ABSTRACT
Molluscum contagiosum virus (MCV) gene MC159 encodes a viral FLICE inhibitory protein (vFLIP) that inhibits caspase-8-mediated apoptosis. The MC159 protein was also reported to inhibit programmed necrosis (necroptosis) and modulate NF-κB activation by interacting with RIP1 and NEMO. The importance of MC159 during MCV infection has remained unknown, as there is no system for propagation and genetic manipulation of this virus. Here we investigated the functions of MC159 during viral infection using murine cytomegalovirus (MCMV) as a surrogate virus. MC159 was inserted into the MCMV genome, replacing M36 or M45, two MCMV genes with functions similar to those reported for MC159. M36 encodes a viral inhibitor of caspase-8-induced apoptosis (vICA) and M45 a viral inhibitor of RIP activation (vIRA), which inhibits RIP1/RIP3-mediated necroptosis. The M45 protein also blocks NF-κB activation by interacting with NEMO. When expressed by MCMV, MC159 blocked tumor necrosis factor alpha (TNF-α)-induced apoptosis of infected cells and partially restored MCMV replication in macrophages. However, MC159 did not fully replace M45, as it did not inhibit necroptosis in murine cells, but it reduced TNF-α-induced necroptosis in MCMV-infected human HT-29 cells. MC159 also differed from M45 in its effect on NF-κB. While MCMV-encoded M45 blocked NF-κB activation by TNF-α and interleukin-1β (IL-1β), MC159 inhibited TNF-α- but not IL-1β-induced NF-κB activation in infected mouse fibroblasts. These results indicate that the spectrum of MC159's functions differs depending on cell type and expression system and that a cell culture system for the propagation of MCV is needed to determine the biological relevance of presumed viral gene functions.
IMPORTANCE MCV is a human-pathogenic poxvirus that cannot be propagated in cell culture or laboratory animals. Therefore, MCV gene products have been studied predominantly in cells expressing individual viral genes. In this study, we analyzed the function of the MCV gene MC159 by expressing it from a different virus and comparing its functions to those of two well-characterized MCMV genes. In this system, MC159 displayed some but not all of the previously described functions, suggesting that the functions of a viral gene depend on the conditions under which it is expressed. Until a cell culture system for the analysis of MCV becomes available, it might be necessary to analyze MCV genes in several different systems to extrapolate their biological importance.
INTRODUCTION
Molluscum contagiosum virus (MCV) is a worldwide-occurring poxvirus that replicates exclusively in humans (1). Seroprevalence ranges from up to 39% in immunocompetent individuals (2, 3), affecting mostly children and young adults, to 91% in HIV-infected patients (2). Molluscum contagiosum (MC) is an infectious disease of the skin characterized by small, benign papular skin lesions that usually regress after a few months. However, in immunocompromised patients MC can become much more severe, with extensive lesions that are difficult to treat (1). This indicates the important role of the host immune system in confining and eliminating the infection.
Despite its importance as a human pathogen, MCV has not been studied extensively due to technical difficulties. There is neither a cell culture system suitable for the propagation of MCV nor an animal model that recapitulates MCV replication. Therefore, individual MCV genes were studied either by using transient transfection or by expression in a recombinant vaccinia virus (VACV) as a surrogate virus (4).
MCV expresses several immune-modulating proteins that are thought to contribute to the persistence of the virus (4, 5). One of these proteins, MC159, has been identified as a viral FLICE-like inhibitory protein (vFLIP) (6) due to its structural and functional similarity to cellular FLIP (cFLIP) (7). Although vFLIPs have not been found in poxviruses other than MCV, they are present in several gammaherpesviruses (6, 8, 9). MC159 interacts with Fas-associated protein with death domain (FADD) and procaspase-8 (formerly known as FADD-like interleukin-1β-converting enzyme [FLICE]) and prevents Fas- and tumor necrosis factor alpha (TNF-α)-induced apoptosis (6, 8–10). However, if caspase-8 is inhibited, receptor interacting protein kinase 1 (RIP1, RIPK1) is not cleaved and recruits RIP3 (RIPK3), initiating a pathway leading to necroptosis, a nonapoptotic form of programmed cell death (11). Next to its antiapoptotic function, cellular FLIP is able to inhibit necroptosis by preventing the association of FADD, RIP1, and RIP3 (12). Two earlier studies showed that MC159 can also block TNF-α- or Fas-induced necroptosis in human Jurkat T cells, but the underlying mechanism remained unclear (13, 14).
In addition to its inhibiting function on programmed cell death, MC159 is able to modulate the activation of transcription factor NF-κB, an important cellular regulator of immediate immune responses and therefore an attractive target for viral modulation (15). It has been shown that overexpression of MC159 inhibits NF-κB activation by several stimuli, including death receptor stimulation or MyD88 overexpression (16–18), and that MC159 acts at the level of the inhibitor of κB kinase (IKK) complex by interacting with the IKKγ subunit (also called NF-κB essential modulator [NEMO]) (18). However, it has also been shown that MC159 can activate NF-κB: transient expression of MC159 in human Jurkat or 293 cells led to a significantly increased NF-κB activity, and the ambivalent effect appeared to depend on MC159 and RIP1 expression levels (19).
Murine cytomegalovirus (MCMV) expresses two proteins whose functions are highly similar to those of MC159 (summarized in Table 1). The MCMV M36 gene is homologous to UL36 of human cytomegalovirus and encodes a viral inhibitor of caspase-8-induced apoptosis (vICA), which binds to procaspase-8 and inhibits its activation (20–23). Deletion of M36 from the MCMV genome rendered infected cells sensitive to apoptosis triggered by death receptor stimulation (21). An MCMV ΔM36 mutant was attenuated in vivo as shown by reduced virus titers in salivary glands and lung (24). Replication of the ΔM36 virus was rescued in cultured macrophages and in vivo by insertion of a dominant negative FADD into the viral genome, suggesting that inhibition of death receptor-dependent apoptosis is important for MCMV replication and dissemination (24, 25).
TABLE 1.
Summary of functions ascribed to MC159/vFLIP, M36/vICA, and M45/vIRA
| Event | Function | Reference(s) ascribing function to: |
||
|---|---|---|---|---|
| MC159 | M36 | M45 | ||
| Cell death | Inhibition of caspase-8-dependent apoptosis | 6, 8–10 | 21, 22 | |
| Inhibition of death receptor-induced necroptosis | 13, 14 | 27, 29 | ||
| NF-κB signaling | Inhibition of NF-κB | 16–18 | 27, 32 | |
| Activation of NF-κB | 19 | 33 | ||
| Interaction with RIP1 | 13, 16 | 27 | ||
| Interaction with NEMO | 18 | 32 | ||
MCMV also expresses a well-characterized necroptosis inhibitor called vIRA (viral inhibitor of RIP activation) that is encoded by gene M45 (26–29). M45 inhibits necroptosis by interacting with RIP1 and RIP3 via a RIP homotypic interaction motif (RHIM), thereby inhibiting formation of the necroptosis-inducing RIP1-RIP3 complex (28, 29). Through the same RHIM-dependent mechanism, M45 also inhibits RIP3 activation by DNA-dependent activator of interferon (IFN)-regulatory factors (DAI), which is activated upon MCMV infection (30). In the absence of M45 or when the M45 RHIM is mutated, MCMV induces rapid cell death and fails to replicate in necroptosis-sensitive cells such as SVEC4-10 endothelial cells (26, 29). The crucial role of M45 for viral fitness in vivo has also been demonstrated (29, 31).
Besides inhibiting necroptosis, M45 also modulates NF-κB activation by interacting with RIP1 and NEMO (27, 32). Importantly, M45 redirects NEMO to autophagosomes, thereby blocking all canonical NF-κB-activating pathways (32). However, M45 also mediates a brief and transient NF-κB activation in fibroblasts immediately after infection: NF-κB is activated during the first few hours of the infection by virion-associated M45 and inhibited at later time points by de novo-synthesized M45, suggesting an M45 concentration-dependent effect on NF-κB (33).
In this study, we analyzed the functions of MCV MC159 in the context of a viral infection and compared them to the well-characterized functions of two other viral proteins, M36/vICA and M45/vIRA of MCMV. Since MCV cannot be propagated in cell culture and there is no system for genetic manipulation of this virus, we decided to express MC159 from recombinant MCMVs lacking either M36 or M45. Using this system, we tested the ability of MC159 to inhibit apoptosis and necroptosis as well as its influence on the NF-κB pathway. This functional analysis of MC159 in the context of a viral infection should provide a refined picture of the diverse functions of this protein and contribute to a better understanding of their biological relevance.
MATERIALS AND METHODS
Plasmids and retroviral vectors.
The coding sequence of MC159 with a C-terminal hemagglutinin (HA) epitope tag was synthesized as a codon-optimized sequence and inserted into pMA-T by GeneArt (ThermoFisher). The MC159 sequence was subcloned in pcDNA3 (Invitrogen). Expression plasmids pcDNA-M45 and pcDNA-M45Ct3 have been described (27, 32). pEGFP-C1 and pNiFty2-SEAP were purchased from Clontech and InvivoGen, respectively. Transfection of NIH 3T3 cells was performed using Polyfect (Qiagen) or Lipofectamine 2000 (Invitrogen), and 293A cells were transfected using polyethylenimine (Sigma).
Retroviral vector plasmids pRetroM45, pRetroM45Ct3, and pRetroGFP have been used in previous studies (27, 32). To generate pRetroMC159, the MC159 sequence was inserted into the PmlI site of the empty retroviral vector plasmid. Production of retrovirus using Phoenix cells and transduction of cells was performed as described previously (34).
Cells and viruses.
NIH 3T3 fibroblasts (CRL-1658), SVEC4-10 endothelial cells (CRL-2181), and RAW 264.7 macrophages (TIB-71) were obtained from ATCC. Human embryonic kidney (HEK) 293A cells were purchased from Invitrogen. 10.1 cells are immortalized mouse embryonic fibroblasts (35). HT-29 cells were kindly provided by Udo Schumacher (University Medical Center Hamburg). Cells were cultured at 37°C and 5% CO2 in Dulbecco's modified Eagle medium (DMEM) or RPMI 1640 medium supplemented with 10% fetal calf serum. NIH 3T3 cells were grown in DMEM with 10% newborn calf serum.
MCMV-M45HA, MCMV-M45Ct3, and the MCMV deletion mutant ΔM45 have been described previously (33). The ΔM36 mutant was constructed essentially in the same way as described earlier for ΔM45 (33). The substitution mutants were generated by en passant mutagenesis (36) of the MCMV Smith bacterial artificial chromosome (BAC). Briefly, the kanamycin resistance gene aphAI and the I-SceI restriction site were PCR amplified from pEPkan-S (37) using primers 5′-AAA GGT ACC GTT ACT GCT ACG CCG CCT CTC CTA GCC TGC CTG TGC GGA CTA GGA TAA CAG GGT AAT CGA TTT-3′ and 5′-AAA GGT ACC GCC AGT GTT ACA ACC AAT TAA CC-3′ and inserted into a KpnI restriction site within pMA-T-MC159. MC159 including the Kanr/I-SceI cassette was PCR amplified using oligonucleotide primers containing 50-nucleotide (nt) homology arms for recombination: 5′-GCT CAT TCT TTC GGG AAA GGG GTG GAG GAG GGT CGT TTG ACA GTG AAA GGA TGA GCG ACA GCA AAG AGG TGC-3′ and 5′-TTT TTT CTC CCC TCA CCC TCT CCG TCC CTT TCT TAT CCG TTT TCC CTC TAT CAG GCG TAG TCG GGC ACG T-3′ for ΔM36::MC159; and 5′-GCT AGA GAA GTT CTA CGT CGA CGT CGG GCC CCT CGT CGA GTT CGC GTG ACA TGA GCG ACA GCA AAG AGG TGC-3′ and 5′-CAC TCG AGC GCC AGA GCA ATA GAA CTC GTT TTT TGG CGA CGA GTT CGC CGT CAG GCG TAG TCG GGC ACG T-3′ for ΔM45::MC159.
For genomic sequencing, 500 ng of BAC DNA was used for library preparation with the NEBNext Ultra DNA Library Prep kit for Illumina according to the manufacturer's recommendations. Diluted libraries were paired-end sequenced (2 × 250 cycles) with an Illumina MiSeq sequencer generating 1.7 to 2.3 million paired reads per sample. Sequences were assembled using SPAdes genome assembler v.3.6.0 (38) and an MCMV BAC reference genome as a guide.
Viruses were propagated and titrated on NIH 3T3 cells. For replication kinetic experiments, cells were infected in triplicate with the virus of interest. Four hours postinfection (hpi), cells were washed and fresh culture medium was added. Supernatants were collected at different time points, and virus titers were determined using the median tissue culture infective dose (TCID50) method.
NF-κB reporter assay.
pNiFty2-SEAP (InvivoGen) is an NF-κB-inducible reporter plasmid expressing secreted embryonic alkaline phosphatase (SEAP). Cells were cotransfected with pNiFty2-SEAP and plasmids encoding the proteins of interest. Thirty hours posttransfection, SEAP activity in supernatants was determined by using the Quanti-Blue detection reagent (InvivoGen) and a FLUOstar Omega (BMG Labtech) photometer. Significance was calculated using analysis of variance (ANOVA).
Antibodies and cytokines.
Mouse monoclonal antibodies against MCMV proteins IE1 (CROMA101), E1/M112-113 (CROMA103), and gB (SN1.01) were kindly provided by Stipan Jonjić (University of Rijeka, Rijeka, Croatia). Monoclonal antibodies against HA (16B12 from Covance or 3F10 from Roche), NEMO (EA2-6; MBL), NF-κB p65 (F-6; Santa Cruz), phospho-p38 (3D7; Cell Signaling), and RIP1 (clone 38; BD Transduction Laboratories) and polyclonal antibodies against HA (H6908; Sigma), IκBα (C-21; Santa Cruz), NEMO (FL-419; Santa Cruz), p38 (C-20; Santa Cruz), RIP1 (H-207; Santa Cruz), and β-actin (AC-74; Sigma) were purchased from suppliers as indicated. Secondary antibodies coupled to horseradish peroxidase (HRP) were purchased from DakoCytomation or Jackson ImmunoResearch. Murine TNF-α (mTNF-α) and interleukin-1β (IL-1β) were purchased from Promokine and Biomol, respectively. Human TNF-α (hTNF-α) was purchased from R&D Systems.
Immunodetection.
For immunoprecipitation, cells were lysed with lysis buffer containing 1% Nonidet P-40 and cOmplete Mini Protease Inhibitor Cocktail (Roche). After preclearing, proteins of interest were precipitated using specific antibodies and protein A-Sepharose (GE Healthcare) or protein A-agarose (Roche). Precipitates were washed six times, eluted by boiling in sample buffer, and subjected to SDS-PAGE and immunoblotting. For all other immunoblot analyses, cells were lysed in boiling SDS-PAGE sample buffer. Immunofluorescence staining and detection were done as described previously (33).
Cell viability assay.
Murine cells were grown in 96-well dishes and infected with MCMV at a multiplicity of infection (MOI) of 3 TCID50/cell. Cells were treated with 20 ng/ml mTNF-α, 0.5 μg/ml cycloheximide (CHX) (Sigma), 75 μM Z-IETD-FMK (BioVision), and 5 μM GSK'872 (BioCat). HT-29 cells were infected at an MOI of 5 TCID50/cell and treated with 30 ng/ml hTNF-α, 2 μg/ml CHX, 50 μM Z-VAD-FMK (R&D Systems), and 10 μM GSK'872. Cell viability was determined by measuring intracellular ATP levels using a Cell Titer-Glo Luminescent Cell Viability Assay kit (Promega) and a FLUOstar Omega (BMG Labtech) luminometer. Significance was calculated using ANOVA.
RESULTS
Characterization of MCMV MC159 substitution mutants.
To examine the functions of the MCV MC159 protein in the context of an MCMV infection and compare them with those of the MCMV M36 and M45 proteins, we constructed substitution mutants expressing MC159 instead of M36 or M45. The mutants were made by en passant mutagenesis of the wild-type (wt) MCMV strain Smith BAC and named ΔM36::MC159 and ΔM45::MC159, respectively (Fig. 1A). Correct insertion of MC159 was verified by restriction analysis and sequencing of the M36 and M45 loci (not shown). To exclude accidental mutations elsewhere in the viral genome, the recombinant BACs were completely sequenced. No unintended mutations were detected (not shown). For optimal expression of the MC159 protein, a codon-optimized MC159 gene sequence was used as it is generally recommended for poxvirus genes (39). Additionally, MC159 was tagged with an HA epitope as it has been done by others in previous studies (10, 40, 41).
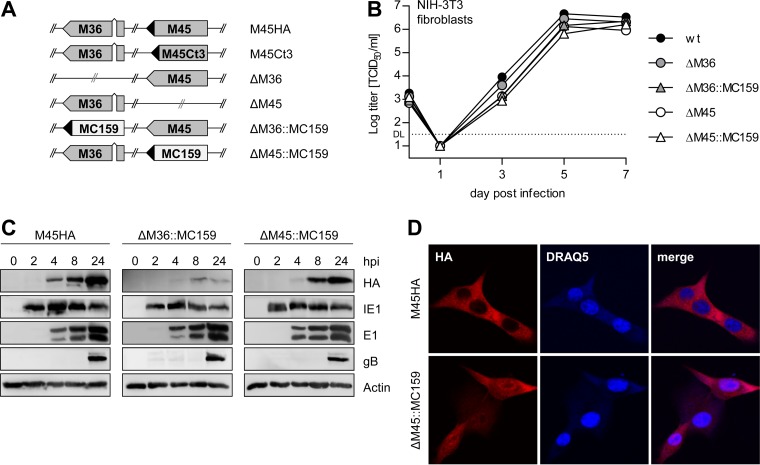
FIG 1.
Characterization of MCMV MC159 substitution mutants. (A) Schematic representation of the viruses used in this study. ◀, HA tag. (B) NIH 3T3 cells were infected with MCMV mutants at an MOI of 0.05 TCID50/cell. Supernatants were collected at the indicated days postinfection, and virus titers were determined. Means and standard errors of the means (±SEM) for experiments done in triplicate are shown. DL, detection limit. (C) NIH 3T3 cells were infected with MCMV M45HA, ΔM36::MC159, or ΔM45::MC159 at an MOI of 5 TCID50/cell. Cells were lysed at the indicated times postinfection, and viral protein expression was analyzed by immunoblotting. (D) NIH 3T3 cells were infected with an MOI of 2 TCID50/cell. Cells were fixed 6 hpi, and the subcellular localization of HA-tagged M45 and MC159 was analyzed by immunofluorescence. Nuclei were stained with DRAQ5.
The substitution mutants replicated to titers similar to those of wt MCMV and ΔM36 and ΔM45 deletion mutants in NIH 3T3 fibroblasts (Fig. 1B). They also expressed the viral immediate early protein IE1, the early protein E1, and the late glycoprotein B (gB) with the same kinetics as MCMV-M45HA, a virus with wt properties expressing an HA-tagged M45 (Fig. 1C). Both substitution mutants expressed HA-tagged MC159, but expression from the M36 locus was lower than from the M45 locus (Fig. 1C). Using immunofluorescence, the MC159 protein was detected in the cytoplasm and the nucleus of infected cells at 6 hpi, whereas the M45 protein was detected only in the cytoplasm (Fig. 1D).
MC159 promotes the activation of NF-κB.
We previously showed that virion-associated M45 induces a rapid and transient activation of NF-κB immediately after infection of fibroblasts, while de novo-synthesized M45 blocks all canonical NF-κB signaling pathways (32, 33). Similarly, M45 expressed transiently by plasmid transfection activates NF-κB (33), whereas prolonged expression (e.g., by retroviral transduction) blocks NF-κB activation (32). For MC159, it was also reported that it is able to both activate and inhibit NF-κB (16–19).
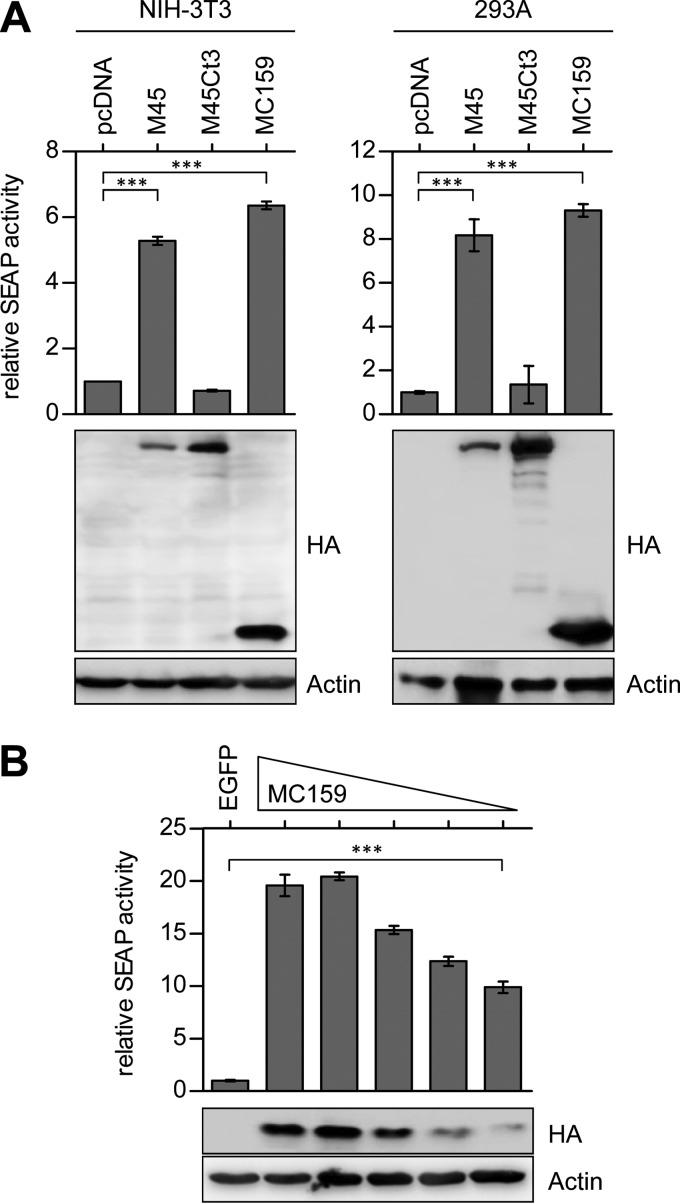
First, we wanted to compare the NF-κB-activating effects of MC159 and M45 using an NF-κB reporter assay. NIH 3T3 fibroblasts and HEK 293A cells were cotransfected with an NF-κB-dependent SEAP reporter plasmid and an MC159 or M45 expression plasmid. MC159 expression induced a significant activation of NF-κB in fibroblasts and HEK cells, similar to the activation induced by M45 expression. In contrast, expression of a C-terminally truncated and therefore inactive M45 did not activate NF-κB (Fig. 2A). As the cellular FLIP modulates NF-κB activity in a concentration-dependent manner (42), we examined whether the viral FLIP MC159 modulates NF-κB activity also in a concentration-dependent manner. We were able to show that the activation of NF-κB by MC159 dropped with decreasing amounts of MC159 (Fig. 2B).
FIG 2.
MC159 induces NF-κB-dependent reporter gene expression in transfected cells. (A) NIH 3T3 cells and 293A cells were cotransfected with expression plasmids as indicated and the reporter plasmid pNiFty-SEAP. Thirty hours posttransfection, Quanti-Blue reagent was added to the supernatant and SEAP activity was quantified colorimetrically. Results were normalized to empty vector (pcDNA). Cells were harvested and proteins analyzed by immunoblotting. (B) NIH 3T3 cells were cotransfected with decreasing amounts of MC159 and constant amounts of the reporter plasmid pNiFty-SEAP. Results were normalized to pEGFP. The experiment was carried out as described for panel A. Means ± SEM for experiments done in triplicate are shown. ***, P < 0.001.
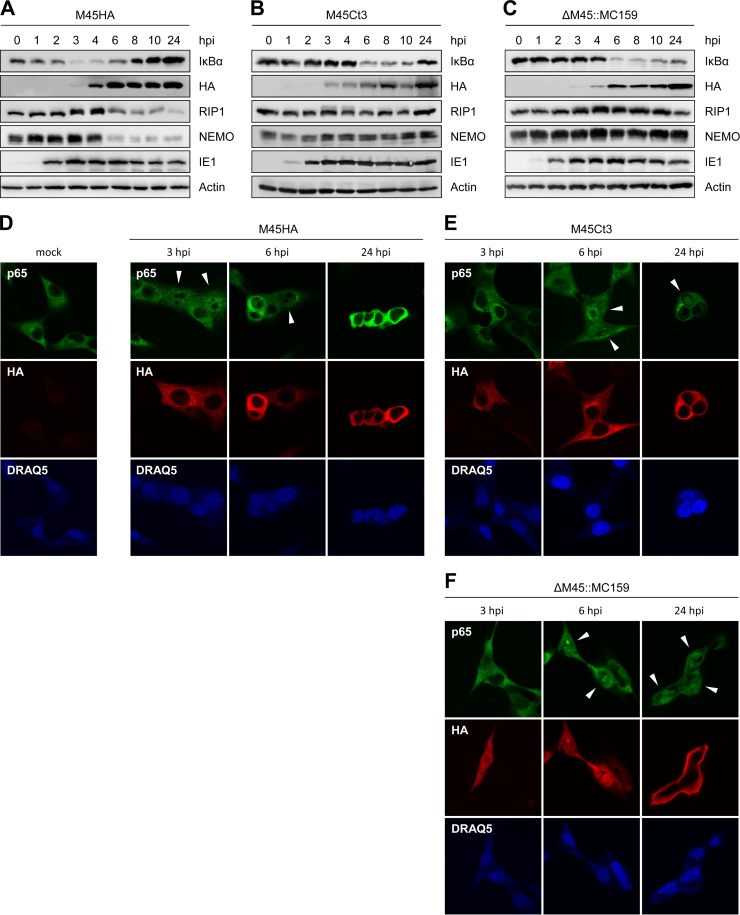
To examine the NF-κB modulating functions of MC159 in the context of an infection and compare them with those of M45, we infected fibroblasts with MCMVΔM45::MC159, MCMV-M45HA, or a virus expressing a C-terminally truncated M45 (MCMV-M45Ct3) (Fig. 1A). Degradation of the NF-κB inhibitor, IκBα, was determined at different times postinfection and served as a measure of NF-κB activation. Infection with MCMV-M45HA induced a decrease of IκBα levels within the first 6 hpi with a maximum reduction at 3 hpi (Fig. 3A). As shown previously, the transient NF-κB activation immediately after infection is mediated by virion-associated M45 protein (33). Recovery of IκBα coincided with an increased expression of M45 and a strong reduction of NEMO levels (Fig. 3A). In cells infected with an MCMV expressing a truncated M45 (Ct3), which neither activates nor inhibits NF-κB (32, 33), IκBα degradation was not induced within the first 4 hpi but was activated at later times as a consequence of viral infection (Fig. 3B). In these cells, a change in NEMO or RIP1 levels was not observed (Fig. 3B) and IκBα levels recovered by 24 hpi, consistent with previous observations (33). In cells infected with the ΔM45::MC159 virus, the kinetics of IκBα levels were similar to those in MCMV-M45Ct3-infected cells (Fig. 3C), suggesting that MC159 does not inhibit MCMV-induced NF-κB activation from approximately 6 hpi onward as M45 does. However, in cells infected with ΔM45::MC159, the IκBα level did not recover by 24 hpi but remained low. The virus did not induce IκBα degradation within the first 4 h, probably because MC159, unlike the viral tegument protein M45, is not packaged into viral particles.
FIG 3.
MCMV ΔM45::MC159 promotes degradation of IκBα and translocation of p65 into the nucleus. (A to C) NIH 3T3 cells were infected with MCMV M45HA, M45Ct3, or ΔM45::MC159 at an MOI of 5 TCID50/cell. Cells were lysed at the indicated times postinfection, and protein levels were analyzed by immunoblotting. (D to F) NIH 3T3 cells were infected with an MOI of 2 TCID50/cell. Cells were fixed at the indicated times postinfection, and the subcellular localization of the NF-κB subunit p65 was analyzed by immunofluorescence. Costaining with HA served as an infection control. Nuclei were stained with DRAQ5. Arrowheads indicate cells with nuclear p65.
In order to confirm that the observed degradation of IκBα actually led to the activation and translocation of NF-κB into the nucleus, we determined the subcellular localization of the NF-κB p65 subunit by immunofluorescence analysis. In MCMV-M45HA-infected cells, p65 was detected in the nucleus at 3 hpi but only in the cytoplasm at 24 hpi (Fig. 3D). In MCMV-M45Ct3- and ΔM45::MC159-infected cells, p65 appeared in the nucleus around 6 hpi and was still detected in the nucleus at 24 hpi (Fig. 3E and F). However, while p65 was found in only a small fraction of nuclei of cells infected with MCMV M45Ct3 by 24 hpi, p65 was present in a high percentage of nuclei of ΔM45::MC159-infected cells (not shown), confirming the previous time course experiment (Fig. 3B and C).
It is noteworthy that M45 and M45Ct3 were found exclusively in the cytoplasm of infected cells (Fig. 3D and E), whereas MC159 changed its localization during the course of infection: at early times, it was distributed throughout the cell, while it accumulated at the periphery at later time points postinfection (Fig. 3F). We observed the latter distribution only in a small fraction of cells transduced with an MC159-expressing retrovirus but in most cells transfected with an MC159-expressing plasmid (data not shown). These observations suggest that MC159 accumulates in the cell periphery when expressed at high levels.
MC159 inhibits TNF-α- but not IL-1β-induced NF-κB activation.
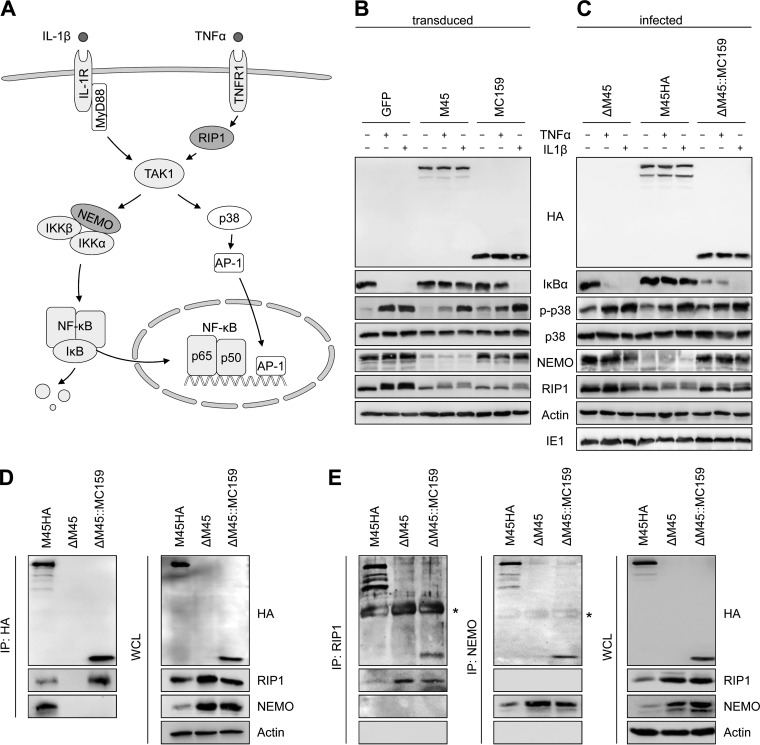
Previous studies have shown that both M45 and MC159 are capable of inhibiting NF-κB activation (16–18, 27, 32). M45 can also inhibit activation (phosphorylation) of p38 MAPK upon TNF-α but not IL-1β stimulation (27, 32). Therefore, we wanted to compare the inhibitory activity of these proteins. First, the two viral proteins were expressed in NIH 3T3 fibroblasts by retroviral transduction. Fibroblasts transduced with a green fluorescent protein (GFP)-expressing retrovirus were used as a control. The cells were stimulated with the cytokines TNF-α and IL-1β, respectively, to examine IκBα degradation as an indicator for NF-κB activation and p38 phosphorylation as a marker for activation of the p38-AP1 signaling pathway (Fig. 4A). In the GFP-expressing control cells, stimulation with TNF-α or IL-1β resulted in IκBα degradation and p38 phosphorylation (Fig. 4B). In M45-expressing fibroblasts, the degradation of IκBα upon TNF-α and IL-1β stimulation was blocked, and p38 phosphorylation upon TNF-α (but not IL-1β) treatment was inhibited. In MC159-expressing fibroblasts, TNF-α-induced IκBα degradation and p38 phosphorylation were blocked, but IL-1β-stimulated activation of these pathways was not inhibited (Fig. 4B).
FIG 4.
MCMV ΔM45::MC159 inhibits TNF-α- but not IL-1β-induced IκBα degradation. (A) Schematic representation of the IL-1 receptor (IL-1R)- and TNF receptor type 1 (TNFR1)-dependent NF-κB- and AP-1-activating pathways. (B and C) NIH 3T3 cells were transduced with retroviral vectors expressing GFP, M45, or MC159 (B) or infected with MCMV M45HA, ΔM45, or ΔM45::MC159 (C). Cells were treated with TNF-α (10 ng/ml) or IL-1β (20 ng/ml). Fifteen minutes after treatment, cells were lysed and protein levels were analyzed by immunoblotting. (D and E) NIH 3T3 cells were infected with MCMV M45HA, ΔM45, or ΔM45::MC159 at an MOI of 3 TCID50/cell. Cells were lysed 15 hpi and subjected to immunoprecipitation (IP) using anti-HA, anti-RIP1, or anti-NEMO antibodies, respectively. Whole-cell lysates (WCL) and immunoprecipitates were analyzed by immunoblotting. *, antibody heavy chain.
Next, we analyzed the same signaling pathways in MCMV-infected cells (Fig. 4C). The results were similar to those in retrovirus-transduced cells, with two notable exceptions: (i) MCMV infection induced a certain level of p38 phosphorylation, which can be further increased by cytokine stimulation; and (ii) IκBα levels were relatively low in ΔM45::MC159-infected cells, even in the absence of cytokine stimulation (Fig. 4C).
The observation that MC159 does not inhibit IκBα degradation upon IL-1β stimulation was somewhat surprising, as MC159 was reported to inhibit the IKK complex by interacting with IKKγ/NEMO (18). Our results would be more consistent with an inhibition at the level of RIP1. Therefore, we tested whether MC159 associates with RIP1 and/or NEMO in NIH 3T3 fibroblasts infected with MCMV ΔM45::MC159. Immunoprecipitation of HA-tagged MC159 resulted in coprecipitation of RIP1 but not NEMO, whereas immunoprecipitation of HA-tagged M45 resulted in coprecipitation of both RIP1 and NEMO (Fig. 4D). However, when RIP1 or NEMO were immunoprecipitated, M45 and MC159 were detected as coprecipitating proteins (Fig. 4E). Taken together, the results of the immunoprecipitation experiments confirm the previously reported interaction with RIP1 (13, 16). However, we could not find convincing evidence for a functional interaction of MC159 with NEMO in MCMV-infected fibroblasts, as NEMO did not coprecipitate with MC159 (Fig. 4D) and IL-1β-induced IκBα degradation was not inhibited (Fig. 4B and C).
MC159 inhibits apoptosis but does not block necroptosis in MCMV-infected mouse cells.
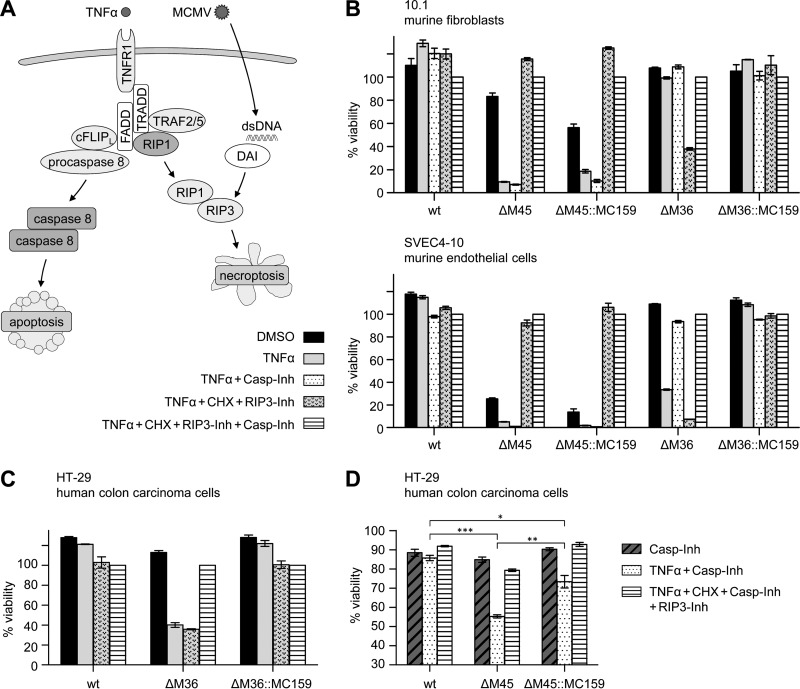
MC159/vFLIP is known to inhibit caspase-8-mediated apoptosis (5, 6, 8, 9). Interestingly, this protein was also reported to inhibit necroptosis in human T cells by an unknown mechanism (13, 14). MCMV, in contrast, inhibits caspase-8-dependent apoptosis and RIP3-dependent necroptosis with two separate viral proteins, M36 and M45, respectively (23, 43). For a functional comparison of the MCMV proteins with MC159, we infected cells with the MC159-expressing MCMV substitution mutants ΔM36::MC159 and ΔM45::MC159, respectively. In the absence of the necroptosis inhibitor M45, MCMV infection by itself (i.e., without additional induction) can trigger necroptosis. As the extent of virus infection-induced necroptosis depends on the cell type, we used 10.1 fibroblasts, which are only moderately sensitive to MCMV-induced necroptosis, and SVEC4-10 endothelial cells, which are highly sensitive. Since TNF-α can trigger both apoptosis and necroptosis (Fig. 5A), TNF-α was combined with inhibitors to discriminate between the different forms of programmed cell death. Infected cells were treated either with TNF-α plus caspase-8 inhibitor Z-IETD-FMK (for the specific induction of necroptosis and inhibition of apoptosis) or with TNF-α plus CHX plus RIP3 kinase inhibitor GSK'872 (for specific induction of apoptosis and inhibition of necroptosis). Cell viability was determined by using an ATP-dependent luminescence assay. Consistent with previous results (26, 29), ΔM45-infected SVEC4-10 cells underwent rapid cell death even without TNF-α treatment. Cell death induced by ΔM45 infection was less pronounced in 10.1 fibroblasts but was strongly enhanced by treatment with TNF-α (Fig. 5B). As expected, ΔM45-infected cells were sensitive to necroptosis but not apoptosis induction. In fact, cell death was completely inhibited by the GSK'872 necroptosis inhibitor (Fig. 5B). Cells infected with the ΔM45::MC159 substitution mutant showed essentially the same phenotype as ΔM45-infected cells, indicating that MC159 does not inhibit programmed necrosis in this setting, neither MCMV infection-induced nor TNF-α-induced necroptosis.
FIG 5.
MC159 expressed by MCMV blocks TNF-α-induced apoptosis in both murine and human cells but inhibits necroptosis only in human cells. (A) Schematic representation of programmed cell death signaling pathways. (B) Murine 10.1 and SVEC4-10 cells were infected with wt and mutant MCMVs and treated 6 hpi with TNF-α, CHX, caspase-8 inhibitor Z-IETD-FMK (Casp-Inh), and RIP3 inhibitor GSK'872 (RIP3-Inh). Cell viability was measured 15 h posttreatment relative to cells treated with both inhibitors. (C) Human HT-29 cells were infected with wt and mutant MCMVs and treated 10 hpi as described above with the exception that Z-VAD-FMK was used as caspase inhibitor. Cell viability was measured 18 h posttreatment. (D) HT-29 cells were infected with wt and mutant MCMVs and treated with Z-VAD-FMK (Casp-Inh), GSK'872 (RIP3-Inh), and TNF-α. Cell viability was measured 20 h posttreatment relative to uninfected cells treated with Z-VAD-FMK. Means ± SEM for experiments done in triplicate are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
As expected, 10.1 and SVEC4-10 cells infected with ΔM36 were sensitive to TNF-α-induced apoptosis. Interestingly, TNF-α treatment alone was not sufficient for apoptosis induction in 10.1 cells but required enhancement by CHX. In contrast, cells infected with the ΔM36::MC159 virus were resistant to apoptosis induction, as were cells infected with wt MCMV (Fig. 5B). This indicates that MC159 inhibits apoptosis induction by death receptor stimulation and compensates for the loss of the caspase-8 inhibitory protein M36.
The inability of MC159 to inhibit necroptosis in MCMV-infected mouse cells (Fig. 5B) was surprising. However, it remained possible that MC159 could inhibit necroptosis in human cells as previously published (13, 14). Although human cells are not permissive for MCMV replication, MCMV-infected human cells usually express viral immediate early and early proteins (44). We decided to use human HT-29 colon carcinoma cells, as these cells have been used previously for analysis of viral necroptosis inhibitors (45–47). First, we infected HT-29 cells with MCMV and analyzed the expression of the immediate early protein IE1 and the early protein M45 at different times postinfection by immunoblotting. The expression kinetics of these two proteins was very similar to the one in NIH 3T3 cells shown in Fig. 1C, indicating that HT-29 cells are infectible and express viral immediate early and early proteins (data not shown). Then, we analyzed the sensitivity of HT-29 cells infected with wt or mutant MCMVs to TNF-α-induced cell death. HT-29 cells infected with MCMVΔM36 were sensitive to TNF-α-induced apoptosis, but cells infected with wt MCMV or the ΔM36::MC159 mutant were resistant (Fig. 5C). Hence, M36 and MC159 are capable of inhibiting death receptor-dependent apoptosis in human and in murine cells. Finally, we infected HT-29 cells with MCMV M45 deletion and substitution mutants to test whether M45 and MC159 inhibit TNF-α-induced necroptosis. As shown in Fig. 5D, HT-29 cells infected with wt MCMV were resistant to necroptosis induction, but cells infected with the ΔM45 mutant were sensitive. HT-29 cells infected with the ΔM45::MC159 substitution mutant were significantly less sensitive to TNF-α-induced necroptosis, although necroptosis inhibition by MC159 was not as robust as by M45.
MC159 partially rescues the replication defect of an MCMV ΔM36 mutant.
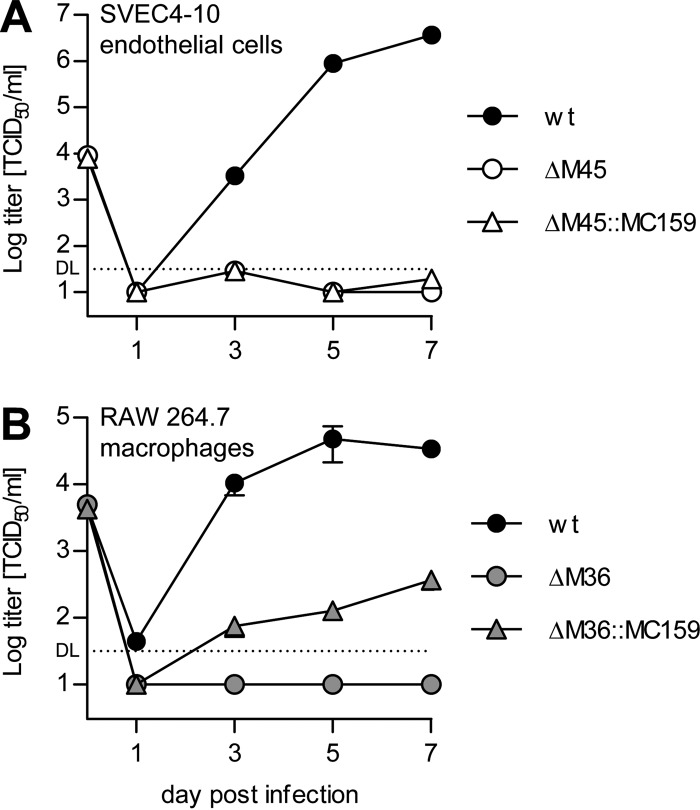
While MCMV M36 and M45 deletion mutants have little or no growth defect in NIH 3T3 fibroblasts (Fig. 1B), it is well established that M45-deficient MCMV mutants do not replicate in SVEC4-10 endothelial cells due to necrosis induction (26, 29). It has also been shown that M36-deficient mutants have a severe replication defect in different macrophage lines (21, 24). Therefore, we tested, using our substitution mutants, whether MC159 could rescue these replication defects. As shown in Fig. 6A, the ΔM45::MC159 virus did not replicate in SVEC4-10 cells. This result was to be expected, since MC159 was not able to compensate for the necroptosis-inhibiting function of M45 in murine cells (Fig. 5B). In contrast, the ΔM36::MC159 substitution mutant did replicate in RAW 264.7 macrophages, although to approximately 100-fold-lower titers than the wt virus (Fig. 6B), indicating that the loss of M36 was only partially compensated by MC159.
FIG 6.
MCMV ΔM45::MC159 does not replicate in endothelial cells, but ΔM36::MC159 replicates in macrophages. SVEC4-10 cells (A) or RAW 264.7 macrophages (B) were infected at an MOI of 0.5 TCID50/cell with the viruses indicated. Supernatants were collected at the indicated days postinfection, and virus titers were determined. Means ± SEM for experiments done in triplicate are shown. DL, detection limit.
DISCUSSION
To date, there is no cell culture or laboratory animal model system to study MCV replication and pathogenesis. Due to this technical barrier, researchers have studied MCV genes either by expressing genes independent of infection (e.g., by plasmid transfection) or by expression from a surrogate virus. In most of the studies published so far, the MCV MC159 gene has been analyzed in cells overexpressing MC159 upon transient transfection with an expression plasmid. Only in a few studies has MC159 been expressed by a surrogate virus, a recombinant VACV with its caspase-8 inhibitor, B13R, deleted (10, 13, 18, 48), and two other studies have examined MC159 functions in transgenic mice expressing the viral protein (19, 49).
In the present study, we used MCMV as a surrogate virus for the analysis of MC159 functions. MCMV has several properties that qualify it as a suitable surrogate virus: (i) both MCV and MCMV are large DNA viruses causing persistent infections, (ii) both viruses express numerous genes involved in viral immune evasion, and (iii) MCMV carries two well-characterized genes, M36 and M45, with functions very similar to those described for MC159, which allows a functional comparison. On the other hand, MCMV replicates its DNA and transcribes its genes in the nucleus, not in the cytoplasm as MCV does, and MCMV is a mouse virus, whereas MCV is a human virus. However, MC159 functions have previously been characterized in transfected mouse cells (18, 50) or in transgenic mice (19, 49), suggesting that MC159 functions are not generally restricted to human cells. Nevertheless, one could argue that VACV is more closely related to MCV and thus should be a better surrogate virus. On the other hand, the fact that VACV carries a substantial number of genes with functions overlapping those of MC159 complicates the interpretation of results (4).
The best-studied function of MC159 is its inhibition of death receptor-dependent apoptosis. We showed here that MC159 blocked apoptosis of MCMV-infected cells upon TNF receptor stimulation and compensated for the lack of the MCMV caspase-8 inhibitor, M36 (Fig. 5B and C). However, the replication defect of an MCMV ΔM36 mutant in RAW 264.7 macrophages was only partially rescued (Fig. 6B). Although both M36 and MC159 have been reported to interact with procaspase 8 (10, 21), differences in the inhibitory mechanisms might be responsible for the partial rescue. While M36 is thought to inhibit caspase-8 activation by binding to procaspase 8 (20, 21), MC159 is thought to exert its inhibitory effect primarily by binding to FADD and preventing recruitment of procaspase 8 to the death-inducing signaling complex (DISC) (51) or oligomerization of the DISC (52). Remarkably, a previous study has shown that the loss of M36 is almost completely rescued by dominant negative FADD, which was overexpressed by using a strong heterologous promoter (24). Our results suggest that the inhibitory activity of MC159 or its expression level is insufficient for a complete reversal of the ΔM36 phenotype in infected macrophages. It is also conceivable that M36 has an additional function (besides inhibiting death receptor-dependent apoptosis) not shared by MC159. This hypothesis, however, seems unlikely, as dominant negative FADD rescued the replication defect in macrophages (24).
MC159 has also been reported to inhibit TNF-α- or Fas-induced programmed necrosis in human Jurkat T cells (13, 14). As TNF-α-induced necroptosis requires RIP1 and RIP3, inhibition by MC159 might occur as a consequence of its interaction with RIP1. We found that MC159 expressed from a recombinant MCMV associated with RIP1 in infected mouse fibroblasts (Fig. 4D and E), but it did not inhibit necroptosis induced by MCMV infection or TNF-α treatment (Fig. 5B), nor did it rescue the replication defect of a MCMV ΔM45 mutant in SVEC4-10 endothelial cells (Fig. 6A). However, MC159 did show a limited inhibitory effect against TNF-α-induced necroptosis in MCMV-infected human HT-29 cells (Fig. 5D). A possible explanation for this discrepancy is that the association of MC159 with RIP1 is sufficient for inhibition of TNF-α-induced necroptosis in human but not in mouse cells. Interestingly, a species-specific necroptosis inhibition has been reported for the herpes simplex virus 1 (HSV-1) ICP6 and the HSV-2 ICP10 proteins: ICP6 and ICP10 inhibit necroptosis in human but not in mouse cells (45, 46, 53). ICP6, ICP10, and M45 are homologous proteins, and all three possess a RHIM (54), which is required for the inhibition of RIP3-dependent necroptosis (29, 45, 46). As MC159 does not appear to have a RHIM, its necroptosis-inhibiting activity is likely to operate through a different mechanism.
Using MCMV as a surrogate virus, we confirmed that MC159 can both activate and inhibit NF-κB, consistent with previous reports (16–19). Although this dual function appears to be similar to the one described for M45 (33), three differences became apparent in our experiments. (i) M45 activates NF-κB rapidly and transiently during the first 6 hpi (Fig. 3A), a function that depends on M45 delivered by the virion (33). In contrast, NF-κB activation in cells infected with ΔM45::MC159 started only around 6 hpi, a time when MC159 was expressed from the viral genome. The most likely explanation for this difference is a lack of MC159 protein in viral particles. (ii) In cells infected with ΔM45::MC159, NF-κB remained activated from 6 hpi onward, whereas M45 expression caused a strong block to NF-κB activation starting 6 hpi. This block is the result of the M45-mediated degradation of NEMO/IKKγ (32). In contrast, MC159 does not induce NEMO degradation (Fig. 3C). (iii) By targeting NEMO for degradation, M45 inhibits all canonical NF-κB activating pathways, including those triggered by TNF-α or IL-1β. In contrast, MC159 inhibited NF-κB activation by TNF-α but not IL-1β (Fig. 4B and C). As MC159 interacts with FADD, RIP1, and TRAF2 (6, 8–10, 16), the most likely explanation is an inhibition at the level or upstream of RIP1 (17). However, an interaction of MC159 with NEMO/IKKγ and an inhibition of the IKK complex have been reported in a previous study (18). In our experiments using the ΔM45::MC159 substitution mutant, we were able to coimmunoprecipitate MC159 with NEMO (Fig. 4E), but NEMO did not coimmunoprecipitate with MC159 (Fig. 4D), suggesting that the interaction might be weak or indirect. In a paper by Randall et al. (18), the association of MC159 with IKKγ was also shown in only one direction. Whether or not the presumed MC159-NEMO interaction results in an inhibition of the IKK complex might depend on the cell type and/or the level of MC159 expression.
In this study, we used MCMV as a surrogate virus for functional analysis of an MCV protein, which has previously been studied predominantly in transfection experiments. Clearly, transfection experiments and surrogate virus infection experiments each have their advantages and disadvantages. For instance, transfection experiments result in an overexpression of the protein of interest, which can lead to exaggerated effects. On the other hand, expression by a surrogate virus might mask real functions of the protein of interest due to the expression of other viral proteins with overlapping or interfering functions. Moreover, the use of different cell types might also account for seemingly conflicting results of different studies. Until a viable cell culture system for MCV becomes available, the use of human keratinocytes, the natural target cells of MCV, and comparison of results from different test systems might be necessary to extrapolate the biological relevance of MCV gene functions.
ACKNOWLEDGMENTS
We thank Daniela Indenbirken and Michael Spohn (HPI Next Generation Sequencing technology platform) for BAC sequencing and Ana Cáceres-Núnez for critical readings of the manuscript.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Birthistle K, Carrington D. 1997. Molluscum contagiosum virus. J Infect 34:21–28. doi: 10.1016/S0163-4453(97)80005-9. [DOI] [PubMed] [Google Scholar]
- 2.Konya J, Thompson CH. 1999. Molluscum contagiosum virus: antibody responses in persons with clinical lesions and seroepidemiology in a representative Australian population. J Infect Dis 179:701–704. doi: 10.1086/314620. [DOI] [PubMed] [Google Scholar]
- 3.Sherwani S, Farleigh L, Agarwal N, Loveless S, Robertson N, Hadaschik E, Schnitzler P, Bugert JJ. 2014. Seroprevalence of Molluscum contagiosum virus in German and UK populations. PLoS One 9:e88734. doi: 10.1371/journal.pone.0088734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shisler JL. 2015. Immune evasion strategies of molluscum contagiosum virus. Adv Virus Res 92:201–252. doi: 10.1016/bs.aivir.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Moss B, Shisler JL, Xiang Y, Senkevich TG. 2000. Immune-defense molecules of molluscum contagiosum virus, a human poxvirus. Trends Microbiol 8:473–477. doi: 10.1016/S0966-842X(00)01838-2. [DOI] [PubMed] [Google Scholar]
- 6.Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer JL, Schroter M, Scaffidi C, Krammer PH, Peter ME, Tschopp J. 1997. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature 386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 7.Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, Steiner V, Bodmer JL, Schroter M, Burns K, Mattmann C, Rimoldi D, French LE, Tschopp J. 1997. Inhibition of death receptor signals by cellular FLIP. Nature 388:190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 8.Bertin J, Armstrong RC, Ottilie S, Martin DA, Wang Y, Banks S, Wang GH, Senkevich TG, Alnemri ES, Moss B, Lenardo MJ, Tomaselli KJ, Cohen JI. 1997. Death effector domain-containing herpesvirus and poxvirus proteins inhibit both Fas- and TNFR1-induced apoptosis. Proc Natl Acad Sci U S A 94:1172–1176. doi: 10.1073/pnas.94.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu S, Vincenz C, Buller M, Dixit VM. 1997. A novel family of viral death effector domain-containing molecules that inhibit both CD-95- and tumor necrosis factor receptor-1-induced apoptosis. J Biol Chem 272:9621–9624. doi: 10.1074/jbc.272.15.9621. [DOI] [PubMed] [Google Scholar]
- 10.Shisler JL, Moss B. 2001. Molluscum contagiosum virus inhibitors of apoptosis: the MC159 v-FLIP protein blocks Fas-induced activation of procaspases and degradation of the related MC160 protein. Virology 282:14–25. doi: 10.1006/viro.2001.0834. [DOI] [PubMed] [Google Scholar]
- 11.Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. 2010. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol 11:700–714. doi: 10.1038/nrm2970. [DOI] [PubMed] [Google Scholar]
- 12.Oberst A, Dillon CP, Weinlich R, McCormick LL, Fitzgerald P, Pop C, Hakem R, Salvesen GS, Green DR. 2011. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature 471:363–367. doi: 10.1038/nature09852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan FK, Shisler J, Bixby JG, Felices M, Zheng L, Appel M, Orenstein J, Moss B, Lenardo MJ. 2003. A role for tumor necrosis factor receptor-2 and receptor-interacting protein in programmed necrosis and antiviral responses. J Biol Chem 278:51613–51621. doi: 10.1074/jbc.M305633200. [DOI] [PubMed] [Google Scholar]
- 14.Thurau M, Everett H, Tapernoux M, Tschopp J, Thome M. 2006. The TRAF3-binding site of human molluscipox virus FLIP molecule MC159 is critical for its capacity to inhibit Fas-induced apoptosis. Cell Death Differ 13:1577–1585. doi: 10.1038/sj.cdd.4401847. [DOI] [PubMed] [Google Scholar]
- 15.Pahl HL. 1999. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene 18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 16.Chaudhary PM, Jasmin A, Eby MT, Hood L. 1999. Modulation of the NF-kappa B pathway by virally encoded death effector domains-containing proteins. Oncogene 18:5738–5746. doi: 10.1038/sj.onc.1202976. [DOI] [PubMed] [Google Scholar]
- 17.Murao LE, Shisler JL. 2005. The MCV MC159 protein inhibits late, but not early, events of TNF-alpha-induced NF-kappaB activation. Virology 340:255–264. doi: 10.1016/j.virol.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 18.Randall CM, Jokela JA, Shisler JL. 2012. The MC159 protein from the molluscum contagiosum poxvirus inhibits NF-kappaB activation by interacting with the IkappaB kinase complex. J Immunol 188:2371–2379. doi: 10.4049/jimmunol.1100136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Challa S, Woelfel M, Guildford M, Moquin D, Chan FK. 2010. Viral cell death inhibitor MC159 enhances innate immunity against vaccinia virus infection. J Virol 84:10467–10476. doi: 10.1128/JVI.00983-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skaletskaya A, Bartle LM, Chittenden T, McCormick AL, Mocarski ES, Goldmacher VS. 2001. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc Natl Acad Sci U S A 98:7829–7834. doi: 10.1073/pnas.141108798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menard C, Wagner M, Ruzsics Z, Holak K, Brune W, Campbell AE, Koszinowski UH. 2003. Role of murine cytomegalovirus US22 gene family members in replication in macrophages. J Virol 77:5557–5570. doi: 10.1128/JVI.77.10.5557-5570.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCormick AL, Skaletskaya A, Barry PA, Mocarski ES, Goldmacher VS. 2003. Differential function and expression of the viral inhibitor of caspase 8-induced apoptosis (vICA) and the viral mitochondria-localized inhibitor of apoptosis (vMIA) cell death suppressors conserved in primate and rodent cytomegaloviruses. Virology 316:221–233. doi: 10.1016/j.virol.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Handke W, Krause E, Brune W. 2012. Live or let die: manipulation of cellular suicide programs by murine cytomegalovirus. Med Microbiol Immunol 201:475–486. doi: 10.1007/s00430-012-0264-z. [DOI] [PubMed] [Google Scholar]
- 24.Cicin-Sain L, Ruzsics Z, Podlech J, Bubic I, Menard C, Jonjic S, Reddehase MJ, Koszinowski UH. 2008. Dominant-negative FADD rescues the in vivo fitness of a cytomegalovirus lacking an antiapoptotic viral gene. J Virol 82:2056–2064. doi: 10.1128/JVI.01803-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ebermann L, Ruzsics Z, Guzman CA, van Rooijen N, Casalegno-Garduno R, Koszinowski U, Cicin-Sain L. 2012. Block of death-receptor apoptosis protects mouse cytomegalovirus from macrophages and is a determinant of virulence in immunodeficient hosts. PLoS Pathog 8:e1003062. doi: 10.1371/journal.ppat.1003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brune W, Ménard C, Heesemann J, Koszinowski UH. 2001. A ribonucleotide reductase homolog of cytomegalovirus and endothelial cell tropism. Science 291:303–305. doi: 10.1126/science.291.5502.303. [DOI] [PubMed] [Google Scholar]
- 27.Mack C, Sickmann A, Lembo D, Brune W. 2008. Inhibition of proinflammatory and innate immune signaling pathways by a cytomegalovirus RIP1-interacting protein. Proc Natl Acad Sci U S A 105:3094–3099. doi: 10.1073/pnas.0800168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Upton JW, Kaiser WJ, Mocarski ES. 2008. Cytomegalovirus M45 cell death suppression requires receptor-interacting protein (RIP) homotypic interaction motif (RHIM)-dependent interaction with RIP1. J Biol Chem 283:16966–16970. doi: 10.1074/jbc.C800051200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Upton JW, Kaiser WJ, Mocarski ES. 2010. Virus inhibition of RIP3-dependent necrosis. Cell Host Microbe 7:302–313. doi: 10.1016/j.chom.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Upton JW, Kaiser WJ, Mocarski ES. 2012. DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA. Cell Host Microbe 11:290–297. doi: 10.1016/j.chom.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lembo D, Donalisio M, Hofer A, Cornaglia M, Brune W, Koszinowski U, Thelander L, Landolfo S. 2004. The ribonucleotide reductase R1 homolog of murine cytomegalovirus is not a functional enzyme subunit but is required for pathogenesis. J Virol 78:4278–4288. doi: 10.1128/JVI.78.8.4278-4288.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fliss PM, Jowers TP, Brinkmann MM, Holstermann B, Mack C, Dickinson P, Hohenberg H, Ghazal P, Brune W. 2012. Viral mediated redirection of NEMO/IKKgamma to autophagosomes curtails the inflammatory cascade. PLoS Pathog 8:e1002517. doi: 10.1371/journal.ppat.1002517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krause E, de Graaf M, Fliss PM, Dölken L, Brune W. 2014. Murine cytomegalovirus virion-associated protein M45 mediates rapid NF-kappaB activation after infection. J Virol 88:9963–9975. doi: 10.1128/JVI.00684-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swift S, Lorens J, Achacoso P, Nolan GP. 2001. Rapid production of retroviruses for efficient gene delivery to mammalian cells using 293T cell-based systems. Curr Protoc Immunol Chapter 10:Unit 10.17C. doi: 10.1002/0471142735.im1017cs31. [DOI] [PubMed] [Google Scholar]
- 35.Harvey DM, Levine AJ. 1991. p53 alteration is a common event in the spontaneous immortalization of primary BALB/c murine embryo fibroblasts. Genes Dev 5:2375–2385. doi: 10.1101/gad.5.12b.2375. [DOI] [PubMed] [Google Scholar]
- 36.Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless red recombination system. Methods Mol Biol 634:421–430. doi: 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- 37.Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 38.Nurk S, Bankevich A, Antipov D, Gurevich A, Korobeynikov A, Lapidus A, Prjibelsky A, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, McLean J, Lasken R, Clingenpeel S, Woyke T, Tesler G, Alekseyev M, Pevzner P. 2013. Assembling genomes and mini-metagenomes from highly chimeric reads, p 158–170. In Deng M, Jiang R, Sun F, Zhang X (ed), Research in computational molecular biology, vol 7821 Springer, Berlin, Germany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barrett JW, Sun Y, Nazarian SH, Belsito TA, Brunetti CR, McFadden G. 2006. Optimization of codon usage of poxvirus genes allows for improved transient expression in mammalian cells. Virus Genes 33:15–26. doi: 10.1007/s11262-005-0035-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garvey TL, Bertin J, Siegel RM, Wang GH, Lenardo MJ, Cohen JI. 2002. Binding of FADD and caspase-8 to molluscum contagiosum virus MC159 v-FLIP is not sufficient for its antiapoptotic function. J Virol 76:697–706. doi: 10.1128/JVI.76.2.697-706.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siegel RM, Martin DA, Zheng L, Ng SY, Bertin J, Cohen J, Lenardo MJ. 1998. Death-effector filaments: novel cytoplasmic structures that recruit caspases and trigger apoptosis. J Cell Biol 141:1243–1253. doi: 10.1083/jcb.141.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kataoka T, Budd RC, Holler N, Thome M, Martinon F, Irmler M, Burns K, Hahne M, Kennedy N, Kovacsovics M, Tschopp J. 2000. The caspase-8 inhibitor FLIP promotes activation of NF-kappaB and Erk signaling pathways. Current Biol 10:640–648. doi: 10.1016/S0960-9822(00)00512-1. [DOI] [PubMed] [Google Scholar]
- 43.McCormick AL, Mocarski ES. 2013. Cell death pathways controlled by cytomegaloviruses, p 264–277. In Reddehase MJ. (ed), Cytomegaloviruses: from molecular pathogenesis to intervention, vol 1 Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 44.Brune W. 2013. Molecular basis of cytomegalovirus host species specificity, p 322–329. In Reddehase MJ. (ed), Cytomegaloviruses: from molecular pathogenesis to intervention, vol 1 Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 45.Guo H, Omoto S, Harris PA, Finger JN, Bertin J, Gough PJ, Kaiser WJ, Mocarski ES. 2015. Herpes simplex virus suppresses necroptosis in human cells. Cell Host Microbe 17:243–251. doi: 10.1016/j.chom.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang Z, Wu SQ, Liang Y, Zhou X, Chen W, Li L, Wu J, Zhuang Q, Chen C, Li J, Zhong CQ, Xia W, Zhou R, Zheng C, Han J. 2015. RIP1/RIP3 binding to HSV-1 ICP6 initiates necroptosis to restrict virus propagation in mice. Cell Host Microbe 17:229–242. doi: 10.1016/j.chom.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 47.Omoto S, Guo H, Talekar GR, Roback L, Kaiser WJ, Mocarski ES. 2015. Suppression of RIP3-dependent necroptosis by human cytomegalovirus. J Biol Chem 290:11635–11648. doi: 10.1074/jbc.M115.646042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gil J, Rullas J, Alcami J, Esteban M. 2001. MC159L protein from the poxvirus molluscum contagiosum virus inhibits NF-kappaB activation and apoptosis induced by PKR. J Gen Virol 82:3027–3034. doi: 10.1099/0022-1317-82-12-3027. [DOI] [PubMed] [Google Scholar]
- 49.Woelfel M, Bixby J, Brehm MA, Chan FK. 2006. Transgenic expression of the viral FLIP MC159 causes lpr/gld-like lymphoproliferation and autoimmunity. J Immunol 177:3814–3820. doi: 10.4049/jimmunol.177.6.3814. [DOI] [PubMed] [Google Scholar]
- 50.Randall CM, Biswas S, Selen CV, Shisler JL. 2014. Inhibition of interferon gene activation by death-effector domain-containing proteins from the molluscum contagiosum virus. Proc Natl Acad Sci U S A 111:E265–272. doi: 10.1073/pnas.1314569111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li FY, Jeffrey PD, Yu JW, Shi Y. 2006. Crystal structure of a viral FLIP: insights into FLIP-mediated inhibition of death receptor signaling. J Biol Chem 281:2960–2968. [DOI] [PubMed] [Google Scholar]
- 52.Yang JK, Wang L, Zheng L, Wan F, Ahmed M, Lenardo MJ, Wu H. 2005. Crystal structure of MC159 reveals molecular mechanism of DISC assembly and FLIP inhibition. Mol Cell 20:939–949. doi: 10.1016/j.molcel.2005.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X, Li Y, Liu S, Yu X, Li L, Shi C, He W, Li J, Xu L, Hu Z, Yu L, Yang Z, Chen Q, Ge L, Zhang Z, Zhou B, Jiang X, Chen S, He S. 2014. Direct activation of RIP3/MLKL-dependent necrosis by herpes simplex virus 1 (HSV-1) protein ICP6 triggers host antiviral defense. Proc Natl Acad Sci U S A 111:15438–15443. doi: 10.1073/pnas.1412767111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lembo D, Brune W. 2009. Tinkering with a viral ribonucleotide reductase. Trends Biochem Sci 34:25–32. doi: 10.1016/j.tibs.2008.09.008. [DOI] [PubMed] [Google Scholar]