Abstract
Background
Insertion sequences (IS) are small DNA segments capable of transposing within and between prokaryotic genomes, often causing insertional mutations and chromosomal rearrangements. Although several methods are available for locating ISs in microbial genomes, they are either labor-intensive or inefficient. Here, we use vectorette PCR to identify and map the genomic positions of the eight insertion sequences (IS1, 2, 3, 4, 5, 30, 150, and 186) found in E. coli strain CGSC6300, a close relative of MG1655 whose genome has been sequenced.
Results
Genomic DNA from strain CGSC6300 was digested with a four-base cutter Rsa I and the resulting restriction fragments ligated onto vectorette units. Using IS-specific primers directed outward from the extreme ends of each IS and a vectorette primer, flanking DNA fragments were amplified from all but one of the 37 IS elements identified in the genomic sequence of MG1655. Purification and sequencing of the PCR products confirmed that they are IS-associated flanking DNA fragments corresponding to the known IS locations in the MG1655 genome. Seven additional insertions were found in strain CGSC6300 indicating that very closely related isolates of the same laboratory strain (the K12 isolate) may differ in their IS complement. Two other E. coli K12 derivatives, TD2 and TD10, were also analyzed by vectorette PCR. They share 36 of the MG1655 IS sites as well as having 16 and 18 additional insertions, respectively.
Conclusion
This study shows that vectorette PCR is a swift, efficient, reliable method for typing microbial strains and identifying and mapping IS insertion sites present in microbial genomes. Unlike Southern hybridization and inverse PCR, our approach involves only one genomic digest and one ligation step. Vectorette PCR is then used to simultaneously amplify all IS elements of a given type, making it a rapid and sensitive means to survey IS elements in genomes. The ability to rapidly identify the IS complements of microbial genomes should facilitate subtyping closely related pathogens during disease outbreaks.
Background
Insertion sequences (IS) are small DNA segments capable of transposing within and between prokaryotic genomes and episomes, often causing insertional mutations and chromosomal rearrangements [1]. Identifying and mapping IS elements in microbial genomes is essential to understand their evolutionary significance [2-5]. So rapidly can IS elements move that even closely related laboratory strains commonly differ in the positions of their IS sequences [6,7]. A swift means to identify IS insertions might therefore allow isolates from specific disease outbreaks to be distinguished from other closely related strains.
Several methods have been used to identify the number and locations of IS elements in bacterial genomes, including Southern hybridization [3] and the inverse polymerase chain reaction (iPCR) [4,8,9]. Southern hybridization is rather time-consuming and requires additional procedures for localizing ISs. Inverse PCR, a commonly used PCR method for recovering unknown flanking sequences of a known target sequence, uses a library of circularized chromosomal DNA fragments as template and two outward primers located in each end of the known fragment for amplification [8]. However, when a target sequence has multiple genomic locations, the variously sized DNA circles formed are difficult to amplify simultaneously. Also, the length of each restriction DNA fragment containing a target sequence must be determined by Southern hybridization followed by sub-genomic fractioning before intramolecular ligation and PCR amplification [4,8,9]. These difficulties render Southern hybridization and iPCR impractical as techniques for quickly surveying repetitive elements in genomes.
Vectorette PCR (vPCR) [10,11] is another method used to amplify unknown sequences flanking a characterized DNA fragment. It involves cutting genomic DNAs with a restriction enzyme, ligating vectorettes to the ends, and amplifying the flanking sequences of a known sequence using primers derived from the known sequence along with a vectorette primer (Fig. 1). This technique has found many applications, including sequencing cosmid insert termini [10], identifying telomeres [12] and microsatellite sequences [13], mapping deletions, insertions, and translocations [14,15], and determining the 5' and 3' ends of mRNAs [16]. Here, we explore the efficiency of vPCR with regards to identifying and mapping IS elements in microbial genomes. We show that multiple copies of an IS are readily amplified using an IS specific primer in combination with a vectorette primer, and that their genomic locations are readily identified from the flanking DNA sequences.
Figure 1.
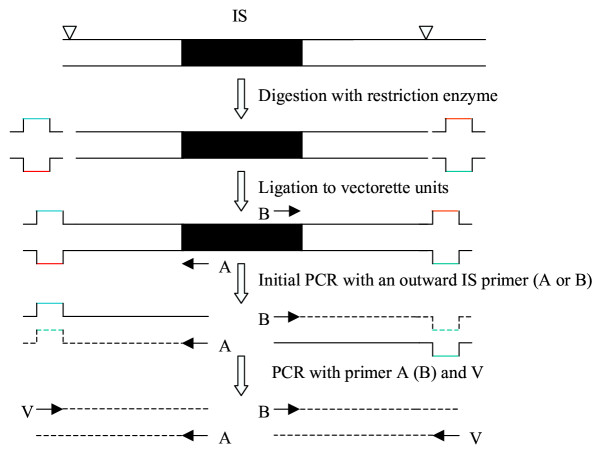
Vectorette PCR for amplification of IS franking sequences. The shadowed area represents the IS sequence. The solid lines indicate the flanking DNA sequences. ∇ indicates the restriction site. A and B are the outward IS-specific primers located at the ends of the IS. V is a vectorette primer.
Results and discussion
The IS insertions of CGSC6300
We used E. coli strain CGSC6300, a close relative of the sequenced strain MG1655, against which to test the efficiency and reliability of vPCR in detecting IS copies. IS insertion sites were identified by sequencing flanking DNA fragments amplified using outward IS-specific primers in combination with the vectorette primer. Based on the whole genome sequence of strain MG1655 [17], there are 37 ISelements, including 7 copies of IS1, 6 copies of IS2, 5 copies of IS3, 1 copy of IS4, 11 copies of IS5, 3 copies of IS30, 1 copy of IS150, and 3 copies of IS186. Our results for each IS in CGSC6300 are summarized in Table 1 and described as follows:
Table 1.
IS elements in closely related E. coli strains MG1655, CGSC6300, TD2 and TD10
| A-side | B-side | Strain | ||||||||
| Element | start (bp) | end (bp) | Flanking Genes | Direction | fragment sizea | fragment sizea | MG1655 | CGSC6300 | TD2 | TD10 |
| Known Elements | ||||||||||
| IS1-1 | 19811 | 20508 | b0020-b0023 | - | 272 | 199* | Pb | P | P | P |
| IS1-2 | 278402 | 279099 | b0263-b0266 | - | 174 | 191* | P | P | P | P |
| IS1-3 | 289873 | 290570 | b0273-b0276 | - | 171* | 577 | P | P | P | P |
| IS1-4 | 1049056 | 1049753 | b0987-b0989 | + | 281* | 139 | P | P | P | P |
| IS1-5 | 1976542 | 1977239 | b1892-b1895 | - | 452 | 226 | P | P | ||
| IS1-6 | 3581114 | 3581811 | b3443-b3446 | + | 712 | 1020 | P | P | P | P |
| IS1-7 | 4516095 | 4516370 | b4293-b4295 | + | 1354 | 575* | P | P | P | P |
| IS2-1 | 380530 | 381803 | b0359-b0362 | + | 308 | 841 | P | P | P | P |
| IS2-2 | 1465945 | 1467218 | b1401-b1405 | - | 194 | 203* | P | P | P | P |
| b1579 transposase | 1649536 | 1650732 | b1578-b1580 | + | 778 | P | P | P | P | |
| IS2-3 | 2066974 | 2068247 | b1995-b1998 | - | 287* | 444 | P | P | P | P |
| IS2-4 | 2994394 | 2995622 | b2859-b2862 | - | 589 | 219 | P | P | P | P |
| IS2-5 | 3184203 | 3185431 | b3043-b3046 | + | 410 | 672 | P | P | P | P |
| IS2-6 | 4495795 | 4497068 | b4271-b4274 | + | 204* | 163 | P | P | P | P |
| IS3-1 | 390963 | 391829 | b0371-b0373 | - | 489* | 142 | P | P | P | P |
| IS3-2 | 566361 | 567227 | b0540-b0542 | + | 787 | 346 | P | P | P | P |
| IS3-3 | 1093498 | 1094364 | b1025-b1027 | - | 1009 | 772 | P | P | P | P |
| IS3-4 | 2168554 | 2169420 | b2088-b2090 | + | 502 | 842 | P | P | P | P |
| IS3-5 | 314811 | 315677 | b0298-b0300 | + | 863 | 158* | P | P | P | P |
| IS4 | 4499671 | 4500999 | b4277-b4279 | - | 629 | 450 | P | P | P | P |
| IS5-1 | 273325 | 274341 | b0258-b0260 | - | 489* | 497 | P | P | P | P |
| IS5-2 | 573960 | 574976 | b0551-b0553 | - | 590 | 1605 | P | P | P | P |
| IS5-3 | 687220 | 688236 | b0655-b0657 | - | 348 | 420* | P | P | P | P |
| IS5-4 | 1394100 | 1395116 | b1330-b1332 | + | 569* | 471* | (P)c | (P) | (P) | (P) |
| IS5-5 | 1425770 | 1426750 | b1369-b1371 | - | 491 | 1543 | P | P | P | P |
| IS5-6 | 2064327 | 2065343 | b1993-b1995 | - | 747 | 1210 | P | P | P | P |
| IS5-7 | 2099917 | 2100933 | b2029-b2031 | - | 306 | 303 | P | P | P | P |
| IS5-8 | 2287085 | 2288101 | b2191-b2193 | - | 314* | 411 | P | P | P | P |
| IS5-9 | 3128193 | 3129209 | b2981-b2983 | + | 1085 | 672 | P | P | P | P |
| IS5-10 | 3363337 | 3364353 | b3217-b3219 | - | 207* | 1126 | P | P | P | P |
| IS5-11 | 3649812 | 3650828 | b3504-b3506 | - | 734* | 1335 | P | P | P | P |
| IS30-1 | 269827 | 270978 | b0255-b0257 | + | 720 | 314 | P | P | P | P |
| IS30-2 | 1467382 | 1468533 | b1401-b1405 | + | 305 | 297 | P | P | P | P |
| IS30-3 | 4505034 | 4506185 | b4283-b4285 | - | 405 | 766 | P | P | P | P |
| IS150 | 3718309 | 3719678 | b3556-b3559 | + | 195 | 176 | P | P | P | P |
| IS186-1 | 15445 | 16557 | b0015-b0017 | + | 503 | 1501 | P | P | P | P |
| IS186-2 | 607288 | 608400 | b0581-b0583 | + | 2204 | 417 | P | P | P | P |
| IS186-3 | 2512345 | 2513463 | b2393-b2395 | + | 764 | 667 | P | P | P | P |
| Additional Elements | ||||||||||
| IS1-a | 257908 | b0240 | - | 1488 | 738 | P | ||||
| IS1-b | 335460 | b0319-b0326 | - | 609 | 309 | P | ||||
| IS1-c | 1871063 | b1786 | - | 1211 | 484 | P | ||||
| IS1-d | 2037484 | b1970-b1971 | + | 617* | 1066 | P | P | |||
| IS1-e | 2623548 | b2502 | + | 282* | 368 | P | P | |||
| IS1-f | 2768501 | b2635 | + | 1608 | 544* | P | P | P | ||
| IS1-g | 3275070 | b3130 | + | 529 | 2117 | P | P | |||
| IS1-h | 4539642 | b4313 | - | 175* | 1119 | P | P | |||
| IS2-a | 1588558 | b1506 | + | 765 | 717 | P | P | |||
| IS2-b | 2927499 | b2796 | + | 138 | 368 | P | P | |||
| IS3-a | 331175 | b0314-b0315 | + | 1002* | 563 | P | ||||
| IS3-b | 838769 | b0805 | + | 450 | 870* | P | P | P | ||
| IS3-c | 2460317 | b2351 | + | 489* | 1642 | P | P | |||
| IS3-d | 3427623 | b3280 | - | 953 | 646 | P | P | |||
| IS3-e | 3776882 | b3604 | - | 1435 | 935* | P | P | P | ||
| IS3-f | 4466303 | b4242 | - | 640 | 935* | P | P | P | ||
| IS5-a | 1102866 | b1040-b1041 | + | 365 | 400 | P | P | |||
| IS30-a | 4115565 | b3927 | - | 692 | 186 | P | P | |||
| IS30-b | 2246187 | b2156 | - | 303* | 445 | P | P | P | ||
| IS186-a | 4541184 | b4314 | - | 606 | 534 | P | P | |||
a * indicates a fragment not recovered. b P, present. c Neither IS5-4 fragment is detected. Conventional PCR of gDNA confirms IS5-4 is present.
IS1
Eight and 6 PCR bands, obtained with primers IS1-A and IS1-B respectively, were observed on ethidium bromide-stained agarose gels (Fig. 2). All 7 IS1 insertion sites in the sequenced genome of MG1655 [17] were successfully identified by isolating and sequencing these fragments. Sequences obtained from both flanking sequences were used to locate 2 IS1 elements (IS1-5 and IS1-6). The remaining 5 IS1 locations were identified from single flanking sequences. Three additional IS1 elements (IS1-a in b0240, IS1-c in b1786, and IS1-f in b2635) were also found in CGSC6300.
Figure 2.
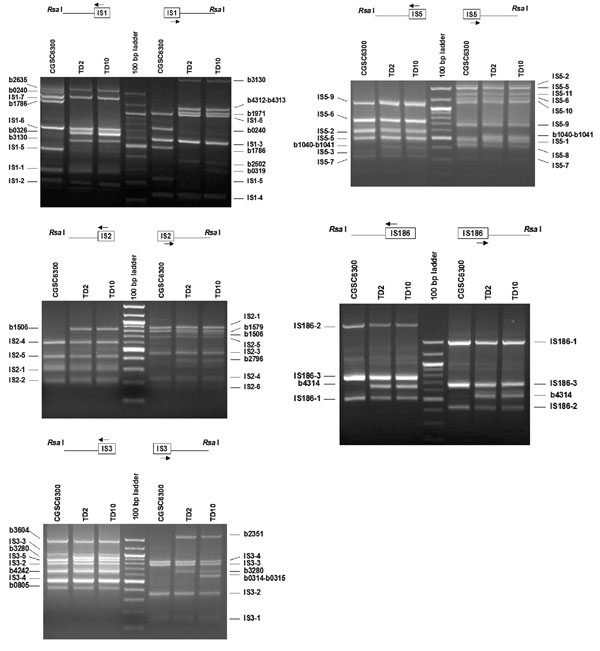
PCR amplification of IS flanking DNA from E. coli strains CGSC6300, TD2 and TD10. Results for IS1, 2, 3, and 5 and 186 are shown. Genomic DNA was digested with Rsa I, ligated with vectorette units and amplified by vPCR. Each panel shows the PCR products generated by two outward IS-specific primers (arrows) of an IS in combination with the vectorette primer. Flanking DNA fragments from both sides of each IS location were amplified. The PCR products were excised, purified, sequenced and identified from the genome sequence of E. coli strain MG1655 [17]. A PCR fragment flanking a known IS site in MG1655 is indicated by the element's name followed by an identifying numeral; for example, IS1-1 is one of 7 IS1 elements in the MG1655 genome. Additional flanking DNAs not found in MG1655 are labeled with the b# of the gene in which the IS is located. PCR products were separated in 1.4% agarose gels and stained with ethidium bromide. Intense bands in the 100 kb ladder correspond to 500 and 1000 bp.
IS2
Primers IS2-A and IS2-B produced 4 bands and 6 bands, respectively (Fig. 2). Three (IS2-1, IS2-4, and IS2-5) were located from both flanking sequences and the remaining 3 (IS2-2, IS2-3, and IS2-6) were located from one flanking sequence. Gene b1579, homologous to the IS2 transposase [17], was also amplified, sequenced and located in CGSC6300.
IS3
Three IS3 elements (IS3-2, -3, and -4) were each located by sequencing amplified flanking DNAs from both sides, and two (IS3-1 and IS3-5) were each located by sequencing a single flanking sequence (Fig. 2). Three additional IS3 elements were found at b0805 (IS3-b),b3604 (IS3-e), b4242 (IS3-f).
IS4
One IS4 was located based on flanking sequences amplified from both sides. No additional IS4 insertions were found.
IS5
Primers IS5-A and IS5-B produced 7 and 9 bands, respectively (Fig. 2). Purification and sequencing of these DNA fragments showed that they correspond to flanking sequences of IS5-2, -3, -5, -6, -7, -9 for the A-side and IS5-1, -2, -5, -6, -7, -8, -9, -10, -11 for the B-side. Fragments flanking either side of IS5-4 were not identified.
IS30
The three known IS30 insertions in MG1655/CGSC6300 were identified based on flanking sequences amplified from both sides, and an additional insertion was identified in b2156.
IS150
The one known IS150 insertion was identified and no other.
IS186
The three known IS186 insertions were identified based on flanking sequences amplified from both sides (Fig. 2).
Additional IS copies in laboratory strains
Several IS elements located in CGSC6300 are not found in the genomic sequence of MG1655 (Table 1). Lyophylized CGSC6300 was obtained from the E. coli Genetic Stock Center, Yale University, and is stored at our laboratory in 15% glycerol at -80°C. It seems likely that the additional IS transpositions arose after separation from the sequenced MG1655, but prior to arrival in our laboratory, probably during storage on agar slants at room temperature, a condition known to promote IS mobilization [6,7]. These results emphasize that the IS complement of each strain should be characterized prior to experimentation.
Two other E. coli K12 derivatives, TD2 and TD10, contain 16 and 18 additional IS insertions (Table 1), respectively. The two additional insertions found in TD10 are: the IS3-a insert between b0314 and b0315 and the IS1-b insert associated with a deletion between b0319 and b0326. Originally, TD2 and TD10 were constructed by P1 transduction of different lac operons into the Δlac of K12 derivative strain DD320 [18]. The IS insertion differences between these two strains probably arose when sequences flanking the lac operon were cotransduced during strain construction.
Reliability of technique
Theoretically, the number of flanking DNA fragments amplified with each IS-specific primer should equal the number of copies of each IS element in the genome. Also, the location of each IS copy should be identifiable from the two flanking DNA sequences. However, some copies of IS elements 1, 2, 3 and 5 were initially located by a single flanking sequence only. DNA fragments not recovered may have been masked by fragments of similar size, amplified from other genomic copies of the IS element. This is evidenced by bands in ethidium-stained agarose gels appearing broader and/or staining more intensely (see Fig. 2). While these bands produce clearly readable sequence in the ISs themselves, their flanking sequences are unreadable or show high noisy background, indicating the presence of multiple fragments of similar size (data not shown). In the case where flanking sequences were readable, we located one of the fragments – presumably the one that was amplified most efficiently.
Despite missing fragments, vectorette PCR provides a reliable estimate of the copy number of elements in a genome. Let the number of copies of the ith IS element be ni, and the number of unidentified flanking sequences be ui. Then the probability that an IS copy is not identified is simply a product of the probabilities of not obtaining either the A-side or the B-side sequences, qi = (ui/ni)A-side·(ui/ni)B-side. The expected number, x, of missing copies is determined by summing over all ni copies of each of the j = 8 elements in MG1655. Our data provide an estimate of  expected missing copies. In fact, only 1 copy was missed entirely. Even when digested by just a single four-cutter restriction enzyme, vectorette methodology is highly reliable with small error rates: 6.8% expected and 2.7% realized.
expected missing copies. In fact, only 1 copy was missed entirely. Even when digested by just a single four-cutter restriction enzyme, vectorette methodology is highly reliable with small error rates: 6.8% expected and 2.7% realized.
The actual error rates are even smaller. Our analysis is restricted to the 37 ISs found in the genomic sequence of MG1655; the 7 additional ISs in CGSC6300 were not used in the calculations even though they may serve to mask fragments and thereby increase the expected and observed error rates.
To determine the reliability of the technique when there are many more than 11 copies of an IS element in a genome requires estimating m, the maximum number of amplified fragments likely to be resolved per lane by agarose minigel electrophoresis. Only a small portion of the resolving power of an agarose gel is actually used because approximately 98% (approximately because the calculation  assumes equal base frequencies) of amplified fragments produced by a 4-base cutter restriction enzyme are less than 1 kb (excluding the IS and the vectorette). Hence, m is less than the maximum number of fragments physically capable of being resolved by agarose minigel electrophoresis.
assumes equal base frequencies) of amplified fragments produced by a 4-base cutter restriction enzyme are less than 1 kb (excluding the IS and the vectorette). Hence, m is less than the maximum number of fragments physically capable of being resolved by agarose minigel electrophoresis.
Consider m as the number of discrete positions that an amplified fragment might occupy. The probability that a particular position is not occupied given ni copies of an IS element i is  . The expected number of unoccupied positions is
. The expected number of unoccupied positions is  and the expected number of occupied positions (i.e. bands visualized) is
and the expected number of occupied positions (i.e. bands visualized) is  . Use fi as an estimate of the number of amplified fragments identified by sequencing. Nonlinear regression of fragments identified,
. Use fi as an estimate of the number of amplified fragments identified by sequencing. Nonlinear regression of fragments identified,  , against the number of known genomic copies, ni, yields an estimate of m = 11.64 ± 1.79 (Fig. 3A). As a practical matter, no more than a dozen amplified fragments is ever likely to be resolved by agarose minigel electrophoresis when a four-cutter restriction enzyme is used to digest genomic DNA.
, against the number of known genomic copies, ni, yields an estimate of m = 11.64 ± 1.79 (Fig. 3A). As a practical matter, no more than a dozen amplified fragments is ever likely to be resolved by agarose minigel electrophoresis when a four-cutter restriction enzyme is used to digest genomic DNA.
Figure 3.
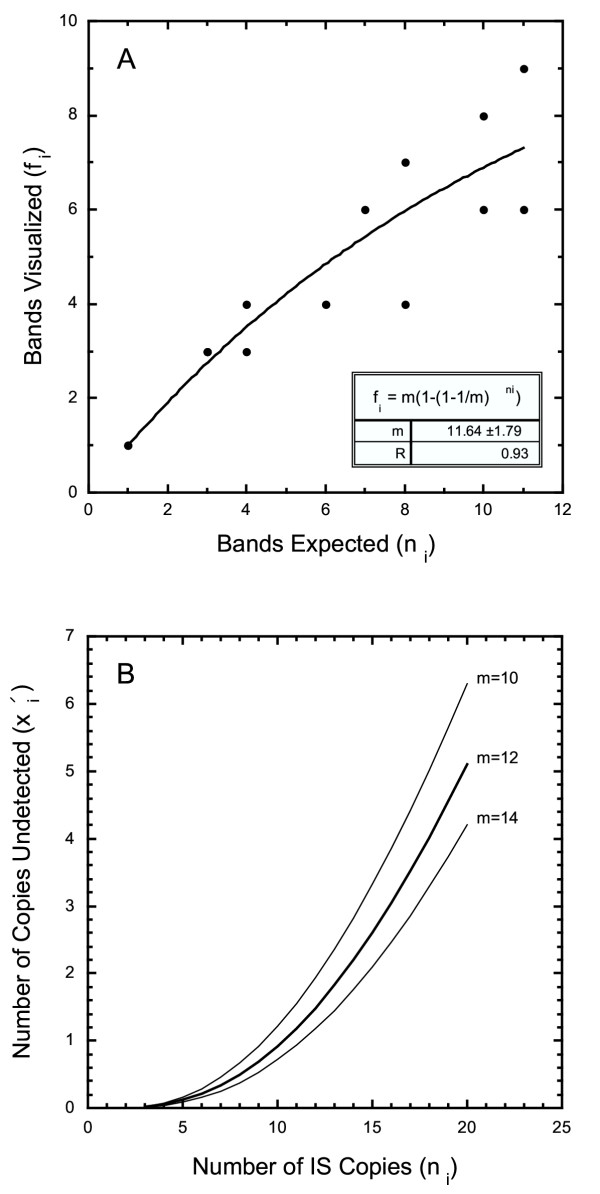
Estimation of IS flanking DNA likely to be resolved and missed. A. The maximum number of fragments likely to be resolved, m, can be estimated by plotting the number of bands observed against the genomic copy number. Only a finite number of bands can be visualized on a gel. Consequently, the likelihood that two amplified fragments comigrate increases with the number of IS copies in the genome. B. The number of amplified flanking sequences likely to be missed rapidly increases when 10 or more bands are visualized. Genomic digests with a single restriction enzyme should be restricted to IS elements with fewer than 10 copies per genome. Genomes with more than 10 copies of an IS element should be screened using high resolution agarose gels and/or using a second restriction enzyme to allow all IS copies to be identified.
Summing the expectations for missing A-side and B-side fragments (i.e. amplified fragments not identified by sequencing) for the j = 8 species of IS elements in MG1655 yields  which is slightly larger than the 17 known masked fragments from MG1655 (each marked with an asterisk in Table 1). The probability that an IS copy is not identified is
which is slightly larger than the 17 known masked fragments from MG1655 (each marked with an asterisk in Table 1). The probability that an IS copy is not identified is  , where the prime designates that this expectation is based on an ability to resolve a maximum of m = 12 fragments per lane. The expected number of missing IS copies is
, where the prime designates that this expectation is based on an ability to resolve a maximum of m = 12 fragments per lane. The expected number of missing IS copies is  , which is only slightly larger than the direct estimate x = 2.54. We conclude that the model provides a robust fit.
, which is only slightly larger than the direct estimate x = 2.54. We conclude that the model provides a robust fit.
A plot of  against ni (Fig. 3B) reveals that the number of missing fragments increases rapidly with the number of genomic copies. With ni = 20
against ni (Fig. 3B) reveals that the number of missing fragments increases rapidly with the number of genomic copies. With ni = 20  = 5 copies (25%,) remain undetected, and even with ni = 10,
= 5 copies (25%,) remain undetected, and even with ni = 10,  = 1 (10%) is expected to be overlooked. To avoid underestimating the number of copies of a highly repeated element, we recommend digesting genomic DNA with a different restriction enzyme and repeating vPCR and sequencing. By using another four-base cutter restriction enzyme Bst UI, we identified all flanking sequences not recovered with the enzyme Rsa I for IS1, IS2, IS3, and IS5, as showed in Fig. 4 for IS2. Larger, temperature controlled high resolution agarose gel electrophoresis apparatus available in some laboratories would also improve resolution of the technique.
= 1 (10%) is expected to be overlooked. To avoid underestimating the number of copies of a highly repeated element, we recommend digesting genomic DNA with a different restriction enzyme and repeating vPCR and sequencing. By using another four-base cutter restriction enzyme Bst UI, we identified all flanking sequences not recovered with the enzyme Rsa I for IS1, IS2, IS3, and IS5, as showed in Fig. 4 for IS2. Larger, temperature controlled high resolution agarose gel electrophoresis apparatus available in some laboratories would also improve resolution of the technique.
Figure 4.
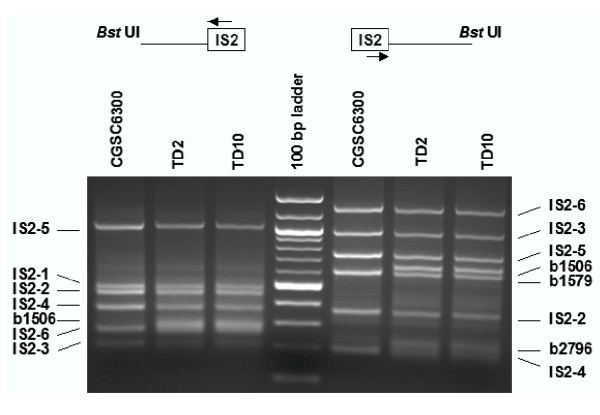
PCR amplification of IS2 flanking DNA from genomic DNA digested with Bst UI. Flanking DNA fragments IS2-3A and IS2-6A (left hand side) and IS2-2B (right hand side), masked by other amplified fragments when genomic DNA was digested with Rsa I (see Fig. 2), were recovered with Bst UI.
Applications
It is apparent that IS complements differ among very closely related laboratory E. coli K12 derivatives MG1655, CGSC6300, TD2 and TD10. The rapidity with which these differences have evolved suggests that ISs may play important roles in experimental evolution. Indeed, adaptation by E. coli to novel laboratory environments is often characterized by IS element mobilization [4,19-22]. Using vPCR will provide these workers with a comprehensive view of genomic reorganization during laboratory evolution. Using this method, we characterized IS elements in 40 isolates which evolved from TD2 and TD10 during chemostats and found a number of IS-mediated gene deletions, duplications and transpositions (unpublished data).
Surveys of natural isolates of E. coli reveal that the numbers and locations of IS elements differ widely among closely related strains, suggesting a brisk turnover of IS elements within and among host lineages [6,23-25]. Comparisons of E. coli genomic sequences confirm that IS elements are commonly associated with chromosomal rearrangements within lineages [17,26,27]. The ability to rapidly and accurately determine the IS complement of the genomes of natural isolates is not only desirable from a population genetic standpoint, but vPCR might also facilitate rapid typing of epidemiological outbreaks of pathogens otherwise indistinguishable from related strains. In this regard it is worth noting that IS sequences are highly conserved compared with most E. coli housekeeping genes [28]. This will greatly aid using vPCR to type strains because only 1 pair of primers is needed for each type of IS element.
Conclusions
This study shows that vPCR is a swift, efficient, reliable method for typing microbial strains and identifying and mapping IS insertion sites present in microbial genomes. Flanking DNA sequences from 36 of the 37IS elements in the E. coli strain MG1655 were recovered by vPCR and confirmed by DNA sequencing. Unlike Southern hybridization and iPCR, our approach involves only one genomic digest and one ligation step. Vectorette PCR is then used to simultaneously amplify all IS elements of a given type, making vPCR a rapid and sensitive means to survey IS elements in genomes.
Methods
Strains
Three derivatives of the K12 isolate were used in this study. Strain CGSC6300, obtained from E. coli genetic Stock Center, Yale University, was used as a control because it is closely related to MG1655 whose entire genome has been sequenced [17]. TD2 and TD10 (derivatives of DD320, itself a K12 derivative) are routinely used in our experiments in molecular evolution [29].
DNA isolation
Genomic DNA was isolated from overnight culture in LB medium using DNAeasy DNA isolation kit (Qiagen, Valencia, CA, USA).
Vectorette unit
The vectorette unit was made using the protocol of Botstein lab http://genome-www.stanford.edu/group/botlab/protocols/vectorette.html[30]. The two anchor bubble primers
![]()
were synthesized by the Advanced Genetic Analysis Center at The University of Minnesota, St. Paul. To anneal bubble primers, 4 μM of each primer (in ddH2O) were combined in a total volume of 100 μl. The mixture was incubated at 65°C for 5 minutes, and then MgCl2 was added to a final concentration of 1–2 mM before cooling down to room temperature.
DNA digestion and ligation of vectorette units
Genomic DNA from each strain was digested using the restriction enzyme Rsa I to produce small, blunt-ended fragments (Fig. 1). The enzyme is a four-base cutter and has 0 to 3 restriction sites within open reading frames (orf) of the eight insertion sequences (IS1, IS2, IS3, IS4, IS5, IS30, IS150, IS186), but does not cut at the extreme ends of each orf. This allows for the design of outward primers to amplify the IS flanking sequence for both sides (see below). Digestion was carried out at 37°C overnight in a 50 μl reaction containing 1 × NEBbuffer (No. 1), 0.5 μg DNA and 10 units of Rsa I. After digestion, 2 μl of anchor bubble unit, 1 μl of 10 mM ATP and 1 unit of T4 DNA ligase (New England Biolabs, Beverly, MA) were added and the reaction was incubated for 5 cycles at 20°C for one hour followed by 37°C for 30 min.
Primers and PCR amplification
Outward primers (Table 2) from each end of the 8 IS sequences were designed and used for PCR amplification in combination with the vectorette primer (5' CGAATCGTAACCGTTCGTACGAGAATCGCT 3') (Fig. 1). The distance between an IS-specific primer position and the extreme end of the IS orf ranged from 16 to 184 bp, which facilitated identifying IS-associated PCR products from DNA sequences. PCR reactions were carried out using Qiagen Multiplex PCR kit (Qiagen, Valencia, CA, USA). Each reaction contained 1 × Qiagen Multiplex PCR Master Mix, 0.2 μM of outward IS primer and vectorette primer and 2 ng of DNA templates (Rsa I-digested DNA ligated with vectorettes). PCR cycling conditions were 95°C for 15 min, 35 cycles of 94°C for 30 s, 60°C for 90 s, 72°C for 2 min, and a final extension step at 72°C for 10 min. The amplified products were separated in 1.4% agarose gel, stained with ethidium bromide and visualized under UV light. DNA bands were excised and purified with Qiagen DNA Gel Extraction Kit (Qiagen, Valencia, CA, USA).
Table 2.
Primers used for identification of ISs using vectorette PCR
| Primer Namea | Sequence (5' to 3')b | Length (bp)c |
| IS1-A | gttacgcacc accccgtcag ta | 22 |
| IS1-B | cggaagtcgc tgtcgttctc aa | 22 |
| IS2-A | ggccc taagacatca atcatctg | 23 |
| IS2-B | tcgctcg ccacgggaat atctg | 22 |
| IS3-A | agccg ctgcgggcca cccggagcac | 25 |
| IS3-B | ggcct cagtccggaa caatttga | 23 |
| IS4-A | cgagagatgag ttcggggtcg agg | 24 |
| IS4-B | aagggccttc ccgagagtgg taa | 23 |
| IS5-A | gccatggca gaatctgctc catgcggg | 27 |
| IS5-B | tgtttcgggc ggaccaaatg ata | 23 |
| IS30-A | ccagctcgt atctcctcgc gctctg | 26 |
| IS30-B | ctagatctgg ttgctgctca gc | 22 |
| IS150-A | cctgacctgg gttcggggga cac | 23 |
| IS150-B | gcgaactgaa ggatgctgtt ac | 22 |
| IS186-A | gggccagaat tgctgaccag ttat | 24 |
| IS186-B | acctgaac tcgcgaaagc gtggata | 25 |
aPrimers are named after the insertion sequence with A and B designating each side. bSequences obtained from MG1655 [17]. cLength of primer in base pairs.
DNA sequencing and analysis
DNA sequencing analysis was carried out on both DNA strands by the AGAC, University of Minnesota, using an IS-specific primer and the vectorette primer. DNA sequences were subjected to BLAST searches against the MG1655 genome sequence.
List of abbreviations
IS: insertion sequences; iPCR: inverse polymerase chain reaction, vPCR: vectorette polymerase chain reaction
Authors' contributions
SZ designed and performed the molecular experiments and prepared the manuscript. AMD provided scientific input and prepared the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We gratefully acknowledge the thorough constructive criticism of an anonymous reviewer that helped improve this manuscript so very much. This study is supported by grants from National Institute of Health (to AMD).
Contributor Information
Shaobin Zhong, Email: zhong020@umn.edu.
Antony M Dean, Email: adean@biosci.umn.edu.
References
- Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blot M. Transposable elements and adaptation of host bacteria. Genetica. 1994;93:5–12. doi: 10.1007/BF01435235. [DOI] [PubMed] [Google Scholar]
- Papadopoulos D, Schneider D, Meier-Eiss J, Arber W, Lenski RE, Blot M. Genomic evolution during a 10,000-generation experiment with bacteria. Proc Natl Acad Sci USA. 1999;96:3807–12. doi: 10.1073/pnas.96.7.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D, Duperchy E, Coursange E, Lenski RE, Blot M. Long-term experimental evolution in Escherichia coli. IX. Characterization of insertion sequence-mediated mutations and rearrangements. Genetics. 2000;156:477–88. doi: 10.1093/genetics/156.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notley-McRobb L, Seeto S, Ferenci T. The influence of cellular physiology on the initiation of mutational pathways in Escherichia coli populations. Proc R Soc Lond B Biol Sci. 2003;270:843–8. doi: 10.1098/rspb.2002.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green L, Miller RD, Dykhuizen DE, Hartl DL. Distribution of DNA insertion element IS5 in natural isolates of Escherichia coli. Proc Natl Acad Sci USA. 1984;81:4500–4. doi: 10.1073/pnas.81.14.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naas T, Blot M, Fitch WM, Arber W. Insertion sequence-related genetic variation in resting Escherichia coli K12. Genetics. 1994;136:721–30. doi: 10.1093/genetics/136.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Gerber AS, Hartl DL. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D, Duperchy E, Depeyrot J, Coursange E, Lenski RE, Blot M. Genomic comparisons among Escherichia coli strains B, K12, and O157:H7 using IS elements as molecular markers. BMC Microbiol. 2002;2:18–26. doi: 10.1186/1471-2180-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley J, Butler R, Ogilvie D, Finniear R, Jenner D, Powell S, Anand R, Smith JC, Markham AF. A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res. 1990;18:2887–2890. doi: 10.1093/nar/18.10.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold C, Hodgson IJ. a novel approach to genomic walking. PCR Methods Appl. 1991;1:39–42. doi: 10.1101/gr.1.1.39. [DOI] [PubMed] [Google Scholar]
- Mao L, Devos KM, Zhu L, Gale MD. Cloning and genetic mapping of wheat telomere-associated sequences. Mol Gen Genet. 1997;254:584–91. doi: 10.1007/s004380050455. [DOI] [PubMed] [Google Scholar]
- Lench NJ, Norris A, Bailey A, Booth A, Markham AF. Vectorette PCR isolation of microsatellite repeat sequences using anchored dinucleotide repeat primers. Nucleic Acids Res. 1996;24:2190–1. doi: 10.1093/nar/24.11.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai M, Huang H, Reed C, Qian C, Smith JS, Alderete B, Jenkins R, Smith DL, Liu W. Genomic organization and mutation analysis of p73 in oligodendrogliomas with chromosome 1 p-arm deletions. Genomics. 1998;51:359–63. doi: 10.1006/geno.1998.5387. [DOI] [PubMed] [Google Scholar]
- Groenen PM, Garcia E, Thoelen R, Aly M, Schoenmakers EF, Devriendt K, Fryns JP, Van de Ven WJ. Isolation of cosmids corresponding to the chromosome breakpoints of a de novo autosomal translocation, t(6;19)(p21;q13.1), in a patient with multicystic renal dysplasia. Cytogenet Cell Genet. 1996;75:210–5. doi: 10.1159/000134485. [DOI] [PubMed] [Google Scholar]
- Chenchik A, Diachenko L, Moqadam F, Tarabykin V, Lukyanov S, Siebert PD. Full-length cDNA cloning and determination of mRNA 5' and 3' ends by amplification of adaptor-ligated cDNA. BioTechniques. 1996;21:526–34. doi: 10.2144/96213pf02. [DOI] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. The complete genome sequence of Escherichia coli K12. Science. 1997;277:1453–74. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Dean AM. A molecular investigation of genotype by environment interactions. Genetics. 1995;139:19–33. doi: 10.1093/genetics/139.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treves DS, Manning S, Adams J. Repeated evolution of an acetate-crossfeeding polymorphism in long-term populations of Escherichia coli. Mol Biol Evol. 1998;15:789–97. doi: 10.1093/oxfordjournals.molbev.a025984. [DOI] [PubMed] [Google Scholar]
- Cooper VS, Schneider D, Blot M, Lenski RE. Mechanisms causing rapid and parallel losses of ribose catabolism in evolving populations of Escherichia coli B. J Bacteriol. 2001;183:2834–41. doi: 10.1128/JB.183.9.2834-2841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riehle MM, Bennett AF, Long AD. Genetic architecture of thermal adaptation in Escherichia coli. Proc Natl Acad Sci USA. 2001;98:525–30. doi: 10.1073/pnas.021448998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser ER, Schneider D, Blot M, Kolter R. Bacterial evolution through the selective loss of beneficial genes. Trade-offs in expression involving two loci. Genetics. 2003;164:1271–7. doi: 10.1093/genetics/164.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SA, Hartl DL. Distribution of transposable elements in prokaryotes. Theor Popul Biol. 1986;30:1–16. doi: 10.1016/0040-5809(86)90021-3. [DOI] [PubMed] [Google Scholar]
- Sawyer SA, Dykhuizen DE, DuBose RF, Green L, Mutangadura-Mhlanga T, Wolczyk DF, Hartl DL. Distribution and abundance of insertion sequences among natural isolates of Escherichia coli. Genetics. 1987;115:51–63. doi: 10.1093/genetics/115.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BG, Parker LL, Betts PW, DuBose RF, Sawyer SA, Hartl DL. IS1 03, a new insertion element in Escherichia coli: characterization and distribution in natural populations. Genetics. 1989;121:423–31. doi: 10.1093/genetics/121.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T, Tanaka M, Tobe T, Iida T, Takami H, Honda T, Sasakawa C, Ogasawara N, Yasunaga T, Kuhara S, Shiba T, Hattori M, Shinagawa H. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- Jin Q, Yuan Z, Xu J, Wang Y, Shen Y, Lu W, Wang J, Liu H, Yang J, Yang F, Zhang X, Zhang J, Yang G, Wu H, Qu D, Dong J, Sun L, Xue Y, Zhao A, Gao Y, Zhu J, Kan B, Ding K, Chen S, Cheng H, Yao Z, He B, Chen R, Ma D, Qiang B, Wen Y, Hou Y, Yu J. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 2002;30:4432–41. doi: 10.1093/nar/gkf566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JG, Ochman H, Hartl DL. The evolution of insertion sequences within enteric bacteria. Genetics. 1992;131:9–20. doi: 10.1093/genetics/131.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunzer M, Natarajan A, Dykhuizen DE, Dean AM. Enzyme kinetics, substitutable resources and competition: from biochemistry to frequency-dependent selection in lac. Genetics. 2002;162:485–99. doi: 10.1093/genetics/162.1.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein lab http://genome-www.stanford.edu/group/botlab/protocols/vectorette.html


