ABSTRACT
We investigated whether there is any association between a native-like conformation and the presence of only the canonical (i.e., native) disulfide bonds in the gp120 subunits of a soluble recombinant human immunodeficiency virus type 1 (HIV-1) envelope (Env) glycoprotein. We used a mass spectrometry (MS)-based method to map the disulfide bonds present in nonnative uncleaved gp140 proteins and native-like SOSIP.664 trimers based on the BG505 env gene. Our results show that uncleaved gp140 proteins were not homogeneous, in that substantial subpopulations (20 to 80%) contained aberrant disulfide bonds. In contrast, the gp120 subunits of the native-like SOSIP.664 trimer almost exclusively retained the canonical disulfide bond pattern. We also observed that the purification method could influence the proportion of an Env protein population that contained aberrant disulfide bonds. We infer that gp140 proteins may always contain a variable but substantial proportion of aberrant disulfide bonds but that the impact of this problem can be minimized via design and/or purification strategies that yield native-like trimers. The same factors may also be relevant to the production and purification of monomeric gp120 proteins that are free of aberrant disulfide bonds.
IMPORTANCE It is widely thought that a successful HIV-1 vaccine will include a recombinant form of the Env protein, a trimer located on the virion surface. To increase yield and simplify purification, Env proteins are often made in truncated, soluble forms. A consequence, however, can be the loss of the native conformation concomitant with the virion-associated trimer. Moreover, some soluble recombinant Env proteins contain aberrant disulfide bonds that are not expected to be present in the native trimer. To assess whether these observations are linked, to determine the extent of disulfide bond scrambling, and to understand why scrambling occurs, we determined the disulfide bond profiles of two soluble Env proteins with different designs that are being assessed as vaccine candidates. We found that uncleaved gp140 forms heterogeneous mixtures in which aberrant disulfide bonds abound. In contrast, BG505 SOSIP.664 trimers are more homogeneous, native-like entities that contain predominantly the native disulfide bond profile.
INTRODUCTION
The first human immunodeficiency virus type 1 (HIV-1) vaccine trial (designated RV144) to yield one statistically significant measure of efficacy was reported in 2009 (1). Since then, attempts to substantially improve on those initial results have been ongoing. Many vaccine design strategies are focused on the HIV-1 Env protein, which is the trimeric antigen present on the viral surface. Some Env immunogens, including proteins used in the RV144 trial, are based on a soluble monomeric form of the gp120 subunit of the trimer (1–7). An alternative approach involves soluble oligomeric gp140 proteins that contain both gp120 and the external component of the gp41 subunit (8–13). The concept underlying the use of gp140 proteins is that they may better mimic the native virion-associated trimer and hence may be superior to the individual gp120 subunits at inducing virus neutralizing antibodies (NAbs) (8–10). The underlying assumption is now known to be invalid in many cases, since the precise design of the soluble gp140 protein is critical for achieving trimer mimicry (11–16).
While one can now infer whether or not a gp140 protein adopts a native-like trimer structure by visualizing it using negative-stain electron microscopy (NS-EM) (11, 12, 14–16), the factors that govern the formation of a native Env protein are only now becoming clear. Increased knowledge of why native and nonnative Env proteins differ, and how those differences arise, may therefore be of substantial value for the further improvement of all Env-based immunogens.
Glycosylation is one modification with regard to which native or native-like Env trimers differ from gp120 monomers or nonnative uncleaved gp140 oligomers: specifically, the native Env proteins are enriched in high-mannose glycans, while the glycans on gp120 monomers and nonnative gp140s are more highly processed (14, 17–21). However, since glycan processing occurs only after the Env protein folds and, when relevant, trimerizes, these differences are likely to be a consequence and not a cause of native trimer formation. In contrast, the generation of intramolecular disulfide bonds is one of the earliest steps in Env protein processing, starting during mRNA translation and continuing as the nascent protein folds. For most genotypes, the gp120 protein (whether a monomer or a trimer subunit) contains nine intramolecular disulfide bonds, and there is also one such bond in the gp41 component of gp140 proteins (22–26). The potential for the formation of aberrant disulfide bonds as the gp120 or gp140 proteins fold and assemble is obvious. Here we have assessed the relationship between the disulfide bond profiles of two soluble Env proteins and their adoption of a native-like structure. Our primary goal was to better understand the interplay between disulfide exchange, protein folding, and glycosylation modifications during the formation of Env trimers.
The canonical disulfide-bonding pattern present in monomeric gp120 was first reported in 1990 (25). The same pattern has since been seen in all high-resolution structures of NAb complexes of gp120 monomers or native-like trimers, including NAbs that recognize trimer-specific epitopes (10, 27–30). The structural data strongly imply that the canonical pattern is the one associated with native, trimeric Env, but the structures do not address whether other Env proteoforms can also be present. Thus, a nonnative form of Env with a different disulfide bond profile probably would not bind the cocrystallizing antibody and hence would be excluded from the crystal.
Noncanonical (i.e., aberrant) disulfide bonds can indeed be present in HIV-1 Env proteins. The formation of aberrant intermolecular disulfide bonds causes the cross-linking of Env proteins into dimers and aggregates (31, 32). There are also multiple reports of recombinant gp120 and gp140 proteins that contain aberrant intramolecular disulfide bonds (6, 15, 23, 24, 33). We show here that the gp120 subunits of native-like BG505 SOSIP.664 trimers contain predominantly the canonical disulfide-bonding profile. In contrast, a nonnative, uncleaved gp140 protein of the same BG505 genotype contains substantial amounts of all the aberrant disulfide bonds that were previously reported to be present in Env proteins of comparable designs (6, 15, 23, 33). The aberrant disulfide bonds were also present in SOSIP.664 gp140 monomers and dimers, implying that the SOSIP trimer-stabilizing modifications do not prevent this problem per se. Instead, we propose that the engineered SOSIP stabilization changes facilitate the production of native-like trimers from which aberrantly folded gp120 subunits are excluded. Taking into account recent knowledge of the glycosylation profiles of native trimers and nonnative Env proteins (11, 14, 19–21), we postulate that the disulfide-bonding profile determines the potential of a proteoform to pack into a native trimer and that stabilizing modifications such as the SOSIP changes then prevent disulfide exchange reactions within the assembled trimer. Thus, there is an inherent interrelationship between native disulfide bonding, native structure, and native glycosylation profile.
Our observations also have practical relevance to vaccine design, by guiding the production and purification of Env proteins with a native disulfide bond profile. Such proteins, be they gp120 monomers or gp140 trimers, may prove to be better immunogens for NAb induction than counterparts containing contaminants in which substantial amounts of aberrant disulfide bonds have formed.
MATERIALS AND METHODS
Reagents.
Trizma base, citric acid, high-performance liquid chromatography (HPLC)-grade acetonitrile (CH3CN), Optima methanol (CH3OH), 4-vinylpyridine, glacial acetic acid, and Optima LC–MS-grade formic acid were purchased from Sigma (St. Louis, MO) and Fisher Scientific (Pittsburgh, PA). Water was purified using a Millipore (Billerica, MA) Direct-Q3 water purification system. Sequencing-grade trypsin was obtained from Promega (Madison, WI). Glycerol-free peptidyl-N-glycosidase F (PNGase F), cloned from Flavobacterium meningosepticum, was bought from New England BioLabs (Ipswich, MA).
Expression and purification of Env proteins.
The BG505 SOSIP.664 and WT.SEKS constructs have been described previously (16, 34). The SOSIP.664 trimers were produced in a stable CHO cell line (35), while the WT.SEKS proteins were made by transient transfection of 293T cells (16). In both cases, the Env proteins were initially isolated via a 2G12 broadly neutralizing antibody (bNAb)-affinity column, followed by size exclusion chromatography (SEC) to isolate the trimer fractions, as described previously (14, 16, 34). The BG505 SOSIP.664-His native-like trimers, which contain a C-terminal His tag, were purified by Ni-nitrilotriacetic acid (NTA) affinity chromatography followed by SEC (14, 15). In some studies, SEC column fractions containing BG505 SOSIP.664 or SOSIP.664-His gp120–gp41 monomers and dimers were also collected (36). As viewed by negative-stain EM, the BG505 SOSIP.664 trimers are native-like, regularly shaped trilobed structures, whereas the uncleaved WT.SEKS gp140 proteins adopt irregular, splayed-out shapes (16, 34).
Deglycosylation of BG505 Env proteins.
Samples containing ∼50 μg of Env were alkylated with a 10-fold molar excess of 4-vinylpyridine for 1 h at room temperature in the dark in order to cap free cysteine residues prior to deglycosylation and thereby prevent disulfide bond shuffling. Deglycosylation was performed by incubating Env samples with 1 μl of a PNGase F solution (500,000 U/ml) in 50 mM Tris citrate buffer (pH 6.5) for 1 week at 37°C, as described previously (24).
Proteolytic digestion of BG505 Env proteins.
The fully deglycosylated and alkylated samples (see above) were digested overnight with trypsin (protein-to-enzyme ratio, 30:1) at 37°C (23, 24). To generate consistent and reproducible Env digests, the complete deglycosylation and subsequent proteolytic digestion procedures were performed at least twice on different days, using samples obtained from the same Env expression batches. Each of these samples was subjected to liquid chromatography (LC)-mass spectrometry (MS) analysis, as described below, and at least two replicate experimental runs were performed on each digest. Thus, in total, four replicate data files were generated for each protein.
Chromatography and mass spectrometry.
High-resolution LC–tandem MS (MS-MS) experiments were performed using an Orbitrap Velos Pro hybrid mass spectrometer (Thermo Scientific, San Jose, CA) coupled to an Acquity ultra-performance liquid chromatography (UPLC) system (Waters, Milford, MA). Chromatography was performed using mobile phases consisting of solvent A (99.9% deionized H2O plus 0.1% formic acid) and solvent B (99.9% CH3CN plus 0.1% formic acid) with a gradient of 3% to 40% solvent B in 50 min. The column was held at 97% solvent B for 10 min before reequilibration. Five microliters of the sample (∼4 to 5 μM) was injected onto a PepMap 300 C18 column (inside diameter [i.d.], 300 μm; length, 15 cm; pore size, 300 Å; Thermo Scientific, Sunnyvale, CA) at a flow rate of 5 μl/min. A short wash and a blank run were performed between every two samples to ensure that there was no sample carryover. All mass spectrometric analysis was performed in a data-dependent mode as described below. The Orbitrap Velos Pro hybrid mass spectrometer was set up to perform experiments by alternating collision-induced dissociation (CID) and electron transfer dissociation (ETD) acquisition. Data-dependent acquisition (DDA) was set up to acquire 10 scan events: for every full high-resolution MS scan in the mass range from 400 to 2,000 m/z, each selected m/z in the MS scan was subjected to three MS-MS events in the ion trap: (i) CID, (ii) ETD, and (iii) CID of the charge-reduced precursor in the previous ETD event. The mass spectrometric parameters used for the experiment were as follows: spray voltage, 3.0 kV; S-lens value, between 45 and 55%; capillary temperature, 250°C; normalized collision energy, 35% for CID. The ion-ion reaction for ETD between the precursor ion and the radical anion, fluoranthene, was set at an automatic gain control (AGC) target value of 2 × 105 and a 100-ms ion-ion reaction time. To improve ETD efficiency, supplemental activation was turned on. The Orbitrap Velos Pro hybrid mass spectrometer was set at a resolution (R) of 30,000 at m/z 400. Under these conditions, the measured R (full width at half maximum [FWHM]) is 22,000 at m/z 1,000 and 16,000 at m/z 1,500.
Peptide identification.
Peptides containing free cysteine residues were identified by searching raw data against a custom HIV database including 157 gp120/gp41 sequences, obtained from the Los Alamos HIV Sequence Database (http://www.hiv.lanl.gov/content/sequence/HIV/mainpage.html), using Mascot (Matrix Science, London, United Kingdom, version 2.5). The peak list was extracted from raw files by use of the MassMatrix conversion tool. Mascot generic format (mgf) files were searched by specifying the following parameters: (i) enzyme, trypsin; (ii) missed cleavages, up to 2; (iii) fixed modification, pyridylethylation; (iv) variable modifications, methionine oxidation and pyro-Glu (Q at N terminus); (v) peptide tolerance, 0.5 Da; (vi) MS-MS tolerance, 0.3 Da. Peptides identified from the Mascot search with 95% confidence were manually validated from tandem mass spectrometry (MS-MS) data to ensure that major fragmentation ions (b, c, y, and z ions) were observed, especially for peptides generated from PNGase F-treated samples that contained Asn-to-Asp conversions.
Disulfide bond analysis.
Raw data acquired from the experiments were analyzed manually. To facilitate the analysis, peptides containing S-pyridylethyl-labeled cysteine residues were first identified using the Mascot search engine as described above. Once the free cysteine residues were identified, the analysis involved using a prediction table containing the sum of the masses of all possible HIV-1 Env disulfide-linked peptide pairs minus 2 Da, at different charge states. From the calculated m/z ratios of the plausible candidates, the elution time profile (or extracted ion chromatogram [XIC]) was constructed. For each plausible candidate, the MS data and corresponding CID and/or ETD spectra of the targeted disulfide-linked peptide that were needed for identification were extracted from the constructed XIC. The observed peptide fragment ions from CID (b/y ions) and ETD (c/z ions) data for each ion identified in the MS scan were manually matched to the theoretical peptide fragment ions generated using MS-Product from ProteinProspector (http://prospector.ucsf.edu/prospector/mshome.htm). Matched fragment ions that were within 0.5 Da of the theoretical value were accepted. In addition to the characteristic c and z fragment ions observed in ETD, ions corresponding to the peptide fragments resulting from cleavage of the disulfide bond during ETD were required to be present in the ETD spectra for the assignment to be unambiguous.
RESULTS
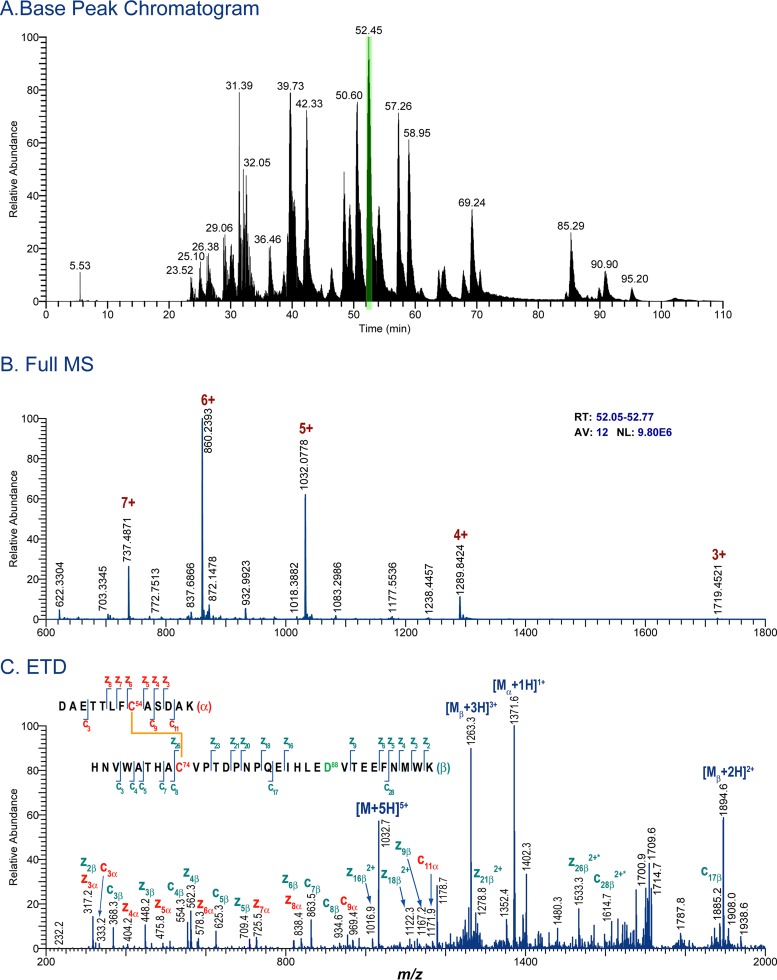
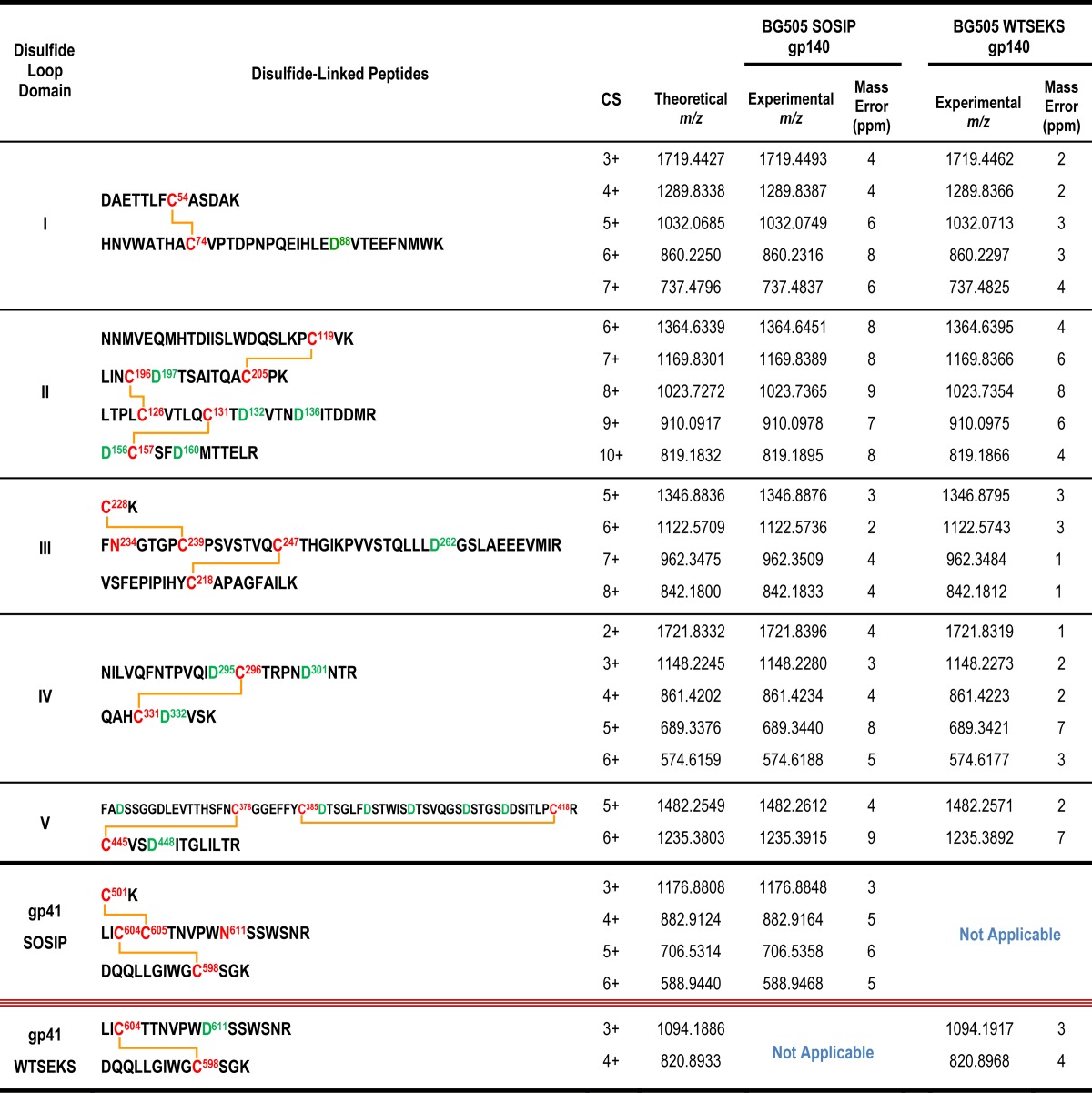
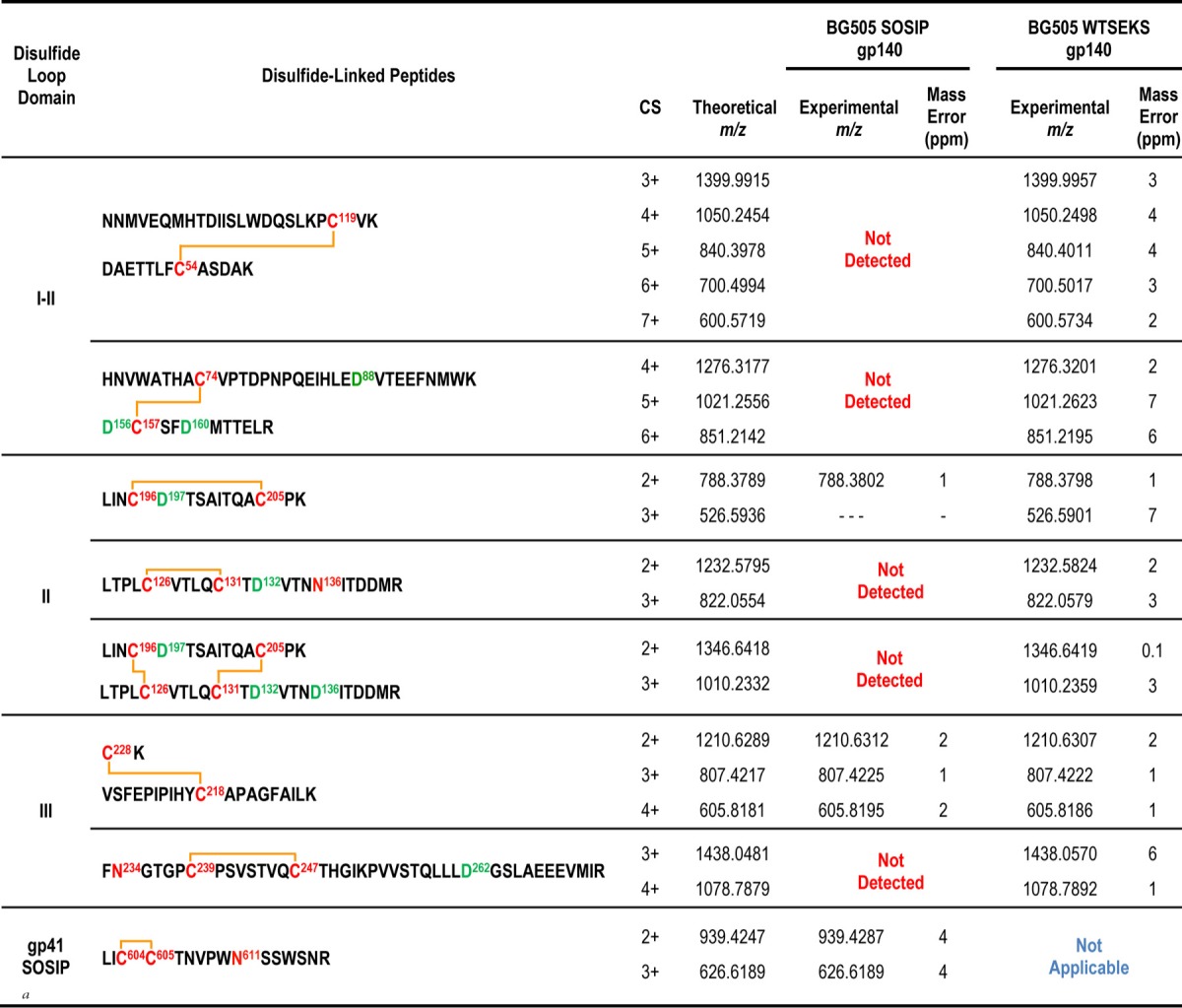
The disulfide bond profiles of the BG505 uncleaved WT.SEKS and cleaved SOSIP.664 trimers were ascertained using MS-based methods for protein disulfide bond determination that have been described previously (6, 23, 24, 33). In brief, the proteins were deglycosylated with PNGase F and were digested with trypsin under acidic conditions that preserve disulfide bonds intact. The disulfide-linked peptides derived were analyzed by liquid chromatography–tandem mass spectrometry (LC–MS-MS) methods. A typical example is shown in Fig. 1A. High-resolution MS data for the portion of the chromatogram containing the first disulfide-bonded peptide in BG505 Env are shown in Fig. 1B. This disulfide-bonded peptide, comprising DAETTLFC54ASDAK and HNVWATHAC74VPTDPNPQEIHLEDVTEEFNMWK, which are linked via the Cys residues at positions 54 and 74 (shown in boldface), has a theoretical m/z of 1,032.0687 in the 5+ charge state and is detected within a 6-ppm mass error in the mass spectrum (Fig. 1B). In addition to the 5+ charge state, the same disulfide-linked peptide is also readily detected in numerous other charge states (Fig. 1B). Its identity is further confirmed by MS-MS experiments; the ETD data used to support this particular assignment are shown in Fig. 1C, where numerous c and z ions consistent with the disulfide-linked peptide can be seen as abundant peaks in the spectrum. Each disulfide-linked peptide that we describe in Tables 1, 2, and 3 was verified using the same approach. Specifically, first, the high-resolution MS data, within a mass error of 10 ppm, were detected for each species, and then the assignment was confirmed by the MS-MS data. All of the canonical disulfide-bonded peptides and all feasible aberrant disulfide-bonded peptides were searched for individually in each data set, as described previously (24, 33). The canonical and aberrant disulfide-bonded peptides detected for each BG505 protein are described in Tables 1 and 2, respectively.
FIG 1.
MS data for tryptic digest of BG505 Env proteins. (A) Base peak chromatogram of the protein digest. (B) High-resolution mass spectrum for a portion of the chromatogram, highlighted in panel A. Several charge states are detected for the disulfide-linked peptide. (C) ETD spectrum of the disulfide-linked peptide detected in panel B.
TABLE 1.
Canonical disulfide bonds in BG505SOSIP.664 and WT.SEKS trimers
CS, charge state.
TABLE 2.
Aberrant disulfide bonds in BG505 SOSIP.664 and WT.SEKS trimers
CS, charge state.
TABLE 3.
Aberrant disulfide-linked peptides in Env proteins purified by various methods
For both the BG505 WT.SEKS and SOSIP.664 trimers, all the canonical disulfide bonds were detected (Table 1). These findings are consistent with all our previous disulfide-bonding analyses of gp140 proteins, since the canonical form is always present to some extent (6, 15, 23, 24, 33). A notable new observation, however, is summarized in Table 2. There it is shown that the trimer fraction of the uncleaved BG505 WT.SEKS gp140 preparation contains proteins with numerous aberrant disulfide bonds in gp120 domains I, II, and III. However, in marked contrast, all of these aberrant forms are almost completely absent from the corresponding SOSIP.664 trimers. Of note is the fact that the comparator proteins were both purified by the same strategy of 2G12 bNAb affinity chromatography followed by SEC to isolate the trimer fraction, so the differences between these proteins are not rooted in the purification process.
We assessed whether the substantial differences between these BG505 WT.SEKS and SOSIP.664 proteins arose because they had been expressed in two different cell lines (293T and CHO cells, respectively). For this purpose, we expressed the SOSIP.664 trimers by transiently transfecting 293T cells, followed by 2G12/SEC purification, and determined their disulfide bond profile. The resulting disulfide-bonded peptides were identical to those found for the BG505 SOSIP.664 trimers expressed in CHO cells and purified by 2G12/SEC (data not shown). The different disulfide bond contents of the SOSIP.664 and WT.SEKS proteins are therefore not attributable to the producer cell.
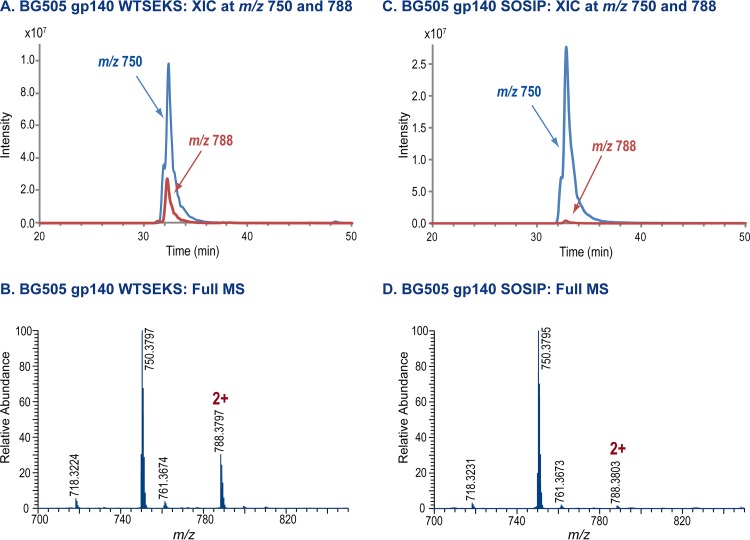
To better assess the magnitude of the differences between the two types of Env protein, we estimated the sizes of the subpopulations that contained the aberrant forms. To do so, we studied the MS data associated with the aberrant disulfide-bonded peptides in greater detail. The MS data for one of the aberrant disulfide-bonded peptides in the V1/V2 region (m/z 788) are shown in Fig. 2, together with a coeluting tryptic peptide (LDVVQINENQGNR; m/z 750) from the protein sequence, which serves as an internal standard (see below). The extracted ion chromatograms of the peptides from the WT.SEKS protein and the corresponding mass spectrum are shown in Fig. 2A and B, respectively. The aberrant disulfide-bonded peptide, m/z 788, is readily detectable in both plots for the WT.SEKS protein, but it is barely detectable above the baseline in the corresponding data plots for the SOSIP.664 trimer (Fig. 2C and D).
FIG 2.
MS data for an aberrant disulfide-linked peptide. (A) Extracted ion chromatograms for the aberrant peptide, at m/z 788, and a coeluting peptide, m/z 750, from the digest of BG505 WT.SEKS gp140. (B) Mass spectrum for the peak shown in panel A. (C) Extracted ion chromatograms for the aberrant peptide, at m/z 788, and a coeluting peptide, m/z 750, from the digest of BG505 SOSIP.664. (D) Mass spectrum for the peak shown in panel C.
A simple visual inspection of the data sets presented above suggests that the aberrant peptide is substantially less abundant in the SOSIP.664 trimer than in its uncleaved, nonnative WT.SEKS counterpart. To provide a more quantitative estimate of the difference, we used the intensity of the coeluting peptide, m/z 750, as an internal standard. The use of coeluting compounds as internal standards to achieve quantitative results is a well-established mass spectrometry technique (37–39). This approach enabled us to estimate that the aberrant disulfide-bonded peptide is ∼20-fold less prevalent in the SOSIP.664 trimer preparation than in the uncleaved WT.SEKS gp140. (In three replicate injections, the estimates of the difference ranged from 17- to 23-fold, with a mean value of 20-fold). While these analyses provide quantitative information on a relative scale, not an absolute scale, it is clear that the prevalence of aberrantly formed disulfide bonds is far greater (∼20-fold) in the uncleaved gp140 protein than in the native-like SOSIP.664 trimer. To put the difference into perspective, it is relevant that we estimate that the biochemical purity of the SOSIP.664 trimers is likely to be in the range of 96 to 99%. Thus, when 2G12/SEC-purified BG505 SOSIP.664 trimers are analyzed on blue native gels, contaminant monomers and dimers typically are not detectable (34). Accordingly, our minimum estimate of trimer purity is ∼96%. It is reasonable to suggest that at least some dimers and aggregates will still be present even after SEC purification, since 100% size homogeneity is unlikely to be achievable by this method alone. Additional information on the purity of trimers after SEC is provided in Fig. S1 in the supplemental material.
The fact that the protein fraction used for these studies is 96 to 99% trimers is relevant because we show below that nontrimeric BG505 SOSIP.664 proteins (i.e., monomers and dimers) are enriched for aberrant disulfide bonds relative to trimers. Thus, if the frequency of the disulfide scrambled V2 domain is in the range of ∼1 to 4% for the overall SOSIP.664 trimer preparation as a result of a low level of nontrimer contaminants, the ∼20-fold-greater prevalence of the aberrant V2 domain in uncleaved WT.SEKS trimer populations implies that 20 to 80% of these Env proteins are misfolded in this way. Irrespective of the exact proportion of the uncleaved gp140 proteins that is aberrantly disulfide bonded, it is clear that the percentage of the total population that is affected is not small but substantial, possibly even the majority. The implications for vaccine design are therefore substantive.
The only other aberrant disulfide-linked peptide that was detectable in both the WT.SEKS and SOSIP.664 trimer preparations was from the gp120 C2 region (Table 2). In both cases, we estimate that this scrambled peptide is present at a low abundance, since the signals for these peaks are about ∼100-fold lower than those for many other peptides. However, based on the peak intensities relative to those of other, coeluting species, the scrambled C2 peptide was, again, less prevalent in the SOSIP.664 trimer than in the WT.SEKS gp140. Several other aberrant disulfide bonds in the C1, C2, and C3 regions were detectable only in the WT.SEKS proteins (Table 2).
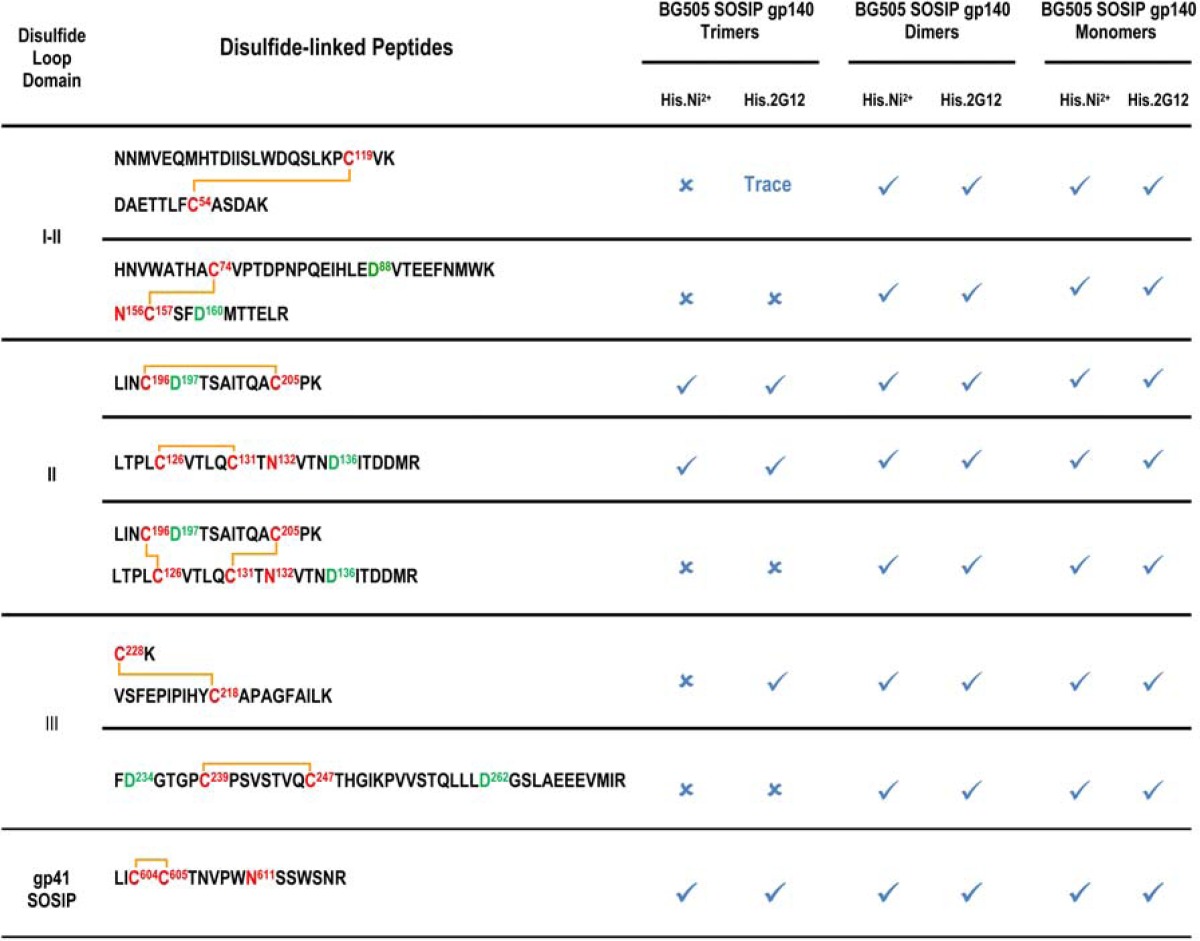
We next addressed whether all secreted Env proteins containing the SOSIP.664 modifications behave in the same way with respect to their disulfide bond content. To do so, we purified His-tagged BG505 SOSIP.664 Env proteins either by 2G12 affinity chromatography, as before, or by Ni2+-NTA affinity chromatography (14, 15). In both cases, we then isolated the gp140 monomer, dimer, and trimer fractions eluting from the SEC column and determined their disulfide bond profiles. An example of an SEC profile showing the pooled fractions can be found in Fig. S2 in the supplemental material, and the results of the disulfide analysis are shown in Table 3. The canonical disulfide bonds were always detected in each protein (not shown) and were accompanied by various numbers of aberrant disulfide-bonded peptides. Of greatest relevance is the fact that the SOSIP.664 gp140 monomers and dimers contained considerably more of the nonnative disulfide bonds than the trimers, irrespective of whether the Env proteins were purified on the 2G12 or the Ni-NTA-column prior to SEC (Table 3).
DISCUSSION
Here we have confirmed and extended earlier reports that HIV-1 Env proteins can contain noncanonical (i.e., aberrant) disulfide bonds (6, 15, 23, 24, 33). The nine disulfide bonds present in the monomeric HIV-1 IIIB gp120 protein were first identified in 1990 (25) and have since been seen in high-resolution structures of native-like trimers (27–30). However, various gp120 and uncleaved gp140 proteins contain subpopulations in which the canonical disulfide bonds have not formed, and alternative configurations are present instead (6, 15, 23, 24, 33). The aberrant disulfide bonds are most prevalent in, but not limited to, the N-terminal half of the protein, and particularly the V1/V2 loop region. The consequence of forming aberrant disulfide bonds is that the resulting subset of gp120 monomers or gp140 subunits is misfolded. In addition, intermolecular disulfide bonds can also form, creating gp120 dimers (31, 32). For the uncleaved gp140 proteins, the problems created by aberrant disulfide bond formation are exacerbated by the formation of dimerizing/aggregating cross-links between different molecules (23, 24, 35). In addition, aberrant cross-links probably also arise between the gp120 subunits within a single gp140 protein, further exacerbating the heterogeneity and nonnative conformation of these entities.
A limitation to the earlier studies of gp120 and uncleaved gp140 proteins is that it was not possible to quantify the proportion of gp120 subunits that were affected by disulfide bond scrambling (6, 23, 24, 33). Thus, whether the problem affected, for example, 1% or 50% of the gp120 moieties was unknown but of major significance to the potential use of these proteins as vaccine immunogens. Now, by conducting comparative studies with native-like SOSIP.664 trimers that we show to be only minimally affected by disulfide bond scrambling, we find that aberrant disulfide bonds are present in a substantial proportion of nonnative gp140 proteins (possibly as high as 40 to 80% for BG505 WT.SEKS). Taking this finding together with a similar outcome of a study based on 92UG037.8 and CZA97.012 Env proteins (15), we can conclude that gp120 monomers and uncleaved gp140 proteins commonly contain a substantial number of noncanonical disulfide bonds (15). Viewing of the uncleaved gp140 proteins by EM and analysis by other biophysical techniques show that this problem contributes to their nonnative configurations (11, 12, 14–16, 20). The implications for the effectiveness of Env proteins with these designs as vaccine components now merit careful consideration. We note that the BG505 WT.SEKS protein was very poorly immunogenic in rabbits, both in absolute terms and relative to the native-like BG505 SOSIP.664 trimers (40).
What underlies the formation of aberrant disulfide bonds in Env proteins? Uncertainties remain, some of which could be explored in future studies on other forms of Env purified in various ways. It is possible that the formation of aberrant disulfide bonds in recombinant forms of Env reflects their overexpression, relative to viral Env expression, in transfection systems. We note, however, that this would not account for the difference between native-like trimers and nonnative gp140 proteins, since in our general experience, total Env expression is comparable in both cases (typically 2 to 5 mg/liter). An alternative possibility is that disulfide heterogeneity is also present in viral Env as part of an immune evasion strategy that involves creating nonnative forms of Env to induce irrelevant, distractive antibodies. Thus, it has long been known that the population of full-length Env proteins present on the surfaces of virus-infected or Env-transfected cells is highly heterogeneous, with both native and nonnative forms readily detectable (41–45). We hypothesize that the nonnative forms of Env will be enriched for gp120 subunits containing aberrant disulfide bonds. The same scenario might also apply in the vaccine context; thus, nonnative, disulfide-scrambled Env protein subpopulations may, at least to some extent, distract the immune response away from any better-folded, more native-like proteins that are also present.
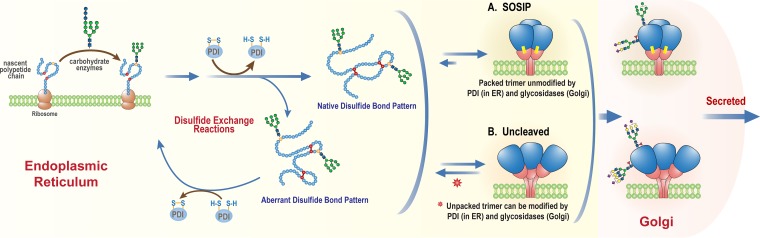
The disulfide heterogeneity problem is common to recombinant soluble Env proteins of multiple designs (6, 15, 23, 24, 33). Thus, it affects SOSIP.664 gp140 monomers and dimers as well as gp120 monomers and uncleaved gp140 proteins that contain or lack Foldon-trimerization domains (15). In each case, we propose that the initially expressed gp120 moieties include subpopulations containing both canonical and aberrant disulfide bonds. We also propose that any gp120 subunits containing aberrant disulfide bonds will be excluded from native-like trimers, because they cannot pack properly against one another in the trimer context. We further propose that once the compact, native-like trimers form, their gp120 subunits are inaccessible to protein disulfide isomerase (PDI), the enzyme in the endoplasmic reticulum (ER) that allows proteins to sample many different disulfide-bonding profiles. In contrast, gp120 monomers and nonnative gp140 proteins that are not subject to the steric constraints imposed by trimerization probably do remain exposed to PDI and thus continue to undergo disulfide exchange until they exit the ER (Fig. 3). The argument here is analogous to that used to account for the higher oligomannose content of native-like trimers than of gp120 and uncleaved gp140 proteins: i.e., steric constraints restrict the access of mannosidase to its glycan substrates on the trimer but not on the other Env forms (14, 15, 19, 20). Thus, in the absence of steric constraints, the glycans on the gp120 subunits of nonnative gp140 proteins can be processed further to complex glycoforms that differ from those found on native, trimeric Env (14, 15, 19–21).
FIG 3.
Protein processing in the ER and Golgi apparatus. The polypeptide chain starts out with no disulfide bonds, and it is homogeneously glycosylated (precursor glycans). A number of disulfide-bonding configurations are sampled as the Env protein folds, assisted by protein disulfide isomerase (PDI). (A) Subunits that contain the canonical disulfide-bonding profile can pack into a native-like trimer, and they are then shielded from the actions of PDI (in the ER) and of glycosidases and glycosyltransferases (in the Golgi apparatus). Accordingly, their glycans remain predominantly in the high-mannose form. (B) In contrast, nonnative trimers in which the gp120 subunits are not packed properly are not subject to the same steric constraints and hence continue to be susceptible to PDI activity until they exit the ER. Their disulfide-bonding profile is therefore heterogeneous, with both canonical and aberrant disulfide bonds present, by the time they exit the ER. In the Golgi apparatus, the glycoforms on these nonnative trimers are trimmed by glycosidases and processed by glycosyltransferases, such that many of their high-mannose glycans are replaced by more highly processed glycoforms. We propose that monomeric gp120 proteins will be processed in the same way as the gp120 subunits of the nonnative trimers.
NS-EM imaging shows how the compact native-like SOSIP.664 trimers differ from the splayed-out, nonnative gp140 proteins, differences reinforced by proteolysis and hydrogen/deuterium exchange studies that directly address the relative accessibility of the protein surfaces to enzymes and solvent (13, 15, 16, 20). The outcomes of these in vitro analyses are entirely consistent with the ways in which the different types of Env protein are produced within the cell; either these Env proteins are subject to trimerization-imposed steric constraints that affect posttranslational modifications by enzymes (e.g., PDI and mannosidase) or they are not (Fig. 3). Accordingly, the canonical disulfide-bonding profile and enrichment for high-mannose glycans are interlinked, and arguably defining, properties of a native-like trimer. Whether gp120 monomer subpopulations with canonical and aberrant disulfide bonds are also differentially glycosylated remains to be determined, but we suspect that the lack of steric constraints on processing enzymes is the dominant factor and that all gp120 proteoforms contain highly processed glycans.
Substantial quantities of aberrant disulfide bonds were identified in the proteins present in the BG505 SOSIP.664 gp140 monomer and dimer fractions isolated from the SEC columns; these aberrant bonds are probably present in Env proteins that either never assembled properly into trimers in the first place or fell apart early enough to be further processed by PDI. A much smaller quantity of aberrant disulfide bonds was also detected in the purified BG505 SOSIP.664 trimer fraction. Here, we think that the nonnative disulfide bond signals arise from a small proportion (∼1 to 4%) of contaminating nonnative forms (e.g., dimers or aggregates) that are incompletely separated from native-like trimers on SEC columns.
The BG505 SOSIP.664 trimers isolated by the 2G12/SEC purification method appear almost completely native-like when viewed by NS-EM (14, 16, 34, 35, 46). BG505 SOSIP.664-His trimers are also fully native-like when purified on Ni-NTA columns via their C-terminal His tags (14). However, such homogeneity is not always seen with SOSIP.664 trimers based on other Env genotypes. Whereas the SOSIP.664-modified BG505, B41, DU422, and ZM197M env genes have a high potential for yielding native-like trimers (34, 46, 47), others (e.g., 92UG037.8 and CZA97.012) have only an intermediate potential (15), and many others have a very low potential (e.g., multiple env genes listed in reference 47). Thus, when CZA97.012 and 92UG037.8 SOSIP.664 trimers are purified by conformationally nonselective methods (lectin, 2G12, or Ni-NTA columns followed by SEC), only ∼50% of the trimers are native-like, while the remainder resemble nonnative uncleaved gp140 trimers (15). The same finding is broadly true of JR-FL and 16055 SOSIP.664 trimers, prior to negative selection to remove the nonnative forms (48). We have shown that the nonnative fraction of CZA97.012 SOSIP.664 trimers is substantially enriched for aberrant disulfide bonds, relative to their fully native-like counterparts (15) We suggest that a similar finding would be made if the nonnative fractions of 92UG037.8, JR-FL, and 16055 SOSIP.664 trimers were similarly analyzed. Conversely, we hypothesize that, when appropriately purified, the recently described uncleaved, native-like SC and NFL gp140 proteins, which include a flexible linker to stabilize the gp120–gp41 interface together with the SOS and/or I559P changes (12, 48), will mimic SOSIP.664 trimers in being predominantly free of scrambled disulfide bonds.
The proportion of native-like trimers that arises when SOSIP.664-modified gp140 proteins are expressed is genotype dependent (15, 47) The underlying reasons are under investigation, but the stability of the gp120–gp41 interface and/or the trimer apex and the efficiency with which the intersubunit SOS bond is formed are probably critical influences on the efficiency of native-like trimer formation. We propose that whether the native trimer assembles properly or not is, itself, the critical determinant of the overall disulfide-bonding profile. Thus, any nonnative gp140 protein in which gp120 has not stably associated with gp41 will remain vulnerable to disulfide bond scrambling; conversely, when gp120 interacts properly with gp41, the resulting native-like trimer is, for steric reasons, resistant to the effects of any PDI family members it encounters, as shown in Fig. 3.
The key to producing properly folded Env proteins that are free of scrambled disulfide bonds therefore lies in the design of the construct and, often, in the purification strategy used. BG505 SOSIP.664 trimers and a few other SOSIP.664 trimers can be purified in fully native form without the use of a bNAb against a trimer-specific epitope (i.e., the 2G12/SEC method used here) (34, 46, 47). However, for SOSIP.664 trimers based on some other genotypes, affinity columns based on quaternary epitope-specific antibodies, such as PGT145 and PGT151, are needed in order to positively select for native-like trimer subpopulations (15, 46). The PGT145 or PGT151 positive-selection columns are not useful for nonnative uncleaved gp140 proteins and gp120 monomers, which predominantly or completely lack these quaternary epitopes. However, it is possible that any subpopulation of gp120 monomers that is free of scrambled disulfide bonds could be purified via other antibody affinity columns. Thus, if there are epitopes, for example, around the CD4-binding site, that are predominantly or exclusively present only on correctly folded gp120 monomers (i.e., those with canonical disulfide bonds), the relevant monoclonal antibodies could be used in positive-selection columns. A practical limitation would be that if the proportion of gp120 subunits with an aberrant disulfide bond profile is too high, the losses during purification could be prohibitive from the yield perspective.
Overall, we conclude that those who design HIV-1 Env proteins for use as vaccine immunogens need to take into account the presence of aberrantly folded proteins that arise because of disulfide bond scrambling during synthesis. The relevant subpopulations can be highly abundant, even predominant. Given the importance of tertiary and often quaternary structure to most Env epitopes, disulfide bond scrambling will inevitably affect antigenicity and may also have a substantial influence on immunogenicity. Here, the presence of aberrantly folded subpopulations may either dilute the responses to the properly folded Env proteins or, in the worst-case scenario, act as an immunological distraction.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michael Golabek for technical assistance.
This work was supported by NIH grants R01 AI094797 (to H.D.), R01 GM103547 (to H.D.), P01 AI082362 (to J.P.M.), and P01 AI110657 to (J.P.M.).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01953-15.
REFERENCES
- 1.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 2.Alam SM, Liao HX, Tomaras GD, Bonsignori M, Tsao CY, Hwang KK, Chen H, Lloyd KE, Bowman C, Sutherland L, Jeffries TL Jr, Kozink DM, Stewart S, Anasti K, Jaeger FH, Parks R, Yates NL, Overman RG, Sinangil F, Berman PW, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Karasavva N, Rerks-Ngarm S, Kim JH, Michael NL, Zolla-Pazner S, Santra S, Letvin NL, Harrison SC, Haynes BF. 2013. Antigenicity and immunogenicity of RV144 vaccine AIDSVAX clade E envelope immunogen is enhanced by a gp120 N-terminal deletion. J Virol 87:1554–1568. doi: 10.1128/JVI.00718-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Du SX, Xu L, Zhang W, Tang S, Boenig RI, Chen H, Mariano EB, Zwick MB, Parren PW, Burton DR, Wrin T, Petropoulos CJ, Ballantyne JA, Chambers M, Whalen RG. 2011. A directed molecular evolution approach to improved immunogenicity of the HIV-1 envelope glycoprotein. PLoS One 6:e20927. doi: 10.1371/journal.pone.0020927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fouda GG, Amos JD, Wilks AB, Pollara J, Ray CA, Chand A, Kunz EL, Liebl BE, Whitaker K, Carville A, Smith S, Colvin L, Pickup DJ, Staats HF, Overman G, Eutsey-Lloyd K, Parks R, Chen H, Labranche C, Barnett S, Tomaras GD, Ferrari G, Montefiori DC, Liao HX, Letvin NL, Haynes BF, Permar SR. 2013. Mucosal immunization of lactating female rhesus monkeys with a transmitted/founder HIV-1 envelope induces strong Env-specific IgA antibody responses in breast milk. J Virol 87:6986–6999. doi: 10.1128/JVI.00528-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fouda GG, Cunningham CK, McFarland EJ, Borkowsky W, Muresan P, Pollara J, Song LY, Liebl BE, Whitaker K, Shen X, Vandergrift NA, Overman RG, Yates NL, Moody MA, Fry C, Kim JH, Michael NL, Robb M, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Liao HX, Haynes BF, Montefiori DC, Ferrari G, Tomaras GD, Permar SR. 2015. Infant HIV type 1 gp120 vaccination elicits robust and durable anti-V1V2 immunoglobulin G responses and only rare envelope-specific immunoglobulin A responses. J Infect Dis 211:508–517. doi: 10.1093/infdis/jiu444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kassa A, Dey AK, Sarkar P, Labranche C, Go EP, Clark DF, Sun Y, Nandi A, Hartog K, Desaire H, Montefiori D, Carfi A, Srivastava IK, Barnett SW. 2013. Stabilizing exposure of conserved epitopes by structure guided insertion of disulfide bond in HIV-1 envelope glycoprotein. PLoS One 8:e76139. doi: 10.1371/journal.pone.0076139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wen Y, Stephenson S, Zambonelli C, Hilt S, Wininger M, Dey A, Barnett S, Carfi A. 2012. Simple, scalable and robust purification of two HIV-1 subtype C gp120 monomer subunit antigens for phase II clinical trial in Republic of South Africa. Retrovirology 9(Suppl 2):P356. doi: 10.1186/1742-4690-9-S2-P356. [DOI] [Google Scholar]
- 8.Mascola JR. 2015. HIV. The modern era of HIV-1 vaccine development. Science 349:139–140. doi: 10.1126/science.aac7800. [DOI] [PubMed] [Google Scholar]
- 9.Mouquet H. 2015. Tailored immunogens for rationally designed antibody-based HIV-1 vaccines. Trends Immunol 36:437–439. doi: 10.1016/j.it.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Ward AB, Wilson IA. 2015. Insights into the trimeric HIV-1 envelope glycoprotein structure. Trends Biochem Sci 40:101–107. doi: 10.1016/j.tibs.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.AlSalmi W, Mahalingam M, Ananthaswamy N, Hamlin C, Flores D, Gao G, Rao VB. 2015. A new approach to produce HIV-1 envelope trimers: both cleavage and proper glycosylation are essential to generate authentic trimers. J Biol Chem 290:19780–19795. doi: 10.1074/jbc.M115.656611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Georgiev IS, Joyce MG, Yang Y, Sastry M, Zhang B, Baxa U, Chen RE, Druz A, Lees CR, Narpala S, Schön A, Van Galen J, Chuang GY, Gorman J, Harned A, Pancera M, Stewart-Jones GB, Cheng C, Freire E, McDermott AB, Mascola JR, Kwong PD. 2015. Single-chain soluble BG505.SOSIP gp140 trimers as structural and antigenic mimics of mature closed HIV-1 Env. J Virol 89:5318–5329. doi: 10.1128/JVI.03451-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guttman M, Lee KK. 2013. A functional interaction between gp41 and gp120 is observed for monomeric but not oligomeric, uncleaved HIV-1 Env gp140. J Virol 87:11462–11475. doi: 10.1128/JVI.01681-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pritchard LK, Vasiljevic S, Ozorowski G, Seabright GE, Cupo A, Ringe R, Kim HJ, Sanders RW, Doores KJ, Burton DR, Wilson IA, Ward AB, Moore JP, Crispin M. 2015. Structural constraints determine the glycosylation of HIV-1 envelope trimers. Cell Rep 11:1604–1613. doi: 10.1016/j.celrep.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ringe RP, Yasmeen A, Ozorowski G, Go EP, Pritchard LK, Guttman M, Ketas TA, Cottrell CA, Wilson IA, Sanders RW, Cupo A, Crispin M, Lee KK, Desaire H, Ward AB, Klasse PJ, Moore JP. 2015. Influences on the design and purification of soluble, recombinant native-like HIV-1 envelope glycoprotein trimers. J Virol 89:12189–12210. doi: 10.1128/JVI.01768-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ringe RP, Sanders RW, Yasmeen A, Kim HJ, Lee JH, Cupo A, Korzun J, Derking R, van Montfort T, Julien JP, Wilson IA, Klasse PJ, Ward AB, Moore JP. 2013. Cleavage strongly influences whether soluble HIV-1 envelope glycoprotein trimers adopt a native-like conformation. Proc Natl Acad Sci U S A 110:18256–18261. doi: 10.1073/pnas.1314351110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonomelli C, Doores KJ, Dunlop DC, Thaney V, Dwek RA, Burton DR, Crispin M, Scanlan CN. 2011. The glycan shield of HIV is predominantly oligomannose independently of production system or viral clade. PLoS One 6:e23521. doi: 10.1371/journal.pone.0023521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doores KJ, Bonomelli C, Harvey DJ, Vasiljevic S, Dwek RA, Burton DR, Crispin M, Scanlan CN. 2010. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc Natl Acad Sci U S A 107:13800–13805. doi: 10.1073/pnas.1006498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Go EP, Herschhorn A, Gu C, Castillo-Menendez L, Zhang S, Mao Y, Chen H, Ding H, Wakefield JK, Hua D, Liao HX, Kappes JC, Sodroski J, Desaire H. 2015. Comparative analysis of the glycosylation profiles of membrane-anchored HIV-1 envelope glycoprotein trimers and soluble gp140. J Virol 89:8245–8257. doi: 10.1128/JVI.00628-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pritchard LK, Spencer DI, Royle L, Bonomelli C, Seabright GE, Behrens AJ, Kulp DW, Menis S, Krumm SA, Dunlop DC, Crispin DJ, Bowden TA, Scanlan CN, Ward AB, Schief WR, Doores KJ, Crispin M. 2015. Glycan clustering stabilizes the mannose patch of HIV-1 and preserves vulnerability to broadly neutralizing antibodies. Nat Commun 6:7479. doi: 10.1038/ncomms8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pritchard LK, Harvey DJ, Bonomelli C, Crispin M, Doores KJ. 2015. Cell- and protein-directed glycosylation of native cleaved HIV-1 envelope. J Virol 89:8932–8944. doi: 10.1128/JVI.01190-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallaher WR, Ball JM, Garry RF, Griffin MC, Montelaro RC. 1989. A general-model for the transmembrane proteins of HIV and other retroviruses. AIDS Res Hum Retroviruses 5:431–440. doi: 10.1089/aid.1989.5.431. [DOI] [PubMed] [Google Scholar]
- 23.Go EP, Zhang Y, Menon S, Desaire H. 2011. Analysis of the disulfide bond arrangement of the HIV-1 envelope protein CON-S gp140 ΔCFI shows variability in the V1 and V2 regions. J Proteome Res 10:578–591. doi: 10.1021/pr100764a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Go EP, Hua D, Desaire H. 2014. Glycosylation and disulfide bond analysis of transiently and stably expressed clade C HIV-1 gp140 trimers in 293T cells identifies disulfide heterogeneity present in both proteins and differences in O-linked glycosylation. J Proteome Res 13:4012–4027. doi: 10.1021/pr5003643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leonard CK, Spellman MW, Riddle L, Harris RJ, Thomas JN, Gregory TJ. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol Chem 265:10373–10382. [PubMed] [Google Scholar]
- 26.Peisajovich SG, Blank L, Epand RF, Epand RM, Shai Y. 2003. On the interaction between gp41 and membranes: the immunodominant loop stabilizes gp41 helical hairpin conformation. J Mol Biol 326:1489–1501. doi: 10.1016/S0022-2836(03)00040-8. [DOI] [PubMed] [Google Scholar]
- 27.Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, Joyce MG, Guttman M, Ma X, Narpala S, Soto C, Terry DS, Yang Y, Zhou T, Ahlsen G, Bailer RT, Chambers M, Chuang GY, Doria-Rose NA, Druz A, Hallen MA, Harned A, Kirys T, Louder MK, O'Dell S, Ofek G, Osawa K, Prabhakaran M, Sastry M, Stewart-Jones GB, Stuckey J, Thomas PV, Tittley T, Williams C, Zhang B, Zhao H, Zhou Z, Donald BR, Lee LK, Zolla-Pazner S, Baxa U, Schön A, Freire E, Shapiro L, Lee KK, Arthos J, Munro JB, Blanchard SC, Mothes W, Binley JM, McDermott AB, Mascola JR, Kwong PD. 2015. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol 22:522–531. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. 2013. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, Stewart-Jones GB, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. 2014. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finzi A, Pacheco B, Zeng X, Kwon YD, Kwong PD, Sodroski J. 2010. Conformational characterization of aberrant disulfide-linked HIV-1 gp120 dimers secreted from overexpressing cells. J Virol Methods 168:155–161. doi: 10.1016/j.jviromet.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Owens RJ, Compans RW. 1990. The human immunodeficiency virus type 1 envelope glycoprotein precursor acquires aberrant intermolecular disulfide bonds that may prevent normal proteolytic processing. Virology 179:827–833. doi: 10.1016/0042-6822(90)90151-G. [DOI] [PubMed] [Google Scholar]
- 33.Clark DF, Go EP, Desaire H. 2013. Simple approach to assign disulfide connectivity using extracted ion chromatograms of electron transfer dissociation spectra. Anal Chem 85:1192–1199. doi: 10.1021/ac303124w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Peña AT, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. 2013. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung NP, Matthews K, Kim HJ, Ketas TJ, Golabek M, de Los Reyes K, Korzun J, Yasmeen A, Sanders RW, Klasse PJ, Wilson IA, Ward AB, Marozsan AJ, Moore JP, Cupo A. 2014. Stable 293 T and CHO cell lines expressing cleaved, stable HIV-1 envelope glycoprotein trimers for structural and vaccine studies. Retrovirology 11:33. doi: 10.1186/1742-4690-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yasmeen A, Ringe R, Derking R, Cupo A, Julien JP, Burton DR, Ward AB, Wilson IA, Sanders RW, Moore JP, Klasse PJ. 2014. Differential binding of neutralizing and non-neutralizing antibodies to native-like soluble HIV-1 Env trimers, uncleaved Env proteins, and monomeric subunits. Retrovirology 11:41. doi: 10.1186/1742-4690-11-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kitamura R, Matsuoka K, Matsushima E, Kawaguchi Y. 2001. Improvement in precision of the liquid chromatographic-electrospray ionization tandem mass spectrometric analysis of 3′-C-ethynylcytidine in rat plasma. J Chromatogr B Biomed Sci Appl 754:113–119. doi: 10.1016/S0378-4347(00)00588-0. [DOI] [PubMed] [Google Scholar]
- 38.Shi G. 2003. Application of co-eluting structural analog internal standards for expanded linear dynamic range in liquid chromatography/electrospray mass spectrometry. Rapid Commun Mass Spectrom 17:202–206. doi: 10.1002/rcm.897. [DOI] [PubMed] [Google Scholar]
- 39.Xu RN, Fan L, Rieser MJ, El-Shourbagy TA. 2007. Recent advances in high-throughput quantitative bioanalysis by LC-MS/MS. J Pharm Biomed Anal 44:342–355. doi: 10.1016/j.jpba.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 40.Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, Arendt H, Kim HJ, Lee JH, Pugach P, Williams M, Debnath G, Moldt B, van Breemen MJ, Isik G, Medina-Ramirez M, Back JW, Koff WC, Julien JP, Rakasz EG, Seaman MS, Guttman M, Lee KK, Klasse PJ, LaBranche C, Schief WR, Wilson IA, Overbaugh J, Burton DR, Ward AB, Montefiori DC, Dean H, Moore JP. 2015. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tong T, Crooks ET, Osawa K, Binley JM. 2012. HIV-1 virus-like particles bearing pure Env trimers expose neutralizing epitopes but occlude nonneutralizing epitopes. J Virol 86:3574–3587. doi: 10.1128/JVI.06938-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pancera M, Wyatt R. 2005. Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology 332:145–156. doi: 10.1016/j.virol.2004.10.042. [DOI] [PubMed] [Google Scholar]
- 43.Herrera C, Spenlehauer C, Fung MS, Burton DR, Beddows S, Moore JP. 2003. Nonneutralizing antibodies to the CD4-binding site on the gp120 subunit of human immunodeficiency virus type 1 do not interfere with the activity of a neutralizing antibody against the same site. J Virol 77:1084–1091. doi: 10.1128/JVI.77.2.1084-1091.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crooks ET, Tong T, Osawa K, Binley JM. 2011. Enzyme digests eliminate nonfunctional Env from HIV-1 particle surfaces, leaving native Env trimers intact and viral infectivity unaffected. J Virol 85:5825–5839. doi: 10.1128/JVI.00154-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blattner C, Lee JH, Sliepen K, Derking R, Falkowska E, de la Peña AT, Cupo A, Julien JP, van Gils M, Lee PS, Peng W, Paulson JC, Poignard P, Burton DR, Moore JP, Sanders RW, Wilson IA, Ward AB. 2014. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity 40:669–680. doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pugach P, Ozorowski G, Cupo A, Ringe R, Yasmeen A, de Val N, Derking R, Kim HJ, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, Julien JP, Burton DR, Wilson IA, Sanders RW, Klasse PJ, Ward AB, Moore JP. 2015. A native-like SOSIP.664 trimer based on an HIV-1 subtype B env gene. J Virol 89:3380–3395. doi: 10.1128/JVI.03473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Julien JP, Lee JH, Ozorowski G, Hua Y, Torrents de la Peña A, de Taeye SW, Nieusma T, Cupo A, Yasmeen A, Golabek M, Pugach P, Klasse PJ, Moore JP, Sanders RW, Ward AB, Wilson IA. 2015. Design and structure of two HIV-1 clade C SOSIP.664 trimers that increase the arsenal of native-like Env immunogens. Proc Natl Acad Sci U S A 112:11947–11952. doi: 10.1073/pnas.1507793112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guenaga J, de Val N, Tran K, Feng Y, Satchwell K, Ward AB, Wyatt RT. 2015. Well-ordered trimeric HIV-1 subtype B and C soluble spike mimetics generated by negative selection display native-like properties. PLoS Pathog 11:e1004570. doi: 10.1371/journal.ppat.1004570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.