ABSTRACT
Hemorrhagic fever arenaviruses (HFAs) pose important public health problems in regions where they are endemic. Concerns about human-pathogenic arenaviruses are exacerbated because of the lack of FDA-licensed arenavirus vaccines and because current antiarenaviral therapy is limited to an off-label use of ribavirin that is only partially effective. We have recently shown that the noncoding intergenic region (IGR) present in each arenavirus genome segment, the S and L segments (S-IGR and L-IGR, respectively), plays important roles in the control of virus protein expression and that this knowledge could be harnessed for the development of live-attenuated vaccine strains to combat HFAs. In this study, we further investigated the sequence plasticity of the arenavirus IGR. We demonstrate that recombinants of the prototypic arenavirus lymphocytic choriomeningitis virus (rLCMVs), whose S-IGRs were replaced by the S-IGR of Lassa virus (LASV) or an entirely nonviral S-IGR-like sequence (Ssyn), are viable, indicating that the function of S-IGR tolerates a high degree of sequence plasticity. In addition, rLCMVs whose L-IGRs were replaced by Ssyn or S-IGRs of the very distantly related reptarenavirus Golden Gate virus (GGV) were viable and severely attenuated in vivo but able to elicit protective immunity against a lethal challenge with wild-type LCMV. Our findings indicate that replacement of L-IGR by a nonviral Ssyn could serve as a universal molecular determinant of arenavirus attenuation.
IMPORTANCE Hemorrhagic fever arenaviruses (HFAs) cause high rates of morbidity and mortality and pose important public health problems in regions where they are endemic. Implementation of live-attenuated vaccines (LAVs) will represent a major step to combat HFAs. Here we document that the arenavirus noncoding intergenic region (IGR) has a high degree of plasticity compatible with virus viability. This observation led us to generate recombinant LCMVs containing nonviral synthetic IGRs. These rLCMVs were severely attenuated in vivo but able to elicit protective immunity against a lethal challenge with wild-type LCMV. These nonviral synthetic IGRs can be used as universal molecular determinants of arenavirus attenuation for the rapid development of safe and effective, as well as stable, LAVs to combat HFA.
INTRODUCTION
Hemorrhagic fever arenaviruses (HFAs) pose important public health problems in regions where they are endemic (1–3). Thus, Lassa virus (LASV) infects several hundred thousand individuals yearly in West Africa, resulting in a high number of Lassa fever (LF) cases, which are associated with high rates of morbidity and significant mortality (4). Notably, increased travel has led to the importation of LF cases into metropolitan areas around the globe where the virus is not endemic (5, 6). Moreover, the regions where LASV is endemic are expanding (4), and the association of the recently identified arenavirus Lujo virus with a recent outbreak of hemorrhagic fever in South Africa (7, 8) has raised concerns about the emergence of novel HFAs. In addition, evidence indicates that the worldwide-distributed prototypic arenavirus lymphocytic choriomeningitis virus (LCMV) is a neglected human pathogen of clinical significance (9–13). Likewise, several arenaviruses pose credible biodefense threats (14). Concerns about human-pathogenic arenaviruses are exacerbated because there are no FDA-licensed arenavirus vaccines (15) and current antiarenaviral therapy is limited to an off-label use of ribavirin that is only partially effective (16–18). Therefore, it is important to develop novel antiviral strategies to combat human-pathogenic arenaviruses. Epidemiological studies indicate that live-attenuated vaccines (LAVs) represent the most feasible approach to the control of HFAs in regions where they are endemic, as LAVs induce long-term robust cellular and humoral immune responses following a single immunization. Moreover, LAVs are very effective for the establishment of herd immunity, which is particularly relevant in developing countries, where it is hard to reach optimal levels of vaccine coverage.
Arenaviruses are enveloped viruses with a bisegmented negative-strand RNA genome (2). Each genome segment, the L and S segments, uses an ambisense coding strategy to direct the synthesis of two proteins in opposite orientation separated by a noncoding intergenic region (IGR) (2). The S-segment RNA encodes the viral nucleoprotein (NP) and surface virion glycoproteins GP1 and GP2, generated by posttranslational cleavage of the glycoprotein precursor (GPC), which mediate virus receptor recognition and cell entry. The L-segment RNA encodes the viral RNA-dependent RNA polymerase (L) and the matrix RING finger protein Z (19, 20). We have documented that the sequences of the IGRs of the L and S segments (L-IGR and S-IGR, respectively) have distinct functional roles in virus multiplication, and we harnessed that knowledge to implement a general molecular strategy for arenavirus attenuation (21). Our approach involved replacement of the IGR of the L genome segment by the IGR of the S genome segment to generate a recombinant LCMV (rLCMV), rLCMV(IGR/S-S), that was highly attenuated in vivo but able to induce protection against a lethal challenge with wild-type (WT) LCMV. Attenuation of rLCMV(IGR/S-S) was associated with the stable alteration of the control of viral gene expression by the IGR at the posttranscriptional level. In the study described in this report, we have further investigated how to utilize the sequence plasticity of the IGR to facilitate the development of attenuated forms of pathogenic arenaviruses. In this work, we demonstrate that rLCMVs where the S-IGR is replaced by the S-IGR of a different arenavirus or by a nonviral S-IGR-like sequence (Ssyn) are viable, supporting the sequence plasticity of a functional IGR. Likewise, rLCMVs whose L-IGRs were replaced by Ssyn or the S-IGR of the distantly related reptarenavirus Golden Gate virus (GGV) (Sggv) (22) were viable and highly attenuated in vivo but able to elicit protective immunity against a lethal challenge with wild-type rLCMV. These results support the feasibility of replacing the L-IGR with a universal Ssyn as a general molecular determinant of arenavirus attenuation.
MATERIALS AND METHODS
Plasmids.
pCAGGS-NP (pC-NP), pCAGGS-L (pC-L), pCAGGS-GPC (pC-GPC), and pCAGGS-LCMV-Z-FLAG (pC-Z-FLAG) have been described previously (23–25). Plasmids pMG/S-CAT/GFP and pMG/S-CAT/GFP-ΔIGR have also been described previously (26). The region containing the S-IGR of pMG/S-CAT/GFP was replaced with the LASV S-IGR (Slas) to generate pMG/S-CAT/GFP w/Slas. Plasmids for rLCMV rescue were generated on the basis of the pPol1S ARM and pPol1L ARM plasmids that direct RNA polymerase I (Pol1)-mediated intracellular synthesis of the S and L RNA genome species of the Armstrong (ARM) strain of LCMV, respectively (27, 28). The IGR of pPol1S ARM was replaced with Slas or synthetic S-IGR like sequences (Ssyn1, Ssyn2, or Ssyn3; Table 1) to generate pPol1S w/Slas or pPol1S w/Ssyn1, pPol1S w/Ssyn2, or pPol1S w/Ssyn3, respectively. The L-IGR of pPol1L ARM was replaced with Ssyn1, Ssyn2, Ssyn3, or the S-IGR of the reptarenavirus GGV (Sggv) to generate pPol1L w/Ssyn1, pPol1L w/Ssyn2, pPol1L w/Ssyn3, or pPol1L w/Sggv, respectively.
TABLE 1.
Synthetic S-IGR-like sequences (viral RNA)b
| IGR | Sequencea (5′–3′) | ΔG (kcal/mol) |
|---|---|---|
| LCMV S-IGR | AGAACAGCGCCUCCCUGACUCUCCACCUCGAAAGAGGUGGAGAGUCAGGGAGGCCCAGAGGGUC | −52.00 |
| Ssyn1 | AGAACAGCcugcaggacugagaggcugcgauuccgcagccucucaguccugcagCCAGAGGGUC | −46.20 |
| Ssyn2 | ucgcggcucugcaggacugagaggcugcgauuccgcagccucucaguccugcaguggaucucag | −47.3 |
| Ssyn3 | ucgcggcuGCCUCCCUGACUCUCCACCUCGAAAGAGGUGGAGAGUCAGGGAGGCuggaucucag | −51.70 |
Substituted nucleotide sequences are shown in lowercase letters.
The GC content of all IGRs was 62%.
Cells.
BHK-21, Vero, A549, and 293T (also known as HEK293T) cells were grown in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) containing 10% fetal bovine serum (FBS), 2 mM l-glutamine, 100 mg/ml streptomycin, and 100 U/ml penicillin at 37°C in a 5% CO2 atmosphere.
Generation of rLCMV.
The different rLCMVs used in this study were generated as described previously (27–29) with minor modifications. BHK-21 cells seeded at 7.5 × 105 cells/6-well plate were cultured overnight and transfected with plasmids pPol1S (0.8 μg) and pPol1L (1.4 μg), respectively, together with plasmids expressing the viral trans-acting factors NP (0.8 μg of pC-NP) and L (1.0 μg of pC-L) using 2.5 μl of the Lipofectamine 2000 reagent (LF2000; Invitrogen)/μg of DNA. After 5 h of transfection, the tissue culture supernatant (TCS) containing the transfection mix was removed and fresh medium was added. At 3 days posttransfection, the TCS was removed, 3 ml of fresh medium was added, and the cells were cultured for another 3 days. The TCS at 6 days posttransfection was designated passage 0 (P0). Rescued viruses were amplified by infection of BHK-21 cells (multiplicity of infection [MOI] = 0.01). At 72 h postinfection (p.i.), the TCS was collected and clarified by centrifugation at 400 × g at 4°C for 5 min to remove cell debris and stored at −80°C. To generate S-IGR mutant viruses rLCMV(IGR/Slas-L), rLCMV(IGR/Ssyn1-L), rLCMV(IGR/Ssyn2-L), and rLCMV(IGR/Ssyn3-L), pPol1S w/Slas, pPol1S w/Ssyn1, pPol1S w/Ssyn2, and pPol1S w/Ssyn3, respectively, were used together with pPol1L ARM. To generate L-IGR mutant viruses rLCMV(IGR/S-Ssyn1), rLCMV(IGR/S-Ssyn2), rLCMV(IGR/S-Ssyn3), and rLCMV(IGR/S-Sggv), pPol1L w/Ssyn1, pPol1L w/Ssyn2, pPol1L w/Ssyn3, and pPol1L w/Sggv, respectively, were used together with pPol1S ARM. Viruses from either passage 1 (P1) or P2 were used for the experiments.
Virus titration.
LCMV titers were determined using an immunofocus assay as described previously (30). Briefly, 10-fold serial virus dilutions were used to infect Vero cell monolayers in a 96-well plate, and at 20 h p.i., cells were fixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS). After cell permeabilization by treatment with 0.3% Triton X-100 in PBS containing 3% bovine serum albumin, the cells were stained using an anti-NP antibody (VL-4; Bio X Cell) and an Alexa Fluor 568-labeled anti-rat IgG secondary antibody.
Infectious VLP assay.
The generation of infectious virus-like particles (VLPs) has been described previously (24, 26). Briefly, BHK-21 cells seeded at 3.5 × 105 cells/12-well plate were cultured overnight and transfected with 0.25 μg of minigenome (MG)-expressing plasmid under the control of murine Pol1, 0.4 μg of pC-NP, 0.5 μg of pC-L, 0.2 μg of pC-GPC, and 50 ng of pC-Z-FLAG using 2.5 μl of LF2000/μg of DNA. Empty pCAGGS instead of pC-L was used as a negative control [L(−)]. Five hours later, the TCS containing the transfection mix was removed and the cells were cultured in 1.5 ml of fresh medium at 37°C in 5% CO2 for 67 h. Then, the TCS was collected and clarified by centrifugation at 400 × g and 4°C for 5 min to remove the cell debris, and 300 μl of clarified TCS was used to infect a fresh monolayer of BHK-21 cells. At 2 h p.i. with VLPs, cells were superinfected with the rLCMV WT (MOI = 2) to supply the trans-acting factors NP and L required for RNA replication and expression of the MG RNA delivered by VLPs. After 90 min of adsorption, the virus inoculum was removed and the cells were cultured in fresh medium for 72 h. Chloramphenicol acetyltransferase (CAT) expression in transfected cells (1st CAT expression level) and VLP-infected cells (2nd CAT expression level) was measured by enzyme-linked immunosorbent assay. VLP infectivity was determined as 2nd/1st CAT expression levels.
Growth kinetics.
Cells were infected with rLCMVs (MOI = 0.01), and at several times p.i., the TCSs were collected and viral titers were determined using an immunofocus assay.
LCMV immunization.
Six-week-old C57BL/6J mice were infected intraperitoneally (i.p.) with rLCMVs (105 focus-forming units [FFU]) or mock infected.
LCMV lethal challenge.
Six-week-old C57BL/6J mice were infected intracranially (i.c.) with rLCMVs (103 FFU). The mice were monitored daily for the development of clinical symptoms and survival. Infections using the i.c. route were done with mice under deep anesthesia. All animal experiments were done under protocol 09-0137, approved by The Scripps Research Institute IACUC.
Stability of rLCMVs.
BHK-21 cells were seeded (7.5 × 105 cells/6-well plate) 18 h prior to infection with rLCMVs (MOI = 0.01). At 72 h p.i., the TCSs were collected and clarified of cell debris by centrifugation at 400 × g at 4°C for 5 min, and virus titers were determined by an immunofocus assay. The TCS from P1 was used to infect a fresh monolayer of BHK-21 cells, and the same process was repeated over 10 serial passages. At P10, total RNA was isolated from infected cells and used in reverse transcription-PCRs to amplify DNA fragments containing the IGR, and their sequences were determined. The data for the rLCMV WT used in Fig. 4B were also used in a previous study (Fig. 3E in reference 21), as the experiment was carried out at the same time.
FIG 4.
In vivo virulence and stability of the rLCMV(IGR/Ssyn-L) constructs. (A) Six-week-old C57BL/6J mice (4 mice/group) were infected (103 FFU i.c.) with the rLCMV WT or an rLCMV(IGR/Ssyn-L) construct or left uninfected (naive). At 28 days p.i. (broken line), mice that survived were subjected to a lethal challenge with the rLCMV WT (103 FFU i.c.). (B) Stability of the rLCMV(IGR/Ssyn-L) constructs. BHK-21 cells were infected with the rLCMV WT or an rLCMV(IGR/Ssyn-L) construct (MOI = 0.01). At 72 h p.i., the TCS was collected and the virus titers in the TCS were determined by an immunofocus assay. Fresh BHK-21 cell monolayers were infected with the TCS (MOI = 0.01); this process was serially repeated throughout P2 to P10. (Panel B includes data republished from reference 21.)
FIG 3.
Growth of rLCMV(IGR/Ssyn-L) in IFN-competent cells. (A) Growth kinetics of the rLCMV(IGR/Ssyn-L) constructs in IFN-competent A549 cells. A549 cells were infected with the rLCMV WT or an rLCMV(IGR/Ssyn-L) construct (MOI = 0.01). Virus titers in the TCS were determined at the indicated times p.i. Data represent the means ± SDs from three independent experiments. (B) Ability of the rLCMV(IGR/Ssyn-L) constructs to induce ISRE-mediated reporter gene expression. A549/ISRE-Fluc cells were infected with the rLCMV WT or an rLCMV(IGR/Ssyn-L) construct (MOI = 0.1) or treated with IFN-α at the concentrations indicated on the x axis, and 17 h later, the levels of Fluc activity were determined. Values of Fluc activity from mock-infected and untreated cells were subtracted as background activity. Data represent the means ± SDs from three independent experiments. RLU, relative light units. (C) Sensitivity of rLCMVs to exogenous IFN-I. Vero cells were infected with the rLCMV WT, rLCMV(IGR/Ssyn2-L), or rLCMV(IGR/Ssyn3-L) (MOI = 0.05) for 90 min and treated with IFN-α (5 or 500 U/ml) or left untreated. At 12 and 48 h p.i., cells were fixed and virus infectivity was determined by IFA.
ISRE reporter assay.
We used an A549 cell line (A549/ISRE-Fluc) that expresses firefly luciferase (Fluc) via interferon (IFN)-stimulated response elements (ISREs) (21). We seeded A549/ISRE-Fluc cells (2.5 × 104 cells/96-well plate) cultured in DMEM containing 10% FBS, 2 mM l-glutamine, 100 mg/ml streptomycin, 100 U/ml penicillin, and 10 μg/ml puromycin at 37°C in 5% CO2 at 20 h prior to infection with rLCMVs (MOI = 0.1) or treatment for 17 h with universal alpha IFN (IFN-α) at the concentrations indicated in the corresponding figures. Firefly luciferase activity was measured using a Steady-Glo luciferase assay kit (Promega) and a luminometer.
Sensitivity of rLCMVs to exogenous type I interferon (IFN-I).
Vero cells were infected with the rLCMV WT, rLCMV(IGR/Ssyn2-L), or rLCMV(IGR/Ssyn3-L) (MOI = 0.05) for 90 min and treated with universal IFN-α at a low (5 U/ml) or a high (500 U/ml) concentration or left untreated. At 12 h and 48 h p.i., infected cells were fixed with 4% PFA in PBS and stained with anti-LCMV-NP antibody (VL-4) followed by Alexa Fluor 568-conjugated anti-rat IgG antibody. Infectivity corresponded to the average percentage of NP-positive cells in three different fields.
RESULTS
Functional consequences of replacing the LCMV S-IGR by the LASV S-IGR.
IGR sequences exhibit a high degree of genetic diversity among arenaviruses, including closely related species like LASV and LCMV (Fig. 1A). However, the predicted S-IGR structures of LCMV and LASV are similar (Fig. 1B), suggesting that the structure, rather than sequence specificity, orchestrates the critical roles played by the IGRs during the arenavirus life cycle. To examine this issue, we generated an LCMV minigenome (MG) construct where the S-IGR of LASV was substituted for the authentic LCMV S-IGR (pMG/S-CAT/GFP w/Slas) and examined the effect of the LASV S-IGR on LCMV MG activity and the production of infectious VLPs. As previously reported (26), the activity of the LCMV MG lacking an IGR (MGΔIGR) was dramatically reduced (Fig. 1C). Replacement of the LCMV S-IGR with the LASV S-IGR caused only a modest reduction in MG activity compared to that of the wild-type MG (Fig. 1C) and did not affect the production of LCMV infectious VLPs, whereas, consistent with previous findings (26), the MGΔIGR was impaired in its ability to form infectious VLPs (Fig. 1D). Based on these observations, we attempted to generate an rLCMV containing the LASV S-IGR instead of the LCMV S-IGR [rLCMV(IGR/Slas-L)]. Even though the LCMV and LASV S-IGRs differed significantly in their sequences, we were able to rescue rLCMV(IGR/Slas-L). However, rLCMV(IGR/Slas-L) showed delayed kinetics, and its peak titer was about 10-fold lower than that of the rLCMV WT in cultured cells (Fig. 1E). These results suggested that the sequence specificity within the S-IGR contributes to optimal viral growth but that the structure of the S-IGR, rather than its specific sequence, is critical for virus viability.
FIG 1.
Sequence plasticity associated with a functional S-IGR. (A, B) Comparison of the predicted structures of the S-IGRs of LASV and LCMV. The sequence alignment (A) and predicted structures (B) of the LCMV and LASV S-IGRs are shown. Predicted RNA secondary structures were determined using the CentroidFold web server (http://rtools.cbrc.jp). Each predicted base pair was colored using a heat color gradation from blue to red that corresponds to a base-pairing probability of from 0 to 1. Hyphens, gaps; asterisks, identical in LCMV and LASV S-IGRs. (C, D) The LASV S-IGR supports the production of LCMV infectious VLPs. The assay for the production of infectious VLPs was performed using the components of the LCMV S-MG, where the MG directed the expression of CAT and contained the S-IGR from either LCMV (LCMV-S) or LASV (LASV-S) or lacked an IGR sequence (ΔIGR). (C) Normalized CAT expression of transfected cells. The value for the WT MG was set to 100%. (D) Normalized VLP infectivity. The level of CAT expression of VLP-infected cells was divided by the level of CAT expression of transfected cells. The level of CAT expression in cells infected with VLPs produced by the WT MG was set to 1. Data represent the means ± SDs from three independent experiments. (E) Growth kinetics of rLCMV(IGR/Slas-L). BHK-21 cells were infected (MOI = 0.01) with either the rLCMV WT or rLCMV(IGR/Slas-L). Virus titers in the TCS were determined at the indicated times p.i. Data represent the means ± SDs from three independent experiments.
Generation of rLCMVs with an Ssyn in the S segment.
The sequences of the LASV and LCMV S-IGRs are quite different, but both S-IGRs are present in bona fide arenaviruses. Therefore, we could not rule out the possibility that both contain a virus-specific motif sequence that is difficult to identify. To address this issue, we generated a series of synthetic S-IGR-like sequences (Ssyns) that are unrelated to any known arenavirus S-IGR sequence but that retained the same GC content as the LCMV S-IGR and had predicted structures similar to that of the LCMV S-IGR (Table 1 and Fig. 2A). To generate Ssyn1, Ssyn2, and Ssyn3, nucleotide substitutions were introduced into the stem-loop region of S-IGR, the entire S-IGR, and the 3′ and 5′ ends but not the stem-loop region of S-IGR, respectively. We substituted each Ssyn for the wild-type S-IGR in the pPol1S ARM plasmid and used the constructs to rescue the corresponding rLCMV-containing Ssyn in its S segment, referred to as rLCMV(IGR/Ssyn-L). We successfully rescued all three rLCMV(IGR/Ssyn-L) constructs: rLCMV(IGR/Ssyn1-L), rLCMV(IGR/Ssyn2-L), and rLCMV(IGR/Ssyn3-L). Both rLCMV(IGR/Ssyn1-L) and rLCMV(IGR/Ssyn2-L) grew more slowly than the rLCMV WT in BHK-21 cells and reached peak titers slightly lower than those of the rLCMV WT (Fig. 2B). In contrast, rLCMV(IGR/Ssyn3-L) showed growth kinetics and peak titers similar to those of the rLCMV WT (Fig. 2B). These results indicated that the structure rather than the overall S-IGR sequence appears to be critical for virus viability but that sequence specificity within the stem-loop region contributes to optimal viral growth.
FIG 2.
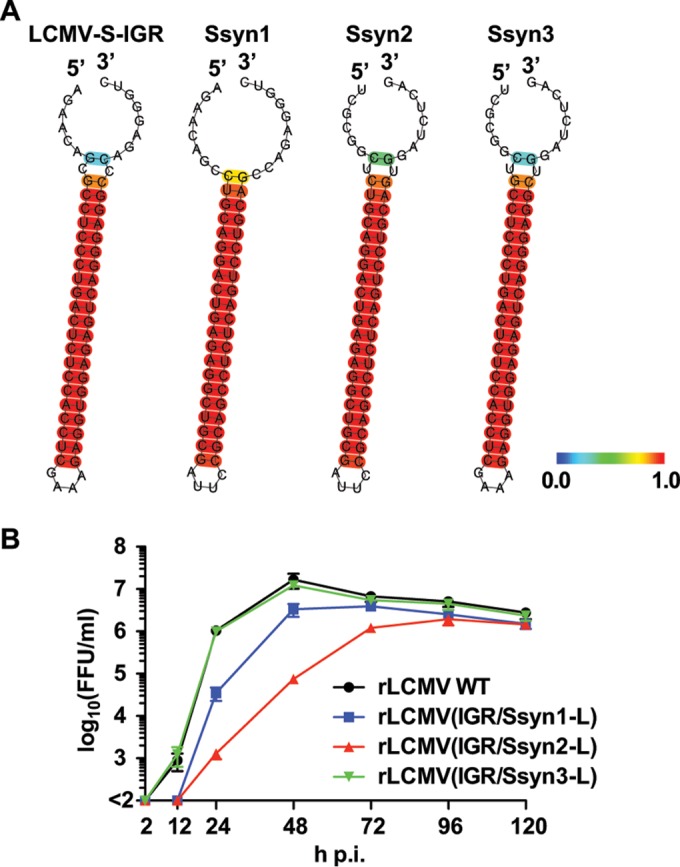
Generation of rLCMVs containing synthetic S-like IGR sequences (Ssyns) in their S segments [rLCMV(IGR/Ssyn-L)]. (A) Predicted RNA secondary structures of Ssyn IGRs were determined using the CentroidFold web server (http://rtools.cbrc.jp). Each predicted base pair was colored using a heat color gradation from blue to red that corresponds to a base-pairing probability of from 0 to 1. (B) Growth kinetics of rLCMV(IGR/Ssyn-L). BHK-21 cells were infected (MOI = 0.01) with the rLCMV WT or the indicated rLCMV(IGR/Ssyn-L) construct. Virus titers in the TCS were determined at the indicated times p.i. Data represent the means ± SDs from three independent experiments.
Ability of rLCMV(IGR/Ssyn-L) to inhibit induction of IFN-I expression.
The IGR plays a critical role in the control of viral gene expression at the posttranscriptional level (21). Replacement of the WT S-IGR with Ssyn could therefore affect viral gene expressions in infected cells, which may result in lower levels of NP expression and, subsequently, compromise the ability to inhibit type I interferon (IFN-I) induction (31). To address this issue, we examined the growth kinetics of rLCMV(IGR/Ssyn-L) in IFN-I-competent A549 cells. rLCMV(IGR/Ssyn3-L) and the rLCMV WT grew to similar titers, whereas rLCMV(IGR/Ssyn1-L) and, especially, rLCMV(IGR/Ssyn2-L) were significantly impaired in their ability to grow in A549 cells compared to that of the rLCMV WT (Fig. 3A). Consistent with this finding, both rLCMV(IGR/Ssyn1-L) and rLCMV(IGR/Ssyn2-L) induced a high level of ISRE-mediated reporter gene expression, while rLCMV WT or rLCMV(IGR/Ssyn3-L) infection resulted in minimal levels of ISRE-mediated reporter gene expression (Fig. 3B). These data also suggested that rLCMV(IGR/Ssyn2-L) might be more sensitive to IFN-I than rLCMV(IGR/Ssyn3-L). To address this issue, we examined the sensitivity of rLCMV(IGR/Ssyn-L) to exogenous IFN-I treatment. For this we treated IFN-I-deficient Vero cells with IFN-α at a low (5 U/ml) or a high (500 U/ml) concentration or left them untreated immediately after infection with rLCMV WT, rLCMV(IGR/Ssyn2-L), or rLCMV(IGR/Ssyn3-L) at a low (0.05) MOI, and 12 h and 48 h later, the numbers of infected cells were determined by an immunofluorescence assay (IFA) to assess virus propagation in the IFN-I-treated cells (Fig. 3C). All three viruses were able to spread efficiently in the presence of exogenous IFN-α, which indicated that rLCMV(IGR/Ssyn2-L) and the rLCMV WT were similarly resistant to IFN-I and that the decreased fitness of rLCMV(IGR/Ssyn2-L) in A549 cells occurred via a mechanism that remains to be determined.
Phenotypic characterization of rLCMV(IGR/Ssyn-L).
To examine whether the decrease in fitness observed for rLCMV(IGR/Ssyn1-L) and rLCMV(IGR/Ssyn2-L) in cultured cells had a significant impact on virulence in vivo, we inoculated 6-week-old WT C57BL/6J mice intracranially (i.c.) with 103 FFU of the rLCMV WT or each of the rLCMV(IGR/Ssyn-L) constructs (Fig. 4A). Consistent with previous findings (32), mice infected with the rLCMV WT developed a fatal case of lymphocytic choriomeningitis (LCM) and succumbed within 8 days of infection. Mice injected with rLCMV(IGR/Ssyn3-L) succumbed by 8 days p.i., which was anticipated, because rLCMV(IGR/Ssyn3-L) exhibited WT-like properties in cultured cells. In contrast, all mice infected with either rLCMV(IGR/Ssyn1-L) or rLCMV(IGR/Ssyn2-L) survived and showed no overt signs of disease throughout the course of the experiment (Fig. 4A). Moreover, rLCMV(IGR/Ssyn1-L)- or rLCMV(IGR/Ssyn2-L)-infected mice subjected 28 days later to a lethal challenge with the rLCMV WT (103 FFU i.c.) were fully protected and did not develop noticeable clinical symptoms (Fig. 4A). These results indicated that rLCMV(IGR/Ssyn1-L) and rLCMV(IGR/Ssyn2-L) exhibit in vivo features characteristic of live-attenuated vaccines.
A key concern with LAVs relates to genetic stability and the possibility that phenotypic revertants with increased virulence may be selected during multiple rounds of virus replication. We therefore examined the stability of rLCMV(IGR/Ssyn-L) during several serial passages in cultured cells (Fig. 4B). The titers of all three rLCMV(IGR/Ssyn-L) virus constructs remained stable over 10 passages in BHK-21 cells. Likewise, sequence analysis showed that all Ssyn constructs retained the initial sequence (Table 1) after 10 serial passages in BHK-21 cells.
Generation of rLCMVs with L-IGR replaced by an Ssyn IGR.
We have shown that rLCMV(IGR/S-S), whose L-IGR was replaced by S-IGR, was highly attenuated in vivo but able to induce protection against a lethal challenge with the LCMV WT (21). These findings motivated us to examine the possibility of using an Ssyn IGR instead of L-IGR as a universal molecular determinant of arenavirus attenuation that could be used for the development of an arenavirus LAV. This approach would have the advantage that Ssyn would not exist in nature and could be used as a molecular marker to distinguish a vaccine strain from field-circulating virus. We successfully rescued all three rLCMV(IGR/S-Ssyn) constructs, which grew to relatively high titers but exhibited some decrease in fitness compared to that of the LCMV WT (Fig. 5A). The growth properties of rLCMV(IGR/Ssyn-L) correlated with the degree of sequence alteration incorporated into the S-IGR, which suggested that the authentic S-IGR sequence is favored for optimal virus growth. In contrast, all rLCMV(IGR/S-Ssyn) constructs exhibited similar growth properties. These findings may reflect the fact that the complete range of functions played by the S-IGR is influenced by its segment location via interactions that remain to be determined.
FIG 5.
Generation of rLCMVs containing an altered L-IGR. (A) Growth kinetics of rLCMVs containing a synthetic S-like IGR in their L segment [rLCMV(IGR/S-Ssyn)]. BHK-21 cells were infected with the rLCMV WT or an rLCMV(IGR/S-Ssyn) construct (MOI = 0.01). Virus titers in the TCS were determined at the indicated times p.i. Data represent the means ± SDs from three independent experiments. (B) The predicted RNA secondary structures of genome (viral RNA [vRNA]) and antigenome (cRNA) S-IGRs of GGV were determined using the CentroidFold web server (http://rtools.cbrc.jp). Each predicted base pair was colored using a heat color gradation from blue to red that corresponds to a base-pairing probability of from 0 to 1. (C) Growth kinetics of rLCMV containing the GGV S-IGR in its L segment [rLCMV(IGR/S-Sggv)]. BHK-21 cells were infected with the rLCMV WT or rLCMV(IGR/S-Sggv) (MOI = 0.01). Virus titers in the TCS were determined at the indicated times p.i. Data represent the means ± SDs from three independent experiments.
Recently, a number of snake arenaviruses have been identified in members of the Boidae and Pythonidae families with a diagnosis of inclusion body disease (IBD) (22, 33–35). Snake arenaviruses are genetically significantly different from the mammalian arenaviruses and constitute the newly proposed genus Reptarenavirus (36). Accordingly, both the sequence and the predicted structure of the S-IGR of the reptarenavirus Golden Gate virus (GGV) (Sggv) are significantly different from those of the LCMV S-IGR (Fig. 5B). We anticipated that Sggv could also be used as a genetic module to attenuate arenaviruses. rLCMV(IGR/S-Sggv) was successfully rescued and grew to titers similar to those of rLCMV(IGR/S-Ssyn1) in BHK-21 cells (Fig. 5C).
Ability of rLCMV with altered L-IGR to inhibit induction of IFN-I expression.
We have previously shown that rLCMV(IGR/S-S) has significantly impaired growth in IFN-I-competent A549 cells and induces a high level of reporter gene expression driven by the ISRE promoter (21). Compared to the fitness of the rLCMV WT, all rLCMVs containing an altered L-IGR exhibited significantly reduced fitness in A549 cells (Fig. 6A and B). Likewise, infection of A549/ISRE-Fluc cells with rLCMV containing an altered L-IGR resulted in ISRE-mediated reporter gene expression at levels higher than those in cells infected with the rLCMV WT (Fig. 6C). We observed differences in the induction of IFN-I among the different rLCMVs with altered L-IGRs. rLCMV(IGR/S-Ssyn3) induced the highest level of reporter gene expression but also showed better growth properties than rLCVM(IGR/S-Ssyn1) and rLCVM(IGR/S-Ssyn2) in A549 cells, suggesting that the reduced fitness of rLCMVs with an altered L-IGR in A549 cells might not be explained only by the magnitude of the IFN-I response that they induced. We have documented that aspartic acid (D) at position 382 of NP is critical for the NP's ability to inhibit IFN-I induction and an rLCMV with the NP mutation D382A [rLCMV(NP/D382A)] lacks the IFN-I-counteracting activity (37). We used rLCMV(NP/D382A) as a benchmark to evaluate the level of IFN-I induced by rLCMV(IGR/S-Ssyn2). The levels of ISRE-mediated reporter gene expression were significantly higher in cells infected with rLCMV(NP/D382A) than in cells infected with rLCMV(IGR/S-Ssyn2) (Fig. 6D). This result could have reflected differences in expression levels rather than differences in the function of NP in cells infected with the different viruses. To investigate this possibility, we infected (MOI = 1) A549 cells with the rLCMV WT, rLCMV(IGR/S-Ssyn2), or rLCMV(IGR/S-Sggv) and examined the kinetics of NP expression (Fig. 6E and F). As with rLCMV(IGR/S-S), NP expression levels in cells infected with rLCMVs containing an altered L-IGR were similar to those in rLCMV WT-infected cells, suggesting that the reduced IFN-I-counteracting activity associated with rLCMV(IGR/S-Ssyn2) cannot be explained on the basis of reduced levels of NP expression.
FIG 6.
(A, B) Growth properties in IFN-competent cells of rLCMVs containing an Ssyn (A) or Sggv (B) in their L segments. A549 cells were infected (MOI = 0.01) with the indicated rLCMV, and the virus titers in the TCS were determined at the indicated times p.i. Data represent the means ± SDs from three independent experiments. (C) Ability of the rLCMV(IGR/S-Ssyn) constructs and rLCMV(IGR/S-Sggv) to induce ISRE-mediated reporter gene expression. A549/ISRE-Fluc cells were infected (MOI = 0.1) with the indicated rLCMV or treated with IFN-α at the concentrations indicated on the x axis. At 17 h p.i., the levels of Fluc activity were determined. Values of Fluc activity from mock-infected and untreated cells were subtracted as background activity. Data represent the means ± SDs from three independent experiments. (D) Comparison of the levels of IFN-I production in cells infected with rLCMV(NP/D382A) and rLCMV(IGR/S-Ssyn2). A549/ISRE-Fluc cells were infected (MOI = 0.1) with the indicated rLCMV or treated with IFN-α at the concentrations indicated on the x axis. At 17 h p.i., the levels of Fluc activity were determined. Values of Fluc activity from mock-infected and untreated cells were subtracted as background activity. Data represent the means ± SDs from three independent experiments. (E, F). Comparison of NP expression levels in cells infected with rLCMV/WT, rLCMV(IGR/S-Ssyn2), and rLCMV(IGR/S-Sggv). A549 cells were infected with rLCMV WT, rLCMV(IGR/S-Ssyn2), or rLCMV(IGR/S-Sggv) (MOI = 1). At 8 and 24 h p.i., infected cells were harvested and NP expression was analyzed by flow cytometry. The percentage of NP-positive cells (E) and the mean signal intensity of the NP-positive cell population (F) are shown. Data represent the means ± SDs from three independent experiments.
Phenotypic characterization of rLCMVs containing an altered S-IGR in their L segments.
We next asked whether, as for rLCMV(IGR/S-S), replacement of the L-IGR with either Ssyn or Sggv could lead to an attenuated phenotype in vivo. For this, we inoculated (i.c.) 6-week-old WT C57BL/6J mice with 103 FFU of the rLCMV WT, rLCMV(IGR/S-Ssyn2), or rLCMV(IGR/S-Sggv). As expected, mice infected with the rLCMV WT developed LCM and succumbed by day 8 p.i. (Fig. 7A). In contrast, all mice infected with rLCMV(IGR/S-Ssyn2) or rLCMV(IGR/S-Sggv) survived without showing overt clinical symptoms and were fully protected from a lethal challenge with the rLCMV WT (103 FFU i.c.), done 30 days after the initial infection (Fig. 7A). These results indicated that both rLCMV(IGR/S-Ssyn2) and rLCMV(IGR/S-Sggv) were highly attenuated in vivo but able to replicate to a sufficient degree to elicit protective immunity against a lethal challenge with the LCMV WT. We also examined whether rLCMV(IGR/S-Ssyn2) and rLCMV(IGR/S-Sggv) could be successfully used in a standard LCMV immunization protocol (i.p. inoculation with 105 FFU). Both rLCMV(IGR/S-Ssyn2) and rLCMV(IGR/S-Sggv) induced protective immunity against a lethal challenge with the rLCMV WT (103 FFU i.c.) (Fig. 7B). Likewise, rLCMV(IGR/S-Ssyn2) and rLCMV(IGR/S-Sggv) exhibited the stable production of infectious progeny during serial passages in cultured cells (Fig. 7C), and sequencing analysis revealed that both rLCMVs were also genetically very stable. Together these results indicated that it is possible to generate rLCMV(IGR/S-Ssyn) that exhibits key features of a safe and effective LAV: (i) growth to high levels in cultured cells, (ii) a highly attenuated phenotype in vivo but an ability to elicit protective immunity against a lethal challenge with wild-type LCMV, and (iii) genetic and, therefore, phenotypic, stability.
FIG 7.
In vivo virulence and stability of rLCMV(IGR/S-Ssyn2) and rLCMV(IGR/S-Sggv). (A) Six-week-old C57BL/6J mice (4 mice/group) were infected (103 FFU i.c.) with the rLCMV WT, rLCMV(IGR/S-Ssyn2), or rLCMV(IGR/S-Sggv) or left uninfected (naive). At 30 days p.i. (broken line), mice that survived were subjected to a lethal challenge with the rLCMV WT (103 FFU i.c.). (B) Immunization with rLCMV(IGR/S-Ssyn2) or rLCMV(IGR/S-Sggv) induces protection against a lethal LCMV challenge. Six-week-old C57BL/6J mice (4 mice/group) were infected (105 FFU i.p.) with the rLCMV WT, rLCMV(IGR/S-Ssyn2), or rLCMV(IGR/S-Sggv) or left uninfected (naive). At 28 days p.i. (broken line), mice were subjected to a lethal LCMV challenge (103 FFU i.c.). (C) Stability of rLCMV(IGR/S-Ssyn2) and rLCMV(IGR/S-Sggv) in cultured cells. BHK-21 cells were infected with the rLCMV WT, rLCMV(IGR/S-Ssyn2), or rLCMV(IGR/S-Sggv) (MOI = 0.01). At 72 h p.i., the TCS was collected and virus titers were determined by an immunofocus assay. Fresh BHK-21 cell monolayers were infected with the TCS (MOI = 0.01); this process was serially repeated throughout P2 to P10.
DISCUSSION
Because LAVs induce robust cellular and humoral immune responses following a single immunization, they represent the most feasible approach to the control of LASV in West Africa and other emergent HFAs in their respective regions of endemicity (38). Accordingly, significant efforts aimed at developing safe and effective vaccines against HFAs have been made (15, 39). The Mopeia virus/LASV reassortant ML29 is a promising LASV LAV candidate (40). However, although the genetic differences that distinguish ML29 from the pathogenic LASV are known, the mechanisms of ML29 attenuation remain unknown, which raises concerns about the phenotypic stability of ML29 in response to additional mutations. The identification of a general strategy to generate recombinant arenaviruses with highly attenuated and stable phenotypes driven by well-defined molecular mechanisms would facilitate the development of safe and effective arenavirus LAVs. Virus attenuation without incorporation of amino acid changes into the gene products encoded by the viral genome has an advantage that the resulting virus would have the same antigenic composition. In this regard, several strategies, including codon deoptimization (41) and gene rearrangement (42), which do not involve amino acid changes in viral gene products, have recently been proposed to develop LAVs for arenaviruses. Similarly, our strategy of generating rLCMVs where the L-IGR is replaced with either Ssyn or Sggv also avoids the introduction of amino acid changes into any of the viral gene products. This alteration of the L-IGR resulted in viruses that had a severely attenuated phenotype in vivo but that were still able to induce protective immunity against a subsequent lethal challenge with wild-type LCMV. Ssyn and Sggv are genetically very distinct from the S-IGRs of any known mammarenavirus, and therefore, they could be used to attenuate currently known as well as potentially newly emerging HFAs to develop LAV strains. Moreover, because of its unique sequence, Ssyn could be also used as a universal molecular marker to distinguish LAV strains from field-circulating virus.
We have shown that the IGR plays critical roles in the virus life cycle. The IGR is necessary for the production of infectious VLPs (26), but the efficiency of production of infectious VLPs appears to be mainly determined by 3′ and 5′ untranslated regions (UTRs) within each genome segment (21). The sequence of Slas is very different from that of the LCMV S-IGR, but the production of LCMV infectious VLPs was minimally affected when Slas was substituted for the LCMV S-IGR. Moreover, we were able to rescue viable rLCMVs containing altered IGRs that nonetheless grew to about 10-fold lower titers than the rLCMV WT. These results indicate that the attenuation of rLCMV containing an altered IGR is unlikely related to a significantly reduced budding efficiency. On the other hand, arenavirus gene expression is exquisitely regulated by the IGR. Both S- and L-IGRs act as bona fide transcriptional termination signals that generate nonpolyadenylated viral mRNA species whose 3′ ends have been mapped to multiple sites within the distal site of the IGR (26, 43). In addition to their role as transcription termination signals, those IGR sequences present at the 3′ ends of mRNA species contribute to the regulation of the efficiency of translation (21). Replacement of L-IGR with Ssyn or Sggv is predicted to alter this orchestrated mechanism of viral gene expression control, thus resulting in an attenuated phenotype.
Arenavirus genome RNA species contain 3′ and 5′ UTRs that are highly conserved within the initial 19 nucleotides (nt) of the 3′ UTR, and the complementary sequence is found in the corresponding 5′ UTR, which results in the formation of predicted panhandle structures that are consistent with results from electron microscopy (2, 44). This terminal complementarity is required for genome RNA encapsidation by NP and subsequent promoter recognition by the L polymerase. Mutations introduced within the highly conserved most 3′-terminal 19 nt disrupt terminal complementarity and abrogate the activity of the genome promoter. Importantly, compensatory mutations within the 5′ end to reestablish terminal complementarity did not rescue promoter activity, indicating that both sequence specificity and structure are strictly required for viral RNA synthesis, mediated by the virus L polymerase (45). In contrast, the data presented in this report indicate that the IGR exhibits a large degree of sequence plasticity, as replacement of the entire IGR with a nonviral synthetic IGR could still support the production of infectious virus, suggesting that the IGR structure, rather than its sequence, is critical for virus viability. We observed a decreased fitness of rLCMVs with altered S-IGRs in cultured cells, indicating that sequence specificity within the S-IGR contributes to optimal viral growth. We were also able to rescue rLCMV(IGR/S-Sggv). Because the structure, in addition to the sequence, of Sggv is significantly different from that of the naturally found LCMV S-IGR, this finding would question the view that the structure of the S-IGR is important for virus viability. It should be noted that Sggv is a naturally found arenavirus S-IGR that can generate the stem-loop structures characteristically predicted for arenavirus S-IGRs. It is plausible that Sggv might contain structural elements that facilitated the rescue of a viable rLCMV.
Notably, none of the mutated IGR sequences incorporated nucleotide substitutions during serial passages in cultured cells. This could partially be explained by the restriction imposed by the need to maintain the stem-loop structure of the IGR, which would require that 2 complementary nucleotides would have to change simultaneously. Therefore, an already viable arenavirus IGR might be intrinsically resistant to the incorporation of mutations, which is consistent with the fact that the IGR exhibits a high degree of conservation within the same arenavirus species. As the rLCMVs containing altered IGRs exhibited different abilities to counteract the induction of the IFN-I response, it would be of great interest to examine the genetic stability of the recombinant viruses during serial passages in IFN-I-competent cells, as well as in both immunocompetent and immunocompromised mice.
We documented that rLCMV(IGR/S-S) lost its ability to efficiently inhibit the induction of IFN-I (21), although the expression levels of NP, a virus-counteracting IFN-I factor (31), in rLCMV(IGR/S-S)-infected cells were not reduced. One possible explanation for this is that newly synthesized RNA species which might be generated as a consequence of the improper termination of viral RNA synthesis or possible RNA complexes formed by the interaction between the identical sequences present at both IGRs in rLCMV(IGR/S-S) could induce IFN-I in rLCMV(IGR/S-S)-infected cells through a pathway that the NP cannot inhibit. Likewise, all rLCMVs with altered IGRs that we have examined in this work were also impaired to different degrees in their ability to prevent the induction of IFN-I expression. Intriguingly, among all three tested rLCMV(IGR/S-Ssyn) constructs, rLCMV(IGR/S-Ssyn3) induced the highest level of reporter gene expression driven by an ISRE promoter. Ssyn3 has the same stem-loop region sequence as wild-type S-IGR. Therefore, it is plausible that RNA complexes generated by the interaction between the identical stem-loop sequences of S- and L-IGRs could be a major cause of IFN-I induction. Alternatively, viral RNAs containing the synthetic IGR might be more immunogenic or somewhat resistant to degradation by the 3′-5′ exoribonuclease activity of NP, and they would thereby provide a more potent or more stable, or both, danger signal. The degree of IFN-I induction by the different rLCMVs with altered IGRs did not necessarily correlate with the defect in growth in A549 cells. Accordingly, these rLCMV constructs and the rLCMV WT exhibited similar degrees of resistance to exogenous IFN-α treatment in Vero cells. These results also indicated that specific sequences within the S-IGR contributed to the significantly reduced fitness of rLCMV(IGR/Ssyn2-L) in A549 cells via a mechanism that remains to be determined.
ACKNOWLEDGMENTS
This research was supported by NIH/NIAID grants AI047140 and AI077719 to J.C.D.L.T. and by the Japan Society for the Promotion of Science, the Daiichi Sankyo Foundation of Life Science, and the KANAE Foundation for the Promotion of Medical Science to M.I.
Footnotes
This is article number 29239 from The Scripps Research Institute.
REFERENCES
- 1.Bray M. 2005. Pathogenesis of viral hemorrhagic fever. Curr Opin Immunol 17:399–403. doi: 10.1016/j.coi.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Buchmeier MJ, Peters CJ, de la Torre JC. 2007. Arenaviridae: the viruses and their replication, p1791–1851. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed, vol 2Lippincott Williams & Wilkins,Philadelphia, PA. [Google Scholar]
- 3.Geisbert TW, Jahrling PB. 2004. Exotic emerging viral diseases: progress and challenges. Nat Med 10:S110–S121. doi: 10.1038/nm1142. [DOI] [PubMed] [Google Scholar]
- 4.Richmond JK, Baglole DJ. 2003. Lassa fever: epidemiology, clinical features, and social consequences. BMJ 327:1271–1275. doi: 10.1136/bmj.327.7426.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freedman DO, Woodall J. 1999. Emerging infectious diseases and risk to the traveler. Med Clin North Am 83:865–883. [PubMed] [Google Scholar]
- 6.Isaacson M. 2001. Viral hemorrhagic fever hazards for travelers in Africa. Clin Infect Dis 33:1707–1712. doi: 10.1086/322620. [DOI] [PubMed] [Google Scholar]
- 7.Briese T, Paweska JT, McMullan LK, Hutchison SK, Street C, Palacios G, Khristova ML, Weyer J, Swanepoel R, Egholm M, Nichol ST, Lipkin WI. 2009. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog 5:e1000455. doi: 10.1371/journal.ppat.1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paweska JT, Sewlall NH, Ksiazek TG, Blumberg LH, Hale MJ, Lipkin WI, Weyer J, Nichol ST, Rollin PE, McMullan LK, Paddock CD, Briese T, Mnyaluza J, Dinh TH, Mukonka V, Ching P, Duse A, Richards G, de Jong G, Cohen C, Ikalafeng B, Mugero C, Asomugha C, Malotle MM, Nteo DM, Misiani E, Swanepoel R, Zaki SR, Outbreak Control and Investigation Teams . 2009. Nosocomial outbreak of novel arenavirus infection, southern Africa. Emerg Infect Dis 15:1598–1602. doi: 10.3201/eid1510.090211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barton LL, Mets MB, Beauchamp CL. 2002. Lymphocytic choriomeningitis virus: emerging fetal teratogen. Am J Obstet Gynecol 187:1715–1716. doi: 10.1067/mob.2002.126297. [DOI] [PubMed] [Google Scholar]
- 10.Fischer SA, Graham MB, Kuehnert MJ, Kotton CN, Srinivasan A, Marty FM, Comer JA, Guarner J, Paddock CD, DeMeo DL, Shieh WJ, Erickson BR, Bandy U, DeMaria A Jr, Davis JP, Delmonico FL, Pavlin B, Likos A, Vincent MJ, Sealy TK, Goldsmith CS, Jernigan DB, Rollin PE, Packard MM, Patel M, Rowland C, Helfand RF, Nichol ST, Fishman JA, Ksiazek T, Zaki SR, LCMV in Transplant Recipients Investigation Team . 2006. Transmission of lymphocytic choriomeningitis virus by organ transplantation. N Engl J Med 354:2235–2249. doi: 10.1056/NEJMoa053240. [DOI] [PubMed] [Google Scholar]
- 11.Jahrling PB, Peters CJ. 1992. Lymphocytic choriomeningitis virus. A neglected pathogen of man. Arch Pathol Lab Med 116:486–488. [PubMed] [Google Scholar]
- 12.Palacios G, Druce J, Du L, Tran T, Birch C, Briese T, Conlan S, Quan PL, Hui J, Marshall J, Simons JF, Egholm M, Paddock CD, Shieh WJ, Goldsmith CS, Zaki SR, Catton M, Lipkin WI. 2008. A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med 358:991–998. doi: 10.1056/NEJMoa073785. [DOI] [PubMed] [Google Scholar]
- 13.Peters CJ. 2006. Lymphocytic choriomeningitis virus—an old enemy up to new tricks. N Engl J Med 354:2208–2211. doi: 10.1056/NEJMp068021. [DOI] [PubMed] [Google Scholar]
- 14.Borio L, Inglesby T, Peters CJ, Schmaljohn AL, Hughes JM, Jahrling PB, Ksiazek T, Johnson KM, Meyerhoff A, O'Toole T, Ascher MS, Bartlett J, Breman JG, Eitzen EM Jr, Hamburg M, Hauer J, Henderson DA, Johnson RT, Kwik G, Layton M, Lillibridge S, Nabel GJ, Osterholm MT, Perl TM, Russell P, Tonat K, Working Group on Civilian Biodefense . 2002. Hemorrhagic fever viruses as biological weapons: medical and public health management. JAMA 287:2391–2405. doi: 10.1001/jama.287.18.2391. [DOI] [PubMed] [Google Scholar]
- 15.Olschlager S, Flatz L. 2013. Vaccination strategies against highly pathogenic arenaviruses: the next steps toward clinical trials. PLoS Pathog 9:e1003212. doi: 10.1371/journal.ppat.1003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bausch DG, Hadi CM, Khan SH, Lertora JJ. 2010. Review of the literature and proposed guidelines for the use of oral ribavirin as postexposure prophylaxis for Lassa fever. Clin Infect Dis 51:1435–1441. doi: 10.1086/657315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damonte EB, Coto CE. 2002. Treatment of arenavirus infections: from basic studies to the challenge of antiviral therapy. Adv Virus Res 58:125–155. doi: 10.1016/S0065-3527(02)58004-0. [DOI] [PubMed] [Google Scholar]
- 18.Hadi CM, Goba A, Khan SH, Bangura J, Sankoh M, Koroma S, Juana B, Bah A, Coulibaly M, Bausch DG. 2010. Ribavirin for Lassa fever postexposure prophylaxis. Emerg Infect Dis 16:2009–2011. doi: 10.3201/eid1612.100994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez M, Craven RC, de la Torre JC. 2003. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc Natl Acad Sci U S A 100:12978–12983. doi: 10.1073/pnas.2133782100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strecker T, Eichler R, Meulen J, Weissenhorn W, Dieter Klenk H, Garten W, Lenz O. 2003. Lassa virus Z protein is a matrix protein and sufficient for the release of virus-like particles. J Virol 77:10700–10705. doi: 10.1128/JVI.77.19.10700-10705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwasaki M, Ngo N, Cubitt B, Teijaro JR, de la Torre JC. 2015. A general molecular strategy for development of arenavirus live-attenuated vaccines. J Virol 89:12166–12177. doi: 10.1128/JVI.02075-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stenglein MD, Sanders C, Kistler AL, Ruby JG, Franco JY, Reavill DR, Dunker F, Derisi JL. 2012. Identification, characterization, and in vitro culture of highly divergent arenaviruses from boa constrictors and annulated tree boas: candidate etiological agents for snake inclusion body disease. mBio 3:e00180–12. doi: 10.1128/mBio.00180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee KJ, Novella IS, Teng MN, Oldstone MB, de La Torre JC. 2000. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J Virol 74:3470–3477. doi: 10.1128/JVI.74.8.3470-3477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee KJ, Perez M, Pinschewer DD, de la Torre JC. 2002. Identification of the lymphocytic choriomeningitis virus (LCMV) proteins required to rescue LCMV RNA analogs into LCMV-like particles. J Virol 76:6393–6397. doi: 10.1128/JVI.76.12.6393-6397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urata S, Yasuda J, de la Torre JC. 2009. The Z protein of the New World arenavirus Tacaribe virus has bona fide budding activity that does not depend on known late domain motifs. J Virol 83:12651–12655. doi: 10.1128/JVI.01012-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinschewer DD, Perez M, de la Torre JC. 2005. Dual role of the lymphocytic choriomeningitis virus intergenic region in transcription termination and virus propagation. J Virol 79:4519–4526. doi: 10.1128/JVI.79.7.4519-4526.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emonet SF, Garidou L, McGavern DB, de la Torre JC. 2009. Generation of recombinant lymphocytic choriomeningitis viruses with trisegmented genomes stably expressing two additional genes of interest. Proc Natl Acad Sci U S A 106:3473–3478. doi: 10.1073/pnas.0900088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flatz L, Bergthaler A, de la Torre JC, Pinschewer DD. 2006. Recovery of an arenavirus entirely from RNA polymerase I/II-driven cDNA. Proc Natl Acad Sci U S A 103:4663–4668. doi: 10.1073/pnas.0600652103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez AB, de la Torre JC. 2006. Rescue of the prototypic arenavirus LCMV entirely from plasmid. Virology 350:370–380. doi: 10.1016/j.virol.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 30.Battegay M. 1993. Quantification of lymphocytic choriomeningitis virus with an immunological focus assay in 24 well plates. ALTEX 10:6–14. [PubMed] [Google Scholar]
- 31.Martinez-Sobrido L, Zuniga EI, Rosario D, Garcia-Sastre A, de la Torre JC. 2006. Inhibition of the type I interferon response by the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol 80:9192–9199. doi: 10.1128/JVI.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cole GA, Gilden DH, Monjan AA, Nathanson N. 1971. Lymphocytic choriomeningitis virus: pathogenesis of acute central nervous system disease. Fed Proc 30:1831–1841. [PubMed] [Google Scholar]
- 33.Bodewes R, Kik MJ, Raj VS, Schapendonk CM, Haagmans BL, Smits SL, Osterhaus AD. 2013. Detection of novel divergent arenaviruses in boid snakes with inclusion body disease in The Netherlands. J Gen Virol 94:1206–1210. doi: 10.1099/vir.0.051995-0. [DOI] [PubMed] [Google Scholar]
- 34.Hetzel U, Sironen T, Laurinmaki P, Liljeroos L, Patjas A, Henttonen H, Vaheri A, Artelt A, Kipar A, Butcher SJ, Vapalahti O, Hepojoki J. 2013. Isolation, identification, and characterization of novel arenaviruses, the etiological agents of boid inclusion body disease. J Virol 87:10918–10935. doi: 10.1128/JVI.01123-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stenglein MD, Jacobson ER, Chang LW, Sanders C, Hawkins MG, Guzman DS, Drazenovich T, Dunker F, Kamaka EK, Fisher D, Reavill DR, Meola LF, Levens G, DeRisi JL. 2015. Widespread recombination, reassortment, and transmission of unbalanced compound viral genotypes in natural arenavirus infections. PLoS Pathog 11:e1004900. doi: 10.1371/journal.ppat.1004900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radoshitzky SR, Bao Y, Buchmeier MJ, Charrel RN, Clawson AN, Clegg CS, DeRisi JL, Emonet S, Gonzalez JP, Kuhn JH, Lukashevich IS, Peters CJ, Romanowski V, Salvato MS, Stenglein MD, de la Torre JC. 2015. Past, present, and future of arenavirus taxonomy. Arch Virol 160:1851–1874. doi: 10.1007/s00705-015-2418-y. [DOI] [PubMed] [Google Scholar]
- 37.Martinez-Sobrido L, Emonet S, Giannakas P, Cubitt B, Garcia-Sastre A, de la Torre JC. 2009. Identification of amino acid residues critical for the anti-interferon activity of the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol 83:11330–11340. doi: 10.1128/JVI.00763-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falzarano D, Feldmann H. 2013. Vaccines for viral hemorrhagic fevers—progress and shortcomings. Curr Opin Virol 3:343–351. doi: 10.1016/j.coviro.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lukashevich IS. 2012. Advanced vaccine candidates for Lassa fever. Viruses 4:2514–2557. doi: 10.3390/v4112514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zapata JC, Poonia B, Bryant J, Davis H, Ateh E, George L, Crasta O, Zhang Y, Slezak T, Jaing C, Pauza CD, Goicochea M, Moshkoff D, Lukashevich IS, Salvato MS. 2013. An attenuated Lassa vaccine in SIV-infected rhesus macaques does not persist or cause arenavirus disease but does elicit Lassa virus-specific immunity. Virol J 10:52. doi: 10.1186/1743-422X-10-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng BY, Ortiz-Riano E, Nogales A, de la Torre JC, Martinez-Sobrido L. 2015. Development of live-attenuated arenavirus vaccines based on codon deoptimization. J Virol 89:3523–3533. doi: 10.1128/JVI.03401-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng BY, Ortiz-Riano E, de la Torre JC, Martinez-Sobrido L. 2015. Arenavirus genome rearrangement for the development of live attenuated vaccines. J Virol 89:7373–7384. doi: 10.1128/JVI.00307-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer BJ, Southern PJ. 1993. Concurrent sequence analysis of 5′ and 3′ RNA termini by intramolecular circularization reveals 5′ nontemplated bases and 3′ terminal heterogeneity for lymphocytic choriomeningitis virus mRNAs. J Virol 67:2621–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Young PR, Howard CR. 1983. Fine structure analysis of Pichinde virus nucleocapsids. J Gen Virol 64(Pt 4):833–842. doi: 10.1099/0022-1317-64-4-833. [DOI] [PubMed] [Google Scholar]
- 45.Perez M, de la Torre JC. 2003. Characterization of the genomic promoter of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol 77:1184–1194. doi: 10.1128/JVI.77.2.1184-1194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]