ABSTRACT
Human cytomegalovirus (HCMV) protein pUL48 is closely associated with the capsid and has a deubiquitinating protease (DUB) activity in its N-terminal region. Although this DUB activity moderately increases virus replication in cultured fibroblast cells, the requirements of the N-terminal region of pUL48 in the viral replication cycle are not fully understood. In this study, we characterized the recombinant viruses encoding UL48(ΔDUB/NLS), which lacks the DUB domain and the adjacent nuclear localization signal (NLS), UL48(ΔDUB), which lacks only the DUB, and UL48(Δ360–1200), which lacks the internal region (amino acids 360 to 1200) downstream of the DUB/NLS. While ΔDUB/NLS and Δ360–1200 mutant viruses did not grow in fibroblasts, the ΔDUB virus replicated to titers 100-fold lower than those for wild-type virus and showed substantially reduced viral gene expression at low multiplicities of infection. The DUB domain contained ubiquitination sites, and DUB activity reduced its own proteasomal degradation in trans. Deletion of the DUB domain did not affect the nuclear and cytoplasmic localization of pUL48, whereas the internal region (360–1200) was necessary for cytoplasmic distribution. In coimmunoprecipitation assays, pUL48 interacted with three tegument proteins (pUL47, pUL45, and pUL88) and two capsid proteins (pUL77 and pUL85) but the DUB domain contributed to only pUL85 binding. Furthermore, we found that the ΔDUB virus showed reduced virion stability and less efficiently delivered its genome into the cell than the wild-type virus. Collectively, our results demonstrate that the N-terminal DUB domain of pUL48 contributes to efficient viral growth by regulating its own stability and promoting virion stabilization and virus entry.
IMPORTANCE HCMV pUL48 and its herpesvirus homologs play key roles in virus entry, regulation of immune signaling pathways, and virion assembly. The N terminus of pUL48 contains the DUB domain, which is well conserved among all herpesviruses. Although studies using the active-site mutant viruses revealed that the DUB activity promotes viral growth, the exact role of this region in the viral life cycle is not fully understood. In this study, using the mutant virus lacking the entire DUB domain, we demonstrate that the DUB domain of pUL48 contributes to viral growth by regulating its own stability via autodeubiquitination and promoting virion stability and virus entry. This report is the first to demonstrate the characteristics of the mutant virus with the entire DUB domain deleted, which, along with information on the functions of this region, is useful in dissecting the functions associated with pUL48.
INTRODUCTION
Human cytomegalovirus (HCMV) belongs to the Betaherpesvirus subfamily. HCMV infection is usually asymptomatic and causes latent or persistent infections in healthy people. However, congenital infection and reactivation from latent infection in immunocompromised individuals can cause severe disease (1). The HCMV virion is composed of an icosahedral capsid containing a 235-kb linear genome, the envelope surrounding the capsid, and the tegument between the capsid and the viral envelope, which contains many viral proteins (1, 2). The tegument proteins are delivered to the cell, and some have been shown to play important roles in both the early and late phases of infection. The early functions of tegument proteins include regulation of viral gene expression and modulation of host cell antiviral responses (3). During the late stages of infection, the tegument proteins are thought to be involved in nuclear egress of the capsid to the cytoplasm and subsequent secondary envelopment (4, 5).
The open reading frame (ORF) UL48-encoded protein of HCMV, pUL48, is the largest inner tegument protein that is closely associated with the capsid (6, 7). A deletion of the UL48 gene is lethal to the virus (8), and viruses containing an insertion of a transposon within the upstream region of the UL48 ORF show severely impaired viral growth (9), suggesting that the function of UL48 is critical for the viral replication cycle to be successful. pUL48 contains deubiquitinating protease (DUB) activity in its N-terminal region (10, 11). The UL48 DUB contains both a ubiquitin-specific carboxyl-terminal hydrolase activity and an isopeptidase activity that cleaves ubiquitin K11, K48, and K64 linkages (11, 12). The growth of active-site mutant virus is reduced by 10-fold in permissive human fibroblast (HF) cells compared to wild-type virus, demonstrating that the DUB activity moderately enhances virus replication in cultured cells (11). The UL48 DUB domain is highly conserved among the pUL48 equivalents of other herpesviruses, including the pUL36 protein (also called VP1-2) of herpes simplex virus 1 (HSV-1) (13, 14).
The function of pUL48 in HCMV replication has not been completely studied. However, studies of the pUL36 proteins of HSV-1 and pseudorabies virus (PRV) have demonstrated that this tegument protein plays important roles in virus entry and maturation. pUL36 is required for capsid transport within the cell by interacting with the microtubule network (15–18). In HSV-1, the nuclear localization signal (NLS) of pUL36 is required for routing of the capsid to the nuclear pore (19, 20) and proteolytic cleavage of pUL36 is necessary for release of HSV-1 DNA into the nucleus (21). Recently, pUL48 was also shown to contain the NLS that is indispensable for viral growth just downstream from the DUB domain (22) and can functionally substitute for the NLS of pUL36 (23). Evidence indicates that the alphaherpesvirus pUL36 proteins are also required for nuclear egress and secondary envelopment. HSV-1 pUL36 has been shown to associate with the capsid (24), while the C-terminal fragment of PRV pUL36 was found to enter the nucleus and enhance nuclear egression of the capsid (25). In cells infected with UL36-deleted HSV-1 and PRV, the newly assembled capsids accumulate in the cytoplasm (26, 27) and this event appears to result from the failure of recruitment of the cytoplasmic capsid to the site of secondary envelopment (28). Recently, it was shown that in the absence of pUL48 expression, development of the cytoplasmic virion assembly complex (cVAC) was abrogated (29).
Although pUL48 of HCMV is thought to play roles similar to those observed for the pUL36 proteins of HSV-1 and PRV, information about the functions associated with the specific domains of this largest tegument protein is limited. In this study, the recombinant HCMV encoding UL48(ΔDUB/NLS), which lacks the entire DUB domain and the NLS, UL48(ΔDUB), which lacks only the DUB, or UL48(Δ360–1200), which lacks the internal region downstream of the DUB/NLS, were produced and their growth patterns were analyzed in permissive HF cells. We also investigated the role of the UL48 DUB and its downstream region in regulating its own stability and intracellular localization. Moreover, we evaluated the requirement of the DUB domain for interactions with other virion proteins, virion stability, and virus entry.
MATERIALS AND METHODS
Cell culture and virus stocks.
Human foreskin diploid fibroblast (HF) and 293T cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml) in a 5% CO2 humidified incubator at 37°C. The HCMV Towne virus stocks used in this study were prepared as previously described (30). Viral titers were determined on HF cells using infectious center assays employing an anti-IE1 antibody (31).
Infectious center assays.
To perform virus infectivity assays, the diluted samples were used to inoculate a monolayer of HF cells (1 × 105) in a 24-well plate. At 24 h postinfection, cells were fixed with 500 μl of cold methanol for 10 min. Cells were then washed three times in phosphate-buffered saline (PBS), incubated with anti-IE1 rabbit polyclonal antibody (PAb) in PBS at 37°C for 1 h, followed by incubation with phosphatase-conjugated anti-rabbit immunoglobulin G (IgG) antibody in PBS at 37°C for 1 h. Finally, the cells were gently washed in PBS and treated with 200 μl of nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate (KPL) at room temperature for 1 h, according to the manufacturer's instructions. The IE1-positive cells were counted in at least three to five separate fields per well under a light microscope (200× magnification).
Transient DNA transfection and electroporation.
293T cells were transfected using polyethylenimine (PEI) (Sigma). The DNA was mixed with PEI in serum-free medium according to the manufacturer's procedure. The mixture was kept at room temperature for 20 min and then added dropwise to cells. HF cells were transfected by electroporation.
Electroporation was used for transfection of HF cells with bacmid DNAs. For each reaction, HF cells (2 × 106) in 200 μl of resuspension buffer were mixed with 3 μg of bacmid DNA, 4 μg of plasmid pCMV71 encoding pp71 to enhance the activation of the major immediate early (MIE) promoter, and 1 μg of pEGFP-C1 to monitor electroporation efficiency. After electroporation at 1,300 V for 40 ms using a Microporator MP-100 (Digital Bio Technology), the cells were plated in T-25 flasks. When they reached confluence, the cells were split into new flasks at a ratio of 1:2. Cultures were grown at 37°C, and the spread of green fluorescent protein (GFP) fluorescence was monitored.
Plasmids.
The wild-type UL48 DNA (Towne strain) and its truncation or deletion mutant versions were PCR amplified from the HCMV (Towne) genome and cloned in the pENTR vector (Invitrogen). Plasmids expressing hemagglutinin (HA)- or Flag-tagged UL48 proteins were produced by transferring the UL48 DNAs from pENTR vectors to pSG5 (32)-based destination vectors using LR Clonase (Invitrogen). Plasmids expressing myc- or glutathione S-transferase (GST)-tagged UL48 proteins were produced on the pCS3-MT (with a 6Myc tag) (33)-based destination vector and pDEST27 (Invitrogen) backgrounds using Gateway technology (Invitrogen). Plasmids expressing the C24S mutant versions of HA, Flag, or GST-tagged UL48 proteins were made using the Stratagene QuikChange site-directed mutagenesis protocol.
pENTR vectors (Invitrogen) containing HCMV ORFs such as UL47, UL45, UL88, and UL77 were described previously (34). Yeast expression was used for GAL4 DNA-binding (DB) domain–UL48 fusion and GAL4 activation (A) domain/HCMV ORF fusion proteins, which were generated on the pAS1-CYH2 and pACTII-based destination vectors (35), respectively, using LR Clonase (Invitrogen). Plasmids expressing myc-tagged HCMV ORFs were produced on the pCS3-MT-based destination vector using LR Clonase.
BAC mutagenesis.
An HCMV (strain Towne) bacterial artificial chromosome (BAC) clone (T-BAC) (36) was used as the template for mutagenesis. In this BAC clone, a 9-kb portion of the genome (US1 to US12) is replaced by a fragment containing both the F plasmid sequence and a GFP expression cassette (36). The T-BAC clones containing the UL48(Δ2–359 [ΔDUB/NLS]), UL48(Δ2–278 [ΔDUB]), or UL48(Δ360–1200) genes were generated by using a counterselection BAC modification kit (Gene Bridges). Briefly, rpsL-neo cassettes were PCR amplified using primers containing homology arms consisting of 50 nucleotides upstream and downstream of the target region plus 24 nucleotides homologous to the rpsL-neo cassette. The primer sets used were LMV794/795, LMV794/1303, and LMV798/799 for UL48(Δ2–359), UL48(Δ2–278), and UL48(Δ360–1200), respectively. The LMV primers used for UL48 mutagenesis are listed in Table 1. The amplified rpsL-neo fragments with homology arms were purified and introduced into E. coli GS243 containing wild-type T-BAC (36) for recombination by electroporation using a Gene Pulser II (Bio-Rad). The intermediate T-BAC constructs containing the rpsL-neo cassette were selected on Luria broth (LB) plates containing kanamycin. Next, the rpsL-neo cassette was replaced by annealed oligonucleotide DNAs consisting of only homology arms (50 nucleotides upstream and downstream of the target region): LMV796/797, LMV1533/1534, and LMV800/801 for UL48(Δ2–359), UL48(Δ2–278), and UL48(Δ360–1200), respectively. The UL48(Δ2–359), UL48(Δ2–278), and UL48(Δ360–1200) T-BACs were selected on LB plates containing streptomycin. The mutated regions were amplified by PCR and sequenced to verify the desired mutations.
TABLE 1.
Primers used for UL48 mutagenesis
| Primer | Sequence |
|---|---|
| LMV794 | 5′-GACCCGCCCGCCGGCTCGACATCGGTGTCCCTGCCGCCGGCCTCGCCATGGGCCTGGTGATGATGGCGGGATCG-3′ |
| LMV795 | 5′-CGTGCAACGTCCAGGCGCTGGTGTTGGCAGGCAACGGCGGCAGCGTCAGTCAGAAGAACTCGTCAAGAAGGCG-3′ |
| LMV796 | 5′-ACCCGCCCGCCGGCTCGACATCGGTGTCCCTGCCGCCGGCCTCGCCATGCTGACGCTGCCGCCGTTGCCTGCCAACACCAGCGCCTGGACGTTGCACGC-3′ |
| LMV797 | 5′-GCGTGCAACGTCCAGGCGCTGGTGTTGGCAGGCAACGGCGGCAGCGTCAGCATGGCGAGGCCGGCGGCAGGGACACCGATGTCGAGCCGGCGGGCGGGTC-3′ |
| LMV798 | 5′-TGGGCCGCTACCAACAGCTGGTCGACGCGGTAGAGCAGGAGTTGAAGGCTGGCCTGGTGATGATGGCGGGATCG-3′ |
| LMV799 | 5′-TGACCCTTGAAGGGTTTCTCGGGTACACGCGTGACGGAGCTGCTCCCGGGTCAGAAGAACTCGTCAAGAAGGCG-3′ |
| LMV800 | 5′-TGGGCCGCTACCAACAGCTGGTCGACGCGGTAGAGCAGGAGTTGAAGGCTCCCGGGAGCAGCTCCGTCACGCGTGTACCCGAGAAACCCTTCAAGGGTCA-3′ |
| LMV801 | 5′-TGACCCTTGAAGGGTTTCTCGGGTACACGCGTGACGGAGCTGCTCCCGGGAGCCTTCAACTCCTGCTCTACCTCGTCGACCAGCTGTTGGTAGCGGCCCA-3′ |
| LMV870 | 5′-GACCCGCCCGCCGGCTCGACA-3′ |
| LMV871 | 5′-GCGTGCAACGTCCAGGCGCTG-3′ |
| LMV872 | 5′-TGGGCCGCTACCAACAGCTGG-3′ |
| LMV873 | 5′-TGACCCTTGAAGGGTTTCTCG-3′ |
| LMV1303 | 5′-CTACCGCTGAGGTCCTTGCGGCGTTTCTCGGGTGTTTTTTTGGGTGAGGATCAGAAGAACTCGTCAAGAAGGCG-3′ |
| LMV1533 | 5′-GACCCGCCCGCCGGCTCGACATCGGTGTCCCTGCCGCCGGCCTCGCCATGACATCCTCACCCAAAAAAACACCCGAGAAACGCCGCAAGGACCTCAGCGGTAG-3′ |
| LMV1534 | 5′-CTACCGCTGAGGTCCTTGCGGCGTTTCTCGGGTGTTTTTTTGGGTGAGGATGTCATGGCGAGGCCGGCGGCAGGGACACCGATGTCGAGCCGGCGGGCGGGTC-3′ |
Revertant T-BACs were also generated from the mutant T-BACs. First, the rps-neo cassettes flanked by homology arms were inserted again into the mutant T-BACs. Next, DNA fragments containing wild-type target genes were PCR amplified from the Towne BAC DNA using LMV870/871 for UL48(1–359) and UL48(1–278) and using LMV872/873 for UL48(360–1200). The amplified genes were then inserted into the T-BACs containing the rps-neo cassette by homologous recombination.
Pulsed-field gel electrophoresis.
The bacmid DNAs were digested with restriction endonucleases and subjected to electrophoresis in a 1% agarose (Bio-Rad) gel containing 0.5× TBE (1× TBE is 89 mM Tris-HCl [pH 7.4], 89 mM boric acid, and 25 mM EDTA [pH 8.0]), using a CHEF-DRII apparatus (Bio-Rad) for 5 h at 6 V/cm, with switching times ramped at 0.1 s. The DNA fragments were stained with ethidium bromide in 0.5× TBE buffer.
qRT-PCR.
Total RNAs were isolated from virus-infected cells using Tri reagent (Molecular Research Center, Inc.) and a MaXtract high-density tube (Qiagen). cDNAs were synthesized using random hexamer/oligo(dT) primers in the QuantiTect reverse transcriptase kit (Qiagen). Quantitative real-time reverse transcription-PCR (qRT-PCR) was performed using Power SYBR green PCR master mix (Applied Biosystems) and the Thermal Cycler Dice real-time system (TaKaRa). PCR was performed using the following primers: 5′-AGAGCTCGCTCTTTGTCAGC-3′ (UL47 forward), 5′-GACGTTCAGTGTCACGTTGC-3′ (UL47 reverse), 5′-GAGGACAAGGCTCCGAAAC-3′ (UL99 forward), 5′-CTTTGCTGATGGTGGTGATG-3′ (UL99 reverse), 5′-ATAAGCGGGAGATGTGGATG-3 (IE1 forward), 5′-TTCATCCTTTTTAGCACGGG-3′ (IE1 reverse), 5′-AGCGGGAAATCGTGCGTG-3′ (β-actin forward), and 5′-CAGGGTACATGGTGGTGCC-3′ (β-actin reverse).
Determination of viral genome copy.
Viral DNAs were isolated from 200 μl of viral stock using the QIAamp DNA minikit (Qiagen). The DNA absorbed on the spin column was eluted with 200 μl of distilled water. qPCR was performed using Power SYBR green PCR master mix (Applied Biosystems) and Thermal Cycler Dice real-time system (TaKaRa). The PCR primers used to amplify IE1 exon 4 were 5′-ATAAGCGGGAGATGTGGATG-3′ (forward) and 5′-TTCATCCTTTTTAGCACGGG-3′ (reverse). Copy number was compared to a standard curve generated from the HCMV Towne BAC DNA. All reactions, including unknown samples and standards were analyzed in triplicate.
Antibodies.
Rabbit polyclonal antibodies (PAb) raised against the synthetic peptide corresponding to UL48 residues 278 to 295 (anti-UL48-N) was described previously (11). Anti-HA mouse monoclonal antibody (MAb) 3F10, anti-myc mouse MAb 9E10 conjugated with horseradish peroxidase (HRP), and fluorescein isothiocyanate (FITC)-conjugated anti-HA rat MAb were purchased from Roche. Anti-GST (B-14) mouse MAb was purchased from Santa Cruz. Mouse MAb 810R, which detects both IE1 and IE2, was purchased from Chemicon. Mouse MAbs against p52 (encoded by UL44) and pp28 (encoded by UL99) were obtained from Virusys. Anti-Flag mouse MAb M2 and anti-β-actin antibody were obtained from Sigma. Anti-UL85 PAb was kindly provided by Wade Gibson (Johns Hopkins University School of Medicine).
IFA.
For the indirect immunofluorescence assays (IFA), cells were fixed in ice-cold methanol and rehydrated in cold phosphate-buffered saline (PBS). All subsequent procedures were described previously (37). Slides were examined and photographed using a Zeiss Axiophot microscope. For confocal microscopy, a Carl Zeiss Axioplan 2 confocal microscope system with LSM510 software (Carl Zeiss) was used.
Immunoblot analysis.
For immunoblot analysis, cells were washed with PBS and total cell extracts were prepared by boiling the cell pellets in sodium dodecyl sulfate (SDS) loading buffer. Equal amounts of the clarified cell extracts were separated on SDS-polyacrylamide gels, and transferred to nitrocellulose membranes (Schleicher & Schuell). The membranes were blocked in PBS-T (PBS plus 0.1% Tween 20 [Sigma]) containing 5% skim milk, washed with PBS-T, and then incubated with the appropriate antibody. The proteins were visualized by the standard procedure using an enhanced chemiluminescence system (Roche).
Ubiquitination assays.
293T cells were cotransfected with plasmids expressing target or effector proteins and plasmids expressing HA-ubiquitin. At 48 h after transfection, cells were treated with 0.5 mM N-ethylmaleimide (NEM) for 30 min before they were harvested. Cell pellets were resuspended with 10% SDS lysis buffer containing protease inhibitors (Sigma) and boiled for 10 min. Cell lysates were diluted 10-fold with coimmunoprecipitation (co-IP) buffer (50 mM Tris-Cl [pH 7.4], 50 mM NaF, 5 mM sodium pyrophosphate, protease inhibitors [Sigma]) and sonicated using a microtip probe (Vibra Cell; Sonics and Materials). The clarified cell lysates were incubated with 30 μl of a 50% slurry of anti-Flag M2 affinity gel (Sigma) for 16 h at 4°C. The mixture was pelleted and washed seven times with co-IP buffer. The bound proteins were boiled and analyzed by SDS-PAGE followed by immunoblot assays.
Co-IP assays.
293T cells (8 × 105) were harvested 2 days after transfection and sonicated in 0.7 ml of co-IP buffer (50 mM Tris-Cl [pH 7.4], 50 mM NaF, 5 mM sodium phosphate, 0.1% Triton X-100, protease inhibitors) by a microtip probe for 10 s (pulse on, l s; pulse off, 3 s). Cell lysates were incubated with the appropriate antibodies at 4°C. After incubation for 16 h, 30 μl of a 50% slurry of protein A- and G-Sepharose (Amersham) was added and then the mixture was incubated for 2 h at 4°C to allow adsorption. The mixture was then pelleted and washed 7 times with co-IP buffer. The beads were resuspended and boiled for 5 min in loading buffer. Each sample was analyzed by SDS-PAGE and immunoblotting.
RESULTS
Production of recombinant viruses encoding the N-terminal variants of pUL48.
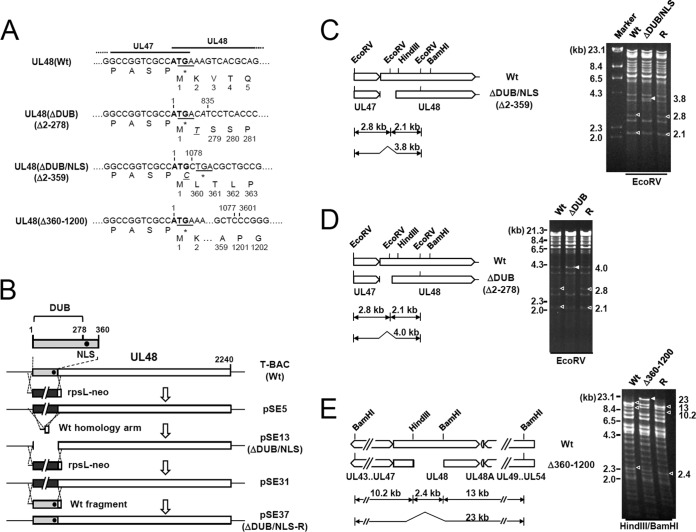
Mutations of the active site of the N-terminal DUB domain of pUL48 only moderately reduce the HCMV growth in permissive HF cells (10, 11). Recently, it was shown that the nuclear targeting of pUL48 via the NLS located just downstream from the DUB domain is essential for viral growth (22). To understand the structural requirements of the UL48 N-terminal domains for viral growth, we produced HCMV (Towne) bacmids (T-BACs) encoding the UL48(Δ2–359 [ΔDUB/NLS]), UL48(Δ2–278 [ΔDUB]), and UL48(Δ360–1200) mutants and their revertants (Fig. 1A and B). The ΔDUB/NLS mutant lacks both the DUB domain and the NLS, whereas the ΔDUB mutant lacks only the DUB domain while retaining the NLS. The Δ360–1200 mutant lacks the internal region downstream from the DUB/NLS. Deletions of the region encompassing amino acids 2 to 359 or 2 to 278 in mutant bacmids and their restoration in revertant bacmids were confirmed by direct sequencing and analysis of the restriction patterns of the EcoRV-digested viral genomes (Fig. 1C and D). Similarly, a deletion of amino acids 360 to 1200 in a mutant and restoration in its revertant were confirmed by sequencing and the HindIII/BamHI restriction patterns (Fig. 1E).
FIG 1.
Construction of the UL48(ΔDUB/NLS), UL48(ΔDUB), or UL48(Δ360–1200) mutant T-BACs and their revertants. (A) Nucleotide and amino acid sequences of the T-BAC clones encoding the wild-type (Wt) or mutant UL48 proteins. The initiation codons for UL48 are indicated in boldface, and the stop codons of UL47 are underlined. The nucleotides and amino acids added during the mutagenesis procedures are indicated in italic. (B) Strategy used to make the UL48 mutant and its revertant BAC clones. The N-terminal region of UL48 (gray boxes) encompassing the DUB domain and the NLS (closed circles) was replaced by an rpsL-neo marker cassette (black boxes) flanked by the 50-nucleotide homologous arm to make pSE5. In a second round of homologous recombination, the rpsL-neo cassette of pSE5 was replaced by the annealed homology arms, resulting in a mutant bacmid with a deletion (pSE13). To construct the revertant T-BAC clone, the mutant gene was replaced with the rpsL-neo cassette (pSE31) and then the wild-type T-BAC clone was rescued by homologous recombination with the wild-type DNA fragment (pSE37). A similar strategy was used to produce the UL48(ΔDUB) and UL48(Δ360–1200) mutant T-BAC clones and their revertants. (C to E) Restriction fragment DNA patterns obtained following EcoRV digestion for the ΔDUB/NLS and ΔDUB mutant T-BACs and their revertants (R) (C and D) or HindIII/BamHI digestion for the Δ360–1200 mutant T-BAC and its revertant (E) were analyzed by agarose gel electrophoresis. Open arrowheads indicate wild-type and revertant T-BACs, and closed arrowheads show mutant T-BACs. The sizes of λ-HindIII are shown by markers. Note that some bands used for diagnosis of the deletion mutants comigrated with other bands.
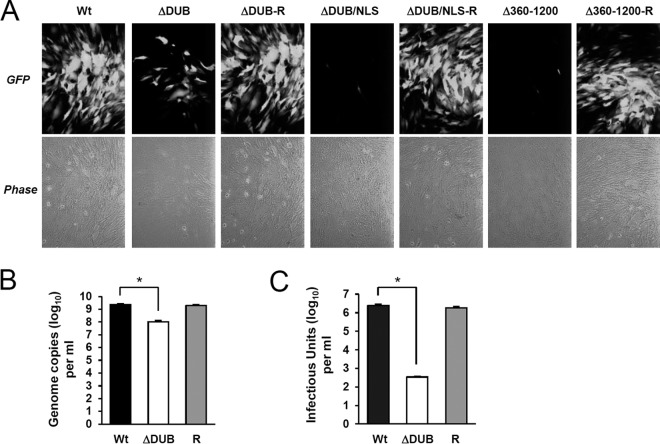
We then tested whether the recombinant viruses can grow when HF cells are transfected with bacmids via electroporation. The results showed that the ΔDUB/NLS and Δ360–1200 mutant viruses did not grow in HF cells and that, while the ΔDUB virus was viable, its growth was severely impaired (Fig. 2A). The viral stocks of wild type, ΔDUB mutant, and its revertant (ΔDUB-R) were prepared by combining the virions produced in cell lysates and in culture supernatants. When the total viral genome copies of the viral stocks were compared, the amount of the ΔDUB virus genome was 20-fold less than those of the wild-type and revertant viruses (Fig. 2B). Furthermore, when the total infectious units in the stocks were measured, the amount of the ΔDUB virus was about 7,000-fold lower than either of those for the wild-type or revertant viruses (Fig. 2C). These results indicate that deletion of both DUB and NLS completely abrogates viral replication, whereas deletion of the DUB domain only with retention of the NLS substantially reduces viral replication. The requirement of the UL48 NLS for viral replication is fully consistent with the results recently reported by Brock et al. (22). Our results demonstrate that the N-terminal region encompassing the DUB domain is also necessary for efficient viral growth independent of the presence of the NLS. Furthermore, these data also demonstrate that the internal region of pUL48 downstream of the DUB/NLS, encompassing amino acids 360 to 1200, is essential for viral growth.
FIG 2.
Comparison of the growth rates of the wild-type and UL48 mutant viruses. (A) HF cells were transfected with wild-type, UL48(ΔDUB), UL48(ΔDUB/NLS), or UL48(Δ360–1200) mutants or their revertant T-BAC clones via electroporation (see Materials and Methods) and were monitored for the propagation of GFP signals. The GFP and phase-contrast images were taken 16 days after electroporation. (B and C) Quantification of viral genomes and infectious units in the stocks of the wild-type, UL48(ΔDUB) mutant, and revertant (R) viruses. After electroporation in panel A, the growth medium was collected and combined with lysates prepared from the cell layer by freezing and thawing three times at 20 days for wild-type and revertant viruses and at 25 days for ΔDUB virus. The viral stocks were prepared after clarification of the samples by centrifugation at 300 × g for 10 min at 4°C. Aliquots of viral stocks were taken, lysates were prepared, and the copy numbers of viral genomes were determined by qPCR (B). The virus titers of wild-type, UL48(ΔDUB) mutant, and revertant viruses in the viral stocks were determined using infectious center assays (C). Statistical significance between samples was determined using Student's t test and is indicated by asterisks (P < 0.01).
Reduced viral gene expression in UL48(ΔDUB) virus infection.
We further investigated the growth property of the ΔDUB mutant virus. Since the ΔDUB virus showed severely reduced infectivity, we infected HF cells with wild-type, ΔDUB, and revertant viruses with equivalent amounts of viral genomes, which correspond to about 0.1 infectious unit (IFU) per cell for wild-type virus. The GFP signals from ΔDUB virus-infected cells appeared and spread much more slowly than the wild-type and revertant virus infections (Fig. 3A). The growth curves of the wild-type, ΔDUB mutant, and revertant viruses were also analyzed after infection at multiplicities of infection (MOI) of 0.1 or 2. The results showed that the ΔDUB virus showed reduced growth at both MOIs and that the peak viral titers of ΔDUB virus were 100-fold lower than those for wild-type virus at an MOI of 0.1, whereas they were 10-fold lower at an MOI of 2 (Fig. 3B). Immunoblot analysis of total cell lysates showed that the reduced growth of the mutant virus correlated with delayed accumulation of viral IE proteins (IE1-p72 and IE2-p86), early proteins [p52(UL44) DNA polymerase processivity factor], and late proteins [pp28(UL99) and pUL48] in cells infected at an MOI of 0.1 (Fig. 3C), although accumulation of viral proteins in mutant virus-infected cells was only slightly delayed at an MOI of 2 (Fig. 3D). These results indicate that the growth defect of ΔDUB virus is more prominent at low MOI.
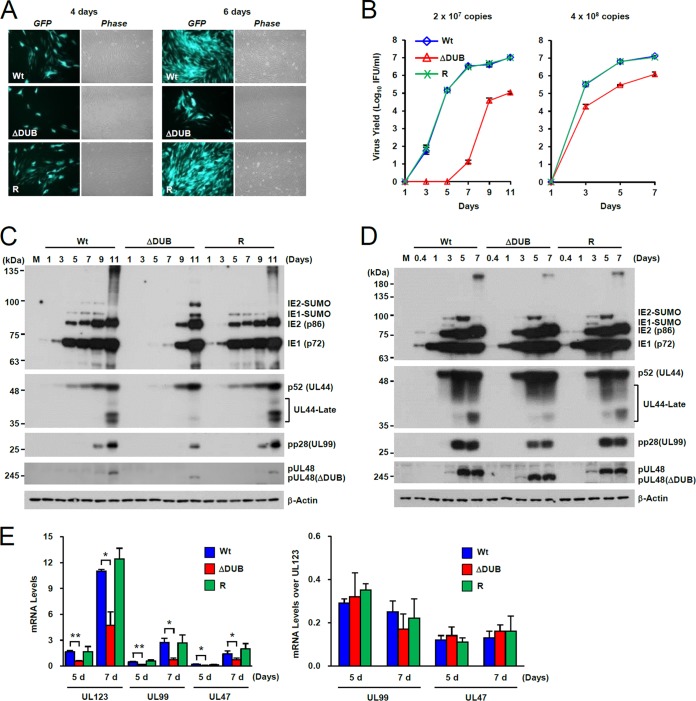
FIG 3.
Growth properties of the UL48(ΔDUB) mutant virus. (A) HF cells (2 × 105) were infected with equivalent amounts of wild-type, UL48(ΔDUB), or revertant (R) viruses (2 × 107 copies of the viral genome), which correspond to an MOI of about 0.1 IFU/cell for wild-type virus. At 4 and 6 days after infection, the GFP images and corresponding phase-contrast images were taken. (B to E) HF cells (2 × 105) were infected with wild-type virus at an MOI of 0.1 or 2 and with equivalent genome copy numbers of UL48(ΔDUB) mutant and revertant viruses. Samples in culture medium were taken at the indicated time points and assayed for virus titer as in Fig. 2D (B). Total cell lysates were prepared from cells infected with MOI of 0.1 (C) and 2 (D) and analyzed by immunoblotting with antibodies for IE1, IE2, p52, pp28, and pUL48 (C and D). M, molecular mass markers. (E) Total RNA samples were prepared from cells infected with an MOI of 0.1 at the indicated time points, and the amounts of UL123 (IE1), UL99, and UL47 transcripts were measured by qRT-PCR. Statistical significance between samples was determined using Student's t test and is indicated by asterisks (*, P < 0.05; **, P < 0.005).
A deletion of the N terminus of the UL48 ORF may affect the transcription of the adjacent UL47 gene, and it may account for the growth defect of UL48(ΔDUB) virus. This possibility is unlikely because the growth of the ΔDUB/NLS virus was more severely impaired than that of the ΔDUB virus, which has a similar deletion in the N-terminal region (Fig. 2A), and UL47 transcription was shown to use the 3′-end polyadenylation signal of UL48 (38). However, to check this possibility, the UL47 mRNA levels were compared with those for wild-type, ΔDUB, and revertant virus infection. The results of qRT-PCR assays showed that the mRNA levels of UL123, UL99, and UL47 were 2- or 3-fold lower in ΔDUB virus infection than those in wild-type and revertant virus infections (Fig. 3E, left). Furthermore, the levels of UL99 or UL47 mRNAs over the UL123 mRNA levels were largely comparable among wild-type, ΔDUB, and revertant virus infections (Fig. 3E, right). Therefore, the growth defect of UL48(ΔDUB) virus cannot be attributed to the defect in UL47 transcription.
The DUB domain-dependent regulation of pUL48 stability.
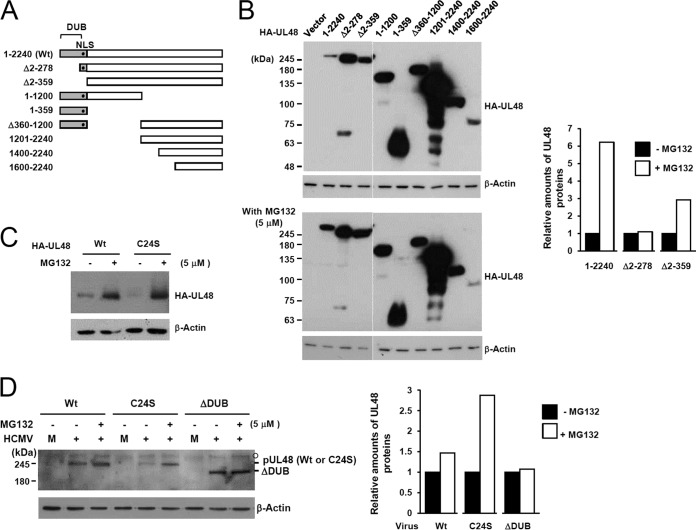
To investigate the structural determinants of pUL48 for protein stability, we produced a series of N- and C-terminal truncation mutants and an internal deletion mutant (Fig. 4A). First, the expression levels of the mutants were compared with that of wild-type pUL48 (2,240 amino acids). We found that the level of pUL48 was much lower than those of other mutant proteins tested, except for the smallest C-terminal fragment (region 1600–2400) and that, in particular, a deletion of the N-terminal DUB domain in Δ2–278 (ΔDUB) and Δ2-359 (ΔDUB/NLS) mutants led to a substantial increase in the protein level (Fig. 4B, top). Treatment with MG132, a proteasome inhibitor, increased the level of wild-type pUL48 while minimally affecting those of the mutant proteins tested (Fig. 4B, bottom). These results suggest that pUL48 is subjected to proteasomal degradation, probably in a manner dependent on the DUB domain.
FIG 4.
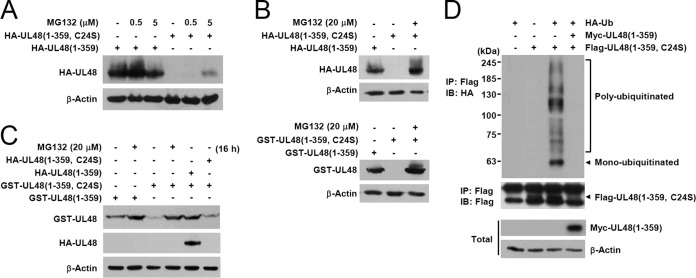
Expression levels of wild-type UL48 and its truncation variants. (A) The structures of wild-type or mutant UL48 proteins expressed in these experiments are illustrated. The DUB region is indicated as gray boxes, while the location of NLS is indicated by closed circles. (B and C) 293T cells were transfected with plasmids encoding HA-tagged wild-type UL48 (2,240 amino acids) or truncated mutant proteins. At 8 h after transfection, culture medium was replaced with fresh medium containing DMSO or 5 μM MG132 in DMSO. After further incubation for 36 h, cell lysates were prepared and subjected to SDS-PAGE (8% in panel B and 6% in panel C) and immunoblotting with anti-HA and anti-β-actin antibodies. (D) HF cells were mock infected (M) or infected with wild-type virus (Wt) or C24S or ΔDUB mutant virus (1 × 108 genome copies per 105 cells) for 9 days. Cells were untreated or treated with 5 μM MG132 for 24 h prior to cell harvest. Total cell lysates were prepared, and immunoblot analysis was performed with anti-UL48 antibody. The relative amounts of UL48 proteins after normalization with β-actin are shown as graphs.
To determine whether the DUB activity affects the stability of pUL48, the levels of wild-type and catalytically inactive C24S mutant proteins were directly compared in transfected cells. The results showed that the C24S mutant protein level was lower than the wild-type pUL48 level, but both were effectively increased to a comparable level by MG132 treatment, suggesting a possible role for the UL48 DUB activity in regulating its own stability (Fig. 4C). Similar stabilization of pUL48 protein after MG132 treatment was observed in virus-infected cells. When HF cells infected with wild-type or mutant (C24S or ΔDUB) viruses were untreated or treated with MG132, the levels of wild-type pUL48 and its C24S mutant proteins were increased by 1.5-fold and 3-fold, respectively, after MG132 treatment; however, the level of the ΔDUB mutant was not significantly affected (Fig. 4D). These results support the notion that the DUB domain is involved in the regulation of pUL48 degradation by proteasomes. In particular, our observation that deletion of the DUB sequence from pUL48 (in the Δ2–278 construct or in ΔDUB virus) increased its metabolic stability, whereas killing the DUB activity with the C24S point mutation made it less stable, suggested that the region of amino acids 2 to 278 may contain ubiquitin attachment sites.
This difference between wild-type and C24S mutant proteins regarding protein stability became more apparent when their stabilities were compared in the N-terminal DUB domain (359-amino-acid) background. The level of the C24S mutant version of HA-UL48(1–359) was much lower than that of the wild-type fragment, but it was partially restored by treatment with 5 μM MG132 for 5 h (Fig. 5A). This restoration was more apparent with MG132 treatment at a higher concentration. When transfected cells were treated with 20 μM MG132 for 16 h, the levels of C24S mutant versions of HA- or GST-UL48(1–359) were increased to levels comparable to those of HA- or GST-UL48(1–359) fragments (Fig. 5B).
FIG 5.
Ubiquitination and deubiquitination of UL48 DUB. (A) 293T cells were transfected with plasmids encoding the HA-tagged UL48(1–359) (wild-type or C24S mutant), as indicated. At 24 h after transfection, cells were untreated or treated with 0.5 or 5 μM MG132 for 5 h before cell harvest. Cell lysates were prepared and subjected to SDS-PAGE (8%) and immunoblotting with anti-HA or anti-β-actin antibodies. (B) Cells were transfected with HA- or GST-tagged UL48(1–359) (wild type or C24S mutant), as indicated. At 24 h, cells were left untreated or treated with 20 μM MG132 for 16 h prior to harvesting. Immunoblotting was performed as in panel A with anti-HA, anti-GST, or anti-β-actin antibodies. (C) Cells were transfected with the HA- or GST-UL48(1–359) (wild-type or C24S mutant) plasmids, as indicated. At 24 h, cells were untreated or treated with MG132 and immunoblotted as in panel B. (D) Cotransfection/ubiquitination assays. Cells were cotransfected with plasmids encoding myc-UL48(1–359), its C24S mutant version, and HA-Ub, as indicated. At 48 h, total cell lysates were prepared (see Materials and Methods). Immunoprecipitation (IP) was performed with anti-Flag antibody, followed by immunoblotting (IB) with anti-HA antibody. The level of myc-UL48(1–359) in total cell lysates was determined by immunoblotting with anti-myc antibody.
We next tested whether the DUB activity of pUL48 can regulate its own stability. In cotransfection assays, the low expression level of the GST-UL48(1–359) C24S mutant was restored by expression of HA-UL48(1–359), but not by expression of its C24S mutant version (Fig. 5C). This result indicates that the UL48 DUB is able to protect against its own degradation by proteasomes in trans. We further investigated whether the DUB domain contains lysine residues that are modified by ubiquitin. In cotransfection/ubiquitination assays, the C24S mutant version of Flag-UL48(1–359) was effectively monoubiquitinated or polyubiquitinated with different lengths and these ubiquitin monomers or polymers could be completely cleaved by expression of myc-UL48(1–359) (Fig. 5D). Taken together, our results demonstrate that the DUB domain of pUL48 contains ubiquitination sites and that its DUB activity can reduce the proteasomal degradation of pUL48 in trans.
N-terminal determinants of pUL48 for intracellular localization.
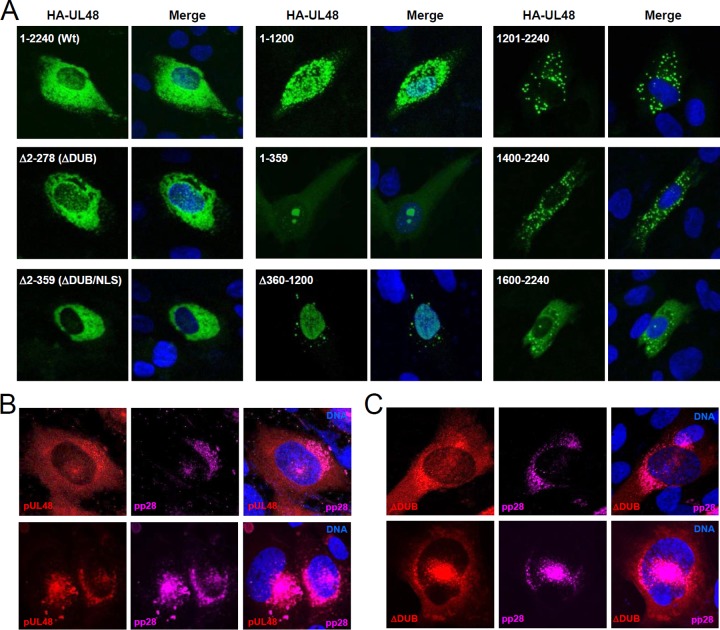
To determine the structural requirement for intracellular localization of pUL48, the HA-tagged mutant UL48 proteins with a truncation or an internal deletion described in Fig. 4A were expressed in HF cells via transfection and their localization patterns were compared with that of wild-type pUL48 (Fig. 6A). By confocal microscopy analysis, we found that wild-type pUL48 is primarily localized in the cytoplasm but is also distributed in the nucleus. The pattern of the Δ2–278 (ΔDUB) mutant was similar to that of the wild type, demonstrating that a deletion of the N-terminal DUB domain does not largely affect the intracellular localization of pUL48. However, as expected, the Δ2–359 (ΔDUB/NLS) mutant, which has a deletion of both DUB and NLS, was strictly cytoplasmic. We also found that the N-terminal half of UL48 (in region 1–1200) lost its typical localization pattern, showing more punctate forms both in the nucleus and in the cytoplasm. The N-terminal 359-amino-acid fragment encompassing the DUB and the NLS largely accumulated in the nucleus as a punctate form and also accumulated in the nucleolus, although it was also found as a cytoplasmic, diffuse form. The UL48(Δ360–1200) protein, which has a deletion of the internal region downstream from the DUB/NLS, lost its typical cytoplasmic distribution and accumulated in the nucleus as a diffuse form. The C-terminal half (in region 1200–2400) and the shorter C-terminal forms (in regions 1400–2400 and 1660–2400) were largely distributed as cytoplasmic punctate forms. Collectively, our results from transfection analysis indicate that both the NLS and the internal region downstream from the NLS are required for the typical nuclear and cytoplasmic localization of pUL48 and that the presence of the DUB domain does not appear to significantly affect the intracellular localization of pUL48.
FIG 6.
Localization patterns of wild-type and mutant UL48 proteins. (A) HF cells were transfected via electroporation with plasmids encoding wild-type UL48 or mutant proteins as indicated. At 48 h, cells were fixed with methanol and labeled with FITC-conjugated anti-HA antibody. A mounting solution containing Hoechst dye was used to stain cell nuclei. The representative images from confocal microscopy are shown. (B and C) HF cells were infected with wild-type HCMV or ΔDUB mutant virus (1 × 108 genome copies) for 72 h. Cells were fixed in methanol, and the confocal double-label IFA for pUL48 and pp28 was performed. Rhodamine Red-X-labeled IgG and Alexa Fluor 647-labeled IgG were used as secondary antibodies to detect UL48 proteins and pp28 proteins, respectively. Two representative images for wild-type virus (B) and for ΔDUB virus (C) are shown.
We also determined the localization patterns of wild-type and DUB-deleted mutant pUL48 proteins in virus-infected cells. Staining of infected cells with anti-peptide UL48 antibody showed that pUL48 proteins were distributed primarily in the cytoplasm as well as the nucleus and that some pUL48 proteins were colocalized with pp28 (Fig. 6B). Consistent with the results from the transfection assays, the ΔDUB mutant protein showed a similar distribution pattern to that of wild-type protein, indicating that the DUB domain does not significantly contribute to the intracellular localization of pUL48 (Fig. 6C). We also observed that the localization pattern of the C24S mutant was similar to that of wild-type pUL48 (data not shown).
Evaluation of the requirement of the DUB domain for the interactions of pUL48 with virion proteins.
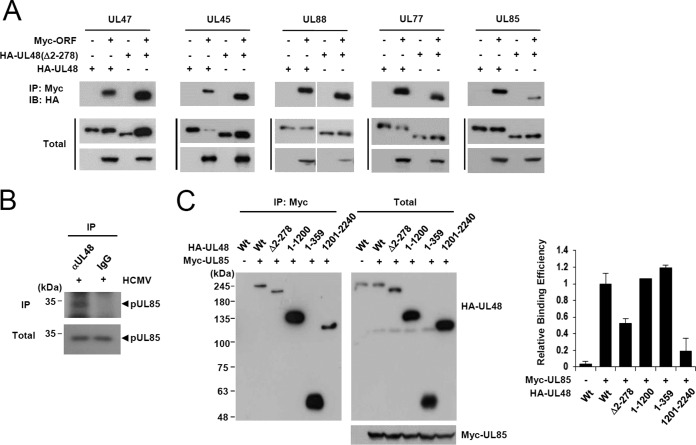
Because pUL48 is the largest tegument protein that is closely associated with the capsid, it is expected to associate with several viral tegument and capsid proteins. It was previously reported that pUL48 interacts with several tegument proteins such as pUL47 (12, 39, 40), pUL45 (41), pUL88 (40, 41), and pUL69 (40). In yeast two-hybrid interaction assays using the GAL4-A/UL48 (full-length) fusion protein as a bait, we also found that pUL48 interacts with pUL47, pUL45, and pUL88 (data not shown). In similar assays, we also newly identified two capsid proteins, pUL77, a capsid vertex-associated protein involved in DNA encapsidation (42, 43), and pUL85, a minor capsid protein, as putative pUL48-interacting proteins (data not shown). To validate these interactions, 293T cells were cotransfected with plasmids encoding HA-UL48 and myc-ORF, and co-IP assays were performed. The ΔDUB UL48 mutant was also employed in co-IP assays to address whether the DUB domain is involved in any of these interactions. The results showed that pUL48 indeed interacts with pUL47, pUL45, pUL88, pUL77, and pUL85, but the DUB domain appears to contribute to only pUL85 binding (Fig. 7A).
FIG 7.
Co-IP assays testing the interactions of wild-type and DUB-deleted mutant pUL48 proteins with other virion proteins. (A) 293T cells were cotransfected with plasmids encoding HA-UL48 (wild type or Δ2–278), myc-UL47, myc-UL45, myc-UL88, myc-UL77, or pUL85 as indicated. At 48 h, total cell lysates were immunoprecipitated with anti-myc antibody, followed by immunoblotting with anti-HA antibody. The levels of UL48 proteins and other HCMV proteins in total cell lysates were also determined by immunoblotting. (B) HF cells were infected with HCMV at an MOI of 3. At 72 h after infection, total cell lysates were prepared and immunoprecipitated with anti-UL48 antibody or IgG (as a control), followed by immunoblotting with anti-UL85 antibody. The levels of pUL85 in total cell lysates are also shown. (C) 293T cells were cotransfected with plasmids expressing HA-UL48 (wild type or mutant) and myc-UL85 as indicated, and co-IP assays were performed as in panel A. The relative binding efficiency of wild-type and mutant pUL48 to pUL85 in panel C is shown as graphs. The results shown are the averages with error bars from two independent experiments.
Since the antibody for pUL85 is available, the interaction of pUL48 with pUL85 was further evaluated by co-IP assays in virus-infected cells. The result confirmed that pUL48 indeed interacts with pUL85 during virus infection (Fig. 7B). The region of pUL48 involved in pUL85 binding was further mapped using cotransfection/co-IP assays. We found that a deletion of the N-terminal half of full-length pUL48 protein reduced its pUL85 binding activity to 20% of that of wild-type protein and that a deletion of the DUB domain reduced its pUL85 binding activity to 50% of that of wild-type protein (Fig. 7C). These results demonstrate that the N-terminal DUB domain of pUL48 promotes its interaction with pUL85.
The DUB domain of pUL48 contributes to virion stabilization and efficient virus entry.
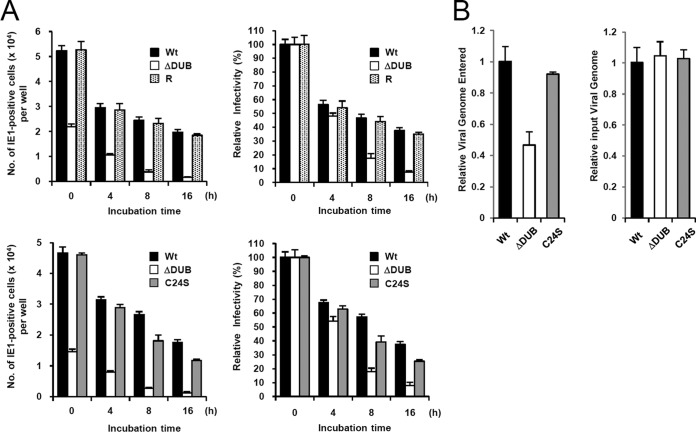
Since pUL48 is tightly associated with virion and is thought to play a role in virus entry, we investigated the role of the DUB domain in virion stability and virus entry. First, HF cells were infected with equivalent amounts of wild-type, ΔDUB mutant, and ΔDUB mutant revertant viruses that were preincubated at 37°C for 4, 8, or 16 h and the levels of IE1-positive cells were counted at 24 h after infection. The results showed that the ΔDUB mutant virus more rapidly lost infectivity than the wild-type and revertant viruses, suggesting that the DUB domain contributes to virion stability (Fig. 8A, top). To determine whether the differences observed in the ΔDUB mutant virus relate to loss of DUB enzymatic activity or to deletion of the amino portion of pUL48, the effects of preincubation at 37°C on the infectivity of the wild-type, ΔDUB mutant, and C24S mutant viruses were directly compared. We found that while the ΔDUB virus lost infectivity more rapidly than wild-type virus, the wild-type and C24S viruses showed similar patterns of infectivity loss (Fig. 8A, bottom). These results indicate that the N-terminal region of pUL48 containing the DUB domain, rather than the DUB catalytic activity, is important for virion stability.
FIG 8.
Comparison of virion stability and virus entry between wild-type and ΔDUB mutant viruses. (A) HF cells (1 × 105) in 24 wells were infected with equivalent amounts of wild-type virus or the ΔDUB mutant and its revertant viruses (5 × 107 copies of the viral genome, corresponding to an MOI of 0.5 for wild-type virus) that were incubated at 37°C for the indicated times (top). HF cells were also similarly infected with equivalent amounts of wild-type, ΔDUB mutant, and C24S mutant viruses (bottom). At 24 h after infection, the number of IE1-positive cells was counted via infectious center assays (left). Relative infectivity is also shown as graphs (right). (B) HF cells (2 × 105) were infected with equivalent amounts of wild-type, ΔDUB, or C24S mutant viruses (2 × 107 copies of the viral genome) in serum-free media for 1 h. Cells were washed with PBS, trypsinized, and collected by centrifugation. Total DNA was isolated from the cells, and the copy numbers of viral genomes were determined by qPCR (left). Relative amounts of input viral genomes were determined by qPCR of the viral inoculum (right).
We also investigated whether deletion of the DUB domain or inactivation of the DUB catalytic activity affects the efficiency of virus entry. HF cells were infected with equivalent amounts of wild-type, ΔDUB mutant, or C24S mutant viruses in serum-free media for 1 h, and the levels of intracellular viral DNA were determined by qPCR. The results showed that the DUB virus less efficiently entered the cell than the wild-type and C23S mutant viruses (Fig. 8B). This result indicates that the DUB region of pUL48 is required for efficient virus entry in a manner independent of its DUB activity.
DISCUSSION
Two functions associated with the N-terminal region of pUL48 tegument protein have been found to influence viral growth: the DUB activity moderately promotes viral growth in cultured fibroblasts (10, 11), and the N-terminal region contains the NLS that is indispensable for viral growth (22). The location of the NLS of pUL48 adjacent to the N-terminal DUB domain is conserved in all herpesvirus subfamilies (23). In this study, we showed that a deletion of the N-terminal 359-amino-acid region encompassing both DUB and NLS (in ΔDUB/NLS virus) completely abrogated viral growth and that a deletion of the DUB domain but not the NLS (as in ΔDUB virus) also substantially reduced viral growth by 100-fold at low MOI, compared to wild-type virus. We previously showed that a DUB catalytic site mutant virus encoding UL48(C24S) replicated to titers about 10-fold lower than that in wild-type virus (11). Therefore, our analysis using the ΔDUB virus indicates for the first time a DUB activity-independent role of the N-terminal region of pUL48 in the viral replication cycle.
Considering a delayed expression of viral IE genes in ΔDUB virus infection, the ΔDUB virus appears to have a defect early in the replication cycle. There are several possible explanations. First, given that deubiquitination is involved in the regulation of diverse immune responses (44), it is possible that the ubiquitin binding activity of pUL48 via the DUB domain, as well as the DUB activity, is important for the regulation of host immune responses by HCMV. Although the targets for the UL48 DUB are not known, several key components of the immune signaling pathways have been identified as cellular targets for other herpesviral DUBs (45): HSV-1 pUL36 inhibits interferon beta production by removing polyubiquitin chains from TRAF3 (46), Epstein-Barr virus (EBV) BPLF1 deubiquitinates TRAF6 to inhibit NF-κB signaling (47), and Kaposi's sarcoma-associated herpesvirus (KSHV) ORF64 suppresses RIG-I-mediated interferon signaling by reducing RIG-I ubiquitination (48). Second, a deletion of the DUB domain adjacent to the NLS may affect the pUL48-associated activity in routing the capsid to the nuclear pore, which requires the functional NLS, as shown in a recent study using HSV-1 pUL36 (20). However, it appears that at least the nuclear transport of the newly synthesized pUL48 protein does not require the DUB domain, since the nuclear and cytoplasmic localization patterns of wild-type and ΔDUB proteins were indistinguishable in our study. Third, it is also conceivable that a deletion of the N-terminal DUB-containing region of pUL48 causes changes in the interaction of pUL48 with other virion proteins or in the composition of tegument proteins during virion assembly and that this may lead to a delay in virus entry or initiation of viral gene expression. Indeed, the DUB domain was necessary for tight binding of pUL48 with a minor capsid protein. It is particularly interesting that deletion of the N-terminal DUB domain, rather than inactivation of its DUB activity, leads to a decrease in virion stability. This finding suggests that the specific interaction of pUL48 with virion proteins through the N-terminal region may be required for optimal virion composition that contributes to virion stability. It cannot be excluded that the N-terminal region of pUL48 may also be involved in the late events of the replication cycle through interactions with the virion proteins involved in nuclear or cytoplasmic egress.
Like the C24S mutant virus (11), the ΔDUB virus displayed an MOI-dependent growth pattern. Both the ΔDUB and C24S mutant viruses showed reduced accumulation of viral IE proteins at low MOI. Our results showed that the C24S mutant virus entered fibroblast cells as efficiently as the wild-type virus. Therefore, in addition to the DUB domain that promotes virus entry, it appears that the DUB activity also contributes to efficient IE gene expression at low MOI after virus entry. At high MOI, the high level of viral tegument proteins may partly compensate for the lack of DUB activity.
Autocatalytic activity of the viral DUB was originally demonstrated in HSV-1 pUL36 (49). In the present study, we also found that the UL48 DUB activity regulates its own stability. We revealed that the N-terminal 399-amino-acid region encompassing both DUB and NLS contains lysine residues that are conjugated by ubiquitin and that the DUB activity can inhibit the formation of polyubiquitin chains in trans. Therefore, the regulation of stability of this large tegument protein by its own DUB activity appears to be conserved among herpesviruses. Although proteasomal degradation is likely dependent on the DUB domain, a C-terminally truncated protein (region 1–1200) containing the DUB was also more stable than the wild-type protein in cotransfected cells. We attribute this to their aberrant intracellular localization, which may interfere with the ubquitination process, as judged by the absence of MG132 effect. How the autocatalytic activity of DUB contributes to viral replication is not clear. However, it is conceivable that the proper maintenance of the pUL48 level in virus-infected cells may be important for productive viral infection. We found that the stability of the UL48(ΔDUB) protein was substantially increased compared to that of the wild-type protein when they were expressed alone by transfection; however, this difference was not observed in virus-infected cells, suggesting that pUL48 is stabilized in infected cells, probably through interactions with other viral or cellular proteins. Studies on the exact role of ubiquitination of pUL48 and its regulation by the DUB in the viral replication cycle are warranted.
Using confocal analysis of HA-tagged pUL48 proteins in transfected cells and pUL48 in virus-infected cells, we observed that a small amount of pUL48 is distributed in the nucleus. A deletion of the DUB domain itself did not largely affect the typical nuclear and cytoplasmic localization of pUL48. Interestingly, a mutant protein with an internal deletion (Δ360–1200) was found to accumulate primarily in the nucleus. Therefore, this internal region of pUL48 may be critical for cytoplasmic distribution. Furthermore, the function associated with the internal region appears to be essential for viral replication, since the Δ360–1200 virus was lethal in our bacmid-based analysis.
Recently, Huffmaster et al. reported that pUL36, a homolog of pUL48, of PRV contains ubiquitin addition sites and that both the DUB activity and modification by ubiquitin at a specific lysine reside play a role in neuroinvasion (50). They demonstrated that the DUB activity was dispensable for neurotropism but essential for neuroinvasion and that ubiquitin addition at a lysine residue (K442) was required for retrograde axonal transport. Although HCMV is not neurovirulent and the K442 in pUL36 does not seem to be conserved in pUL48, the results in the present study demonstrate that pUL48 is also ubiquitinated and the DUB activity can control its ubiquitination level. Whether ubiquitin attachment of pUL48 and its homologs in other beta- and gammaherpesviruses plays a role in viral pathogenesis warrants further investigation.
In this study, we demonstrate that the N-terminal DUB domain-containing region of pUL48 is required for the early phase of infection, independent of the DUB activity, that the DUB activity regulates its own protein stability, and that the internal region downstream from the NLS is essential for proper cytoplasmic localization of pUL48 and viral growth. Our results also demonstrate that the DUB domain of pUL48 is necessary for virion stabilization and efficient virus entry. Given that pUL48 is the largest HCMV protein with multiple functions, fine dissection of the functions associated with specific regions using several mutant viruses will be necessary.
ACKNOWLEDGMENTS
We thank Wade Gibson and Gary S. Hayward (Johns Hopkins University School of Medicine) for providing the UL48 antibody and the HCMV ORF library, respectively.
REFERENCES
- 1.Mocarski ES, Shenk T, Griffiths PD, Pass RF. 2013. Cytomegaloviruses, p 1960–2014. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Gibson W. 2008. Structure and formation of the cytomegalovirus virion. Curr Top Microbiol Immunol 325:187–204. [DOI] [PubMed] [Google Scholar]
- 3.Kalejta RF. 2008. Functions of human cytomegalovirus tegument proteins prior to immediate early gene expression. Curr Top Microbiol Immunol 325:101–115. [DOI] [PubMed] [Google Scholar]
- 4.Kalejta RF. 2008. Tegument proteins of human cytomegalovirus. Microbiol Mol Biol Rev 72:249–265. doi: 10.1128/MMBR.00040-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tandon R, Mocarski ES. 2012. Viral and host control of cytomegalovirus maturation. Trends Microbiol 20:392–401. doi: 10.1016/j.tim.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradshaw PA, Duran-Guarino MR, Perkins S, Rowe JI, Fernandez J, Fry KE, Reyes GR, Young L, Foung SK. 1994. Localization of antigenic sites on human cytomegalovirus virion structural proteins encoded by UL48 and UL56. Virology 205:321–328. doi: 10.1006/viro.1994.1648. [DOI] [PubMed] [Google Scholar]
- 7.Yu X, Shah S, Lee M, Dai W, Lo P, Britt W, Zhu H, Liu F, Zhou ZH. 2011. Biochemical and structural characterization of the capsid-bound tegument proteins of human cytomegalovirus. J Struct Biol 174:451–460. doi: 10.1016/j.jsb.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunn W, Chou C, Li H, Hai R, Patterson D, Stolc V, Zhu H, Liu F. 2003. Functional profiling of a human cytomegalovirus genome. Proc Natl Acad Sci U S A 100:14223–14228. doi: 10.1073/pnas.2334032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu D, Silva MC, Shenk T. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc Natl Acad Sci U S A 100:12396–12401. doi: 10.1073/pnas.1635160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Loveland AN, Kattenhorn LM, Ploegh HL, Gibson W. 2006. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: mutant viruses altered in its active-site cysteine or histidine are viable. J Virol 80:6003–6012. doi: 10.1128/JVI.00401-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim ET, Oh SE, Lee YO, Gibson W, Ahn JH. 2009. Cleavage specificity of the UL48 deubiquitinating protease activity of human cytomegalovirus and the growth of an active-site mutant virus in cultured cells. J Virol 83:12046–12056. doi: 10.1128/JVI.00411-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tullman JA, Harmon ME, Delannoy M, Gibson W. 2014. Recovery of an HMWP/hmwBP (pUL48/pUL47) complex from virions of human cytomegalovirus: subunit interactions, oligomer composition, and deubiquitylase activity. J Virol 88:8256–8267. doi: 10.1128/JVI.00971-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kattenhorn LM, Korbel GA, Kessler BM, Spooner E, Ploegh HL. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol Cell 19:547–557. doi: 10.1016/j.molcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 14.Schlieker C, Korbel GA, Kattenhorn LM, Ploegh HL. 2005. A deubiquitinating activity is conserved in the large tegument protein of the Herpesviridae. J Virol 79:15582–15585. doi: 10.1128/JVI.79.24.15582-15585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luxton GW, Lee JI, Haverlock-Moyns S, Schober JM, Smith GA. 2006. The pseudorabies virus VP1/2 tegument protein is required for intracellular capsid transport. J Virol 80:201–209. doi: 10.1128/JVI.80.1.201-209.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Radtke K, Kieneke D, Wolfstein A, Michael K, Steffen W, Scholz T, Karger A, Sodeik B. 2010. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog 6:e1000991. doi: 10.1371/journal.ppat.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shanda SK, Wilson DW. 2008. UL36p is required for efficient transport of membrane-associated herpes simplex virus type 1 along microtubules. J Virol 82:7388–7394. doi: 10.1128/JVI.00225-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaichick SV, Bohannon KP, Hughes A, Sollars PJ, Pickard GE, Smith GA. 2013. The herpesvirus VP1/2 protein is an effector of dynein-mediated capsid transport and neuroinvasion. Cell Host Microbe 13:193–203. doi: 10.1016/j.chom.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abaitua F, O'Hare P. 2008. Identification of a highly conserved, functional nuclear localization signal within the N-terminal region of herpes simplex virus type 1 VP1-2 tegument protein. J Virol 82:5234–5244. doi: 10.1128/JVI.02497-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abaitua F, Hollinshead M, Bolstad M, Crump CM, O'Hare P. 2012. A nuclear localization signal in herpesvirus protein VP1-2 is essential for infection via capsid routing to the nuclear pore. J Virol 86:8998–9014. doi: 10.1128/JVI.01209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jovasevic V, Liang L, Roizman B. 2008. Proteolytic cleavage of VP1-2 is required for release of herpes simplex virus 1 DNA into the nucleus. J Virol 82:3311–3319. doi: 10.1128/JVI.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brock I, Kruger M, Mertens T, von Einem J. 2013. Nuclear targeting of human cytomegalovirus large tegument protein pUL48 is essential for viral growth. J Virol 87:6005–6019. doi: 10.1128/JVI.03558-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hennig T, Abaitua F, O'Hare P. 2014. Functional analysis of nuclear localization signals in VP1-2 homologues from all herpesvirus subfamilies. J Virol 88:5391–5405. doi: 10.1128/JVI.03797-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bucks MA, O'Regan KJ, Murphy MA, Wills JW, Courtney RJ. 2007. Herpes simplex virus type 1 tegument proteins VP1/2 and UL37 are associated with intranuclear capsids. Virology 361:316–324. doi: 10.1016/j.virol.2006.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leelawong M, Lee JI, Smith GA. 2012. Nuclear egress of pseudorabies virus capsids is enhanced by a subspecies of the large tegument protein that is lost upon cytoplasmic maturation. J Virol 86:6303–6314. doi: 10.1128/JVI.07051-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desai PJ. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J Virol 74:11608–11618. doi: 10.1128/JVI.74.24.11608-11618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuchs W, Klupp BG, Granzow H, Mettenleiter TC. 2004. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J Virol 78:11879–11889. doi: 10.1128/JVI.78.21.11879-11889.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schipke J, Pohlmann A, Diestel R, Binz A, Rudolph K, Nagel CH, Bauerfeind R, Sodeik B. 2012. The C terminus of the large tegument protein pUL36 contains multiple capsid binding sites that function differently during assembly and cell entry of herpes simplex virus. J Virol 86:3682–3700. doi: 10.1128/JVI.06432-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das S, Ortiz DA, Gurczynski SJ, Khan F, Pellett PE. 2014. Identification of human cytomegalovirus genes important for biogenesis of the cytoplasmic virion assembly complex. J Virol 88:9086–9099. doi: 10.1128/JVI.01141-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee HR, Kim DJ, Lee JM, Choi CY, Ahn BY, Hayward GS, Ahn JH. 2004. Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J Virol 78:6527–6542. doi: 10.1128/JVI.78.12.6527-6542.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huh YH, Kim YE, Kim ET, Park JJ, Song MJ, Zhu H, Hayward GS, Ahn JH. 2008. Binding STAT2 by the acidic domain of human cytomegalovirus IE1 promotes viral growth and is negatively regulated by SUMO. J Virol 82:10444–10454. doi: 10.1128/JVI.00833-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green S, Issemann I, Sheer E. 1988. A versatile in vivo and in vitro eukaryotic expression vector for protein engineering. Nucleic Acids Res 16:369. doi: 10.1093/nar/16.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roth MB, Zahler AM, Stolk JA. 1991. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J Cell Biol 115:587–596. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim ET, Kim YE, Kim YJ, Lee MK, Hayward GS, Ahn JH. 2014. Analysis of human cytomegalovirus-encoded SUMO targets and temporal regulation of SUMOylation of the immediate-early proteins IE1 and IE2 during infection. PLoS One 9:e103308. doi: 10.1371/journal.pone.0103308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahn JH, Xu Y, Jang WJ, Matunis MJ, Hayward GS. 2001. Evaluation of interactions of human cytomegalovirus immediate-early IE2 regulatory protein with small ubiquitin-like modifiers and their conjugation enzyme Ubc9. J Virol 75:3859–3872. doi: 10.1128/JVI.75.8.3859-3872.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchini A, Liu H, Zhu H. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J Virol 75:1870–1878. doi: 10.1128/JVI.75.4.1870-1878.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahn JH, Jang WJ, Hayward GS. 1999. The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10). J Virol 73:10458–10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hyun JJ, Park HS, Kim KH, Kim HJ. 1999. Analysis of transcripts expressed from the UL47 gene of human cytomegalovirus. Arch Pharm Res 22:542–548. doi: 10.1007/BF02975323. [DOI] [PubMed] [Google Scholar]
- 39.Roby C, Gibson W. 1986. Characterization of phosphoproteins and protein kinase activity of virions, noninfectious enveloped particles, and dense bodies of human cytomegalovirus. J Virol 59:714–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.To A, Bai Y, Shen A, Gong H, Umamoto S, Lu S, Liu F. 2011. Yeast two hybrid analyses reveal novel binary interactions between human cytomegalovirus-encoded virion proteins. PLoS One 6:e17796. doi: 10.1371/journal.pone.0017796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phillips SL, Bresnahan WA. 2011. Identification of binary interactions between human cytomegalovirus virion proteins. J Virol 85:440–447. doi: 10.1128/JVI.01551-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cockrell SK, Sanchez ME, Erazo A, Homa FL. 2009. Role of the UL25 protein in herpes simplex virus DNA encapsidation. J Virol 83:47–57. doi: 10.1128/JVI.01889-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coller KE, Lee JI, Ueda A, Smith GA. 2007. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J Virol 81:11790–11797. doi: 10.1128/JVI.01113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun SC. 2008. Deubiquitylation and regulation of the immune response. Nat Rev Immunol 8:501–511. doi: 10.1038/nri2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwon KM, Ahn JH. 2013. Herpesvirus-encoded deubiquitinating proteases and their roles in regulating immune signaling pathways. J Bacteriol Virol 43:244–252. doi: 10.4167/jbv.2013.43.4.244. [DOI] [Google Scholar]
- 46.Wang S, Wang K, Li J, Zheng C. 2013. Herpes simplex virus 1 ubiquitin-specific protease UL36 inhibits beta interferon production by deubiquitinating TRAF3. J Virol 87:11851–11860. doi: 10.1128/JVI.01211-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saito S, Murata T, Kanda T, Isomura H, Narita Y, Sugimoto A, Kawashima D, Tsurumi T. 2013. Epstein-Barr virus deubiquitinase downregulates TRAF6-mediated NF-kappaB signaling during productive replication. J Virol 87:4060–4070. doi: 10.1128/JVI.02020-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inn KS, Lee SH, Rathbun JY, Wong LY, Toth Z, Machida K, Ou JH, Jung JU. 2011. Inhibition of RIG-I-mediated signaling by Kaposi's sarcoma-associated herpesvirus-encoded deubiquitinase ORF64. J Virol 85:10899–10904. doi: 10.1128/JVI.00690-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bolstad M, Abaitua F, Crump CM, O'Hare P. 2011. Autocatalytic activity of the ubiquitin-specific protease domain of herpes simplex virus 1 VP1-2. J Virol 85:8738–8751. doi: 10.1128/JVI.00798-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huffmaster NJ, Sollars PJ, Richards AL, Pickard GE, Smith GA. 2015. Dynamic ubiquitination drives herpesvirus neuroinvasion. Proc Natl Acad Sci U S A 112:12818–12823. doi: 10.1073/pnas.1512559112. [DOI] [PMC free article] [PubMed] [Google Scholar]