ABSTRACT
Due to high viral diversity, an effective HIV-1 vaccine will likely require Envs derived from multiple subtypes to generate broadly neutralizing antibodies (bNAbs). Soluble Env mimics, like the native flexibly linked (NFL) and SOSIP trimers, derived from the subtype A BG505 Env, form homogeneous, stable native-like trimers. However, other Env sequences, such as JRFL and 16055 from subtypes B and C, do so to a lesser degree. The high-resolution BG505 SOSIP crystal structures permit the identification and redesign of Env elements involved in trimer stability. Here, we identified structure trimer-derived (TD) residues that increased the propensity of the subtype B JRFL and subtype C 16055 Env sequences to form well-ordered, homogenous, and highly stable soluble trimers. The generation of these spike mimics no longer required antibody-based selection, positive or negative. Using the redesigned subtype B and C trimer representatives as respective foundations, we further stabilized the NFL TD trimers by engineering an intraprotomer disulfide linkage in the prebridging sheet, I201C-A433C (CC), that locks the gp120 in the receptor nontriggered state. We demonstrated that this disulfide pair prevented CD4 induced-conformational rearrangements in NFL trimers derived from the prototypic subtype A, B, and C representatives. Coupling the TD-based design with the engineered disulfide linkage, CC, increased the propensity of Env to form soluble highly stable spike mimics that are resistant to CD4-induced changes. These advances will allow testing of the hypothesis that such stabilized immunogens will more efficiently elicit neutralizing antibodies in small-animal models and primates.
IMPORTANCE HIV-1 displays unprecedented global diversity circulating in the human population. Since the envelope glycoprotein (Env) is the target of neutralizing antibodies, Env-based vaccine candidates that address such diversity are needed. Soluble well-ordered Env mimics, typified by NFL and SOSIP trimers, are attractive vaccine candidates. However, the current designs do not allow most Envs to form well-ordered trimers. Here, we made design modifications to increase the propensity of representatives from two of the major HIV subtypes to form highly stable trimers. This approach should be applicable to other viral Envs, permitting the generation of a repertoire of homogeneous, highly stable trimers. The availability of such an array will allow us to assess if sequential or cocktail immune strategies can overcome some of the vaccine challenges presented by HIV diversity.
INTRODUCTION
The HIV-1 envelope glycoproteins (Envs), which sparsely decorate the viral surface, are the sole targets of host-elicited broadly neutralizing antibodies (bNAbs). A robust antibody response to Env will likely be required to generate a broadly effective HIV vaccine. To generate Env-specific neutralizing antibody responses, soluble mimics have been developed as candidate immunogens with the objective of recapitulating the viral spike. Soluble mimics of Env are difficult to produce, in large part due to the labile nature of the normally noncovalent interaction between gp120 and gp41 subunits (1–11). However, Env modifications stabilize subunit interactions by engineered disulfides, resulting in the so-called SOSIP.664 trimers. These trimers are well-ordered native-spike mimics, requiring cleavage for proper quaternary packing (12–15). The recent high-resolution structure of the BG505 SOSIP trimer reinvigorated efforts to develop an HIV vaccine as it elicits tier 2 neutralizing serum antibodies in preclinical models (16–21). We developed two other SOSIP trimers, the HIV subtype B JRFL- and subtype C 16055-derived trimers which require negative selection to yield homogenous, well-ordered trimers (22), and other clade B and C SOSIPs are also now available (23, 24). Subsequently, we designed a different means to covalently link the subunits, creating cleavage-independent native flexibly linked (NFL) trimers that do not require precursor cleavage. The NFL trimers display a native-like conformation while obviating the need for cleavage by cellular furins required by the SOSIP trimers (25).
BG505, JRFL, and 16055 NFL and SOSIP trimers increase the expanding arsenal of soluble Env mimics to assess immunogenicity in vivo as preclinical vaccine candidates. Subtype A HIV-1 BG505-derived NFL and SOSIP designs form highly homogeneous and thermostable trimers that are purified by an initial affinity step (lectin or antibody), followed by size exclusion chromatography (SEC) (15, 25). JRFL- and 16055- or B41-derived trimers, however, are less homogenous and require antibody affinity negative or positive selection to generate trimer homogeneity (22, 23, 25). In this study, we used BG505 structural information and sequence alignments to generate improved variants of the 16055 and JRFL uncleaved NFL trimers. We demonstrate that substitution of selected residues, defined by the BG505 SOSIP structure, in the 16055 and JRFL NFL contexts results in more homogenous and thermostable trimers, comparable to the trimer of the BG505 NFL or SOSIP. The transfer of these dispersed residues overcomes the need for antibody-based positive or negative selection, yielding subtype B and C well-ordered NFL trimers directly from lectin affinity/SEC purification. This analysis reveals three regions of stability of Env: the gp120-gp41 interface, the prebridging sheet, and the V2/V3 interface. These improvements allowed us to add an additional element of trimer stability, an intraprotomer disulfide bond, without negatively impacting trimer yields. This internal cysteine pair (I201C and A433C, or CC) was designed to prevent primate CD4-induced conformational changes on the trimer, limiting exposure of nonneutralizing epitopes. It does so while still permitting the generation of well-ordered trimeric spike mimics. The lack of CD4 induction is desirable as the well-ordered trimers advance into preclinical vaccine testing in nonhuman primates (NHPs), which possess CD4 that binds gp120 with high affinity. The property of increased stability, along with furin-independence and increased yields, position the improved NFL trimers for future preclinical and clinical utility for assessing vaccine immunogenicity.
MATERIALS AND METHODS
Design of NFL TD trimer constructs.
The 16055 (GenBank accession number EF117268) and JRFL (GenBank accession number U63632) primary sequences were modified as follows to express fully uncleaved but well-ordered soluble gp140 trimers. The furin cleavage motif at the C terminus of gp120, REKR, was genetically deleted and replaced with two copies of the G4S (GGGGS) flexible linker, covalently joining the C terminus of gp120 to the N terminus of gp41. A proline substitution at residue 559 was introduced to facilitate trimer formation (14). The sequence of the membrane-proximal external region (MPER) was deleted from the constructs for better expression (26), and the native signal sequence was replaced by the CD5 leader sequence to increase secretion (27). The gp140 sequence was terminated at D664, followed by a G4S linker, His6 tag, and stop codon. For JRFL, an E168K mutation was introduced to restore PG9/PG16 binding, and K334S was introduced in 16055 to restore the N-glycan at position 332. The gene constructs were codon optimized for mammalian expression and synthesized (GenScript). The gp140s designed here are designated native flexibly linked (NFL) Envs, containing two copies of a G4S linker and designated wild-type (wt) NFL. The NFL BG505 trimer-derived (TD) variants contained the additional substitutions derived from the BG505 HIV Env sequences as follows: for 16055 NFL TD8 variants, E47D, K49E, V65K, E106T, I165L, E429R, R432Q, and A500R; for JRFL NFL TD15 variants, E47D, T49E, V65K, E106T, S164E, I165L, E172V, H308R, E429R, K432Q, K500R, L543N, N553S, G588R, and E662A. The TD CC variants contained the TD modifications and the I201C and A433C substitutions.
Site-directed mutagenesis.
The TD and CC substitutions were introduced via site-directed mutagenesis PCR using a QuikChange Lightning Multi Site-Directed Mutagenesis kit (Agilent Technologies) into NFL (or SOSIP) expression plasmids (CMV-R, where CMV is cytomegalovirus). In brief, single primers were designed for each mutation. We used up to five primers in each reaction mixture to knock in multiple substitutions simultaneously. Reaction products were transformed into competent bacteria and plated onto Luria broth agar plates for colony selection, subsequent plasmid DNA isolation, and sequencing.
Expression and purification of soluble trimeric proteins.
The NFL (or SOSIP) trimeric proteins were transiently expressed as soluble glycoproteins in 293F cells from expression plasmids using 293fectin transfection reagent (Invitrogen). Cell culture supernatants were harvested at 5 to 6 days posttransfection, and the Env-derived proteins were purified by affinity chromatography using a Galanthus nivalis lectin-agarose (Vector Laboratories) column. Bound glycoproteins were eluted with phosphate-buffered saline (PBS) containing 500 mM NaCl and 500 mM methyl-α-d-mannopyranoside and then concentrated with an Amicon filter (100-kDa) to 1 ml. The lectin-purified proteins were subsequently purified by size exclusion chromatography (SEC) using a HiLoad Superdex 200 16/60 column to isolate the trimer fractions.
DSC.
The thermal melting profiles of the 16055 and JRFL trimers were determined by differential scanning calorimetry (DSC) using a MicroCal VP-Capillary differential scanning calorimetry instrument (General Electric). Prior to the DSC melting scan, the protein samples were extensively dialyzed in PBS, pH 7.4, and the concentrations were adjusted to 0.25 mg/ml. The dialysis buffer was used as the reference solution. The DSC experiments were done at a scanning rate of 1 K/min under 3.0 atm. DSC data were analyzed after buffer correction, normalization, and baseline subtraction using CpCalc software provided by the manufacturer.
EM sample preparation.
The purified trimers were analyzed by negative staining electron microscopy (EM). A 3-μl aliquot containing ∼0.03 mg/ml of the sample was applied for 15 s onto a carbon-coated 400-mesh Cu grid that had been glow discharged at 20 mA for 30 s and then negatively stained with 2% uranyl formate for 30 s. Data were collected using an FEI Tecnai Spirit electron microscope operating at 120 kV, with an electron dose of ∼30 e−/Å2 and a magnification of ×52,000 that resulted in a pixel size of 2.05 Å at the specimen plane. Images were acquired with a Tietz 4,000-by-4,000 pixel TemCam-F416 complementary metal-oxide semiconductor (CMOS) camera using a nominal defocus of 1,000 nm and the Leginon package (28).
Data processing.
Particles were picked automatically using DoG Picker and put into a particle stack using the Appion software package (29, 30). Reference-free, two-dimensional (2D) class averages were calculated using particles binned by two via the iterative multivariate statistical analysis/multireference alignment (MSA/MRA) Clustering 2D alignment (31) and IMAGIC software systems (32) and sorted into classes. To analyze the quality of the trimers (closed, open, or nonnative-like trimers), the reference-free 2D class averages were examined by eye using the same metrics that were previously described (23).
Enzyme-linked immunosorbent assay (ELISA).
MaxiSorp plates (Thermo) were coated overnight at 4°C with 1.5 μg/ml of a mouse anti-His tag monoclonal antibody (MAb) (R&D Systems) in PBS, pH 7.5. The next day the plates were incubated at 4°C in blocking buffer (2% [wt] milk powder–5% [vol] fetal bovine serum in PBS, pH 7.5) for 2 h and then washed three times in 0.05% Tween 20-PBS, pH 7.4. The Env-derived soluble trimer was added to the plate (100 μl per well) at a concentration of 2 μg/ml in blocking buffer and incubated at 4°C for 1 h. The plates were washed three times in 0.05% Tween 20-PBS, pH 7.4. The primary antibodies (human anti-HIV Env bNAbs) were added to the plates at a maximum concentration of 5 μg/ml and serially diluted 1:5 in blocking buffer. The plates were incubated at 4°C for 1 h and then washed three times in 0.05% Tween 20-PBS, pH 7.4. A secondary antibody (peroxidase-conjugated goat anti-human IgG antibody; Jackson ImmunoResearch Labs) diluted 5,000 times in blocking buffer was added to the plates and incubated at 4°C for 30 min. Then the plates were washed three times in 0.05% Tween 20-PBS, pH 7.4. The substrate solution (3,3′,5,5′-tetramethylbenzidine [TMB chromogen solution; Invitrogen) was added to the plates (100 μl per well) and incubated at room temperature (RT) for 5 min. A total of 100 μl per well of 1N sulfuric acid was added to stop the reaction, and the plates were read at 450 nm.
Biolayer interferometry (BLI) binding analysis and kinetics.
Kinetic measurements were obtained with an Octet Red instrument immobilizing IgGs on previously hydrated (in PBS pH 7.4) anti-human IgG Fc sensors (Fortebio, Inc.). The NFL trimers were analyzed as free analytes in solution (PBS, pH 7.4). Briefly, the biosensors were immersed in PBS, pH 7.4, containing IgGs at a concentration of 10 μg/ml for 2 min and at 1,000 rpm prior to the encounter with the analyte. The NFL trimer analytes were concentrated to 800 nM and then serially diluted 1:2 to 12.5 nM. The IgG-immobilized sensor was in contact with the analyte in solution for 2 min at 1,000 rpm and then removed from the analyte solution and placed into PBS for another 2 min. These time intervals generated the association and dissociation binding curves reported in this study. Data analysis was done using the ForteBio analysis software, version 7.1 (ForteBio), and the kinetic parameters were calculated using a global fit 1:1 model for applicable MAbs.
RESULTS
Dispersed Env elements increase stability of the NFL trimers.
The BG505 Env-derived NFL glycoprotein readily forms trimers that adopt a native-like conformation. These trimers can be purified in a two-step scheme of lectin affinity chromatography, followed by size exclusion chromatography (SEC). JRFL and 16055 NFL trimers result in a mixture of native and aberrant trimers that are not resolved by this process and require the additional step of negative selection to remove aberrant trimers, contaminating monomers, and dimers (22, 25). Consequently, yields of these trimers are lower than those of BG505 NFL trimers. Also, the JRFL and 16055 NFL trimers display thermostability profiles inferior to those of the BG505 NFLs, possessing lower thermal transition midpoints (Tm) (Fig. 1A). We reasoned that the BG505 Env sequence contained elements of stability, absent in the other two Env sequences, that increase the propensity to form well-ordered stable trimers in both the NFL and SOSIP platforms. By alignment of the three Env sequences of BG505, JRFL, and 16055, we identified BG505 residues potentially involved in enhanced trimer formation. We selected residues that differed in both the 16055 and JRFL Env sequences from those of the BG505 Env sequence. Given the variability of Env sequences, this analysis yielded tens of residues. To interpret the analysis contextually, we annotated these differences in the high-resolution structure of BG505 SOSIP and selected residues proximal to the trimer axis, primarily those near the gp120-gp41 interface. We reasoned that it was at the semiconserved gp120-gp41 interface where the major elements of enhanced BG505 stability would likely reside due to the high diversity between all Envs in the variable regions. This approach allowed us to eliminate many hypervariable residues exposed on the surface of the trimer, reducing the complexity of the analysis.
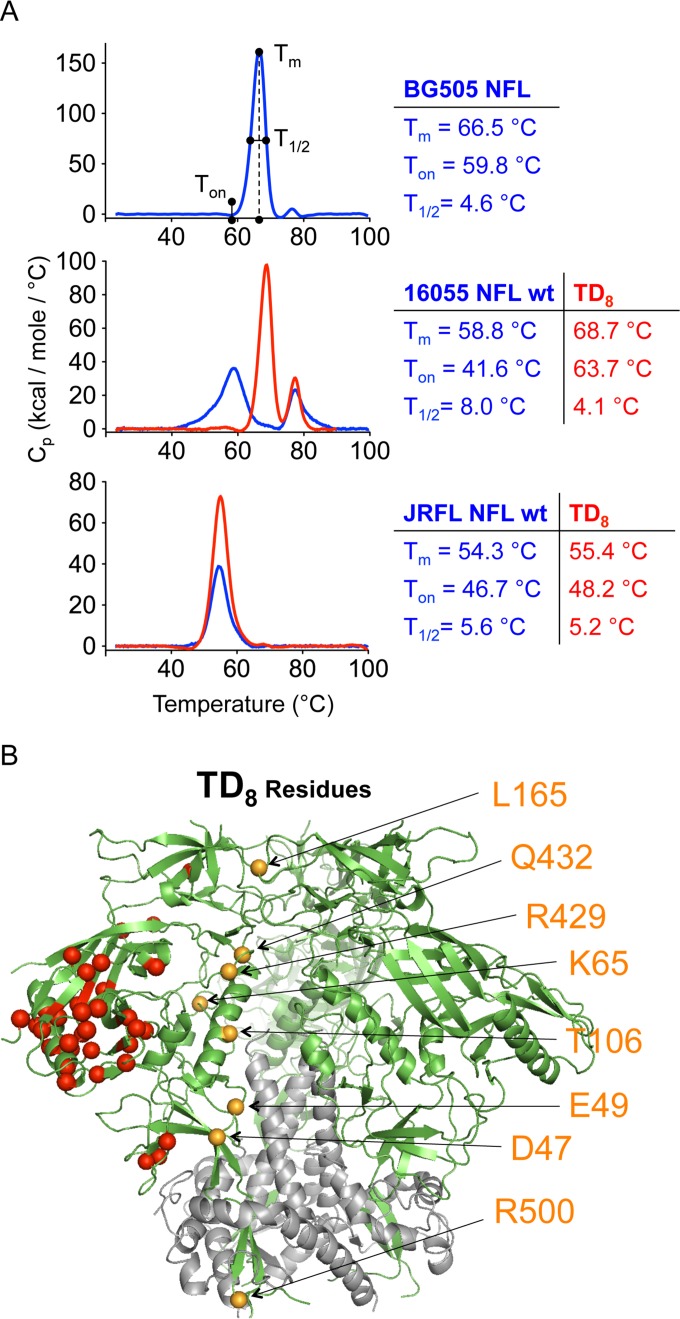
FIG 1.
Modifications and thermostability of NFL trimers. (A) DSC analysis of NFL trimers before and after BG505 trimer-derived (TD) substitutions. Clade A BG505, clade B JRFL, and clade C 16055 NFL wt DSC thermal transition curves are shown in blue while those corresponding to the NFL TD8 versions are shown in red. DSC parameters (Tm, Ton, and T1/2) are displayed next to the curves in corresponding colors. (B) Ribbon representation of the BG505 SOSIP structure (derived from PDB entry 4TVP) where gp120 is shown in green and gp41 is shown in gray. The eight BG505 TD residues located proximal to the trimer axis that were substituted in JRFL and 16055 NFL to generate the TD8 variants are annotated by orange spheres. Red spheres represent residues that were identified in the sequence alignments but that were not evaluated due to their distal location to the trimer axis. Cp, heat capacity at constant pressure.
We identified eight gp120 residues in the BG505 sequence, 47D, 49E, 65K, 106T, 165L, 429R, 432Q, and 500R, that differed in both JRFL and 16055. We termed these amino acids BG505 trimer-derived (TD) residues (Fig. 1B). We transferred these eight residues (TD8) as a full set to 16055 and JRFL NFL coding sequences and evaluated their impact on trimer formation and stability. Following lectin affinity chromatography and SEC purification, we assessed stability of the engineered 16055 and JRFL NFL TD8 variant trimers by differential scanning calorimetry (DSC), revealing positive effects on stability. For the 16055 NFL trimers, the selected TD8 mutations increased the Tm of the 16055 NFL trimers relative to those of the unmodified wild-type (wt) trimers from 58.8°C to 68.7°C. The latter value actually exceeded that of the BG505 NFL trimer (66.5°C). These mutations also increased the Tm of the JRFL Env-derived NFL TD8 trimers (from 54.3°C to 55.4°C), but the effect was less profound. Other DSC parameters, such as the width of the thermal transition at half-height of the DSC peak (T1/2) and the melting starting onset temperature (Ton), also improved for the NFL TD8 variants in comparison with the values for the parental wt variants (Fig. 1A). A lower T1/2 is associated with structural homogeneity, and a higher Ton indicates increased resistance of the trimer to disassemble.
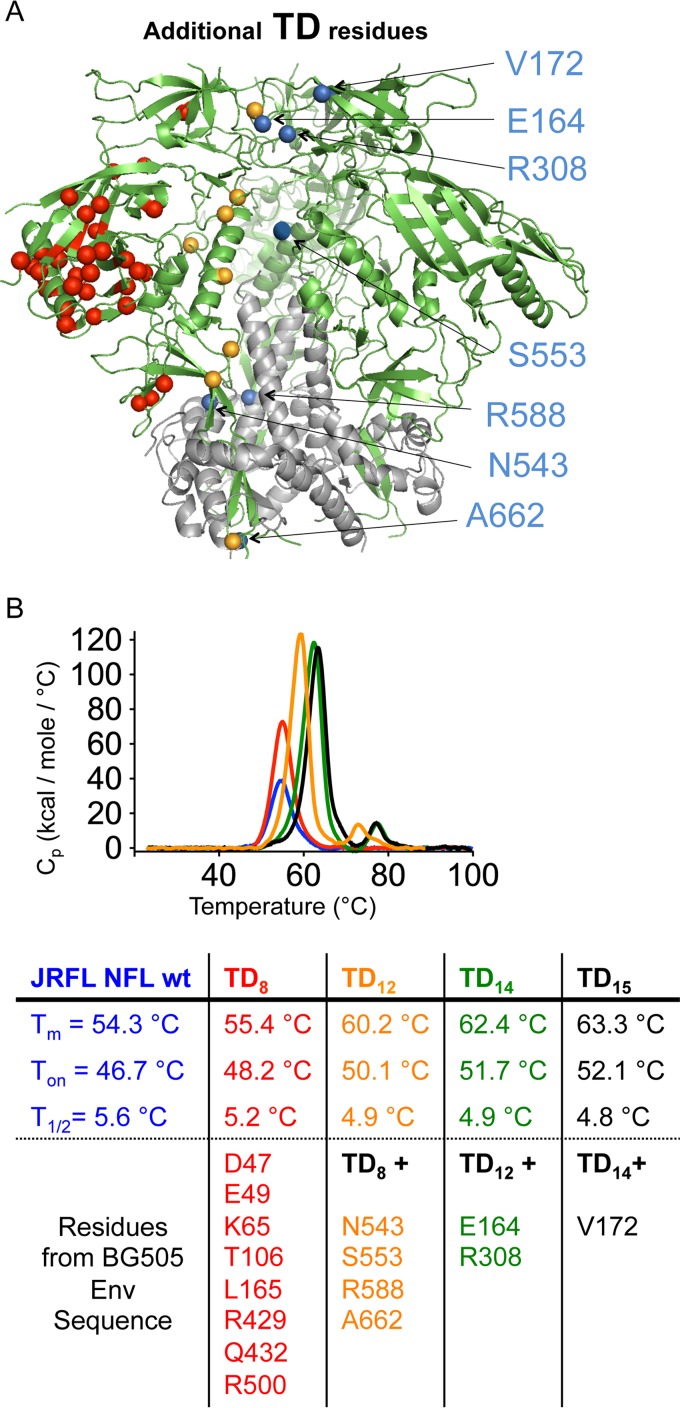
Since the stability of 16055-derived trimers, but not that of the JRFL-Env derived trimers, reached that of the reference BG505 Env-derived trimers, we reasoned that there might be additional elements of Env stability that remained to be identified from our first screen that might further improve the JRFL NFL TD8 variants. By now comparing the JRFL to the composite 16055 and BG505 Env sequences, we identified additional residues that potentially contribute to the superior stability of both BG505 and 16055 TD8 NFL trimers relative to those derived from JRFL. These newly identified residues were located in gp41, at the gp41-gp120 interface (543N, 553S, 588R, and 662A), and in the variable cap loops of gp120 (164E, 172V, and 308R). We transferred a total of 15 residues from BG505/16055 to JRFL to generate JRFL NFL TD15 trimers. These modifications included the original gp120 TD8 residues and the seven newly identified gp41/variable region cap residues (Fig. 2A). DSC analysis of the newly generated JRFL NFL TD15 trimers revealed a considerable enhancement of stability as Tm increased from 54.3°C to 63.3°C with the newly identified residues, consistent with their overall contribution to increased JRFL NFL TD15 stability (Fig. 2B). Along the pathway to TD15 design, we also generated intermediate TD12 and TD14 variants that demonstrated stepwise improvement in trimer stability (Fig. 2B). When interpreted in the context of the SOSIP high-resolution structure, the TD residues clustered in three distinct, but dispersed, regions of Env, the gp120-gp41 interface (residues 47, 49, 65, 106, 500, 543, 553, 588, and 662), the variable loops (residues 164, 165, 172, and 308), and the prebridging sheet (residues 429 and 432) (Fig. 3A). The bridging sheet substitutions (residues 429 and 432) suggested additional modifications that would further stabilize this region, such as covalently anchoring the β21 to β3 via a disulfide linkage that might further increase stability, which we investigated below.
FIG 2.
Additional TD15 modifications and thermostability of JRFL NFL variants. (A) Additional TD substitutions beyond the original TD8 residues (orange) from Fig. 1 are indicated as blue spheres in the context of the BG505 SOSIP structure (PDB accession number 4TVP). A total of 15 residues were transferred from the BG505 envelope glycoprotein sequence to the JRFL NFL trimer to generate the more stable TD15 trimer variant with increased thermostability. (B) DSC thermal transition curves and derived parameters of the JRFL HIV-1 sequence-derived NFL trimers, with the wt in blue, TD8 in red, TD12 in orange, TD14 in green, and TD15 in black, associated with progressive improvement of the JRFL NFL trimer thermal stability.
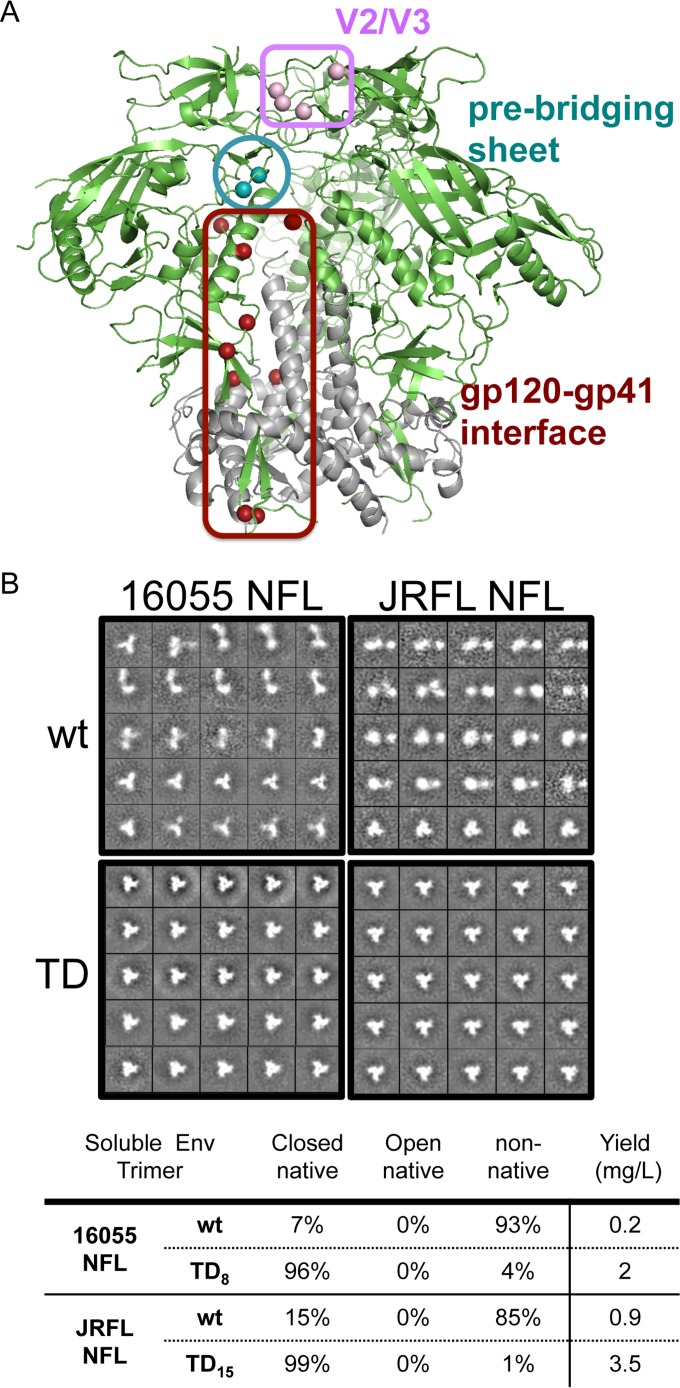
FIG 3.
Regions of stability of soluble Env and representative 2D class averages of NFL trimers by negative staining EM. (A) BG505 TD residues transferred to the JRFL and 16055 HIV sequences to generate more stable NFL TD variant trimers highlight three regions critical for soluble Env stability, namely, the variable region V2/V3 (lavender), the prebridging sheet (teal), and the gp120-gp41 interface (brown). (B) Comparison of representative 2D class averages of the 16055 and JRFL NFL wt trimers (top panels) versus those of the corresponding TD variants (bottom panels) by negative staining EM. Indicated below the EM images are the corresponding calculated proportions of native trimers and nonnative trimers and the final yields of protein in milligrams per liter of transfected cell supernatants.
TD modifications increased NFL trimer formation propensity and yields.
Following lectin affinity chromatography/SEC, we investigated the conformational state of both unmodified (wt) and modified (TD) trimers by negative staining EM. Visual inspection of the EM images of the wt 16055 NFL trimers showed a preponderance of aggregates with the detection of few well-ordered trimers. Computational analysis of the EM images revealed <10% closed, native-like trimers. In contrast, analysis of the more stable 16055 NFL TD8 trimers revealed >95% well-ordered native-like trimers (Fig. 3B). Similarly, EM images of the wt JRFL NFL trimers displayed a low level of well-ordered trimers (∼15%), whereas micrographs of the stabilized JRFL NFL TD15 variants revealed a preponderance of well-ordered trimers, exceeding 95% (Fig. 3B).
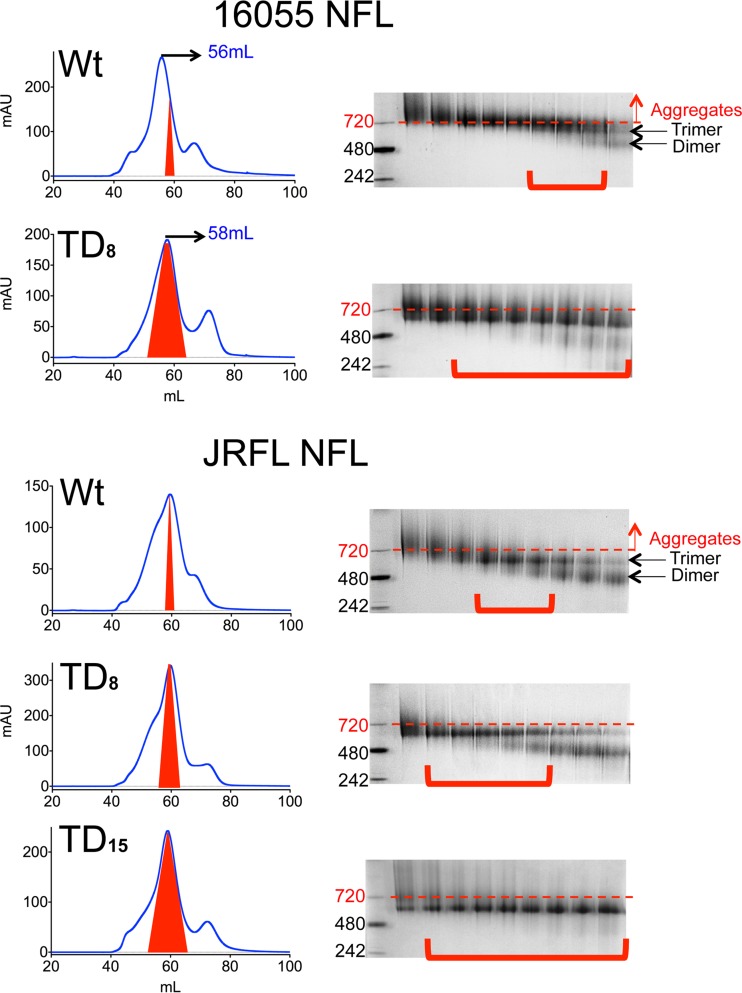
By retrospective analysis, the enhanced propensity of the NFL TD variants to form well-ordered homogenous trimers as determined by DSC and EM was also manifested on SEC profiles and blue native (BN) gels of the trimer fractions collected during purification as follows. The wt 16055 NFL trimer peak is normally detected at an elution volume of 56 ml. However, with the TD modifications, the major peak elution volume shifted to the right on the graph, from 56 to 58 ml, indicating a smaller Stokes radius of the TD trimers (Fig. 4). The collected SEC trimer fractions were analyzed by BN gel electrophoresis, where the more compact nature was corroborated. While the wt 16055 NFL protein showed a preponderance of bands at higher molecular mass aggregates (∼720 kDa), the TD8 variant displayed a majority of bands at the expected molecular size of a well-ordered trimeric soluble Env mimic (∼640 kDa) (Fig. 4). Similarly, a narrower SEC trimer peak was observed for the final JRFL NFL TD15 variant, suggesting an increased propensity to form homogenous trimers, as computed by the EM analysis (Fig. 3B and 4). Accordingly, BN gels revealed a decreased in aggregate, dimer, and monomer bands with the full set of TD15 mutations (Fig. 4). Overall, the negative staining EM analysis, SEC profiles, and BN gels indicated that the TD trimer variants were conformationally more homogeneous and native-like than the parental wt versions. This general improvement in trimer homogeneity and stability indicates that the TD mutations increased the propensity of 16055 and JRFL Envs to form well-ordered trimers, favorably increasing final trimer yields (Fig. 3B).
FIG 4.
SEC profiles and BN gels of lectin affinity-purified trimers. SEC profiles of lectin affinity-purified wt and TD 16055 NFL and JRFL NFL trimer variants are shown. The shaded red area indicates the native-like trimer fractions. These fractions are indicated with red brackets in the BN-PAGE gel images. AU, arbitrary units.
TD trimers are efficiently recognized by bNAbs and inefficiently by nonneutralizing antibodies.
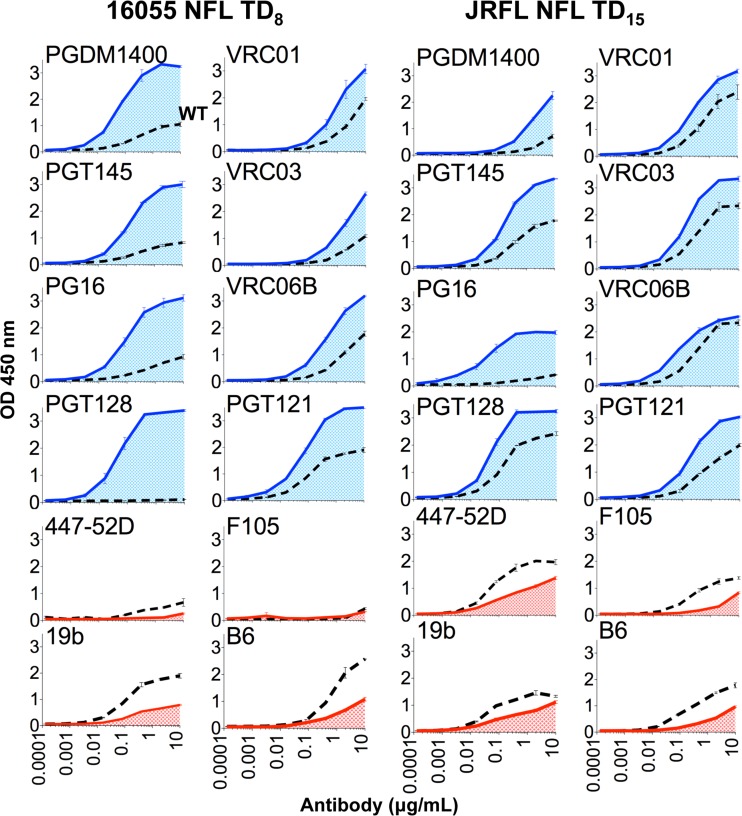
Following EM and DSC analysis, we sought to verify that the favorable antigenic profile of the well-ordered trimers was maintained following the TD modifications. Anti-HIV antibodies targeting different regions of Env provide valuable information about the quaternary conformation of the soluble trimers, as demonstrated in several recent studies (20, 22, 23, 25, 33, 34). The antibodies PGDM1400, PGT145, and PG16 are potent broadly neutralizing antibodies (bNAbs) targeting the variable cap of the spike and are considered to be “trimer preferring” since they selectively bind the native spike and well-ordered native-like soluble trimers (35–37). Efficient recognition by the trimer-preferring antibodies indicates a native-like assembly of the soluble trimers. In contrast, the antibodies 447-52D and 19b, also targeting the V3 region, are considered nonneutralizing antibodies (non-NAbs) since they neutralize only HIV-1 tier 1A or 1B isolates that are not representative of most circulating clinical isolates. Efficient trimer binding by these antibodies is not desired since efficient recognition of these non-NAbs likely indicates an open conformation of Env. Such global opening occurs in disordered trimers lacking proper quaternary packing or, for full-length functional Env and soluble, ordered timers, in the CD4-triggered state. Similarly, at the conserved CD4 binding site (CD4bs), efficient recognition by the CD4-binding site-directed bNAbs, VRC01, VRC03, and VRC06b, is desired, as is inefficient binding by the nonneutralizing CD4bs-directed b6 and F105 MAbs (38, 39). Accordingly, we performed ELISA binding avidity experiments to assess recognition of the NFL TD trimer variants by a panel of bNAbs and non-NAbs. Both 16055 NFL TD8 and JRFL NFL TD15 trimers were well recognized by bNAbs and poorly recognized by non-NAbs at both the variable cap and the CD4 binding regions of Env (Fig. 5). The 16055 NFL TD8 trimer was better recognized by the variable cap trimer-preferring bNAbs PGDM1400, PGT145, and PG16 than JRFL NFL TD15, while the JRFL NFL TD15 trimer was better recognized by the CD4bs-directed bNAbs VRC01, VRC03, and VRC06b (Fig. 5). This recognition pattern is likely explained in part since the “PG” bNAbs were isolated from patients infected with subtype C HIV, whereas the “VRC” bNAbs were isolated from a patient infected with subtype B HIV (35, 36, 38–40). In general, within a given Env context, the bNAbs recognized the TD trimers with higher avidity than non-NAbs, suggesting native presentation of these two sites and quaternary assembly of the TD trimers. The bNAbs PGT121 and PGT128, targeting the N332 supersite, bound with high avidity to both 16055 and JRFL NFL TD trimers. The N332 glycan is naturally absent in the 16055 Env but was introduced in the 16055 NFL TD version to accommodate binding of bNAbs targeting this site of vulnerability (Fig. 5).
FIG 5.
ELISA binding of selected antibodies to the NFL trimers. (A) ELISA binding of selected bNAbs (in blue) and non-NAbs (in red) targeting the variable region cap and CD4bs regions to 16055 NFL TD8 (left) and JRFL NFL TD15 (right) trimers purified by lectin affinity followed by SEC. Black discontinuous lines represent reference values for the wt NFL trimers.
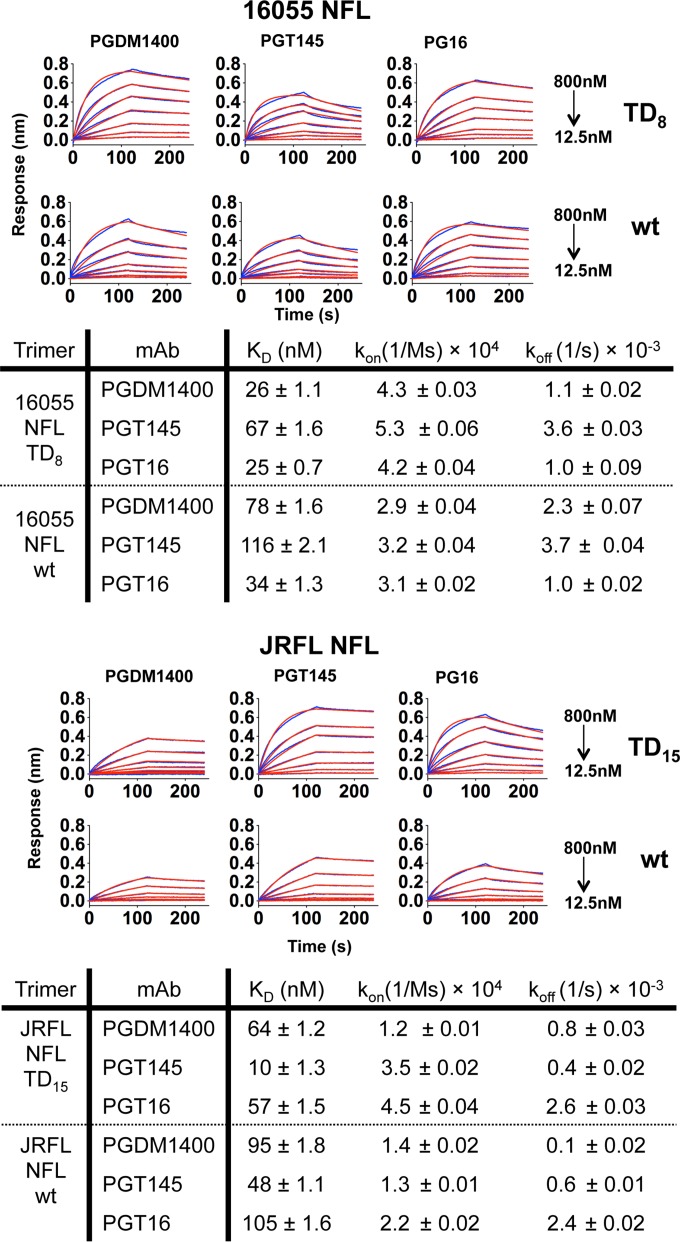
Because PGDM1400, PGT145, and PG16 bind only one site per spike, we were able to determine bona fide binding kinetics and affinities for these trimer-preferring bNAbs by BLI (Octet platform). All of these cap-directed bNAbs recognized the TD trimers with nanomolar affinities, with PGDM1400 and PG16 displaying highest affinities for 16055 NFL TD trimers while PGT145 displayed the highest affinity for JRFL NFL TD trimers (Fig. 6).
FIG 6.
Biolayer interferometry (Octet) kinetic measurements for trimer-preferring bNAbs targeting the V2 cap of Env. Kinetic parameters were derived by biolayer light interferometry using three trimer-preferring bNAbs as ligands immobilized on the sensor surface and the NFL trimers as analytes in solution. KD, equilibrium dissociation constant; kon, association rate constant; koff, dissociation rate constant.
An engineered intraprotomer disulfide (I201C-A433C) link prevents CD4-induced NFL trimer rearrangements.
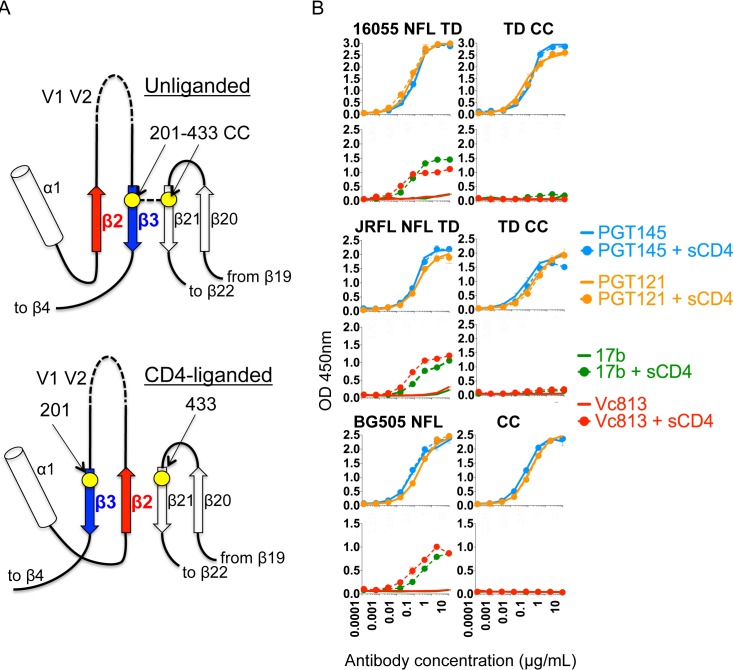
During natural infection, Env on the surface of the virus engages the primary receptor, CD4, on the target T cell, leading to large conformational changes that form the HIV Env coreceptor binding site. Because in the context of vaccination these Env-soluble mimics will encounter CD4 in NHPs and humans, elimination of CD4-induced changes is desirable if other aspects of the ordered trimers are maintained. Consequently, we sought to further stabilize the NFL trimers beyond the TD modifications so that they would not respond to CD4 engagement, preventing CD4-induced conformational changes that would alter quaternary packing and expose nonneutralizing epitopes. For this purpose, we engineered a disulfide bond between gp120 residues 201 on β3 and 433 on β21 of the gp120 subunit (Fig. 7A). A similar approach has recently been reported in the context of the BG505 SOSIP trimer (20). These two β-strands are situated adjacent to each other and in parallel in the pre-receptor-engaged spike structure (17). In contrast, published soluble CD4-liganded gp120 structures reveal β2 rearrangements relative to β3, indicating that CD4 binding influences β20-β21, allowing β2-β3 to rearrange, and formation of the bridging sheet (Fig. 7A) (41). β2-β3 transition forces V1/V2 to pivot 180°, freeing the underlying V3 loop to spring open for coreceptor interaction as part of the viral entry process. This open-trimer conformation is recognized by non-NAbs such as 17b and Vc813 that target the receptor-triggered bridging sheet epitope and in a vaccine context may hinder the elicitation of neutralizing antibodies (39, 42). We can infer formation of the Cys-Cys pair through trimer binding experiments using the antibodies 17b and Vc813 in the absence or presence of soluble CD4 (sCD4). The bridging sheet epitope is not formed in the pre-receptor-engaged spike and becomes accessible only after CD4 engagement. The antibodies 17b and Vc813 efficiently recognized the 16055 NFL TD8, JRFL NFL TD15, and BG505 NFL wt trimers after incubation with sCD4, indicating the formation of the bridging sheet following CD4-induced conformational changes. In contrast, the disulfide-stabilized trimer variants were not recognized by 17b or Vc813 in the absence or presence of sCD4, demonstrating that the bridging sheet was not induced in these additionally stabilized trimers (Fig. 7B). To confirm quaternary packing, we demonstrated that the disulfide-engineered NFL trimers were efficiently recognized by bNAbs, including the trimer-preferring bNAbs, and inefficiently by the nonneutralizing antibodies (Fig. 8A).
FIG 7.
Effect of the stabilizing disulfide (I201C-A433C) on CD4-induced trimer antigenicity with and without sCD4. (A) Schematic model of the prebridging sheet region of the HIV trimer depicting the location of the residues implicated in the formation of the stabilizing intraprotomer disulfide (CC), marked here as yellow circles. The residues 201 and 433 are located within disulfide bond distances on adjacent β-strands (β3 and β21, respectively) in the unliganded HIV trimer while they are separated by a third β-strand, β2, when the trimer is CD4 liganded (from PDB accession number 3J5M). (B) Antigenic changes detected on NFL trimers following CD4 induction by ELISA. Solid lines represent trimer recognition by antibodies in the absence of sCD4, whereas dotted lines represent binding of antibodies following sCD4-induced conformational changes.
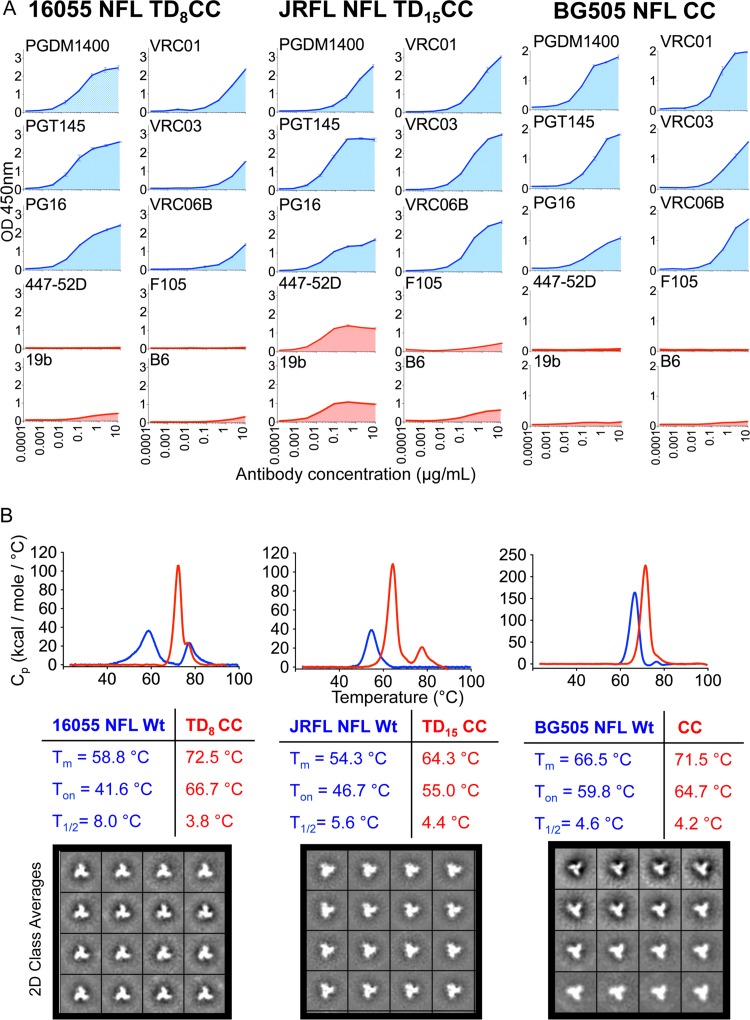
FIG 8.
Biophysical characterization of the disulfide CC-stabilized NFL trimers. (A) ELISA binding curves of selected bNAbs (blue) and non-NAbs (red) to the three disulfide-stabilized NFL trimers representing the three major HIV subtypes, BG505 NFL CC from subtype A, JRFL NFL TD15 CC from subtype B, and 16055 NFL TD8 CC from subtype C. (B) DSC thermal transition curves comparing the wt NFL trimers to the stabilized trimer variants 16055 NFL TD8 CC, JRFL NFL TD15 CC, and BG505 NFL CC. DSC parameters are shown next to the curves in blue for wt NFLs and in red for the stabilized NFL trimers. Representative negative staining EM 2D class averages corresponding to the disulfide-stabilized NFL trimer variants 16055 NFL TD8 CC, JRFL NFL TD15 CC, and BG505 NFL CC are also shown. OD450, optical density at 450 nm.
We then investigated the effect of the engineered disulfide linkage (I201C-A433C) on overall trimer thermostability. Trimer stability may be relevant to maintain structural integrity in adjuvant at 37°C in serum or lymph and for antigen presentation in the context of vaccination, improvements of which may lead to qualitatively better antibody responses. We assessed the Tm of the TD CC trimers by DSC. The 16055 NFL TD8 CC construct displayed a Tm of 72.5°C, significantly higher than that of the original wt 16055 NFL trimer (58°C) (Fig. 8B). Similarly, the JRFL NFL TD15 CC variant displayed a Tm of 64.3°C, significantly higher than that of the wt JRFL NFL at 54.3°C, and the BG505 NFL CC variant experienced a significant increase in the Tm, to 71.5°C from the wt BG505 NFL level of 66.5°C. The T1/2 parameters were narrower, indicating increased molecular homogeneity (Fig. 8B). Analysis of the TD CC trimers by negative staining EM confirmed that the majority of the trimers adopted a native-like conformation (Fig. 8B).
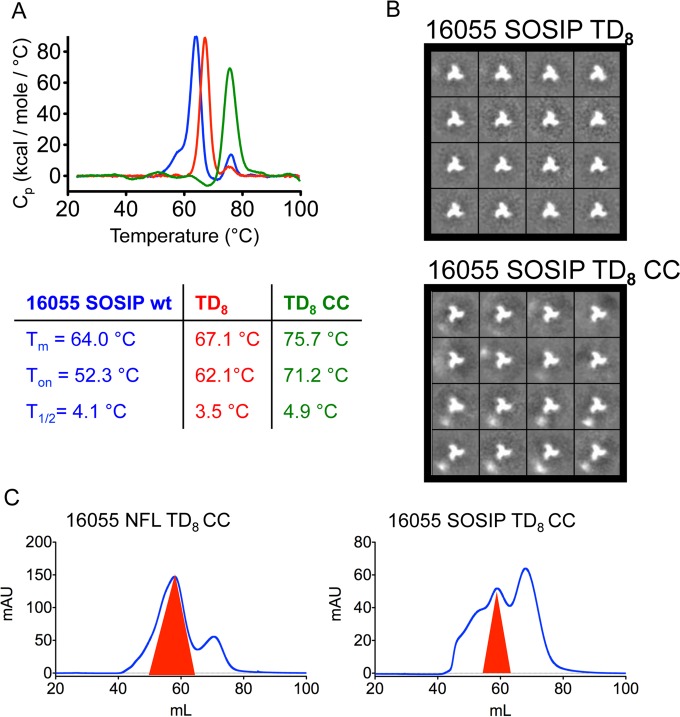
To investigate the general applicability of these modifications, we introduced the same TD8 and CC modifications in the context of 16055 SOSIP. The resulting SOSIP trimer variants displayed increased stability. The Tm increased from 64.0°C for the wt to 67.1°C for the TD8 variant and 75.7°C for the TD8 CC variant (Fig. 9A). Unlike the NFLs, the T1/2 of the 16055 SOSIP TD8 CC variant was increased compared to that of the wt or TD8 trimer, suggesting increased trimer heterogeneity, although EM images and 2D class averages showed predominantly well-ordered trimers (Fig. 9B). The apparent increased heterogeneity may be due to the two unnatural disulfides introduced into 16055 SOSIP TD8 CC (501-605CC [A501C-T605C] and I201C-A433C). Compared to the NFLs, the well-ordered 16055 TD8 SOSIPs, with and without I201C-A433C, displayed a less predominant trimer peak by SEC and consequently lower yields (Fig. 9C).
FIG 9.
Biophysical characterization of stabilized 16055 SOSIP trimer variants. (A) DSC transition melting curves corresponding to 16055 SOSIP wt (blue), TD8 (orange), and TD8 CC (red) and derived DSC parameters. (B) Negative staining EM micrographs and derived 2D class averages for 16055 NFL TD8 and 16055 NFL TD8 CC variants. (C) SEC profiles comparing 16055 NFL versus SOSIP with both the TD and the CC modifications.
DISCUSSION
In this study, we generated improved variants of the soluble NFL trimers derived from the prototypic clade C 16055 and clade B JRFL. We also identified dispersed elements of stability of Env that contributed to the enhancement in homogeneity and stability of the improved soluble NFL trimers. To accomplish these improvements, we identified a set of trimer-associated residues via sequence alignment, followed by structure-guided selection on the BG505 SOSIP high-resolution structure (19). Transfer of selected BG505 TD residues, distributed in dispersed and distinct regions of Env, was sufficient to increase the propensity of 16055 and JRFL Env sequences to form well-ordered native-like NFL trimers, decreasing aggregation and dissociation into dimers and monomers. Furthermore, the stability of the NFL TD trimer variants, as judged by their thermostability profiles, increased substantially, achieving stability parameters similar to those of the BG505-derived trimers, demonstrating that the residues identified in this study accurately pinpointed important Env stability elements. Transfer of the BG505 TD residues to the 16055 and JRFL NFL trimers also enhanced homogeneity and ordered trimer yields. These elements of stability clustered in three different areas of Env: the gp120-gp41 interface, the prebridging sheet, and the variable region cap. In the 16055 Env context, eight single-residue substitutions were sufficient to increase both homogeneity and stability, while JRFL Env required 15 to achieve levels of homogeneity and stability approaching those of BG505-derived well-ordered trimers. It is possible that although the specific residues may somewhat differ depending on the specific HIV Env sequence under investigation, the premise and method of identification of the stability residues should be applicable to many other Env sequences, thereby extending options and our capacity to generate more diverse, HIV Env-soluble spike mimics. We transferred these improvements as groups of substitutions, making strategic intermediates, but we did not determine the contribution of each individual residue. Therefore, at this juncture, we do not know the absolute contribution of each of these individual residues, a subject of further study. The JRFL and 16055 Env sequences were selected because they display a weak trimer-generating propensity and are phylogenetically distant from the BG505 subtype A sequence, within the major HIV subtypes of clades B and C. Why some Env sequences easily adopt the NFL or SOSIP trimer platform while others do not is unknown but may be related to the stability of the various subdomains of the trimeric Env. While the BG505 Env sequence readily forms NFL or SOSIP trimers, others, such as JRFL and 16055, show a weaker trimer-forming propensity in these contexts or none at all. The elements of stability pinpointed in this study helped those sequences with a weak trimer-forming propensity to be increased in this aspect. The data suggest that trimer formation and stability as assessed by DSC are intimately associated and that a threshold of stability may need to be reached in order for the TD approach to be expandable to Env sequences with little inherent propensity to form well-ordered NFL trimers.
In addition to this transferable trimer-enhancing propensity BG505 signature, we engineered an intraprotomer disulfide linkage between residues I201 and A433 of the gp120 subunit to stabilize these soluble spike mimics in the pre-receptor-engaged conformation. This unnatural covalent bond linking β3 and β21, if formed, would prevent the formation of the bridging sheet by not allowing the positional receptor triggered to switch between β3 and β2. This swap is inferred from the structures of the BG505 SOSIP, which is presumably in the pre-receptor-engaged state, in which β3 aligns in parallel to β21. By inference and using the CD4-bound gp120 core high-resolution structure, β2 aligns in an antiparallel manner with respect to β21, reflecting β2- and β3-focused conformational changes following CD4 engagement. Essentially, CD4 interacts with β21-β22, allowing a repositioning of β2 relative to β3 of nearly 180 degrees; this pivot triggers torquing of the V1 stem, allowing V1/V2 to swing open from the closed prereceptor state, forming the full coreceptor binding site (CoRbs) with the distal bridging sheet, and now V1/V2 is released and V3 is extended.
The antibodies 17b and Vc813 recognized the 16055 NFL TD8, JRFL NFL TD15, and BG505 NFL trimers after CD4 induction while they did not recognize the disulfide-stabilized CC trimer variants. These results are consistent with the formation of the engineered linkage between β3 and β21 and with an inability to form the receptor-induced bridging sheet. The stabilized (CC) trimers displayed a superior thermostability profile compared to that of the TD and wt variants, with higher Tm and narrower T1/2s, providing supporting evidence that the engineered disulfide was formed. EM analysis revealed that the trimers remained well ordered in the presence of the specific substitutions. In the absence of a high-resolution structure, formation of the bridging sheet disulfide pair is indirect, but the experimental data described here are consistent with Cys-Cys pair formation and the subsequent inability of CD4 to induce 17b-sensing conformational changes in the CC variant compared to the ability of the TD trimer alone.
Higher thermal transition points show a resistance of the trimeric NFL protein to disassemble into its monomeric components under a defined stress test. We believe that this thermal resistance, as measured by DSC, may be surrogate measurements of in vivo durability of the integrity of the quaternary conformation of these immunogens. Overall enhanced trimer stability is desired to maintain conformational integrity in vivo so that during the immune response/GC reaction, B cells will encounter only the preferentially exposed neutralizing epitopes on the trimeric immunogens, thereby preferentially driving B cells sensing these targets. B cells recognizing nonneutralizing determinants, exposed when the trimer loses structural integrity, should be disfavored by this approach. In addition, engineered stable and well-ordered trimers that are not conformationally altered by soluble CD4 interaction may be important when such spike mimics are inoculated into NHPs or humans that possess CD4+ cells displaying CD4 with high affinity for Env. Immunogen-CD4 interaction could potentially disrupt the quaternary packing of the novel well-ordered HIV-1 soluble spike mimics and expose undesirable nonneutralizing epitopes to the primate humoral immune system. The stepwise designs described here provide insight into key regions of Env involved in trimer integrity. These designs also provide improved tools to interrogate the immune response to test the hypothesis of whether structure-based Env trimer stability will alter the immunogenic outcome or if eliminating induction by CD4 in primates will better elicit vaccine-induced HIV-1 neutralizing antibodies.
ACKNOWLEDGMENTS
This work was funded by the HIVRAD program (grant number P01 AI104722 to V.D. and R.T.W.), a CHAVI-ID grant (AI100663 to A.B.W. and R.T.W.), and the International AIDS Initiative (IAVI) and its generous donors (to J.G., S.K.S., B.C., A.B.W., and R.T.W.). IAVI's work is made possible by generous support from many donors, including the Bill & Melinda Gates Foundation, the Ministry of Foreign Affairs of Denmark, Irish Aid, the Ministry of Finance of Japan, the Ministry of Foreign Affairs of the Netherlands, the Norwegian Agency for Development Cooperation, the United Kingdom Department for International Development, and the U.S. Agency for International Development (USAID). The full list of IAVI donors is available at www.iavi.org. This study is made possible by the generous support of the Bill & Melinda Gates Foundation Collaboration for AIDS Vaccine Discovery and the American people through USAID.
The contents are the responsibility of the International AIDS Vaccine Initiative and do not necessarily reflect the views of USAID or the Government of the United States. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Earl PL, Broder CC, Long D, Lee SA, Peterson J, Chakrabarti S, Doms RW, Moss B. 1994. Native oligomeric human immunodeficiency virus type 1 envelope glycoprotein elicits diverse monoclonal antibody reactivities. J Virol 68:3015–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Earl PL, Sugiura W, Montefiori DC, Broder CC, Lee SA, Wild C, Lifson J, Moss B. 2001. Immunogenicity and protective efficacy of oligomeric human immunodeficiency virus type 1 gp140. J Virol 75:645–653. doi: 10.1128/JVI.75.2.645-653.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao F, Weaver EA, Lu Z, Li Y, Liao HX, Ma B, Alam SM, Scearce RM, Sutherland LL, Yu JS, Decker JM, Shaw GM, Montefiori DC, Korber BT, Hahn BH, Haynes BF. 2005. Antigenicity and immunogenicity of a synthetic human immunodeficiency virus type 1 group m consensus envelope glycoprotein. J Virol 79:1154–1163. doi: 10.1128/JVI.79.2.1154-1163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kovacs JM, Noeldeke E, Ha HJ, Peng H, Rits-Volloch S, Harrison SC, Chen B. 2014. Stable, uncleaved HIV-1 envelope glycoprotein gp140 forms a tightly folded trimer with a native-like structure. Proc Natl Acad Sci U S A 111:18542–18547. doi: 10.1073/pnas.1422269112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forsell MN, Li Y, Sundback M, Svehla K, Liljestrom P, Mascola JR, Wyatt R, Karlsson Hedestam GB. 2005. Biochemical and immunogenic characterization of soluble human immunodeficiency virus type 1 envelope glycoprotein trimers expressed by Semliki Forest virus. J Virol 79:10902–10914. doi: 10.1128/JVI.79.17.10902-10914.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang X, Florin L, Farzan M, Kolchinsky P, Kwong PD, Sodroski J, Wyatt R. 2000. Modifications that stabilize human immunodeficiency virus envelope glycoprotein trimers in solution. J Virol 74:4746–4754. doi: 10.1128/JVI.74.10.4746-4754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spearman P, Lally MA, Elizaga M, Montefiori D, Tomaras GD, McElrath MJ, Hural J, De Rosa SC, Sato A, Huang Y, Frey SE, Sato P, Donnelly J, Barnett S, Corey LJ, HIV Vaccine Trials Network of NIAID . 2011. A trimeric, V2-deleted HIV-1 envelope glycoprotein vaccine elicits potent neutralizing antibodies but limited breadth of neutralization in human volunteers. J Infect Dis 203:1165–1173. doi: 10.1093/infdis/jiq175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Srivastava IK, Stamatatos L, Kan E, Vajdy M, Lian Y, Hilt S, Martin L, Vita C, Zhu P, Roux KH, Vojtech L, D CM, Donnelly J, Ulmer JB, Barnett SW. 2003. Purification, characterization, and immunogenicity of a soluble trimeric envelope protein containing a partial deletion of the V2 loop derived from SF162, an R5-tropic human immunodeficiency virus type 1 isolate. J Virol 77:11244–11259. doi: 10.1128/JVI.77.20.11244-11259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang X, Farzan M, Wyatt R, Sodroski J. 2000. Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J Virol 74:5716–5725. doi: 10.1128/JVI.74.12.5716-5725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang X, Lee J, Mahony EM, Kwong PD, Wyatt R, Sodroski J. 2002. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J Virol 76:4634–4642. doi: 10.1128/JVI.76.9.4634-4642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang X, Wyatt R, Sodroski J. 2001. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J Virol 75:1165–1171. doi: 10.1128/JVI.75.3.1165-1171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, Moore JP. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol 74:627–643. doi: 10.1128/JVI.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Binley JM, Sanders RW, Master A, Cayanan CS, Wiley CL, Schiffner L, Travis B, Kuhmann S, Burton DR, Hu SL, Olson WC, Moore JP. 2002. Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. J Virol 76:2606–2616. doi: 10.1128/JVI.76.6.2606-2616.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, Lu M, Moore JP. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol 76:8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. 2013. A next-generation cleaved, soluble HIV-1 Env Trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. 2013. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartesaghi A, Merk A, Borgnia MJ, Milne JL, Subramaniam S. 2013. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat Struct Mol Biol 20:1352–1357. doi: 10.1038/nsmb.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, Stewart-Jones GB, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. 2014. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, Joyce MG, Guttman M, Ma X, Narpala S, Soto C, Terry DS, Yang Y, Zhou T, Ahlsen G, Bailer RT, Chambers M, Chuang GY, Doria-Rose NA, Druz A, Hallen MA, Harned A, Kirys T, Louder MK, O'Dell S, Ofek G, Osawa K, Prabhakaran M, Sastry M, Stewart-Jones GB, Stuckey J, Thomas PV, Tittley T, Williams C, Zhang B, Zhao H, Zhou Z, Donald BR, Lee LK, Zolla-Pazner S, Baxa U, Schon A, Freire E, Shapiro L, Lee KK, Arthos J, Munro JB, Blanchard SC, Mothes W, Binley JM, et al. 2015. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol 22:522–531. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, Arendt H, Kim HJ, Lee JH, Pugach P, Williams M, Debnath G, Moldt B, van Breemen MJ, Isik G, Medina-Ramirez M, Back JW, Koff WC, Julien JP, Rakasz EG, Seaman MS, Guttman M, Lee KK, Klasse PJ, LaBranche C, Schief WR, Wilson IA, Overbaugh J, Burton DR, Ward AB, Montefiori DC, Dean H, Moore JP. 2015. HIV-1 vaccines. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 349:aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guenaga J, de Val N, Tran K, Feng Y, Satchwell K, Ward AB, Wyatt RT. 2015. Well-ordered trimeric HIV-1 subtype B and C soluble spike mimetics generated by negative selection display native-like properties. PLoS Pathog 11:e1004570. doi: 10.1371/journal.ppat.1004570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pugach P, Ozorowski G, Cupo A, Ringe R, Yasmeen A, de Val N, Derking R, Kim HJ, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, Julien JP, Burton DR, Wilson IA, Sanders RW, Klasse PJ, Ward AB, Moore JP. 2015. A native-like SOSIP.664 trimer based on a HIV-1 subtype B env gene. J Virol 89:3380–3395. doi: 10.1128/JVI.03473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Julien JP, Lee JH, Ozorowski G, Hua Y, Torrents de la Pena A, de Taeye SW, Nieusma T, Cupo A, Yasmeen A, Golabek M, Pugach P, Klasse PJ, Moore JP, Sanders RW, Ward AB, Wilson IA. 2015. Design and structure of two HIV-1 clade C SOSIP.664 trimers that increase the arsenal of native-like Env immunogens. Proc Natl Acad Sci U S A 112:11947–11952. doi: 10.1073/pnas.1507793112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma SK, de Val N, Bale S, Guenaga J, Tran K, Feng Y, Dubrovskaya V, Ward AB, Wyatt RT. 2015. Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep 11:539–550. doi: 10.1016/j.celrep.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klasse PJ, Depetris RS, Pejchal R, Julien JP, Khayat R, Lee JH, Marozsan AJ, Cupo A, Cocco N, Korzun J, Yasmeen A, Ward AB, Wilson IA, Sanders RW, Moore JP. 2013. Influences on trimerization and aggregation of soluble, cleaved HIV-1 SOSIP envelope glycoprotein. J Virol 87:9873–9885. doi: 10.1128/JVI.01226-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haas J, Park EC, Seed B. 1996. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr Biol 6:315–324. doi: 10.1016/S0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- 28.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. 2005. Automated molecular microscopy: the new Leginon system. J Struct Biol 151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 29.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. 2009. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol 166:205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, Lyumkis D, Potter CS, Carragher B. 2009. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol 166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sorzano CO, Bilbao-Castro JR, Shkolnisky Y, Alcorlo M, Melero R, Caffarena-Fernandez G, Li M, Xu G, Marabini R, Carazo JM. 2010. A clustering approach to multireference alignment of single-particle projections in electron microscopy. J Struct Biol 171:197–206. doi: 10.1016/j.jsb.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. 1996. A new generation of the IMAGIC image processing system. J Struct Biol 116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 33.Blattner C, Lee JH, Sliepen K, Derking R, Falkowska E, de la Pena AT, Cupo A, Julien JP, van Gils M, Lee PS, Peng W, Paulson JC, Poignard P, Burton DR, Moore JP, Sanders RW, Wilson IA, Ward AB. 2014. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity 40:669–680. doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Georgiev IS, Joyce MG, Yang Y, Sastry M, Zhang B, Baxa U, Chen RE, Druz A, Lees CR, Narpala S, Schon A, Van Galen J, Chuang GY, Gorman J, Harned A, Pancera M, Stewart-Jones GB, Cheng C, Freire E, McDermott AB, Mascola JR, Kwong PD. 2015. Single-chain soluble BG505.SOSIP gp140 trimers as structural and antigenic mimics of mature closed HIV-1 Env. J Virol 89:5318–5329. doi: 10.1128/JVI.03451-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Protocol GPI, Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sok D, van Gils MJ, Pauthner M, Julien JP, Saye-Francisco KL, Hsueh J, Briney B, Lee JH, Le KM, Lee PS, Hua Y, Seaman MS, Moore JP, Ward AB, Wilson IA, Sanders RW, Burton DR. 2014. Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proc Natl Acad Sci U S A 111:17624–17629. doi: 10.1073/pnas.1415789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, O'Dell S, Wilson R, Wu X, Schmidt SD, Hogerkorp CM, Louder MK, Longo NS, Poulsen C, Guenaga J, Chakrabarti BK, Doria-Rose N, Roederer M, Connors M, Mascola JR, Wyatt RT. 2012. HIV-1 neutralizing antibodies display dual recognition of the primary and coreceptor binding sites and preferential binding to fully cleaved envelope glycoproteins. J Virol 86:11231–11241. doi: 10.1128/JVI.01543-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y, Migueles SA, Welcher B, Svehla K, Phogat A, Louder MK, Wu X, Shaw GM, Connors M, Wyatt RT, Mascola JR. 2007. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med 13:1032–1034. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang CC, Venturi M, Majeed S, Moore MJ, Phogat S, Zhang MY, Dimitrov DS, Hendrickson WA, Robinson J, Sodroski J, Wyatt R, Choe H, Farzan M, Kwong PD. 2004. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc Natl Acad Sci U S A 101:2706–2711. doi: 10.1073/pnas.0308527100. [DOI] [PMC free article] [PubMed] [Google Scholar]