyme-1/site-1 protease inhibitor PF-429242. Virology 423:14–22. doi: 10.1016/j.virol.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kunz S, Edelmann KH, de la Torre JC, Gorney R, Oldstone MB. 2003. Mechanisms for lymphocytic choriomeningitis virus glycoprotein cleavage, transport, and incorporation into virions. Virology 314:168–178. doi: 10.1016/S0042-6822(03)00421-5. [DOI] [PubMed] [Google Scholar]