Abstract
Functional genomics approaches that leverage the RNA interference (RNAi) pathway have been applied in vivo to examine the roles of hundreds or thousands of genes, mainly in the context of cancer. Here we discuss principles guiding the design of RNAi screens, parameters that determine success and recent developments that have improved accuracy and expanded the applicability of these approaches to other in vivo settings, including the immune system. We review recent studies that have applied in vivo RNAi screens to examine the networks of factors that drive the development and function of the immune system, and in this context, we put forward an argument as to why RNAi approaches in vivo are likely to provide particularly salient insight into immunology.
Introduction
Most factors that regulate immune cell development, activation and function are still uncharacterized. The first step in a widely used approach has been to identify genes controlling these processes based on differential gene expression analyses. Based on the results, hypotheses about these factors are generated and tested in genetic experiments, generally involving application of gene-disrupted mice, examining one factor at a time. From this perspective, it is easy to see that much of our knowledge has coalesced around relatively few factors that are expressed in a cell-type specific fashion, which might bias understanding toward the concept of “master regulators” [1]. However, the gene expression programs that potentiate cell development are governed by a much more complicated milieu of factors that function cooperatively, many of which are expressed in multiple cell types, and might differ in expression at the RNA level so minimally that they are not readily discerned using basic genome-wide differential expression profiling, or they undergo posttranscriptional, translational or posttranslational modifications that alter their activity [2–5]. To comprehensively identify these factors and clarify their roles, functional approaches are necessary.
The discovery RNA interference (RNAi) pathway, followed closely by the development of approaches that co-opted the endogenous RNAi machinery to conduct gene-specific loss-of-function experiments have ushered in a new era of functional screening [6, 7]. Despite the initial promise that early RNAi tools and screening approaches provided, the burden of caveats and complications inherent to conducting large-scale screens using RNAi became more apparent, and these realities seemed to curtail excitement, or at least raised skepticism, for using these new and otherwise powerful tools [8, 9]. However, in the last five years substantial improvements in tools for inducing prolonged (i.e., stable) RNAi combined with high-throughput approaches to quantify and analyze DNA sequences using next generation sequencing has reinvigorated the field and prompted a shift from conducting screens by assaying thousands of individual genes in series, to pooled screens in which thousands of genes are interrogated in parallel in single tubes [6, 7]. Most recently, these tools have been adapted to studying the immune response to tumors and viral infections by conducting pooled RNAi screens in T cells in mice [10, 11]. These studies suggest the field is poised to make larger strides toward more comprehensive identification of factors that control the immune system.
Here we review RNAi approaches, tools and critical parameters that have contributed to the success of multiple pooled RNAi screens in mice. We discuss basic principles that set the framework for using RNAi for functional screens, including recent developments that have improved accuracy and expanded the applicability of these approaches to in vivo settings. In this context, we put forward an argument as to why RNAi approaches in vivo are likely to provide particularly salient insight into immunology.
Learning to co-opt the endogenous RNAi machinery for functional screens
The canonical endogenous RNAi pathway (reviewed in [12]) derives from genes that are transcribed by RNA Pol II into pri-microRNAs (primary transcripts) that are processed into short stem-loop structures called pre-microRNAs, by the microprocessor complex in the nucleus. Pre-microRNAs are subsequently exported to the cytosol via Exportin 5 where they are further cleaved by the class II RNAse III enzyme Dicer into ~22nt mature microRNAs. The two strands of mature microRNA duplexes are subsequently treated unequally. One strand from the duplex is loaded onto one of several Argonaute-family proteins to form the RNA Induced Silencing Complex (RISC), and serves as the “guide” that directs target gene-silencing, whereas the other “passenger” strand is discarded. The mechanisms that determine guide strand selection are still not fully understood, but involve sensing thermodynamic stability of the miRNA’s 5′-ends. Guide RNAs loaded into the RISC target the complex to complementary RNAs via base pairing, which can cause either cleavage or translational repression of the targeted RNA, resulting in gene silencing.
The natural RNAi pathway can be co-opted experimentally. The earliest approaches involved transfecting cells with small interfering RNAs (siRNAs), 20–24nt duplexes, which results in one strand of the duplex being loaded into the RISC as the RNA “guide” [13]. The effects of siRNAs can be potent because they are introduced in high molar amounts, but they are transient [6, 14]. siRNAs are suitable for conducting screens that assay conditions soon after introducing siRNAs, but are limited to cells that are easily transfected, and are not appropriate for screens that demand stable target gene silencing [6].
The utility of RNAi in screens was expanded by configuring siRNAs into stem loop structures termed short hairpin RNAs (shRNAs) [15] that could be expressed continuously from Pol III promoters [15, 16] and were later adapted to both retroviral and lentiviral vectors to induce sustained gene-silencing [17, 18]. These simple stem loops mimic pre-microRNAs that must be cleaved by Dicer before entering the RISC and, thus, enter the RNAi biogenesis pathway at a step before exogenously introduced siRNAs. However, imprecise processing of the pre-microRNA form of these shRNAs by Dicer has been demonstrated, leading to both passenger strand and altered-guide strand derived off-target effects [19]. Furthermore, strong expression of these basic shRNA stem loops has been associated with dose dependent toxicities, resulting from saturation of the endogenous RNAi machinery and displacement of endogenous microRNAs [20]. Both of these complications are likely to have contributed to difficulties in early screening approaches.
The use of isolated stem loop shRNAs as RNAi triggers was improved substantially by embedding the stem loops within the sequence of an endogenous pri-microRNA to form “shRNAmirs”. This results in shRNAs expressed in a pri-microRNA form, a substrate for Drosha processing that enters the RNAi pathway in the nucleus and that might facilitate more accurate downstream processing by Dicer. In fact, this strategy increased shRNA maturation substantially compared to the basic stem loop pre-microRNA form, and correlated with enhanced target-specific RNAi [18]. Importantly, shRNAmirs exhibit potent target gene silencing when expressed from single copy genomic integrations and have been validated extensively for this effect [21–23]. In contrast, aspects of most basic stem loop shRNA tools have not been optimized recently, or been validated extensively for their function from single copy insertions, suggesting that shRNAmirs are likely to have benefits over basic stem loop shRNAs for pooled screens. Multiple endogenous microRNA sequence contexts have proven effective for shRNAmirs, although most studies have employed human miR30-based sequences [21–26]. Additional improvements to this approach have included expressing shRNAmirs using Pol II promoters [27] and preserving conserved nucleotides from the endogenous sequences of the microRNA surrounding the embedded shRNA stems [21]. New computational tools to predict highly effective on-target shRNA sequences have been developed by training them on high-throughput data, and this has enhanced the efficacy of the latest generation of available shRNAmirs [22, 23]. Notably, sequences of shRNAmirs designed using these algorithms are ranked for potential functional effectiveness, which might prove to offer benefits in terms of aiding in selection the most highly effective shRNAmirs for particular genes prior to experimental validation [23].
Looking ahead, multiple additional improvements in the potency and on-target effects of experimentally triggered RNAi are likely to emerge, as key aspects of endogenous RNAi pathways continue to be elucidated. The structures of Argonaute proteins in complex with guide RNAs [28, 29], and Dicer in conjunction with Dicer-associated RNA binding proteins [30], are providing insight into the structural basis for guide strand selection into the RISC, target recognition and target inhibition. In addition, it is clear that primary sequence plays a role in governing pri-microRNA recognition and processing in bilitareans [31]; clarifying these rules is likely to provide guidance in selecting or engineering the most appropriate precursor context in which to express desired shRNA stem loops. Furthermore, not all endogenous RNA triggers develop through the canonical biogenesis pathway [32, 33]. For example, a small fraction have been identified that are transcribed directly into 5′-capped pre-microRNAs that result in Dicer processing and specific loading of the mature microRNA’s 3p strand into the RISC [33], and are the basis for developing new shRNA-tools that capitalize on highly specific utilization of the intended guide strand. Thus, as understanding of the rules that underlie natural RNAi biogenesis and target gene-silencing continue to be resolved, more sophisticated and potent experimental RNAi tools, with fewer off-target problems, will be developed.
Approaches to shRNA-mediated screens in vitro and in vivo
The availability of genome-scale shRNA-based libraries (A description of available tools has been reviewed recently [6]) has propelled development of approaches to conduct high-throughput screens in cultured mammalian cells. Arrayed in vitro screens using shRNAs have a number of advantages. However, the high-throughput tools and resources required to analyze thousands of constructs individually in these screens, and the extent of physiological information that can be gleaned from in vitro systems can limit their potential.
More recently, RNAi screening both in vitro and in vivo has trended toward applying shRNAs in pooled formats. The concept of pooled shRNA-mediated screens is built upon the idea that by creating a pool of cells in which each cell carries a distinct shRNA, genes required for distinct cellular states can be inferred based on quantifying alterations in the representation of the shRNAs between the starting populations of cells and the cells at the end of the experiment (Figure 1). Cells carrying an shRNA whose sequence suppresses expression of a gene that is required for a particular phenotype are diluted from the pool of cells with the desired phenotype. Reciprocally, enrichment of particular shRNAs in the pool of cells with the desired phenotype suggests their cognate gene product normally restrains cells from accumulating or acquiring the desired phenotype. Pooled screens capitalize on lentiviral or retroviral shRNA delivery vectors that indelibly mark cells upon transduction with each shRNA, by virtue of the DNA provirus integrating into the host cell genome. In effect, each cell carrying an shRNA is barcoded by the shRNA’s gene-specific sequence. Thus, the frequency of shRNA sequences from a population of cells carrying many different shRNAs can be measured and differential enrichment of the different shRNAs between experimental populations can be discerned [34]. Early studies used microarray approaches for detecting shRNA representation. However, recent advances and flexibility in next generation sequencing approaches has increased the reliability of pooled shRNA screens compared to microarray analysis, owing to greater dynamic range, the ability to assay shRNA guide or passenger strand sequences directly without requiring additional barcodes in shRNA libraries, and no need to generate custom arrays matched to the particular shRNA libraries being screened.
Figure 1. In vivo screen approaches using pooled shRNA DNAs delivered to cells.
(A) Screens based on pooling shRNA DNAs. DNA encoding tens to tens-of-thousands of viral (or transposon) delivered shRNAs is pooled.
(B) Pooled shRNA DNA is transfected into viral packaging cells to generate viral supernatants, and used to transduce query cells (tumors, or tissue progenitors), or is transferred in utero to transduce developing fetal tissue progenitor cells [51], at a low multiplicity of infection (MOI) to promote integration of one shRNA clone per cell. Alternatively, pooled DNA is injected (transposon-mediated) into recipient mice to directly deliver shRNAs to target tissues (e.g., liver) [36].
(C) Sequencing methods are used to quantify representation of proviral DNA encoding shRNA sequences in libraries prepared from DNA extracted from the input pool at time zero, as compared to libraries prepared from DNA extracted from tumors, regenerated tissue, or normal tissue to determine differential selection of cells bearing shRNAs that induce functional effects.
Pooled screens provide a number of advantages over arrayed screens. Most importantly they have opened the door for conducting screens in vivo [10, 11, 35–38]. In addition, pooled screens have the potential to drastically minimize certain aspects of technical variability compared to arrayed screens, because cells carrying all shRNAs are combined and are assayed simultaneously in single tubes for each different experimental condition (e.g., treated versus untreated), and are thus internally controlled. Finally, pooled approaches reduce or virtually eliminate the need for large-scale liquid-handling and high-throughput sampling, imaging and assaying tools. Thus, thoughtfully designed pooled screens in vivo have opened the door to efficiently interrogate the function of a large number of genes in physiological settings.
Lessons from in vivo pooled RNAi screens of genes in oncogenesis: Divide and Conquer
The applicability of in vitro and in vivo pooled RNAi screens has grown out of studies aimed at identifying genes that regulate development, growth and survival of normal or cancerous cells. These studies capitalize on strong selective forces to enrich for shRNAs that produce an advantage from within a pool. Thus, tissue development and tumor models have proven ideal systems and have begun outlining the parameters necessary to conduct cogent analyses of hundreds or thousands of genes in parallel in vivo using pooled RNAi screens.
The number of distinct shRNAs that can be screened simultaneously in a pool is influenced by multiple factors. In practical terms, each shRNA must be represented in many cells in the input pool. In this way, each shRNA has the potential to influence the fate of multiple individual cells during the developmental process. The number of different shRNAs that can be screened is a function of how frequently each shRNA can be represented in the pool at experimental inception, and is therefore constrained by the frequency of precursor cells in a developmental system. Tumors and normal tissues derive and maintain homeostasis from a limited number precursor cells [39–42], and this establishes the baseline. In addition, the diversity of the pool that can be screened successfully is reduced by the natural stochasticity that is inherent to cell fate decisions [39, 40, 43, 44], the low probability of delivering distinct shRNAs evenly among all precursor cells during transduction, and the fact that even the most current libraries of shRNA designs are likely to contain a substantial number of molecules with weak or absent gene-silencing activity [6, 22, 23].
Thus, a general scheme in the setting of cell development and tumors has been to transduce tumor cells, or their progenitors, with focused pools of shRNAs, and to assess shRNA representation in the resulting tumors or tissue at time points after implantation into mice (Figure 1). Nearly all in vivo pooled RNAi screens to date have been geared toward interrogating focused gene sets, rather than surveying all potential genes genome-wide. Such approaches maximize representation of individual shRNAs and are likely to reveal genes from across the regulatory continuum including both strong and weak modulators [45]. In addition, dividing libraries into smaller focused sets by prioritizing genes with potential functional roles based on correlative data (e.g., oncogenomics, or differential gene expression analysis), constitutes a hypothesis-driven approach [46]. In one of the earliest approaches, a reconstitution system was developed in which liver cell progenitors were rendered tumor-prone (but not committed to malignancy) by virtue of p53 deficiency combined with ectopic expression of myc [47]. To identify potential tumor suppressor genes, these progenitors were transferred into recipient mice after transduction with pools of shRNAs targeting genes that map in focal deletions associated with hepatocellular carcinoma [38]. Whereas mice receiving progenitors transduced only with negative control shRNAs produced no tumors, many tumors developed in mice reconstituted with cells carrying the experimental shRNA pools. Thus, the shRNAs found in the resulting tumors indicated that genes targeted by these shRNAs normally suppressed oncogenic transition (in the context of combined p53 deficiency and myc-overexpression), and could be inferred as tumor suppressors.
Hematologic malignancies have been investigated in conceptually similar ways. Using the well-characterized Eμ-myc transgenic model of lymphoma, multiple candidate genes that regulate lymphoma progression in vivo have been identified after adoptive transfer of cells transduced with focused sets of thousands of shRNAs targeting cancer-related genes (the “cancer 1000”) [45, 48]. A similar adoptive transfer approach is likely possible for multiple different hematologic tumors as the tumorigenic precursor frequency in many hematologic malignancies is quite high, and the ability to transfer disease into animals is robust [40].
To sidestep the requirement of initial in vitro manipulation of target cells, direct delivery of shRNA pools to cells in vivo may be advantageous and has been developed (Figure 1A–C). Both the potential for liver regeneration, and the resistance of liver carcinomas to sorafenib treatment have been probed using this approach. In the first case, factors that regulate tissue regeneration could be studied upon complementing the lethality associated with defective liver function in FAH-deficient mice by directly delivering to the liver (using hydrodynamic injection) transposable DNA that co-expressed a normal FAH gene and shRNAmirs [36]. Thus, the strong positive selection for cells that took up the pooled shRNA DNA and expressed FAH could be used to infer the genes necessary for liver regeneration based on the loss of particular shRNAs in regenerated livers (Figure 1A–C). In the second case, genes residing in known amplified genomic regions of hepatocellular carcinomas that might confer resistance of hepatic carcinomas to sorafenib treatment were interrogated by delivering focused sets of shRNAmirs on transposable elements that also expressed NrasG12V into livers of p19Arf-deficient mice (this genetic combination induces aggressive tumors)[49]. By comparing representation of shRNAs in the resulting tumors from mice that were either treated or untreated with sorafenib, genes that were responsible for resistance to this therapy were identified.
Although most in vivo screens might be most informative when using smaller pools of shRNAs, a pioneering genome-wide screen of normal and oncogenic growth in the epidermis has been accomplished using shRNA pools, indicating that large screens in vivo are possible in some settings [35, 50]. Using a direct in vivo approach in which ~ 70,000 lentiviral shRNAs were delivered to epidermal precursor cells in fetuses in utero [35, 51] (Figure 1A–C), approximately 1,800 genes were identified as potential regulators of normal epidermal growth, and an additional 160 genes that were specifically involved in HrasG12V-dependent tumorigenic expansion of cells. A clear strength of the approach was the use of two computational strategies, one of which is extremely stringent (DESeq, [52]), in a complementary fashion to classify differentially represented shRNAs statistically. An important question to be addressed in the future is how closely the genes identified in this approach approximates the actual comprehensive set of genes controlling epidermal growth. Future development of validated whole-genome shRNA libraries in which constructs with low potency and/or unreliable off-target effects have been “weeded-out”, effectively reducing the size of pools that are necessary to screen, are likely to improve the feasibility and reliability of genome-wide screens.
T-cell shRNA screens in a tumor microenvironment
The analysis of T cells is an ideal starting point for conducting in vivo pooled RNAi screens. A number of very well characterized TCR transgenic models and adoptive transfer systems matched with model pathogens and tumors expressing defined antigens are available. The precursor frequencies of endogenous naïve T cells have been estimated, and it has been determined how this impacts the development of resulting T cell populations during infections, including lineage-tracing experiments using barcodes to follow responding T cell “families” during primary and secondary immune responses [39, 43, 53–55]. Furthermore, the in vivo phenotypes of TCR transgenic CD4 and CD8 T cells after manipulation in vitro by transduction with retroviruses expressing cDNAs or shRNAs and adoptive transfer have been validated in these systems for single genes [56–59]. Thus, many of the tools and critical knowledge that are important for designing large-scale screens in vivo are available.
CD8+ cytotoxic T lymphocytes (CTLs) can provide immune defense against tumors. Adoptive transfer of “re-directed”, tumor-specific and/or, chimeric antigen receptor (CAR) expressing CTLs holds promise for treating some cancer patients [60]. However, the tumor microenvironment can resist immune attack by inhibiting T cell function [61]. Using a creative pooled RNAi approach in vivo, CD8 T cell intrinsic genes encoding factors that might underlie this process have been identified. The antitumor CD8 T cell screen capitalized by focusing on two gene target lists: 255 genes encoding factors associated with T-cell dysfunction and 1,307 genes encoding kinases or phosphatases. OT-I TCR transgenic CD8 T cells, specific for the model antigen Ova, were transduced in vitro with a pool of lentivirally-expressed shRNAs at a relatively low MOI to favor single integrations, and were transferred to multiple mice bearing B16-Ova melanoma tumors. One week after adoptive transfer, shRNA representation in T cells re-isolated from tumors (where antigen was presented directly by the tumor) and spleens (in which a contribution to antigen cross-presentation was excluded) was quantified by next generation sequencing. This revealed a substantial number of shRNAs that were differentially enriched between the tissues. Candidate genes that were likely to restrain T cell accumulation were inferred from shRNAs that were enriched in tumor T cells over splenic T cells, and were reanalyzed in a secondary screen using ~15 shRNAs per gene. This resulted in identification of 17 candidate genes from the T cell dysfunction list, and 32 candidate genes from the kinase/phosphatase list, that restrained T cell accumulation in the tumor microenvironment.
Suppression of the gene Ppp2r2d had the strongest effect in the screen, and validation showed its suppression promoted very strong T cell accumulation in tumors, an increased effector phenotype and enhanced clearance of tumors. Identification of Ppp2r2d appears to tie results from the in vivo screen to other known negative regulators of T cell function during chronic infection and tumors. Ppp2r2d encodes a regulatory subunit of the PP2A phosphatase complex, which regulates cell cycle entry and exit; reduced PP2A activity is associated with mitosis. A number of receptors, including PD-1 and CTLA-4 inhibit T cell activation and have pathological roles during chronic infections and tumors [62–64]. Tumor patients treated with molecules that block PD-1 and CTLA-4 have seen positive clinical outcomes [64]. The normal inhibitory function of CTLA-4 on phosphotidylinostitol 3 kinase (PI3K) activity in activated T cells is dependent upon PP2A [63]. Thus, genetic evidence from the pooled RNAi screen in vivo might provide a molecular explanation for a CD8 T cell intrinsic model of downstream events upon CTLA-4 blockade during tumor therapy.
On the one hand, this screen was likely to be robust, because it allowed transferring a large number of T cells and both the spleen and the tumor offer a sufficiently large repository to allow seeding, accumulation and recovery of a large number of cells during the experiment. These features are likely to relax constraints on the size of an RNAi pool that can be analyzed simultaneously. Accordingly, multiple genes related to the TCR, inhibitory cytokine signaling, phosphoinositide metabolism, cell cycle regulation and more were identified. On the other hand, it is not yet clear whether additional possible weak modulators were missed. The approach relied on a very strong selection for T cell outgrowth in the tumor, which could have favored strong modulators (or more effective shRNAs) to outcompete less strong regulators. In addition, it is unclear whether initial recruitment and retention in both the spleen and the tumor was equivalent. Indeed, some shRNAs could act quickly enough to affect recruitment into these different compartments upon adoptive transfer. Thus, some observed differences could relate to initial ingress into the tumor. An approach that utilizes a conditional system to activate shRNA expression in vivo [27, 65], after cells have equilibrated into target tissues, should aid in resolving these unknowns and identify new important regulators of antitumor immunity.
Pooled RNAi screens using shRNAmirs in CD4 and CD8 T cells during viral infection
To functionally interrogate factors responsible for the differentiation of effector CD4 T helper cells (Th1 and T follicular helper (Tfh) cells), as well as, effector and memory precursor CD8 T cells, a pooled in vivo RNAi screening approach was devised in the context of the immune response to acute lymphocytic choriomeningitis virus (LCMV) infection [10]. This approach constitutes the first example of an in vivo pooled RNAi screen to assay cell differentiation based primarily on phenotype, rather than positive or negative selection based on growth or survival fitness (Figure 2).
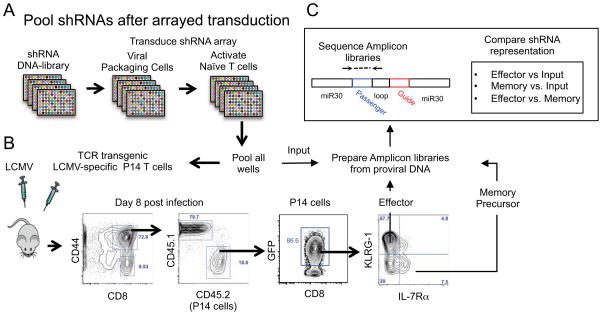
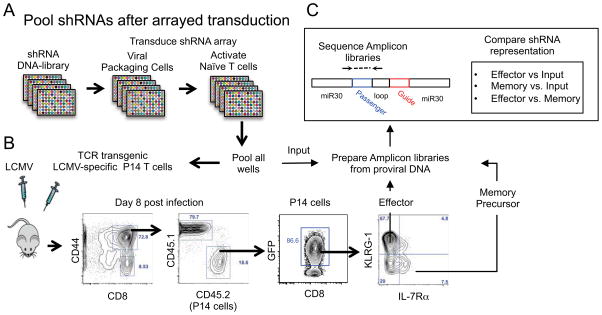
Figure 2. A pooled approach to in vivo T cell screens using arrayed transduction of shRNAs.
(A) Screens based on pooling cells after transduction with unique shRNAs in arrays. DNA for viral shRNA constructs is used to package virus and transduce query cells at high-efficiency (high MOIs) in arrayed format and cells are pooled before transfer in vivo [10].
(B) FACS-based recovery of all transferred cells after subsetting based on differentiation-associated phenotypes.
(C) Next generation sequencing libraries amplified from proviral DNA encoding shRNA sequences are prepared from genomic DNA samples of input and sorted cell populations and differential shRNA representation is quantified after high-throughput sequencing.
T cell responses naturally derive from a limited number of naïve precursor cells [53–55]. Therefore, a focused set of ~110 shRNAmirs targeting ~34 candidate genes encoding transcriptional regulatory factors were selected to minimize cell transfer numbers and maximize shRNA representation. These shRNAs were cloned into an optimized retroviral delivery vector that does not cause rejection of transduced T cells after adoptive transfer. Pools of shRNA-transduced TCR transgenic, LCMV GP61–80 specific (SMARTA) CD4 T cells and LCMV GP33–41 specific (P14) CD8 T cells were transferred into separate groups of multiple mice that were subsequently infected with LCMV. Approximately 1 week following infection, the transferred T cells were re-isolated. In one experiment, Th1 and Tfh cells were sorted based on expression of CXCR5 and SLAM [58, 66]. In another experiment, effector and memory precursor CD8 T cells were sorted on the basis of KLRG-1 and IL-7Rα expression [59]. Quantitation of shRNA representation in each population confirmed the known requirements for a number of genes (e.g., Tbx21, Id2, Id3, Prdm1 and Itch) [58, 59, 67–71].
The screens in both cell types suggested a role for transcription elongation [72] in controlling the selective generation of Th1 cells and effector CD8 T cells. Multiple shRNAmirs targeting Ccnt1 (Cyclin T1), the regulatory subunit of positive transcription elongation factor b (P-TEFb), were strongly depleted from Th1 cells and effector CD8 T cells, relative to Tfh and memory precursor CD8 T cells, respectively. Notably, these shRNAmirs did not appear to affect negatively the accumulation of either CD4 or CD8 T cells in vitro, or in vivo. The impact of Ccnt1 shRNAmirs were validated individually in both cell types, and a role for P-TEFb was bolstered by demonstrating that shRNAmirs targeting Cdk9, which encodes the kinase subunit that dimerizes with Cyclin T1 to form P-TEFb, resulted in a similar phenotype as Ccnt1 inhibition. P-TEFb components are not strongly differentially expressed between effector and memory CD8 T cells, or Th1 and Tfh cells. Thus, mechanisms outside of differential RNA expression are likely to explain the selective effects of P-TEFb. Identifying factors such as these emphasizes the importance of functional screens for identifying factors that do not demonstrate differential expression between distinct cell lineages, but nevertheless have cell type-specific roles.
The CD4 and CD8 T cell screens in the LCMV system differ fundamentally from previously reported pooled in vivo shRNA screens. An arrayed format was used to maximize transduction efficiency of shRNAmirs. To create pools of shRNAs for analysis in vivo, all cells from each well in the transduced arrays were combined before transfer (Figure 2A and B). The increased transduction efficiency using this approach raises the potential for target gene suppression by each examined shRNA and might be beneficial because mature lymphocytes may be less competent for RNAi than other cell types [73]. Indeed, validation of single shRNAs indicated target gene suppression and T cell phenotype was strongest in the most highly transduced cells, based on reporter protein fluorescence intensity [10] (and unpublished data, SC and MEP). Nevertheless, outcomes from the screens did not suggest that high transduction efficiency (and presumably multiple integrations) and the array-pooling approach amplified any commonly expected off-target issues [6, 10]. Thus, highly efficient delivery of shRNAs to cells in arrayed format followed by pooling is suitable for screening in vivo, and might present advantages in some screens.
Additional modifications to these approaches might improve their significance and utility. In the current screens, T cell activation in vitro was required for delivery of the retrovirally expressed shRNAs [10, 11], which is a potential limitation. It is formally possible that some genes whose products function early during T cell activation might not have been silenced soon enough to observe effects, and requirements for these genes missed in this setting, even though all T cells apparently experienced additional TCR stimulation after transduction and adoptive transfer during the infection or tumor encounter in vivo. Thus, approaches to generate naïve T cells bearing shRNAs will be critical to be sure that factors which regulate the early aspects of naïve T cell activation and differentiation to effector cells are identified. With this in mind, inducible or reversible shRNA systems [27, 65, 74, 75] applied to T cell precursors would be important. Such systems would also allow one to generate effector or memory T cells from naïve T cells in the absence of shRNA activity, followed by inducible target gene silencing, to examine specifically, the transition to memory [76]. While vector systems to conduct these experiments exist [27, 65, 74], they may confer immune rejection to transduced T cells upon adoptive transfer into infected mice, and shRNA libraries in the vectors are not widely available (although they are not problematic to create). Finally, it would be beneficial to increase the number of shRNAmirs that can be surveyed simultaneously. However, even though the differentiation of individual CD4 and CD8 T cells can recapitulate all differentiated subsets observed on the population level upon infection [77, 78], lineage-tracing experiments in CD8 T cells indicates that only a very small number of clones dominate the response [39, 43]. Thus, the complexity of RNAi pools that can be interrogated in T cells will have to be determined carefully in each experimental system to account for the natural stochasticity in these processes.
Concluding remarks
RNAi should remain a mainstay in the repertoire of tools to perform loss-of-function analyses in vivo, despite powerful new systems employing CRISPR-Cas9 machinery, which can be used to rapidly engineer null alleles in mice, and that have been adapted to conduct large scale loss-of-function screens in somatic cells [7, 79]. Although gene-disruption in mice has been the gold-standard in genetic loss-of-function studies, they are subject to a number caveats, and it is difficult to know whether null alleles model biology very accurately [80, 81]. Even using conditional inactivation approaches, organisms with null alleles may invoke complicated compensatory mechanisms for survival, lack entire cellular lineages, or fail to develop beyond embryonic stages. The complete absence of a gene might represent an entirely different system, rather than simply reflect the wild-type system minus one of its cognate factors.
Cell fate “decisions” result from fluctuations in gene expression noise that become amplified, ultimately reinforcing these initial “decisions” and driving lineage commitment [82]. Variations in factor concentrations, such as cytokine or transcription factor “gradients”, amplify (or dampen) these fluctuations and mediate cell differentiation in a deterministic fashion [4, 44]. Thus, the biology of cell development is determined by quantitative, rather than all-or-none, changes in gene expression via feedback networks that culminate in qualitatively distinct cell states. With this in mind, the physiology of most factors likely resides in a range between its maximal expression (or function) and some very low level of expression (or function), and it is reasonable to propose that experimental application of RNAi might be one of the most suitable systems to probe the biology of cellular function. The natural function of RNAi appears to titrate gene expression within a physiological window during cell activation, differentiation and function, rather than inhibiting targets completely [83, 84]. In like fashion, experimental target gene suppression induced by shRNAmirs is frequently incomplete [22, 23]. As a result, the effects induced by experimental RNAi likely resemble conditions of altered factor amounts that cells might encounter naturally when genes are modulated during otherwise normal cell-state transitions. Such approaches are likely to provide cogent understanding of how biological systems are controlled, and in turn may facilitate incisive approaches to manipulate cells therapeutically.
Highlights.
Tools for stable RNAi have improved and facilitate pooled screens in vivo
Applying pooled RNAi screens in vivo emerged from approaches in tumors
Pooled RNAi screens in T cells in tumor microenvironments and during infection
RNAi phenotypes may inform biological understanding better than null alleles
Acknowledgments
This work was supported by NIH grant R01 072543 to S.C.; NIH U19 AI109976 to S.C. and M.E.P.; NIH R01 AI095634 to M.E.P and funding from the Frenchman’s Creek Women for Cancer Research to M.E.P. We would like to thank Dr. Gustavo Martinez for suggestions on the manuscript, and apologize to other investigators for the inability to include discussion of many other important contributions that have moved the RNAi screening field forward.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Oestreich KJ, Weinmann AS. Master regulators or lineage-specifying? Changing views on CD4+ T cell transcription factors. Nat Rev Immunol. 2012;12:799–804. doi: 10.1038/nri3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Best JA, et al. Transcriptional insights into the CD8(+) T cell response to infection and memory T cell formation. Nat Immunol. 2013;14:404–412. doi: 10.1038/ni.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin YC, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pipkin ME, Rao A. SnapShot: effector and memory T cell differentiation. Cell. 2009;138:606 e601–602. doi: 10.1016/j.cell.2009.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ravasi T, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fellmann C, Lowe SW. Stable RNA interference rules for silencing. Nat Cell Biol. 2014;16:10–18. doi: 10.1038/ncb2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohr SE, et al. RNAi screening comes of age: improved techniques and complementary approaches. Nature reviews. Molecular cell biology. 2014;15:591–600. doi: 10.1038/nrm3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Echeverri CJ, et al. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat Methods. 2006;3:777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- 9.Sigoillot FD, King RW. Vigilance and validation: Keys to success in RNAi screening. ACS chemical biology. 2011;6:47–60. doi: 10.1021/cb100358f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen R, et al. In vivo RNA interference screens identify regulators of antiviral CD4(+) and CD8(+) T cell differentiation. Immunity. 2014;41:325–338. doi: 10.1016/j.immuni.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou P, et al. In vivo discovery of immunotherapy targets in the tumour microenvironment. Nature. 2014;506:52–57. doi: 10.1038/nature12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annual review of biophysics. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elbashir SM, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 14.Sharma S, Rao A. RNAi screening: tips and techniques. Nat Immunol. 2009;10:799–804. doi: 10.1038/ni0809-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brummelkamp TR, et al. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 16.McManus MT, et al. Gene silencing using micro-RNA designed hairpins. Rna. 2002;8:842–850. doi: 10.1017/s1355838202024032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 18.Silva JM, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 19.Gu S, et al. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell. 2012;151:900–911. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimm D, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 21.Fellmann C, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 22.Fellmann C, et al. Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Mol Cell. 2011;41:733–746. doi: 10.1016/j.molcel.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knott SR, et al. A Computational Algorithm to Predict shRNA Potency. Mol Cell. 2014;56:796–807. doi: 10.1016/j.molcel.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung KH, et al. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res. 2006;34:e53. doi: 10.1093/nar/gkl143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu YP, et al. Inhibition of HIV-1 by multiple siRNAs expressed from a single microRNA polycistron. Nucleic Acids Res. 2008;36:2811–2824. doi: 10.1093/nar/gkn109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng Y, et al. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 27.Dickins RA, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 28.Schirle NT, MacRae IJ. The crystal structure of human Argonaute2. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schirle NT, et al. Gene regulation. Structural basis for microRNA targeting. Science. 2014;346:608–613. doi: 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson RC, et al. Dicer-TRBP Complex Formation Ensures Accurate Mammalian MicroRNA Biogenesis. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Auyeung VC, et al. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013;152:844–858. doi: 10.1016/j.cell.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maurin T, et al. RNase III-independent microRNA biogenesis in mammalian cells. Rna. 2012;18:2166–2173. doi: 10.1261/rna.036194.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie M, et al. Mammalian 5′-capped microRNA precursors that generate a single microRNA. Cell. 2013;155:1568–1580. doi: 10.1016/j.cell.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sims D, et al. High-throughput RNA interference screening using pooled shRNA libraries and next generation sequencing. Genome biology. 2011;12:R104. doi: 10.1186/gb-2011-12-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beronja S, et al. RNAi screens in mice identify physiological regulators of oncogenic growth. Nature. 2013;501:185–190. doi: 10.1038/nature12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wuestefeld T, et al. A Direct in vivo RNAi screen identifies MKK4 as a key regulator of liver regeneration. Cell. 2013;153:389–401. doi: 10.1016/j.cell.2013.03.026. [DOI] [PubMed] [Google Scholar]
- 37.Zender L, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zender L, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buchholz VR, et al. Disparate individual fates compose robust CD8+ T cell immunity. Science. 2013;340:630–635. doi: 10.1126/science.1235454. [DOI] [PubMed] [Google Scholar]
- 40.Magee JA, et al. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer cell. 2012;21:283–296. doi: 10.1016/j.ccr.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mascre G, et al. Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature. 2012;489:257–262. doi: 10.1038/nature11393. [DOI] [PubMed] [Google Scholar]
- 42.Simons BD, Clevers H. Strategies for homeostatic stem cell self-renewal in adult tissues. Cell. 2011;145:851–862. doi: 10.1016/j.cell.2011.05.033. [DOI] [PubMed] [Google Scholar]
- 43.Gerlach C, et al. Heterogeneous differentiation patterns of individual CD8+ T cells. Science. 2013;340:635–639. doi: 10.1126/science.1235487. [DOI] [PubMed] [Google Scholar]
- 44.Losick R, Desplan C. Stochasticity and cell fate. Science. 2008;320:65–68. doi: 10.1126/science.1147888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bric A, et al. Functional identification of tumor-suppressor genes through an in vivo RNA interference screen in a mouse lymphoma model. Cancer cell. 2009;16:324–335. doi: 10.1016/j.ccr.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McFadden DG, et al. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell. 2014;156:1298–1311. doi: 10.1016/j.cell.2014.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zender L, et al. Generation and analysis of genetically defined liver carcinomas derived from bipotential liver progenitors. Cold Spring Harb Symp Quant Biol. 2005;70:251–261. doi: 10.1101/sqb.2005.70.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meacham CE, et al. In vivo RNAi screening identifies regulators of actin dynamics as key determinants of lymphoma progression. Nat Genet. 2009;41:1133–1137. doi: 10.1038/ng.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rudalska R, et al. In vivo RNAi screening identifies a mechanism of sorafenib resistance in liver cancer. Nat Med. 2014;20:1138–1146. doi: 10.1038/nm.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beronja S, Fuchs E. RNAi-mediated gene function analysis in skin. Methods Mol Biol. 2013;961:351–361. doi: 10.1007/978-1-62703-227-8_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beronja S, et al. Rapid functional dissection of genetic networks via tissue-specific transduction and RNAi in mouse embryos. Nat Med. 2010;16:821–827. doi: 10.1038/nm.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Badovinac VP, et al. Initial T cell receptor transgenic cell precursor frequency dictates critical aspects of the CD8(+) T cell response to infection. Immunity. 2007;26:827–841. doi: 10.1016/j.immuni.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moon JJ, et al. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27:203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Obar JJ, et al. Endogenous naive CD8+ T cell precursor frequency regulates primary and memory responses to infection. Immunity. 2008;28:859–869. doi: 10.1016/j.immuni.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Araki K, et al. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu JK, et al. Expression of chemokine receptor CXCR3 on T cells affects the balance between effector and memory CD8 T-cell generation. Proc Natl Acad Sci U S A. 2011;108:E118–127. doi: 10.1073/pnas.1101881108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnston RJ, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Joshi NS, et al. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Restifo NP, et al. Adoptive immunotherapy for cancer: harnessing the T cell response. Nat Rev Immunol. 2012;12:269–281. doi: 10.1038/nri3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tumeh PC, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barber DL, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 63.Parry RV, et al. CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol Cell Biol. 2005;25:9543–9553. doi: 10.1128/MCB.25.21.9543-9553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wolchok JD, et al. Nivolumab plus ipilimumab in advanced melanoma. The New England journal of medicine. 2013;369:122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zuber J, et al. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotechnol. 2011;29:79–83. doi: 10.1038/nbt.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Choi YS, et al. Bcl6 Expressing Follicular Helper CD4 T Cells Are Fate Committed Early and Have the Capacity To Form Memory. J Immunol. 2013 doi: 10.4049/jimmunol.1202963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cannarile MA, et al. Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nat Immunol. 2006;7:1317–1325. doi: 10.1038/ni1403. [DOI] [PubMed] [Google Scholar]
- 68.Kallies A, et al. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity. 2009;31:283–295. doi: 10.1016/j.immuni.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 69.Rutishauser RL, et al. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31:296–308. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang CY, et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat Immunol. 2011;12:1221–1229. doi: 10.1038/ni.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xiao N, et al. The E3 ubiquitin ligase Itch is required for the differentiation of follicular helper T cells. Nat Immunol. 2014;15:657–666. doi: 10.1038/ni.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 73.Oberdoerffer P, et al. Efficiency of RNA interference in the mouse hematopoietic system varies between cell types and developmental stages. Mol Cell Biol. 2005;25:3896–3905. doi: 10.1128/MCB.25.10.3896-3905.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Godec J, et al. Inducible RNAi in vivo reveals that the transcription factor BATF is required to initiate but not maintain CD8+ T-cell effector differentiation. Proc Natl Acad Sci U S A. 2015;112:512–517. doi: 10.1073/pnas.1413291112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kaech SM, et al. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111:837–851. doi: 10.1016/s0092-8674(02)01139-x. [DOI] [PubMed] [Google Scholar]
- 77.Becattini S, et al. T cell immunity. Functional heterogeneity of human memory CD4(+) T cell clones primed by pathogens or vaccines. Science. 2015;347:400–406. doi: 10.1126/science.1260668. [DOI] [PubMed] [Google Scholar]
- 78.Stemberger C, et al. A single naive CD8+ T cell precursor can develop into diverse effector and memory subsets. Immunity. 2007;27:985–997. doi: 10.1016/j.immuni.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 79.Wang T, et al. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gingrich JR, Roder J. Inducible gene expression in the nervous system of transgenic mice. Annual review of neuroscience. 1998;21:377–405. doi: 10.1146/annurev.neuro.21.1.377. [DOI] [PubMed] [Google Scholar]
- 81.Matthaei KI. Genetically manipulated mice: a powerful tool with unsuspected caveats. The Journal of physiology. 2007;582:481–488. doi: 10.1113/jphysiol.2007.134908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chang HH, et al. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trifari S, et al. MicroRNA-directed program of cytotoxic CD8+ T-cell differentiation. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1317191110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pipkin ME, Monticelli S. Genomics and the immune system. Immunology. 2008;124:23–32. doi: 10.1111/j.1365-2567.2008.02818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]