ABSTRACT
DNAJC14, a heat shock protein 40 (Hsp40) cochaperone, assists with Hsp70-mediated protein folding. Overexpressed DNAJC14 is targeted to sites of yellow fever virus (YFV) replication complex (RC) formation, where it interacts with viral nonstructural (NS) proteins and inhibits viral RNA replication. How RCs are assembled and the roles of chaperones in this coordinated process are largely unknown. We hypothesized that chaperones are diverted from their normal cellular protein quality control function to play similar roles during viral infection. Here, we show that DNAJC14 overexpression affects YFV polyprotein processing and alters RC assembly. We monitored YFV NS2A-5 polyprotein processing by the viral NS2B-3 protease in DNAJC14-overexpressing cells. Notably, DNAJC14 mutants that did not inhibit YFV replication had minimal effects on polyprotein processing, while overexpressed wild-type DNAJC14 affected the NS3/4A and NS4A/2K cleavage sites, resulting in altered NS3-to-NS3-4A ratios. This suggests that DNAJC14's folding activity normally modulates NS3/4A/2K cleavage events to liberate appropriate levels of NS3 and NS4A and promote RC formation. We introduced amino acid substitutions at the NS3/4A site to alter the levels of the NS3 and NS4A products and examined their effects on YFV replication. Residues with reduced cleavage efficiency did not support viral RNA replication, and only revertant viruses with a restored wild-type arginine or lysine residue at the NS3/4A site were obtained. We conclude that DNAJC14 inhibition of RC formation upon DNAJC14 overexpression is likely due to chaperone dysregulation and that YFV probably utilizes DNAJC14's cochaperone function to modulate processing at the NS3/4A site as a mechanism ensuring virus replication.
IMPORTANCE Flaviviruses are single-stranded RNA viruses that cause a wide range of illnesses. Upon host cell entry, the viral genome is translated on endoplasmic reticulum (ER) membranes to produce a single polyprotein, which is cleaved by host and viral proteases to generate viral proteins required for genome replication and virion production. Several studies suggest a role for molecular chaperones during these processes. While the details of chaperone roles have been elusive, in this report we show that overexpression of the ER-resident cochaperone DNAJC14 affects YFV polyprotein processing at the NS3/4A site. This work reveals that DNAJC14 modulation of NS3/4A site processing is an important mechanism to ensure virus replication. Our work highlights the importance of finely regulating flavivirus polyprotein processing. In addition, it suggests future studies to address similarities and/or differences among flaviviruses and to interrogate the precise mechanisms employed for polyprotein processing, a critical step that can ultimately be targeted for novel drug development.
INTRODUCTION
Molecular chaperones belong to a class of proteins that help polypeptides reach a properly folded conformation, assemble into oligomeric complexes or disassemble, transport across membranes, or undergo degradation (1). A wide range of cellular activities involve chaperones (2). Chaperone-mediated folding relies on ATP-driven conformational changes and is assisted by a variety of partner cochaperones. A well-known class of chaperones is represented by heat shock protein 70 (Hsp70), whose function is regulated by cochaperone proteins of the DNAJ family. Cochaperone proteins are recruited to bind Hsp70 and stimulate its ATPase activity (3, 4). Given the numerous cellular activities that involve chaperones and the dependence of viruses on the host apparatus for their propagation, it is expected that molecular chaperones play important roles in virus life cycles. Indeed, the functions of cellular chaperones, such as Hsp70, Hsp40, and Hsp90, have been observed in the context of viral infections (reviewed in references 3 and 5). Since chaperones are upregulated during cellular stress, it is problematic to discern if chaperones are induced by and required for viral replication or are a result of the stress response caused by viral infection. Only recently, Taguwa and colleagues (6) have elucidated the role of the Hsp70 machinery in multiple steps of the dengue virus (DENV) life cycle. Interestingly, the specific activity of Hsp70 in each of these steps is determined by different DNAJ proteins (6). In a previous study, we showed that overexpression of an Hsp40 family member, DNAJC14, conferred resistance to several members of the Flaviviridae family. Viruses inhibited by overexpression of DNAJC14 included, among the members of the Flavivirus genus, yellow fever virus (YFV), DENV, the Kunjin strain of West Nile virus (WNV), and the Langat strain of tick-borne encephalitis virus; the hepacivirus hepatitis C virus (HCV) was also inhibited. DNAJC14 was previously implicated in the life cycle of bovine viral diarrhea virus (BVDV), a pestivirus also belonging to the Flaviviridae family (8–10).
Flaviviruses are enveloped, positive-strand RNA viruses. Like other flaviviruses, the YFV genome encodes a single large polyprotein that is cotranslationally inserted into the endoplasmic reticulum (ER) membrane and processed by viral and host cell proteases to yield three structural proteins (the core [C], premembrane, and envelope proteins) and seven nonstructural (NS) proteins, including NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5 (reviewed in reference 11). Viral protein expression results in the formation of characteristic membrane structures on which viral replication complexes (RCs) are assembled (12). NS proteins also perform various enzymatic functions, including protease, helicase, methyltransferase, and RNA-dependent RNA polymerase activities. NS3 is a multifunctional protein. The N-terminal region of NS3 and its cofactor, NS2B, fold into a serine protease that cleaves the viral polyprotein after dibasic residues present at the C terminus of the C protein and at NS2A/2B, NS2B/3, NS3/4A, and NS4B/5 sites. It also cleaves within NS4A to generate the 2K peptide derived from its C terminus. The C-terminal domain of NS3 possesses nucleoside triphosphatase/RNA helicase activities and is involved in viral RNA replication and virus particle formation. In addition, the NS2B-3 complex has also been shown to modulate viral pathogenesis and the host immune response (13–16).
This study focuses on the molecular mechanism by which DNAJC14 overexpression inhibits YFV infection. Our recent reports (7, 17) demonstrated the inhibitory effect of DNAJC14 overexpression on YFV replication, and the data suggested that DNAJC14 overexpression exerts its antiviral effects by inhibiting the steps required for RC formation. This led us to hypothesize that DNAJC14 overexpression likely alters the chaperone-client stoichiometry (7), disrupting the folding process and altering the protein network needed for productive RC formation (17). Given DNAJC14's critical role in the folding of multipass transmembrane proteins in the ER, such as members of the G-protein-coupled receptor (GPCR) family (18), we hypothesize that during translation DNAJC14 helps with the proper folding of the YFV polyprotein. While we previously showed that cleavage at the NS2B/3 site was unaffected (7), we wondered if improper polyprotein folding due to DNAJC14 overexpression could affect cleavage events at other sites. Here, our results demonstrate specific inhibition of NS3/4A/2K cleavage events, resulting in a marked reduction in the levels of the NS3-4A, NS3, and NS4B proteins. We employed a variety of assays to show that inhibition of these cleavage events has a significant effect on YFV fitness. Therefore, we propose that normal DNAJC14 chaperone activity is required for YFV NS3/4A cleavage, which is a highly regulated rate-limiting step occurring primarily in cis (19) to ensure production of the proper amount of the YFV NS proteins for RC formation and viral RNA replication.
MATERIALS AND METHODS
Plasmid constructs.
YFV polyprotein expression vectors, derivatives of the pET-8C system, sig2A-5356, sig2A-5356*, NS3-5356, and NS3-5356*** were previously described (19). The proteinase construct pET-8c-2B-3181 is described in reference 20.
The vectors pV1, pV1-GFP, pV1-DNAJC14 FL, pV1-DNAJC14 CT1, and pV1-DNAJC14 NT5 H471Q were previously described (7). Viral plasmid pACNR-FLYF17Dx containing the sequences of YFV 17D downstream of an SP6 promoter with XhoI as a runoff site is described in reference 21. Yellow fever virus replicon plasmid pYF-R.luc2A-RP (22) was provided by R. J. Kuhn (Purdue University). Plasmid pYF-R.luc2A-RP delta DD was described previously (7). Additional plasmids were constructed by standard cloning methods. All constructs were checked by diagnostic restriction digestion, and the PCR products were verified by sequencing. Descriptions of the constructs and the cloning strategy are presented below; all sequences and primers used are available upon request.
pNS4A-GFP.
The YFV NS4A sequence was cloned into the pEGFP-N1 expression vector with a green fluorescent protein (GFP) tag on the C terminus, resulting in pNS4A-GFP. A methionine codon for translation initiation was added to the NS4A amino terminus normally obtained through cleavage of the polyprotein.
NS3/4A P1 substitutions.
Mutations were originally created in the pET-BS(+) expression vector backbone and are described in reference 23. The SapI and NgoMIV restriction sites were used to introduce the region containing the mutations from individual pET-BS(+) constructs into pACNR-FLYF17Dx and pYF-R.luc2A-RP for mutant virus and replicon generation, respectively. The presence of the desired mutations was confirmed by nucleotide sequencing.
pACNR-YF17D NS3-4A SPLIT.
The pACNR-YF17D NS3-4A SPLIT construct was generated by introducing an expression cassette—stop codon, the encephalomyocarditis virus internal ribosome entry site (IRES) sequence, and the ubiquitin monomer—at the NS3/4A junction by performing standard overlapping PCRs, using the pACNR-FLYF17Dx and J6/JFH NS2-IRES-nsGluc2AUb (24) plasmids as the templates. The SapI and NgoMIV restriction sites were used to swap in the cassette. This construct did not generate virus. Inadvertent mutations in other sites of the viral genome were ruled out by reintroducing the correct NS3/4A junction into the bicistronic NS3-4A split genome. All data collected from this reconstituted genome were comparable to those for the wild type. This demonstrated that the observations from the SPLIT genome are solely due to the insertion of the IRES-ubiquitin segment placed between NS3 and NS4A.
Cell lines and virus growth.
SW13 cells (human adrenal carcinoma cells; CCL-105; ATCC) were cultured in minimum essential medium (MEM) alpha medium (MEMa; Invitrogen) supplemented with 10% fetal bovine serum (FBS; Invitrogen). 293T cells (CRL-3216; ATCC) were cultured in Dulbecco modified Eagle medium supplemented with 10% FBS. BHK-J cells, a previously described (25) line of BHK-21 hamster kidney cells, were cultured in MEM supplemented with 7.5% FBS. Virus growth was measured by plaque assay on BHK-J cells as described previously (7, 26).
Antibodies.
Anti-myc mouse monoclonal antibody (clone 9E10; catalog number SC-40; Santa Cruz) was used for immunoprecipitation at 3 μg of antibody/30 μl protein G Dynabeads (Invitrogen). YFV NS3-4A and NS4B rabbit polyclonal antisera were previously described (27), recognize NS3- and NS4B-containing sequences, respectively, and were utilized for immunoprecipitation at 1:500 and 1:1,000 dilutions, respectively. YFV NS3 polyclonal serum was diluted 1:10,000 for Western blotting. Rabbit polyclonal anti-GFP antiserum was generated as described previously (28) and utilized at a 1:20,000 dilution for Western blotting. Rabbit polyclonal anti-DNAJC14 antibody (catalog number HPA017653; Sigma) was used at a 1:2,000 dilution for Western blotting. Horseradish peroxidase (HRP)-conjugated secondary anti-rabbit IgG antibodies (catalog number 474-15160; KLP) were utilized at a 1:20,000 dilution.
Western blot analysis.
Protein samples in 2× sodium dodecyl sulfate (SDS) loading buffer were separated on a NuPAGE 4 to 12% bis-Tris gel (Novex; Thermo Fisher) and transferred to a nitrocellulose membrane (GE Healthcare Life Sciences), as described previously (7). The membrane was blocked with 5% skim milk in phosphate-buffered saline (PBS), 0.05% Tween 20. After incubation with primary antibodies or sera, an appropriate HRP-conjugated secondary antibody and chemiluminescence reagent (SuperSignal West Pico chemiluminescent substrate; Thermo Fisher) were applied for signal detection.
Transient expression, metabolic labeling, immunoprecipitation, and gel electrophoresis.
The expression of YFV NS polyprotein regions (by use of the pET/BS series of plasmids) in SW13 cells, metabolic labeling, and immunoprecipitation were performed essentially as described previously (23). Cells were infected (multiplicity of infection [MOI] = 20) with vTF7-3, a recombinant vaccinia virus expressing the bacteriophage T7 RNA polymerase, and were then transfected with plasmid DNA using the Lipofectamine 2000 reagent (Invitrogen) to drive RNA synthesis from the T7 promoter upstream of the YFV NS proteins (19). Transfection was continued for 2.5 h, and then metabolic labeling of proteins with Express protein-labeling mix (PerkinElmer), lysis with 0.5% SDS lysis buffer containing a mammalian protease inhibitor cocktail (Roche), and immunoprecipitation with anti-YFV antiserum were performed as described previously (19). Protein G Dynabeads were used to collect the immunoprecipitated proteins. For pulse-chase experiments, medium with 1 mM unlabeled methionine and cysteine was added after washing and cells were harvested at the times indicated on the figures. Proteins were separated on NuPAGE 4 to 12% bis-Tris gels, fixed in 40% methanol and 10% acetic acid, incubated in enlightening solution (Autofluor; National Diagnostics), and dried. Dried gels were placed in contact with Kodak Biomax XAR autoradiography film overnight or placed in contact with a phosphorimage screen for a few hours before the screen was scanned using an Amersham Biosciences Typhoon 9400 variable imager. Image analysis for protein band quantification was performed using ImageQuant (v5.2) software.
Proteinase K protection experiments.
SW13 cells transduced or not to express DNAJC14 were infected with vTF7-3 (29) and transfected to express sig2A-5536* or sig2A-5536. Alternatively, cells were infected with YFV (MOI = 10). After metabolic labeling for 4 h, cells were permeabilized by digitonin at 50 μg/ml (0.005%) and treated with or without proteinase K at 10 μg/ml. Proteinase K digestion was terminated 30 min later as reported previously (19). For some samples, prior to proteinase K digestion the membranes were solubilized by the addition of digitonin, and differential centrifugation was used to separate the soluble (S) and insoluble (P) fractions as previously described (7).
Immunoprecipitation.
To demonstrate a DNAJC14 interaction with the NS4A protein, 293T cells were seeded 16 h before transfection onto a 6-well plate at a density of 3 × 105 cells/well. The cells were cotransfected with 1 μg each of pNS4A-GFP and pV1-hDNAJC14-FL-myc (7) using Lipofectamine and Plus reagent (Invitrogen) according to the manufacturer's recommendation. Forty-eight hours later, cells were scraped into ice-cold PBS, harvested by centrifugation, and solubilized with 300 μl lysis buffer (10 mM HEPES, pH 7.5, 150 mM KCl, 3 mM MgCl2, 0.5% NP-40, proteinase inhibitor cocktail [Roche]). After disruption by passing through a 26-gauge 3/8-in. needle and clarification by centrifugation at 15,000 × g for 10 min at 4°C, 270 μl of the soluble fraction was incubated overnight at 4°C with anti-myc or anti-GFP antibody. Preequilibrated protein G Dynabeads were then added, and after 2 h of incubation, the beads were collected by a DynaMag magnetic stand (Invitrogen) and washed four times with 600 μl washing buffer (10 mM HEPES, pH 7.5, 150 mM KCl, 3 mM MgCl2, 0.05% NP-40). The bound proteins were eluted by boiling in SDS sample buffer and were subjected to Western blot analysis.
Preparation of lentiviral stocks and cell transduction.
The production of vesicular stomatitis virus G glycoprotein (VSV-G)-pseudotyped lentiviral particles was performed essentially as described in reference 7. After transduction, 70 to 95% of the cells were positive for protein expression.
Flow cytometry.
SW13 cells were harvested, fixed, permeabilized with Cytofix/Cytoperm (BD Biosciences), and stained with anti-myc for 40 min at room temperature, followed by incubation with Alexa Fluor 488-conjugated anti-mouse IgG antibody (Invitrogen) at a 1:1,000 dilution for 30 min. Cells were resuspended in PBS containing 2% FBS, and data were acquired using a BD LSR II flow cytometer. Data were analyzed with FlowJo software.
RNA interference (RNAi)-mediated silencing.
Triplicate wells of SW13 cells were seeded in 24-well plates at 1 × 105 cells per well in the presence of 80 nM Stealth RNAi siRNA Negative Control Med GC (catalog number 12935-300; Invitrogen) or DNAJC14-targeting Stealth siRNA and Lipofectamine RNAiMAX (Invitrogen) as described previously (7). The transfection of small interfering RNA (siRNA) was repeated 2 days later, using forward transfection with 80 nM siRNA. After an additional 2 days of incubation, the cells were ready to be used in downstream metabolic labeling experiments. For each condition, cells from one of the triplicate wells were harvested for Western blot analysis.
RNA electroporation, virus production, and viral assays.
A preparation of vTF7-3 (23, 29) was made, and titers on BSC-40 cells were determined. The YFV 17D strain was made by electroporation of 6 × 106 to 8 × 106 BHK-J cells with in vitro-transcribed RNA (3 to 5 μg) generated from XhoI-linearized pACNR-FLYF17Dx as described previously (26, 30). The titers of the stocks were determined on BHK-J and SW13 cells after serial dilution as previously described (7). Plaques were enumerated by crystal violet staining after 72 h. For YFV infections, the MOI was based on the titers obtained on BHK-J or SW13 cells, depending on the cell type used in the experiment. For the infectious center assay (31), a sample of the electroporated cells was first diluted 1:100 and 1,000 μl was added to a BHK-J cell monolayer (5 × 105 cells/35-cm dish) and then further diluted 1:10, and 2, 20, 200, and 1,000 μl each was added to a BHK-J cell monolayer. After 4 h of incubation, the cells were overlaid with 0.6% agarose in MEM containing 2% FBS and incubated at 37°C. After 72 h, infectious centers were enumerated by crystal violet staining as described above. For replicon experiments, BHK-J cells were electroporated with RNA transcribed in vitro from YF-R.luc2A-RP-based replicon constructs; electroporated cells were seeded in 24-well plates at 3 × 105 cells/well, and luciferase activity was measured from cell lysates at different time points using a Renilla luciferase assay system (Promega) and a FLUOstar Omega luminometer (BMG Labtech).
Statistical analysis.
The graphical representation of the results, expressed as the mean with the range from two independent experiments (see Fig. 4E and 6C and D) or including the standard deviation (SD; from two independent experiments each conducted in triplicate; see Fig. 6F), was performed using Prism software (version 5.0b; GraphPad Software Inc.).
FIG 4.
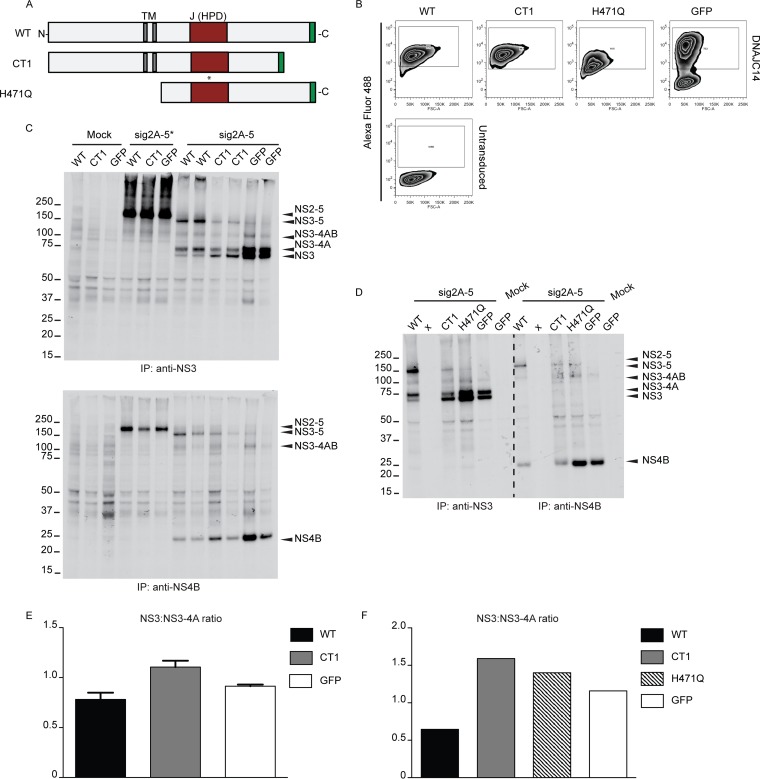
Alterations of YFV NS cleavage events are minimal upon overexpression of noninhibitory DNAJC14 mutants. (A) Diagram of DNAJC14 protein mutants. The transmembrane (TM) segments (gray) and the J domain (red) containing the HPD sequence are indicated. Each mutant contained a C-terminal myc epitope tag, depicted in green. (B) Intracellular levels of DNAJC14 protein mutants measured by fluorescence-activated cell sorting analysis after staining with anti-myc antibody and Alexa Fluor 488-conjugated secondary antibody. (Bottom) Untransduced SW13 cells; (top) cells transduced to express the indicated protein. GFP expression was measured by direct determination of the fluorescent signal. (C and D) Cleavage assays after transient expression of sig2A-5* and sig2A-5 polyproteins were performed in SW13 cells overexpressing wild-type DNAJC14, CT1, and the H471Q mutant, as indicated. GFP served as a negative control. Mock, cells left untransduced and infected with vTF7-3 without YFV plasmid DNA transfection; x, an empty lane. NS3- or NS4B-containing proteins were immunoprecipitated and subjected to autoradiography as described in the legend to Fig. 1A. Identically labeled lanes indicate samples obtained from replicate wells. (E and F) Individual protein bands from the experiments whose results are presented in panels C and D were quantified by phosphorimaging, and NS3-to-NS3-4A protein ratios are displayed in panels E and F, respectively. The bar graphs in panel E represent the means and ranges of the measurements obtained from the duplicate samples analyzed in the experiment whose results are presented in C. The numbers to the left of the gels in panels C and D are molecular masses (in kilodaltons). WT, wild type.
FIG 6.
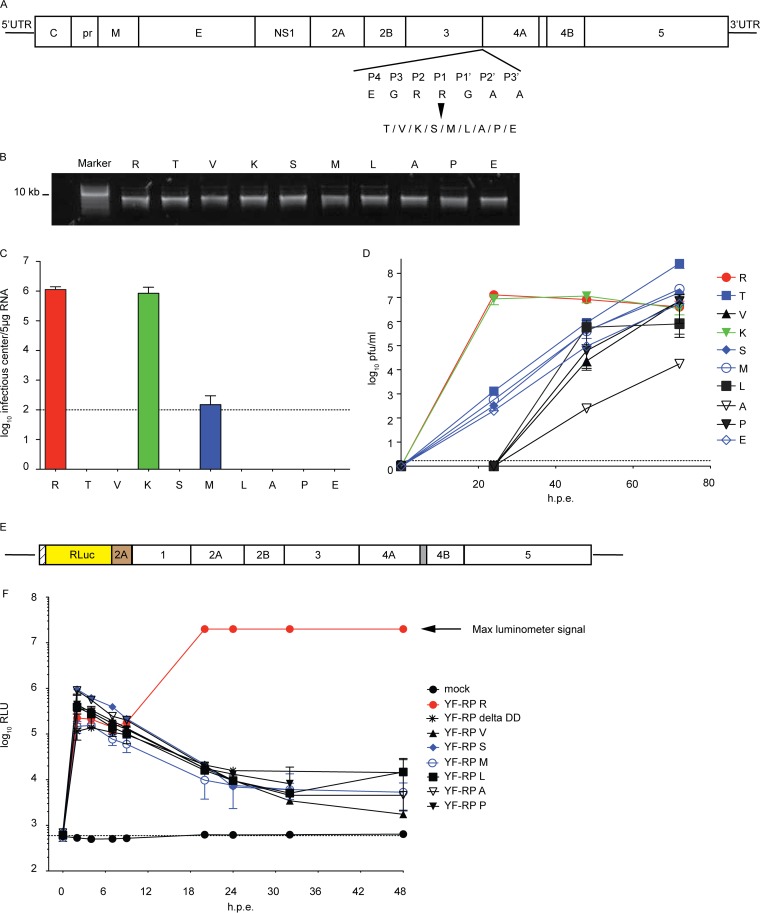
YFV fitness correlates with NS3/4A cleavage efficiency: impaired NS3/4A cleavage delays YFV growth kinetics. (A) Diagram of the YFV molecular organization and the amino acid substitutions at the NS3/4A junction. UTR, untranslated region; C, core; pr, pre-; M, membrane; E, envelope. (B) RNA transcripts for YFV NS3/4A mutants were synthesized in vitro by SP6 RNA polymerase. RNA integrity was assessed by agarose gel electrophoresis. (C) BHK-J cells were electroporated with equal amounts of viral RNA to launch infection. For each viral genome, the efficiency of initiation of infection and spread was determined by an infectious center assay with staining at 72 h postelectroporation. Dashed horizontal line, the detection limit of the assay. The mean infectious center titers from two independent experiments are plotted; error bars indicate ranges. (D) Viral titers were measured in the supernatant by plaque assay at the indicated times (in hours) postelectroporation. Mean titers from two independent experiments are plotted; error bars indicate ranges. (E) Schematic of the YFV replicon. RLuc, Renilla luciferase. (F) BHK-J cells were electroporated with YFV replicon RNAs containing the indicated substitutions at P1 at the NS3/4A site or with a replication-incompetent replicon RNA (delta DD). Luciferase activity was measured at the indicated time points from cell lysates derived from triplicate wells. Mock indicates cells without replicon RNA. Results from two independent experiments, each performed in triplicate, are shown; error bars represent SDs. h.p.e., hours postelectroporation; RLU, relative light units.
RESULTS
The ER cochaperone DNAJC14 alters YFV NS protein maturation.
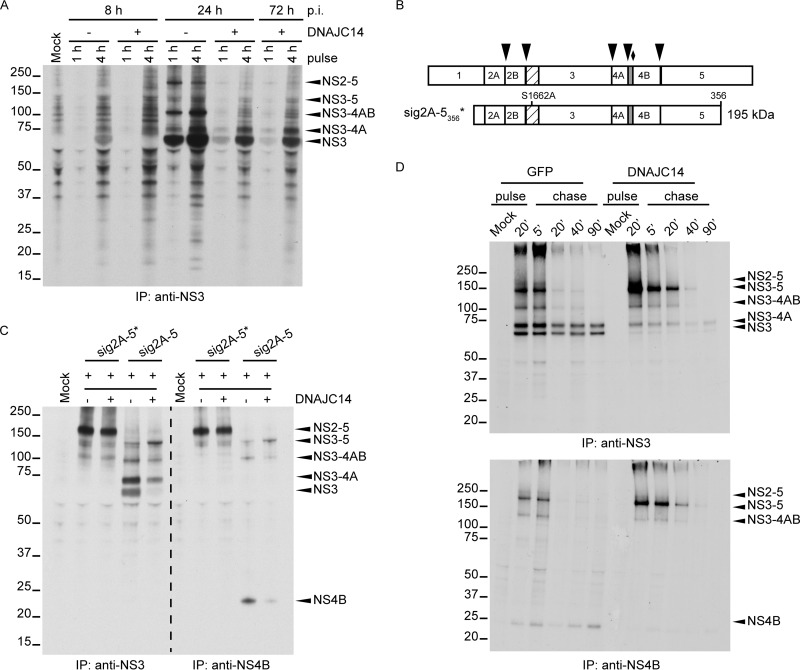
To test whether processing of the YFV polyprotein is affected by DNAJC14 overexpression, we expressed DNAJC14 in SW13 cells and infected them with YFV. At different times of infection, the cells were radioactively labeled for 1 h or 4 h, and the levels of NS3 and its precursor proteins were examined by immunoprecipitation with anti-NS3 antiserum and autoradiography. As shown in Fig. 1A, we observed that at 8 h postinfection (p.i.), cells that overexpressed DNAJC14 exhibited reduced levels of NS3 protein compared to untransduced cells, and this was accompanied by a moderate accumulation of NS3-4A. At 24 h p.i., NS3 was efficiently generated in untransduced cells. In DNAJC14-transduced cells, the NS3 protein became detectable at 24 or 72 h p.i., although processing was less efficient. We concluded that NS3/4A cleavage was impaired in the presence of overexpressed DNAJC14.
FIG 1.
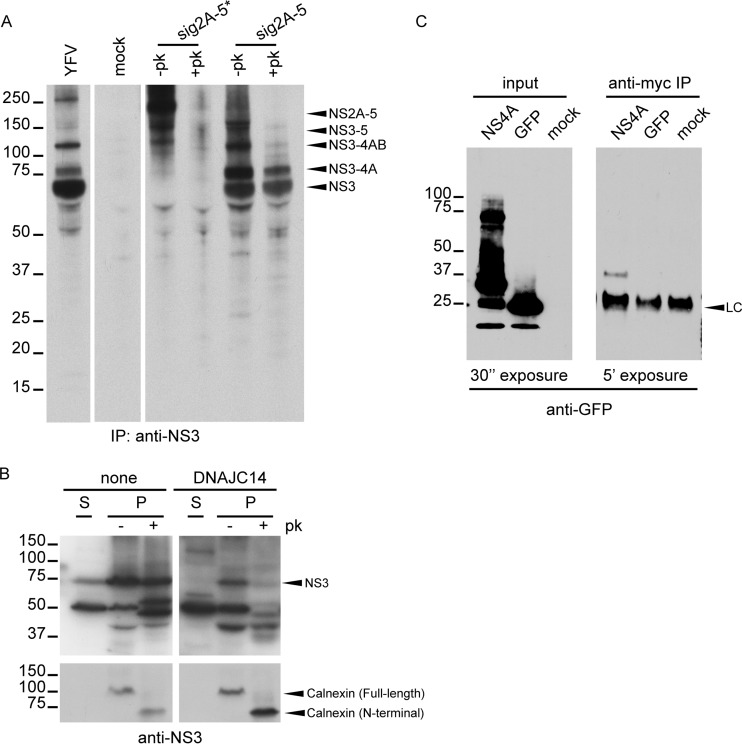
DNAJC14 overexpression affects processing of YFV NS proteins, particularly at the NS3/4A site. (A) Polyprotein processing during YFV infection. SW13 cells were transduced or not with lentiviral pseudoparticles expressing DNAJC14. Cells were then infected with YFV (MOI = 10) and labeled with [35S]methionine for 1 or 4 h at the indicated times. Cell lysates were prepared as described in Materials and Methods, and NS3-precipitated proteins were analyzed by polyacrylamide gel electrophoresis and autoradiography. Mock, immunoprecipitates from uninfected cells. The migration of the size standards is indicated on the left (in kilodaltons); positions of relevant YFV proteins are indicated by arrowheads on the right. (B) Schematic of YFV NS regions used in vaccinia virus expressing bacteriophage T7-based cleavage assays. The entire NS region is depicted at the top. Arrowheads, cleavage sites mediated by NS2B-3 protease; diamond, the cellular signalase cleavage site. The N-terminal protease domain of NS3 (hatched) and 2K peptide portion of NS4A (gray) are indicated. sig2A-5356* represents an NS polypeptide containing the E-derived signal peptide and short NS1-derived linker (open box) fused to NS2-5356 and containing the S1662A protease active-site mutation in NS3 that abolishes proteolytic capacity. The wild-type equivalent is not shown. The predicted molecular mass of sig2A-5356 is shown on the right. (C) Cells transduced or not to express DNAJC14 were infected with the recombinant vaccinia virus vTF7-3 and transfected to transiently express the sig2A-5* or sig2A-5 region of the YFV polyprotein. As a control (Mock), cells were left untransduced and were infected with vTF7-3 without YFV plasmid DNA transfection. [35S]methionine labeling was conducted for 4 h, and solubilized NS3 or NS4B proteins were immunoprecipitated (IP) and analyzed as described in the legend to panel A. (D) Cells transduced to express DNAJC14 or GFP were infected with vTF7-3 and transfected to transiently express the sig2A-5 region of the YFV polyprotein. Pulse-chase analysis of NS3- and NS4B-containing proteins was performed. After 20 min of metabolic labeling, cells were harvested and used directly for immunoprecipitation (pulse) or were incubated with medium enriched in unlabeled methionine and cysteine amino acids (chase) for the indicated times prior to lysis and immunoprecipitation. The data presented in panels A and C are representative of those from at least two independent experiments. The experiment whose results are presented in panel D was performed once. The numbers to the left of the gels in panels A, C, and D are molecular masses (in kilodaltons).
Because the inhibitory effect of DNAJC14 on YFV replication complicates comparisons of NS protein levels, we used a transient-expression system that is independent of YFV infection and replication. The system consists of a recombinant vaccinia virus expressing the T7 RNA polymerase (vTF7-3) to drive expression of YFV sequences from plasmids containing NS regions under the control of the T7 promoter (20, 23, 29). Figure 1B is a schematic representation of the portions of the YFV NS polyprotein expressed in the following experiment. SW13 cells were infected with vTF7-3 and transfected with plasmids carrying the YFV NS region. NS3- and NS4B-containing products were analyzed after metabolic labeling and immunoprecipitation. We expressed the autocatalytic polyprotein sig2A-5 and its corresponding mutant, sig2A-5*, which is incapable of self-cleaving at the NS2A/2B, NS2B/3, NS3/4A, NS4A/2K, and NS4B/5 sites (23) (Fig. 1C). The mutant polyprotein was detected as an uncleaved product of about 195 kDa at similar levels in both mock- and DNAJC14-transduced cells (Fig. 1C). Upon transfection of cells with the wild-type sig2A-5 construct, no full-length sig2A-5 was detected under either condition and NS3-containing proteins were detected, indicating that, as we previously found (7), the NS2B/3 site is readily cleaved in the presence of endogenous or overexpressed DNAJC14 levels. NS3- and NS4B-containing proteins of ∼150 and 115 kDa, whose molecular masses matched the predicted molecular masses for NS3-4-5 and NS3-4AB, respectively, were observed. NS3-4A and NS3 proteins of ∼75 and 70 kDa, respectively, and NS4B were also detected in mock-transduced cells. However, in cells overexpressing DNAJC14, the levels of NS3-5 were increased and the levels of NS3-4A, NS3, and NS4B were drastically reduced. In particular, mature NS3 and NS4B were almost absent when DNAJC14 was overexpressed. Accumulation of the NS3-5 precursor under conditions of DNAJC14 overexpression suggested that cleavage events in this region of the polyprotein were altered.
NS3-4AB levels were similar irrespective of DNAJC14 levels, and we concluded that cleavage at the NS4B/5 site was also (like that at the NS2B/3 site) unaffected by DNAJC14. Since the levels of both NS3 and NS3-4A, as well as the level of NS4B, were reduced following DNAJC14 overexpression, cleavage at both the NS3/4A site and either the NS4A/2K or the 2K/4B (mediated by signalase) site was affected. We were unable to discriminate NS3-4A-2K from NS3-4A, since we lacked antiserum specific for NS4A or 2K. However, it has been reported that cleavage at NS4A/2K is a prerequisite for cleavage at the downstream signalase 2K/4B site (19). All these observations, coupled with the cytosolic localization of the DNAJC14 J domain (17) and of the NS4A/2K site, suggest that DNAJC14 overexpression inhibits cleavage activity at the NS3/4A and NS4A/2K sites, with possible subsequent inhibition at the 2K/4B signalase site.
Next, to understand in greater detail the effect of DNAJC14 overexpression on the kinetics of YFV polyprotein processing, we analyzed the maturation of NS3- and NS4B-containing proteins in a traditional pulse-chase experiment (Fig. 1D). SW13 cells were transduced to express DNAJC14 or GFP as a negative control, infected with vTF7-3, and transfected with the YFV sig2A-5-expressing plasmid. Pulse-labeling was conducted for 20 min, and the chase was started by removal of the labeling medium and replenishment with excess amounts of unlabeled methionine and cysteine. Cells were lysed at different times, and NS3- and NS4B-containing species were immunoprecipitated (Fig. 1D, top and bottom, respectively). NS2-5 was not detected in either control cells or DNAJC14-overexpressing cells, confirming that cleavage at the NS2B/3 site was active and insensitive to DNAJC14 levels. In control cells, uncleaved NS3-5 and NS3-4AB were detected only until 5 min of chase, while NS3-4A and NS3 were synthesized immediately, and their levels remained stable over time and did not increase as the levels of NS3-5 and NS3-4AB decreased (Fig. 1D, top). These data demonstrate a lack of a precursor-product relationship between NS3 and all its potential precursors, as has previously been documented for NS3-4A and NS3 (23). There was also no obvious generation of NS3-4AB from NS3-5. Similarly, NS4B was generated during the pulse and was relatively stable during the chase (Fig. 1D, bottom), again demonstrating a lack of a precursor-product relationship between NS3-5 or NS3-4AB and mature NS4B. The presence of the multiple NS3- and NS4B-containing forms at the end of the pulse, coupled with the lack of an obvious subsequent precursor-product relationship, suggests that in control cells, cleavage of the YFV polyprotein occurs cotranslationally and via several distinct pathways that lead to the production of either NS3 or NS3-4A. Likely polyprotein precursors that fail to cleave at the NS3/4A or NS4A/2K sites are targeted for degradation. In cells overexpressing DNAJC14, NS3-5 accumulated during the pulse, whereas it did not accumulate in control cells, supporting the data in Fig. 1C and indicating that cleavage sites in the NS3-5 region of the polyprotein were affected by DNAJC14 overexpression (Fig. 1D, top). NS3-4AB levels were minimally reduced. However, DNAJC14 overexpression resulted in a marked reduction in mature NS3-4A, demonstrating once more that cleavage at the NS4A/2K or the 2K/4B (signalase) site was affected. The most striking effect upon DNAJC14 overexpression was the drastic reduction in NS3 and NS4B proteins (Fig. 1D), indicating that DNAJC14 overexpression altered proteolytic cleavage in cis at the NS3/4A and downstream NS4A/2K and 2K/4B sites. Quantification of the band intensities in Fig. 1D demonstrated that NS3 levels in the presence of GFP overexpression were reduced by 9% after 5 min of chase and 35% after 90 min of chase compared to the levels at the end of the 20-min pulse. Similarly, NS3 levels in the presence of DNAJC14 overexpression were reduced 11% and 39% at the same time points. Thus, NS3 degradation was not markedly altered in the presence of DNAJC14 overexpression.
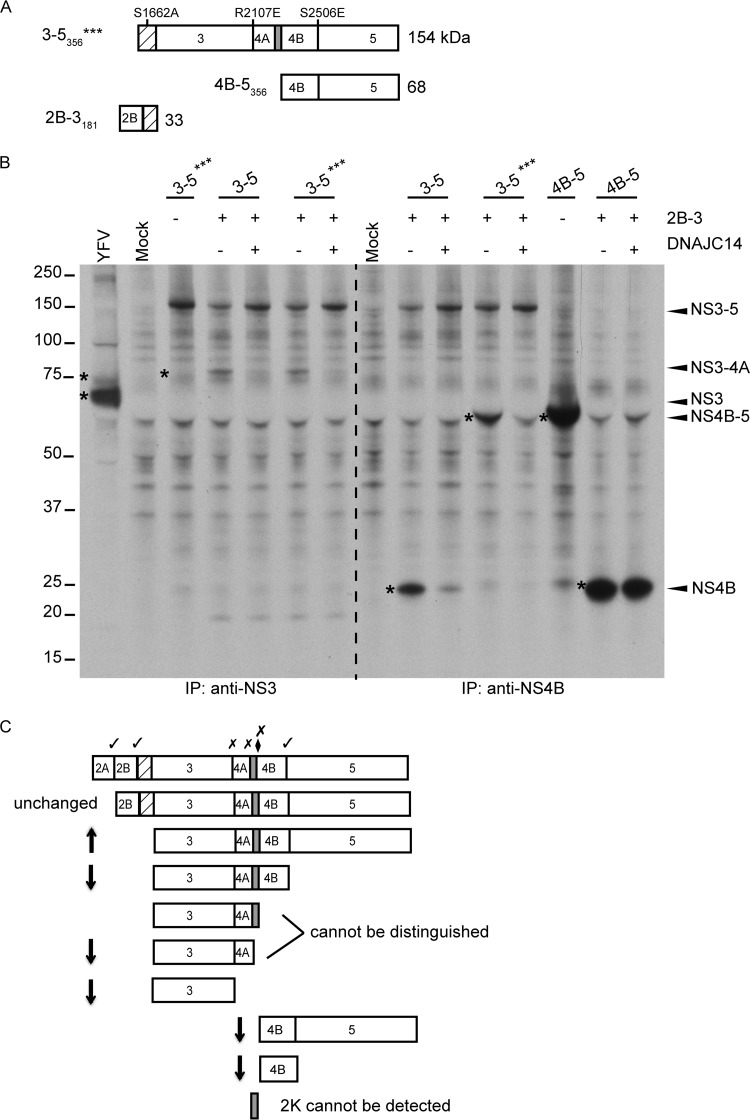
To directly examine the effect of DNAJC14 on cleavage at specific sites in the NS3-4-5 polyprotein region, we utilized a trans-cleavage assay where the NS2B-3 protease activity was provided by an additional DNA plasmid (in trans) that expresses the minimal protease domain, NS2B-3181 (19). Figure 2A is a schematic illustration of the YFV NS polyprotein regions used. For the experiment whose results are shown in Fig. 2B, we used as the substrate both NS3-5 and the triple mutant (NS3-5***). This mutant polyprotein contains an inactive NS3 protease and mutations abrogating cleavage at the NS3/4A and 4B/5 sites (Fig. 2A), allowing analysis of cleavage at the NS4A/2K site. In untransduced cells, NS2B-3 expressed in trans was able to cleave both NS3-5 and NS3-5*** at the NS4A/2K site, as demonstrated by NS3-4A production. As shown in Fig. 2B (left), in DNAJC14-overexpressing cells, the substrate form of NS3-5 (and NS3-5***) accumulated slightly compared to its level of accumulation in control untransduced cells, while NS3-4A levels were reduced, suggesting potential effects on the NS4A/2K site. NS3 was undetectable in both control and DNAJC14-overexpressing cells, an observation consistent with the findings of previous work demonstrating that the NS2B-3 protease functions very poorly in trans at the NS3/4A site (20). To evaluate cleavage events at the NS4B/5 site, NS4B forms were examined (Fig. 2B, right). Using the NS3-5*** substrate, where NS4B/5 cleavage is blocked, the NS4B-5 protein was readily detected in control cells, demonstrating cleavage at the NS4A/2K or 2K/4B site. The levels of NS4B-5 were markedly reduced in DNAJC14-overexpressing cells, indicating that cleavage at NS4A/2K or 2K/4B was affected. As expected, due to the mutation at the NS4B/5 site, no NS4B was detected under either condition. Using wild-type NS3-5, NS4B-5 was not detectable in control cells, suggesting that cleavage at the NS4B/5 site was occurring normally. Indeed, NS4B was readily detectable. In the presence of overexpressed DNAJC14, NS4B-5 was also not detected and the levels of NS4B were reduced, consistent with a reduction in cleavage events at the NS4A/2K site.
FIG 2.
DNAJC14 overexpression affects cleavage at the 4A/2K site but not the NS4B/5 site. (A) Schematic of YFV NS regions used in the vaccinia virus expressing bacteriophage T7-based trans-cleavage assays. (Top) The NS3-5*** (3-5***) construct encodes NS3-5356 and also contains the protease active-site mutation along with the indicated mutations at the NS3/4A and NS4B/5 cleavage sites, which block cleavage. The wild-type version is not shown. (Bottom) The NS4B-5356 substrate used in trans cleavage assays along with the active NS2B-3181 (2B-3181) protease. For additional details, see Fig. 1B. (B) trans-Cleavage analyses were conducted in vTF7-3-infected SW13 cells that had previously been transduced or not to express DNAJC14. YFV proteins were coexpressed by transfection of a combination of plasmids carrying the parental NS3-5 polyprotein, the triple mutant NS3-5***, the NS4B-5 (4B-5) region, and the cofactor protease NS2B-3, as indicated. Cells were labeled with [35S]methionine for 4 h. Labeled NS3- or NS4B-containing proteins were analyzed as described in the legend to Fig. 1A. The data are representative of those from two independent experiments. The numbers to the left of the gel are molecular masses (in kilodaltons). Asterisks indicate bands of interest. (C) Schematic summary of the effect of DNAJC14 overexpression on cleavage events. Arrows, relative accumulation or reduction in protein levels.
To examine the effects of DNAJC14 on the NS4B/5 site directly, we expressed NS4B-5356 (Fig. 2A) as a substrate for the NS2B-3181 protease, which is capable of trans cleavage at the NS4B/5 site (19). As seen on the far right of Fig. 2B, we observed equivalent amounts of NS4B protein in control and DNAJC14-transduced cells, demonstrating that trans cleavage at the NS4B-5 site was unaffected by DNAJC14 overexpression. All together, the data collected during the course of YFV infection or in a transient-expression system for the YFV polyprotein indicate that NS3/4A/2K cis cleavage events but not NS2/3 or NS4B/5 cleavage events are altered by DNAJC14 overexpression. Figure 2C provides a schematic summary of the effects of DNAJC14 overexpression on cleavage events in the polyprotein.
NS3-4A-2K region cleavage events correlate with changes in the physical properties of NS3 and NS3-4A.
NS4A/2K cleavage was shown to regulate RC formation for DENV and WNV (32, 33). Therefore, we investigated the possible relationship between cleavage of the NS3-4A-2K region and YFV RC formation. Since in vitro replicase activity and viral RNA in the RC are fully resistant to nuclease and protease treatment (34), we examined the proteinase K sensitivity of YFV NS3-containing proteins. SW13 cells were infected with YFV as a control or were infected with vTF7-3 and transfected to express sig2A-5 polyprotein or the corresponding protease active-site mutant, sig2A-5*. After metabolic labeling, the cells were permeabilized with digitonin and treated or not with proteinase K. Upon proteinase K treatment, cytosolic proteins would be digested, while those inside membrane structures would be protected. In Fig. 3A, we observed that the uncleaved sig2A-5* form was sensitive to protease digestion. However, in cells expressing the sig2A-5 polyprotein and treated with proteinase K, the NS3-4A and NS3 protein cleavage products were resistant to digestion. This, coupled with the fact that larger polyproteins containing NS3-4A (NS2-5, NS3-5, and NS3-4AB) were sensitive to digestion, suggested that cleavage events at the NS3/4A or 4A/2K site were associated with a change in conformation and/or the subcellular localization of the NS3-4A and NS3 proteins. Conformational changes occurring upon cleavage might alter protease sensitivity due to changes in the folding of these domains of the polyprotein and could also alter their localization with respect to that of other proteins in the polyprotein. Moreover, invagination of the ER membrane into the luminal space, which occurs during RC formation, could render these components protected from protease digestion. In any of these cases, this would involve a conformational change for NS3 and NS3-4A. In the experiment whose results are shown in Fig. 3B, the sensitivity of NS3 was examined in the context of YFV infection of DNAJC14-overexpressing cells. Note that in infected cells (Fig. 3A, YFV), the primary NS3-containing protein is NS3, although NS3-4A is also produced. Soluble proteins were extracted with digitonin, and the remaining insoluble (membrane) fraction was treated or not with proteinase K and analyzed by Western blotting using anti-NS3 serum. In control cells, most of the NS3 protein associated with the membrane fraction was proteinase K resistant, suggesting that it had undergone a conformational change. In contrast, in cells overexpressing DNAJC14, lower levels of NS3 were present, as expected, due to viral inhibition, and the small amount of NS3 produced was more sensitive to proteinase K digestion, suggesting a failure to achieve a new conformation or new subcellular localization because of a chaperone process dysregulation. We also reason that normal cleavage to generate NS3 or NS3-4A is likely correlated with a conformational and/or subcellular localization change, compatible with RC formation.
FIG 3.
NS3/4A/2K cleavage events correlate with physical changes to NS3 and NS3-4A. The proteinase K sensitivity of YFV polyprotein products was used to assess physical changes. (A) SW13 cells were infected with vTF7-3 and transfected to express the sig2A-5* or sig2A-5 polyprotein. After 4 h of metabolic labeling, cells were permeabilized by digitonin and then treated with or without proteinase K (+pk and –pk, respectively). NS3-containing products were analyzed as described in the legend to Fig. 1A. (B) SW13 cells transduced or not to express DNAJC14 were infected with YFV (MOI = 10) and 8 h later permeabilized with digitonin to separate the soluble (S) and insoluble (P) fractions. Proteins precipitated from the insoluble fraction were treated or not with proteinase K. Western blot analysis was performed using NS3- and calnexin-specific antibodies. Digestion of calnexin and protection of the N-terminal luminal fragment served as controls for protease activity and membrane integrity, respectively. (C) DNAJC14 interacts with NS4A. 293T cells were cotransfected with plasmids expressing myc-tagged DNAJC14 along with the YFV NS4A-GFP fusion protein. Expression of GFP served as a control. Total cell lysates and anti-myc-immunoprecipitated products were blotted with anti-GFP antibody. LC, antibody light chain. The results presented in each panel in panels B and C are representative of those from three independent experiments. The numbers to the left of the gels are molecular masses (in kilodaltons).
DNAJC14, also known as DRIP78 (35), is an essential factor for proper folding and cell surface transport of several G-protein-coupled receptors (35–38) and interacts with an amphipathic helix, designated helix 8 (H8), in the cytoplasmically localized C termini of the receptors (39). The DENV NS4A protein has been shown to have an N-terminal amphipathic helix (40), and similar properties are predicted for YFV NS4A. Given the effects of DNAJC14 on regions of the polyprotein in the vicinity of NS4A, we hypothesized that DNAJC14 might bind to an amphipathic helix in NS4A and modulate polyprotein cleavage. To test this, cells were cotransfected with plasmids expressing myc-tagged DNAJC14 and NS4A containing a carboxyl-terminal GFP tag or GFP as a negative control. We specifically detected NS4A-GFP and not GFP in DNAJC14 (myc) immunoprecipitates (Fig. 3C), consistent with the predicted interaction of DNAJC14 with an amphipathic region of the polyprotein. These findings together indicate that the effect of DNAJC14 overexpression on NS3/4A/2K cleavage events correlates with changes in NS3 and NS3-4A, likely due to the DNAJC14 interaction with NS4A.
DNAJC14 mutants have different effects on YFV NS3/4A/2K cleavage events.
To better understand DNAJC14 function, we tested the effects of various deletions and point mutations (17) on NS3/4A/2K processing events. Figure 4A shows a schematic of the different mutant forms. Full-length DNAJC14 was previously shown to have an inhibitory effect on YFV replication (7). DNAJC14 with a 77-amino-acid, C-terminal deletion that was designated CT1 and that was not inhibitory for YFV replication (7) was selected for this study. Since the carboxyl terminus is involved in the DNAJC14-DNAJC14 interaction, this mutant was predicted to be incapable of cochaperone activity. To confirm our findings with CT1 (see below), we also utilized another noninhibitory mutant, designated H471Q (7), which contains an N-terminal truncation deleting the transmembrane domains and also contains a histidine-to-glutamine amino acid substitution at position 471 within the conserved HPD sequence in the DNAJC14 J domain. Since the HPD motif is involved in the interaction with Hsp70 and the stimulation of Hsp70 ATPase activity (41), this mutant is also predicted to be incapable of cochaperone activity. The wild type and each mutant form expressed a C-terminal myc epitope tag, which allowed assessment of intracellular expression levels (Fig. 4B).
To investigate the effect of the DNAJC14 mutants on processing, we utilized the vaccinia virus system to express the YFV sig2A-5 polyprotein. SW13 cells that expressed wild-type DNAJC14, the CT1 mutant, or GFP (as a control) were infected with vTF7-3 and transfected with plasmids expressing mutated NS3 protease (sig2A-5*) or the wild-type sig2A-5 polyprotein (Fig. 4C). As expected, sig2A-5* remained uncleaved in all samples (Fig. 4C, top and bottom). In contrast, sig2A-5 exhibited different cleavage patterns with wild-type and mutant DNAJC14. While wild-type DNAJC14 caused the accumulation of NS3-5 and a reduction of NS3 levels relative to the levels of NS3-4A, the CT1 noninhibitory mutant did not and facilitated the accumulation of NS3 relative to the levels of NS3-4A (Fig. 4C, top). Similar results were obtained for NS4B and its uncleaved forms (Fig. 4C, bottom). Wild-type DNAJC14 caused the accumulation of NS3-5 and a reduction of NS4B levels, while the CT1 mutant resulted in intermediate levels of NS4B compared to the levels obtained with wild-type DNAJC14 or GFP. To confirm that overexpression of a noninhibitory mutant did not have the effect of overexpressed wild-type DNAJC14, the H471Q mutant was also tested along with the wild-type and CT1 proteins (Fig. 4D). The levels of NS3-containing products (Fig. 4D, left) observed upon wild-type and CT1 overexpression were similar to those obtained in the experiment whose results are presented in Fig. 4C. Interestingly, the H471Q mutant did not induce the marked alterations observed for the wild-type and CT1 proteins. In the presence of the H471Q mutant, the NS3/4A/2K cleavage products were more abundant and were present at protein ratios similar to those seen with GFP overexpression. The increased levels are consistent with those presented in the published literature on mutations in this region of the J domain (see Discussion). The gels presented in Fig. 4C and D were quantified by phosphorimaging, and NS3-to-NS3-4A protein ratios were plotted for easier visualization (Fig. 4E and F, respectively). Overall, the results presented in Fig. 4 demonstrate that DNAJC14 protein mutants, previously shown to lack inhibitory activity on the YFV life cycle, produced minimal alterations to NS3/4A/2K cleavage events. The fact that the overexpression of mutant forms of DNAJC14 did not produce the same alteration in processing as wild-type DNAJC14 suggests that the results are not due to a nonspecific or off-target effect. In particular, the C-terminal deletion mutant CT1 slightly promoted the formation of NS3 as the final product, opposite the results seen with wild-type DNAJC14. The H471Q N-terminal deletion mutant, which also contained an amino acid substitution in the J domain, increased the levels of NS3-4A and NS3 protein accumulation, highlighting the importance of Hsp70 ATPase activity stimulation in the mechanism by which DNAJC14 overexpression alters polyprotein processing.
Reduction of DNAJC14 levels minimally affects YFV polyprotein processing.
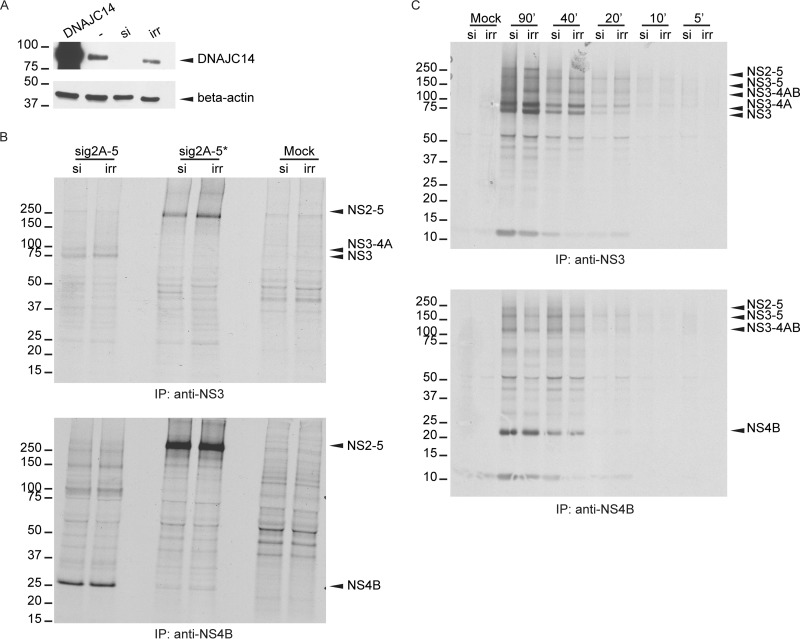
To examine a possible requirement for endogenous DNAJC14 during YFV polyprotein processing, we silenced the expression of DNAJC14. SW13 cells were transfected with irrelevant siRNA or siRNA directed against DNAJC14, and reduced levels of DNAJC14 protein expression were confirmed by Western blotting (Fig. 5A), using DNAJC14 overexpression for unambiguous protein identification. We analyzed the processing of the sig2A-5 protein, following 4 h of labeling, in cells that contained endogenous levels of DNAJC14 and compared it to that in cells with reduced levels of DNAJC14 (Fig. 5B). In DNAJC14-knockdown cells, the levels of NS3-containing proteins (Fig. 5B, top) were similar to those observed in control cells. Similarly, the amounts of NS4B-containing products were unchanged by DNAJC14 silencing (Fig. 5B, bottom). To test these preliminary observations in greater detail, we examined, using shorter labeling periods, the early processing of the sig2A-5 polyprotein in cells that had previously been treated with irrelevant or DNAJC14-specific siRNA (Fig. 5C). The effects of DNAJC14 silencing on NS3/4A/2K cleavage events were minimal over time. Early during labeling (20 min), NS3 levels were similar in the presence of normal or reduced DNAJC14 levels, while after 90 min of labeling, NS3 levels were slightly decreased in the presence of reduced DNAJC14 levels. NS4B levels did not change upon specific siRNA treatment (Fig. 5C, bottom). Overall, YFV polyprotein processing in the NS3-4A-2K region was minimally affected by a reduction of DNAJC14 levels, although we cannot exclude the possibility of the existence of a subtle kinetic effect that we were unable to measure.
FIG 5.
Reduction of DNAJC14 levels by siRNA transfection has minimal effects on NS3/4A processing. (A) Cells were transfected with a plasmid expressing DNAJC14 to allow identification of the band corresponding to endogenous DNAJC14. Actin served as a loading control. SW13 cells were transfected with irrelevant siRNA (irr) or with an siRNA pool targeting DNAJC14 (si), before being collected and analyzed by Western blotting using anti-DNAJC14 antibody. (B) Cells that were transfected twice with DNAJC14 or irrelevant siRNA were subsequently infected with vTF7-3 and transfected to express the sig2A-5* or sig2A-5 protein. As a control (Mock), cells were treated with siRNA and infected with vTF7-3 without YFV plasmid transfection. [35S]methionine labeling was conducted for 4 h, NS3 (top) or NS4B (bottom) antiserum was used for immunoprecipitation, and labeled products were visualized by autoradiography. (C) SW13 cells treated with siRNA, infected with vTF7-3, and transfected to express the sig2A-5 or sig2A-5* polyprotein were metabolically labeled for the indicated times (in minutes) prior to harvest. NS3- or NS4B-precipitated proteins were analyzed as described in the legend to Fig. 1A. Control samples (Mock) were prepared as described in the legend to panel B. The numbers to the left of the gels in panels B and C are molecular masses (in kilodaltons).
YFV growth kinetics correlate with NS3/4A cleavage efficiency.
To further confirm the effect of DNAJC14 overexpression on NS3/4A site cleavage, we engineered a bicistronic YFV genome with a stop-codon-IRES-ubiquitin cassette cloned between NS3 and NS4A (NS3-4A SPLIT). Since replication of this genome would not require cleavage between NS3 and NS4A, we hypothesized that DNAJC14 should not inhibit viral replication. Analysis of cells and quantification of virus in the culture supernatant revealed that no detectable polyprotein or infectious virus was produced by the bicistronic NS3-4A split genome (data not shown). While the reasons why this viral genome failed to replicate remain unknown, it is possible that incomplete NS3/4A processing is required for YFV RNA replication and/or infectious virion formation, suggesting a crucial role for the uncleaved NS3-4A protein. This possibility is not easily tested, for example, through trans-complementation of the NS3-4A SPLIT genome with NS3-4A, due to the fact that the cis-cleavage activity of the flavivirus NS3 protease is tightly coupled with viral genome translation and RNA replication (42–45). Because the construct did not yield detectable polyprotein or virions, the NS3-4A SPLIT construct could not be used to directly test whether viral inhibition by DNAJC14 was due to inhibition of NS3/4A cleavage.
Therefore, as an alternative approach to probe whether the inhibition of YFV replication that we observed upon DNAJC14 overexpression (7) was related to alteration of NS3/4A/2K cleavage events (Fig. 1 and 2), we altered the cleavage efficiency independently of DNAJC14. Early studies on YFV polyprotein processing conducted by Lin et al. (23) examined the effect on polyprotein processing of a number of amino acid substitutions in the conserved dibasic NS3/4A cleavage site consisting of Arg (P2)-Arg (P1)/Gly (P1′). Using the expression of P1-site mutants in the context of the sig2A-5 polyprotein, the authors reported that replacement of the P1 residue with uncharged or hydrophobic residues reduced the cleavage efficiency, while replacement of the P1 residue with Glu or Pro drastically impaired cleavage. We introduced these substitutions into the YFV infectious molecular clone to evaluate the effects of altered NS3/4A cleavage efficiency on YFV replication independently of alterations in DNAJC14 levels (Fig. 6A). Table 1 lists the amino acid properties of the P1 substitutions tested in this study.
TABLE 1.
Summary of the properties of YFV NS3/4A P1-site mutants
| YFV NS3/4A site mutation (aaa 2107) | Amino acid property | DNA codon | Titerb at: |
Polyprotein processingc | Revertant substitution at NS3/4A site (aa 2107) | Mutation typed | |
|---|---|---|---|---|---|---|---|
| 24 h p.e. | 48 h p.e. | ||||||
| R (Arg) wild type | Basic | AGG | 1.3 x 107 | 8.0 × 106 | +++ | None | None |
| T (Thr) | Polar uncharged | ACG | 1,300 | 9.0 × 105 | ++ | AGG (Arg) | Tv |
| V (Val) | Nonpolar hydrophobic | GTG | <50 | 2.0 × 105 | +/− | AGG (Arg) | Ts/Tv |
| K (Lys) | Basic | AAG | 8.7 × 106 | 1.1 × 107 | +++ | None | None |
| S (Ser) | Polar uncharged | TCG | 325 | 4.0 × 105 | ++ | AGG (Arg) | Tv/Tv |
| M (Met) | Nonpolar hydrophobic | ATG | 575 | 4.0 × 105 | + | AGG (Arg) | Tv |
| L (Leu) | Nonpolar hydrophobic | TTG | <50 | 5.7 × 105 | + | AAG (Lys) | Tv/Tv |
| A (Ala) | Nonpolar hydrophobic | GCG | <50 | 250 | ++ | AAG (Lys) | Ts/Tv |
| P (Pro) | Nonpolar hydrophobic | CCG | <50 | 6.0 × 104 | − | CGG (Arg) | Tv |
| E (Glu) | Acidic | GAG | 200 | 9.0 × 104 | − | AAG (Lys) | Ts |
aa, amino acid.
Titers are in PFU per milliliter and are the means from two independent experiments. p.e., postelectroporation.
Data are from Fig. 5 of Lin et al. (19).
Ts, transition; Tv, transversion
For each NS3/4A mutant, infectious YFV RNA was transcribed in vitro and RNA integrity was assessed by agarose gel electrophoresis (Fig. 6B). The capacity of the viral mutants to initiate replication and make virus progeny was examined by an infectious center assay, where electroporated cells were serially diluted and plated onto a permissive monolayer. As seen in Fig. 6C, only virus containing the Lys (K) substitution formed infectious centers similarly to the wild type, while the number of infectious centers formed by the Met (M) mutant was near the lower limit of detection for this assay. The remaining viral mutants were unable to form plaques in the infectious center assay. In parallel, we assessed virus replication and spread by quantification of the virions in the medium. Culture supernatants collected at different times from cells electroporated with RNA were evaluated by a plaque assay (Fig. 6D), revealing three phenotypes. Virus with the Lys substitution was confirmed to be as efficient as wild-type virus, producing overlapping viral growth curves. Substitutions that included Thr, Ser, Met, and Glu resulted in lower levels of virus accumulation that eventually reached wild-type levels, while substitutions that included Val, Leu, Ala, and Pro resulted in markedly decreased levels of viral accumulation. Of note, the titers of the Ala mutant at 48 h postelectroporation were 30,000-fold lower than those of the wild type. Table 1 reports the viral titers measured for each clone at two different time points after RNA electroporation. Overall, the observed viral kinetics suggested that P1 substitutions at the NS3/4A cleavage site affected viral production to different extents.
Given the delay in infectious virion production by the mutants, we wondered if reversion of the P1 residue or other mutation at the NS3/4A site might have occurred to allow eventual robust replication. To test this, equivalent numbers of infectious virions (equivalent numbers of PFU) obtained at 48 h postelectroporation (Fig. 6D) were used to infect BHK-J cells and RNA was harvested when cytopathic effects started to appear. After cDNA generation, the NS3-4A region was amplified by PCR and the product was sequenced. Table 1 reports the sequence and amino acid changes that were identified for each viral clone. Sequencing quality allowed the unambiguous verification of the region amplified from nucleotide 6412 to nucleotide 6635 of the YFV genome, and we observed no changes outside the nucleotides encoding the P1 residue. Only the sequence of the mutant with the Lys substitution at the NS3/4A P1 position remained unchanged at the P1 site. Characterization of the sequences indicated two classes of amino acid substitutions associated with reversion (Table 1). Reversion to the original parental amino acid, Arg, occurred for mutants with Thr, Val, Ser, Met, and Pro at the NS3/4A P1 site. Mutations to Lys rather than the parental Arg were observed for the remaining clones (Table 1). These changes were achieved through nucleotide transition, transversion, or both. Overall, these observations reinforce a requirement for a basic residue at the P1 position of the NS3/4A cleavage site in YFV (23).
Altered cleavage efficiency at the NS3/4A site could affect RC formation, RNA replication, and/or subsequent virion production. To evaluate whether P1 substitutions and the NS3/4A cleavage efficiency affected YFV translation and RNA replication, the NS3/4A P1 mutations were introduced in a YFV replicon which expressed a luciferase reporter gene in place of the structural proteins (22). A schematic representation of the YFV replicon is depicted in Fig. 6E. As a negative control, we used a replication-incompetent replicon that contained a delta DD mutation abolishing RNA-dependent RNA polymerase activity (7) and for which the luciferase signal is derived completely from translation of the input RNA. As shown in Fig. 6F, luciferase expression at early time points after electroporation was similar for the control (delta DD), the wild type, and all the NS3/4A mutant replicons tested. This indicates that the mutations did not affect RNA stability or translation. The gradual decline in signal seen with the delta DD mutant reflects the gradual loss of RNA that was unable to replicate. For the wild-type replicon, a significantly increased luciferase signal was detected at 8 h after electroporation, reflecting functional RC formation and active RNA replication with subsequent translation of the newly generated RNA. In contrast, all the tested NS3/4A mutant replicons exhibited a gradual decline in luciferase expression, consistent with RNA decay and a failure of RC formation and RNA replication. Therefore, the replacement of the NS3/4A P1 Arg residue with nonpolar hydrophobic or polar uncharged residues inhibited the YFV RNA replication efficiency. All together, the data collected revealed that the cleavage efficiency at the NS3/4A site significantly affected YFV replication fitness, as measured by polyprotein processing, RNA replication, and virus production.
The finding of uncleaved NS3-4A in YFV-infected cells (Fig. 1A, 2B, and 3A), along with the cleaved products NS3 and NS4A, suggests an important role for NS3-4A, although its function in the virus life cycle is unknown (27, 46, 47). Our work demonstrates that cleavage at the NS3/4A junction is a critical step in YFV replication and determines YFV sensitivity to DNAJC14 overexpression. We propose a model where DNAJC14's chaperone activity is required for YFV NS3/4A cleavage, which occurs primarily in cis, and is a highly regulated, rate-limiting step that ensures the production of the proper amount of final YFV proteins, including NS3, NS4A, and NS3-4A.
DISCUSSION
The proper folding of proteins within cells is mediated by the activities of molecular chaperone and other folding enzymes that stabilize nascent polypeptides to prevent incorrect folding or aggregation and that allow the polypeptide chain to acquire its correct conformation. Recent work studying the role of chaperones in DENV infection revealed a role for Hsp70 chaperone activity at multiple life cycle steps. Moreover, the chaperone process was found to involve distinct DNAJ cochaperone family members to facilitate viral entry, RNA replication, and virion production (6). DNAJC14 is an Hsp40 cochaperone that assists a wide range of cellular folding processes mediated by Hsp70. Overexpression of DNAJC14 was shown to inhibit several members of the Flaviviridae family. Here, we hypothesize that DNAJC14 affects processing of the NS region of the YFV polyprotein and alters downstream RC assembly. While previous studies have been elusive in addressing a detailed role of chaperones in virus replication (reviewed in reference 3), in our work, we monitored processing of the YFV NS2A-5 polyprotein in DNAJC14-overexpressing cells. We found that NS3/4A/2K cleavage events were particularly affected, with an overall effect on uncleaved NS3-5 accumulation and reduced levels of NS3-4A, NS3, and NS4B being seen. The cleavage at the NS3/4A site was most significantly inhibited. We anticipated that downstream cleavage events in the NS2-5 polyprotein could be altered as a consequence of inefficient processing at the NS3/4A junction (19). A correlation between the folding function of a molecular chaperone and viral polyprotein cleavage events was established, suggesting that these cellular molecules are diverted from their cellular functions to play a complementary role during virus infection of the host cell. At present, the host requirements in the coordinated process of viral replication and production are not yet fully understood, but several prior studies suggest a role for molecular chaperones (48, 49). Several reports, particularly on positive-strand RNA viruses of plants, examined the key role of cellular chaperones and folding enzymes. During plant virus infection, several Hsp proteins are involved in the formation of membrane-bound RCs for certain members of the Tombusvirus, Tobamovirus, Potyvirus, and Potexvirus genera (50–53). There are several cochaperones, including Hsp40, that associate with the bromovirus RC (54–56). There are also examples of plant viruses that rely on chaperone systems in the ER to support cell-to-cell movement (57). It is possible that some viral proteins have a common fold that can be recognized by Hsp70, Hsp40, or Hsp90 partners; however, how these cellular chaperones participate in the replication of a wide range of unrelated viruses was one of our driving questions. From the data presented here, we propose that the chaperone machinery remains intact but is diverted to recognize alternative substrates, which may or may not impact normal cellular chaperone functions.
Hsp40 family members can be classified into three groups (58) depending on the J-domain localization within the protein. Type I proteins contain the J domain at the N terminus and a C-terminal dimerization domain. Type II proteins also contain the J domain at the N terminus but a peptide binding sequence at the C terminus. Type III proteins, like DNAJC14, are variable, with the J domain being localized anywhere in the protein. Structural and functional studies of different classes of Hsp40 demonstrated that the C terminus mediates chaperone dimerization (59). The different classes of Hsp40 are thought to interact with the Hsp70 folding machinery separately from each other, chaperoning different types of client proteins (4). However, if and how a combination of proteins of different DNAJ classes might act in tandem has only recently started to be addressed (60). In our previously published report (7), a DNAJC14 truncation mutant (CT1) lacking the C-terminal 77 amino acids did not exhibit antiviral activity. In the same study, we showed that DNAJC14 carrying a mutation in the conserved HPD motif within the J domain, which is important for binding to and accelerating the ATPase activity of Hsp70 (61), was noninhibitory for YFV when overexpressed.
In the present study, we evaluated these noninhibitory DNAJC14 mutants in the context of YFV polyprotein processing and showed that they caused minimal alterations to YFV NS3/4A/2K cleavage events. Consistent with the notion that cochaperone activity stimulates the Hsp70 folding cycle and ATP hydrolysis, we verified that the H471Q mutant form that contained an amino acid substitution in the HPD motif did not cause any YFV cleavage alteration; on the contrary, it seemed to stabilize the protein products (Fig. 3). A similar observation has been made for ERdj5, an ER chaperone involved in low-density lipoprotein (LDL) receptor biogenesis (62), where coexpression of the LDL receptor with a J-domain mutant of ERdj5 increased the relative abundance of the ER form of the LDL receptor. Confirmation of this interesting finding for DNAJC14 and the YFV polyprotein requires further investigation.
In both yeasts and animals, Hsp proteins assist with protein folding and prevent the formation of aggregates by binding to hydrophobic stretches of amino acids in the target protein. Hsp70, for example, together with J proteins, captures unfolded, newly synthesized proteins or misfolded polypeptides and maintains them in a soluble state to ensure proper conformational changes, oligomeric assembly, and transport competence through different cell compartments (63). We anticipated that DNAJC14 interacted with an amphipathic portion of the viral polyprotein region, since it was previously shown to interact with the amphipathic helix of the C-terminal tail of several GPCRs (35, 36). YFV NS4A, like DENV NS4A, contains a putative amphipathic helix in its amino terminus (40). As predicted, the YFV NS4A protein was coimmunoprecipitated with DNAJC14 in our experiments (Fig. 2B). The amphipathic helix in the N terminus of DENV NS4A was shown to mediate NS4A oligomerization and be required for viral replication (40). The parallel between the DNAJC14 interaction with a likely amphipathic helix in NS4A, which is involved in oligomerization, and the DNAJC14 interaction with the amphipathic H8 of GPCRs and its possible role in oligomerization (40) is very attractive, suggesting a role of YFV NS4A in coordinating viral RC assembly, as reported for other members of the Flaviviridae family (64).
It was surprising to observe that polyprotein processing was not dramatically affected by using a pool of three siRNAs to knock down DNAJC14 (Fig. 4), since we previously reported (7) an approximately 1-log decrease in YFV replication after DNAJC14 knockdown. Several considerations are needed to interpret the findings of the DNAJC14 silencing experiments. (i) In the Escherichia coli system, it has been shown that DnaJ has a relatively low-affinity association with ATP-bound DnaK, the bacterial Hsp70 equivalent, leading to a transient and dynamic association (65). A transient association is optimal in the cellular context, as it prevents the nonproductive hydrolysis of ATP in the absence of client proteins. This model has also been verified for the Hsp70 folding cycle in yeast and mammalian cells (66, 67). Furthermore, optimal refolding activity occurs at substoichiometric levels of J proteins in different experimental systems (68–70). It is reasonable to speculate that our experimental siRNA knockdown conditions were not sufficient to achieve 100% protein knockdown and low levels of DNAJC14 expression would be sufficient to actively drive the Hsp70-DNAJC14 folding cycle. Slight alterations of the highly regulated processing in the NS3/4A/2K region might still impart deleterious effects on YFV fitness, with the ultimate effect being dependent on the knockdown efficiency. (ii) Changes in the abundance and relative levels of chaperones and cochaperones can generate novel combinations of folding proteins, which, in turn, could determine the overall effect on cellular functions. While some cellular responses may become favored because of an increase in the level of a particular cochaperone, other cellular pathways might be suppressed or even constitutively activated by the lack of critical chaperone components. DNAJC14 may be important for YFV polyprotein processing, but upon DNAJC14 silencing, cells do not demonstrate a phenotype due to the presence of other cellular chaperones. (iii) The overall effect of changes in chaperone or cochaperone levels on cellular and organismal phenotypes likely depends on which chaperone or cochaperone is affected (71). Moreover, chaperone and cochaperone molecules generally have pleiotropic effects on a variety of cellular functions (protein folding, trafficking, stress sensing, cell growth, and cell death). In this study, DNAJC14 knockdown did not cause a dramatic defect in YFV polyprotein processing but could have affected other viral or cellular functions that have not yet been investigated.
Our data shed light on the molecular mechanism by which DNAJC14 overexpression inhibits YFV infection and highlight the biological significance of NS3/4A/2K cleavage events to ensure viral fitness. For noncytopathic BVDV, which establishes persistent infection, DNAJC14 serves as a cofactor for the NS2 protease to allow early but limited proteolytic action and RNA replication. At later times, cleavage at the NS2/3 site no longer occurs, limiting new RC formation and providing uncleaved NS2-3 important for virion production. Interestingly, some cytopathic biotypes of BVDV contain insertions of DNAJC14 sequences within the NS2-3 region, which serve to stimulate NS2/3 cleavage, RNA replication, and cytopathogenicity (72). It is not surprising that cellular chaperones actively regulate viral polyprotein processing. Virus proliferation depends on the successful recruitment of host cellular components for their own replication, protein synthesis, and virion assembly. In the course of infection, a large number of proteins are synthesized in a relatively short time, with proper protein folding being a requirement to ensure downstream events, such as polyprotein processing, RNA replication, and ultimately, virus assembly. Most viruses therefore need cellular chaperones during their life cycle. Hsp70 proteins, as central components of the cellular chaperone network, are indeed frequently recruited by viruses (6, 56, 73–75).
Because of the essential functions that the NS2B-3 complex plays in the flavivirus life cycle, it represented an important subject for further investigation in our study on DNAJC14 and YFV cleavage events. Therefore, we introduced amino acid substitutions at the P1 position of the NS3/4A site in the YFV genome to alter NS3/4A/2K cleavage events and the NS3-to-NS3-4A ratio. Analysis of translation, replication, and virus production indicated that viral fitness positively correlates with processing efficiency (Fig. 6 and Table 1), revealing a critical biological significance of the NS3/4A cleavage site in the context of the YFV life cycle. Previous studies showed that polyprotein processing at the NS3/4A site is incomplete in cells infected with YFV or with other flaviviruses. (27, 46, 47). We also observed that NS3-4A was quickly generated from the sig2A-5 polyprotein, remained stable over time, and was not a precursor to NS3 (Fig. 1D). This finding indicates (i) the existence of at least two alternative pathways that independently generate NS3 or NS3-4A from different precursor molecules. (ii) It also indicates that the unprocessed form of YFV NS3-4A might have an as-yet-unknown function in YFV replication. Studies on BVDV suggested that uncleaved NS2-3 is needed for virus assembly (76, 77). It is likely that the YFV NS3-4A protein is functionally distinct from NS3, as it contains additional NS4A-derived sequences with potential amphipathic properties as well as transmembrane domains that could be important for protein oligomerization and/or changes in membrane curvature required for competent RC formation (40, 78). In this regard, Roosendaal et al. reported that cleavage by NS3 of the Kunjin virus polyprotein at the NS4A/4B site initiates the induction of membrane rearrangement and that NS4A, containing the C-terminal transmembrane 2K domain, plays a role in membrane rearrangements (33). (iii) Furthermore, it indicates that the YFV NS3/4A site is tightly regulated to have a controlled rate of processing in order to generate appropriate levels of uncleaved NS3-4A and cleaved products, NS3 and NS4A, similar to what has been described for cleavage of NS2/3 of BDVD and classical swine fever virus (CSFV) and NS3/4A of Murray Valley encephalitis virus (46, 77, 79). In this report, we propose that DNAJC14 plays a role in controlling this specific YFV cleavage site, similar to what has been described for BVDV NS2/3 cleavage, although the mechanism and biological relevance may be different (72, 77, 80). Indeed, to probe for a role of NS3-4A in YFV infection, we abolished the uncleaved NS3-4A form by molecularly engineering the YFV genome to encode the polyprotein in a bicistronic manner with no possibility of NS3-4A expression (NS3-4A SPLIT). We hoped to evaluate whether the abolishment of uncleaved NS3-4A affected viral RNA replication and infectious virus production. Moreover, if virus progeny were generated, then we further hypothesized that this virus would not be susceptible to inhibition by DNAJC14, since the generation of NS3 and NS4A proteins would be independent of the normal NS3/4A cleavage event and DNAJC14's chaperone activity. Unfortunately, the NS3-4A SPLIT YFV genome did not encode a viable virus, precluding further investigation. We cannot exclude the possibility, among other plausible explanations for this result, that uncleaved NS3-4A is an essential requirement for viral replication and virion production.
YFV has two basic residues at the P1 and P2 positions within the NS3/4A cleavage site (Arg-Arg/Gly-Glu). Mutant polyproteins containing various P1 substitutions at the NS3/4A site exhibited various grades of efficiency outside the context of viral replication (23). In particular, substitution with Glu or Pro completely abolished cleavage (23). We introduced these substitutions in the full-length YFV genome and in a subgenomic replicon to evaluate the effects of altered NS3/4A cleavage efficiency on virus replication independently of DNAJC14 overexpression. All of these substitutions, expect Lys, abrogated the ability of the virus to replicate (Fig. 6C and D), confirming the requirement for a basic residue at the P1 position. Interestingly, all escape mutants had reverted to either Arg or Lys at the NS3/4A P1 position (Table 1). Whether the viable revertant viruses that were eventually selected and appeared in the culture supernatant were derived from rare RNA molecules containing nucleotide substitutions introduced during RNA transcription in vitro or were the result of low-level replication and mutation due to the error-prone YFV RNA-dependent RNA polymerase is unknown but could be investigated in future studies. Overall, these findings further support a requirement for a basic residue, Arg or Lys, at the NS3/4A junction (Arg-Gly), as previously reported (20). When we introduced several of the NS3/4A P1 substitutions in a YFV replicon system (Fig. 6E), we found that efficiency at the NS3/4 site is a critical step for RNA replication, since all mutants tested were replication incompetent, a finding similar to that for a control replicon bearing a deletion in the RNA-dependent RNA polymerase protein (delta DD; Fig. 6F). All together these data show a correlation between YFV fitness, polyprotein cleavage at the NS3/4A site, and the observed sensitivity of YFV to DNAJC14's overexpression.
In conclusion, evidence supports a model wherein the DNAJC14 chaperone function modulates the viral NS protein conformation to regulate NS2B-3-mediated polyprotein cleavage, specifically, at the site between NS3 and NS4A, which occurs in cis. Notably, the modulation of processing at the NS3/4A/2K sites with the generation of distinct amounts of NS3-containing species is revealed here to be an important mechanism to ensure virus replication. DNAJC14's overexpression inhibits NS3/4A cleavage due to a dysregulation of proper Hsp70-mediated chaperone events, which normally ensure well-regulated cleavages to promote successful viral replication.
ACKNOWLEDGMENTS
We acknowledge all members of the M. R. MacDonald and C. M. Rice labs for critical discussions and for contributing reagents and technical help. In particular, we are grateful to H. H. Hoffmann, M. Gu, P. Ambrose, and Y. Yu. We thank W. M. Schneider, M. Saeed, and M. A. Scull for critical readings of the manuscript.
Funding Statement
This work was supported in part by The Starr Foundation, the Greenberg Medical Research Institute, and CTSA RUCCTS grant UL1 TR000043 from the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health, Clinical and Translational Science Award (CTSA) program. The funding agencies had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Stirling PC, Lundin VF, Leroux MR. 2003. Getting a grip on non-native proteins. EMBO Rep 4:565–570. doi: 10.1038/sj.embor.embor869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mayer MP, Bukau B. 2005. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci 62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sullivan CS, Pipas JM. 2001. The virus-chaperone connection. Virology 287:1–8. doi: 10.1006/viro.2001.1038. [DOI] [PubMed] [Google Scholar]
- 4.Kampinga HH, Craig EA. 2010. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol 11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geller R, Taguwa S, Frydman J. 2012. Broad action of Hsp90 as a host chaperone required for viral replication. Biochim Biophys Acta 1823:698–706. doi: 10.1016/j.bbamcr.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taguwa SM, Maringer K, Li X, Bernal-Rubio D, Rauch JN, Gestwicki JE, Andino R, Fernandez-Sesma A, Frydman J. 2015. Defining Hsp70 subnetworks in dengue virus replication reveals key vulnerability in flavivirus infection. Cell 163:1108–1123. doi: 10.1016/j.cell.2015.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yi Z, Sperzel L, Nurnberger C, Bredenbeek PJ, Lubick KJ, Best SM, Stoyanov CT, Law LM, Yuan Z, Rice CM, MacDonald MR. 2011. Identification and characterization of the host protein DNAJC14 as a broadly active flavivirus replication modulator. PLoS Pathog 7:e1001255. doi: 10.1371/journal.ppat.1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rinck G, Birghan C, Harada T, Meyers G, Thiel HJ, Tautz N. 2001. A cellular J-domain protein modulates polyprotein processing and cytopathogenicity of a pestivirus. J Virol 75:9470–9482. doi: 10.1128/JVI.75.19.9470-9482.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muller A, Rinck G, Thiel HJ, Tautz N. 2003. Cell-derived sequences in the N-terminal region of the polyprotein of a cytopathogenic pestivirus. J Virol 77:10663–10669. doi: 10.1128/JVI.77.19.10663-10669.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, Huang Y, Wu H, Ni X, Cheng H, Fan J, Gu S, Gu X, Cao G, Ying K, Mao Y, Lu Y, Xie Y. 2003. Molecular cloning and characterization of a novel human J-domain protein gene (HDJ3) from the fetal brain. J Hum Genet 48:217–221. doi: 10.1007/s10038-003-0012-8. [DOI] [PubMed] [Google Scholar]
- 11.Lindenbach BD, Murray CL, Thiel H-J, Rice CM. 2013. Flaviviridae, p 712–746. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 12.Mackenzie JM, Westaway EG. 2001. Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J Virol 75:10787–10799. doi: 10.1128/JVI.75.22.10787-10799.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samuel MA, Diamond MS. 2006. Pathogenesis of West Nile virus infection: a balance between virulence, innate and adaptive immunity, and viral evasion. J Virol 80:9349–9360. doi: 10.1128/JVI.01122-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu WJ, Chen HB, Khromykh AA. 2003. Molecular and functional analyses of Kunjin virus infectious cDNA clones demonstrate the essential roles for NS2A in virus assembly and for a nonconservative residue in NS3 in RNA replication. J Virol 77:7804–7813. doi: 10.1128/JVI.77.14.7804-7813.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu WJ, Wang XJ, Mokhonov VV, Shi PY, Randall R, Khromykh AA. 2005. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J Virol 79:1934–1942. doi: 10.1128/JVI.79.3.1934-1942.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu WJ, Wang XJ, Clark DC, Lobigs M, Hall RA, Khromykh AA. 2006. A single amino acid substitution in the West Nile virus nonstructural protein NS2A disables its ability to inhibit alpha/beta interferon induction and attenuates virus virulence in mice. J Virol 80:2396–2404. doi: 10.1128/JVI.80.5.2396-2404.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yi Z, Yuan Z, Rice CM, MacDonald MR. 2012. Flavivirus replication complex assembly revealed by DNAJC14 functional mapping. J Virol 86:11815–11832. doi: 10.1128/JVI.01022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ritter SL, Hall RA. 2009. Fine-tuning of GPCR activity by receptor-interacting proteins. Nat Rev Mol Cell Biol 10:819–830. doi: 10.1038/nrm2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin C, Amberg SM, Chambers TJ, Rice CM. 1993. Cleavage at a novel site in the NS4A region by the yellow fever virus NS2B-3 proteinase is a prerequisite for processing at the downstream 4A/4B signalase site. J Virol 67:2327–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chambers TJ, Grakoui A, Rice CM. 1991. Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J Virol 65:6042–6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bredenbeek PJ, Kooi EA, Lindenbach B, Huijkman N, Rice CM, Spaan WJ. 2003. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J Gen Virol 84:1261–1268. doi: 10.1099/vir.0.18860-0. [DOI] [PubMed] [Google Scholar]
- 22.Jones CT, Patkar CG, Kuhn RJ. 2005. Construction and applications of yellow fever virus replicons. Virology 331:247–259. doi: 10.1016/j.virol.2004.10.034. [DOI] [PubMed] [Google Scholar]
- 23.Lin C, Chambers TJ, Rice CM. 1993. Mutagenesis of conserved residues at the yellow fever virus 3/4A and 4B/5 dibasic cleavage sites: effects on cleavage efficiency and polyprotein processing. Virology 192:596–604. doi: 10.1006/viro.1993.1076. [DOI] [PubMed] [Google Scholar]
- 24.Jones CT, Murray CL, Eastman DK, Tassello J, Rice CM. 2007. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J Virol 81:8374–8383. doi: 10.1128/JVI.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindenbach BD, Rice CM. 1997. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J Virol 71:9608–9617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chambers TJ, McCourt DW, Rice CM. 1989. Yellow fever virus proteins NS2A, NS2B, and NS4B: identification and partial N-terminal amino acid sequence analysis. Virology 169:100–109. doi: 10.1016/0042-6822(89)90045-7. [DOI] [PubMed] [Google Scholar]
- 27.Chambers TJ, McCourt DW, Rice CM. 1990. Production of yellow fever virus proteins in infected cells: identification of discrete polyprotein species and analysis of cleavage kinetics using region-specific polyclonal antisera. Virology 177:159–174. doi: 10.1016/0042-6822(90)90470-C. [DOI] [PubMed] [Google Scholar]
- 28.Cristea IM, Carroll JW, Rout MP, Rice CM, Chait BT, MacDonald MR. 2006. Tracking and elucidating alphavirus-host protein interactions. J Biol Chem 281:30269–30278. doi: 10.1074/jbc.M603980200. [DOI] [PubMed] [Google Scholar]
- 29.Fuerst TR, Niles EG, Studier FW, Moss B. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A 83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amberg SM, Rice CM. 1999. Mutagenesis of the NS2B-NS3-mediated cleavage site in the flavivirus capsid protein demonstrates a requirement for coordinated processing. J Virol 73:8083–8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kummerer BM, Rice CM. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J Virol 76:4773–4784. doi: 10.1128/JVI.76.10.4773-4784.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller S, Kastner S, Krijnse-Locker J, Buhler S, Bartenschlager R. 2007. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J Biol Chem 282:8873–8882. doi: 10.1074/jbc.M609919200. [DOI] [PubMed] [Google Scholar]
- 33.Roosendaal J, Westaway EG, Khromykh A, Mackenzie JM. 2006. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J Virol 80:4623–4632. doi: 10.1128/JVI.80.9.4623-4632.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quinkert D, Bartenschlager R, Lohmann V. 2005. Quantitative analysis of the hepatitis C virus replication complex. J Virol 79:13594–13605. doi: 10.1128/JVI.79.21.13594-13605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bermak JC, Li M, Bullock C, Zhou QY. 2001. Regulation of transport of the dopamine D1 receptor by a new membrane-associated ER protein. Nat Cell Biol 3:492–498. doi: 10.1038/35074561. [DOI] [PubMed] [Google Scholar]
- 36.Kuang YQ, Charette N, Frazer J, Holland PJ, Attwood KM, Dellaire G, Dupre DJ. 2012. Dopamine receptor-interacting protein 78 acts as a molecular chaperone for CCR5 chemokine receptor signaling complex organization. PLoS One 7:e40522. doi: 10.1371/journal.pone.0040522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leclerc PC, Auger-Messier M, Lanctot PM, Escher E, Leduc R, Guillemette G. 2002. A polyaromatic caveolin-binding-like motif in the cytoplasmic tail of the type 1 receptor for angiotensin II plays an important role in receptor trafficking and signaling. Endocrinology 143:4702–4710. doi: 10.1210/en.2002-220679. [DOI] [PubMed] [Google Scholar]
- 38.Dupre DJ, Robitaille M, Richer M, Ethier N, Mamarbachi AM, Hebert TE. 2007. Dopamine receptor-interacting protein 78 acts as a molecular chaperone for Ggamma subunits before assembly with Gbeta. J Biol Chem 282:13703–13715. doi: 10.1074/jbc.M608846200. [DOI] [PubMed] [Google Scholar]
- 39.Bermak JC, Zhou QY. 2001. Accessory proteins in the biogenesis of G protein-coupled receptors. Mol Interv 1:282–287. [PubMed] [Google Scholar]
- 40.Stern O, Hung YF, Valdau O, Yaffe Y, Harris E, Hoffmann S, Willbold D, Sklan EH. 2013. An N-terminal amphipathic helix in dengue virus nonstructural protein 4A mediates oligomerization and is essential for replication. J Virol 87:4080–4085. doi: 10.1128/JVI.01900-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wall D, Zylicz M, Georgopoulos C. 1994. The NH2-terminal 108 amino acids of the Escherichia coli DnaJ protein stimulate the ATPase activity of DnaK and are sufficient for lambda replication. J Biol Chem 269:5446–5451. [PubMed] [Google Scholar]
- 42.Herod MR, Schregel V, Hinds C, Liu M, McLauchlan J, McCormick CJ. 2014. Genetic complementation of hepatitis C virus nonstructural protein functions associated with replication exhibits requirements that differ from those for virion assembly. J Virol 88:2748–2762. doi: 10.1128/JVI.03588-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pijlman GP, Kondratieva N, Khromykh AA. 2006. Translation of the flavivirus Kunjin NS3 gene in cis but not its RNA sequence or secondary structure is essential for efficient RNA packaging. J Virol 80:11255–11264. doi: 10.1128/JVI.01559-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu WJ, Sedlak PL, Kondratieva N, Khromykh AA. 2002. Complementation analysis of the flavivirus Kunjin NS3 and NS5 proteins defines the minimal regions essential for formation of a replication complex and shows a requirement of NS3 in cis for virus assembly. J Virol 76:10766–10775. doi: 10.1128/JVI.76.21.10766-10775.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khromykh AA, Varnavski AN, Sedlak PL, Westaway EG. 2001. Coupling between replication and packaging of flavivirus RNA: evidence derived from the use of DNA-based full-length cDNA clones of Kunjin virus. J Virol 75:4633–4640. doi: 10.1128/JVI.75.10.4633-4640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lobigs M. 1992. Proteolytic processing of a Murray Valley encephalitis virus non-structural polyprotein segment containing the viral proteinase: accumulation of a NS3-4A precursor which requires mature NS3 for efficient processing. J Gen Virol 73(Pt 9):2305–2312. doi: 10.1099/0022-1317-73-9-2305. [DOI] [PubMed] [Google Scholar]
- 47.Klemens O, Dubrau D, Tautz N. 2015. Characterization of the determinants of NS2-3-independent virion morphogenesis of pestiviruses. J Virol 89:11668–11680. doi: 10.1128/JVI.01646-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verchot J. 2012. Cellular chaperones and folding enzymes are vital contributors to membrane bound replication and movement complexes during plant RNA virus infection. Front Plant Sci 3:275. doi: 10.3389/fpls.2012.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiao A, Wong J, Luo H. 2010. Viral interaction with molecular chaperones: role in regulating viral infection. Arch Virol 155:1021–1031. doi: 10.1007/s00705-010-0691-3. [DOI] [PubMed] [Google Scholar]
- 50.Aparicio F, Thomas CL, Lederer C, Niu Y, Wang D, Maule AJ. 2005. Virus induction of heat shock protein 70 reflects a general response to protein accumulation in the plant cytosol. Plant Physiol 138:529–536. doi: 10.1104/pp.104.058958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Z, Zhou T, Wu X, Hong Y, Fan Z, Li H. 2008. Influence of cytoplasmic heat shock protein 70 on viral infection of Nicotiana benthamiana. Mol Plant Pathol 9:809–817. doi: 10.1111/j.1364-3703.2008.00505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mathioudakis MM, Veiga R, Ghita M, Tsikou D, Medina V, Canto T, Makris AM, Livieratos IC. 2012. Pepino mosaic virus capsid protein interacts with a tomato heat shock protein cognate 70. Virus Res 163:28–39. doi: 10.1016/j.virusres.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 53.Wang RY, Stork J, Nagy PD. 2009. A key role for heat shock protein 70 in the localization and insertion of tombusvirus replication proteins to intracellular membranes. J Virol 83:3276–3287. doi: 10.1128/JVI.02313-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tomita Y, Mizuno T, Diez J, Naito S, Ahlquist P, Ishikawa M. 2003. Mutation of host DnaJ homolog inhibits brome mosaic virus negative-strand RNA synthesis. J Virol 77:2990–2997. doi: 10.1128/JVI.77.5.2990-2997.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weeks SA, Miller DJ. 2008. The heat shock protein 70 cochaperone YDJ1 is required for efficient membrane-specific flock house virus RNA replication complex assembly and function in Saccharomyces cerevisiae. J Virol 82:2004–2012. doi: 10.1128/JVI.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weeks SA, Shield WP, Sahi C, Craig EA, Rospert S, Miller DJ. 2010. A targeted analysis of cellular chaperones reveals contrasting roles for heat shock protein 70 in flock house virus RNA replication. J Virol 84:330–339. doi: 10.1128/JVI.01808-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen MH, Tian GW, Gafni Y, Citovsky V. 2005. Effects of calreticulin on viral cell-to-cell movement. Plant Physiol 138:1866–1876. doi: 10.1104/pp.105.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vos MJ, Hageman J, Carra S, Kampinga HH. 2008. Structural and functional diversities between members of the human HSPB, HSPH, HSPA, and DNAJ chaperone families. Biochemistry 47:7001–7011. doi: 10.1021/bi800639z. [DOI] [PubMed] [Google Scholar]
- 59.Li J, Qian X, Sha B. 2009. Heat shock protein 40: structural studies and their functional implications. Protein Pept Lett 16:606–612. doi: 10.2174/092986609788490159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nillegoda NB, Kirstein J, Szlachcic A, Berynskyy M, Stank A, Stengel F, Arnsburg K, Gao X, Scior A, Aebersold R, Guilbride DL, Wade RC, Morimoto RI, Mayer MP, Bukau B. 2015. Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature 524:247–251. doi: 10.1038/nature14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hennessy F, Nicoll WS, Zimmermann R, Cheetham ME, Blatch GL. 2005. Not all J domains are created equal: implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci 14:1697–1709. doi: 10.1110/ps.051406805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oka OB, Pringle MA, Schopp IM, Braakman I, Bulleid NJ. 2013. ERdj5 is the ER reductase that catalyzes the removal of non-native disulfides and correct folding of the LDL receptor. Mol Cell 50:793–804. doi: 10.1016/j.molcel.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ellgaard L, Helenius A. 2003. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol 4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 64.Murray CL, Jones CT, Rice CM. 2008. Architects of assembly: roles of Flaviviridae non-structural proteins in virion morphogenesis. Nat Rev Microbiol 6:699–708. doi: 10.1038/nrmicro1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Russell R, Wali Karzai A, Mehl AF, McMacken R. 1999. DnaJ dramatically stimulates ATP hydrolysis by DnaK: insight into targeting of Hsp70 proteins to polypeptide substrates. Biochemistry 38:4165–4176. doi: 10.1021/bi9824036. [DOI] [PubMed] [Google Scholar]
- 66.Li Z, Hartl FU, Bracher A. 2013. Structure and function of Hip, an attenuator of the Hsp70 chaperone cycle. Nat Struct Mol Biol 20:929–935. doi: 10.1038/nsmb.2608. [DOI] [PubMed] [Google Scholar]
- 67.Kanelakis KC, Morishima Y, Dittmar KD, Galigniana MD, Takayama S, Reed JC, Pratt WB. 1999. Differential effects of the hsp70-binding protein BAG-1 on glucocorticoid receptor folding by the hsp90-based chaperone machinery. J Biol Chem 274:34134–34140. doi: 10.1074/jbc.274.48.34134. [DOI] [PubMed] [Google Scholar]
- 68.Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. 1991. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci U S A 88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Laufen T, Mayer MP, Beisel C, Klostermeier D, Mogk A, Reinstein J, Bukau B. 1999. Mechanism of regulation of Hsp70 chaperones by DnaJ cochaperones. Proc Natl Acad Sci U S A 96:5452–5457. doi: 10.1073/pnas.96.10.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kirschke E, Goswami D, Southworth D, Griffin PR, Agard DA. 2014. Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell 157:1685–1697. doi: 10.1016/j.cell.2014.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nollen EA, Morimoto RI. 2002. Chaperoning signaling pathways: molecular chaperones as stress-sensing ‘heat shock’ proteins. J Cell Sci 115:2809–2816. [DOI] [PubMed] [Google Scholar]
- 72.Lackner T, Muller A, Konig M, Thiel HJ, Tautz N. 2005. Persistence of bovine viral diarrhea virus is determined by a cellular cofactor of a viral autoprotease. J Virol 79:9746–9755. doi: 10.1128/JVI.79.15.9746-9755.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Inoue T, Tsai B. 2015. A nucleotide exchange factor promotes endoplasmic reticulum-to-cytosol membrane penetration of the nonenveloped virus simian virus 40. J Virol 89:4069–4079. doi: 10.1128/JVI.03552-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mayer MP. 2005. Recruitment of Hsp70 chaperones: a crucial part of viral survival strategies. Rev Physiol Biochem Pharmacol 153:1–46. [DOI] [PubMed] [Google Scholar]
- 75.Zhang C, Kang K, Ning P, Peng Y, Lin Z, Cui H, Cao Z, Wang J, Zhang Y. 2015. Heat shock protein 70 is associated with CSFV NS5A protein and enhances viral RNA replication. Virology 482:9–18. doi: 10.1016/j.virol.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 76.Agapov EV, Murray CL, Frolov I, Qu L, Myers TM, Rice CM. 2004. Uncleaved NS2-3 is required for production of infectious bovine viral diarrhea virus. J Virol 78:2414–2425. doi: 10.1128/JVI.78.5.2414-2425.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lackner T, Muller A, Pankraz A, Becher P, Thiel HJ, Gorbalenya AE, Tautz N. 2004. Temporal modulation of an autoprotease is crucial for replication and pathogenicity of an RNA virus. J Virol 78:10765–10775. doi: 10.1128/JVI.78.19.10765-10775.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hung YF, Schwarten M, Hoffmann S, Willbold D, Sklan EH, Koenig B. 2015. Amino terminal region of dengue virus NS4A cytosolic domain binds to highly curved liposomes. Viruses 7:4119–4130. doi: 10.3390/v7072812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lamp B, Riedel C, Roman-Sosa G, Heimann M, Jacobi S, Becher P, Thiel HJ, Rumenapf T. 2011. Biosynthesis of classical swine fever virus nonstructural proteins. J Virol 85:3607–3620. doi: 10.1128/JVI.02206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lackner T, Thiel HJ, Tautz N. 2006. Dissection of a viral autoprotease elucidates a function of a cellular chaperone in proteolysis. Proc Natl Acad Sci U S A 103:1510–1515. doi: 10.1073/pnas.0508247103. [DOI] [PMC free article] [PubMed] [Google Scholar]