Abstract
Rationale
Inactivating mutations in the FOXF1 gene locus are frequently found in patients with Alveolar Capillary Dysplasia with Misalignment of Pulmonary Veins (ACD/MPV), a lethal congenital disorder, which is characterized by severe abnormalities in the respiratory, cardio-vascular and gastro-intestinal systems. In mice, haploinsufficiency of the Foxf1 gene causes alveolar capillary dysplasia and developmental defects in lung, intestinal and gall bladder morphogenesis.
Objective
While FOXF1 is expressed in multiple mesenchyme-derived cell types, cellular origins and molecular mechanisms of developmental abnormalities in FOXF1-deficient mice and ACD/MPV patients remain uncharacterized due to lack of mouse models with cell-restricted inactivation of the Foxf1 gene. In the present study, the role of FOXF1 in endothelial cells was examined using a conditional knockout approach.
Methods and Results
A novel mouse line harboring Foxf1-floxed alleles was generated by homologous recombination. Tie2-Cre and Pdgfb-CreER transgenes were used to delete Foxf1 from endothelial cells. FOXF1-deficient embryos exhibited embryonic lethality, growth retardation, polyhydramnios, cardiac ventricular hypoplasia and vascular abnormalities in the lung, placenta, yolk sac and retina. Deletion of FOXF1 from endothelial cells reduced endothelial proliferation, increased apoptosis, inhibited VEGF signaling and decreased expression of endothelial genes critical for vascular development, including VEGF receptors Flt1 and Flk1, Pdgfb, Pecam1, CD34, integrin β3, ephrin B2, Tie2 and the non-coding RNA Fendrr. ChIP assay demonstrated that Flt1, Flk1, Pdgfb, Pecam1 and Tie2 genes are direct transcriptional targets of FOXF1.
Conclusions
FOXF1 is required for formation of embryonic vasculature by regulating endothelial genes critical for vascular development and VEGF signaling.
Keywords: FOXF1, endothelial cells, VEGF, Flk1, vascular development, angiogenesis, pulmonary circulation, developmental biology, gene regulation, genetically altered mice
INTRODUCTION
Development of the embryonic vasculature depends on vasculogenesis (de novo formation of blood vessels) and angiogenesis (branching of preexisting blood vessels) in a process requiring appropriate levels of Vascular Endothelial Growth Factor (VEGF). Targeted disruption of the Vegf gene produces an embryonic lethal phenotype displaying impaired blood-island formation and delayed endothelial cell differentiation, leading to abnormal blood vessel development1, 2. VEGF is the ligand for tyrosine kinase receptors Flk1 and Flt1, both of which are expressed in endothelial cells and their mesenchymal precursors. Flk1−/− mice die in utero due to inhibition of vasculogenesis and formation of angioblast cells in the blood islands3, whereas Flt1−/− embryos fail to form mature blood vessels4. Other signaling pathways involved in formation of embryonic vasculature include Angiopoietin/Tie2, PDGF, PI3K/ AKT, TGF-β, Shh, Wnt and Notch, as well as transcription factors Etv2, Hand1, MEF2c, Prox1, Hey1/2, COUP-TFII, Tbx4, Snail, FOXC2, GATA, Sox and KLF (reviewed in5–7). Identification of additional proteins that regulate embryonic vascular development will provide information regarding pathogenesis of human vascular disorders.
Alveolar Capillary Dysplasia with Misalignment of Pulmonary Veins (ACD/MPV) is a congenital disorder of neonates and infants, which is characterized by severe defects in development of pulmonary capillaries, hypoxemia, pulmonary hypertension and thickening of small pulmonary arteries, malposition of pulmonary veins, lung edema and impaired lobular development8. Structural abnormalities of the genitourinary, gastrointestinal and cardiovascular systems are also common. Due to the severity of developmental defects and progressive respiratory insufficiency in ACD/MPV infants, the survival after the first month of birth is rare8. While genetic factors associated with ACD/MPV are not fully characterized, heterozygous deletions and point mutations in the Forkhead Box F1 (FOXF1) gene locus account for approximately 40% of ACD/MPV cases9. In addition, genomic deletions in FOXF1 gene were recently found in prenatal cystic hygroma10, a congenital vascular defect which can result in fetal hydrops (tissue edema) and embryonic death. These clinical data illustrate a critical role of FOXF1 in vascular development.
FOXF1 protein (previously known as HFH-8 or Freac-1) is a member of the Forkhead Box (Fox) family of transcription factors that share homology in the Winged helix/Forkhead DNA binding domain. FOXF1 is expressed in extraembryonic mesoderm, allantois, splanchnic mesoderm and septum transversum mesenchyme11, 12. Foxf1−/− mice die by E8.5 due to severe abnormalities in development of the yolk sac and allantois13. While FOXF1 haploinsufficiency causes alveolar capillary dysplasia, fusion of the lung lobes and various developmental defects in mesenchyme of the gallbladder, esophagus and trachea12, 14, 15, Foxf1+/− mice do not recapitulate all histopathological features of human ACD/MPV. Approximately half of Foxf1+/− mice survived past birth14 but these mice exhibited severe pulmonary hemorrhage in response to lung injury16 and abnormal liver regeneration after liver injury17.
FOXF1 is activated by the Shh signaling pathway through a direct binding of Gli transcription factors to the Foxf1 promoter region15, 18. Deletions of Gli-binding sites were found in the FOXF1 gene locus of ACD/MPV patients19. Shh−/− mouse embryos exhibit a reduction in Foxf1 mRNA15, implicating Shh/Gli signaling in regulation of Foxf1 gene expression. FOXF1 induces migration of mesenchymal cells through direct transcriptional activation of Integrin β3 and Notch-2 genes20, 21. While FOXF1 is expressed in multiple mesenchyme-derived cell types, including fibroblasts, peribronchial smooth muscle, endothelial and hepatic stellate cells, cellular origins and molecular mechanisms of developmental abnormalities in FOXF1-deficient mice and ACD/MPV patients remain uncharacterized due to the lack of mouse models with cell-restricted inactivation of the Foxf1 gene. In the present study, we generated mice harboring Foxf1-floxed alleles and used Tie2-Cre and Pdgfb-CreER transgenes to investigate the role of FOXF1 in endothelial cells. We demonstrated that FOXF1 is critical for formation of embryonic vasculature by stimulating endothelial proliferation and promoting the VEGF, PDGF and Angpt/Tie2 signaling pathways in endothelial cells through direct transcriptional activation of Flk1, Flt1, Pdgfb and Tie2 genes.
METHODS
Generation of Foxf1-floxed mice and deletion of FOXF1 from endothelial cells
Foxf1-targeting vector contained a LoxP site inserted into the Foxf1 promoter and PGK-gb2 LoxP/FRT-flanked Neomycin (neo) cassette placed into the first intron (Fig. 2A). The PGK promoter-driven herpes simplex virus-thymidine kinase (HSV-TK) gene was placed outside of the Foxf1 gene homology region for negative selection of non-homologous recombination in ES cells. The Foxf1fl-targeting vector was used for electroporation of mouse ES cells (C57Bl/6 × 129/SVEV), which were selected for neo (G418) and HSV-TK resistance (ganciclovir). ES cells with the appropriate Foxf1fl-targeted locus were used to generate chimeric mice by injecting Foxf1fl ES cells into mouse blastocysts. Mice containing the Foxf1fl-targeted allele were determined by PCR amplification with primers flanking the LoxP sequence located in the Foxf1 promoter (P1 and P2) and primers located in the 3’ region of the Foxf1fl allele (P3 and P4) (Fig. 2A–B and Online Table I). To produce Foxf1fl/+ mice, chimeric mice were bred with C57Bl/6 mice in the animal facility of Cincinnati Children’s Hospital Medical Center. The Neo cassette was deleted by breeding of Foxf1fl/+ mice with ACT-FLP1 mice (Jackson Lab.) (Fig. 2A). The loss of Neo in Foxf1fl/+ mice was confirmed by PCR using P5 and P6 primers (Fig. 2A and Online Table I). Foxf1fl/+ mice were backcrossed to generate viable Foxf1fl/fl mice that were bred into the C57Bl/6 background for ten generations. Deletion of the Foxf1fl alleles from endothelial cell lineage was accomplished through breeding with Tie2-Cre (C57Bl/6, Jackson Lab.) and Pdgfb-CreER (C57Bl/6,22) transgenic mice. To activate Pdgfb-CreER, tamoxifen was given in food (200mg of tamoxifen citrate with 24.8g of sucrose per kg of diet, Harlan Lab.) at E9.5. For postnatal activation of Cre, tamoxifen was injected i.p. (20µg/ day) at postnatal days P0, P1 and P2. Deletion of FOXF1 was confirmed by breeding FOXF1-deficient mice with LoxP-stop-LoxP-β-gal (R26R) and LoxP-tdTomato-LoxP-GFP (mT/mG) reporter mice (both from Jackson Lab.). Flk1-null mutant mice were previously described23. Animal studies were approved by the Animal Care and Use Committee of Cincinnati Children’s Hospital Research Foundation.
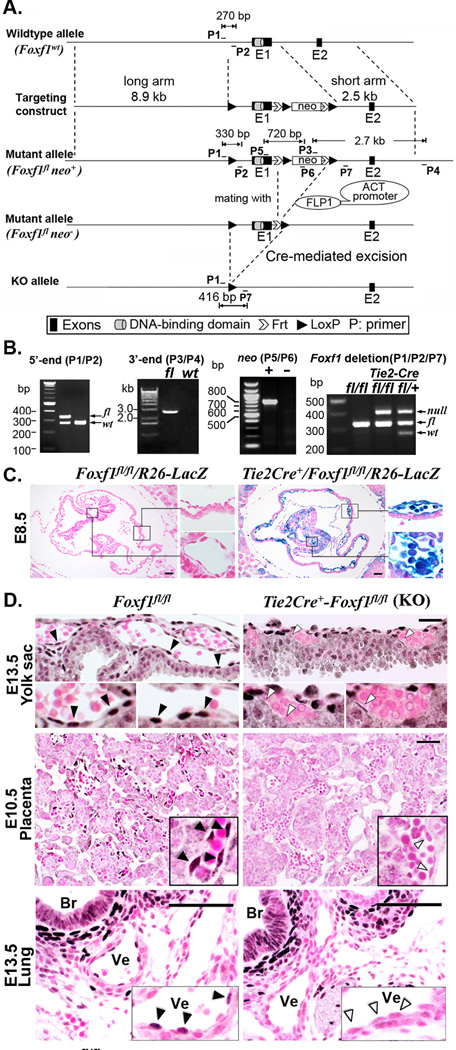
Figure 2. Generation of Foxf1fl/fl mice and conditional deletion of FOXF1 from endothelial cells.
(A) Schematic diagram of Foxf1 gene-targeting construct with two Frt sites (white arrows) and three LoxP sites (black arrowheads) that surround the Neomycin (neo) gene and exon 1 (E1), encoding the DNA-binding domain of the FOXF1 protein. The Neo cassette was removed after breeding of Foxf1-floxed mice with β-actin (ACT)-FLP1 mice. Endothelial deletion of FOXF1 was achieved by breeding with Tie2-Cre mice. (B) PCR of mouse tail DNA using primers (P1–P7). Locations of primers are indicated in panel A. (C) β-gal activity (blue staining) is detected in both endothelial and hematopoietic cells in the yolk sac of Tie2-Cre Foxf1fl/fl /R26R E8.5 embryos. Slides were counterstained with nuclear fast red (red nuclei). Inserts show blood vessels in the yolk sac (top insert) and embryo proper (bottom insert). (D) Immunostaining shows the presence of FOXF1 protein in endothelial cells (black arrowheads) of control Foxf1fl/fl embryos. FOXF1 staining is absent from majority of endothelial cells of Tie2-Cre Foxf1fl/fl embryos (white arrowheads). Abbreviations: Br, bronchiole; Ve, blood vessel. Scale bars are 50µm.
RNA preparation and quantitative real-time RT-PCR (qRT-PCR)
Total RNA was prepared from MFLM-91U cells, mouse tissue and flow-sorted endothelial cells using RNeasy micro kit (Qiagen). qRT-PCR analysis was performed using a StepOnePlus Real-Time PCR system (Applied Biosystems) as described24. Samples were amplified using inventoried TaqMan primers (Online Table II). Reactions were analyzed in triplicates and expression levels were normalized to β-actin mRNA.
siRNA transfection, Western blot and matrigel angiogenesis assay
MFLM-91U cells20 were cultured in serum-free UltraCULTURE medium (Lonza, Walkersville, MD). To inhibit FOXF1, we transfected either non-targeting siRNA or siRNA specific to mouse Foxf1 (Dharmacon) using Lipofectamine™ 2000 reagent (Invitrogen) as described20, 25. Cells were harvested 48 hours after transfection and used for matrigel angiogenesis assay (BD Biosciences). VEGF 165 (20 ng/ ml, Millipore) was added to matrigel for 14 hr. Cells in matrigel were stained with calcein AM fluorescent viability dye which is transported through the cellular membrane into live cells. Confocal 3D images were quantitated using IMARIS software (Bitplane, CT). Western Blot analysis was performed using antibodies described in Supplemental Material. Detection of the immune complex was accomplished by using secondary antibodies directly conjugated with HRP followed by the Supersignal chemiluminescence substrate (Pierce, Rockford, IL).
Immunohistochemical staining and Flow cytometry
Paraffin sections were stained with hematoxylin and eosin (H&E) or used for immunohistochemical staining as described26, 27. Primary antibodies and detection systems are listed in Supplemental Material. For co-localization experiments, secondary antibodies conjugated with Alexa Fluor 488 or Alexa Fluor 594 (Invitrogen) were used as previously described28, 29. Slides were counterstained with DAPI (Vector Lab). Fluorescent images were obtained using a Zeiss Axioplan2 microscope equipped with an AxioCam MRm digital camera and AxioVision 4.3 Software (Carl Zeiss Microimaging, Thornwood, NY). Flow cytometry was performed using cells isolated from yolk sacs and lungs as described27, 30. Antibodies used for flow cytometry are listed in Supplemental Materials. BrdU was injected i.p. into pregnant females 2 hr prior to embryo harvest. Annexin V kit was from eBioscience. Stained cells were separated using cell sorting (Five-laser FACSAria II, BD Biosciences). Purified cells were used for RNA preparation and qRT-PCR analysis.
Chromatin Immunoprecipitation (ChIP) assay
ChIP assay was performed using in situ cross-linked MFLM-91U cells as described24, 27. Antibodies used for ChIP were: FOXF120 and control rabbit IgG (Vector Lab). Sense (S) and antisense (AS) PCR primers that were used to amplify mouse promoter DNA fragments in ChIP assay are provided in Online Table III.
Statistical analysis
ANOVA and Student’s T-test was used to determine statistical significance. P values less then 0.05 were considered significant. Values for all measurements were expressed as the mean ± standard deviation (SD).
RESULTS
FOXF1 is expressed in mesenchyme and endothelial cells during embryogenesis
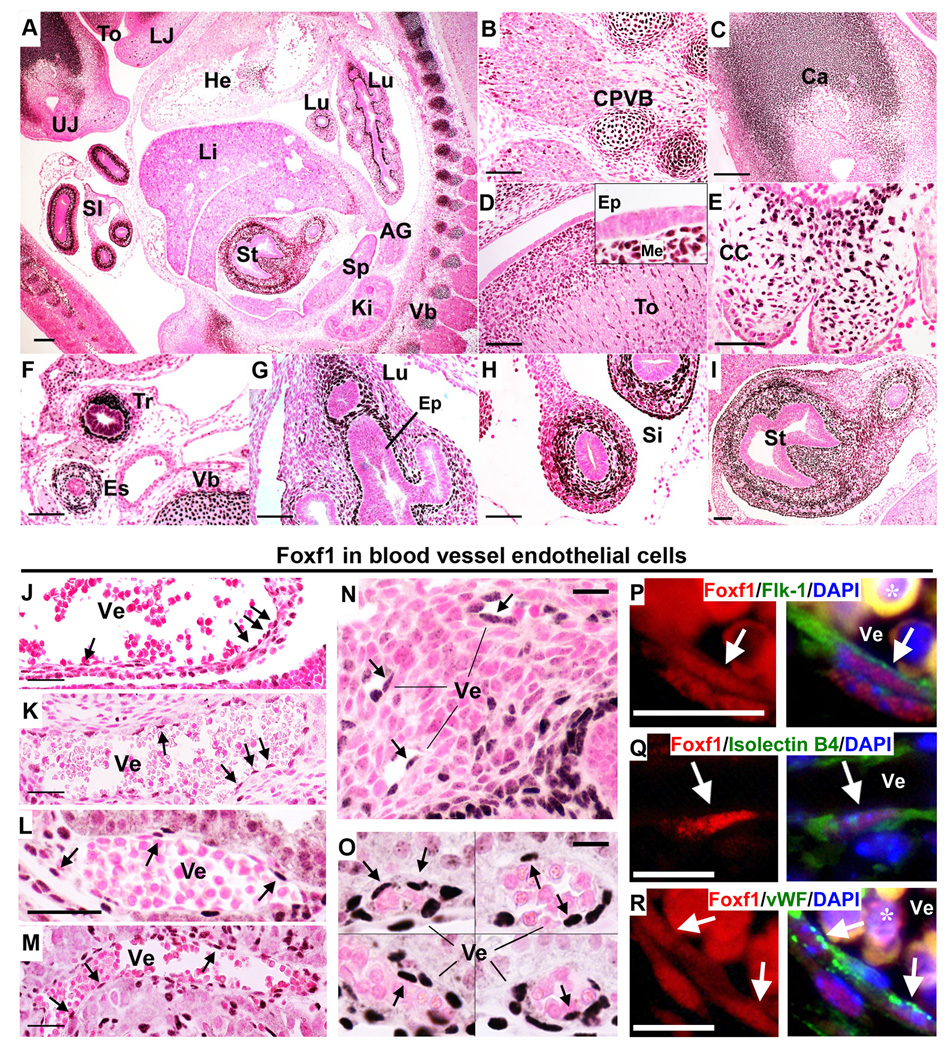
Immunostaining with FOXF1 antibodies was used to visualize FOXF1-expressing cells in E13.5 mouse embryos. FOXF1 protein was found in mesenchyme of the lung, trachea, esophagus, stomach, intestine, oral cavity, tongue and cartilage (Fig. 1A–I), which is consistent with previous in situ hybridization studies11, 13. Additional site of FOXF1 expression was found in the embryonic heart where FOXF1 was present in mesenchyme of cardiac cushion but absent from myocardium and endocardial cells (Fig. 1E and data not shown). FOXF1 was detected in hemangioblasts of the yolk sac but was absent from hematopoietic cells at E8.5 – E12.5 (Fig. 1O and data not shown). FOXF1 was also detected in nuclei of endothelial cells of the lung, yolk sac and embryonic regions of the placenta (Fig. 1J–N). FOXF1 protein co-localized with endothelial marker proteins Flk1, Isolectin B4 and vWF in lung tissue (Fig. 1P–R). Thus, FOXF1 is expressed in endothelial cells during embryogenesis.
Figure 1. FOXF1 is expressed in mesenchymal and endothelial cells of the developing embryo.
(A–I) Immunostaining shows that FOXF1 is expressed in mesenchyme-derived cells in E13.5 wild type mouse embryos (A). FOXF1 is detected in cartilage primordiums of vertebral bodies (CPVB in B), oral cavity (Ca in C), tongue (To in D), atrioventricular cardiac cushions (CC in E), trachea (Tr) and esophagus (Es) (F), lung (Lu in G), small intestine (Si in H), stomach (St in I). (J–O) FOXF1 is expressed in endothelial cells. Low levels of FOXF1 were detected in a subset of endothelial cells (shown with arrows) located in the inferior vena cava (J) and pulmonary vein (K). FOXF1 is detected in blood vessels of the yolk sac (L and O), lung (N) and the fetal part of the placenta (M). A composite of four different images is shown in O. FOXF1 protein was not found in hematopoietic cells. (P–R) FOXF1 co-localizes with endothelial markers Flk1 (P), Isolectin B4 (Q) and vWF (R) in E12.5 lungs. Location of FOXF1 in nuclei of endothelial cells is shown with white arrows. Autofluorescence of red blood cells is denoted with *. Abbreviations: ep, epithelial cells; me, mesenchyme; Ve, blood vessel; AG, adrenal gland; He, heart; Ki, kidney; Li, liver; LJ, lower jaw; UJ, upper jaw; Sp, spleen; Vb, vertebrae. Scale bars: A, 100µm; B–M, 50µm; N–O, 20µm; P–R, 10µm.
Generation of Foxf1-floxed mice and deletion of Foxf1 from endothelial cells
Given the importance of FOXF1 in pathogenesis of ACD/MPV in humans, we determined FOXF1 requirements in endothelial cells using a conditional knockout approach. A triple-LoxP Foxf1-floxed (fl) - targeting vector containing the Neo cassette and two Frt sites was constructed (Fig. 2A) and used for electroporation of mouse ES cells. After mouse blastocyst injection of the Neo-resistant Foxf1fl/+ ES cells, chimeric mice with germ line transmission were obtained and bred to generate a stable Foxf1fl/+ mouse line. The Neo cassette was deleted by breeding Foxf1fl/+ mice with ACT-FLP1 mice (Fig. 2A). PCR amplification of mouse tail genomic DNA was used to distinguish between Foxf1-floxed and Foxf1-wild type (wt) alleles (Fig. 2B). The Foxf1fl/fl mice were bred with Tie2-Cre mice to delete the first exon of the Foxf1 gene encoding the DNA-binding domain and part of the transcriptional activation domain of the FOXF1 protein (Fig. 2A and Online Fig. I), both of which are required for FOXF1 transcriptional activity9, 21. Cre-mediated recombination and the loss of FOXF1 protein in Tie2-Cre Foxf1fl/fl endothelial cells were confirmed by Rosa26-LacZ-reporter (Fig. 2C) and immunostaining with FOXF1 antibodies (Fig. 2D).
Embryonic lethality and cardiovascular defects in Tie2-Cre Foxf1fl/fl embryos
Tie2-Cre Foxf1fl/fl embryos were present in Mendelian ratio prior to E13.5 (Table 1). The number of Tie2-Cre Foxf1fl/fl embryos progressively decreased from E13.5 to E16.5, consistent with embryonic lethality during this period (Table 1). Histological examination of the Tie2-Cre Foxf1fl/fl embryos revealed severe growth retardation as demonstrated by decreased embryo size and body weight (Fig. 3A). Liver size was also decreased in FOXF1 mutants (Fig. 3B). Deletion of FOXF1 caused ventricular hypoplasia and an interventricular septal defect in the embryonic heart (Online Fig. IIA–B). Furthermore, FOXF1 mutants exhibited accumulation of fluid in the amniotic cavity (polyhydramnios) and pericardial cavity (pericardial efflux) (Fig. 3A), a common finding in embryos with various cardiovascular abnormalities5, 6. Thus, deletion of FOXF1 resulted in embryonic lethality due to severe growth retardation and cardiovascular defects.
Table 1.
Genotype frequency of offspring from the breeding of Tie2-Cre+/− Foxf1fl/wt males and Foxf1fl/fl females.
| Total embryos | Foxf1fl/wt | Foxf1fl/fl |
Tie2-Cre+/− Foxf1fl/wt |
Tie2-Cre+/− Foxf1fl/fl |
|
|---|---|---|---|---|---|
| Theoretical | 25% | 25% | 25% | 25% | |
| Experimental | |||||
| E10.5 | 55 | 14 (25%) | 15 (27%) | 14 (25%) | 13 (24%) |
| E11.5 | 39 | 14 (36%) | 8 (21%) | 7 (18%) | 10 (26%) |
| E12.5 | 60 | 20 (33%) | 9 (15%) | 12 (20%) | 19 (32%) |
| E13.5 | 74 | 21 (28%) | 20 (27%) | 20 (27%) | 13 (18%)* |
| E14.5 | 21 | 6 (29%) | 7 (33%) | 6 (29%) | 2 (10%)* |
| E15.5-16.5 | 17 | 2 (12%) | 7 (41%) | 7 (41%) | 1 (6%)* |
shows significant differences between experimental and theoretical ratios.
Figure 3. Embryonic abnormalities in Tie2-Cre Foxf1fl/fl embryos.
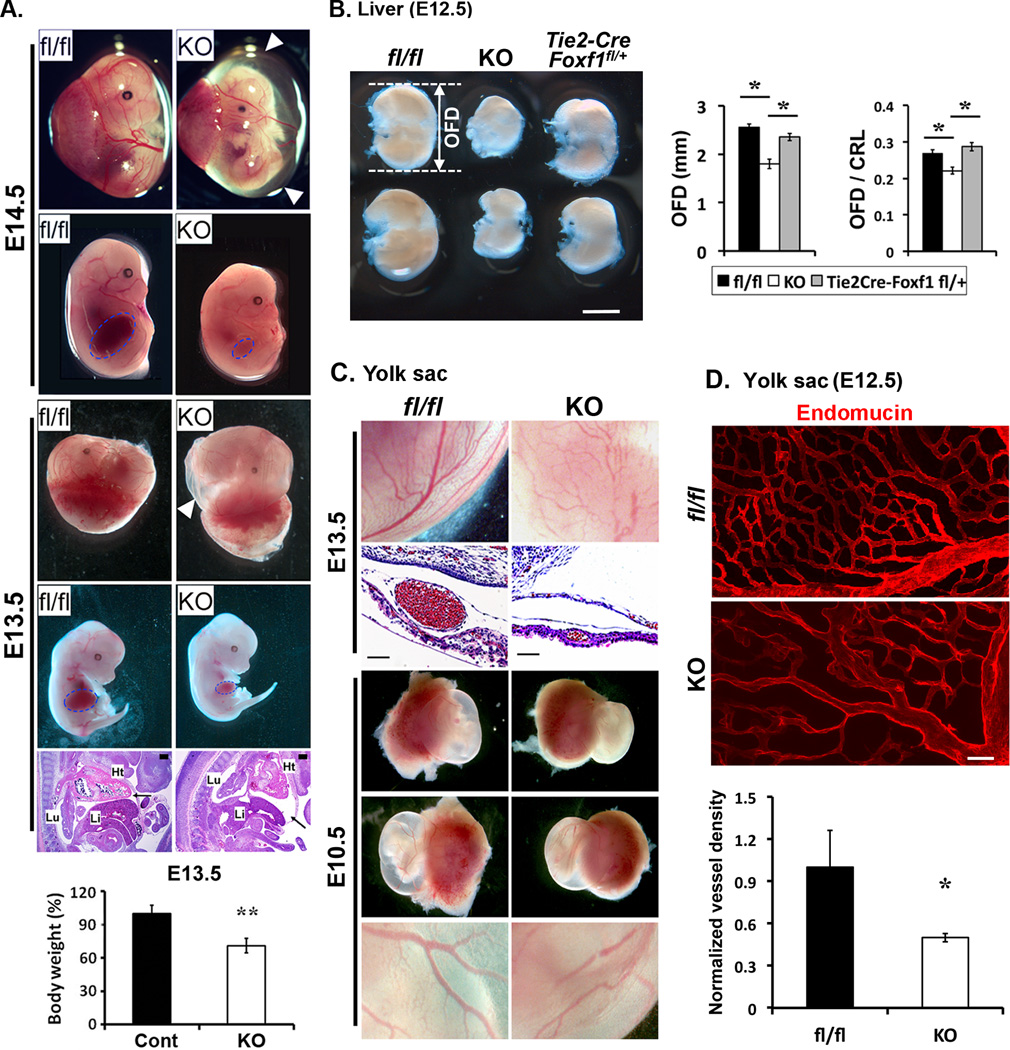
(A) Tie2-Cre Foxf1fl/fl embryos (KO) exhibit growth retardation and severe polyhydramnios (white arrowheads) at E13.5–14.5. Foxf1fl/fl littermates (fl/fl) are shown for comparison. Boundary of the liver is indicated by blue line. Pericardial efflux is shown with a black arrow on H&E-stained section of Tie2-Cre Foxf1fl/fl embryos. Body weight is decreased in Tie2-Cre Foxf1fl/fl E13.5 embryos (n=10) compared to control Foxf1fl/fl littermates (n=43). (B) Liver size was decreased in Tie2-Cre Foxf1fl/fl embryos. Occipital frontal diameter (OFD) of the liver and the ratio between OFD and the Crown rump length (CRL) were significantly reduced after deletion of FOXF1 (n=5). (C–D) Diminished vascular branching in the yolk sac of FOXF1 deficient embryos. The whole mount immunoistaining was performed using endomucin Abs. Confocal microscopy was used to quantitate the vessel density as a ratio between endothelial (endomucin) and epithelial (E-cadherin) staining (bottom panel in D). p<0.05 is *, p<0.01 is **. Abbreviations: Ht, heart; Li, liver; Lu, lung. Scale bars: A, 50µm; B, 1mm; C, 50µm, D, 100µm.
FOXF1 deletion impairs the formation of embryonic vasculature in the yolk sac and placenta
At E13.5, vascular branching was reduced in the yolk sac and placenta of Tie2-Cre Foxf1fl/fl embryos when compared to control Foxf1fl/fl embryos (Fig. 3C and Online Fig. IIIA). Reduced vascular branching was confirmed by whole mount immunostaining of yolk sacs for endothelial-specific endomucin (Fig. 3D). Decreased vascular branching and polyhydramnios were also found in the yolk sac of Foxf1+/− embryos (Online Fig. IIIB–C), findings consistent with published studies14, 15. Vascular phenotypes in Tie2-Cre Foxf1fl/fl and Foxf1+/− embryos were less severe compared to Flk1-null mutant mice that exhibited growth retardation, a near complete loss of yolk sac vasculature and embryonic lethality at E10.5 (Online Fig. IIID).
Vascular defects in Tie2-Cre Foxf1fl/fl yolk sacs were associated with reduced Foxf1 mRNA (Fig. 4A) and decreased expression of Flt1, Flk1 and angiopoietin-1 (Angpt1) (Fig. 4B), all of which are critical for angiogenesis and VEGF signaling in endothelial cells3, 4, 23, 31. Angpt2 and Nrp1 mRNAs were increased in FOXF1 mutants (Fig. 4B). Expression of Ephrin B2 was reduced in arteries of Tie2-Cre Foxf1fl/fl embryos (Online Fig. IVA) as well as in isolated Pecam1+ endothelial cells (Online Fig. IVB). There were no differences in the number of lymphatic vessels stained for LYVE1 (Online Fig. IVA). Ephrin b4, Sox-18, Foxc1 and Foxc2 mRNAs were unaltered (Online Fig. IVB–C). Interestingly, when the Tie2-Cre Foxf1fl/fl embryos were examined at E10.5, diminished branching of blood vessels was still evident in the yolk sac and placenta (Fig. 3C and data not shown) but cardiac abnormalities and polyhydramnios were absent (Fig. 3C and Online Fig. IIC). The number of Pecam1+/ Tie2+/ CD45− endothelial cells was reduced in Tie2-Cre Foxf1fl/fl E10.5 yolk sacs (Online Fig. VB), whereas the number of Pecam1−/ CD45+ and Pecam−/ CD41+ hematopoietic cells was normal (Online Fig. VA). Thus, vascular insufficiency in the yolk sac and placenta occurs earlier than other embryonic defects and is likely to be a primary cause of growth retardation and embryonic lethality in Tie2-Cre Foxf1fl/fl embryos. Altogether, FOXF1 deletion from endothelial cells impairs heart development and decreases vascular branching in the yolk sac and placenta.
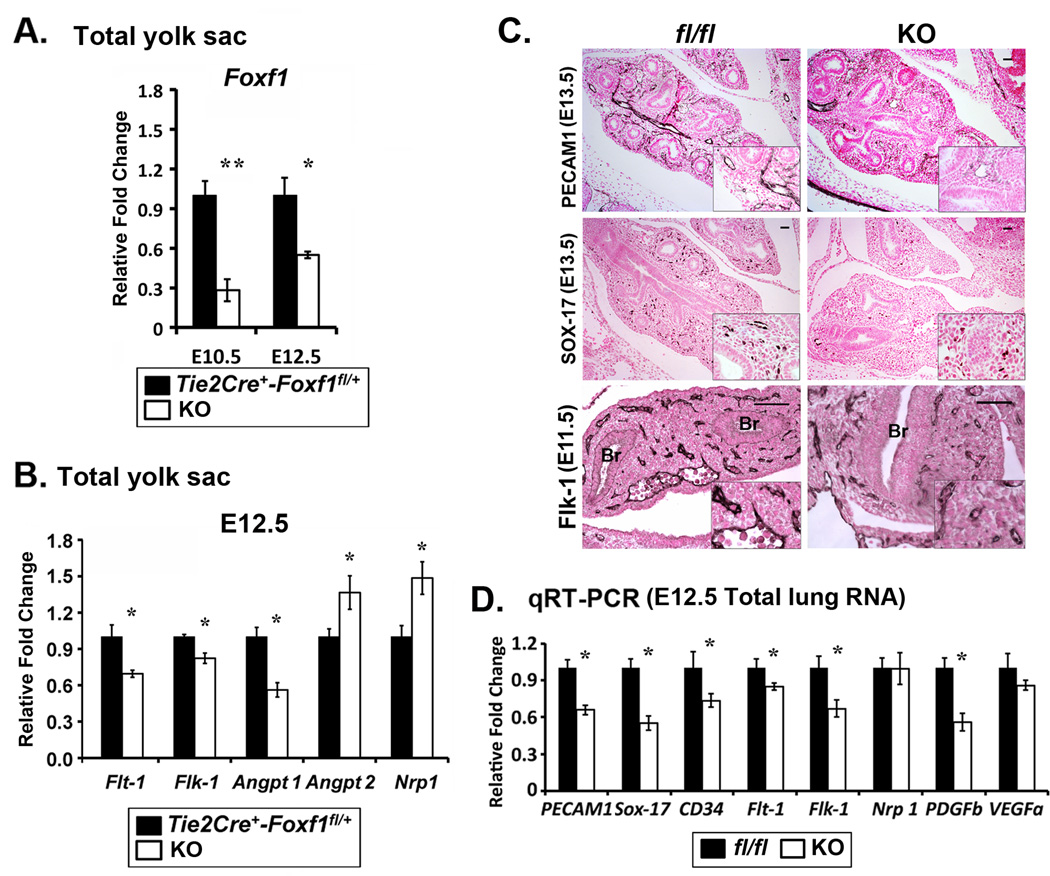
Figure 4. Altered expression of endothelial genes in the yolk sac and lung of Tie2-Cre Foxf1fl/fl embryos.
(A–B) Total RNA was prepared from yolk sacs of Tie2-Cre Foxf1fl/fl (KO) and control Tie2-Cre Foxf1fl/+ E12.5 embryos and analyzed by qRT-PCR. Decreased mRNAs of Foxf1, Flt-1, Flk-1 and Angpt 1 were found in Tie2-Cre Foxf1fl/fl yolk sacs. Angpt2 and Nrp1 mRNAs were increased (n=5). Expression levels were normalized to β-actin mRNA. (C) Immunohistochemical staining shows reduced PECAM1 and SOX-17 in lungs of FOXF1 KO embryos. The intensity of Flk1 staining was also reduced in FOXF1 KO lungs. (D) Decreased mRNAs of Pecam1, Sox-17, CD34, Flt1, Flk1 and Pdgfb were found in whole lung RNA from E12.5 KO embryos (n=6). p<0.05 is *, p<0.01 is **. Scale bars are 50µm.
FOXF1 deletion impairs the formation of pulmonary vascular plexus
Since FOXF1 deficiency is associated with reduced numbers of pulmonary capillaries in ACD/MPV infants and Foxf1+/− embryos8, 14, we examined vasculature in Tie2-Cre Foxf1fl/fl lungs. Similar to the yolk sac and placenta, diminished number of blood vessels was observed in the lung of Tie2-Cre Foxf1fl/fl embryos, as demonstrated by reduced staining for endothelial-specific markers PECAM-1 and SOX-17 (Fig. 4C). Despite impaired vasculature, epithelial tubules were still present in Tie2-Cre Foxf1fl/fl lungs (Fig. 4C). Flk1 staining and Flk1 mRNA were reduced after deletion of FOXF1 (Fig. 4C–D). Flt1, Pecam-1, Sox-17, CD34 and Pdgfb mRNAs were decreased in whole lung RNA from Tie2-Cre Foxf1fl/fl embryos (Fig. 4D), confirming the loss of pulmonary vascular plexuses.
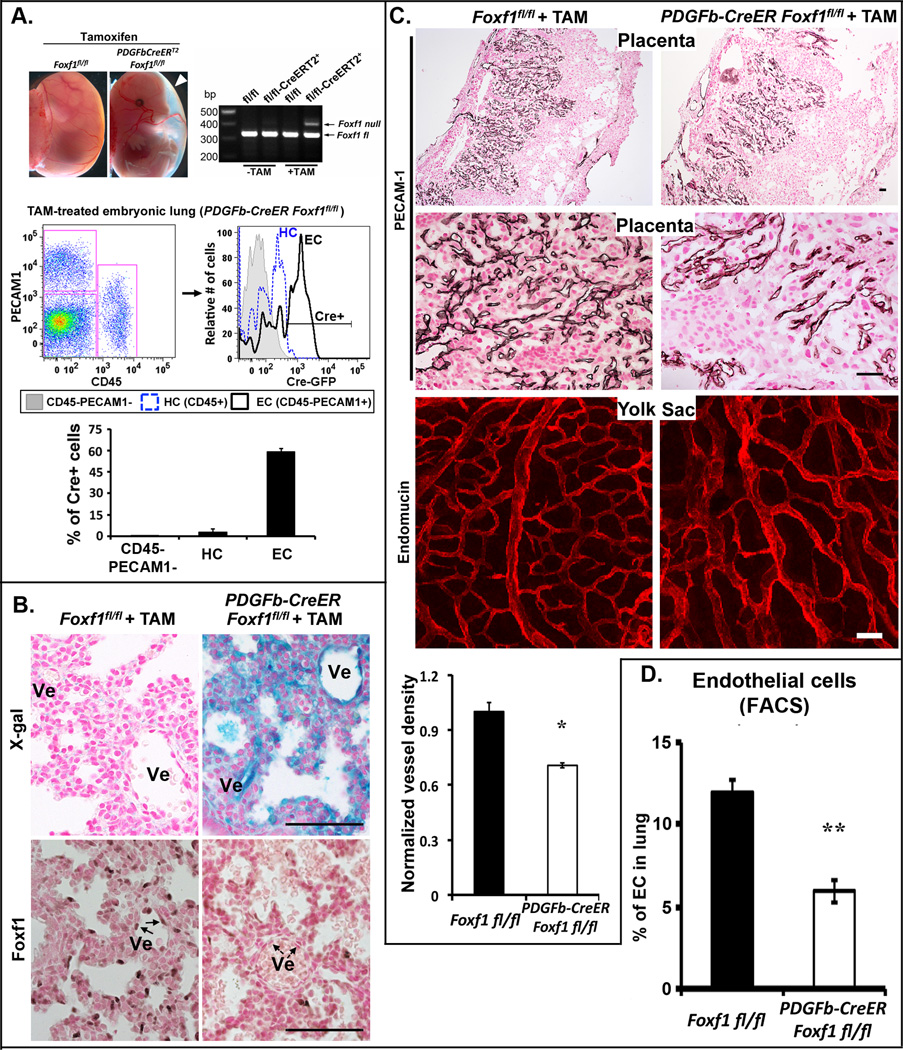
Reduced vascular branching in Pdgfb-CreER Foxf1fl/fl embryos
Pdgfb-CreER Foxf1fl/fl mouse line was generated to achieve an inducible deletion of Foxf1 from endothelial cells without targeting hematopoietic cells. To activate Cre, tamoxifen was given to pregnant females at E9.5 and embryos were harvested at E12.5. The Foxf1-null allele was detected in tamoxifen-treated Pdgfb-CreER Foxf1fl/fl mice by PCR (Fig. 5A), a finding consistent with activation of Cre by tamoxifen. Cre-mediated recombination in lung tissue was confirmed by β-gal reporter and diminished FOXF1 immunostaining (Fig. 5B). Flow cytometry showed that approximately 60% of PECAM-1+ endothelial cells and only 2% of CD45+ hematopoietic cells in the yolk sac were positive for Cre (Fig. 5A). Pdgfb-CreER Foxf1fl/fl embryos exhibited polyhydramnios (Fig. 5A), reduced vascular branching in the placenta and yolk sac (Fig. 5C) and decreased numbers of endothelial cells in the lung (Fig. 5D). Deletion of FOXF1 during the postnatal period (P0–P2) impaired retinal angiogenesis in Pdgfb-CreER Foxf1fl/fl mice (Online Fig. VI). Thus, FOXF1 stimulates angiogenesis in the developing lung, eye, placenta and yolk sac.
Figure 5. Reduced vascular branching in Pdgfb-CreER Foxf1fl/fl embryos.
(A) Pdgfb-CreER Foxf1fl/fl embryos were treated with tamoxifen at E9.5 and harvested at E12.5. Polyhydramnios in FOXF1-deficient embryos is indicated with white arrowhead. PCR of genomic tail DNA shows Cre-mediated recombination. Flow cytometry shows GFP fluorescence, which is associated with the Pdgfb-CreER transgene, in 60% of Pecam1+ endothelial cells (EC) isolated from the yolk sac. GFP is not detected in a majority of CD45+ hematopoietic cells (HC). (B) Increased β-gal activity and decreased FOXF1 staining is observed in lungs of Pdgfb-CreER Foxf1fl/fl/ R26R E17.5 embryos. (C) Immunostaining for Pecam1 and endomucin shows reduced vessel (Ve) density in the placenta and yolk sac of FOXF1-deficient embryos. Quantification is shown in bottom panel. (D) Decreased numbers of Pecam1+ endothelial cells are found in FOXF1-deficient lungs by flow cytometry (n=5). p<0.05 is *, p<0.01 is **. Scale bars are 50µm.
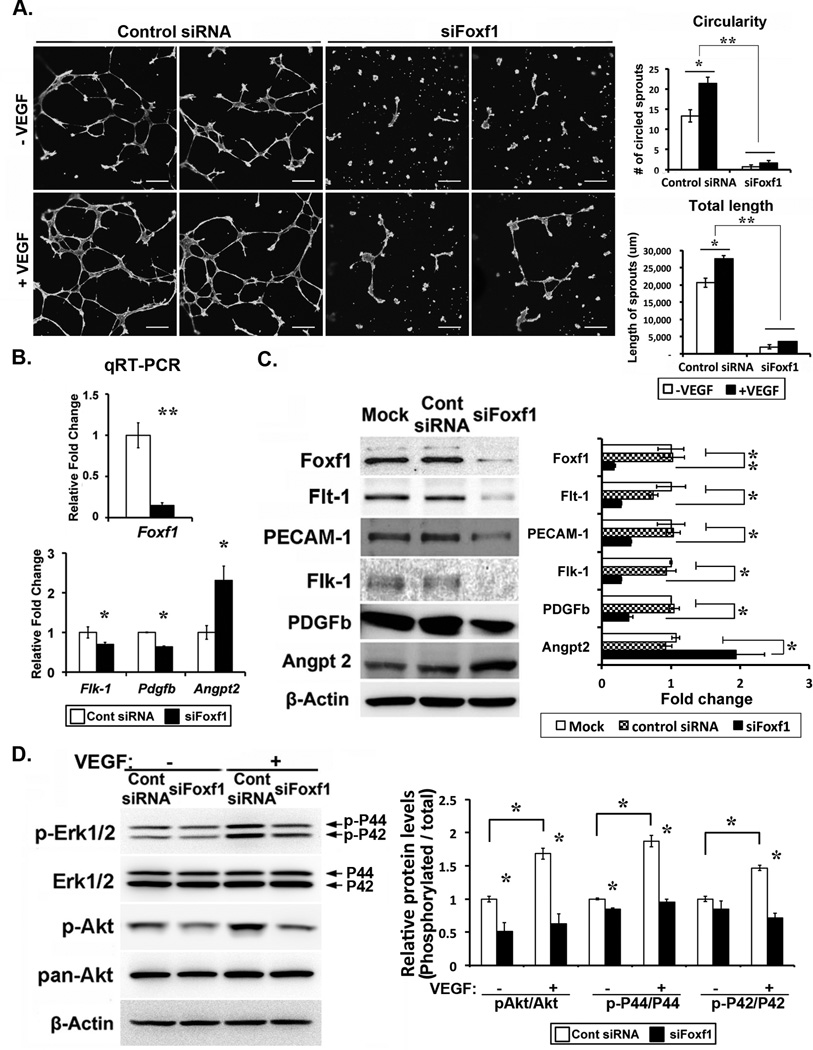
FOXF1 stimulates angiogenesis in vitro
The ability of FOXF1 to stimulate angiogenesis in endothelial cells was directly tested in vitro. siRNA transfection was used to deplete FOXF1 mRNA and protein in endothelial MFLM-91U cells (Fig. 6B–C). Depletion of FOXF1 reduced the ability of MFLM-91U cells to form vessel-like sprouts in matrigel (Fig. 6A), a common model of angiogenesis in vitro. Diminished angiogenesis in FOXF1-depleted cells was associated with reduced proteins and mRNAs of Flk1, Flt1, Pecam-1 and Pdgfb, whereas expression of Angpt2 was increased (Fig. 6B–C). After depletion of FOXF1, phosphorylation of ERK and AKT was decreased (Fig. 6D), indicating reduced VEGF signaling. Interestingly, VEGF-A ligand did not rescue the FOXF1-mediated decrease in angiogenesis (Fig. 6A and 6D), a finding consistent with reduced expression of VEGF receptors in FOXF1-depleted cells. Thus, FOXF1 promotes VEGF signaling and increases angiogenesis in vitro.
Figure 6. siRNA-mediated depletion of FOXF1 impairs angiogenesis in matrigel and inhibits VEGF signaling in cultured MFLM-91U cells.
(A) MFLM-91U cells were transfected with either FOXF1 siRNA or control non-targeting siRNA. Forty-eight hours after siRNA transfection, matrigel angiogenesis assay was performed in the presence and absence of VEGF-A(165). FOXF1 knockdown reduced the number and total length of endothelial sprouts (n=5). (B) qRT-PCR shows reduced Foxf1, Flk1 and Pdgfb mRNAs at 24 hrs after siRNA transfection. Angp2 mRNA was increased after depletion of FOXF1. (C) Altered protein levels of FOXF1, Flk1, Flt1, PECAM-1, PDGFb and Angpt2 at 48 hrs after siRNA transfection are shown by Western blots. (D) FOXF1-depleted MFLM-91U cells are resistant to VEGF-A(165) stimulation as indicated by decreases in phosphorylated Erk1/2 (pErk1/2) and phosphorylated Akt (pAkt). Quantification of Western blots is shown in right panels in C and D. p<0.05 is *, p<0.01 is **. Scale bars are 500µm.
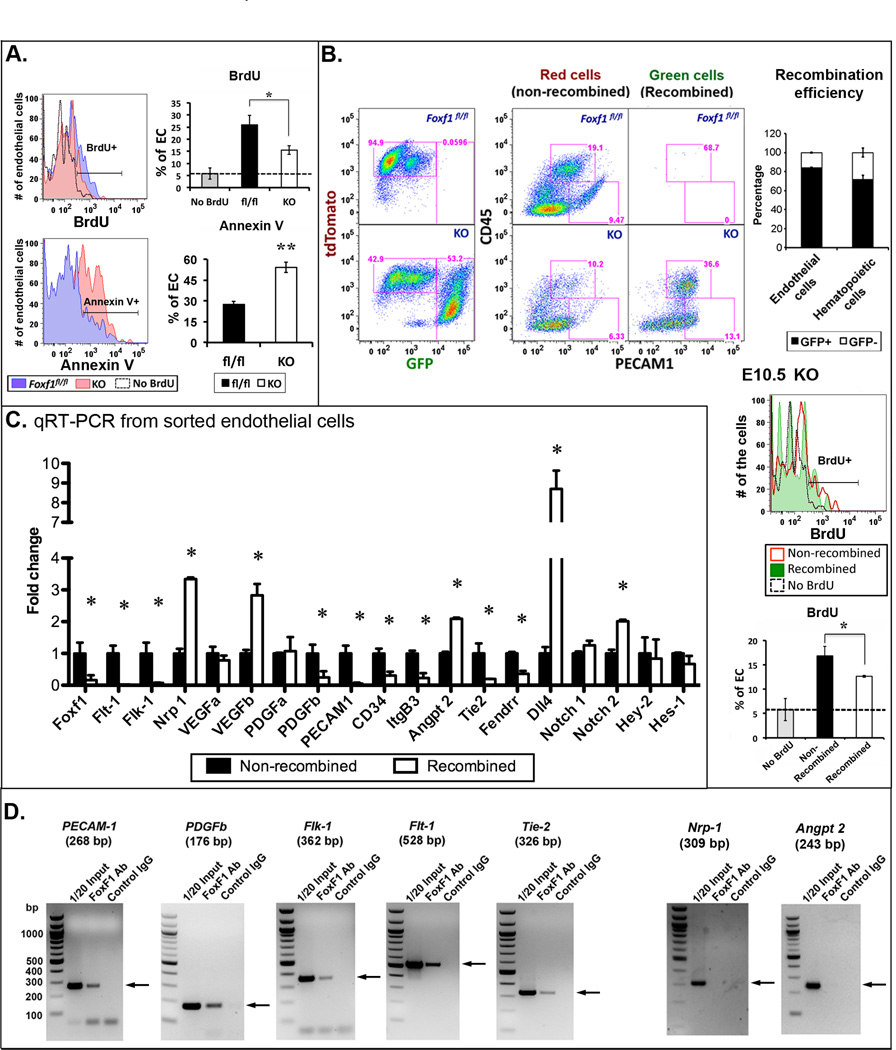
Decreased proliferation and increased apoptosis in endothelial cells of Tie2-Cre Foxf1fl/fl embryos
VEGF induces cellular proliferation and inhibits apoptosis by activating the tyrosine kinase receptor Flk1, which is present on the surface of endothelial cells3, 6. Given that Flk1 mRNA and protein were decreased in FOXF1-deficient mice (Fig. 4B–D) and cultured MFLM-91U cells (Fig. 6B–C), we next determined whether FOXF1 affects endothelial proliferation and apoptosis in vivo. Yolk sacs from E12.5 embryos were enzymatically digested and cells were stained for the endothelial marker Pecam-1 and the hematopoietic marker CD45 followed by flow cytometry. Consistent with reduced VEGF/ Flk1 signaling, DNA replication was decreased in Pecam-1+/ CD45− endothelial cells of Tie2-Cre Foxf1fl/fl yolk sacs as demonstrated by reduced BrdU incorporation into DNA (Fig. 7A). In addition, apoptosis of FOXF1-deficient endothelial cells was increased as shown by annexin V staining (Fig. 7A). Thus, reduced proliferation and increased apoptosis of endothelial cells can contribute to reduced angiogenesis and embryonic lethality in Tie2-Cre Foxf1fl/fl mice.
Figure 7. FOXF1 directly regulates expression of endothelial genes critical for angiogenesis and VEGF signaling.
(A) Reduced BrdU incorporation and increased annexin V staining is detected by flow cytometry in endothelial cells of Tie2-Cre Foxf1fl/fl E12.5 embryos. The dotted line represents non-specific staining with BrdU Abs. (B) GFP and tdTomato fluorescence were measured in endothelial (CD45−PECAM-1+) and hematopoietic cells (CD45+PECAM-1−) isolated from Tie2-Cre Foxf1fl/fl mT/mG yolk sacs. GFP is not detected in cells from control Foxf1fl/fl mT/mG yolk sacs. Efficiency of Cre-mediated recombination is shown as a percentage of GFP+ cells among total cells isolated from the yolk sac (n=5) (right panel). BrdU incorporation is decreased in GFP+ endothelial cells at E10.5 (right middle panels). (C) Endothelial cells were flow sorted from E12.5 yolk sacs. qRT-PCR was used to examine mRNAs. (D) ChIP assay shows that FOXF1 binds to promoter regions of Pecam-1, Pdgfb, Tie-2, Flt1 and Flk1 genes. FOXF1 does not bind to Nrp1 and Angpt2 promoters. Fetal endothelial MFLM-91U cells were used for ChIP. p<0.05 is *, p<0.01 is **.
FOXF1 directly regulates expression of endothelial genes critical for angiogenesis and VEGF signaling
To label endothelial cells that underwent Cre-mediated recombination, Tie2-Cre Foxf1fl/+ mice were crossed with mT/mG reporter mice that contain the LoxP-tdTomato-LoxP-GFP cassette knocked into the Rosa26 locus. The use of mTmG reporter enabled us to distinguish between Cre-targeted (GFP+) and non-targeted (tdTomato+) endothelial cells in Tie2-Cre Foxf1fl/fl/ mT/mG embryos. In control Foxf1fl/fl/ mTmG embryos, both endothelial (Pecam-1+/ CD45−) and hematopoietic cells (Pecam-1−/ CD45+) were negative for GFP but positive for tdTomato reporter, indicating the lack of Cre-mediated recombination (Fig. 7B). In contrast, 84.1 ± 0.5% of endothelial cells and 71.6 ± 4.6% of hematopoietic from Tie2-Cre Foxf1fl/fl/ mTmG embryos were positive for GFP (Fig. 7B). We next used flow cytometry-based cell sorting to isolate GFP+ and tdTomato+ endothelial cells and use these cells for qRT-PCR. Loss of Foxf1 mRNA and decreased expression of the FOXF1 target gene, integrin β320, were specifically found in GFP+ endothelial cells compared to control tdTomato+ endothelial cells (Fig. 7C). Deletion of FOXF1 efficiently reduced mRNAs of Flk1, Flt1, Pdgfb, Pecam-1, CD34, Tie2 and the non-coding RNA Fendrr, all of which are critical for embryonic vascular development5–7, 32, 33. In contrast, Angpt2, Nrp1, Dll4, Notch2 and VEGFb mRNAs were increased (Fig. 7C). There was no difference in expression levels of Notch1 and Notch target genes Hey2 and Hes1 (Fig. 7C). Finally, Chromatin Immunoprecipitation assay (ChIP) demonstrated that FOXF1 protein directly bound to promoter DNAs of Flk1, Flt1, Pdgfb, Pecam-1 and Tie2 (Fig. 7D), a finding consistent with a direct transcriptional regulation of these genes by FOXF1. FOXF1 protein did not bind to Nrp1 and Angpt2 promoters (Fig. 7D). Thus, FOXF1 directly regulates expression of endothelial genes critical for angiogenesis and VEGF signaling.
DISCUSSION
Although various vascular abnormalities were previously reported for Foxf1−/− and Foxf1+/− mouse embryos as well as for ACD/MPV infants with FOXF1 mutations9, 13, 14, cellular origins and molecular mechanisms of these developmental defects remain uncharacterized. In the present study, we showed that FOXF1 transcription factor is expressed in endothelial cells and that endothelial deletion of FOXF1 causes a variety of developmental defects, including impaired vasculature in the yolk sac, placenta, lung and retina. These data demonstrate that FOXF1 functions in a cell-autonomous manner to induce formation of embryonic vasculature. This hypothesis is consistent with diminished angiogenesis in FOXF1-deficient endothelial MFLM-91U cells in vitro. Interestingly, during embryogenesis FOXF1 is abundantly expressed in endothelial cells of capillaries and small blood vessels of the yolk sac, placenta and lung, but found only in a subset of endothelial cells of vena cava and pulmonary vein. In adult mice, FOXF1 was absent from endothelium of large pulmonary vessels but present in pulmonary capillaries34. Therefore, FOXF1 is not a marker of endothelial cells. It is possible that FOXF1 expression in endothelial cells depends on proliferation or differentiation status, or reflects endothelial responses to various stimuli.
While Tie2-Cre Foxf1fl/fl mutant mice exhibited a complex developmental phenotype, we believe that vascular insufficiency in the yolk sac and placenta were primary causes of growth retardation and embryonic lethality in Tie2-Cre Foxf1fl/fl embryos. This conclusion is based on the fact that diminished branching of blood vessels was found at E10.5, whereas as other developmental abnormalities, such as cardiac defects and polyhydramnios, occurred after E13.5. Vascular insufficiency in the yolk sac and placenta may alter embryonic circulation in FOXF1 mutants, causing secondary heart defects and polyhydramnios and contributing to embryonic death. Interestingly, FOXF1 is expressed in the cardiac cushion and important for mesenchyme migration (20 and this manuscript). Therefore, interventricular septal defect in Tie2-Cre Foxf1fl/fl embryos can be a direct consequence of FOXF1 deletion from mesenchymal cells of cardiac cushion, which is critical for formation of interventricular septum and cardiac valves. FOXF1 was not detected in hematopoietic cells that are targeted by the Tie2-Cre transgene in our mouse model and therefore, it is unlikely that FOXF1 deletion in hematopoietic cell lineages contributed to the vascular phenotype in FOXF1 mutant mice. However, we can not exclude the possibility that FOXF1 is expressed in rare population(s) of hematopoietic progenitors and targeting these cells contributed to the phenotype in Tie2-Cre Foxf1fl/fl embryos. Interestingly, the Pdgfb-CreER transgene, which is more specific to endothelial cells compared to the Tie2-Cre22, caused similar defects in vascular development, suggesting that endothelial cells are the main cellular targets of FOXF1. It is also possible that FOXF1 deletion reduces the number of circulating endothelial progenitors or mesenchymal stem cells, contributing to vascular insufficiency in FOXF1-deficient embryos. Previous studies demonstrated that Foxc2 and Foxc1 genes are critical for vascular development in zebrafish and mice5, 32. We found no significant differences in expression of FOXC2 and FOXC1 after deletion of FOXF1. Since FOXC proteins and FOXF1 have high homology in their DNA binding domains, it is possible that FOXC1/2 can compensate for the loss of FOXF1 from endothelial cells by regulating similar target genes.
Published studies reported a direct correlation between FOXF1 levels and the number of pulmonary capillaries during lung development14 and lung injury16. In the present study, we used in vivo and in vitro models to demonstrate that FOXF1 induces angiogenesis, endothelial proliferation and VEGF signaling through transcriptional activation of VEGF receptors Flk1 and Flt1 (Fig. 7). Reduction of Flk1 and Flt1 in FOXF1 mutants is consistent with the role of these genes in stimulating proliferation and differentiation of endothelial cells, leading to formation and maturation of blood vessels3, 4. In addition to VEGF signaling, FOXF1 may influence other signaling pathways critical for endothelial development, such as PDGF and Angpt/ Tie2 pathways. Reduced expression of Pdgfb and Tie2 may account for decreased proliferation and increased apoptosis of endothelial cells in FOXF1-deficient embryos. Our data also suggest that FOXF1 stimulates angiogenesis through transcriptional activation of integrin-β3, pecam-1 and lncRNA Fendrr, all of which are critical for vascular development5–7, 33. Altogether, disruption of several key EC regulators can contribute to the FOXF1 phenotype. Since FOXF1 is a downstream target of the Shh signaling pathway15, 18, 19, it is possible that FOXF1 mediates cross-talk between the Shh and VEGF pathways during development of embryonic vasculature. Our results suggest that inability of FOXF1-deficient endothelial cells to respond to VEGF, PDGF and Angpt/Tie2 signaling is a key mechanism in development of alveolar capillary dysplasia in ACD/MPV fetuses and infants harboring inactivating mutations in the FOXF1 gene locus.
Supplementary Material
Novelty and Significance.
What Is Known?
Inactivating mutations in the FOXF1 gene are found in 40% of patients with Alveolar Capillary Dysplasia with Misalignment of Pulmonary Veins (ACD/MPV).
Haploinsufficiency of the Foxf1 gene causes alveolar capillary dysplasia and developmental defects in lung, intestinal and gall bladder morphogenesis in mice.
FOXF1 is a transcription factor, which is found in multiple cell types, including endothelial cells.
What New Information Does This Article Contribute?
Disruption of Foxf1 gene in endothelial cells caused embryonic lethality, growth retardation and cardiovascular abnormalities.
Disruption of Foxf1 reduced cell proliferation, increased apoptosis and inhibited the Vascular Endothelial Growth Factor (VEGF) signaling pathway in endothelial cells.
FOXF1 induces transcription of VEGF receptor genes Flk1 and Flt1.
Inactivating mutations in the FOXF1 gene were recently found in 40% of human patients with ACD/MPV. The molecular mechanisms by which these mutations cause vascular defects remain unknown. In the present study, we used transgenic mice with endothelial-specific inactivation of the Foxf1 gene to demonstrate that FOXF1 is critical for formation of embryonic vasculature. FOXF1 stimulates endothelial proliferation and promotes the VEGF signaling pathway in embryonic endothelial cells through direct transcriptional activation of VEGF receptor genes. Our results suggest that inability of FOXF1-deficient endothelial cells to respond to VEGF signaling is a key mechanism in development of alveolar capillary dysplasia in ACD/MPV fetuses and infants harboring inactivating mutations in the FOXF1 gene. Pharmacological agents that stimulate or stabilize FOXF1 protein might serve as promising therapeutic agents in patients with ACD/MPV caused by heterozygous loss-of-function mutations in FOXF1 gene.
Acknowledgments
We thank Y. Zhang for excellent technical assistance.
SOURCES OF FUNDING
This work was supported by NIH Grants HL 84151 (to V. V. K.) and CA 142724 (T. V. K.).
Nonstandard Abbreviations and Acronyms
- Cre
Cre recombinase
- E
Embryonic day
- FOXF1
Forkhead Box transcription factor F1
- VEGF
Vascular Endothelial Growth Factor
- ACD/MPV
Alveolar Capillary Dysplasia with Misalignment of Pulmonary Veins
- HSV-TK
Herpes simplex virus-thymidine kinase
- EC
Endothelial cells
Footnotes
DISCLOSURES
No relationships to disclose
REFERENCES
- 1.Carmeliet P, Ferreira V, Breier G, Pollefeyt S, Kieckens L, Gertsenstein M, Fahrig M, Vandenhoeck A, Harpal K, Eberhardt C, Declercq C, Pawling J, Moons L, Collen D, Risau W, Nagy A. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature. 1996;380:435–439. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- 2.Ferrara N, Carver-Moore K, Chen H, Dowd M, Lu L, O'Shea KS, Powell-Braxton L, Hillan KJ, Moore MW. Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature. 1996;380:439–442. doi: 10.1038/380439a0. [DOI] [PubMed] [Google Scholar]
- 3.Shalaby F, Rossant J, Yamaguchi TP, Gertsenstein M, Wu XF, Breitman ML, Schuh AC. Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature. 1995;376:62–66. doi: 10.1038/376062a0. [DOI] [PubMed] [Google Scholar]
- 4.Fong GH, Rossant J, Gertsenstein M, Breitman ML. Role of the Flt-1 receptor tyrosine kinase in regulating the assembly of vascular endothelium. Nature. 1995;376:66–70. doi: 10.1038/376066a0. [DOI] [PubMed] [Google Scholar]
- 5.De Val S, Black BL. Transcriptional control of endothelial cell development. Developmental cell. 2009;16:180–195. doi: 10.1016/j.devcel.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arora R, Papaioannou VE. The murine allantois: a model system for the study of blood vessel formation. Blood. 2012;120:2562–2572. doi: 10.1182/blood-2012-03-390070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tiozzo C, Carraro G, Al Alam D, Baptista S, Danopoulos S, Li A, Lavarreda-Pearce M, Li C, De Langhe S, Chan B, Borok Z, Bellusci S, Minoo P. Mesodermal Pten inactivation leads to alveolar capillary dysplasia- like phenotype. The Journal of clinical investigation. 2012;122:3862–3872. doi: 10.1172/JCI61334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bishop NB, Stankiewicz P, Steinhorn RH. Alveolar capillary dysplasia. American journal of respiratory and critical care medicine. 2011;184:172–179. doi: 10.1164/rccm.201010-1697CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stankiewicz P, Sen P, Bhatt SS, Storer M, Xia Z, Bejjani BA, Ou Z, Wiszniewska J, Driscoll DJ, Maisenbacher MK, Bolivar J, Bauer M, Zackai EH, McDonald-McGinn D, Nowaczyk MM, Murray M, Hustead V, Mascotti K, Schultz R, Hallam L, McRae D, Nicholson AG, Newbury R, Durham-O'Donnell J, Knight G, Kini U, Shaikh TH, Martin V, Tyreman M, Simonic I, Willatt L, Paterson J, Mehta S, Rajan D, Fitzgerald T, Gribble S, Prigmore E, Patel A, Shaffer LG, Carter NP, Cheung SW, Langston C, Shaw-Smith C. Genomic and genic deletions of the FOX gene cluster on 16q24.1 and inactivating mutations of FOXF1 cause alveolar capillary dysplasia and other malformations. Am J Hum Genet. 2009;84:780–791. doi: 10.1016/j.ajhg.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garabedian MJ, Wallerstein D, Medina N, Byrne J, Wallerstein RJ. Prenatal Diagnosis of Cystic Hygroma related to a Deletion of 16q24.1 with Haploinsufficiency of FOXF1 and FOXC2 Genes. Case reports in genetics. 2012;2012:490408. doi: 10.1155/2012/490408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peterson RS, Lim L, Ye H, Zhou H, Overdier DG, Costa RH. The winged helix transcriptional activator HFH-8 is expressed in the mesoderm of the primitive streak stage of mouse embryos and its cellular derivatives. Mech. Dev. 1997;69:53–69. doi: 10.1016/s0925-4773(97)00153-6. [DOI] [PubMed] [Google Scholar]
- 12.Kalinichenko VV, Zhou Y, Bhattacharyya D, Kim W, Shin B, Bambal K, Costa RH. Haploinsufficiency of the Mouse Forkhead Box f1 Gene Causes Defects in Gall Bladder Development. The Journal of biological chemistry. 2002;277:12369–12374. doi: 10.1074/jbc.M112162200. [DOI] [PubMed] [Google Scholar]
- 13.Mahlapuu M, Ormestad M, Enerback S, Carlsson P. The forkhead transcription factor Foxf1 is required for differentiation of extra-embryonic and lateral plate mesoderm. Development. 2001;128:155–166. doi: 10.1242/dev.128.2.155. [DOI] [PubMed] [Google Scholar]
- 14.Kalinichenko VV, Lim L, Beer-Stoltz D, Shin B, Rausa FM, Clark J, Whitsett JA, Watkins SC, Costa RH. Defects in Pulmonary Vasculature and Perinatal Lung Hemorrhage in Mice Heterozygous Null for the Forkhead Box f1 transcription factor. Developmental biology. 2001;235:489–506. doi: 10.1006/dbio.2001.0322. [DOI] [PubMed] [Google Scholar]
- 15.Mahlapuu M, Enerbäck S, Carlsson P. Haploinsufficiency of the forkhead gene Foxf1, a target for sonic hedgehog signaling, causes lung and foregut malformations. Development. 2001;128:2397–2406. doi: 10.1242/dev.128.12.2397. [DOI] [PubMed] [Google Scholar]
- 16.Kalinichenko VV, Zhou Y, Shin B, Beer-Stoltz D, Watkins SC, A WJ, Costa RH. Wild Type Levels of the Mouse Forkhead Box f1 Gene are Essential for Lung Repair. Am J Physiol Lung Cell Mol Physiol. 2002;282:L1253–L1265. doi: 10.1152/ajplung.00463.2001. [DOI] [PubMed] [Google Scholar]
- 17.Kalinichenko VV, Bhattacharyya D, Zhou Y, Gusarova GA, Kim W, Shin B, Costa RH. Foxf1+/− Mice Exhibit Defective Stellate Cell Activation and Abnormal Liver Regeneration Following CCl4 Injury. Hepatology. 2003;37:107–117. doi: 10.1053/jhep.2003.50005. [DOI] [PubMed] [Google Scholar]
- 18.Madison BB, McKenna LB, Dolson D, Epstein DJ, Kaestner KH. FoxF1 and FoxL1 link hedgehog signaling and the control of epithelial proliferation in the developing stomach and intestine. The Journal of biological chemistry. 2009;284:5936–5944. doi: 10.1074/jbc.M808103200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szafranski P, Dharmadhikari AV, Brosens E, Gurha P, Kolodziejska KE, Zhishuo O, Dittwald P, Majewski T, Mohan KN, Chen B, Person RE, Tibboel D, de Klein A, Pinner J, Chopra M, Malcolm G, Peters G, Arbuckle S, Guiang SF, Hustead VA, 3rd, Jessurun J, Hirsch R, Witte DP, Maystadt I, Sebire N, Fisher R, Langston C, Sen P, Stankiewicz P. Small noncoding differentially methylated copy-number variants, including lncRNA genes, cause a lethal lung developmental disorder. Genome research. 2013;23:23–33. doi: 10.1101/gr.141887.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malin D, Kim IM, Boetticher E, Kalin TV, Ramakrishna S, Meliton L, Ustiyan V, Zhu X, Kalinichenko VV. Forkhead box F1 is essential for migration of mesenchymal cells and directly induces integrin-beta3 expression. Molecular and cellular biology. 2007;27:2486–2498. doi: 10.1128/MCB.01736-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalinichenko VV, Gusarova GA, Kim I-M, Shin B, Yoder HM, Clark J, Sapozhnikov AM, Whitsett JA, Costa RH. Foxf1 Haploinsufficiency Reduces Notch-2 Signaling during Mouse Lung Development. Am J Physiol Lung Cell Mol Physiol. 2004;286:L521–L530. doi: 10.1152/ajplung.00212.2003. [DOI] [PubMed] [Google Scholar]
- 22.Claxton S, Kostourou V, Jadeja S, Chambon P, Hodivala-Dilke K, Fruttiger M. Efficient, inducible Cre-recombinase activation in vascular endothelium. Genesis. 2008;46:74–80. doi: 10.1002/dvg.20367. [DOI] [PubMed] [Google Scholar]
- 23.Sandell LL, Iulianella A, Melton KR, Lynn M, Walker M, Inman KE, Bhatt S, Leroux-Berger M, Crawford M, Jones NC, Dennis JF, Trainor PA. A phenotype-driven ENU mutagenesis screen identifies novel alleles with functional roles in early mouse craniofacial development. Genesis. 2011;49:342–359. doi: 10.1002/dvg.20727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalin TV, Wang IC, Meliton L, Zhang Y, Wert SE, Ren X, Snyder J, Bell SM, Graf L, Jr, Whitsett JA, Kalinichenko VV. Forkhead Box m1 transcription factor is required for perinatal lung function. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19330–19335. doi: 10.1073/pnas.0806748105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang IC, Snyder J, Zhang Y, Lander J, Nakafuku Y, Lin J, Chen G, Kalin TV, Whitsett JA, Kalinichenko VV. Foxm1 Mediates Cross Talk between Kras/Mitogen-Activated Protein Kinase and Canonical Wnt Pathways during Development of Respiratory Epithelium. Molecular and cellular biology. 2012;32:3838–3850. doi: 10.1128/MCB.00355-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalin TV, Meliton L, Meliton AY, Zhu X, Whitsett JA, Kalinichenko VV. Pulmonary mastocytosis and enhanced lung inflammation in mice heterozygous null for the Foxf1 gene. American journal of respiratory cell and molecular biology. 2008;39:390–399. doi: 10.1165/rcmb.2008-0044OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ren X, Shah TA, Ustiyan V, Zhang Y, Shinn J, Chen G, Whitsett JA, Kalin TV, Kalinichenko VV. FOXM1 promotes allergen-induced goblet cell metaplasia and pulmonary inflammation. Molecular and cellular biology. 2013;33:371–386. doi: 10.1128/MCB.00934-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang IC, Zhang Y, Snyder J, Sutherland MJ, Burhans MS, Shannon JM, Park HJ, Whitsett JA, Kalinichenko VV. Increased expression of FoxM1 transcription factor in respiratory epithelium inhibits lung sacculation and causes Clara cell hyperplasia. Developmental biology. 2010;347:301–314. doi: 10.1016/j.ydbio.2010.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ustiyan V, Wang IC, Ren X, Zhang Y, Snyder J, Xu Y, Wert SE, Lessard JL, Kalin TV, Kalinichenko VV. Forkhead box M1 transcriptional factor is required for smooth muscle cells during embryonic development of blood vessels and esophagus. Developmental biology. 2009;336:266–279. doi: 10.1016/j.ydbio.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren X, Zhang Y, Snyder J, Cross ER, Shah TA, Kalin TV, Kalinichenko VV. Forkhead box M1 transcription factor is required for macrophage recruitment during liver repair. Molecular and cellular biology. 2010;30:5381–5393. doi: 10.1128/MCB.00876-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suri C, Jones PF, Patan S, Bartunkova S, Maisonpierre PC, Davis S, Sato TN, Yancopoulos GD. Requisite role of angiopoietin-1, a ligand for the TIE2 receptor, during embryonic angiogenesis. Cell. 1996;87:1171–1180. doi: 10.1016/s0092-8674(00)81813-9. [DOI] [PubMed] [Google Scholar]
- 32.De Val S, Chi NC, Meadows SM, Minovitsky S, Anderson JP, Harris IS, Ehlers ML, Agarwal P, Visel A, Xu SM, Pennacchio LA, Dubchak I, Krieg PA, Stainier DY, Black BL. Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell. 2008;135:1053–1064. doi: 10.1016/j.cell.2008.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, Macura K, Blass G, Kellis M, Werber M, Herrmann BG. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Developmental cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalinichenko VV, Lim L, Shin B, Costa RH. Differential Expression of Forkhead Box Transcription Factors Following Butylated Hydroxytoluene Lung Injury. American journal of physiology. 2001;280:L695–L704. doi: 10.1152/ajplung.2001.280.4.L695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.