ABSTRACT
More than 80 cases of lethal hemorrhagic disease associated with elephant endotheliotropic herpesviruses (EEHVs) have been identified in young Asian elephants worldwide. Diagnostic PCR tests detected six types of EEHV in blood of elephants with acute disease, although EEHV1A is the predominant pathogenic type. Previously, the presence of herpesvirus virions within benign lung and skin nodules from healthy African elephants led to suggestions that African elephants may be the source of EEHV disease in Asian elephants. Here, we used direct PCR-based DNA sequencing to detect EEHV genomes in necropsy tissue from five healthy adult African elephants. Two large lung nodules collected from culled wild South African elephants contained high levels of either EEHV3 alone or both EEHV2 and EEHV3. Similarly, a euthanized U.S. elephant proved to harbor multiple EEHV types distributed nonuniformly across four small lung nodules, including high levels of EEHV6, lower levels of EEHV3 and EEHV2, and a new GC-rich branch type, EEHV7. Several of the same EEHV types were also detected in random lung and spleen samples from two other elephants. Sanger PCR DNA sequence data comprising 100 kb were obtained from a total of 15 different strains identified, with (except for a few hypervariable genes) the EEHV2, EEHV3, and EEHV6 strains all being closely related to known genotypes from cases of acute disease, whereas the seven loci (4.0 kb) obtained from EEHV7 averaged 18% divergence from their nearest relative, EEHV3. Overall, we conclude that these four EEHV species, but probably not EEHV1, occur commonly as quiescent infections in African elephants.
IMPORTANCE Acute hemorrhagic disease characterized by high-level viremia due to infection by members of the Proboscivirus genus threatens the future breeding success of endangered Asian elephants worldwide. Although the genomes of six EEHV types from acute cases have been partially or fully characterized, lethal disease predominantly involves a variety of strains of EEHV1, whose natural host has been unclear. Here, we carried out genotype analyses by partial PCR sequencing of necropsy tissue from five asymptomatic African elephants and identified multiple simultaneous infections by several different EEHV types, including high concentrations in lymphoid lung nodules. Overall, the results provide strong evidence that EEHV2, EEHV3, EEHV6, and EEHV7 represent natural ubiquitous infections in African elephants, whereas Asian elephants harbor EEHV1A, EEHV1B, EEHV4, and EEHV5. Although a single case of fatal cross-species infection by EEHV3 is known, the results do not support the previous concept that highly pathogenic EEHV1A crossed from African to Asian elephants in zoos.
INTRODUCTION
Elephant endotheliotropic herpesvirus 1 (EEHV1) is the prototype species of the novel Proboscivirus genus that has evidently evolved separately from all other mammalian herpesviruses over the past 100 million years within the ancestors of modern elephants (1–5). Small 250-bp terminase U60(TERex3) and DNA polymerase U38(POL) gene segments of the first two EEHV types discovered were identified originally in samples from 11 zoo elephants with lethal hemorrhagic disease by PCR (1). Since then, five more species of EEHV have been detected in blood or necropsy tissue samples from captive zoo elephants with systemic disease (3, 6–8). DNA from predominantly EEHV1, but also from EEHV3, EEHV4, or EEHV5, has been found in Asian elephant cases (3–5, 9–11), whereas EEHV2 and EEHV6 DNAs have been found only occasionally in the very few African elephant cases (1, 3).
Acute EEHV disease, which has a fatality rate of 80%, affects primarily Asian elephant calves between 1 and 8 years of age (1, 8, 12, 13). Histopathological examination at necropsy shows damaged microvascular endothelial cells containing typical herpesvirus nuclear inclusion bodies, with both blood and necropsy tissue being proven to contain extremely high levels of EEHV DNA by standard PCR DNA analysis (3, 14). In contrast, samples from six Asian elephant calves in the United States that survived symptomatic EEHV1 DNA-positive viremic disease (3, 14–17) as well as those with only mild early-stage symptomatic disease contain several orders of magnitude less viral DNA in blood and none in serum. A series of real-time PCR-based tests developed for quantitative diagnosis of different EEHV types also detected asymptomatic shedding of either EEHV1A, EEHV1B, EEHV4, EEHV5A, or EEHV5B in trunk wash samples from a significant number of monitored U.S. elephants (9, 14, 15, 18). The latter episodes appear to represent normal low-grade primary infections that did not lead to fulminant systemic disease.
Forty-one cases of acute EEHV disease have been confirmed by DNA analysis in North America, 12 have been confirmed in Europe, and 16 have been confirmed in Asian countries (19). PCR-based DNA sequencing of multiple genetic loci from samples from 58 cases, including 9 in India, revealed that lethal disease is associated predominantly with EEHV1, including >37 distinct strains of EEHV1A and 6 strains of EEHV1B (11, 19). Just one or two known lethal cases each have been associated with EEHV3, EEHV4, and EEHV5 in Asian elephants (8, 10, 20), there have been just two known deaths caused by EEHV2 in African elephants (1), and one surviving African calf had mild EEHV6 viremic disease (3).
Although EEHVs have not yet been propagated in cell culture, a great deal of genetic analysis has been carried out directly on blood samples or necropsy tissue from cases of hemorrhagic disease, including the prototype strains for each of the six known major EEHV types and two other partially chimeric subgroups (3–6, 21–24). The results showed that, compared to all other known mammalian herpesviruses, they form a single novel clade, in which EEHV1 and EEHV6 plus EEHV2 and EEHV5 cluster together into two distinct lineages of an AT-rich branch, whereas EEHV3 and EEHV4 form a separate, highly diverged GC-rich branch. Most conserved core EEHV genes harbored in common with orthologues from other herpesvirus subfamilies fall into an intermediate position between the Betaherpesvirinae and Gammaherpesvirinae in phylogenetic trees (3–5). EEHV1A, EEHV1B, and EEHV5 each have a total genome size of close to 180 kb, encompassing 118 genes, including 60 that are novel and not found in any other herpesvirus groups (22, 24). Only EEHV1 has been officially given species status as yet by the International Committee on Taxonomy of Viruses (ICTV), where it was designated the prototype species of the novel Proboscivirus genus within the Betaherpesvirinae subfamily (2). However, based on evidence provided from phylogenetic trees, the many unique features of their biology, as well as the overall gene content and genomic organization, we have proposed that all six EEHVs be assigned as independent species within the Proboscivirus genus and placed together within the newly designated Deltaherpesvirinae subfamily within the family Herpesviridae, order Herpesvirales (4, 5, 25).
Before acute systemic EEHV disease was identified, morphologically recognizable herpesvirus intracellular virions were found originally in 1971 by microscopic techniques within typical nuclear inclusion bodies in syncytial alveolar cells present in lymphoid lung nodules found in up to 75% of 50 healthy culled adult African elephants in Kruger National Park, South Africa (26). Similar inclusion bodies were also observed in epithelial cells from an outbreak of skin nodules on the trunk and head of a subset of orphan African elephant calves imported from Zimbabwe to Florida and Texas in the 1980s (27). From preliminary evidence by PCR amplification of EEHV1 DNA from several of these African elephant skin nodules from archival paraffin blocks, an initial plausible model that EEHV1 may have crossed from African to Asian elephants in zoos was proposed (28). On the other hand, one of two tested lung nodules from culled adult South African elephants in the same study proved to contain EEHV2 DNA instead. However, subsequent unsuccessful attempts to replicate the skin nodule results, the finding of occasional asymptomatic shedding of EEHV1 in healthy adult Asian elephants, as well as confirmation of numerous cases of EEHV1-positive disease throughout Asian countries have all cast serious doubts about the concept of cross-species infection.
To obtain a more definitive picture of which of these herpesvirus types are harbored as apparent quiescent passengers in healthy asymptomatic African elephants, we used highly sensitive and specific targeted PCR-based DNA sequencing approaches to test lung nodules and other clinical samples obtained from five adult African elephants from around the world, including three with lung nodules, that died from a variety of random unrelated causes.
MATERIALS AND METHODS
EEHV-positive pathological samples from healthy adult African elephants. (i) Frozen lung nodule tissue from South Africa.
DNA was extracted from two large (1.5-cm-diameter) frozen lung lesion samples (AfLng54 and AfLng47) collected from two healthy adult wild African bull elephants that were culled in 1997 at Kruger National Park, Republic of South Africa (Table 1). Several sets of archival paraffin blocks from these and additional necropsied lung nodules collected at that time were also available.
TABLE 1.
Summary of the 15 EEHV genomes detected in African elephant lung and spleen samplesa
| Virusb | Strain or case | Host animal sex, age (yr)d | Location | Pathology description | Tissue type or source | Total amt of sequenced DNA (bp) |
|---|---|---|---|---|---|---|
| 1 EEHV2 | AfLng54 | ?, adult | South Africa | Culled | Large lung nodule | 29,699 |
| 2 EEHV3 | AfLng47 | ?, adult | South Africa | Culled | Large lung nodule | 3,707 |
| 3 EEHV3 | AfLng54 | ?, adult | South Africa | Culled | Large lung nodule | 4,019 |
| 4 EEHV6 | NAP42 | F, 28 | USA | Necropsy | Small lung nodule Dc | 31,532 |
| 5a EEHV3v1 | NAP42 | F, 28 | USA | Necropsy | Small lung nodule B | 2,902 |
| 5b EEHV3v1 | Small lung nodule D | 3,061 | ||||
| 5c EEHV3v1 | Small lung nodule A | 287 | ||||
| 5d EEHV3v1 | Lung bronchiole E | 233 | ||||
| 5a EEHV3v3 | Lung bronchiole H | 234 | ||||
| 6a EEHV3v2 | NAP42 | F, 28 | USA | Necropsy | Small lung nodule C | 1,994 |
| 6b EEHV3v2 | Small lung nodule D | 2,572 | ||||
| 7a EEHV7 | NAP42 | F, 28 | USA | Necropsy | Small lung nodule D | 4,008 |
| 7b EEHV7 | Small lung nodule B | 340 | ||||
| 8a EEHV2v1 | NAP42 | F, 28 | USA | Necropsy | Small lung nodule B | 449 |
| 8b EEHV2v1 | Small lung nodule D | 464 | ||||
| 8c EEHV2v1 | Lung bronchiole H | 419 | ||||
| 8d EEHV2v2 | Lung bronchiole H | 419 | ||||
| 9 EEHV2 | SAM7 | F, mature | Kenya | Necropsy | Lung 2 | 581 |
| 10a EEHV3 | SAM6 | F, mature | Kenya | Necropsy | Lung 1 | 3,965 |
| 10b EEHV3 | SAM7 | Lung 2 | 1,657 | |||
| 11a EEHV6 | SAM6 | F, mature | Kenya | Necropsy | Lung 1 | 2,227 |
| 11b EEHV6 | SAM7 | Lung 2 | 1,186 | |||
| 12 EEHV7 | SAM7 | F, mature | Kenya | Necropsy | Lung 2 | 581 |
| 13 EEHV2 | EP26 | M, 18 | Spain | Necropsy | Spleen | 1,398 |
| 14 EEHV3 | EP26 | M, 18 | Spain | Necropsy | Spleen | 1,121 |
| 15 EEHV6 | EP26 | M, 18 | Spain | Necropsy | Spleen | 574 |
| Total | 99,394 |
Detailed ORF and gene locus identifiers, GenBank accession numbers, and matching map coordinates for plus divergence values from EEHV1A(Kimba) for each virus and sample are listed in Section S1 in the supplemental material.
Row numbers refer to each of the 15 different strains involved and match the strain numbering system in Table S1 in the supplemental material. v1, v2, and v3 refer to multiple strains or variants of the same EEHV type, all found within lung samples from elephant NAP42.
Additional EEHV6 sequences of the identical strain were also found in elephant NAP42 nodules A, B, and C and in bronchiole sample H.
All animals were Loxodonta africana elephants. M, male; F, female.
(ii) Necropsy lung tissue from an adult African elephant housed in the United States.
A variety of clinical samples were collected at necropsy from a 28-year-old wild-born female African elephant (case NAP42) housed at a North American zoo that was euthanized because of severe degenerative joint disease in December 2010. To attempt to obtain additional data about potential quiescent EEHV infections, we requested fresh lung necropsy samples that may have lung nodules present. After bread-loafing of the whole lung into 1-cm layers, a small “palpable” section of bronchiolar tissue (3 cm3) that proved to include four shiny white spherical lymphoid “mininodules,” each with a diameter of 2 to 3 mm, within the bronchiolar area or embedded in the upper airway wall. After shipping on ice, DNA was extracted separately from all four dissected nodules (labeled samples A, B, C, and D) as well as from a surrounding segment of parenchymal tissue (sample E) plus similar-sized segments from three other “normal” lung tissue samples from the same animal (samples F, G, and H). In addition to the fresh necropsy lung tissue samples, blood and trunk wash samples intended to be tested for possible evidence of EEHV systemic infection or asymptomatic shedding (14) were also collected (Table 1).
This animal was born in Zimbabwe, rescued as a 3-year-old calf during culling, and then imported to Florida with a large group of other orphan elephants in the mid-1980s. Soon after arrival, a large subset of juveniles from this group were reported to exhibit localized clusters of skin nodules attributed to reactivated EEHV infections (27). These skin nodules were transient and regressed after several weeks or months, although there have been no reports that the nodules recurred as the animals became adults. It is not known whether this particular elephant was included among those that suffered from skin nodules. She was subsequently transferred several times to various other private zoo facilities over the next 25 years but is not known to have ever been in contact with any Asian elephants. In consultations between the zoo veterinarian and owners and the International Elephant Foundation (IEF), a team of veterinarians and two veterinary pathologists familiar with elephant necropsy samples was assembled and agreed to follow newly revised recommended euthanasia and investigative necropsy protocols developed by the IEF and the American Zoological Association Taxon Advisory Group (AZA TAG). The North American proboscivirus case number used here for this animal (NAP42) is part of a larger series encompassing all EEHV DNA-positive pathological samples identified among the collection at the National Elephant Herpesvirus Laboratory (NEHL) at the Smithsonian's National Zoo in Washington, DC.
(iii) Lung necropsy samples from an African elephant in Kenya.
After EEHVs were found at high levels in several lung nodules as described above, a general request for additional samples from elephant lung nodules from routine necropsy tissue was issued. One such response occurred while searching for evidence of potential herpesvirus-containing skin nodules among wild elephants at Samburu National Park in Kenya. One of the authors (V. R. Pearson) working together with the Kenya Wildlife Service and Save the Elephants was provided an opportunity to collect fresh lung tissue from a recently deceased young adult female that died of gunshot wounds. Two random lung samples (samples 1 [SAM6] and 2 [SAM7]) were immersed immediately in Qiagen RNAlater reagent and then subsequently subjected to DNA extraction, followed by PCR amplification and comparative DNA genotyping analysis, as reported previously (29).
(iv) Spleen necropsy tissue from an African elephant at a European zoo.
Frozen blood and tissue samples from an 18-year-old male African elephant (elephant EP26) that died suddenly of an unexplained illness at a zoo in Spain after transfer from another facility were sent to the NEHL diagnostic laboratory in Washington, DC, for EEHV diagnostic testing. A case report (30) suggested that the pathology, which included hemorrhagic symptoms, was consistent with Citrobacter freundii infection. Diagnostic blood tests for EEHV were negative, as were toxicology tests for possible ingested poisons as well as assays for other potential bacterial pathogens, viral pathogens, and parasites (including encephalomyocarditis [EMC] virus). However, despite the negative blood tests, the only other tissue available for testing from this elephant (spleen) proved to contain very low levels of three different EEHVs, as presented in detail here.
GenomiPhi amplification, DNA sequencing, and phylogenetic analysis.
The total DNA amplification and DNA sequencing procedures were all described previously in detail by Richman et al. (4). Molecular phylogenetic analyses by the Bayesian maximum likelihood method with the Kimura 2-parameter model for nucleotide sequences or by the Jones-Thornton-Taylor (JTT) matrix-based model for amino acid sequences were conducted with MEGA5 (31). Bootstrap values (100 replicates) and distance scales (number of substitutions per site) are included directly on the diagrams.
PCR amplification and sequencing primers used.
A list of selected multiround PCR amplification and sequencing primers for 13 of the most relevant and significant gene loci described here, including the new U48.5(TK) and U81(UDG) loci for genomes of the GC-rich branch, are given in Section S2 in the supplemental material. Details of the numerous additional PCR primer sets used were either reported previously (3, 9, 11, 14) or listed in previous papers by Richman et al. (4) and Zong et al. (5) or can be obtained upon request.
Nucleotide sequence accession numbers.
DNA sequence data files representing all 71 new targeted-PCR-sequenced loci from a total of 15 distinct EEHV2, EEHV3A, EEHV6, and EEHV7 genomes found within necropsy tissues from the five African elephants described above (and summarized in Table 1) have been deposited in the NCBI GenBank database under the following accession numbers: JF692766, JQ300034 to JQ300087, JX011086 to JX011088, KC854730 to KC854739, KC854747, KT832514 to KT832517, and KU321582 (AfLng47 and AfLng54 and elephant NAP42); KT832496 to KT832512, KU147226, and KU147227 (SAM6 and SAM7); as well as KM282198 to KM282203 (elephant EP26). Full details of individual loci and their associated accession numbers are listed in Section S1 in the supplemental material.
RESULTS
Detection of both EEHV2 and EEHV3 DNAs in lung nodules from two culled adult African elephants.
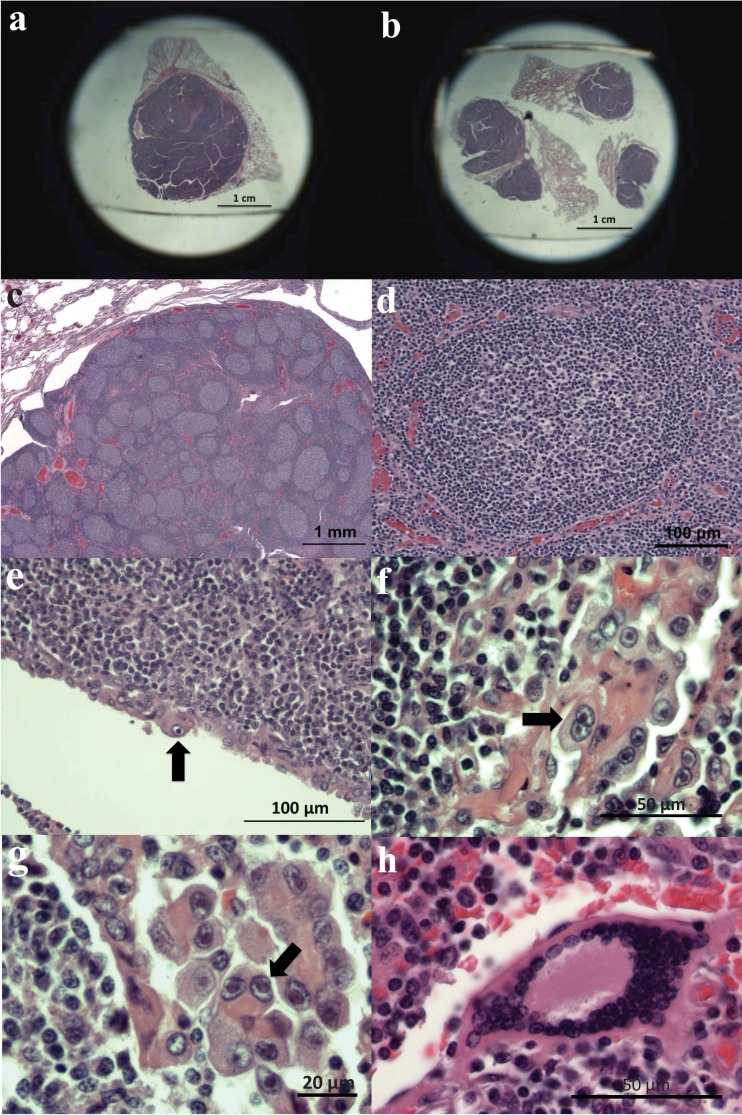
To revisit the question of which EEHV types might be present as quiescent infections within asymptomatic wild African elephants, we prepared DNA samples from the only two frozen lung nodules known to still be available that had been collected at necropsy from a group of culled African elephants in Kruger National Park in 1996. Pathologically, as shown in Fig. 1, the lung nodule tissue in parallel archival formalin-fixed and paraffin-embedded (FFPE) sections from the same collection closely resembled those described in a previous study by McCully et al. (26), including displaying syncytial alveolar epithelial cells and large follicle-like areas consisting predominantly of hyperplasic or reactive lymphoid cells. Low-power images of intact nodules (Fig. 1a and b) and of the internal follicular areas (Fig. 1c and d) as well as higher-magnification images of two types of abnormal alveolar epithelial cells show clustered giant cells (Fig. 1f and g) and multinucleated syncytia (Fig. 1h). Both the giant cells and some scattered single epithelial cells also appear to exhibit typical herpesvirus nuclear inclusion bodies and an enlarged cytoplasm indicative of active lytic infection (Fig. 1e to g, arrows). Unfortunately, there were no samples properly prepared for electron microscopy (EM) investigation available.
FIG 1.
Examples of EEHV-associated pathology in lung nodules from culled adult South African elephants. Shown are photomicrographs from hematoxylin-and-eosin-stained sections of formalin-fixed, paraffin-embedded archival blocks prepared at necropsy from healthy adult African elephants culled at Kruger National Park in 1997. (a and b) Low-power images of four examples of large lymphoid nodules in lung tissue; (c) cross-sectional image of a large lung nodule showing multiple lymphocyte-containing follicular areas; (d) more detailed higher-power image of a region with extensive follicular hyperplasia; (e) presumed viral nuclear inclusion body within an isolated epithelial cell; (f and g) higher-power images of representative clusters of enlarged alveolar epithelial cells in the area surrounding the follicular mass; (h) higher-power image of a representative syncytial cell cluster flanking the follicular mass. Arrows indicate likely nuclei with viral inclusion bodies.
One of these two nodules had previously proven positive for a small U38(POL) DNA segment of EEHV2 but was negative for EEHV1 by using redundant panherpesvirus PCR primers in the original studies by Richman et al. (28), whereas the second nodule was negative for both EEHV1 and EEHV2. Here, we used a new set of more sensitive diagnostic PCR primers designed specifically for the EEHV2 DNA POL gene, which yielded a strong 500-bp first-round band from AfLng54 DNA (Fig. 2, lane 1) but was again negative with the second lung nodule DNA sample (AfLng47) (Fig. 2, lane 2). Two parallel control samples of random biopsy specimen DNA from a captive U.S. African elephant (Fig. 2, lane 4) and a positive heart DNA sample from an Asian elephant case, designated EEHV1(NAP11) (Fig. 2, lane 3), were negative. The DNA sequence of EEHV2(AfLng54) differed from that of the prototype strain EEHV(NAP12) by 2 out of 497 bp.
FIG 2.
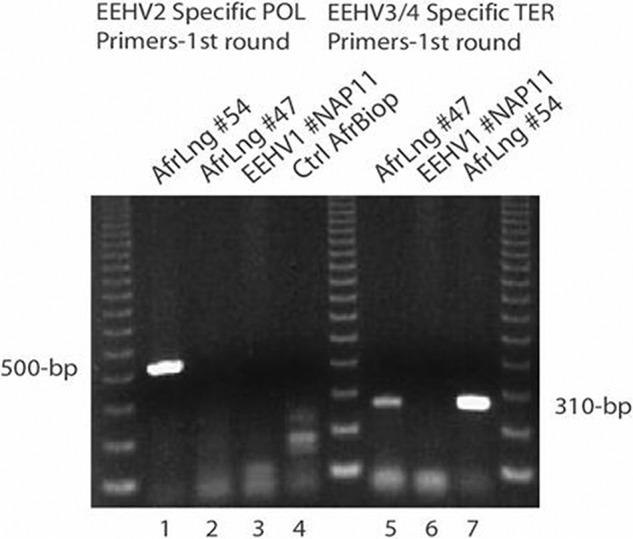
Detection of both EEHV2 and EEHV3 DNAs in lung nodules from two culled adult South African elephants by PCR amplification. Shown is agarose gel electrophoretic separation of ethidium bromide-stained first-round PCR products obtained by using EEHV2-specific POL primers LGH6525 and LGH7440 (500 bp) (lanes 1 to 4) and EEHV3- and EEHV4-specific TER primers LGH6707 and LGH6708 (310 bp) (lanes 5 to 7). Lanes: 1, AfLng47; 2, AfLng54; 3, Asian elephant EEHV1A case NAP11 heart; 4, negative-control African elephant vestibular lesion biopsy specimen; 5, AfLng47; 6, Asian elephant EEHV1A case NAP11 heart; 7, AfLng54. Three size marker lanes contain multimerized 123-bp ladders.
Interestingly, examination of the same two African elephant lung nodule DNA samples with primers specific for the EEHV3/4 TERex3 gene locus also produced strong 310-bp first-round PCR bands (Fig. 2, lanes 5 and 7). Again, these primers failed to detect the known EEHV1A prototype genome in control heart tissue DNA from Asian elephant case NAP11 (Fig. 2, lane 6) and produced negative results with two other unrelated captive African elephant biopsy samples tested in parallel (not shown). The two African elephant lung nodule samples proved to differ from the prototype Asian case of EEHV3A(NAP27) sample as well as from each other by 2 bp within a 390-bp POL locus PCR product (3). The three EEHV genomes found within these two large Kruger lung nodules are just the second example of EEHV2 identified and just the second and third examples of EEHV3 ever examined. Additional tests with multiple-round PCR primers specific for EEHV1, EEHV5, and EEHV6 (and later EEHV7) failed to detect any evidence of these other EEHV types.
Characterization of EEHV2 genomic DNA sequences from the South African lung nodule sample.
All attempts to propagate EEHV derived from blood samples from acute disease or necropsy tissue in cell culture in primary elephant endothelial cells and other cell types have failed so far. However, the challenging process of de novo assembly of the entire 180-kb genomes has been accomplished for three strains of EEHV1 and one strain of EEHV5 (22–24) from very-high-quality DNA samples by high-throughput next-generation approaches. In comparison, for the prototype EEHV2(NAP12) sample, a total of 58 kb of DNA sequence in eight noncontiguous segments was determined with a set of phage lambda clones as well as by direct PCR-based sequencing of necropsy tissue DNA (4). Because those prototype data came from a case of lethal hemorrhagic disease in a young zoo calf (21), we also generated of total of 29.7 kb of Sanger DNA sequence data for 17 nonadjacent PCR loci from the EEHV2(AfLng54) genome amplified directly from lung nodule DNA using both EEHV2-specific and EEHV1-EEHV2-common redundant primers. An abbreviated summary of the locations and nomenclature for all 32 partial or complete open reading frames (ORFs) obtained are listed in order in Table 2. Full details of the positions and sizes of the individual ORFs obtained at each PCR locus, together with GenBank accession numbers and the equivalent genomic coordinates and nucleotide divergence levels for each locus from EEHV1A(Kimba) (22), are listed in Section S1 in the supplemental material. Nucleotide differences between the EEHV2(AfLng54) genome and the prototype EEHV2(NAP12) genome within each of 20 selected ORF segments that have been analyzed in common are presented in Table 3, along with the matching positional coordinates for orthologous protein regions derived from the EEHV1A(Kimba) genome. Almost all ORFs from the two strains evaluated proved to be very closely related, with the total nucleotide differences between the two strains reaching 361 bp (1.25%). However, the U51(vGPCR1) locus proved to be the only major exception, displaying nucleotide differences in 171 out of 1,042 bp (16.4%) as well as 8% divergence at the protein level. Excluding this locus, there were a total of 190 nucleotide polymorphisms found across the remaining 28.5 kb of matching sequence, representing just 0.76% overall variation between the viremic U.S. strain and the quiescent South African lung nodule strain, showing that they both clearly belong to the same independent EEHV2 species.
TABLE 2.
Summary of sequenced African elephant EEHV gene coding regions
| Gene (ORF) | Protein | Presence of coding regiona |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild African elephant lung nodules |
Elephant NAP42 lung nodules (USA) |
SAM6/7 lung (Kenya) |
Elephant EP26 spleen (Spain) |
||||||||||||
| EEHV2 (AfLng54) |
EEHV3 (AfLng47) |
EEHV3 (AfLng54) |
EEHV2 | EEHV3v1 and -v2 | EEHV6 | EEHV7 | EEHV2 | EEHV3 | EEHV6 | EEHV7 | EEHV2 | EEHV3 | EEHV6 | ||
| U39 | gB | + | + | ||||||||||||
| U38 | POL | + | + | + | + | + | + | + | + | + | + | + | − | ||
| U33 | Cys-R | + | + | ||||||||||||
| U28 | RRA | + | + | ||||||||||||
| U27.5 | RRB | + | + | ||||||||||||
| U27 | PPF | + | + | ||||||||||||
| U45.7 | ORF-J | + | + | ||||||||||||
| U46 | gN | + | + | ||||||||||||
| U47 | gO | + | + | ||||||||||||
| U48 | gH | + | + | ||||||||||||
| U48.5 | TK | + | + | + | + | + | + | + | − | + | − | − | |||
| U49 | + | + | + | − | − | ||||||||||
| U50 | PAC2 | + | + | ||||||||||||
| U51 | vGPCR1 | + | + | ||||||||||||
| U57 | MCP | + | + | ||||||||||||
| U60ex3 | TERex3 | + | + | + | + | + | + | + | + | + | − | + | − | ||
| U62 | + | + | |||||||||||||
| U63 | + | ||||||||||||||
| U66ex2 | TERex2 | + | + | ||||||||||||
| U66ex1 | TERex1 | + | + | ||||||||||||
| U70 | EXO | + | + | + | + | + | + | ||||||||
| U71 | MyrTeg | + | + | + | + | + | + | + | + | − | − | − | |||
| U72 | gM | + | + | + | + | + | + | + | + | ||||||
| U73 | OBP | + | + | + | + | + | + | + | + | + | − | − | + | − | |
| U76 | POR | + | + | + | + | + | − | − | + | ||||||
| U77 | HEL | + | + | + | + | + | + | + | + | + | + | − | − | + | |
| U77.5 | ORF-M | + | + | ||||||||||||
| U80.5 | ORF-N | + | + | ||||||||||||
| U81 | UDG | + | + | + | + | + | + | ||||||||
| U82 | gL | + | + | ||||||||||||
| U82.5 | ORF-O | + | + | ||||||||||||
| U85.5 | ORF-K | + | + | ||||||||||||
| U86.5 | ORF-L | + | + | ||||||||||||
+, present and amplified; −, presumed to be present but not able to be amplified.
TABLE 3.
Differences in African lung nodule strains from the EEHV2 and EEHV6 prototypes
| PCR locus | No. of nucleotide differences/total no. of nucleotides (%) across selected locia |
||||||
|---|---|---|---|---|---|---|---|
| EEHV2 |
EEHV6 |
||||||
| AfLng54 vs NAP12 | NAP42 vs NAP12 | SAM7 vs NAP12 | EP26 vs NAP12 | NAP42 vs NAP35 | SAM6 vs NAP35 | EP26 vs NAP35 | |
| U39(gB) | 1/2,313 | 16/2,354 | |||||
| U38(POL) | 11/2,313 | 12/419 | 1/487 | 8/2,364 | 6/467 | ||
| U33(Cys-R) | 20/851 | 10/916 | |||||
| U28-27.5(RRA-B) | 14/2,043 | 4/880 | |||||
| U27-U46-47(PPF-gN-gO) | 16/2,558 | 31/2,516 | |||||
| U48(gH) | 1/501 | 74/539 | |||||
| U48.5(TK)-U49 | 3/1,195 | 53/922 | 30/1,029 | ||||
| U49-50(PAC2) | 11/1,702 | 11/1,105 | |||||
| U51(vGPCR1) | 171/1,042 | 3/832 | |||||
| U57(MCP) | 15/2,767 | 5/2,244 | |||||
| U60-62(TERex3) | 6/816 | 10/874 | 2/347 | ||||
| U66(TERex1) | 10/914 | 2/325 | |||||
| U71-72(MyrTeg-gM) | 18/2,674 | 3/667 | |||||
| U73(OBP) | 0/568 | 1/181 | 1/832 | 0/470 | 14/558 | ||
| U76-77(POR-HELn) | 24/1,864 | 12/1,867 | 9/943 | ||||
| U77-U78.5(HELc-ORF-Mn) | 4/1,082 | 2/1,300 | |||||
| U77.5-U80.5(ORF-Mc-vCXCL1) | 4/970 | 53/900 | |||||
| U81-U82(UDG) | 9/954 | 22/957 | |||||
| U82(gL) | 9/1,001 | 30/819 | |||||
| U85.5(ORF-Kex3) | 11/1,431 | 16/1,371 | |||||
| U86.5(ORF-L) | 6/748 | 22/2,253 | |||||
| Total | 363/29,523 (1.25) | 12/214 | 1/581 | 55/1,990 | 367/27002 (1.45) | 17/2,227 | 14/558 |
Data in boldface type represent ORF segments that diverge by >3% between the two strains.
Partial sequence characterization of two EEHV3 genomes from South African lung nodules.
The first example of a second major branch of the Proboscivirus genus, designated EEHV3(NAP27), came from blood and necropsy tissues of a 7-year-old Asian elephant calf that died of acute hemorrhagic disease in a U.S. zoo (3, 8). Because of the highly divergent and very-GC-rich nature of this genome, we were able to identify only five small genomic DNA loci totaling 3.9 kb and encompassing parts of the U38(POL), U60(TERex3), U71-U72(gM), U73(OBP), and U76(POR)-U77(HEL) genes (5). Many of the same primer sets were used here, although because of the high levels of EEHV2 DNA present in the AfLng54 DNA sample, the generic pan-EEHV primers for both U73(OBP) and U76(POR)-U77(HEL) had to be redesigned to be specific for the viruses of the GC-rich branch (the successful new primer sets employed are listed in Section S2 in the supplemental material). Initially, we were able to obtain totals of 3,330 and 3,144 bp of DNA sequence each across these same five loci for both South African lung nodule genomes (Table 2; see also Section S1 in the supplemental material). Their sequences proved to differ from one another by 56 bp (1.6%) and from the previously reported prototype EEHV3A(NAP27) sequence at 39 (1.05%) and 42 (1.1%) polymorphic positions, respectively (Table 4). In comparison, they differed from the prototype EEHV4(NAP22) genome by 11.5% and from the equivalent EEHV1, EEHV2, and EEHV5 segments by between 21 and 35%.
TABLE 4.
African lung nodule strain differences from the EEHV3A, EEHV3B, and EEHV4 prototypes
| PCR locus | No. of nucleotide differences/total no. of nucleotides (%)a |
|||||||
|---|---|---|---|---|---|---|---|---|
| EEHV3A (NAP27) vs EEHV3A (AfLng47) |
EEHV3A (NAP27) vs EEHV3A (AfLng54) |
EEHV3A (NAP27) vs EEHV3A (NAP42v1) |
EEHV3A (NAP27) vs EEHV3A (NAP42v2) |
EEHV3A (NAP27) vs EEHV3A (SAM6) |
EEHV3B (NAP62) vs EEHV3A (AfLng54)b |
EEHV3A (NAP27) vs EEHV7 (NAP42-D) |
EEHV4 (NAP22) vs EEHV7 (NAP42-D) |
|
| U38(POL) | 5/996 | 7/1,001 | 1/382 | 1/382 | 1/397 | 5/360 | 94/660 | 91/630 |
| U60(TERex3) | 0/311 | 0/309 | 0/290 | 0/290 | 0/279 | 9/310 | 25/310 | 22/310 |
| U48.5(TK) | ND | ND | ND | ND | ND | 31/315 | NDc | 75/423 |
| U71-72(MyrTeg-gM) | 18/650 | 10/749 | 13/732 | 16/726 | 2/665 | 120/696 | 291/698 | 290/764 |
| U73(OBP) | 12/697 | 9/525 | ND | ND | 44/885 | 14/286 | 161/621 | 145/637 |
| U76-77(POR-HEL) | 2/576 | 3/552 | 2/640 | 2/640 | 3/903 | 14/553 | 62/911 | 64/877 |
| U81(UDG) | 2/449 | 13/449 | ND | ND | 11/359 | 18/338 | 44/353 | 39/353 |
| Total | 39/3,698 (1.05) | 42/3,695 (1.1) | 16/2,024 (0.8) | 19/2,018 (0.85) | 61/3,488 (0.7) | 211/2,858 (7.4) | 677/3,553 (19.1) | 726/3,994 (18.2) |
ND, no data available.
ORF and GenBank information for EEHV3B (Bronson et al., unpublished) is included in Section S1 in the supplemental material.
However, the number of nucleotide differences/total number of nucleotides between EEHV3A(AfLng54) and EEHV7(NAP42-D) was 28/226 for U48.5(TK).
While this work was in progress, the first known example of symptomatic viremic infection of a young African elephant calf in a U.S. zoo by EEHV3 occurred. This infection was associated with mild clinical symptoms of acute hemorrhagic disease, with moderately high levels of viral DNA being detected in the blood of a surviving 5-year-old male Loxodonta africana elephant. A clinical case report, including a time course evaluation of viremia and subsequent virus shedding in trunk wash and saliva samples from this calf and his adult herd mates during the time course of the calf's illness and recovery, will be presented elsewhere (E. Bronson, M. McClure, J. Sohl, E. Weidner, E. M. Latimer, G. S. Hayward, V. R. Pearson, and P. D. Ling, unpublished data). Data from comparative genotype analysis of this case, involving 4,050 bp of DNA sequence obtained across all seven PCR loci for EEHV3(NAP62), are included in Tables 4 and 5 and in Section S1 in the supplemental material. EEHV3(NAP62) displayed a total of 211 differences (7.4%) from the prototype EEHV3(NAP27) versions across 2,858 bp of common areas, especially within the TK and U71-gM loci, with 9.8 and 17% differences, respectively, but closely resembles another recently analyzed sample, EEHV3(SAM5) (29) (see Section S1 in the supplemental material). Therefore, we have designated the new viremic case genome the second known example of a distinctive EEHV3B subtype. However, all four of the EEHV3 genomes from the African elephant lung nodules described here clearly belong to the EEHV3A rather than the EEHV3B subtype at all loci tested.
TABLE 5.
DNA- and protein-level divergences in ORFs across EEHV7, EEHV3, EEHV4, and EEHV1Ab
| Gene locus | EEHV1A (Kimba) positionsa |
Protein size (bp) | % divergence |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide level |
Amino acid level |
|||||||||||
| EEHV7/ EEHV3 |
EEHV7/ EEHV4 |
EEHV3A/ EEHV3B |
EEHV3/ EEHV4 |
EEHV7/ EEHV1 |
EEHV7/ EEHV3 |
EEHV7/ EEHV4 |
EEHV3A/ EEHV3B |
EEHV3/ EEHV4 |
EEHV7/ EEHV1 |
|||
| U38(POL) | 78384–79040 | 220 | 13 | 14 | 1.6 | 7 | 34 | 12 | 11 | 1.8 | 7.5 | 26 |
| U48.5(TK) | 106674–106294 | 126 | 18 | 16 | 9.8 | 12 | 38 | 19 | 16 | 7.6 | 14 | 48 |
| U60(TERex3) | 123731–124037 | 105 | 8 | 7 | 2.9 | 3.8 | 22 | 1 | 1 | 2 | 1 | 5 |
| U71(MyrTeg) | 132954–133241 | 96 | 37 | 39 | 15.3 | 26 | 50 | 42 | 45 | 20 | 29 | 71 |
| U72(gM) | 133320–133554 | 78 | 24 | 23 | 9.8 | 14 | 50 | 24 | 31 | 15 | 13 | 53 |
| U73(OBP) | 134670–135287 | 207 | 26 | 23 | 4.9 | 17 | 41 | 27 | 24 | 4 | 22 | 40 |
| U76(POR) | 140967–141295 | 104 | 6 | 5.8 | ND | 3.3 | 29 | >1 | >2 | ND | >1 | 23 |
| U77(HEL) | 141246–141878 | 189 | 7.2 | 8 | 2.7 | 3.5 | 28 | 9 | 7 | 3.3 | 2.5 | 21 |
| U81(UDG) | 146566–146168 | 133 | 11 | 12 | 5.3 | 7.3 | 35 | 7 | 6 | 0 | 3 | 26 |
Based on data reported previously by Ling et al. (22).
ORF and GenBank data for EEHV3B are included for comparison in Section S1 in the supplemental material (Bronson et al., unpublished). The prototype strains used for these comparisons were EEHV1A(Kimba, NAP23). EEHV3A(AfrLng54), EEHV3B(NAP62), EEHV4A(NAP22), and EEHV7(NAP42-D).
Finally, based on the recently completed intact 206-kb genomic DNA sequence of EEHV4(NAP69) (GenBank accession number KT832477 [P. D. Ling, S. Y. Long, A. Feury, R.-S. Peng, S. Y. Heaggans, X. Qin, K. C. Worley, S. Dugan, and G. S. Hayward, submitted for publication]), we successfully designed PCR primers that amplified two more EEHV3 PCR loci involving the U48.5(TK) and U81(UDG) gene coding regions from EEHV3B(NAP62). By using these primers (listed in Section S2 in the supplemental material), additional comparative data were then also obtained for the 315-bp TK locus for EEHV3A(AfLng54) and for the 367-bp UDG locus for EEHV3A(NAP27), EEHV3A(AfLng54), and EEHV3A(AfLng47), which are all included in Tables 2 and 4 and in Section S1 in the supplemental material. Again, EEHV3A(AfLng54) proved to differ from the EEHV3B(NAP62) subtype at the TK locus by 9.8% and at the UDG locus by 6%. However, whereas EEHV3A(AfLng47) had only 2 bp of polymorphisms compared to EEHV3A(NAP27) in UDG, the EEHV3A(AfLng54) version differed by 4.2% from them both, potentially indicative of another novel, third EEHV3 subtype at this locus.
Detection of high levels of EEHV6 in lung nodules from an asymptomatic adult African elephant.
Each of the eight lung DNA samples from African elephant NAP42 were screened extensively with a large panel of first-, second-, and third-round PCR primer sets that were either known to detect multiple EEHV species at moderate efficiencies (pan-EEHV primers) or designed to detect just single specific viruses or subsets of EEHV types at high efficiency (3, 5). Initially, the standard highly redundant, diagnostic pan-EEHV POL primer set produced correct-sized first-round PCR DNA products from nodule D and second-round bands from nodules B and C but produced negative results for nodule A and the other four samples. DNA sequencing of these bands revealed an identical EEHV6 POL gene in each case, with differences in just 2 out of 480 bp from the single previously known prototype viremic strain, EEHV6(NAP35) (3). Pan-EEHV primer sets for U73(OBP) and U76(POR)-U77(HEL) also gave identical first-round EEHV6 products from nodules B, C, and D, and the OBP set also detected it in nodule A after second-round PCR. EEHV1/6-common second-round U38(POL), U71(gM), and U51(vGPCR1) primer sets also detected the same EEHV6 sequence in all four lung nodules, as well as at lower levels in sample E, but not in sample F, G, or H.
Because the EEHV6(NAP42) genome was only the second example of EEHV6 encountered, we again carried out extensive direct targeted PCR sequencing of nodule D to obtain a total of 30.2 kb of DNA sequence distributed over 16 loci for comparison with the known 27.7 kb of sequence for the viremic EEHV6(NAP35) strain (5). A summary of the location, nomenclature, and core gene status of all 32 partial or complete ORFs identified according to their orthologues from related herpesviruses is presented in Table 2, and details of the GenBank accession numbers as well as equivalent genomic coordinates for and levels of nucleotide divergence from EEHV1A(Kimba) for each locus are listed in Section S1 in the supplemental material.
Comparative data spread across 20 selected matching ORF locations within EEHV6(NAP42-D) and the prototype EEHV6(NAP35) genome are also presented in Table 3. The results revealed a total of 367 nucleotide differences (1.45%), with nearly half being localized within just four small segments with ≥3% nucleotide divergence (Table 3). The most significant of these variable regions was the N terminus of the U48(gH) protein, where the two strains differed by 15% at the protein level. This compares with 20% protein divergence within the same segment of both EEHV6 strains from EEHV1A(Kimba). The next most variable locus fell within a contiguous 4-kb block encompassing parts of ORF-M plus all of the CXCL1(ORF-N), UDG, and gL ORFs, where there were 18, 13, 22, and 30 polymorphisms, respectively, giving amino acid differences of 4.5, 5.1, 3.2, and 4.0%, respectively. Overall, when the U48(gH) hypervariable locus is omitted, the sum of the nucleotide differences totals 288 bp (1.15%), and when all ORFs that vary by >3% are omitted, it drops to just 181 bp (0.8%) across the other 16 remaining ORFs evaluated (Table 3). Again, these results make a strong case for considering both samples to belong to the same genotype representing an independent EEHV6 species.
Identification of EEHV2 and two distinct strains of EEHV3 in the same U.S. elephant lung samples.
Further examination of the NAP42 lung nodule samples also revealed low levels of EEHV2 DNA in 460-bp bands obtained in the second round with EEHV2-specific POL primers from nodule D and also in the third round from nodule B. These sequences proved to be identical but showed seven and four unique DNA polymorphisms compared to the matching loci from EEHV2(NAP12) and EEHV2(AfLng54). Further analysis with a third-round EEHV2-specific POL primer set also revealed the presence of very low levels of two closely related variants of EEHV2 within lung tissue sample H. One of these variants (variant 1 [v1]) was identical to EEHV2 in nodules B and D, whereas the second (v2) retained all of the same polymorphisms but also differed from it at three more positions, but we considered this to be insufficient evidence to definitively indicate the presence of two independent strains.
Despite the presence of much higher levels of EEHV6, EEHV3 genomes were also detected in elephant NAP42 nodules B, C, and D as well as at very low levels in lung samples E and H but not in sample F or G. Overall, sequence data totaling 11.3 kb derived from all of the EEHV3A(NAP42) lung tissue PCR products obtained showed that, unlike EEHV6, which was identical in all five positive tissue samples, there were at least two distinct strains of EEHV3 distributed nonuniformly among both the nodules and adjacent nonnodular lung samples. In particular, the EEHV3 U71-gM locus gave a clean unique sequence from nodule B (v1) that differed at 16 out of 726 nucleotide positions (2.1%) from that from nodule C (v2), whereas nodule D contained an equal mixture of both. Similar levels of polymorphisms were also found when these two sequences were compared with African EEHV3A(AfLng47) and EEHV3A(AfLng54) or with the prototype Asian EEHV3A(NAP27) strain sequence. Indeed, all five known strains of EEHV3A differ from one another by between 1.6% and 3.2% at this locus (Table 4). Importantly, the exact same 16-bp differences between nodules B and C were also reproduced twice more with independent PCR sets starting with additional new, completely different primer pairs, thus ruling out any possible type of sequencing artifact. In contrast, the 230-bp EEHV3 POL locus for lung bronchiolar sample H showed two nucleotide differences from EEHV3A(NAP27) and one difference from the EEHV3A(NAP42) nodules B, C, and D as well as from lung sample E. Unfortunately, the more variable EEHV3 U71-gM locus could not be amplified from sample E or H to provide sufficient evidence to designate this third variant a separate strain.
Many unsuccessful attempts to detect EEHV1A, EEHV1B, or EEHV5 in all of the same lung nodule samples by three-round POL- or TER-specific PCR were also carried out. Trace amounts of EEHV6 POL DNA, but none of the other types, were also occasionally amplified from whole-blood samples from elephant NAP42, but because the sequence was the same as that in the nodules, we could not validate this result. Based on our current threshold limits for reliable three-round PCR detection, the maximum possible levels of EEHV1 present were estimated to be below 120 viral genomes per million host cells in both blood and tissue. Similarly, searches for both EEHV1 and EEHV6 in trunk wash samples from this elephant by real-time PCR approaches carried out at Baylor College of Medicine were negative (P. D. Ling, personal communication).
Detection of novel DNA sequences from another GC-rich Proboscivirus species, EEHV7.
Remarkably, in addition, a 230-bp DNA sequence obtained from elephant NAP42 lung nodules B and D with the second-round EEHV3-specific POL primer set proved to contain an equal mixture of two obviously related but distinct sequences. This segment of POL lies within the Codehops domain (32) used for robust panherpesvirus diagnostic PCR tests using highly redundant primers that detect nearly all known types of herpesviruses if sufficiently abundant (although the standard Codehops set does not usually work very well for EEHV species). Because these two sequences differed from each other by between 10 and 15% at the nucleotide level (i.e., more than twice as much as EEHV3 differs from EEHV4 here), we interpreted this result to indicate the presence of a likely new virus species, designated EEHV7. To obtain clean POL DNA sequence data from both of these GC-rich genomes, new primers were designed from inside this locus, in which several nucleotide positions (including the extreme 3′ nucleotide) were altered to match either only EEHV3 or only the putative EEHV7 sequence. When paired individually with several outside pan-EEHV POL primers and used for additional second-round PCR amplification, these primers successfully resolved the mixtures over the full standard 490-bp Codehops region. The resulting nucleotide sequence data revealed that one of the two genomes amplified was identical to the previous EEHV3v1 and -v2 strains, whereas the other was confirmed to be a new and distinctive Proboscivirus type that differed at 63 and 67 nucleotide positions (13%) and by 21 amino acids (aa) (13%) from the prototype EEHV3A, EEHV3B, and EEHV4 strains at this locus. In comparison, EEHV3A and EEHV4 themselves differ over this same region by just 26 out of 490 bp (5.3%) and 16 out of 165 aa (9.7%).
Furthermore, while searching for larger segments of the DNA POL region with a new set of EEHV2-specific primers, we obtained a 700-bp second-round PCR product from nodule D that evidently contained EEHV7 sequences only, without any EEHV2 or EEHV3 DNA being simultaneously amplified. In contrast, many other primer sets amplified only one of the other EEHV genomes present there. A 342-bp EEHV7 POL sequence was also obtained from the nodule B sample, which proved to be identical to that in nodule D. To further validate these results, we eventually obtained data for a total of 4,008 bp of DNA sequence for the new EEHV7 genome from across all seven PCR loci now known for EEHV3. To do so in the presence of similar levels of EEHV3 DNA plus much higher levels of EEHV6 required multiple different approaches to identify new sets of specific primers that either incorporated common redundant features of both the EEHV3 and EEHV4 versions or maintained the high overall GC-rich characteristics but with deliberately introduced mismatches against just the EEHV3 version. The results are summarized in Tables 1 and 2 and in Section S1 in the supplemental material, and the combined EEHV7(NAP42-D) data proved to differ overall by 19.1% and 18.2% from the equivalent matching EEHV3A(NAP27) and EEHV4(NAP22) loci, respectively (Table 4), compared to an overall difference of 12.6% between EEHV3A and EEHV4 themselves across the same regions.
Percent differences in both DNA and protein levels across each of the seven partial ORFs in EEHV7 compared to the matching positions in EEHV1A, EEHV3A, EEHV3B, and EEHV4 are listed in Table 5. The catenated EEHV7 sequences proved to have a GC content of 57.6%, compared to 61.3% for the matching regions of EEHV3 (5). Similarly, the extremely high GC content found at the third (wobble) codon position in EEHV3 and EEHV4 was not quite as high in EEHV7 but still reached 86, 83, 87, 89, 92, and 88% for those parts of the POL, TK, TER, OBP, POR-HEL, and UDG coding regions, respectively, that were sequenced. In contrast, EEHV1, EEHV2, EEHV5, and EEHV6 all have an overall GC content of just 42 to 43% across the same loci, with between 40 and 50% GC contents at their third codon positions (4, 5).
Detection of EEHV2, EEHV3, EEHV6, and EEHV7 in lung tissue from a wild Kenyan elephant.
Although no nodules were observed in the lung of this young adult female Loxodonta africana elephant at field necropsy, two random segments of fresh parenchymal tissue were immediately immersed in RNAlater reagent, and the DNA was then subsequently extracted under laboratory conditions and couriered to the United States under appropriate USDA and Convention on International Trade in Endangered Species (CITES) permits. Diagnostic PCR tests with appropriate standard pan-EEHV POL, TER, U71-gM, and HEL primers produced positive results after either second- or third-round amplification for both EEHV3A and EEHV6 in lung sample 1 (SAM6) and for EEHV2, EEHV3A, and EEHV6 in lung sample 2 (SAM7). Subsequently, several more specific follow-up primer sets for U71-gM, TK, OBP, and UDG also gave positive results for EEHV3A in one or both samples (Tables 1 and 2). Note that in this case, initial PCR and sequencing were all carried out at a separate institution and laboratory that had never previously handled any elephant samples of this type. Importantly, the detailed PCR sequence data obtained (see Section S1 in the supplemental material) confirmed that although all three Kenya strains were very similar to the EEHV2, EEHV3, and EEHV6 species prototypes identified in the United States, they also revealed several unique and reproducible polymorphisms compared to all other strains of these species that we have encountered previously (Tables 2, 4, and 5; see also Section S1 in the supplemental material). In particular, the six loci from EEHV3A(SAM6) displayed differences from the matching loci for EEHV3A(NAP27) in 61 out of 3,488 bp (0.7%). Finally, further studies also detected a new, second example of EEHV7 at the U77(HEL) locus from SAM7 that differed from the prototype U.S. NAP42-D lung nodule example in 1 out of 575 bp, but there was evidently not enough virus present to be detected with any other EEHV7 primers tested.
Additional EEHV2, EEHV3, and EEHV6 strains found in the spleen of an African elephant at a European zoo.
Finally, spleen (but not blood) samples from a fifth, different adult African elephant (EP26) that died of unrelated causes but was sent to the NEHL for evaluation (30) again proved to contain multiple EEHV types present at low borderline levels in which just a subset but not all of the PCR loci tested produced visible second-round PCR bands (Tables 1 and 2). However, once again, the positive loci obtained included parts of each of the same three typical African elephant types, EEHV2, EEHV3A, and EEHV6, as those found in the U.S., South African, and Kenyan examples. Furthermore, for EEHV2 here (see Section S1 in the supplemental material), although the POL locus was very similar to that of the standard EEHV genotypes examined previously, the positive U48.5(TK) locus proved to be almost 20% different at the DNA level from the two EEHV2 TK genes evaluated previously or nearly as far diverged as is EEHV5 TK from the standard EEHV2 version (Table 3). Potentially, this novel TK gene could represent a segment from a new independent EEHV2 subtype, but we currently interpret that it most likely represents just a hypervariable segment from an otherwise standard EEHV2 strain, similar to the situation found here for the vGPCR1 gene of EEHV2(AfLng54) and for the U48(gH) genes of EEHV1 and EEHV6 (4, 5, 11). Although the studies in this case were carried out in our regular diagnostic laboratory, all three EEHV3A loci and the single EEHV6 locus that were PCR amplifiable were again slightly but reproducibly different from those of all other strains of these EEHV types studied previously (Table 3).
Phylogenetic tree comparisons of the African elephant lung nodule viruses and those from hemorrhagic disease cases.
A DNA-level linear phylogram for the well-conserved combined U76(POR)-U77(HEL) PCR locus for many of these asymptomatic African elephant samples, compared to prototype examples from both the GC-rich and AT-rich EEHV branches and the other selected mammalian herpesvirus subfamilies, is presented in Fig. 3. As expected, this phylogram shows that all the EEHV samples fall into one of two linked major branches of a cladal Proboscivirus cluster that are both quite distinct from the primate Cytomegalovirus genus and Roseolovirus genus branches as well as from the Gammaherpesvirus cluster. Whereas the African elephant lung nodule EEHV2, EEHV3, and EEHV6 strains are barely distinguishable from the hemorrhagic disease prototypes, the novel African elephant lung nodule EEHV7 genome instead proved to be closely related to but also extensively diverged from those of both of the other known GC-rich branch prototypes, EEHV3 and EEHV4.
FIG 3.
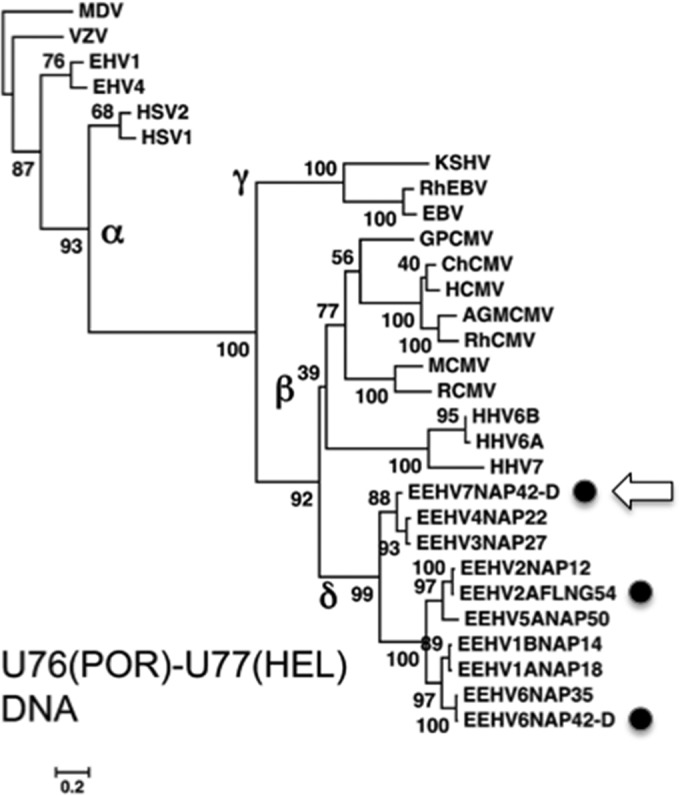
DNA-level phylogenetic tree for African elephant lung nodule strains at the combined POR-HEL gene locus. The diagram presents linear distance-based Bayesian nucleotide dendrograms generated in MEGA5 with MUSCLE illustrating the genetic relationships at the U76(POR)-U77(HEL) PCR gene locus (628 bp) among five of the EEHV2, EEHV6, and EEHV7 strains (solid circles) from African elephant lung nodules compared to their previously described Proboscivirus orthologues EEHV1, EEHV2, EEHV3, EEHV4, EEHV5, and EEHV6 from hemorrhagic disease cases as well as selected key herpesviruses from all three other mammalian subfamilies (alpha-, beta-, and gammaherpesviruses). In comparison, the probosciviruses belong to the proposed Deltaherpesvirus subfamily (δ). The position of the novel EEHV7 version is also indicated with an arrow. Marek's disease alphaherpesvirus (MDV) was used as the outgroup. Bootstrap values (percentages from 1,000 reiterations) and a distance scale of 0.2 are shown. Alphaherpesvirinae (α) include Marek's disease virus (MDV), varicella-zoster virus (VZV), equine herpesvirus 1 (EEHV1), EEHV4, herpes simplex virus 1 (HSV-1), and HSV-2; Gammaherpesvirinae (γ) include Kaposi's sarcoma-associated herpesvirus (KSHV), Epstein-Barr virus (EBV), and rhesus EBV (RhEBV); and Betaherpesvirinae (β) include human cytomegalovirus (HCMV), chimpanzee CMV (ChCMV), RhCMV, African green monkey CMV (AgmCMV), guinea pig CMV (GPCMV), mouse CMV (MCMV), rat CMV (RCMV), and the Roseolovirus species human herpesvirus 6A (HHV-6A), HHV-6B, and HHV-7.
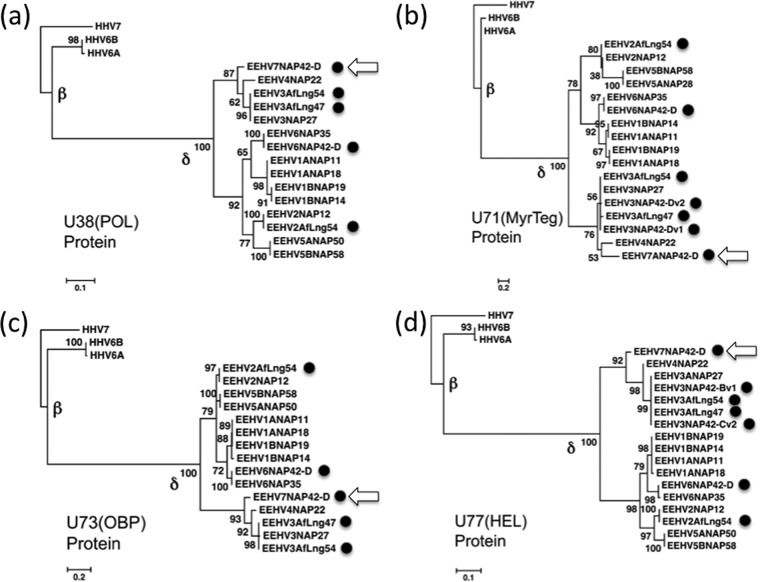
In addition, Fig. 4 presents four protein-level linear phylogenetic trees focused on much shorter segments of U38(POL), U71(MyrTeg), U73(OBP), and U77(HEL) that again allow comparisons to be made using data for all three GC-rich branch types among the quiescent African elephant viral genomes, including EEHV7, as well as for the EEHV2 and EEHV6 strains. To focus on relationship comparisons among the EEHVs, these data include only the three human Roseolovirus species for outgroup comparisons, together with between 15 and 17 different Proboscivirus examples. In each case, the three predominant branches of the EEHVs are very obvious, with the GC-rich arm being very distinct from the AT-rich arm and the latter splitting cleanly into the two major sublineages of EEHV1/6 and EEHV2/5. The POL and MyrTeg trees especially also illustrate the sometimes highly significant differences between the EEHV1A and EEHV1B or between the EEHV5A and EEHV5B subtype branches (4, 5), whereas in the OBP and HEL trees, they are nearly indistinguishable. On the other hand, in all four protein trees, the EEHV7 versions (Fig. 4, arrows) are highly diverged from the EEHV3 versions, with EEHV4 being intermediate, whereas the various EEHV3 strains from multiple African elephant lung nodule sources are nearly identical to the prototype hemorrhagic disease strain EEHV3A(NAP27).
FIG 4.
Protein-level phylogenetic trees comparing POL, MyrTeg, OBP, and HEL from African elephant lung nodules with all other known EEHV types. The diagrams present distance-based linear Bayesian amino acid dendrograms generated in MEGA5 with MUSCLE illustrating the genetic relationships among four proteins each from the EEHV2, EEHV3, EEHV6, and EEHV7 strains detected in African elephant lung nodules (solid circles) compared to their Proboscivirus (EEHV) orthologues from other Asian and African elephant hemorrhagic disease cases. The positions of the novel EEHV7 versions are also indicated with arrows. The Proboscivirus genus belongs to the proposed Deltaherpesviridae family (δ). The HHV-6A, HHV-6B, and HHV-7 orthologues within the Roseolovirus genus of the Betaherpesviridae (β) are the only non-Proboscivirus examples included for reference comparisons here. The HHV-7 version is used as the outgroup in all panels. Bootstrap values (percent) and distance scales of 0.1 or 0.2 are shown. (a) U38(POL), a DNA polymerase of 128 aa (18 examples). (b) U71(MyrTeg), a myristylated tegument protein of 48 aa (20 examples). (c) U73(OBP), an origin binding protein of 125 aa (18 examples). (d) U77(HEL), a helicase subunit of 163 aa (20 examples).
Minimal patterns of strain variability within EEHV2, EEHV3, and EEHV6.
In summary, both the EEHV2(AfLng54) and EEHV6(NAP42) genomes proved to differ only minimally (by 363 bp and 367 bp or 1.25% and 1.45%, respectively) from the matching regions in the prototypes from viremic blood samples of two zoo African elephant calves with either lethal or mild acute disease (5). In both cases, the measured variability dropped to nearly half when the one or two known hypervariable loci in each genome were omitted. This residual range of intraspecies variability closely matches that found among most conserved genes in parallel studies of 36 independent strains of EEHV1A. However, EEHV1A strains display clustering into multiple subtype patterns within several hypervariable genes, including those encoding the U51(vGPCR1) and U48(gH) proteins (4, 11, 24), which appears to parallel the observed exceptions here of divergence among the three strains of EEHV2 within vGPCR1 (14%) and TK (20%) and between the two strains of EEHV6 within gH (16%). Neither the EEHV2(AfLng54) nor the EEHV6(NAP42) strain shows any other evidence of complicating major chimeric patterns of the type found consistently among all known EEHV1B strains compared to EEHV1A strains within chimeric domain I (CD-I), CD-II, and CD-III (4, 22, 24) and similarly within EEHV5B compared to several EEHV5A strains (5). Therefore, we consider that they should all be unambiguously assigned as members of the same species as the prototypes.
Although it was possible to analyze only a subset of up to seven small PCR segments for each of the six new EEHV3 strains found here, they all differed slightly among themselves and from the previously described prototype EEHV3A(NAP27) strain that killed a young Asian elephant in the United States (3, 8). Again, the overall observed genomic variation represents just 1.2% over a total of 3 to 4 kb each (Table 4), clearly placing them all within the boundaries of the EEHV3A subtype, which differs by 211 out of 2,858 bp (7.4%) from the more recently recognized EEHV3B subtype from a surviving U.S. African elephant calf with acute disease symptoms (Bronson et al., unpublished). Nevertheless, ∼40% of these differences occurred just within the U71(MyrTeg)-U72(gM) locus, which seems to represent a specific “hot spot” for variation within EEHV3A, and this level is greater than that found at this locus among multiple strains of EEHV1A. Thus, the latter anomalous region leads to an exaggeration of the overall levels of polymorphism among these several distinct EEHV3A strains, which measures just 0.6% when the U71 region is omitted.
DISCUSSION
Multiple species of EEHV found in African elephant lung nodules.
Virtually all samples from previous analyses of EEHV genomic DNA have been obtained from cases of acute disease or trunk wash shedding in Asian elephants. However, herpesvirus-like infections of an unknown type were first detected as inclusion bodies or enveloped virions in localized lung and skin nodules from healthy adult African elephants by microscopic techniques several decades ago (26, 27). To determine which, if any, of the now many known types of EEHV may be present in this quiescent form in asymptomatic African elephants, we sought all available samples of pulmonary tissue that contained “herpes-like” lung nodules. PCR sequencing analysis of the six examined lung nodules from three different African elephants revealed moderate to very high levels of EEHV DNA, including multiple EEHV genomes within five of the six nodules and with a total of four different types of EEHV (including at least eight different strains) being present among them.
In particular, targeted PCR sequencing using DNA samples obtained from two 20-year-old frozen lung nodules still available from culled wild elephants from South Africa revealed high levels of a single distinct strain each of EEHV3A plus even higher levels of a single strain of EEHV2 in one of them. In addition, the only lung nodules from a U.S. zoo African elephant reported so far proved to harbor four different EEHV types. The highest levels by far were obtained for a single strain of EEHV6, which was present in all four small lymphoid nodules within a localized “palpable” segment of bronchiolar tissue. Other EEHV types, including two distinct EEHV3A strains, were distributed nonrandomly at moderately high levels within three of these same nodules, and low levels of a single EEHV2 strain were found in two of them. Furthermore, evidence for another previously unknown genome from a GC-rich Proboscivirus branch, designated EEHV7, was also found in two of the same small lung nodules. Finally, much smaller amounts of the same EEHV2, EEHV3A, and EEHV6 strains were also occasionally detected in the parenchyma surrounding the nodules.
Samples from the other two EEHV-positive necropsy cases studied here came as freshly collected random segments of lung or spleen tissue, but even these samples yielded low levels of multiple EEHV types, including EEHV2, EEHV3A, EEHV6, and EEHV7 in the wild Kenyan elephant and EEHV2, EEHV3A, and EEHV6 in the Spanish zoo elephant. These results contrast with data for all of the numerous positive clinical samples of either necropsy tissue or blood from elephants with acute viremic disease, as well as occasional positive trunk wash samples from asymptomatic animals, that we have evaluated previously, which have always yielded DNA sequence data for just a single type and strain of EEHV, even in samples from multiple organs or from sequential collections from the same shedding episode. Several surviving mildly viremic elephants as well as a few asymptomatic trunk wash shedders have yielded different virus species or subtypes during different illness or shedding episodes but never from the same sample or from sequential blood or trunk wash samples collected over a time period of <6 months (9, 14, 15).
The most surprising aspect of these results was our evidence for multiple distinct strains and types of EEHV found within just single (even very small) lung nodules from otherwise healthy asymptomatic elephant case NAP42. The most extreme example was nodule D, containing five different viral genomes, including EEHV2, two different strains of EEHV3A, plus both EEHV6 and EEHV7, although other different distribution patterns of the same set of viruses occurred across the other three nearby small nodules. For humans, detection of multiple types of herpesvirus within the same clinical specimen, even in blood, saliva, or cerebrospinal fluid (CSF) samples, is quite rare in healthy individuals although well documented in immunosuppressed patients. One might anticipate that each nodule begins from a single “reactivated” virus-infected cell, followed by either the proliferation of virus-carrying cells themselves or infiltration of circulating reactive inflammatory cells targeted to the nodules, but this result must imply either that the different viruses are carried there by multiple responding inflammatory cells or that several nearby, initially distinct micronodules coalesce into larger ones. The positive results for all four genome types, EEHV2, EEHV3, EEHV6, and EEHV7, seem to be completely valid to the extent that none of them match any of the prototype strains examined previously in our laboratory or each other and because the same unique sequence signatures have been reproducibly obtained from multiple independent PCRs as well as from multiple separate nodules or tissue samples and DNA preparations. In addition, multiple gene loci were identified for most of the viruses detected here, except where there were only trace amounts of EEHV2 or the second EEHV7 strain.
Evidence for a new, seventh type of elephant proboscivirus.
Confirmation of the presence of a previously unidentified novel GC-rich EEHV genome within two of the lung nodules from the euthanized African elephant required considerable effort, partly because of the nearly 20% nucleotide divergence but also especially uniquely because of the presence of three other related EEHV genomes within the same samples, including EEHV6 at a >50-fold-higher abundance. However, the combined total of 4,008 bp of EEHV7 DNA sequence obtained from seven different PCR loci is close to the totals obtained by conventional Sanger PCR sequencing for the prototype EEHV3A (4.1-kb) and EEHV4 (5.7-kb) genomes from viremic Asian elephant calves as described previously by Garner et al. (8) and Zong et al. (5). Most importantly, the seven EEHV7 loci identified all proved to diverge by between 8 and 32% (average, 18%) from those of all three of its closest known relatives EEHV3A, EEHV3B, and EEHV4. This value is significantly higher than the levels of divergence of EEHV3A from EEHV4 (12%) and of EEHV3A from EEHV3B (7.4%). From the phylogenetic data presented in Fig. 3 and 4 and by the same criteria and reasoning presented previously by Zong et al. (5) for the other EEHV types, this level of nucleotide divergence implies that EEHV7 likely initially underwent an evolutionary split from both EEHV3 and EEHV4 ∼15 million to 20 million years ago.
Considering that all seven sets of GC branch-specific redundant primers that could be tested were successful in amplifying PCR loci from EEHV7(NAP42-D), the most parsimonious interpretation is that they represent portions of a presumed intact infectious viral genome. The fact that at least three other members of the GC-rich branch of the probosciviruses, EEHV3A, EEHV3B, and EEHV4, are infectious entities was already apparent because of the sudden and symptomatic high-level viremias that they caused, with other herd mates also becoming infected with the same identical strains at about the same time in two of these instances (18; Bronson et al., unpublished). Furthermore, the complete 206-kb genome of EEHV4B(Baylor) from a high-quality trunk wash DNA sample from an Asian elephant recovering from a moderate viremic episode was recently determined (P. D. Ling et al., submitted for publication). The fact that we have also identified a least one locus, U77(HEL), from a second example of EEHV7 in a wild adult African elephant also tends to further validate EEHV7 as a distinct species that has so far been found only at low levels in asymptomatic animals. Finally, the discovery of this apparent third member of the GC branch within the Proboscivirus genus also raises the question of whether there might be yet another, fourth, undiscovered virus of this highly GC-rich type that might be an Asian elephant equivalent of EEHV7.
Criteria for assignment of distinct species or subtype status to novel EEHV genomes.
In searching for and evaluating rarely studied and often uncultured herpesviruses from numerous wild-animal host species, most evolutionary analyses have used just a small common 250- or 480-bp segment of the DNA polymerase gene (referred to as Codehops) for which highly redundant PCR primers can be used to detect previously unknown species in clinical samples (32). In making assessments about whether different herpesviruses are likely to be distinct species or not, samples in which the DNA POL Codehops region shows <1 to 2% variation have always been judged to be of the same species, whereas related viruses with variations beyond the 5 to 10% level have always turned out to be distinct species that are often (but not always) native to a different host species. When even just the POL Codehops region data are considered, all seven EEHVs that we have described here as different types (or species) have >7% consistent nucleotide differences (and often between 15 and 35%), and this same yardstick also applies to almost all other well-conserved genes evaluated that are not among the hypervariable subset.
However, genomic subtype PCR sequence scanning across numerous strains of EEHV1 (4) as well as complete genome sequencing of three high-quality representative prototypes (22, 24) have led to the recognition of two major subgroups, designated EEHV1A and EEHV1B, that differ overall by 7.3% when measured by the EMBOSS Stretcher program. Furthermore, these subgroups display a mosaic combination of nearly identical sequence blocks plus subtype-common nucleotide variations of ∼3% in blocks encompassing loci such as POL, U71-gM, and HEL as well as large characteristic subgroup-specific variations (17 to 32%) within at least three scattered multigene loci, known as CD-I, CD-II, and CD-III, that comprise a total of 15 kb (<8% of the genome) (4). EEHV1A strains can also be subdivided further into between four and six distinct clusters in several known hypervariable gene loci, of which the two best characterized so far are U51(vGPCR1) and U48(gH) (4, 11).
Similarly, for the other EEHV types studied here, which also display complex genomic chimerism and cannot be isolated or propagated in cell culture, it has proven necessary to go far beyond just the POL Codehops sequence data to assess whether any of these types are distinct virus species rather than subtypes of the same species. In addition, by now having several independent examples each of EEHV2, EEHV3A, and EEHV6 that proved to be almost exactly the same as the prototypes across multiple genomic scanning loci (except for occasional hypervariable regions) greatly strengthens the evidence that these three types all represent independent virus species. Although all five newly identified EEHV3A strains from asymptomatic adult African elephants that we have studied here clearly fall within the jurisdiction of a single species and subtype, the situation for two other strains, now identified as EEHV3B(NAP62) and EEHV3B(SAM5), is not yet fully resolved, with the overall divergence from EEHV3A across all seven PCR loci being 4.5% but with two loci, U71-gM and TK, showing divergences of between 8 and 9%, whereas for the POL locus, it is only 1.5%. Accordingly, similarly to EEHV1B and EEHV5B, we judge that while the EEHV3B strains clearly have a common pattern of divergence from EEHV3A, they are probably not sufficiently different to be designated a distinct species.
Finally, the two examples of EEHV7 identified also differ from both EEHV3 and EEHV4 by nearly 20% across the POL Codehops region, with the total 4,008 bp accumulated for EEHV7(NAP42-D) also differing by 19.1% and 18.2% from the equivalent regions in the EEHV3A and EEHV4 prototypes, respectively. Individually, the nine EEHV7(NAP42-D) genes identified so far differ from the EEHV3A versions by 13% (POL), 18% (TK), 8% (TER), 37% (U71), 24% (gM), 26% (OBP), 6% (POR), 7.2% (HEL), and 7% (UDG). Therefore, presuming (as expected) that this represents an intact genome, these results clearly justify separate-species status for EEHV7 as well, although additional examples and more genomic loci will be needed to fully validate this interpretation. On the other hand, for the strains that we have determined to be of the EEHV1B and EEHV5B subtypes, although there are several gene loci with consistent strain variations well above the 5 to 10% level compared to their EEHV1A and EEHV5A counterparts, this does not continue throughout the whole length of the genome, and in particular, they differ by <3 to 5% across the intact POL genes.
Considering the evidence that at least five highly diverged types of elephant gammaherpesvirus, designated elephant gammaherpesvirus 1 (EGHV1) to EGHV5, can also infect and be shed asymptomatically from otherwise healthy captive Asian and African elephants (3, 33, 34), the new Proboscivirus genotype designated EEHV7 now represents the 12th known major type of elephant herpesvirus, all of which have been detected within North American elephants and all of which would be judged worthy of species status according to the standard genetic criteria described above.
Lack of support for the concept of cross-species transmission as the cause of EEHV1 hemorrhagic disease.
Richman et al. (1, 28) originally suggested that cross-species transmission of EEHV1 infection from African elephant herd mates to Asian elephant calves in captivity may be the reason for the devastating nature of the acute hemorrhagic disease observed. This interpretation was based on preliminary PCR evidence for EEHV1 being present in archival paraffin block sections from several skin nodules found among African elephant calves imported to Florida in the mid-1980s (27). However, we have not been able to confirm this result with more extensive DNA sequence analysis of the same archival pathological samples of the Florida skin nodules. Furthermore, as described here, several other EEHV species instead, but not yet any examples of EEHV1, have now been found within necropsied lung nodules from five different adult African elephants. Another major problem with the old concept that EEHV1 may be of African elephant origin is the confirmation by PCR-based DNA sequencing that nine examined cases of lethal disease with exactly the same hemorrhagic pathology occurring in orphan and wild Asian elephant calves in India have also involved high-level disseminated infections with seven different highly diverged strains of EEHV1A and one strain of EEHV1B (11), with at least four more cases in Thailand, Laos, and Cambodia being associated with either EEHV1A or EEHV4 (20, 35, 36). Therefore, it is now quite clear that EEHV1 has been an established endogenous virus in wild Elephas maximus elephants throughout Asian countries for a very long time.
There is also no question that a considerable fraction of healthy monitored adult Asian elephant herd mates housed in American and European elephant facilities carry and occasionally shed either EEHV1A, EEHV1B, or both and that at least a subset of them also harbor either EEHV4, EEHV5, or both, implying that these individuals (many of them wild born) survived primary infections with all of these viruses, presumably as young calves (9, 14–16, 18, 37, 38). Possibly, the 20% of Asian elephant calves that are susceptible to EEHV disease (whether in zoos or in the wild) become exposed to a herd mate shedding levels of EEHV1 high enough for successful transmission at a much later age than usual in the absence of maternal antibodies and without potentially protective prior infections by the other less pathogenic EEHV types. Unfortunately, specific serological assays that can discriminate among antibodies against the various EEHV species are not yet available to further address these issues.
Likely universality of simultaneous infections by multiple EEHV types in asymptomatic adult African elephants.
Our analysis here clarifies some aspects of previous, largely circumstantial evidence for the presence of localized nonsystemic EEHV infection in lung or skin nodules of healthy adult African elephants. In particular, the results confirm and expand on the original observation of EEHV2 DNA in one of the same lung nodules from a culled African elephant but make highly suspect the similar original report of PCR detection (without detailed sequence analysis) of small EEHV1 TER or POL PCR products in DNA extracted from archival biopsied skin nodules collected in the 1980s from several African elephant calves in Florida (27, 28). Although there is still no solid evidence for any EEHV type being present within the latter archival skin nodule samples, we have reported here the detection of multiple examples of EEHV2, EEHV3, EEHV6, and EEHV7 in five of the six lung nodule samples examined as well as occasionally in fresh necropsied lung or spleen samples from a total of five different adult African elephants.
The lung nodules from the first three asymptomatic adult African elephants studied here represented the only known examples of lung nodules that have yet been available for testing for herpesvirus DNA. Therefore, considering that the original report by McCully et al. (26) described the presence of similar large lung nodules in 75% of culled South African elephants evaluated, we conclude that most wild-born African elephants are likely to harbor similar inapparent infections with one or more of the EEHV2, EEHV3, EEHV6, and EEHV7 types, which evidently target otherwise healthy parenchymal lung tissue to produce localized multifocal nodular reactivation. However, the lower levels of the same set of viruses detected in random, freshly collected lung and spleen samples from the other two positive animals also imply either that micronodules may also be commonly present in lung or spleen tissue or that some other form of localized reactivation can occur. Combined with previously reported evidence for occasional shedding of low levels of EEHV1A or EEHV1B (14, 15) and later also of EEHV5 DNA (9) and EEHV4 DNA (18) in trunk wash samples collected from many healthy adult Asian zoo elephants, this tends to suggest that probably all of the known Proboscivirus species occur commonly as asymptomatic quiescent infections of either Asian or African elephant hosts. We suspect that this will be a nearly universal occurrence in the wild, as well as in wild-born animals in zoos, and suggest that there may be a logical biological connection between these lung nodules, especially at sites of apparent pulmonary reactivation, and viral DNA-positive nasal wash secretions as a source of virus shedding and transmission.
Another obvious prediction from these results is that we would now expect that nearly all captive African elephants, especially wild-born adults, carry quiescent infections with several different Proboscivirus species. On the other hand, no such lung or skin nodules have yet become available from routine pathology examinations of Asian elephants at necropsy, whether captive or wild, despite the high levels of concern over the problem of EEHV hemorrhagic disease since 1995. Admittedly, there was only a vaguely palpable mass (and visible nodules were not observed) during the very thorough initial gross examination of the euthanized U.S. zoo elephant NAP42 studied here.
Clarification of EEHV species that cause natural infections of African versus Asian elephant hosts.
Overall, we interpret that African elephants are most likely to be the natural hosts for EEHV2, EEHV3, EEHV6, and EEHV7. For EEHV2 and EEHV3 especially, these findings provide direct evidence that both intraspecies and interspecies disease scenarios can occur. Thus, the death of an African elephant calf from acute hemorrhagic disease that was confirmed to have been caused by EEHV2 (the index case NAP12 in 1998) would have been an intraspecies event, whereas the death of an Asian elephant calf from acute EEHV3 hemorrhagic disease (the index case NAP27 in 2007) would now be the only known example of an apparently very rare cross-species event. Based on the finding of multiple different EEHV types in four of the five adult African elephants studied here, it seems clear that prior infection by one EEHV species does not necessarily provide protection against the establishment of persistent infection by one or more of the other EEHV types, possibly even including infection by a second strain of the same EEHV type, but it could mitigate the pathological symptoms and disease severity of subsequent primary infections by other EEHV strains or types. On the other hand, the majority of lethal disease cases in young Asian elephants, whether under human care in North America and Europe (19) or within Asian countries such as India (11), Thailand (20), Cambodia (35), and Laos (36), have been caused by a variety of different strains of EEHV1A or EEHV1B (3, 7), together with two examples of EEHV4 (18) and one example of EEHV5 (10, 23). This suggests that the two subgroups of EEHV1 and probably also EEHV4 and EEHV5 originated in Asian elephants (9, 18, 19).
Supplementary Material
ACKNOWLEDGMENTS
We particularly thank Mitch Bush (Smithsonian National Zoo), Kruger National Park and the Department of Pathology, Pretoria University, for the collection of lung nodules from the Republic of South Africa, as well as Ed Ramsey (University of Tennessee), Scott Terrell (Disney, Orlando, FL), and Dalen Agnew (Michigan State University), members of the pathology team that was organized in association with AZA TAG protocols and with kind permission of Chuck and Rise Pankow and the Nashville Zoo, Grassmere, TN.
We also thank Lynn W. Enquist, Department of Molecular Biology, Princeton University (Princeton, NJ), for his generous support of the PCR and sequencing costs for V.R.P.
J.-C.Z. carried out the majority of the PCR amplification and sequencing studies. L.K.R. initiated the project at the NEHL and organized the collection of the South African elephant lung nodule samples. E.B. and M.C. collected and provided the elephant samples from the Maryland Zoo and from Spain. E.M.L. carried out initial PCR sequencing work with the samples from South Africa, Spain, and the Maryland Zoo. V.R.P. organized the collection and importation of the Kenyan samples and carried out the PCR sequencing work on them. V.R.P. and S.Y.L. performed additional PCR sequence analyses of the Maryland Zoo DNA samples. S.Y.L. generated the photomicrographs in Fig. 1 and carried out long-range PCR work on the EEHV6(NAP42) genome. M.D.F. was the principal investigator for the Morris Animal Fund research grant, and S.A.N. assisted in organizing the necropsy team and the search for lung nodules from the euthanized U.S. elephant. S.Y.H. extracted most of the DNA samples. G.S.H. and S.Y.H. carried out all of the DNA sequence difference analyses and comparisons for identifying ORFs, generated the annotated GenBank files as well as the phylogenetic trees, and prepared the tables and Fig. 2 through 5. G.S.H. conceived of and coordinated the project, designed primers, and wrote multiple drafts and revisions of the manuscript. All authors read and provided input for the manuscript.
We declare no competing financial interests.
Funding Statement
Studies at the Smithsonian National Zoo in Washington, DC, were funded by Research grants to L.K.R. from the Smithsonian Institution, the Morris Animal Fund, the Fort Worth Zoo, the International Elephant Foundation, and Ringling Bros. and Barnum and Bailey Center for Elephant Conservation. Studies at Johns Hopkins School of Medicine were funded through NIH research grant R01 AI24576 to G.S.H., by a grant from the Morris Animal Foundation in association with Fort Worth Zoo and the International Elephant Foundation, and by a subcontract within a Collections Stewardship Leadership grant from the Institute of Museums and Library Services to Lauren Howard at the Houston Zoo in Texas.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02936-15.
REFERENCES
- 1.Richman LK, Montali RJ, Garber RL, Kennedy MA, Lehnhardt J, Hildebrandt T, Schmitt D, Hardy D, Alcendor DJ, Hayward GS. 1999. Novel endotheliotropic herpesviruses fatal for Asian and African elephants. Science 283:1171–1176. [DOI] [PubMed] [Google Scholar]
- 2.Davison AJ, Eberle R, Ehlers B, Hayward GS, McGeoch DJ, Minson AC, Pellett PE, Roizman B, Studdert MJ, Thiry E. 2009. The order Herpesvirales. Arch Virol 154:171–177. doi: 10.1007/s00705-008-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Latimer E, Zong J-C, Heaggans SY, Richman LK, Hayward GS. 2011. Detection and evaluation of novel herpesviruses in routine and pathological samples from Asian and African elephants: identification of two new probosciviruses (EEHV5 and EEHV6) and two new gammaherpesviruses (EGHV3B and EGHV5). Vet Microbiol 147:28–41. doi: 10.1016/j.vetmic.2010.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richman LK, Zong J-C, Latimer E, Lock J, Fleischer RC, Heaggans SY, Hayward GS. 2014. Elephant endotheliotropic herpesviruses EEHV1A, EEHV1B, and EEHV2 from cases of hemorrhagic disease are highly diverged from other mammalian herpesviruses and may form a new subfamily. J Virol 88:13523–13546. doi: 10.1128/JVI.01673-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zong J-C, Latimer EM, Long SY, Richman LK, Heaggans SY, Hayward GS. 2014. Comparative genome analysis of four elephant endotheliotropic herpesviruses, EEHV3, EEHV4, EEHV5, and EEHV6, from cases of hemorrhagic disease or viremia. J Virol 88:13547–13569. doi: 10.1128/JVI.01675-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehlers B, Dural G, Marschall M, Schregel V, Goltz M, Hentschke J. 2006. Endotheliotropic elephant herpesvirus, the first betaherpesvirus with a thymidine kinase gene. J Gen Virol 87:2781–2789. doi: 10.1099/vir.0.81977-0. [DOI] [PubMed] [Google Scholar]
- 7.Fickel J, Lieckfeldt D, Richman LK, Streich WJ, Hildebrandt TB, Pitra C. 2003. Comparison of glycoprotein B (gB) variants of the elephant endotheliotropic herpesvirus (EEHV) isolated from Asian elephants (Elephas maximus). Vet Microbiol 91:11–21. doi: 10.1016/S0378-1135(02)00264-X. [DOI] [PubMed] [Google Scholar]
- 8.Garner MM, Helmick K, Ochsenreiter J, Richman LK, Latimer E, Wise AG, Maes RK, Kiupel M, Nordhausen RW, Zong J-C, Hayward GS. 2009. Clinico-pathologic features of fatal disease attributed to new variants of endotheliotropic herpesviruses in two Asian elephants (Elephas maximus). Vet Pathol 46:97–104. doi: 10.1354/vp.46-1-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atkins L, Zong J-C, Tan J, Mejia A, Heaggans SY, Nofs SA, Stanton JJ, Flanagan JP, Howard L, Latimer E, Stevens MR, Hoffman DS, Hayward GS, Ling PD. 2013. EEHV-5, a newly recognized elephant herpesvirus associated with clinical and subclinical infections in captive Asian elephants (Elephas maximus). J Zoo Wildl Med 44:136–143. doi: 10.1638/1042-7260-44.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denk D, Stidworthy MF, Redrobe S, Latimer E, Hayward GS, Cracknell J, Claessens A, Steinbach F, McGowan S, Dastjerdi A. 2012. Fatal elephant endotheliotropic herpesvirus type 5 infection in a captive Asian elephant. Vet Rec 171:380–381. doi: 10.1136/vr.e6833. [DOI] [PubMed] [Google Scholar]
- 11.Zachariah A, Zong J-C, Long SY, Latimer EM, Heaggans SY, Richman LK, Hayward GS. 2013. Fatal herpesvirus (EEHV) hemorrhagic disease in wild and orphan Asian elephants in Southern India. J Wildl Dis 49:381–393. doi: 10.7589/2012-07-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ossent P, Guscetti F, Metzler AE, Lang EM, Rübel A, Hauser B. 1990. Acute and fatal herpesvirus infection in a young Asian elephant (Elephas maximus). Vet Pathol 27:131–133. doi: 10.1177/030098589002700212. [DOI] [PubMed] [Google Scholar]
- 13.Richman LK, Montali RJ, Cambre RC, Schmitt D, Hardy D, Hildbrandt T, Bengis RG, Hamzeh FM, Shahkolahi A, Hayward GS. 2000. Clinical and pathological findings of a newly recognized disease of elephants caused by endotheliotropic herpesviruses. J Wildl Dis 36:1–12. doi: 10.7589/0090-3558-36.1.1. [DOI] [PubMed] [Google Scholar]
- 14.Stanton JJ, Zong J-C, Latimer E, Tan J, Herron A, Hayward GS, Ling PD. 2010. Detection of pathogenic elephant endotheliotropic herpesvirus in routine trunk washes from healthy adult Asian elephants (Elephas maximus) by use of a real-time quantitative polymerase chain reaction assay. Am J Vet Res 71:925–933. doi: 10.2460/ajvr.71.8.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stanton JJ, Zong J-C, Eng C, Howard L, Flanagan J, Stevens S, Schmitt D, Wiedner E, Graham D, Junge R, Weber MA, Fischer M, Mejia A, Tan J, Latimer E, Herron A, Hayward GS, Ling PD. 2013. Kinetics of viral loads and genotypic analysis of elephant endotheliotropic herpesvirus-1 infection in captive Asian elephants (Elephas maximus). J Zoo Wildl Med 44:42–54. doi: 10.1638/1042-7260-44.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuery A, Tan J, Peng RS, Flanagan JP, Tocidlowski ME, Howard LL, Ling PD. Clinical infection of two captive Asian elephants (Elephas maximus) with elephant endotheliotropic herpesvirus 1B. J Zoo Wildl Med, in press. [DOI] [PubMed] [Google Scholar]
- 17.Schmitt DL, Hardy DA, Montali RJ, Richman LK, Lindsay WA, Isaza R, West G. 2000. Use of famciclovir for the treatment of endotheliotrophic herpesvirus infections in Asian elephants (Elephas maximus). J Zoo Wildl Med 31:518–522. doi: 10.1638/1042-7260(2000)031[0518:UOFFTT]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 18.Fuery A, Browning GR, Ling PD, Tan J, Long SY, Hayward GS, Cox SK, Flanagan JP, Tocidlowski ME, Howard LL, Ling PD. Clinical infection of captive elephants (Elephas maximus) with elephant endotheliotropic herpesvirus 4. J Zoo Wildl Med, in press. [DOI] [PubMed] [Google Scholar]
- 19.Hayward GS. 2012. Conservation: clarifying the risk from herpesvirus to captive Asian elephants. Vet Rec 170:202–203. doi: 10.1136/vr.e1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sripiboon S, Tankaew P, Lungka G, Thitaram C. 2013. The occurrence of elephant endotheliotropic herpesvirus in captive Asian elephants (Elephas maximus): first case of EEHV4 in Asia. J Zoo Wildl Med 44:100–104. doi: 10.1638/1042-7260-44.1.100. [DOI] [PubMed] [Google Scholar]
- 21.Richman LK. 2003. Pathological and molecular aspects of fatal endotheliotropic herpesviruses of elephants. PhD thesis. Johns Hopkins University, Baltimore, MD. [Google Scholar]
- 22.Ling PD, Reid JG, Qin X, Muzny DM, Gibbs R, Petrosino J, Peng R, Zong J-C, Heaggans SY, Hayward GS. 2013. Complete genome sequence of elephant endotheliotropic herpesvirus 1A. Genome Announc 1(2):e0010613. doi: 10.1128/genomeA.00106-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilkie GS, Davison AJ, Kerr K, Stidworthy MF, Redrobe S, Steinbach F, Dastjerdi A, Denk D. 2014. First fatality associated with elephant endotheliotropic herpesvirus 5 in an Asian elephant: pathological findings and complete viral genome sequence. Sci Rep 4:6299. doi: 10.1038/srep06299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilkie GS, Davison AJ, Watson M, Kerr K, Sanderson S, Bouts T, Steinbach F, Dastjerdi A. 2013. Complete genome sequences of elephant endotheliotropic herpesviruses 1A and 1B determined directly from fatal cases. J Virol 87:6700–6712. doi: 10.1128/JVI.00655-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pellett PE. 2014. Trunkloads of viruses. J Virol 88:13520–13522. doi: 10.1128/JVI.02359-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCully RM, Basson PA, Pienaar JG, Erasmus BJ, Young E. 1971. Herpes nodules in the lung of the African elephant (Loxodonta africana (Blumebach, 1792)). Onderstepoort J Vet Res 38:225–235. [PubMed] [Google Scholar]
- 27.Jacobson ER, Sundberg JP, Gaskin JM, Kollias GV, O'Banion MK. 1986. Cutaneous papillomas associated with a herpesvirus-like infection in a herd of captive African elephants. J Am Vet Med Assoc 189:1075–1078. [PubMed] [Google Scholar]
- 28.Richman LK, Montali RJ, Hayward GS. 2000. Review of a newly recognized disease of elephants caused by endotheliotropic herpesviruses. Zoo Biol 19:383–392. doi:. [DOI] [PubMed] [Google Scholar]
- 29.Pearson VR. 2012. Finding EEHVs in wild Kenyan elephants. Elephant Managers Association, Lake Buena Vista, FL: https://elephantmanagers.com/uploads/ElephantManagersArticleKWS__1_.doc. [Google Scholar]
- 30.Ortega J, Corpa JM, Orden JA, Blanco J, Carbonell MD, Gerique AC, Latimer E, Hayward GS, Roemmelt A, Kraemer T, Romey A, Kassimi LB, Casares M. 2015. Acute death associated with Citrobacter freundii infection in an African elephant (Loxodonta africana). J Vet Diagn Invest 27:632–636. doi: 10.1177/1040638715596034. [DOI] [PubMed] [Google Scholar]
- 31.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rose TM. 2005. CODEHOP-mediated PCR—a powerful technique for the identification and characterization of viral genomes. Virol J 2:20. doi: 10.1186/1743-422X-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wellehan JF, Johnson AJ, Childress AL, Harr KE, Isaza R. 2008. Six novel gammaherpesviruses of Afrotheria provide insight into the early divergence of the Gammaherpesvirinae. Vet Microbiol 127:249–257. doi: 10.1016/j.vetmic.2007.08.024. [DOI] [PubMed] [Google Scholar]
- 34.Masters NJ, Stidworthy MF, Everest DJ, Dastjerdi A, Baulmer S. 2011. Detection of EGHV-5 in a self-limiting papilloma-like lesion in the trunk of an Asian elephant (Elephas maximus). Vet Rec 169:209. doi: 10.1136/vr.d4226. [DOI] [PubMed] [Google Scholar]
- 35.Reid CE, Hildebrandt TB, Marx N, Hunt M, Thy N, Reynes JM, Schaftenaar W, Fickel J. 2006. Endotheliotropic elephant herpes virus (EEHV) infection. The first PCR-confirmed fatal case in Asia. Vet Q 28:61–64. doi: 10.1080/01652176.2006.9695209. [DOI] [PubMed] [Google Scholar]
- 36.Bouchard B, Xaymountry B, Thongtip N, Lertwatcharasarakul P, Wajjwalku W. 2014. First reported case of elephant endotheliotropic herpes virus infection in Laos. J Zoo Wildl Med 45:704–707. doi: 10.1638/2013-0264R1.1. [DOI] [PubMed] [Google Scholar]
- 37.Schaftenaar W, Reid C, Martina B, Fickel J, Osterhaus AD. 2010. Nonfatal clinical presentation of elephant endotheliotropic herpes virus discovered in a group of captive Asian elephants (Elephas maximus). J Zoo Wildl Med 41:626–632. doi: 10.1638/2009-0217.1. [DOI] [PubMed] [Google Scholar]
- 38.Hardman K, Dastjerdi A, Gurrala R, Routh A, Banks M, Steinbach F, Bouts T. 2012. Detection of elephant endotheliotropic herpesvirus type 1 in asymptomatic elephants using TaqMan real-time PCR. Vet Rec 170:205. doi: 10.1136/vr.100270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.