ABSTRACT
Influenza virus mRNA synthesis by the RNA-dependent RNA polymerase involves binding and cleavage of capped cellular mRNA by the PB2 and PA subunits, respectively, and extension of viral mRNA by PB1. However, the mechanism for such a dynamic process is unclear. Using high-throughput mutagenesis and sequencing analysis, we have not only generated a comprehensive functional map for the microdomains of individual subunits but also have revealed the PA linker to be critical for polymerase activity. This PA linker binds to PB1 and also forms ionic interactions with the PA C-terminal channel. Nearly all mutants with five-amino-acid insertions in the linker were nonviable. Our model further suggests that the PA linker plays an important role in the conformational changes that occur between stages that favor capped mRNA binding and cleavage and those associated with viral mRNA synthesis.
IMPORTANCE The RNA-dependent RNA polymerase of influenza virus consists of the PB1, PB2, and PA subunits. By combining genome-wide mutagenesis analysis with the recently discovered crystal structure of the influenza polymerase heterotrimer, we generated a comprehensive functional map of the entire influenza polymerase complex. We identified the microdomains of individual subunits, including the catalytic domains, the interaction interfaces between subunits, and nine linkers interconnecting different domains. Interestingly, we found that mutants with five-amino-acid insertions in individual linkers were nonviable, suggesting the critical roles these linkers play in coordinating spatial relationships between the subunits. We further identified an extended PA linker that binds to PB1 and also forms ionic interactions with the PA C-terminal channel.
INTRODUCTION
Influenza A virus is a major public health problem, infecting as many as 500 million people a year worldwide and causing more than 500,000 deaths (1). Its RNA-dependent RNA polymerase (RdRp) consists of the PA (polymerase acidic), PB1 (polymerase basic 1), and PB2 (polymerase basic 2) subunits and has been the focus of a great deal of research (2–6). The nearly complete crystal structure of the bat influenza virus polymerase was recently published, providing a physical map of the subunits (7). Although it is a promising drug target, its multiple functions and how they relate to specific structures within the heterotrimer have not yet been fully elucidated.
Once the virus has invaded the host cell nucleus, the RdRp transcribes the vRNA genome into capped and polyadenylated mRNAs. The cap-binding domain of the PB2 subunit binds the 5′ 7-methyl-guanosine cap structure of a cellular pre-mRNA (8, 9), and subsequently the N-terminal region of the PA subunit endonuclease (PAn) is thought to cleave off and snatch the 10 to 15 nucleotides (nt) downstream of the cap (10, 11), allowing them to serve as a primer for mRNA synthesis on the vRNA template through a conformational change in PB2. The ends of the single-stranded vRNAs then reassociate to restore the dsRNA promoter structure, leading to PB1 initiating polyadenylation of the viral mRNA (8, 12). The N terminus of PB2 (PB2n) also is responsible for binding with the C-terminal region of PB1 (PB1c) and the replication of the vRNA genome, using cRNA intermediates as templates (8, 13, 14). The C terminus of PB2 (PB2c) carries a bipartite nuclear localization signal (NLS), which is thought to be responsible for α5 importin binding (15, 16). However, the functions of many microdomains of the influenza virus's heterotrimeric replication machinery remain unclear.
Our approach has involved using a transposon mutagenesis system to generate short in-frame insertions and create ∼105 different insertion clones from a single reaction for each subunit gene. This nonbiased high-throughput analysis provides a rapid and effective method to pinpoint microdomains of individual subunits for further functional studies. Here, we have shown the first structure-function analysis of the entire influenza polymerase complex, including catalytic regions and nine linkers connecting different microdomains. Information we acquired from this approach has led us to discover the function of an extended PA linker, which binds to PB1 and also forms ionic interactions with the PA C-terminal (PAc) channel. Our data also have highlighted the critical and previously uncharacterized role influenza A virus linkers play in coordinating the spatial interactions and functions of the polymerase complex.
The elucidation of the structure-function relationship of the influenza virus replication machinery is indispensable not only for a profound mechanistic understanding of the virus's infectivity, activity, and pathogenesis, but also for designing more effective therapeutics against this recurring pathogen that has caused and will continue to cause significant morbidity and mortality.
MATERIALS AND METHODS
Cells and plasmids.
Human embryonic kidney 293 cells (HEK293T) and Madin-Darby canine kidney cells (MDCK) were purchased from the ATCC and maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (FBS; Gibco) and penicillin-streptomycin (100 U/ml and 50 μg/ml, respectively). All cells were incubated at 37°C and 5% CO2. Plasmid pPolI-CAT-RT and influenza A/WSN/33 virus-derived plasmids pcDNA-PB2/PB1/PA/NP were described previously (6).
Generation of mutagenesis library of PA, PB1, and PB2.
All three gene segments (PA, PB1, and PB2) of influenza virus A/WSN/33 underwent Mu transposon-mediated mutagenesis (MGS kit; Finnzymes) according to the manufacturer's instructions. This resulted in three mutant libraries, with each gene containing a randomly positioned 15-nt insertion that includes the 10-nt unique sequence 5′-TGCGGCCGCA-3′. The mutated segment and seven wild-type (WT) plasmids were cotransfected into HEK293T cells for virus packaging (17), and virus then was collected and used to infect MDCK cells for 24 to 48 h for each of the three consecutive passages. The 2nd and 3rd passages in MDCK cells were achieved through subsequent inoculation of the viral pool at a multiplicity of infection (MOI) of 0.1 from the previous passage. Treatment at an MOI of 0.1 still maintains genetic diversity and does not introduce artificial stochastic bottlenecks. To ensure that there was no contamination with DNA used for transfection, the medium was replaced 12 h postinfection. Total RNA of the mutant pool from all passages of virus was isolated using TRIzol and converted to cDNA with iScript (Life Technology). The mutants were genotyped by PCR amplification with gene-specific forward primers and a Vic-labeled reverse primer against the 10-nt insertion. To minimize PCR bias, we designed forward primers to hybridize 300 to 500 nt apart to ensure complete coverage of each gene (see Table S1 in the supplemental material). The final product was sequenced using a 96-capillary DNA Analyzer with the size standard Liz 500 (3730xl DNA Analyzer; Applied Biosystems) at the UCLA GenoSeq Core facility. Sequencing data were analyzed for clarity using the ABI software with the following criteria: (i) all data passed the standard default detection level; (ii) the first 70 nt were removed due to nonspecific background noise; (iii) all data were aligned to the nearest nucleotide or amino acid position in the specific gene; and (iv) all genotyping experimental data were normalized with WT WSN, nontransfected cells, and a different gene library as controls. This eliminated nonspecific data from the PCR, primers, and DNA analyzer.
Viral packaging assay.
A packaging reporter was constructed by replacing nt 67 to 2158 of the PB1 coding sequence (in the PB1 plasmid from the 8-plasmid reverse genetics system) with the open reading frame of green fluorescent protein (GFP). The resulting construct contains GFP flanked by the minimal signal for efficient packaging of the PB1 segment (i.e., the untranslated regions [UTRs] plus 66 nt and 50 nt of the PB1 coding sequence at the 3′ and 5′ ends, respectively) (18). Mutants containing 15-nt insertions at the indicated positions were generated by site-directed mutagenesis. For the viral packaging assay, 293T cells were transfected with the 8-plasmid system plus the packaging reporter. Forty-eight hours later, supernatants from transfected cells were collected, cleared by centrifugation, and used for infection on fresh target cells. At 16 h postinfection, cells were stained with an anti-NP antibody (AA5H) to quantify total infection. A GFP+ NP+ double-positive population indicates infection by viruses that have packaged the GFP packaging reporter. Packaging efficiency was calculated by dividing the percentage of GFP+ NP+ double-positive cells by the percentage of NP+ cells.
Construction of the polymerase complex model.
The influenza virus polymerase complex functional map was constructed using the published crystal structure of the complete polymerase complex (7). We adapted the influenza A virus polymerase complex structure (PDB code 4WSB), and all mappings were made using UCSF Chimera (19).
Primer extension assay.
Plasmid pPolI-CAT-RT and A/WSN/33 virus-derived plasmids pcDNA-PB2/PB1/PA/NP were described previously. The primer extension assay has also been described (6).
Minireplicon system for influenza A virus.
HEK293T cells were transfected with Lipo2000 (Invitrogen) in 6-well plates. A/WSN/33-derived PB2-, PB1-, PA-, and NP-expressing plasmids and a luciferase reporter plasmid, pPolI-NP-luc, were cotransfected into 293T cells. Renilla luciferase activity was used for normalizing variation in transfection efficiency. Twenty-four hours posttransfection, firefly luciferase and renilla luciferase of cell lysates were detected by the Dual-Glo luciferase assay system (Promega) using a microplate reader (GENios Plus; Tecan). The activity detected with samples containing WT PA was set to 100%.
Polymerase assay and strand-specific real-time PCR.
A minigenome reporter for influenza virus polymerase activity was constructed. Briefly, the Gaussia luciferase (GLuc) gene flanked by the UTRs from the PA segment of the WSN virus was inserted between the human polymerase I (Pol I) promoter and mouse Pol I terminator in the reverse-sense orientation. Cotransfection of WSN PB1, PB2, PA, and NP expression constructs results in the expression of GLuc protein, which was used as an indicator of polymerase activity. The mRNA, vRNA, and cRNA transcript levels were quantified using a strand-specific real-time RT-PCR method developed by Kawakami et al., which has been shown to be able to distinguish between the three different types of RNA with high specificity. The same tags as those described by Kawakami et al. were used, and primers were designed to target the GLuc gene (20).
Bimolecular luminescence complementation assay (BiLC) assay.
The GLuc gene was divided at site 109 into C- and N-terminal fragments labeled GC and GN, respectively. PB1 was cloned upstream of GN, and PA was cloned upstream of GC to create fusion products. Additional constructs containing truncated fragments of PA (residues 1 to 195, 196 to 250, 196 to 270, 220 to 250, and 220 to 270) or with a 5-amino-acid addition (ALGST) at position 196 or 270 and with a 5-amino-acid deletion from position 266 to 270 also were created. The fused proteins were coexpressed in 293T cells for 24 h. Cells were lysed and protein interactions were assessed with the Gaussia luciferase kit (Promega) using a microplate reader.
Protein immunoblotting.
PB1 and PA were detected by rabbit polyclonal serum (1:1,500), and alpha-tubulin was detected by anti-tubulin antibody (1:2,000). Blots were incubated with primary antibodies for 2 h at room temperature and then incubated for 1 h at room temperature with horseradish peroxidase (HRP)-conjugated secondary antibodies (1:10,000; Beijing ZSBio).
TAP pulldown of influenza virus polymerase complex.
Recombinant PB2-tandem affinity purification (TAP)-tagged influenza virus polymerase containing PA and PB1 were purified and then separated by SDS-PAGE and silver staining as described previously (21).
ApG-primed transcription.
Reactions were conducted using recombinant heterotrimer polymerase and a model vRNA promoter consisting of short 15- and 14-nt synthetic 5′ and 3′ ends in vitro (37°C for 1 h) as described previously (21).
RESULTS
Generation of high-throughput mutagenesis profiles of the PA, PB1, and PB2 proteins.
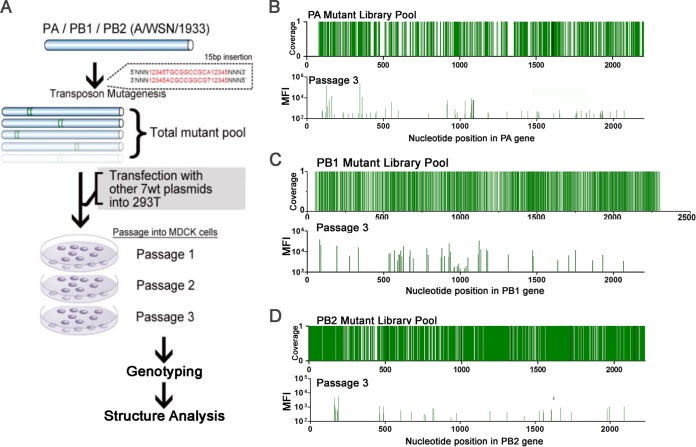
We created three mutant libraries comprised of >105 random in-frame insertions of a 15-nucleotide sequence, 5′-NNNNNTGCGGCCGCA-3′, in the PB1, PB2, and PA gene segments of influenza A/WSN/1933 H1N1 (WSN) using a Mu-transposon mutagenesis method previously described (22). We used the reverse genetics system of influenza virus to transfect mutant plasmid libraries of each polymerase gene segment with the complementary 7 segments of WT WSN into HEK293T cells, creating a mutant virus library (Fig. 1A; also see Fig. S1 and S2 in the supplemental material). High-throughput genotyping of each library shows a low level of insertion tolerance, which is expected given the vital role of polymerase genes. However, we found that ∼50% of the clones that were observed in the passage 1 population remained in passage 3 (Fig. 1B to D, peaks represent mean fluorescence intensity; also see Table S5). One advantage of using Mu-transposon mutagenesis is that it generates random insertions of 5 amino acids in all 3 reading frames without producing stop codons. To verify this, we selected 152 single insertion mutant clones from all 3 libraries and examined polymerase activity (see Table S2 to 4). We found that the viability and the polymerase activity of the single clones strongly supported the high-density mutagenesis results (see Fig. S3 and S4), and 40/42 single clones could express protein at the WT level (data not shown). These data provide the fundamental justification that our insertional mutation method can be used as a high-throughput loss-of-function analysis tool.
FIG 1.
High-throughput mutagenesis profiling of the PA, PB1, and PB2 proteins. (A) The flowchart for the generation of a PB1, PA, and PB2 mutant library pool. Peaks from the PA (B), PB1 (C), and PB2 (D) plasmid library pool demonstrate the total coverage of each subunit, which is cotransfected with 7 WT WSN plasmids in 293T cells and 3 passages in MDCK cells. The in vitro replication efficiency of the entire mutant pool was determined by genotyping of the cell lysates as mean fluorescence intensity (MFI). The list of all viable mutants of each mutant library from passage 1 to 3 can be found in Table S5 in the supplemental material.
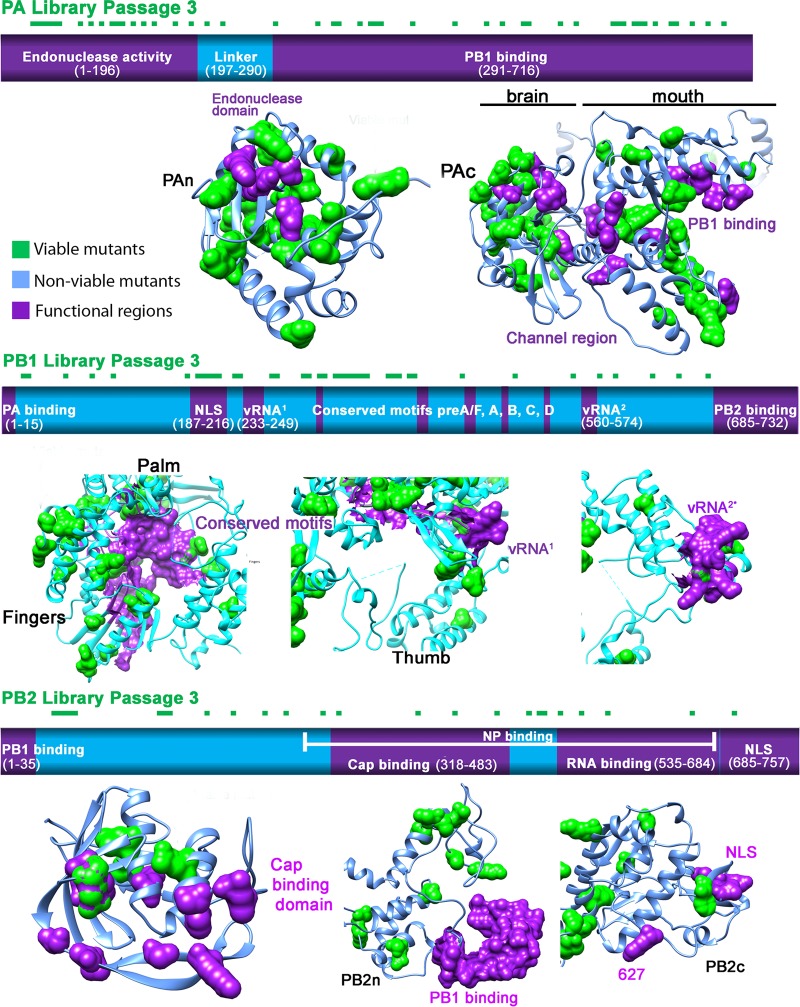
After three rounds of passaging in MDCK cells, we mapped the viable clones of the PA, PB1, and PB2 libraries onto a linear schematic and the published three-dimensional (3D) protein structure (Fig. 2A to C) (7, 23). The regions we identified where viable insertion mutants could not be recovered matched the previously identified functional domains, confirming their importance. For instance, we confirmed that the PB1 library recovered no viable mutations from the vRNA-binding regions (233-249 and 560-574) or the conserved motifs preA to preD (225-252, 300-317, 403-426, 440-452, and 475-487) (24–26). Several mutants in the PB1c-PB2n-binding region were recovered from passage 1, but only one remained by passage 3. The newly discovered PB1 priming site and the β hairpin loop (7), which were suggested to interact with the template and vRNA, tolerated only one insertion mutant. However, we found many viable mutations outside those functional regions (see Fig. S4B in the supplemental material). Several mutants in the single-partite nuclear localizing signal (NLS) regions at amino acid positions 205, 206, 207, and 210 survived through all passages.
FIG 2.
Structure and functional characterization of the influenza virus polymerase complex. Graphical illustration of PA (A), PB1 (B), and PB2 (C) functional domains (based on previous literature; shown in purple) overlapping the passage 3 mutant library as square dots on top of the bar (green). Using the same color scheme, we mapped all of the viable and nonviable (light blue) mutants from passage 3 into the recently identified influenza crystal structure (PDB code 4WSB) (7, 23). The viable and functional regions were highlighted using the surface display.
In the PB2 schematic shown in Fig. 2C, the PB1c- and NP-binding sites both are located at PB2n and overlap. No viable mutants were discovered in the PB2n-PB1c-binding domain. The newly discovered Lid domain, which may open to allow template exiting, tolerates few mutations at the nonflexible α helixes (7). No mutants were recovered within the two suggested cap-binding regions. Mutations in close proximity to amino acid 627, which plays a crucial role in viral transmission, also failed to produce virus. In contrast to the PB1 NLS region, no viable mutants were found in the bipartite NLS regions in PB2c (736-739 and 752-755, respectively).
In the PA schematic shown in Fig. 2A, PAn, which is responsible for endonuclease activity, surprisingly can tolerate multiple insertions, but not at position K102, D108, or K134. What is known is that the function of the endonuclease relies on a negatively charged sequence in these residues (11, 21). The high tolerance we observed also correlates with Hiromoto et al.'s finding that PAn has been the most mutated region in the PA subunit over the course of evolution (27). In contrast to the many viable mutants in the endonuclease domain, we found no mutants in the “mouth” region, particularly the α11, α21, and α22 helixes, or the “brain” of PAc (291-716), which consists of the channel domain and the RNA-binding groove (28). These findings demonstrate that genome-wide insertion mutagenesis can provide important clues in identifying functionally important regions in less characterized proteins.
Characterization of critical PB1 microdomains using insertion mutagenesis.
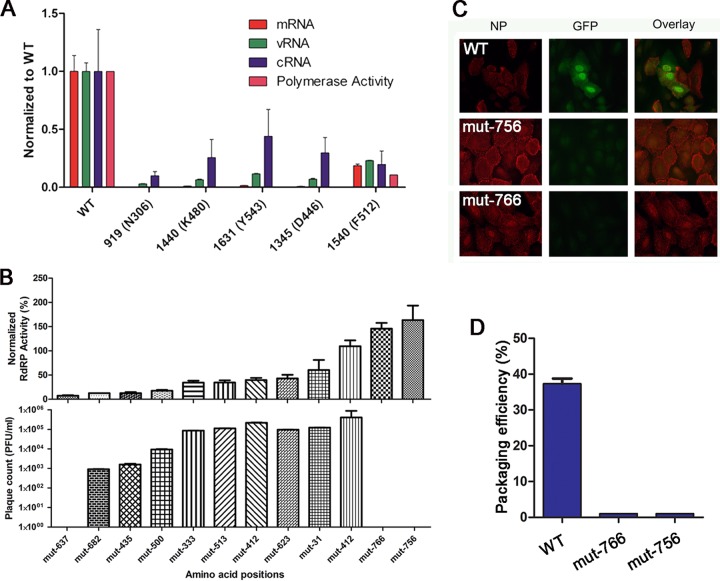
The template-binding and catalytic channel (TBC) is crucial for binding template, substrates, and cofactors, as well as catalyzing the nucleotidyltransferase reactions (26, 29–31). The third passage of the PB1 library revealed that residues inside the TBC could not tolerate insertions, in contrast to residues on the surface away from the TBC (data not shown). We hypothesized that the TBC is involved in RNA synthesis. Therefore, we isolated five individual mutant clones from the PB1 mutant plasmid library (see Table S2 in the supplemental material) with insertions in the TBC and tested their RdRp activity and mRNA, vRNA, and cRNA synthesis efficiency using a minireplicon assay (Fig. 3A). We found that these mutant clones lacked any RdRp activity, were unable to proceed with any step in RNA synthesis, and were unable to produce viable virions when transfected, supporting the critical role of this channel.
FIG 3.
Characterization of PB1 TBC region and viral packaging signals. (A) The effects of individual insertion mutants on the m/c/vRNA synthesis of influenza virus detected using qPCR. (B) The polymerase activity and PFU value of individual PB1 mutants. (C) Viral packaging assay. (D) Packaging signal detection using fluorescence microscopy.
In addition, our mutational analysis uncovered two interesting nonviable PB1 mutants with insertions at amino acid positions 756 and 766, which are at the end and outside the coding region, respectively (Fig. 3B). When we isolated these mutants and tested their RdRp activity, we found levels similar to that of WT PB1, suggesting that polymerase functions related to heterotrimeric interactions and RNA binding were unaffected. However, results from a viral packaging assay showed that these mutants had defects in viral assembly (Fig. 3C and D). This additional involvement in later stages of viral replication suggests that the PB1 3′ UTR plays a more crucial role in viral packaging than previously thought (32, 33).
The heterotrimeric interfaces of the influenza virus polymerase complex.
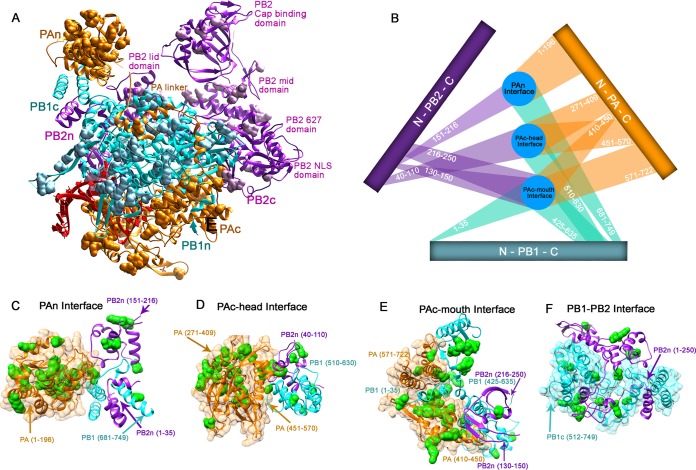
To elucidate the interactions between the subunits of the RdRp, we mapped the viable mutants of PA, PB1, and PB2 recovered from passage 3 onto the polymerase complex protein structure (Fig. 4A). There are several locations where all three subunits share the same interface (Fig. 4C to E). These encompass the entire PA subunit (PAn, PAc-head, and PAc-mouth), the N terminus of PB2 (40-256), and the C terminus of PB1 (425-749; see Fig. S4 in the supplemental material). Our mutational studies were able to recover only a few viable mutants at these sites. PB1 and the entire PB2-N1 domain (1-250) also share an interface. Functional analysis of individual insertion mutants we isolated from these domains showed that they lacked any polymerase activity (see Fig. S4 and Table S3).
FIG 4.
Heterotrimeric interfaces of the influenza polymerase complex. (A) Mapping of all viable mutants onto the crystal structure of the influenza polymerase complex. (B) Schematics of the heterotrimer interfaces. (C to F) The interface locations labeled on the polymerase structure. The viable mutants from passage 3 are labeled in green.
Critical role of the interdomain linkers in the dynamics and functions of influenza polymerase complex.
The structure of the influenza virus RdRp reveals a tightly interlocking architecture between the PA, PB1, and PB2 subunits. The gaps between domains are spanned by numerous interdomain linkers. Using genome-wide profiling, we found that most of the linker regions in all three polymerase proteins do not tolerate any insertional mutations (Fig. 5A), which is unusual given the flexible and nonconserved role typically observed in other proteins with linkers of this length. We hypothesize that the RdRp's linkers play an indispensable role in achieving multiple polymerase functions.
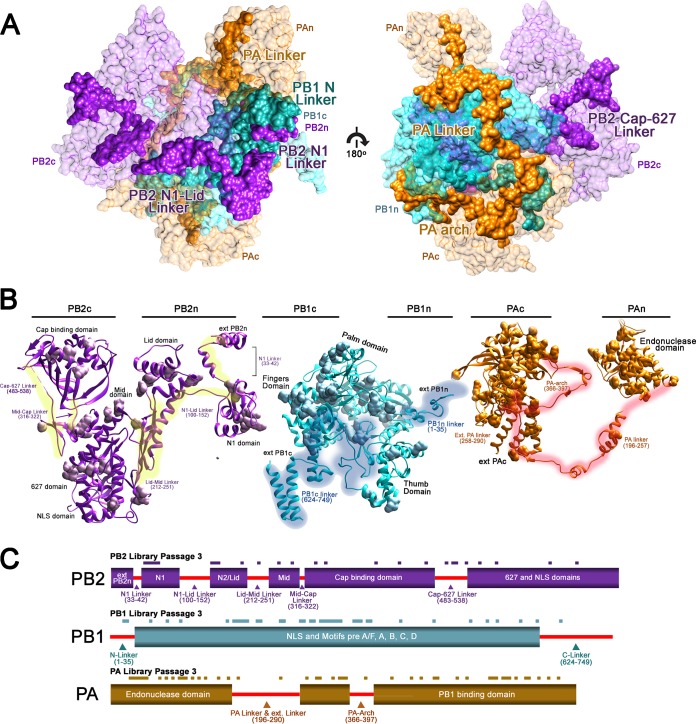
FIG 5.
Critical role of the interdomain linkers in the dynamics and functions of influenza polymerase complex. (A) The locations of each linker from the subunit of the entire RdRp complex are highlighted. (B) Each identified linker for PB2, PB1, and PA are highlighted in yellow, cyan, and red, respectively. The viable mutants from passage 3 were labeled using the surface display. (C) Schematics of the linker and viable mutants from passage 3 from each subunit are shown.
Although they are spread far apart from each other, the domains of PB2 are connected by an interdomain linker (highlighted in yellow in Fig. 5A). The N1 linker (33-42) and the N1-Lid linker (100-152) of the PB2 subunit connect the N1 and Lid domains and are packed closely against PB1c as well as PAn. No viable mutants were discovered in any of these regions, and insertion at nucleotide position 115 completely abolished the polymerase activity (see Fig. S4C in the supplemental material). The Lid-Mid linker (212-251) and Cap-627 linker (483-538) were highly intolerant of mutation, as was the Mid-Cap linker (316-322), which was proposed to facilitate the conformational change in PB2 (7, 23). None of the single clones found in the linker regions had polymerase activity (see Fig. S4A and Table S3). The PB1 subunit is the central subunit of the influenza polymerase complex. We found two distinct linker regions (highlighted in blue in Fig. 5B) at PB1c and PB1n, which are mainly involved in binding PB2 and PA, respectively, and are very intolerant to mutation. The PAc and PAn domains are spread far apart and are proximal to different sides of PB1. The PA linker (196-257; highlighted in red in Fig. 5C) that bridges them, of which a portion has been identified previously and studied (34), circles around PB1 (7). Here, we also have analyzed a region of PB1 (315-345) which directly interacts with the PA linker α7, α8, and α9 helixes (201-257). Using randomly selected single clones, we found that insertions in the PB1-PA linker interaction sites greatly reduce polymerase activity (see Fig. S4A and Table S2). Furthermore, we highlighted two additional linkers, the extended PA linker (258-290) and the PA arch (366-397), which may interact with PAc and vRNA, respectively. None of the three linkers in the PA subunit are very tolerant of insertion mutations. Linear schematics of the mutants recovered from passage 3 of each library mapped onto the linker positions are depicted in Fig. 5C. Our data suggest that genome-wide insertion mutagenesis also can be used to highlight key protein-protein interaction domains for further studies.
Critical ionic interaction between the channel and linker of the PA protein.
Genome-wide mutagenesis of the PA subunit confirmed the importance of the PA linker and channel regions, which have been poorly understood. To further clarify why almost all of the insertion mutants in these regions were nonviable, we generated specific mutations in the PA linker region.
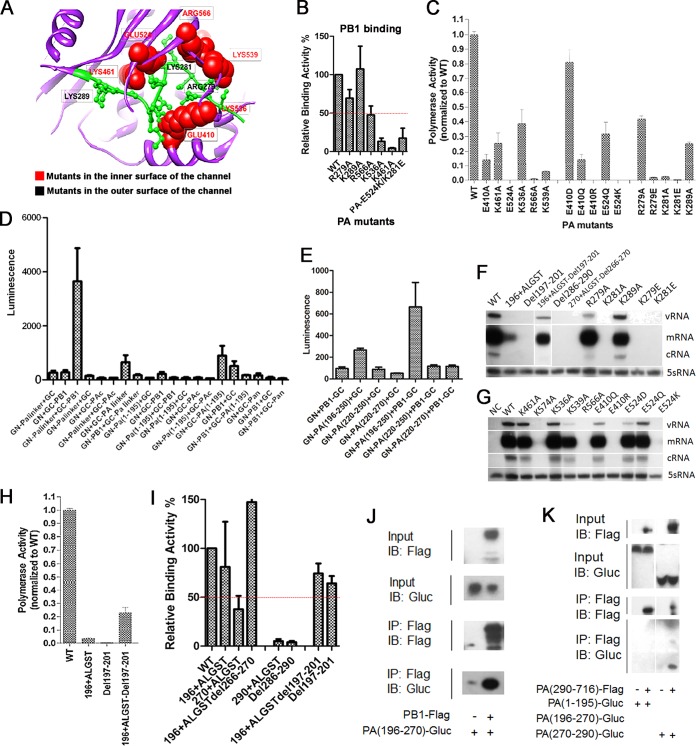
Previous research on the crystal structure of the PAc domain has revealed the presence of a channel-like structure in proximity to the putative RNA-binding groove (28). Although several amino acid residues around this area are known to affect polymerase activity and/or viral growth (6, 35), the biological significance of this channel has yet to be demonstrated and its function remains elusive. Therefore, we systematically mutated the amino acid residues that line the inner surface of the channel (K461, E524, K536, and E410) as well as those that connect the channel to the RNA-binding groove (R566 and K539) (Fig. 6A). Alanine substitution of any of these amino acids resulted in a significant reduction of both binding affinity to PB1 and polymerase activity, with E524A showing the most dramatic effect (Fig. 6B and C).
FIG 6.
Critical ionic interaction between the linker and channel of PA protein. (A) Residues within the PAc channel region (K461, E524, K536, and E410), as well as those that connect the channel to the RNA-binding groove (R566 and K539), are labeled in the PA protein structure. (B) We used BiLC to examine protein binding affinity. The luminescence of Gaussia luciferase (GLuc)-fused PA linker is shown for the indicated mutants. (C) The RdRp activity was measured using the minireplicon assay. (D and E) PA linker truncations were generated to examine their aversion to PB1. Relative binding aversion to PB1 using BiLC was shown. Additional PA linker mutants were generated. The negative control is labeled NC. (F and G) To analyze the level of m/c/vRNA synthesis in the PA mutants, A/WSN/33 virus-derived PB1, PB2, NP, and the indicated PA mutants were coexpressed in 293T cells with pPolI-NA, which can produce NA vRNA. Total RNA was analyzed using a primer extension assay. The relative polymerase activity (H) and PB1 binding affinity (I) of the PA mutant were shown using the minireplicon assay and BiLC, respectively. (J and K) PB1-PA linker and PA linker-PAc interactions between the truncated PA linker mutants and PA channel region were shown using coimmunoprecipitation. IB, immunoblot; IP, immunoprecipitation.
We used a bimolecular luminescence complementation assay (BiLC) to examine protein-binding affinity. The PA linker fused with the N-terminal half of Gaussia luciferase (GN), when expressed together with the full-length PB1 protein fused with the C-terminal half, showed strong luciferase activity. We found that a fragment of the PA linker region (196-250; α7-α9 helices of PA shown in Fig. S5 in the supplemental material) could bind to PB1 but not 220-250 or 220-270 (Fig. 6D and E). We made a series of deletion and insertion mutants at three different locations in the PA linker (amino acids 196, 265, and 285, respectively) to further explore its functions. We found that insertion or deletion of five random amino acids (Ala-Leu-Gly-Ser-Thr) at PA-196 did not affect PB1-binding affinity but did abolish polymerase activity and RNA synthesis (Fig. 6F, H, and I). Deleting or inserting at PA-265 and PA-285 severely impaired its binding affinity to PB1 as well as RdRp polymerase activity. Remarkably, simultaneous insertion and deletion in the original sequence to maintain the length of the linker completely or partially restored binding affinity, polymerase activity, and RNA synthesis. This finding supports our model that the lengths of the linkers are critical for polymerase binding and function.
A short peptide comprised of residues 279 to 289 in the extended PA linker occupies the channel. The structural model suggests that two positively charged amino acids in this peptide (R279 and K281) can interact with the two negatively charged amino acids on the inner surface of the channel (E410 and E524, respectively). Although alanine substitution in R279 and K289 did not alter the binding affinity to PB1, the polymerase activities of R279A, K281A, and K289A were significantly reduced. We confirmed that R279A and K289A retain the heterotrimeric form of the influenza virus polymerase complex using TAP pulldown of the influenza virus polymerase complex (see Fig. S6 in the supplemental material). We performed additional mutagenesis studies to further explore the nature of these interactions. It appears that electrostatic interaction is critical, since changing the residues to the opposite charge completely abolished PA function, and neutralizing the negative charges of E410 and E524 through E-to-Q mutations also severely compromised polymerase activities (Fig. 6C). We also showed that positively charged E410R and E524K mutations abolish the heterotrimeric assembly of the purified influenza virus polymerase proteins (see Fig. S6). Furthermore, we performed an in vitro replication assay using ApG-primed transcription and found that ionic interaction also is functionally important for RNA synthesis (see Fig. S7). We showed that oppositely charged but not similarly or uncharged mutations in residues E410, E524, K279, and K281 could completely abolish RNA synthesis (Fig. 6F and G). The PA linker coimmunoprecipitation supports the model in which the linker is not only crucial for binding with PB1 but also mediates specific interactions with PAc via charged residues (channel region; Fig. 6J and K).
DISCUSSION
Here, we took an unbiased functional genomics approach to generate a comprehensive functional map of the microdomains of the influenza RdRp heterotrimer, which allow us to rapidly pinpoint microdomains of each individual subunit. Guided by these data, we have identified critical domains in the PA linker and channel regions. We showed the critical role these linkers play in coordinating the spatial interactions of the polymerase complex. We also found that the extended PA linker forms ionic interactions with the PA C-terminal channel.
Transposon-based mutagenesis is a valuable and cost-effective tool that can identify critical segments of uncharacterized proteins, guiding further functional analysis (22, 36, 37). We acknowledge that 15-nt insertions are large enough to potentially cause significant changes to the structure or stability of the protein, which could impact our assessment of the function of that specific targeted region. Although we do not have the tools to characterize the global conformation changes caused by these insertions, we were able to verify that subunit expression was only rarely affected in the hundreds of mutant clones that we screened, as the vast majority had a protein level comparable to that of WT samples (data not shown). Given the extensive coverage of our mutant libraries, the loss of a few individual clones within each functional region, which span tens to hundreds of amino acids long, would not be expected to affect our overall observations about those regions. In addition, functional assays of polymerase activity from these clones were consistently in agreement with the viability profile seen in the initial genotyping screen. This confirms that the mutagenesis resulted in the loss of function of these critical subunits.
Guided by the recently solved crystal structure, we highlighted three heterotrimeric interfaces, and mutations at these locations almost always resulted in nonviable virus with partially or fully abolished polymerase activity. The PB1c and PB2n domains are important for their interactions with each other. However, they also may play a much larger role in the complex as a whole, as they share an interface with the entire PA subunit.
A result of our unbiased functional genomics analysis was the surprise discovery of the critical importance of a sophisticated web of linkers connecting the different subunit domains. We identified a total of nine linkers, most of which could not tolerate any insertional mutations and resulted in abolished polymerase activity. We conclude that the linkers play a critical role in the spatial arrangement required to achieve a functional polymerase complex.
In addition, our data demonstrate the importance of the linker length, as the insertion of five amino acids at almost any position in the entire linker region led to nonviable virus. We have also shown that the deletion of five amino acids at different positions abolished the polymerase activity, whereas the addition of five amino acids near the deletion site could partially rescue it. Our mutational studies have demonstrated the essential function of the PA linker in supporting polymerase activity, especially when it affects the interactions between PB1 and PA.
Our comprehensive functional map of the influenza polymerase complex will help in the future design and screening of novel anti-influenza drugs. One potential target is the TBC of the PB1 protein, in which we have shown that almost all of the surrounding residues are conserved. Another strategy is to use small molecules to disrupt the ionic interactions between the linker and the channel of the PA protein. The size of the channel could permit small molecules to enter, and we have shown that every ionic interaction is essential for RNA polymerase activity. Finally, targeting the critical PA linker itself could be the means of fully disrupting influenza virus transcription.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yuying Liang and David Sanchez for providing the 8-plasmid reverse genetics system. We thank Ren Sun for his help in the mutagenesis assay.
We confirm that there are no known conflicts of interest associated with this publication, and there has been no significant financial support for this work that could have influenced its outcome.
L.W., A.W., and Y.E.W. designed and performed most of the experiments. L.W. and A.W. collected the data. H.-W.C., S.L., Y.E.W., J.W., C.L., and F.X.-F.Q. developed the methodology and performed the analysis. T.J. provided structural and bioinformatic analysis. L.W., A.W., Y.E.W., N.Q., and G.C. wrote the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02400-15.
REFERENCES
- 1.WHO. 2003. Influenza: fact sheets. WHO, Geneva, Switzerland. [Google Scholar]
- 2.Kolpashchikov DM, Honda A, Ishihama A. 2004. Structure-function relationship of the influenza virus RNA polymerase: primer-binding site on the PB1 subunit. Biochemistry 43:5882–5887. doi: 10.1021/bi036139e. [DOI] [PubMed] [Google Scholar]
- 3.Kabsch W, Sander C. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 4.Buchan DW, Ward SM, Lobley AE, Nugent TC, Bryson K, Jones DT. 2010. Protein annotation and modelling servers at University College London. Nucleic Acids Res 38:W563–W568. doi: 10.1093/nar/gkq427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, Lipman D. 2008. The influenza virus resource at the National Center for Biotechnology Information. J Virol 82:596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fodor E, Crow M, Mingay LJ, Deng T, Sharps J, Fechter P, Brownlee GG. 2002. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J Virol 76:8989–9001. doi: 10.1128/JVI.76.18.8989-9001.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pflug A, Guilligay D, Reich S, Cusack S. 2014. Structure of influenza A polymerase bound to the viral RNA promoter. Nature 516:355–360. doi: 10.1038/nature14008. [DOI] [PubMed] [Google Scholar]
- 8.Fodor E. 2013. The RNA polymerase of influenza a virus: mechanisms of viral transcription and replication. Acta Virol 57:113–122. doi: 10.4149/av_2013_02_113. [DOI] [PubMed] [Google Scholar]
- 9.Guilligay D, Tarendeau F, Resa-Infante P, Coloma R, Crepin T, Sehr P, Lewis J, Ruigrok RW, Ortin J, Hart DJ, Cusack S. 2008. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat Struct Mol Biol 15:500–506. doi: 10.1038/nsmb.1421. [DOI] [PubMed] [Google Scholar]
- 10.Dias A, Bouvier D, Crepin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RW. 2009. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458:914–918. doi: 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- 11.Yuan P, Bartlam M, Lou Z, Chen S, Zhou J, He X, Lv Z, Ge R, Li X, Deng T, Fodor E, Rao Z, Liu Y. 2009. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature 458:909–913. doi: 10.1038/nature07720. [DOI] [PubMed] [Google Scholar]
- 12.Pritlove DC, Poon LL, Devenish LJ, Leahy MB, Brownlee GG. 1999. A hairpin loop at the 5′ end of influenza A virus virion RNA is required for synthesis of poly(A)+ mRNA in vitro. J Virol 73:2109–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gastaminza P, Perales B, Falcon AM, Ortin J. 2003. Mutations in the N-terminal region of influenza virus PB2 protein affect virus RNA replication but not transcription. J Virol 77:5098–5108. doi: 10.1128/JVI.77.9.5098-5108.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatakeyama D, Shoji M, Yamayoshi S, Hirota T, Nagae M, Yanagisawa S, Nakano M, Ohmi N, Noda T, Kawaoka Y, Kuzuhara T. 2014. A novel functional site in the PB2 subunit of influenza A virus essential for acetyl-CoA interaction, RNA polymerase activity, and viral replication. J Biol Chem 289:24980–24994. doi: 10.1074/jbc.M114.559708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarendeau F, Boudet J, Guilligay D, Mas PJ, Bougault CM, Boulo S, Baudin F, Ruigrok RW, Daigle N, Ellenberg J, Cusack S, Simorre JP, Hart DJ. 2007. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat Struct Mol Biol 14:229–233. doi: 10.1038/nsmb1212. [DOI] [PubMed] [Google Scholar]
- 16.Resa-Infante P, Jorba N, Zamarreno N, Fernandez Y, Juarez S, Ortin J. 2008. The host-dependent interaction of alpha-importins with influenza PB2 polymerase subunit is required for virus RNA replication. PLoS One 3:e3904. doi: 10.1371/journal.pone.0003904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A 97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang Y, Hong Y, Parslow TG. 2005. Cis-acting packaging signals in the influenza virus PB1, PB2, and PA genomic RNA segments. J Virol 79:10348–10355. doi: 10.1128/JVI.79.16.10348-10355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 20.Kawakami E, Watanabe T, Fujii K, Goto H, Watanabe S, Noda T, Kawaoka Y. 2011. Strand-specific real-time RT-PCR for distinguishing influenza vRNA, cRNA, and mRNA. J Virol Methods 173:1–6. doi: 10.1016/j.jviromet.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hara K, Schmidt FI, Crow M, Brownlee GG. 2006. Amino acid residues in the N-terminal region of the PA subunit of influenza A virus RNA polymerase play a critical role in protein stability, endonuclease activity, cap binding, and virion RNA promoter binding. J Virol 80:7789–7798. doi: 10.1128/JVI.00600-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arumugaswami V, Remenyi R, Kanagavel V, Sue EY, Ngoc Ho T, Liu C, Fontanes V, Dasgupta A, Sun R. 2008. High-resolution functional profiling of hepatitis C virus genome. PLoS Pathog 4:e1000182. doi: 10.1371/journal.ppat.1000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reich S, Guilligay D, Pflug A, Malet H, Berger I, Crepin T, Hart D, Lunardi T, Nanao M, Ruigrok RW, Cusack S. 2014. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature 516:361–366. doi: 10.1038/nature14009. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez S, Ortin J. 1999. Characterization of influenza virus PB1 protein binding to viral RNA: two separate regions of the protein contribute to the interaction domain. J Virol 73:631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li ML, Ramirez BC, Krug RM. 1998. RNA-dependent activation of primer RNA production by influenza virus polymerase: different regions of the same protein subunit constitute the two required RNA-binding sites. EMBO J 17:5844–5852. doi: 10.1093/emboj/17.19.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biswas SK, Nayak DP. 1994. Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. J Virol 68:1819–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiromoto Y, Saito T, Lindstrom SE, Li Y, Nerome R, Sugita S, Shinjoh M, Nerome K. 2000. Phylogenetic analysis of the three polymerase genes (PB1, PB2 and PA) of influenza B virus. J Gen Virol 81:929–937. doi: 10.1099/0022-1317-81-4-929. [DOI] [PubMed] [Google Scholar]
- 28.He X, Zhou J, Bartlam M, Zhang R, Ma J, Lou Z, Li X, Li J, Joachimiak A, Zeng Z, Ge R, Rao Z, Liu Y. 2008. Crystal structure of the polymerase PA(C)-PB1(N) complex from an avian influenza H5N1 virus. Nature 454:1123–1126. doi: 10.1038/nature07120. [DOI] [PubMed] [Google Scholar]
- 29.Poch O, Sauvaget I, Delarue M, Tordo N. 1989. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J 8:3867–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C, Wu A, Peng Y, Wang J, Guo Y, Chen Z, Zhang H, Wang Y, Dong J, Wang L, Qin FX, Cheng G, Deng T, Jiang T. 2014. Integrating computational modeling and functional assays to decipher the structure-function relationship of influenza virus PB1 protein. Sci Rep 4:7192. doi: 10.1038/srep07192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferrer-Orta C, Arias A, Escarmis C, Verdaguer N. 2006. A comparison of viral RNA-dependent RNA polymerases. Curr Opin Struct Biol 16:27–34. doi: 10.1016/j.sbi.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Marsh GA, Rabadan R, Levine AJ, Palese P. 2008. Highly conserved regions of influenza a virus polymerase gene segments are critical for efficient viral RNA packaging. J Virol 82:2295–2304. doi: 10.1128/JVI.02267-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hutchinson EC, von Kirchbach JC, Gog JR, Digard P. 2010. Genome packaging in influenza A virus. J Gen Virol 91:313–328. doi: 10.1099/vir.0.017608-0. [DOI] [PubMed] [Google Scholar]
- 34.Guu TS, Dong L, Wittung-Stafshede P, Tao YJ. 2008. Mapping the domain structure of the influenza A virus polymerase acidic protein (PA) and its interaction with the basic protein 1 (PB1) subunit. Virology 379:135–142. doi: 10.1016/j.virol.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 35.Regan JF, Liang Y, Parslow TG. 2006. Defective assembly of influenza A virus due to a mutation in the polymerase subunit PA. J Virol 80:252–261. doi: 10.1128/JVI.80.1.252-261.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heaton NS, Sachs D, Chen CJ, Hai R, Palese P. 2013. Genome-wide mutagenesis of influenza virus reveals unique plasticity of the hemagglutinin and NS1 proteins. Proc Natl Acad Sci U S A 110:20248–20253. doi: 10.1073/pnas.1320524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu NC, Young AP, Al-Mawsawi LQ, Olson CA, Feng J, Qi H, Chen SH, Lu IH, Lin CY, Chin RG, Luan HH, Nguyen N, Nelson SF, Li X, Wu TT, Sun R. 2014. High-throughput profiling of influenza A virus hemagglutinin gene at single-nucleotide resolution. Sci Rep 4:4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.