ABSTRACT
Retargeting of gammaretroviral envelope proteins has shown promising results in the isolation of novel isolates with therapeutic potential. However, the optimal conditions required to obtain high-affinity retargeted envelope proteins with narrow tropism are not understood. This study highlights the advantage of constrained peptides within receptor binding domains and validates the random library screening technique of obtaining novel retargeted Env proteins. Using a modified vector backbone to screen the envelope libraries on 143B osteosarcoma cells, three novel and unique retargeted envelopes were isolated. The use of complex disulfide bonds within variable regions required for receptor binding is found within natural gammaretroviral envelope isolates. Interestingly, two of the isolates, named AII and BV2, have a pair of cysteines located within the randomized region of 11 amino acids similar to that identified within the CP Env, an isolate identified in a previous Env library screen on the human renal carcinoma Caki-1 cell line. The amino acids within the randomized region of AII and BV2 envelopes that are essential for viral infection have been identified in this study and include these cysteine residues. Through mutagenesis studies, the putative disulfide bond pairs including and beyond the randomized region were examined. In parallel, the disulfide bonds of CP Env were identified using mass spectrometry. The results indicate that this pair of cysteines creates the structural context to position key hydrophobic (F and W) and basic (K and H) residues critical for viral titer and suggest that AII, BV2, and CP internal cysteines bond together in distinct ways.
IMPORTANCE Retargeted gammaretroviral particles have broad applications for therapeutic use. Although great advances have been achieved in identifying new Env-host cell receptor pairs, the rules for designing optimal Env libraries are still unclear. We have found that isolates with an additional pair of cysteines within the randomized region have the highest transduction efficiencies. This emphasizes the importance of considering cysteine pairs in the design of new libraries. Furthermore, our data clearly indicate that these cysteines are essential for viral infectivity by presenting essential residues to the host cell receptor. These studies facilitate the screening of Env libraries for functional entry into target cells, allowing the identification of novel gammaretroviral Envs targeting alternative host cell receptors for gene and protein delivery.
INTRODUCTION
The specificity of retroviral vectors is initially conferred by the envelope (Env) protein present on the surface of virus particles. This Env protein is responsible for binding to a host cell receptor, thereby initiating virus entry. In gammaretroviruses, the Env protein is composed of a transmembrane protein (TM) and a surface protein (SU). For murine leukemia virus (MLV) and feline leukemia virus (FeLV) SUs, primary receptor binding is localized to the N-terminal half of SU (1, 2, 12). Critical residues for specificity are encoded within the N-terminal variable regions, VRA and VRB. Secondary receptor binding sites that promote viral infection have also been identified within the SU C terminus (3–5).
FeLV Envs are classified as A, B, and C, originally based on interference assays (6, 7). The receptors for these three major subgroups have been identified as THTR1 (8), PiT1 (9), and FLVCR1 (10, 11), respectively, molecularly confirming the distinct receptor usage for each subgroup. For FeLV-A, 19 residues in the VRA can be replaced with 16 residues of FeLV-C VRA to sufficiently alter the binding specificity, thus switching receptor usage (12). In FeLV-A, the VRA and VRB each include a pair of cysteines, which have the potential to form intramolecular disulfide bonds (5). The cysteine contents of MLV and FeLV-B SUs are more complex; ecotropic MLV SU has six cysteines in the VRA and two in the VRB (2, 13), while amphotropic MLV and FeLV B SUs have four cysteines in both VRA and VRB (1, 2, 14, 15).
Targeted entry of retroviral particles is a key interest in gene and protein delivery (16, 17). Insertion of ligand binding domains, single-chain antibody (Ab) regions, and other conjugates into the Env SU of MLV and FeLV has been explored, with relative success (reviewed in reference 16). However, proteins bearing these modifications may fail to be incorporated into viral particles and/or the addition of such domains may interfere with postbinding processes. Binding to the host receptor(s) is the first step in a series of complex conformational changes required to deliver the viral core particle into the cells. The localization of the receptor recognition sequence to the VRA loop in FeLV-A, coupled with the simplicity of the disulfide bonding potential, made the FeLV-A/C Env backbone more suitable to generate randomized Env libraries for receptor retargeting (18–21). Generation of the random Env libraries within the context of the viral SU while also screening for gene delivery ensures maintenance of a functional entry process.
The use of randomized Env libraries within the FeLV-A/C backbone has yielded a variety of novel Env isolates (18–21). For two of these isolates, A5 and constrained peptide (CP), the corresponding receptors have been identified (22, 23). The receptor for the A5 isolate is the SLC35F2 protein, a multitransmembrane spanning protein. The receptor for the CP isolate is GPR172A (riboflavin transporter; HuPAR-1), which also functions as a receptor for porcine endogenous retrovirus A (PERV-A) (24). Interestingly, the VRA region of CP Env shows little homology to that of PERV-A, despite binding to the same extracellular loop of the receptor (25). The CP Env, selected in tissue culture, maintained high viral titers and could be readily concentrated. These features were attributed to the presence of an internal cysteine pair located within the randomized VRA region (22, 25).
While a variety of retargeted Env proteins have been obtained, the overall yield of high-titer isolates remains low. This study optimized the selection of high-affinity gammaretroviral Env with defined specificities, with the goal of identifying novel retroviral Env-host cell receptor pairs to facilitate targeted viral delivery. Surprisingly, two isolates identified require a complex disulfide bonding pattern within the VRA to present the residues essential for viral binding and entry into the host cell. A better understanding of the gammaretroviral receptor binding domain (RBD) is obtained through characterization of these isolates, which can be applied toward the design of next-generation Env libraries.
MATERIALS AND METHODS
Cell lines.
Cells were all cultured in Dulbecco's modified Eagle medium (DMEM) containing 10% heat-inactivated fetal bovine serum and 1% antibiotic-antimycotic (Gibco) unless mentioned otherwise. Human 143B osteosarcoma cells were used as the target cell for library screening (19), TE671 rhabdomyosarcoma cells were used for binding studies (22), and HEK293T cells were used for virus production. CeB (22)-expressing cell lines were maintained in 9 μg/ml of blasticidin S (InvivoGen). Virus-packaging 293TCeB (18) and 143BCeB cell lines were grown in DMEM and minimum essential medium, respectively. TELCeB and all other cell lines were maintained as previously described (22, 26).
Oligonucleotides.
The sequences of the oligonucleotides used in this study will be provided upon request.
Generation of alternative bicistronic vectors.
Five alternative bicistronic vectors were generated to eliminate aberrant splicing detected within pRVL (22). In order to incorporate the M1 mutation into pRVL-CP (22), the 2,099-bp BamHI-ClaI fragment from pRVL (22) was subcloned into pBABE, generating pBABE + 2KB. The M1 mutation in the simian virus 40 (SV40) region was amplified using primers FSVE-M1/RSVE-M1 and SVE-core M1 plasmid as the template (27). The fragment was digested with HindIII/NcoI and ligated with the 1.1-kb HindIII/ClaI fragment of pRVL into NcoI/ClaI-digested pBABE + 2KB. The 4.9-kb BamHI/ClaI fragment from pRVL encoding the cytomegalovirus (CMV) promoter replaced the pBABE fragment. Finally, the BamHI CP Env cassette was inserted into the BamHI site to generate the pRVL-M1-CP. pRVL-CP-NWPRE was constructed by digesting pRVL-CP with BamHI and inserting the 620-bp woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) BamHI-BglII fragment previously described (28). Env was subsequently reinserted back using the BamHI site. The pRVL-CP-CWPRE construct was built by overlapping PCR. Primer pairs FNeo/RWPREneoX, FneoXWPRE/RDtWPREp4, and FDtWPREp5/RpRVLClaI were used to amplify neo, WPRE, and the long terminal repeat (LTR), respectively. All PCR templates used were the same as described below for construction of plasmid SD-env-SA-neo-CWPRE. The final PCR fragment was BssHII digested and inserted into BssHII-digested pRVL-CP.
To build our SD-env-SA plasmid, we introduced an AgeI and PacI site into pRVL using overlapping PCR of fragments amplified with primer pairs FSpRVL1/RAPpRVL2 and FAPKpRVL3/RKpRVL. The PCR product was digested with SacII and KpnI, and a three-piece ligation was performed with the pRVL 4,107-bp KpnI-KpnI and 2,543-bp KpnI-SacII fragments, generating pRVL-AgeI/PacI. The splice site acceptor (SA) site was amplified from the pMX plasmid using primers FBpMX1 and RPpMX1. The PCR product was digested and ligated into pRVL-AgeI/PacI using the BamHI and PacI sites. The neomycin resistance gene was PCR amplified from pRVL (18) using primers FNeo/RLTR, digested with PacI/ClaI, and ligated using the PacI/ClaI sites of pRVL-AgeI/PacI. The CP env was introduced using the BamHI sites as previously described (22). In order to create the library vector SD-env-SA-neo-CWPRE, a point mutation was introduced to destroy a BbsI site in the WPRE sequence, using primers FneoXWPRE/RWPRE560 and FWPRE539/RDtWPREp4. The parental template used, HIV2gtmtWPRE (28), encoded a substitution of the ATG start codon in the WPRE sequence, to abrogate translation of the woodchuck hepatitis virus X protein (29). The BbsI− WPRE sequence was introduced into the plasmid vector downstream of the neomycin resistance gene by overlapping PCR. The neomycin resistance gene was amplified from SD-env-SA-neo using FNeo7730/RWPREneoX primers, the BbsI− WPRE region was amplified using FneoXWPRE/RDtWPREp4 primers, and the LTR from the pRVL plasmid was amplified with FDtWPREp5/RpRVLCla primers. The final overlapping PCR product was exchanged into the pSD-CP-SA-neo vector using ClaI and PacI. The cloned product was confirmed using multiple restriction digests and DNA sequencing. Finally, using the BamHI restriction site, we exchanged the CP Env with the library Env containing the BbsI-Stop-BbsI cassette previously described (22).
Library screening.
A library of oligonucleotides was inserted at the BbsI site of the plasmid vector as previously described (22). Briefly, the library of retroviral particles was produced transiently in GP2-293 cells (Clontech) by cotransfecting 70 μg of both the Env library plasmid and pHIT-G (30) using Lipofectamine 2000 (Life Technologies). The following day, transfected cells were incubated with 10 mM sodium butyrate for ∼6 h. Two days after transfection, ∼70 ml of supernatant containing the library of virus particles was collected, filtered through a 0.45-μm filter, and divided among three HYPERflasks (Corning) seeded with 2 × 107 143BCeB cells per flask. All infections were performed in the presence of 8 μg/ml of Polybrene and at a multiplicity of infection (MOI) of 0.6. The medium was replaced after 12 h, and infected positive cells were selected with 400 μg/ml of G418 after 48 h. The G418r cells constitutively produced the library of virus particles, with each particle expressing a unique Env on its surface.
To screen the library, viral supernatant was collected from confluent producer cells grown in the absence of G418. Filtered supernatant (0.45-μm filter) was used to infect 143BCeB target cells in three 15-cm plates in the presence of 8 μg/ml of Polybrene. Two days after infection, target cells were subjected to G418 selection. Nine clones were identified from the screen. To further characterize the clones, each individual colony was isolated and expanded into a larger plate. The genomic DNA was extracted, and the env gene was PCR amplified and sequenced using primers FeLV1/2.
Viral titer and receptor interference assays.
A total of 2 × 105 cells were seeded 1 day prior to infection. Cells were infected with 1 ml of viral supernatant in the presence of 8 μg/ml Polybrene, and the medium was changed 12 h later. Forty-eight hours postinfection, cells were grown in the presence of 400 μg/ml of G418. Fourteen days later, colonies were counted. In the case of viral particles carrying the lacZ gene, infected cells were stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) at 48 h postinfection. The LacZ-positive cells were counted 48 h later.
For viral receptor interference studies, 1 ml of viral supernatant was used to infect 2 × 105 HEK293T cells or HEK293T cells constitutively producing the secreted CP-SU1-235.
Statistical analyses.
All statistical analyses were performed using MINITAB 17 software. All experimental means represent the results of at least three independent experiments. Significant differences between the means were analyzed using analysis of variance (ANOVA) with a post hoc Dunnet test. The respective wild-type (WT) Env protein was used as a control. The level of significance was set at an α value of 0.05 unless otherwise indicated.
Mutagenesis of AII and BV2.
Specific mutations in the AII and BV2 Env proteins were generated within the pSD-Env-SA-neo-CWPRE vector at the back-to-back BbsI sites using three oligonucleotides, similar to that used to generate the Env libraries. After sequencing, individual mutations were amplified with In-Fusion primers, Fprvltopsin and Rprvltopsin, cloned into BamHI-digested pSinEF1α-IRES-Puro using In-Fusion HD enzyme (Clontech). pSinEF1α-IRES-Puro was generated by digesting pSin-EF2-Oct4-Pur (Addgene) with EcoRI and SpeI, treated with Klenow large fragment enzyme (NEB M0210S) to generate blunt ends, and ligated with T4 ligase (NEB M0202S). To establish producer cell lines expressing the individual Env mutants, 2 μg of each plasmid (pSinEF1α-mutantEnv-IRES-Puro, pMD2.G [Addgene], and psPAX2 [Addgene]) were transfected into 293T cells overnight using FuGENE (Promega) as per the manufacturer's protocol. The following day, cells were induced with sodium butyrate for 5 h. Viral supernatant was collected after a minimum of 20 h and used to infect TELCeB in the presence of 8 μg/ml of Polybrene. The medium was changed the next day, and cells were selected with puromycin (2.5 μg/ml) 48 h postinfection.
MLV-based viral particles expressing the mutant AII and BV2 Envs were collected from the TELCeB supernatants, normalized by capsid (CA) levels (31), and used to determine viral titers by lacZ transfer as described above.
Western blot analysis.
Western blotting was performed using Any kD (Bio-Rad) SDS-PAGE; proteins were then transferred onto Trans-Blot Turbo Mini polyvinylidene difluoride (PVDF; Bio-Rad). Each lane was normalized to contain 100 ng of CA. Membranes were probed with C11D8 mouse anti-GP70 (AbD Serotec; 1:1,000 dilution) and secondary 31430 goat anti-mouse horseradish peroxidase (HRP) conjugate (1:5,000 dilution). Signals were detected using SuperSignal West maximum-sensitivity substrate (Life Technologies). To detect the presence of the SU in whole-cell extracts, 5 × 104 producer cells were pelleted, resuspended, and lysed in 20 μl of 2× lithium dodecyl sulfate (LDS) buffer. The entire sample was analyzed by Western blotting for SU Env as described above for viral proteins.
CP-SU1-235 protein A domain B fusion construct.
The Env SU was PCR amplified from amino acid position 1 to position 235 using pRVL-CP (22) as a template and primers FprvltopPRO and RprvltopPRO. The PCR product was digested with SacI and BamHI and ligated into pPro (32) digested with SacI and BamHI using T4 ligase. Ligation into pPro positions the SU upstream of three tandem repeats of the protein A B domain followed by a 3× FLAG tag at the C-terminal domain. The tagged SU1-235 region was PCR amplified using primers FpJG and RpJG and cloned into lentiviral expression vector pJG (33) digested with PmeI, using In-Fusion HD (Clontech).
Two micrograms each of plasmids pJG, pMD2.G (Addgene), and psPAX2 (Addgene) was transfected into 293T cells overnight using FuGENE (Promega) as per the manufacturer's protocol. The medium was changed the following day, and sodium butyrate was added for 5 h (23). Viral supernatant was collected the following day after a minimum of 20 h and used to transduce HEK293T cells overnight in the presence of Polybrene (8 μg/ml). Cell medium was changed the next day, and cells were selected in the presence of puromycin (2.5 μg/ml) at 48 h postinfection.
Binding assay by flow cytometry.
Supernatant from 293T-CP-SU1-235 cells was collected and filtered. The secreted CP-SU1-235 protein was purified on an IgGFF-Sepharose 6 Fast Flow column (GE Healthcare Life Sciences). Binding studies were performed on TE671 human rhabdomyosarcoma cells, which support high titers of virus bearing CP Env (22). A total of 2.5 × 106 TE671 cells were centrifuged at 228 × g and exposed to 10 ml of supernatant containing CP viral particles or 1 ml with 400 nM purified CPSU1-235 on a tube rotator at 4°C for 1.5 h. Env was probed using either the C11D8 mouse anti-gp70 Ab (1:50 dilution) for virus-associated Env or mouse anti-3XFLAG (1:50) for the secreted CP-SU1-235. Sigma PE goat anti-mouse Ab (P9287) was used as a secondary antibody. TE671 cells in the absence of antigen were used as a negative control. The binding analysis was performed on a Beckman Coulter Cytomics FC500 at the Cytometry Core Facility at the Environmental and Occupational Health Sciences Institute of Rutgers University in Piscataway, NJ.
Tryptic digestion of CP-SU1-235.
Free cysteines of CP-SU1-235 were blocked with 10 mM iodoacetamide (IAM) in 200 mM Tris-HCl, pH 8.0, and 6 M urea in the dark at room temperature for 60 min, followed by SDS-PAGE. The CP-SU1-235 protein band was excised and subjected to in-gel tryptic digestion at a ratio of 10:1 (wt/wt, CP-SU1-235 to trypsin) in 50 mM ammonium acetate buffer (pH 6.0) at 37°C overnight. Peptides were extracted, dried under vacuum, and then solubilized in 50 mM ammonium acetate buffer (pH 6.0). The digested peptides were then divided into two aliquots. One aliquot was reduced with 10 mM dithiothreitol (DTT) in ammonium bicarbonate buffer (pH 8.0) at 37°C, followed by alkylation with 20 mM N-ethylmaleimide (NEM) in the dark at room temperature for 30 min. For the second aliquot, water was used in the place of DTT and NEM as a control. After the treatment, the samples were acidified with trifluoroacetic acid for liquid chromatography-tandem mass spectrometry (LC-MS/MS).
LC-MS/MS.
Peptides were analyzed by nano-LC-MS/MS using a rapid-separation liquid chromatography (RSLC) system (Dionex, Sunnyvale CA) interfaced with a Velos-LTQ-Orbitrap (Thermo Fisher, San Jose, CA). Samples were loaded onto a self-packed 100-μm by 2-cm trap packed with Magic C18AQ (5 μm, 200 ˚; Michrom Bioresources Inc., Auburn, CA) and washed with buffer A (0.2% formic acid) for 5 min with a flow rate of 10 μl/min. The trap was brought in-line with the homemade analytical column (Magic C18AQ; 3 μm, 200 ˚, 75 μm by 50 cm), and peptides were fractionated at 300 nl/min with a multistep gradient (4 to 15% buffer B [0.16% formic acid and 80% acetonitrile] in 10 min and 15 to 55% buffer B in 35 min). Mass spectrometry data were acquired using a data-dependent acquisition procedure with a cyclic series of a full scan acquired in Orbitrap with resolution of 60,000 followed by MS/MS scans of the 20 most intense ions in the ion trap (CID) with a repeat count of two and the dynamic exclusion duration of 60 s.
Disulfide bond analysis.
LC-MS/MS data were first analyzed by an automatic database search against a custom FASTA database composed of target protein sequences and a random collection of ∼2,000 other nonrelated protein sequences as well as a cRAP database with common contaminants using X!tandem (SLEDGEHAMMER [2013.09.01]; http://thegpm.org). Oxidation of methionine and deamidation on asparagine were set as variable modifications, and 10 ppm and 0.4 Da were set as precursor ion and fragment ion tolerances, respectively. For the purpose of identification of free cysteines, +57.02 Da (carbomidal) was set as a variable modification; −1.0079 Da was set as a variable modification on cysteine to identify internal disulfide bonds. All theoretical masses of tryptic sequences containing cysteine minus two hydrogens were also used as modifications on cysteine for automatic identification of interpeptide disulfide bonds. Any inter- or intrapeptide disulfide bonds identified by MS/MS through a database search as described above were manually verified. Finally, a mass list of all possible combinations of disulfide-bonded tryptic peptide pairs at different charge states was generated, and extracted ion chromatograms were compared for reducing and nonreducing LC-MS/MS runs. The peptide pairs showing significant reduction in reducing LC-MS/MS runs were further confirmed by manual inspection of MS/MS.
RESULTS
Modification of the vector backbone for improved library screening.
One critical factor toward achieving successful gene and drug delivery lies in the specificity of the cell targeting mechanism. Our studies have shown that viral gene delivery can be altered using alternative host proteins as target receptors by randomizing the receptor binding domains within the gammaretroviral Env (18–23, 25). Previous techniques produced a limited number of high-affinity Env isolates in each round of screening (18–22). Additional studies indicated that the pRVL vector (22) used to build the library of Envs had a high frequency of aberrant splicing into the internal SV40 promoter region, eliminating the packaging of the RNA into viral particles (28).
In order to address this limitation, a panel of modified bicistronic vectors (Fig. 1A) was tested for optimal delivery and drug selection of the CP Env construct into the target 143B cells. Variations aimed at eliminating the aberrant splice acceptor site, replacing the internal promoter with the natural MLV splice site acceptor (SA) to express the neomycin resistance gene, and inserting a woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) reported to increase the half-life of the viral RNA transcripts (34). Vectors expressing the secondary neo gene through the MLV splice site acceptor in the presence of the WPRE downstream of the neomycin resistance gene (3.4 × 105 neomycin-resistant colony-forming units [NCFU]/ml) resulted in titers 15-fold higher than with the parental pRVL-CP Env constructs (2.3 × 104 NCFU/ml) (Fig. 1A). Therefore, the pSD-Env-SA-neo-CWPRE vector was used for subsequent Env library studies (Fig. 1B).
FIG 1.
Generation of bicistronic vectors for Env library screening. (A) Schematic of the various vector backbones (left) and their corresponding viral titers (neomycin CFU [NCFU]/milliliter). The internal SV40 promoter was replaced with the M1 mutant variant (27) or the MLV SA sequence. Arrows mark the transcriptional start sites. Positions of the WPRE are indicated. ANOVA and a post hoc Dunnet test using pRVL-CP as a control were used to calculate significance. **, P < 0.05; *, P < 0.1. Error bars indicate the SEMs for at least three independent experiments. Ψ, MLV packaging signal. (B) Positions of the viral long terminal repeat (LTR), splice donor (SD), splice acceptor (SA), packaging signal (Ψ), env, and WPRE elements are indicated. The arrow marks the transcriptional start site. The position of the randomized 11 amino acids (X11) within the VRA of the FeLV-A/C Env backbone is indicated.
Figure 1B shows the schematic of the pSD-Env-SA-neo-CWPRE vector used in the current library screen. The Env cassette is expressed from the MLV promoter, the neomycin resistance gene is expressed as a spliced message, and the WPRE is downstream of the neomycin resistance gene. The BbsI site within the WPRE element was mutated to facilitate the generation of the Env library, which requires back-to-back BbsI sites.
Isolation and characterization of novel Env isolates capable of infecting 143B cells.
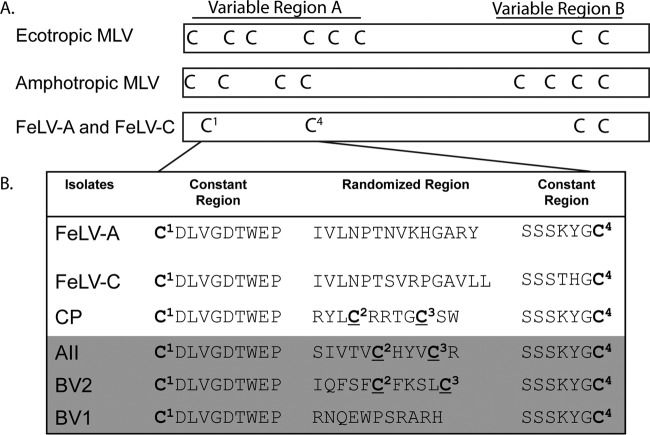
Using the pSD-Env-SA-neo-CWPRE vector (Fig. 1B), a constitutive Env library randomized at 11 positions within the FeLV-A VRA (Fig. 2A) (complexity of 106 inserts) was generated and screened for its ability to infect the human osteosarcoma cell line 143BCeB (26). After sequencing of the neomycin-resistant isolates, supernatants were collected from the corresponding expanded cell cultures and tested for neomycin transfer into naive 143B cells for verification of the Env function. Figure 2B highlights the sequences of three isolates with confirmed neomycin resistance gene transfer, AII, BVI, and BV2. These three isolates encode unique insertions in the randomized VRA region that are distinct from each other and from the parental FeLV-A and FeLV-C (18–20, 22, 23).
FIG 2.
Comparison of the Env variable regions A and B. (A) Localization of cysteines within the VRA and VRB of three closely related gammaretroviral Envs. (B) Primary sequence of the VRA from FeLV A and C with four Env library isolates. Constrained peptide (CP) Env was isolated from an Env library screen on Caki1 cells (22). Isolates identified with the highest transduction efficiencies using the pSD-Env-SA-neo-CWPRE vector (AII, BV2, and BVI) are highlighted in dark gray. Cysteines (superscripted) are numbered based on their order within the AII, BV2, and CP isolates. Cysteines within the randomized regions of VRA are underlined.
Interestingly, both AII and BV2 encode two cysteines (Fig. 2B, labeled as C2 and C3) within the randomized VRA region and thus have the potential to constrain the conformation of the receptor binding domain by forming disulfide bonds. The FeLV CP Env isolate (Fig. 2B) previously shown to maintain high viral titers on 143B cells (105 LacZ staining unit [LSU]/ml), recognizes the PAR1/2 host cell receptor (22) and also contains cysteines at 2 out of the 11 randomized positions. This was surprising, since we selected to build our library of Envs in the FeLV Env backbone due to a single cysteine loop within both the VRA and VRB, resulting in a less complex secondary structure in comparison to those of the ecotropic and amphotropic MLV Envs (Fig. 2A). The two novel envelope isolates, AII and BV2, were further investigated in order to understand the role of cysteine pairs within the RBD in viral entry. Studies of BVI Env will be described independently.
Host range analysis of AII and BV2 Envs.
A dual-expression construct is necessary for the library screening in order to link the packaging of the unique Env isolate with the selectable marker. For subsequent analysis, an alternative vector system that independently expresses the Env construct is preferred, providing the flexibility to vary visual and selectable markers to optimize viral titers. The FeLV Env isolates described here were expressed from the internal EF1α promoter within the self-inactivated (SIN) lentiviral vector pSinEF1α-IRES-puro. Vectors were introduced into TELCeB cells (26), which constitutively express the MLV Gag/Pol proteins and package the retroviral vector expressing the lacZ gene. Expression of Env from pSinEF1α-Env-IRES-puro in the TELCeB cells resulted in titers of 1.63 × 103 LSU/ml for AII and 6.6 × 103 LSU/ml for BV2 in 143B osteosarcoma cells and up to 105 LSU/ml in HEK293T cells (Fig. 3). Using this system, we examined the host range of AII and BV2. Analysis of the host range and tissue specificity of AII and BV2 confirmed that their receptor usage is distinct from that of the parental FeLV-A and -C isolates due to their inability to infect feline AH927 cells (Fig. 3).
FIG 3.
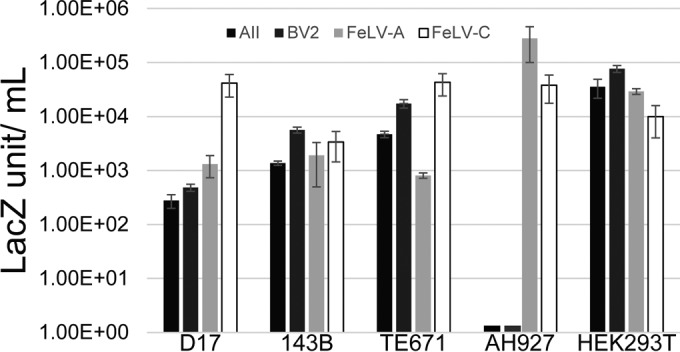
Defining the host range of virus bearing the AII and BV2 Env. A total of 2 × 105 cells were infected with viral supernatant containing the corresponding Env proteins and packaging the lacZ gene. Viral titers are LacZ staining units/milliliter of virus. Individual cell lines are indicated, with the color key at the top. Error bars indicate the SEMs for at least three independent experiments.
Mutagenesis studies reveal that the cysteines within the randomized region of AII are essential for viral titer.
The role of individual residues within the RBD of FeLV Env AII was investigated by mutating each of the 11 amino acids to alanine in HEK293T cells. Figure 4A shows the results of lacZ transfer into HEK293T cells infected with retroviral particles bearing the respective mutant Env proteins. Viral infections were normalized to equivalent levels of capsid (CA) (31).
FIG 4.

AII Env mutagenesis studies identify residues critical for viral titers. (A) HEK293T cells were exposed to viral particles carrying the respective Env mutant protein and quantified for lacZ gene transfer, normalized to 200 ng of capsid. Error bars indicate the SEMs for at least three independent experiments. Asterisks indicate P values of <0.05 compared to the value for the parental AII. (B) Western blot analysis of virus-associated SU. Viral supernatant was collected and lanes were normalized to analyze 100 ng of CA. Env was detected using C11D8 mouse anti-GP70 (AbD Serotec; 1:1,000 dilution). (C) Western blot analysis of whole-cell extracts. SU was detected using C11D8 mouse anti-GP70 (AbD Serotec; 1:1,000 dilution). Positions of protein size markers are indicated on the left in panels B and C.
For AII Env mutants, five individual positions—V3A, C6A, H7A, Y8A, and C10A—decreased viral titers at least 1,000-fold (Fig. 4A). While statistically significant variations in lacZ titers were also observed for S1A, I2A, T4A, V5A, V9A, and R11A mutants compared to viral particles bearing the WT AII Env, this group of residues is more tolerant to alanine substitutions but still necessary for optimal viral titer (Fig. 4A).
Western blot analysis of the viral particles was performed to determine whether the reduction in titer was due to incorrect assembly of the mutant Env onto viral particles (Fig. 4B). SU bearing the mutant AII V3A Env was not detected on viral particles, although SU was observed in whole-cell extracts from the TELCeB AII V3A producer cell line (Fig. 4C), indicating a block in transport or assembly. The H7A and Y8A AII SU proteins were detected on viral particles, albeit at lower levels for the H7A mutant. The presence of SU on viral particles indicates a loss of SU function for viral entry (Fig. 4B).
Of particular interest is the role of C6 and C10 in receptor binding. Individual mutation of each resulted in loss of viral titer and decreased association of SU on viral particles. Interestingly, mutation of both C6A and C10A, reestablishing an even number of cysteine residues within the VRA region, restored assembly of SU onto the viral particles (Fig. 4B) but did not restore viral infectivity. More significantly, the double mutant C6E:C10K, which introduces the potential for a salt bridge to mimic a cysteine bond, restored the viral titer to 1 × 103 LSU/200 ng of CA, with levels of SU similar to those of the parental AII Env (Fig. 4A). The requirement for either the C6:C10 pair or a salt bridge between these residues strongly suggests that C6 and C10 form a disulfide bond in order to present the essential residues H7 and Y8 to the host-cell receptor. Since the V3A mutant affects the assembly of the Env onto viral particles, we cannot examine its effect on host cell receptor binding and entry.
The cysteines within the randomized region of the BV2 isolate are essential for viral titers.
Mutational analysis of the BV2 Env isolate was performed, again with a particular interest in the role of the pair of cysteines selected within the randomized region. In BV2 Env, the spacing between the cysteines is four residues, similar to that of the CP Env, while the AII Env isolate has three amino acids between the cysteines (Fig. 2B). Strikingly, BV2 Env C6A, F7A, K8A, and C11A mutants resulted in a total loss of infectivity (Fig. 5A), highlighting the importance of the two cysteines and a limited number of hydrophobic and charged residues within this intervening region. An interesting feature of BV2 is the presence of three phenylalanines at positions 3, 5, and 7. Complete loss of infectivity was observed only for F7A, while F5A resulted in a reproducible 10-fold reduction of titer and F3A Env was similar to the parental BV2 Env. Titers of virus bearing BV2 Env I1A, Q2A, F3A, S4A, S9A, and L10A mutants were not significantly lower than those of the parental virus and may therefore be considered nonessential for viral entry.
FIG 5.
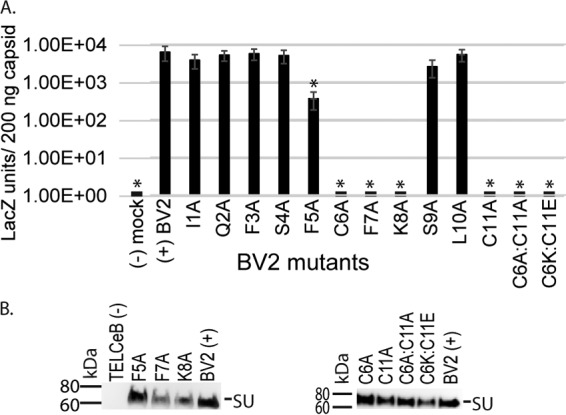
BV2 mutagenesis studies revealed that the pair of cysteines and residues in between are critical for viral titers. (A) Viral titers (LacZ units/200 ng of CA) were determined as for Fig. 4. (B) Western blot analysis of virus-associated SU. Viral samples were normalized to 100 ng of CA. Env was detected using C11D8 mouse anti-GP70 (AbD Serotec; 1:1,000 dilution).
For BV2 Env, loss of a single cysteine (C6A or C11A) or both cysteine residues did not affect the association of SU on viral particles. The substitution C6K:C11E maintained SU on the viral particles but did not restore viral titer. Similarly, substituting a potential salt bridge for the cysteines within the CP SU VRA did not restore functionality of the Env protein (22). Western blot analysis indicated the presence of Env on viral particles of all mutants with significantly lower titers in HEK293T cells (Fig. 5B). This indicates that the low titers cannot be attributed to an absence of Env on the surface of the viral particle but rather that these residues are essential for high titers. Taken together, these data indicate that for the BV2 Env, the presentation of F7 and K8 within a loop formed between C6 and C11 is essential for viral titers.
The purified CP-SU1-235 monomer has binding and interference activity and does not block AII and BV2 infection.
Since insertion of a salt bridge did not restore the function of the BV2 (Fig. 5A) or the CP (22) Env, the use of mass spectrometry was applied to identify the putative disulfide bonds. The amino acid sequence within the CP Env randomized region encodes three arginines (Fig. 2B), making it suitable for trypsin digestion and for mapping of the disulfide bonds using liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. In order to obtain sufficient purified material for the analysis, the N-terminal residues from 1 to 235 of CP SU were expressed in 293T cells as a secreted protein, based on the construct used to determine the crystal structure of FeLV-B SU (14). The schematic of the construct within the vector pJG is shown in Fig. 6A. The construct was modified to replace the prolactin signal peptide with the natural FeLV signal peptide, which resulted in increased secretion of the CP-SU1-235. To ensure that any mass spectrometry results reflected data from a functional CP-SU1-235 fusion protein, we performed receptor binding and interference assays. The ability of the secreted CP-SU1-235 protein to maintain receptor binding was confirmed using a FACS binding assay on permissive TE671 cells. Binding of CP-SU1-235 was detected, although it was less efficient than for retroviral particles bearing the full-length CP Env protein (Fig. 6B). This can be attributed to the fact that the secreted CP-SU1-235 is a monomer, whereas the virus-associated CP Env, like its parental FeLV-A/C Env, is likely to form a trimer that binds more efficiently to its host cell receptor. Alternatively, it is possible that domains outside the SU RBD region also function in receptor binding, similar to what has been shown for the SU C-terminal domain of FeLV-C (5).
FIG 6.

CP-SU1-235 monomer has binding and interference activity and does not block AII and BV2 infection. (A) Schematic of the vector pJG expressing CP-SU1-235. The black box in the LTR indicates SIN vector. Individual components are as indicated. (B) SU binding studies on HEK293T cells. Secreted CP-SU1-235 from HEK293T (1 ml of medium, red line) and CP viral particles (blue line) produced in TELCeB6 (1 ml) was incubated with HEK293T (control green line), and binding was verified by flow cytometry as described on the methodology. (C) CP-SU1-235 interference assay, with comparison of infection of 2 × 105 HEK293T cells or HEK293T cells constitutively producing CP-SU1-235. LacZ staining units/milliliter of virus were determined for virus expressing CP, AII, or BV2 Env, as indicated. Error bars indicate the SEMs for at least three independent experiments.
The ability of secreted CP-SU1-235 to block exogenous infection of CP retroviral particles packaging the β-galactosidase gene was tested in receptor interference assays. Expression of the secreted CP-SU1-235 lowered the titer of the challenge virus bearing the CP Env greater than 3 orders of magnitude (Fig. 6C), similar to previous results with CP retroviral particles (25). In contrast, constitutive expression of the secreted CP-SU1-235 in 293T cells did not block infection of virus bearing the AII or BV2 Envs. This indicates that AII and BV2 Envs use a host cell receptor other than PAR1 and PAR2, in addition to being distinct from the receptors of FeLV-A and -C (Fig. 3). The recombinant CP-SU1-235 monomer has activity sufficient to block the infection of the Env trimer on the retroviral particles, indicating the presence of proteins folded in a functional conformation.
Disulfide bonding within the randomized region of CP.
In order to determine if the cysteines within the randomized region of CP-SU1-235 form a disulfide bond, we performed mass spectrometry studies with the purified CP-SU1-235 protein. The protein sample was alkylated using iodoacetamide (IAM) in the absence of a reducing agent to label free cysteines, whereas cysteines within a disulfide bond were unavailable for this modification. After SDS-PAGE and in-gel tryptic digestion, the peptides were extracted, solubilized, and divided into two samples. One sample was treated with a reducing agent (DTT) to break the disulfide bonds, followed by alkylation with N-ethylmaleimide (NEM) to label the newly formed free cysteines. The second sample was treated with NEM in the absence of DTT, leaving the disulfide bonds intact. The results are summarized in Fig. 7 and Table 1. Two disulfide linkage peptides corresponding to C2 and C3 were identified (Table 1). Figure 7A shows the data for the peptide with a 498.5557 m/z and a retention time of 27.42 min (shown schematically in Fig. 7B) that is absent in the presence of DTT treatment. This corresponds to the oxidized form of YLC2R disulfide linked to TGC3SWSSSK, with a 3+ charge and a spectral count of 5 (Table 1). The fragmentation pattern (Fig. 7C) of the 498.5557 m/z peak in Fig. 7A further confirmed that the peak identified indeed corresponded to C2-C3 forming a disulfide bond. The ions y and b correspond to the fragmentation resulting at the amide bond. The y ion contains the C-terminal fragment, and the b ion contains the N-terminal fragment. We have named and labeled the smaller and the larger peptide fragments α and β, respectively, in superscripts in Fig. 7B to D and F to H. The subscript numbers correspond to the amino acid relative to the terminal that the fragment contains. Sixteen y and b ion peptides corresponding to the fragmentation of the parental peptide were identified (Fig. 7D). y and b ion 738.58 m/z could be assigned to two different peptides yielding the same theoretical result (Fig. 7D). We also observed the YLC2R disulfide linked to RTGC3SWSSSK peptide with an m/z of 413.1936 (Table 1) with a retention time of 25.44 min and a spectral count of 32, which was further confirmed in the final fragmentation patterns (data not shown).
FIG 7.
Identification of CP-SU1-235 cysteine bonds using mass spectrometry. (A to D) Identification of the C2-C3 bond; (E to H) identification of the C3-C4 bond. (A and E) Analysis of products in the presence and absence of DTT); (B and F) schematic of the peptide structure corresponding to the identified products; (C and G) fragmentation pattern of the identified peptides containing a disulfide bond; (D and H) assignment of the experimental mass of peaks in panels C and G with the y and b fragment ions, defined in panels B and F. M, full peptide mass.
TABLE 1.
Summary of mass spectrometry data for cysteines within the randomized loop of CP
| Cysteine no. | Sequencea | Spectral count | Charge | m/z | RTb (min) | Peak area |
|
|---|---|---|---|---|---|---|---|
| Without DTT | With DTT | ||||||
| C2-C3 | YLCR RTGCSWSSSK | 32 | 4+ | 413.1941 | 25.44 | 145,006,359 | 0 |
| C2-C3 | YLCR TGCSWSSSK | 5 | 3+ | 498.5558 | 27.42 | 17,050,837 | 0 |
| C3-C4 | TGCSWSSSK YGCK | 37 | 3+ | 470.5320 | 24.33 | 56,186,750 | 0 |
| C9-C10 | GSSQDNNCEGKCNPLILQFTQK | 50 | 3+ | 808.0429 | 44.99 | 366,697,279 | 31,759,741 |
Cysteines linked by disulfide bonds are underlined and in bold type. Spaces indicate separate tryptic peptides.
RT, retention time.
A second peptide, with a spectral count of 37 and a predicted C3-C4 cysteine bond, was identified. This corresponded to the peptide TGC3SWSSSK disulfide linked to YGC4K with a retention time of 24.33 min (Fig. 7E). Seventeen y and b ion peptides corresponding to the parental peptide were identified (Fig. 7G and H). Three of the y and b ions obtained experimentally could be assigned to two different theoretical fragments, at 408.3, 632.49 and 1,263.60 m/z (Fig. 7H). Additionally, the peptide GSSQDNNC9EGKC10NPLILQFTQK, with a retention time of 44.99 min and a spectral count of 50 (Table 1), was identified in the LC-MS/MS. While spectral count is considered a semiquantitative measure, peptides C2-C3, C3-C4, and C9-C10 were identified as the predominant species, with total spectral counts of 37, 37, and 50, respectively, and peak areas over 106. These data strongly suggest that C2-C3, C3-C4, and C9-C10 are cysteine bonds of a functional CP-SU and indicate the presence of an unexpected heterogeneity in the disulfide bond pattern within the VRA. Interestingly, generating the potential to form salt bridges at the C1-C2, C1-C4, and C3-C4 positions in CP by introducing Lys and Glu pairs did not rescue viral titers (data not shown).
DISCUSSION
The success of screening random libraries is greatly influenced by the vector backbone and the library design. Viruses have evolved multiple mechanisms for efficient expression of bicistronic RNAs. Herein we show that a bicistronic vector that uses the wild-type MLV SA to express the second gene results in viral titers that are significantly higher than with our previous vector, which contained an internal SV40 promoter to express the selectable marker (Fig. 1A and B). This is not surprising given that MLV has evolved an efficient mechanism to balance the transcription and export of unspliced mRNA encoding the gag-pol gene as well as the spliced mRNA expressing the env gene (35). With the incorporation of the WPRE element downstream of the neomycin resistance gene in the SD-env-SA backbone, we obtained 15-fold-higher titers (Fig. 1A). Using this vector three novel retargeted Env isolates were identified on 143B osteosarcoma cells (Fig. 2B).
A major goal of this study was to optimize the expression and screening of new Env libraries to obtain high-affinity gammaretroviral isolates in 143B osteosarcoma cells. The two isolates with the highest titers are constrained by the presence of a pair of cysteines within the randomized loop. Interestingly, from all of our Env libraries that were screened, the construct with the highest titer, CP Env (22), was also constrained by the presence of two internal cysteines. Therefore, we examined the role of these cysteines and nearby residues within the randomized loop. Our data clearly indicate that the internal cysteines are indispensable for viral titer within the identified Env isolates (Fig. 4 and 5).
Comparison of the role of the internal cysteines from the three Env isolates CP, AII, and BV2 did not result in a uniform model. In AII Env, substitution of a Glu-Lys salt bridge for the C2-C3 disulfide bond restored viral titers, whereas replacement of C6E:C11K in BV2 Env resulted in complete loss of titers (Fig. 5A). In both AII and BV2 Envs, however, the specific residues identified as critical for viral entry localize to the loop between the C2 and C3 residues. Because the AII isolate contains only 3 amino acid residues between C2 and C3, while the BV2 isolate contains 4 amino acid residues between C2 and C3, it is possible that the spacing and amino acid side chain organization of the BV2 isolate are not conducive to the formation of a salt bridge at these positions. Since the salt bridge substitutes for the AII Env C6:C10 disulfide bond, it is reasonable to expect that an isolate with a salt bridge might have been selected in the initial screen. However, our randomized library was not saturating, with only 106/1012 possible combinations.
In CP Env, like BV2, the C2-C3 residues are separated by 4 amino acids. Again, providing residues with the potential to form a salt bridge in CP Env did not substitute for a disulfide bond (Fig. 5A) (22). However, mass spectrometry analysis of a secreted CP Env construct shown to be functional for receptor binding indicated the presence of disulfide-linked peptides at C2-C3 and C3-C4, suggesting the existence of heterogeneity in the VRA bonding pattern. It is interesting that in CP, only two other residues within the randomized region in addition to C2 and C3 were critical for viral titer (22). The first was a Gly between C2 and adjacent to C3, providing flexibility to position C3. The other residue indispensable for infectivity was the Trp at position 11. It is possible that formation of either C2-C3 or C3-C4 is required to position the essential Trp11 for interaction with the host cell receptor (Fig. 8).
FIG 8.

Summary of disulfide bonds and mutagenesis studies. Critical amino acids required for viral entry are indicated in red for each isolate. Cysteines that form intramolecular bonds are indicated via bars (solid and dashed). For AII, the linkage is based on functional substitution of a putative salt bridge. For CP, disulfide bonds were identified by mass spectrometry.
These results are similar to what has been observed in the study of HIV Env trimers, where heterogeneity of the disulfide bonding pattern has been found for the N-terminal variable regions 1 and 2 (V1 and V2). The V1/V2 region is implicated in exposure of the variable region 3 (V3) to HIV host cell receptors (36). Conversely, the lack of a pair of cysteines present in the V2 region of HIV-2 and SIV strains but not in the HIV-1 Env glycoprotein has been suggested as a factor contributing to the high virulence of HIV-1 (37).
The heterogeneity in the disulfide bond patterns does not appear to be limited to members of the Retroviridae family. Recently, two crystal structures were solved for the E2 Env protein of hepatitis C virus (HCV) of the Flaviviridae family. These structures revealed two possible cysteine arrangements of a functional E2 Env glycoprotein (38). These cysteines are known to be conserved across different HCV genotypes, implying an important role in structure and/or function of the Env. However, the presence of this heterogeneity in cysteine bonding on HCV viral particles is not known. It is possible that certain cysteines are important to hold the structure in place, while other cysteines can be easily rearranged by the virus as a way to evade the immune system, without having major conformational changes.
The protein expression system utilized in this study was identical to that used to express and solve the HCV E2 Env protein crystal structure (33). Interestingly, to achieve successful secretion of the soluble CP-SU1-235, we had to exchange the prolactin secretion signal for that of the native FeLV Env signal peptide. Surprisingly, expression of the CP-SU35-235 with the prolactin signal peptide resulted in minimal secretion of the SU protein, with detectable perinuclear and nuclear signals by immunostaining and confocal imaging (data not shown). These results suggest that the signal peptide of the FeLV Env may have additional unknown properties, leading to correct transport and secretion of the viral Env.
Ironically, we initially selected the FeLV-A/C backbone for the library screen due to the absence of complex cysteine bonding within variable regions A and B. Selecting for the additional cysteine pairs generates variable loops that are more similar to the complex loops found in related gammaretroviral Envs. The studies presented here will assist in the design of third-generation Env libraries. Future studies will aim at constructing libraries holding in place C2 and C3 at the locations found in AII, BV2, and CP Envs and randomizing nine residues to screen for new retargeted Env proteins.
ACKNOWLEDGMENTS
We thank Haiyan Zheng, Meiqian Qian, and Peter Lobel, Rutgers Biological Mass Spectrometry Facility of Robert Wood Johnson Medical School, for their discussions on interpreting the mass spectrometry data.
This work was supported by grant R01 CA49932 to M.J.R. and P50GM103368 and R01 GM111959 to J.M. NIH grants T32AI007403, 3R01CA049932-19S1, and 2R25GM05839 supported Leonardo Valdivieso-Torres.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Battini J-L, Heard JM, Danos O. 1992. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J Virol 66:1468–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fass D, Davey RA, Hamson CA, Kim PS, Cunningham JM, Berger JM. 1997. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science 277:1662–1666. doi: 10.1126/science.277.5332.1662. [DOI] [PubMed] [Google Scholar]
- 3.Gemeniano M, Mpanju O, Salomon DR, Eiden MV, Wilson CA. 2006. The infectivity and host range of the ecotropic porcine endogenous retrovirus, PERV-C, is modulated by residues in the C-terminal region of its surface envelope protein. Virology 446:108–117. [DOI] [PubMed] [Google Scholar]
- 4.Lavillette D, Boson B, Russell SJ, Cosset FL. 2001. Activation of membrane fusion by murine leukemia viruses is controlled in cis or in trans by interactions between the receptor-binding domain and a conserved disulfide loop of the carboxy terminus of the surface glycoprotein. J Virol 75:3685–3695. doi: 10.1128/JVI.75.8.3685-3695.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rey MA PR, Tailor CS. 2008. The C domain in the surface envelope glycoprotein of subgroup C feline leukemia virus is a second receptor-binding domain. Virology 370:273–284. doi: 10.1016/j.virol.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 6.Sarma PS, Log T, Jain D, Hill PR, Huebner RJ. 1975. Differential host range of viruses of feline leukemia-sarcoma complex. Virology 64:438–446. doi: 10.1016/0042-6822(75)90121-X. [DOI] [PubMed] [Google Scholar]
- 7.Sarma PS, Log T. 1971. Viral interference in feline leukemia-sarcoma complex. Virology 44:352–358. doi: 10.1016/0042-6822(71)90266-2. [DOI] [PubMed] [Google Scholar]
- 8.Mendoza R, Anderson MM, Overbaugh J. 2006. A putative thiamine transport protein is a receptor for feline leukemia virus subgroup A. J Virol 80:3378–3385. doi: 10.1128/JVI.80.7.3378-3385.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rudra-Ganguly N, Ghosh AK, Roy-Burman P. 1998. Retrovirus receptor PiT-1 of the Felis catus. Biochim Biophys Acta 1443:407–413. doi: 10.1016/S0167-4781(98)00241-3. [DOI] [PubMed] [Google Scholar]
- 10.Tailor CS, Willett BJ, Kabat D. 1999. A putative cell surface receptor for anemia-inducing feline leukemia virus subgroup C is a member of a transporter superfamily. J Virol 73:6500–6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quigley JG, Burns CC, Anderson MM, Lynch ED, Sabo KM, Overbaugh J, Abkowitz JL. 2000. Cloning of the cellular receptor for feline leukemia virus subgroup C (FeLV-C), a retrovirus that induces red cell aplasia. Blood 95:1093–1099. [PubMed] [Google Scholar]
- 12.Rigby MA, Rojko JL, Stewart MA, Kociba GJ, Cheney CM, Rezanka LJ, Mathes LE, Hartke JR, Jarrett O, Neil JC. 1992. Partial dissociation of subgroup C phenotype and in vivo behaviour in feline leukaemia viruses with chimeric envelope genes. J Gen Virol 73:2839–2847. doi: 10.1099/0022-1317-73-11-2839. [DOI] [PubMed] [Google Scholar]
- 13.Linder M, Wenzel V, Linder D, Stirm S. 1994. Structural elements in glycoprotein 70 from polytropic Friend mink cell focus-inducing virus and glycoprotein 71 from ecotropic Friend murine leukemia virus, as defined by disulfide-bonding pattern and limited proteolysis. J Virol 68:5133–5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barnett AL, Wensel DL, Li W, Fass D, Cunningham JM. 2003. Structure and mechanism of a coreceptor for infection by a pathogenic feline retrovirus. J Virol 77:2717–2729. doi: 10.1128/JVI.77.4.2717-2729.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tailor CS, Kabat D. 1997. Variable regions A and B in the envelope glycoproteins of feline leukemia virus subgroup B and amphotropic murine leukemia virus interact with discrete receptor domains. J Virol 71:9383–9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazari P, Roth M. 2013. Library screening and receptor-directed targeting of gammaretroviral vectors. Future Microbiol 8:107–121. doi: 10.2217/fmb.12.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu D-T, Roth MJ. 2014. MLV based viral-like-particles for delivery of toxic proteins and nuclear transcription factors. Biomaterials 35:8416–8426. doi: 10.1016/j.biomaterials.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bupp K, Roth M. 2002. Altering retroviral tropism using a random display envelope library. Mol Ther 5:329–335. doi: 10.1006/mthe.2002.0546. [DOI] [PubMed] [Google Scholar]
- 19.Bupp K, Roth MJ. 2003. Targeting a retroviral vector in the absence of a known cell-targeting ligand. Hum Gene Ther 14:1557–1564. doi: 10.1089/104303403322495061. [DOI] [PubMed] [Google Scholar]
- 20.Bupp K, Sarangi A, Roth MJ. 2005. Probing sequence variation in the receptor-targeting domain of feline leukemia virus envelope proteins with peptide display libraries. J Virol 79:1463–1469. doi: 10.1128/JVI.79.3.1463-1469.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bupp K, Sarangi A, Roth MJ. 2006. Selection of feline leukemia virus envelope proteins from a library by functional association with a murine leukemia virus envelope. Virology 351:340–348. doi: 10.1016/j.virol.2006.03.040. [DOI] [PubMed] [Google Scholar]
- 22.Mazari PM, Linder-Basso D, Sarangi A, Chang Y, Roth MJ. 2009. Single-round selection yields a unique retroviral envelope utilizing GPR172A as its host receptor. Proc Natl Acad Sci U S A 106:5848–5853. doi: 10.1073/pnas.0809741106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarangi A, Bupp K, Roth MJ. 2007. Identification of a retroviral receptor used by an Envelope protein derived by peptide library screening. Proc Natl Acad Sci U S A 104:11032–11037. doi: 10.1073/pnas.0704182104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ericsson TA, Takeuchi Y, Templin C, Quinn G, Farhadian SF, Wood JC, Oldmixon BA, Suling KM, Ishii JK, Kitagawa Y, Miyazawa T, Salomon DR, Weiss RA, Patience C. 2003. Identification of receptors for pig endogenous retrovirus. Proc Natl Acad Sci U S A 100:6759–6764. doi: 10.1073/pnas.1138025100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazari PM, Argaw T, Valdivieso L, Zhang X, Marcucci KT, Salomon DR, Wilson CA, Roth MJ. 2012. Comparison of the convergent receptor utilization of a retargeted feline leukemia virus envelope with a naturally-occurring porcine endogenous retrovirus A. Virology 427:118–126. doi: 10.1016/j.virol.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cosset FL, Takeuchi Y, Battini JL, Weiss RA, Collins MK. 1995. High-titer packaging cells producing recombinant retroviruses resistant to human serum. J Virol 69:7430–7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gendra E, Colgan DF, Meany B, Konarska MM. 2007. A sequence motif in the simian virus 40 (SV40) early core promoter affects alternative splicing of transcribed mRNA. J Biol Chem 282:11648–11657. doi: 10.1074/jbc.M611126200. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Sarangi A, Wu DT, Kanduri J, Roth MJ. 2013. Gene delivery in a mouse xenograft of a retargeted retrovirus to a solid 143B osteosarcoma. Virol J 10:194. doi: 10.1186/1743-422X-10-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zanta-Boussif MA, Charrier S, Brice-Ouzet A, Martin S, Opolon P, Thrasher AJ, Hope TJ, Galy A. 2009. Validation of a mutated PRE sequence allowing high and sustained transgene expression while abrogating WHV-X protein synthesis: application to the gene therapy of WAS. Gene Ther 16:605–619. doi: 10.1038/gt.2009.3. [DOI] [PubMed] [Google Scholar]
- 30.Fouchier RAM, Meyer BE, Simon JHM, Fischer U, Malim MH. 1997. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J 16:4531–4539. doi: 10.1093/emboj/16.15.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu DT, Aiyer S, Villanueva RA, Roth MJ. 2013. Development of an enzyme-linked immunosorbent assay based on the murine leukemia virus p30 capsid protein. J Virol Methods 193:332–336. doi: 10.1016/j.jviromet.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whidby J, Mateu G, Scarborough H, Demeler B, Grakoui A, Marcotrigiano J. 2009. Blocking hepatitis C virus infection with recombinant form of envelope protein 2 ectodomain. J Virol 83:11078–11089. doi: 10.1128/JVI.00800-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khan AG, Whidby J, Miller MT, Scarborough H, Zatorski AV, Cygan A, Price AA, Yost SA, Bohannon CD, Jacob J, Grakoui A, Marcotrigiano J. 2014. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature 509:381–384. doi: 10.1038/nature13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zufferey R, Donello JE, Trono D, Hope TJ. 1999. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J Virol 73:2886–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trubetskoy AM, Okenquist SA, Lenz J. 1999. R region sequences in the long terminal repeat of a murine retrovirus specifically increase expression of unspliced RNAs. J Virol 73:3477–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Go EP, Zhang Y, Menon S, Desaire H. 2011. Analysis of the disulfide bond arrangement of the HIV-1 envelope protein CON-S gp140 DeltaCFI shows variability in the V1 and V2 regions. J Proteome Res 10:578–591. doi: 10.1021/pr100764a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bohl C, Bowder D, Thompson J, Abrahamyan L, Gonzalez-Ramirez S, Mao YD, Sodroski J, Wood C, Xiang SH. 2013. A twin-cysteine motif in the V2 region of gp120 is associated with SIV envelope trimer stabilization. PLoS One 8:e69406. doi: 10.1371/journal.pone.0069406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khan AG, Miller MT, Marcotrigiano J. 2015. HCV glycoprotein structures: what to expect from the unexpected. Curr Opin Virol 12:53–58. doi: 10.1016/j.coviro.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]