Abstract
We investigated naturally occurring variation within the major (L1) and minor (L2) capsid proteins of human papillomavirus genotype 45 (HPV45). Pseudoviruses (PsVs) representing HPV45 sublineages A1, A2, A3, B1, and B2 exhibited comparable particle-to-infectivity ratios and morphologies but demonstrated both increased (A2, A3, and B1) and decreased (B2) sensitivities to cross-neutralization by HPV vaccine antibodies compared to that of the A1 sublineage. Mutant PsVs identified HI loop residue 357 as being critical for conferring this differential sensitivity.
TEXT
The evolutionary mutation rate of the human papillomavirus (HPV) double-stranded DNA genome is low at ca. 2 × 10−8 base substitutions per site per year (1, 2), yet distinct genotypes and intragenotype variant lineages have arisen over time (3). HPV genotype 45 (HPV45) is closely related to HPV18 within the Alpha-7 species group and is associated with ca. 5% of cervical cancer cases worldwide (4, 5). Whole-genome sequence analysis of HPV45 strains has led to the delineation of distinct variant lineages (A and B) and sublineages (A1, A2, A3, B1, and B2) (3, 6, 7), with the possibility of a lineage C suggested from subgenomic sequences (8). Although firm data on their contribution to the risk of cervical disease progression are lacking, in part due to the low relative prevalences of individual lineages and sublineages in the population, current evidence does support some lineage-specific bias such that sublineage variant B2 (and possibly A3) appears to be overrepresented in patients with high-grade disease compared to controls (8–10). There may also be some geographical bias to the distribution of HPV45 sublineages (9). Intragenotypic variation occurs throughout the HPV genome, but the consequences of these polymorphisms on the functions of the resulting gene products are uncertain.
The HPV structural genes encode the major (L1) and minor (L2) capsid proteins. The L1 protein multimerizes to form the nonenveloped icosahedral viral capsid (comprising 72 L1 pentameric capsomers) that mediates attachment to host cells (11), while the L2 protein is essential for viral infectivity (12). Structural alterations of the external surface topography of L1 can be conferred by minor sequence differences between genotypes (13), supporting observations that almost all neutralizing monoclonal antibodies (MAbs) that target these external surfaces are type specific (14–17). Nevertheless, functional antibody cross-reactivity is a common feature of sera from recipients of the Cervarix (bivalent) and Gardasil (quadrivalent) vaccines (18–22) and may be responsible for conferring HPV vaccine-induced cross-protection (23).
It is reasonable to consider that lineage-specific variation in surface-exposed domains (7, 24, 25) may influence capsid recognition by HPV vaccine-derived antibodies. Single-cycle replication-incompetent pseudoviruses (PsVs) representing HPV16 L1, but not L2, variants (26) appear to exhibit little difference in their susceptibilities to type-specific antibodies elicited by HPV16 virus-like particles (VLP). We recently demonstrated that although PsVs incorporating HPV31 L1 and L2 lineage variants (A, B, and C) were susceptible to cross-neutralizing antibodies elicited by the Cervarix and Gardasil HPV vaccines, there were lineage-specific differences in sensitivity (27). Here we examine the potential impact of lineage-specific L1 and L2 HPV45 variation on sensitivity to cross-neutralizing antibodies elicited by the Cervarix and Gardasil HPV vaccines.
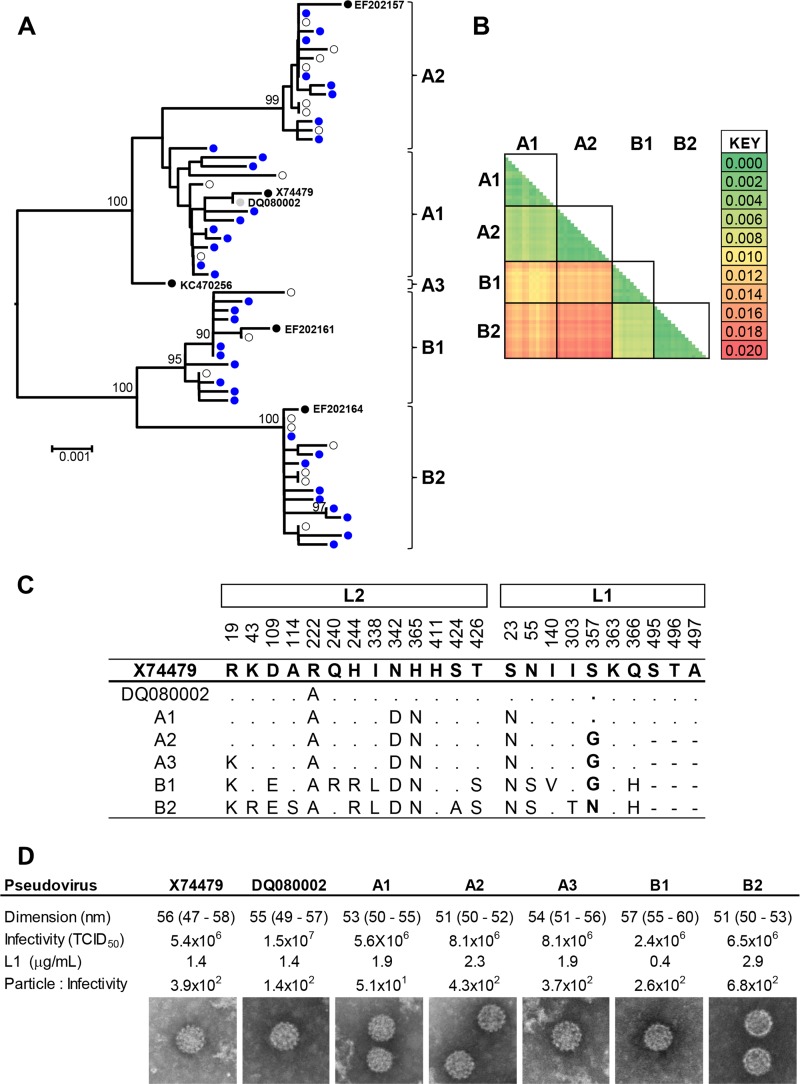
To improve estimates of the sublineage consensus sequences, we first generated 35 L1 (bp 5608 to 7149, numbered according to the HPV45 reference sequence [X74479]) and L2 (bp 4236 to 5627) sequences from samples collected from 16- to 24-year-old females previously confirmed as HPV45 DNA positive by the Linear Array HPV genotyping test (Roche) (28). Additional HPV45 L1 and L2 sequences were downloaded from the National Center for Biotechnology Information (NCBI [http://www.ncbi.nlm.nih.gov/] accession numbers X74479 [29], DQ080002 [30], EF202156 to EF202167 [6], and KC470250 to KC470260 [7]). X74479 is considered to be the reference sequence for the HPV45 genotype (3), while DQ080002 (30) was used as the basis of the HPV45 pseudovirus. These sequences are not identical, so for clarity, we refer to X74479 for sequence-based comparisons and to DQ080002 for comparison of biological data. The concatenated L2 L1 (2.9-kb) fragment contained sufficient numbers of diagnostic motifs to allow segregation of sequences into the sublineages A1, A2, A3, B1, and B2 defined (3, 6, 7) by whole-genome sequence analysis (Fig. 1A). Mean intralineage sequence diversity was 0.14% (standard deviation [SD], 0.09%), while mean interlineage sequence diversity was 1.27% (SD, 0.14%) (Fig. 1B). A consensus sequence for each sublineage was determined (Fig. 1C), and bicistronic psheLL vectors (31) containing codon-optimized HPV45 L1 and L2 genes representing these consensus sublineage variants were generated (20) (Fig. 1D). All HPV45 variant PsVs displayed similar particle sizes (median, 55 nm; interquartile range [IQR], 53 to 57 nm) and particle-to-infectivity ratios (27) (median, 2.9 × 102; IQR, 2.0 × 102 to 4.7 × 102).
FIG 1.
HPV45 L1 and L2 variation. (A) Neighbor-joining tree constructed (MEGA v6 [42]) from concatenated L1 and L2 nucleotide sequences. Sublineage (A1, A2, A3, B1, and B2) attribution is based upon whole-genome sequencing (representative sequences are included [3, 6, 7]) and is supported by bootstrap values of ≥90%. (B) Inter- and intralineage sequence diversity. A3 was omitted from this analysis due to a low representation of sequences. (C) Site-specific amino acids within the consensus L1 and L2 protein sequences used to generate PsV. (D) PsV preparations were characterized for particle dimension in nanometers (median [IQR]), infectivity, L1 concentration, and the resultant particle-to-infectivity ratio. TCID50, 50% tissue culture infective dose.
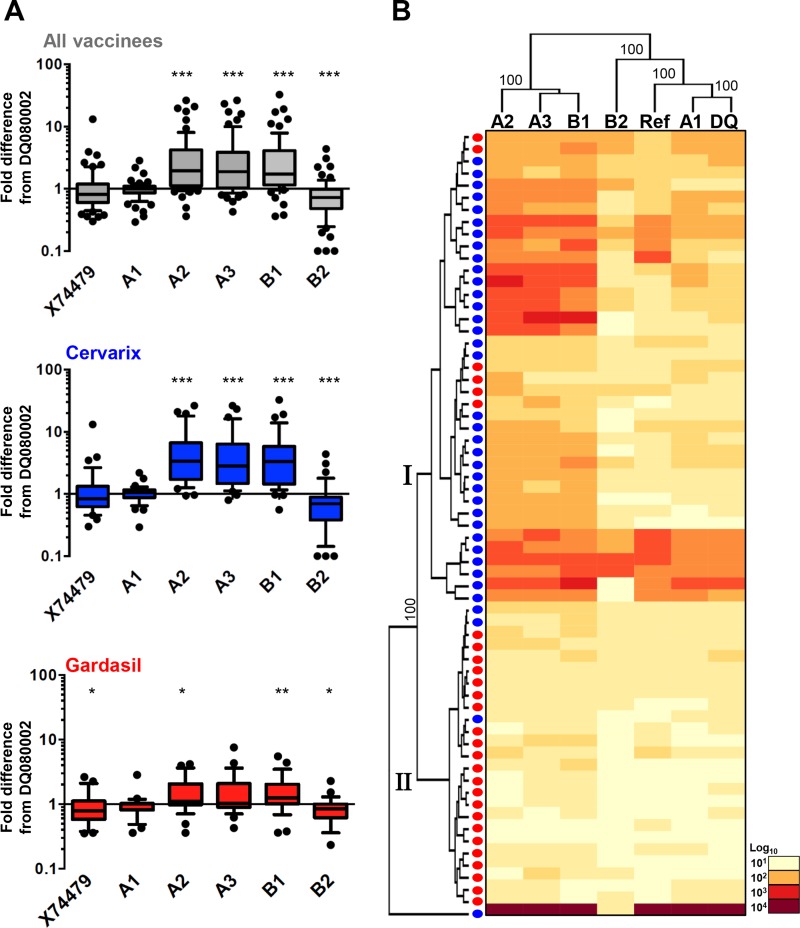
Sera from 12- to 15-year-old girls, collected following three doses of Cervarix or Gardasil HPV vaccine (22), were used to assess HPV45 sublineage variant sensitivity to cross-neutralizing antibodies (Fig. 2A) in a PsV-based neutralization assay (20, 32). PsVs based upon the reference sequence (A1, X74479) and a consensus A1 sequence displayed neutralization sensitivities similar to that of the commonly used HPV45 PsV (A1, DQ080002 [30]). These data suggest that L2 (R222A, N342D, and H365N) and L1 (S23N) variant residues (Fig. 1C) have no discernible impact on sensitivity to cross-neutralizing antibodies elicited by the HPV vaccines. PsV variants A2, A3, and B1 exhibited ca. 3-fold-increased sensitivity (P < 0.001; Wilcoxon signed-rank test) to cross-neutralizing antibodies, while variant B2 displayed slightly decreased sensitivity (P < 0.001), compared to that of the DQ080002 HPV45 PsV.
FIG 2.
Neutralization sensitivities of sublineage HPV45 L1 and L2 pseudoviruses. (A) Box (median, IQR) and whisker (10th and 90th percentiles) plots of the fold difference in neutralization titer from that of the DQ080002 PsV for each PsV (reference sequence A1 [X74479] and consensus constructs for A1, A2, A3, B1, and B2) for all (n = 65) vaccine sera tested and separately for Cervarix (n = 37) and Gardasil (n = 28) sera. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Wilcoxon paired signed-rank test). (B) Hierarchical clustering of log10 PsV neutralization titers (center, heat map) reordered according to serological (left) and PsV (top) dendrograms constructed from the resulting Euclidean distance matrices, with clusters supported by bootstrap values as indicated. Blue dots, Cervarix sera; red dots, Gardasil sera.
The source of the antibodies appeared to influence the magnitude of these sensitivity differences for some sublineage variants. For example, sublineage variant A2 was 3.4 (IQR, 1.8 to 6.4)-fold more sensitive than the DQ080002 PsV to sera from Cervarix recipients, compared to 1.1 (1.0 to 1.9)-fold for the sera from Gardasil recipients (P < 0.001) (Fig. 2A). As HPV45 antibody titers generated by the Gardasil vaccine are generally lower than those generated by the Cervarix vaccine (19, 22), we also compared the data from a subset of sera with low titers (<50). Median antibody titers neutralizing the DQ080002 HPV45 PsV were 37 (IQR, 34 to 42; n = 14) and 32 (28 to 42; n = 18) for the sera from Cervarix and Gardasil recipients, respectively (Mann-Whitney U test, P = 0.203), while titers neutralizing the A2 variant PsV were 172 (IQR, 89 to 205) and 39 (IQR, 27 to 50), respectively (P < 0.001). Similar differences were apparent for PsV variants A3 and B1 (data not shown).
We next subjected the log10-transformed PsV neutralization assay data to hierarchical clustering (http://www.hiv.lanl.gov/content/sequence/HEATMAP/heatmap.html) and identified two serum clusters; cluster I contained predominantly sera from Cervarix recipients (n = 33; Gardasil n = 6), while cluster II contained mostly sera from Gardasil recipients (n = 22; Cervarix n = 4) (Fisher's exact test, P < 0.001) (Fig. 2B). The variant PsV formed three distinct branches: one containing the A1 PsV (including the consensus A1 sequences [DQ080002 and X74479]), another containing PsVs A2, A3, and B1, and another containing PsV B2. These data support distinct profiles for HPV45 sublineage variant sensitivity to cross-neutralizing antibodies generated by the HPV vaccines and suggest that the antibody repertoires generated by the Cervarix and Gardasil HPV vaccines are not identical.
Although there were multiple amino acid residue differences between the L1 and L2 variant lineage sequences (Fig. 1C), residue 357 in the HI loop appeared to track differences in neutralizing antibody sensitivities displayed by the variant PsVs (Fig. 2). We next constructed three mutant HPV45 PsVs (A1 S357G, A2 G357S, and B2 N357G) to examine the potential impact of this HI loop residue on sensitivity to cross-neutralizing antibodies (Fig. 3). These mutant PsVs displayed particle-to-infectivity ratios similar to those of the variant PsVs (data not shown). These contextual substitutions support a strong influence of residue 357 in the HI loop on PsV sensitivity to cross-neutralizing antibodies elicited by the current HPV vaccines; specifically, a glycine at position 357 renders the PsV more sensitive to such antibodies, followed in decreasing order by serine and asparagine. For example, the S357G substitution in the context of PsV A1 increased sensitivity to sera from Cervarix recipients from a median of 182 (IQR, 94 to 288) to 1,018 (IQR, 774 to 1,155) (P < 0.001), while replacement of the glycine with serine in the context of PsV A2 (G357S) reduced sensitivity from 906 (IQR, 796 to 1,138) to 174 (IQR, 73 to 295) (P < 0.001).
FIG 3.
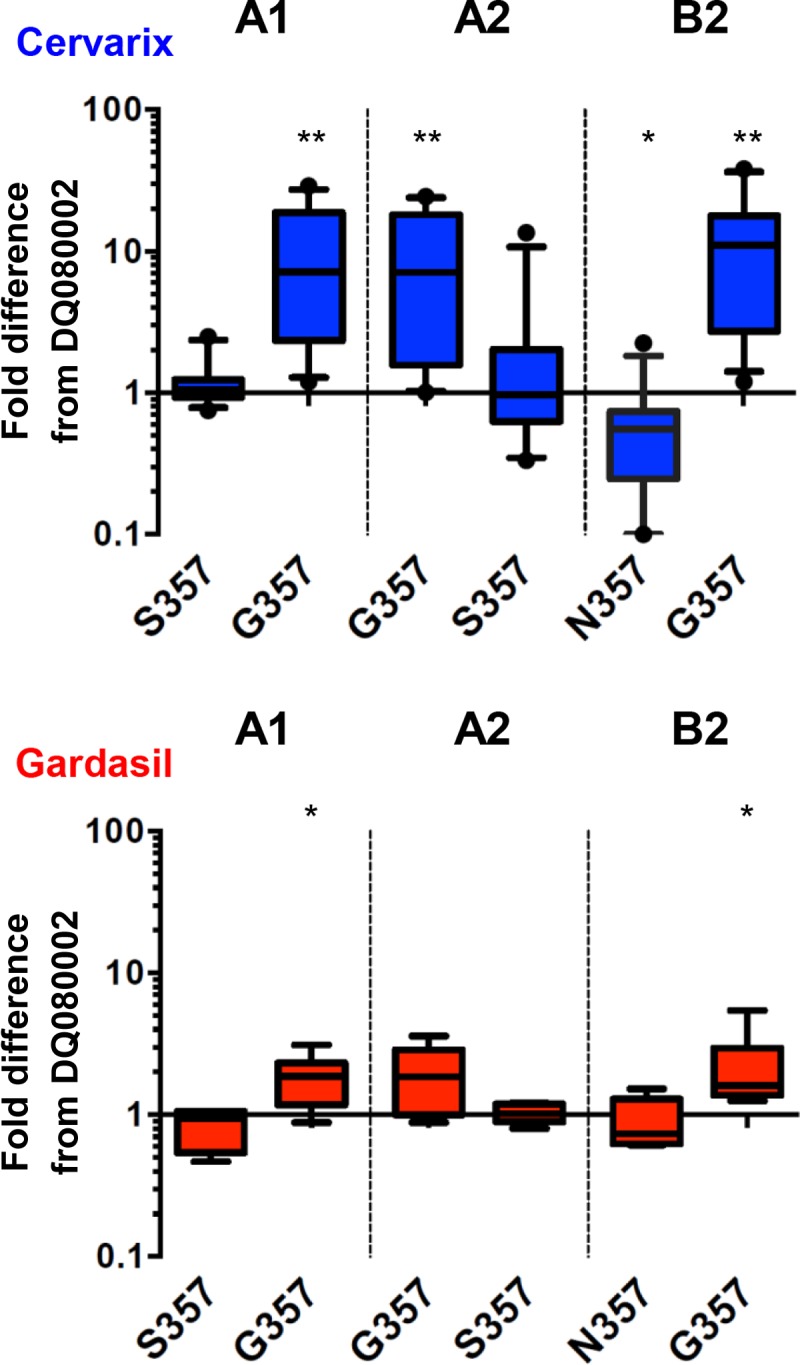
Neutralization sensitivities of mutant HPV45 L1 and L2 PsVs. Box (median, IQR) and whisker (10th and 90th percentiles) plots of fold differences in neutralization titer from that of the DQ080002 PsV for PsVs A1, A2, and B2 and their indicated mutants in Cervarix (n = 12) and Gardasil (n = 6) sera. *, P < 0.05; **, P < 0.01 (Wilcoxon paired sign-rank test).
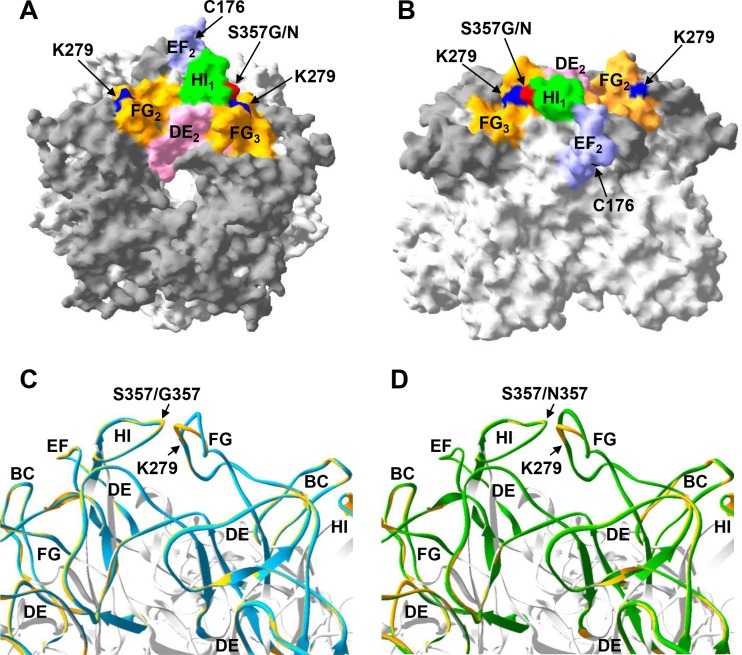
We next used the crystal structure of the HPV18 L1 pentamer (13) to create homology models (33, 34) of HPV45 A1, A2, and B2 L1 variants (http://swissmodel.expasy.org/), which were visualized using Swiss-PDP viewer v4.0 software (Fig. 4A and B). Pairwise model comparisons of HPV45 A2 and B2 with A1 were performed by superimposition (root mean square deviations of 0.20 Å and 0.16 Å, respectively) (Fig. 4C and D). The models predicted that substitution at HI residue 357 may influence local topography by shifting the adjacent FG loop by a mean ± standard error [SE] of 3.0 ± 1.2 Å (HPV45 A2) or 1.5 ± 0.8 Å (HPV45 B2) relative to its position in the HPV45 A1 model.
FIG 4.
Homology modeling of HPV45 sublineage variants. HPV45 sublineage variant homology models are based upon the HPV18 L1 pentamer crystal (Protein Databank [PDB] accession number 2R5I.1), with top (A) and side surface-filled (B) views shown for HPV45 A1 and with external loops indicated by dark-gray shading. External loops adjacent to the indicated HI loop (in this case, the HI loop of monomer 1 [HI1]) are indicated. Residues C176, K279, and S357 are also indicated for context. Superimposition models for HPV45 A2 (blue) (C) and HPV45 B2 (green) (D) relative to HPV45 A1 (gold) are used to indicate relative shifts in the position of the FG loop. HI loop residue 357 and FG loop residue K279 are indicated.
Notably, K279 in the FG loop is within 3.5 Å of residue 357, and the corresponding residue in HPV16 and HPV18 is involved in HPV binding to heparin sulfate (35). Recent cryo-electron microscopy studies have significantly improved the resolution of the antigenic domains of two classes of HPV16-neutralizing MAbs, exemplified by H16.U4 (36) and H16.V5 (15, 37). The H16.U4 epitope encompasses residues in the C-terminal portion of L1, which is involved in forming intercapsomer contacts via the “invading arm” disulfide bridge between Cys 428 and Cys 175 on adjacent capsomers of HPV16 L1. The epitopes of H16.V5-like MAbs include residues primarily within the DE and FG loops of the external surface of the capsomer, with contribution from residues within the BC, EF, and HI loops. The model predictions made herein for HPV45 suggest that the subtle structural alterations conferred by substitution of HI 357 occur within a domain proximal to the type-specific immunogenic domain on the external surface of HPV16 L1. These data suggest that variation within this region may influence the presentation of cross-neutralizing antibody epitopes in a way similar to the observation that subtle structural differences between genotypes in this region may bestow type-specific susceptibility to neutralizing MAbs (13).
The quality of the predicted models was demonstrated by their Qualitative Model Energy Analysis (QMEAN4) (38) Z-scores, which were −2.81, −2.91, and −2.93 for the A1, A2, and B2 models, respectively, and by their global model quality estimation (GMQE) score, which was 0.99 for all three models. HPV45 contains an insertion (S282, A283) within the FG loop relative to the HPV18 template, and although this is common to all HPV45 sequence variants examined here, it may nevertheless introduce a certain degree of measurement error into these model predictions.
Taken together, these data suggest that HPV45 lineage variants differ in their sensitivities to cross-neutralizing antibodies induced by the HPV vaccines through subtle alteration of L1's topography. HPV PsVs have been used widely to monitor antibody responses to vaccines and natural infection (18, 22, 32, 39, 40), as well as to elucidate steps in the entry process (41). Nevertheless differences between how PsVs behave in vitro and how authentic HPV45 lineage variants behave in vivo are uncertain, although this is a limitation of all PsV-based systems. Whether differences in PsV variant sensitivities noted here will influence the prevalence of individual variant lineages over time in countries that have introduced national vaccination programs will require further study. These data inform our understanding of the antigenicity of the HPV structural proteins and may be useful in guiding impact modeling of the current HPV vaccines and informing postvaccine surveillance programs.
Nucleotide sequence accession numbers.
The L1 and L2 sequences (KU049723 to KU049757) generated in this report have been deposited in GenBank under the indicated accession numbers.
ACKNOWLEDGMENTS
The sera from young girls who received Gardasil or Cervarix came from a study funded in part by the United Kingdom Department of Health Policy Research Programme (National Vaccine Evaluation Consortium, 039/0031; E.M.). A.F. was in receipt of traveling fellowships from the ExTRA and Erasmus Programs of the University of Milan-Bicocca, Monza, Italy.
We declare no conflicts of interest.
The views expressed in this publication are those of the authors and not necessarily those of the United Kingdom Department of Health.
We are indebted to John T. Schiller and Chris Buck (National Cancer Institute, Bethesda, MD) for access to the p45sheLL clone.
Funding Statement
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
REFERENCES
- 1.Rector A, Lemey P, Tachezy R, Mostmans S, Ghim SJ, Van Doorslaer K, Roelke M, Bush M, Montali RJ, Joslin J, Burk RD, Jenson AB, Sundberg JP, Shapiro B, Van Ranst M. 2007. Ancient papillomavirus-host co-speciation in Felidae. Genome Biol 8:R57. doi: 10.1186/gb-2007-8-4-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duffy S, Shackelton LA, Holmes EC. 2008. Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet 9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- 3.Burk RD, Harari A, Chen Z. 2013. Human papillomavirus genome variants. Virology 445:232–243. doi: 10.1016/j.virol.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li N, Franceschi S, Howell-Jones R, Snijders PJ, Clifford GM. 2011. Human papillomavirus type distribution in 30,848 invasive cervical cancers worldwide: variation by geographical region, histological type and year of publication. Int J Cancer 128:927–935. [DOI] [PubMed] [Google Scholar]
- 5.de Sanjose S, Quint WG, Alemany L, Geraets DT, Klaustermeier JE, Lloveras B, Tous S, Felix A, Bravo LE, Shin HR, Vallejos CS, de Ruiz PA, Lima MA, Guimera N, Clavero O, Alejo M, Llombart-Bosch A, Cheng-Yang C, Tatti SA, Kasamatsu E, Iljazovic E, Odida M, Prado R, Seoud M, Grce M, Usubutun A, Jain A, Suarez GA, Lombardi LE, Banjo A, Menendez C, Domingo EJ, Velasco J, Nessa A, Chichareon SC, Qiao YL, Lerma E, Garland SM, Sasagawa T, Ferrera A, Hammouda D, Mariani L, Pelayo A, Steiner I, Oliva E, Meijer CJ, Al-Jassar WF, Cruz E, Wright TC, Puras A, et al. 2010. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol 11:1048–1056. doi: 10.1016/S1470-2045(10)70230-8. [DOI] [PubMed] [Google Scholar]
- 6.Chen Z, DeSalle R, Schiffman M, Herrero R, Burk RD. 2009. Evolutionary dynamics of variant genomes of human papillomavirus types 18, 45, and 97. J Virol 83:1443–1455. doi: 10.1128/JVI.02068-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z, Schiffman M, Herrero R, DeSalle R, Anastos K, Segondy M, Sahasrabuddhe VV, Gravitt PE, Hsing AW, Burk RD. 2013. Evolution and taxonomic classification of alphapapillomavirus 7 complete genomes: HPV18, HPV39, HPV45, HPV59, HPV68 and HPV70. PLoS One 8:e72565. doi: 10.1371/journal.pone.0072565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xi LF, Schiffman M, Koutsky LA, Hughes JP, Winer RL, Mao C, Hulbert A, Lee SK, Shen Z, Kiviat NB. 2014. Lineages of oncogenic human papillomavirus types other than type 16 and 18 and risk for cervical intraepithelial neoplasia. J Natl Cancer Inst 106:dju270. doi: 10.1093/jnci/dju270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen AA, Heideman DA, Boon D, Gheit T, Snijders PJ, Tommasino M, Franceschi S, Clifford GM. 2014. Human papillomavirus 45 genetic variation and cervical cancer risk worldwide. J Virol 88:4514–4521. doi: 10.1128/JVI.03534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schiffman M, Rodriguez AC, Chen Z, Wacholder S, Herrero R, Hildesheim A, Desalle R, Befano B, Yu K, Safaeian M, Sherman ME, Morales J, Guillen D, Alfaro M, Hutchinson M, Solomon D, Castle PE, Burk RD. 2010. A population-based prospective study of carcinogenic human papillomavirus variant lineages, viral persistence, and cervical neoplasia. Cancer Res 70:3159–3169. doi: 10.1158/0008-5472.CAN-09-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buck CB, Day PM, Trus BL. 2013. The papillomavirus major capsid protein L1. Virology 445:169–174. doi: 10.1016/j.virol.2013.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang JW, Roden RB. 2013. L2, the minor capsid protein of papillomavirus. Virology 445:175–186. doi: 10.1016/j.virol.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bishop B, Dasgupta J, Klein M, Garcea RL, Christensen ND, Zhao R, Chen XS. 2007. Crystal structures of four types of human papillomavirus L1 capsid proteins: understanding the specificity of neutralizing monoclonal antibodies. J Biol Chem 282:31803–31811. doi: 10.1074/jbc.M706380200. [DOI] [PubMed] [Google Scholar]
- 14.Christensen ND, Cladel NM, Reed CA, Budgeon LR, Embers ME, Skulsky DM, McClements WL, Ludmerer SW, Jansen KU. 2001. Hybrid papillomavirus L1 molecules assemble into virus-like particles that reconstitute conformational epitopes and induce neutralizing antibodies to distinct HPV types. Virology 291:324–334. doi: 10.1006/viro.2001.1220. [DOI] [PubMed] [Google Scholar]
- 15.Lee H, Brendle SA, Bywaters SM, Guan J, Ashley RE, Yoder JD, Makhov AM, Conway JF, Christensen ND, Hafenstein S. 2015. A cryo-electron microscopy study identifies the complete H16.V5 epitope and reveals global conformational changes initiated by binding of the neutralizing antibody fragment. J Virol 89:1428–1438. doi: 10.1128/JVI.02898-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleury MJ, Touze A, Maurel MC, Moreau T, Coursaget P. 2009. Identification of neutralizing conformational epitopes on the human papillomavirus type 31 major capsid protein and functional implications. Protein Sci 18:1425–1438. doi: 10.1002/pro.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roth SD, Sapp M, Streeck RE, Selinka HC. 2006. Characterization of neutralizing epitopes within the major capsid protein of human papillomavirus type 33. Virol J 3:83. doi: 10.1186/1743-422X-3-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kemp TJ, Hildesheim A, Safaeian M, Dauner JG, Pan Y, Porras C, Schiller JT, Lowy DR, Herrero R, Pinto LA. 2011. HPV16/18 L1 VLP vaccine induces cross-neutralizing antibodies that may mediate cross-protection. Vaccine 29:2011–2014. doi: 10.1016/j.vaccine.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Einstein MH, Baron M, Levin MJ, Chatterjee A, Fox B, Scholar S, Rosen J, Chakhtoura N, Lebacq M, van der Most R, Moris P, Giannini SL, Schuind A, Datta SK, Descamps D. 2011. Comparison of the immunogenicity of the human papillomavirus (HPV)-16/18 vaccine and the HPV-6/11/16/18 vaccine for oncogenic non-vaccine types HPV-31 and HPV-45 in healthy women aged 18–45 years. Hum Vaccin 7:1359–1373. doi: 10.4161/hv.7.12.18282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Draper E, Bissett SL, Howell-Jones R, Edwards D, Munslow G, Soldan K, Beddows S. 2011. Neutralization of non-vaccine human papillomavirus pseudoviruses from the A7 and A9 species groups by bivalent HPV vaccine sera. Vaccine 29:8585–8590. doi: 10.1016/j.vaccine.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith JF, Brownlow M, Brown M, Kowalski R, Esser MT, Ruiz W, Barr E, Brown DR, Bryan JT. 2007. Antibodies from women immunized with Gardasil® cross-neutralize HPV 45 pseudovirions. Hum Vaccin 3:109–116. doi: 10.4161/hv.3.4.4058. [DOI] [PubMed] [Google Scholar]
- 22.Draper E, Bissett SL, Howell-Jones R, Waight P, Soldan K, Jit M, Andrews N, Miller E, Beddows S. 2013. A randomized, observer-blinded immunogenicity trial of Cervarix® and Gardasil® human papillomavirus vaccines in 12–15 year old girls. PLoS One 8:e61825. doi: 10.1371/journal.pone.0061825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehtinen M, Dillner J. 2013. Clinical trials of human papillomavirus vaccines and beyond. Nat Rev Clin Oncol 10:400–410. doi: 10.1038/nrclinonc.2013.84. [DOI] [PubMed] [Google Scholar]
- 24.Ahmed AI, Bissett SL, Beddows S. 2013. Amino acid sequence diversity of the major human papillomavirus capsid protein: implications for current and next generation vaccines. Infect Genet Evol 18:151–159. doi: 10.1016/j.meegid.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Z, Schiffman M, Herrero R, Desalle R, Anastos K, Segondy M, Sahasrabuddhe VV, Gravitt PE, Hsing AW, Burk RD. 2011. Evolution and taxonomic classification of human papillomavirus 16 (HPV16)-related variant genomes: HPV31, HPV33, HPV35, HPV52, HPV58 and HPV67. PLoS One 6:e20183. doi: 10.1371/journal.pone.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pastrana DV, Vass WC, Lowy DR, Schiller JT. 2001. NHPV16 VLP vaccine induces human antibodies that neutralize divergent variants of HPV16. Virology 279:361–369. doi: 10.1006/viro.2000.0702. [DOI] [PubMed] [Google Scholar]
- 27.Bissett SL, Godi A, Fleury MJ, Touze A, Cocuzza C, Beddows S. 2015. Naturally occurring capsid protein variants of human papillomavirus genotype 31 represent a single L1 serotype. J Virol 89:7748–7757. doi: 10.1128/JVI.00842-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howell-Jones R, de Silva N, Akpan M, Oakeshott P, Carder C, Coupland L, Sillis M, Mallinson H, Ellis V, Frodsham D, Robinson TI, Gill ON, Beddows S, Soldan K. 2012. Prevalence of human papillomavirus (HPV) infections in sexually active adolescents and young women in England, prior to widespread HPV immunisation. Vaccine 30:3867–3875. doi: 10.1016/j.vaccine.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 29.Delius H, Hofmann B. 1994. Primer-directed sequencing of human papillomavirus types. Curr Top Microbiol Immunol 186:13–31. [DOI] [PubMed] [Google Scholar]
- 30.Buck CB, Thompson CD, Roberts JN, Muller M, Lowy DR, Schiller JT. 2006. Carrageenan is a potent inhibitor of papillomavirus infection. PLoS Pathog 2:e69. doi: 10.1371/journal.ppat.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buck CB, Thompson CD. 2007. Production of papillomavirus-based gene transfer vectors. Curr Protoc Cell Biol Chapter 26:Unit 26.21. [DOI] [PubMed] [Google Scholar]
- 32.Pastrana DV, Buck CB, Pang YY, Thompson CD, Castle PE, FitzGerald PC, Kruger Kjaer S, Lowy DR, Schiller JT. 2004. Reactivity of human sera in a sensitive, high-throughput pseudovirus-based papillomavirus neutralization assay for HPV16 and HPV18. Virology 321:205–216. doi: 10.1016/j.virol.2003.12.027. [DOI] [PubMed] [Google Scholar]
- 33.Schwede T, Kopp J, Guex N, Peitsch MC. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. 2009. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc 4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 35.Dasgupta J, Bienkowska-Haba M, Ortega ME, Patel HD, Bodevin S, Spillmann D, Bishop B, Sapp M, Chen XS. 2011. Structural basis of oligosaccharide receptor recognition by human papillomavirus. J Biol Chem 286:2617–2624. doi: 10.1074/jbc.M110.160184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guan J, Bywaters SM, Brendle SA, Lee H, Ashley RE, Christensen ND, Hafenstein S. 2015. The U4 antibody epitope on human papillomavirus 16 identified by cryo-electron microscopy. J Virol 89:12108–12117. doi: 10.1128/JVI.02020-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guan J, Bywaters SM, Brendle SA, Lee H, Ashley RE, Makhov AM, Conway JF, Christensen ND, Hafenstein S. 2015. Structural comparison of four different antibodies interacting with human papillomavirus 16 and mechanisms of neutralization. Virology 483:253–263. doi: 10.1016/j.virol.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benkert P, Biasini M, Schwede T. 2011. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krajden M, Cook D, Yu A, Chow R, Su Q, Mei W, McNeil S, Money D, Dionne M, Palefsky J, Karunakaran K, Kollmann T, Ogilvie G, Petric M, Dobson S. 2014. Assessment of HPV 16 and HPV 18 antibody responses by pseudovirus neutralization, Merck cLIA and Merck total IgG LIA immunoassays in a reduced dosage quadrivalent HPV vaccine trial. Vaccine 32:624–630. doi: 10.1016/j.vaccine.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 40.Einstein MH, Baron M, Levin MJ, Chatterjee A, Edwards RP, Zepp F, Carletti I, Dessy FJ, Trofa AF, Schuind A, Dubin G. 2009. Comparison of the immunogenicity and safety of Cervarix and Gardasil human papillomavirus (HPV) cervical cancer vaccines in healthy women aged 18–45 years. Hum Vaccin 5:705–719. doi: 10.4161/hv.5.10.9518. [DOI] [PubMed] [Google Scholar]
- 41.Raff AB, Woodham AW, Raff LM, Skeate JG, Yan L, Da Silva DM, Schelhaas M, Kast WM. 2013. The evolving field of human papillomavirus receptor research: a review of binding and entry. J Virol 87:6062–6072. doi: 10.1128/JVI.00330-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]