Abstract
Recent reports suggest that human genogroup II genotype 17 (GII.17) noroviruses are increasing in prevalence. We analyzed the evolutionary changes of three GII.17 capsid protruding (P) domains. We found that the GII.17 P domains had little cross-reactivity with antisera raised against the dominant GII.4 strains. X-ray structural analysis of GII.17 P domains from 2002 to 2014 and 2015 suggested that surface-exposed substitutions on the uppermost part of the P domain might have generated a novel 2014-2015 GII.17 variant.
TEXT
Human noroviruses are the dominant cause of outbreaks of acute gastroenteritis. In the past decade, genogroup II genotype 4 (GII.4) norovirus strains were those mostly responsible for epidemic outbreaks (1–3). However, a GII.17 variant norovirus was found recently to cause an alarming number of outbreaks in certain parts of Asia in 2014 to 2015 (4–8). Before this time, the GII.17 norovirus was only a minor cause of infections, although it was first described in 1978 (9). Researchers are now reporting that the GII.17 variant is emerging in other parts of the world, and molecular epidemiologists have warned that the GII.17 norovirus might replace the predominant GII.4 norovirus (10).
Noroviruses have a single-stranded, positive-sense RNA genome of 7.5 to 7.7 kb. The genome contains three open reading frames (ORFs). The first ORF (ORF1) encodes nonstructural proteins, including the RNA-dependent RNA polymerase (RdRp), ORF2 encodes capsid protein (VP1), and ORF3 encodes a minor capsid protein (VP2) (11). The X-ray crystal structure of the prototype (GI.1) virus-like particles (VLPs) identified two domains, the shell (S) domain and the protruding (P) domain, which can be further subdivided into P1 and P2 subdomains (12). The S domain surrounds the viral RNA, whereas the P domain contains the determinants for cell attachment and antigenicity. Human noroviruses are known to bind histo-blood group antigens (HBGAs), and the interaction is thought to be important for infection (13–16). Two recent reports indicated that, similarly to other GII noroviruses, the recent GII.17 strains bind a panel of different HBGA types (4, 8).
Human noroviruses are believed to evolve in a manner similar to that seen with influenza viruses, where new norovirus genotype variants emerge every other year. Evolving strains with an ∼5% amino acid change can reinfect the same individual (17). Data on short- and long-term immunity to human norovirus are still unclear, although vaccines are currently been tested in clinical trials (18, 19). Unfortunately, the vaccines, which can include VLPs or P domains (20, 21), may not protect from antigenically divergent strains (18–21). Here, we report the first X-ray crystal structure of GII.17 norovirus P domains and describe the cross-reactivities with antibodies (Abs) raised against GII.4 strains, which are targeted by the current vaccines in clinical trials.
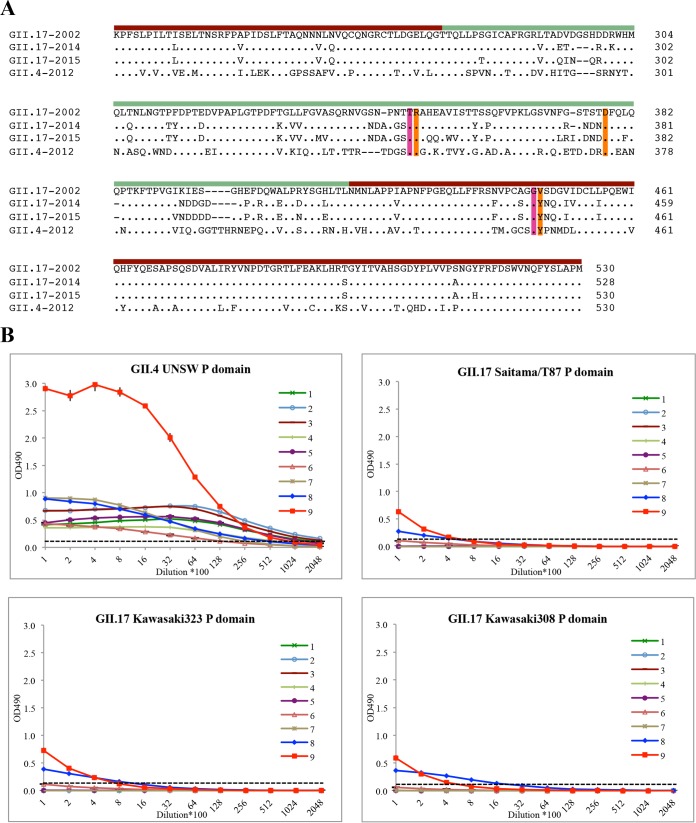
Three different GII.17 norovirus strains were selected for antibody binding and structural analysis: a nonprevalent 2002 strain (Saitama/T87; GenBank accession number KJ196286), a prevalent 2014 variant strain (Kawasaki323; AB983218), and a prevalent 2015 variant strain (Kawasaki308; LC037415) (Fig. 1A). The GII.17 P domains (Fig. 1A) were expressed and purified as previously described (22). An antigen enzyme-linked immunosorbent assay (ELISA) was used to determine the cross-reactivities of GII.17 P domains with eight different monoclonal antibodies (MAbs) and one polyclonal antibody (PAb) raised against GII.4 strains using an established method (23). The titer was expressed as the reciprocal of the highest dilution of antiserum giving an absorbance value at 490 nm (OD490) of >0.15, which was three times the blank value (Fig. 1B). The antibodies reacted to the GII.4 P domains at high titers (>12,800 dilution). Seven of nine antibodies (numbers 1 to 7) showed no cross-reactivity against the GII.17 P domains. Two antibodies (numbers 8 and 9) weakly cross-reacted to the GII.17 P domains, i.e., at about 400 to 800 dilutions. The weak cross-reactivities with the two antibodies indicated that the GII.4 and GII.17 P domains are antigenically distinct. Considering that GII.4 and GII.17 P domains had ∼60% amino acid identity, this result is similar to those of an earlier cross-reactivity study that showed that most GII genotypes were antigenically distinct (23). Based on these findings, it is likely that the current vaccines might not provide protection for the GII.17 strains.
FIG 1.
An amino acid alignment of norovirus GII.17 and GII.4 P domains and antigen ELISA of GII.4 and GII.17 P domains. (A) The P domain amino acid sequences of three GII.17 variants (isolated in 2002, 2014, and 2015) and GII.4 Sydney2012 were aligned using Clustal Omega. Among the GII.17 P domains, the P1 subdomain (ruby bar) was more conserved than the P2 subdomain (pale green bar). The GII.4 main-chain (magenta) and side-chain (orange) residues interacting with the fucose moiety of HBGAs were mostly conserved in the GII.17 P domains. (B) Plates were coated with 10 μg/ml of Saitama/T87, Kawasaki323, Kawasaki308, or Sydney2012 (University of New South Wales [UNSW]) (24) P domains, and detection was performed with serially diluted antibodies. The antibodies used in this study were as follows: 1 (MAb 2H2, raised against Minerva GII.4 VLPs; starting concentration, 2.4 mg/ml), 2 (MAb 4933, raised against Minerva GII.4 VLPs; 3.4 mg/ml), 3 (MAb 564, raised against Minerva GII.4 VLPs; 2.6 mg/ml), 4 (MAb 10E2, raised against Minerva GII.4 VLPs; 2.4 mg/ml), 5 (MAb 6C10, raised against Minerva GII.4 VLPs; 2.0 mg/ml), 6 (MAb SM875 antigen, raised against Sydney2012 P domain; 1.4 mg/ml), 7 (MAb SM875 antigen, raised against Sydney2012 P domain; 0.6 mg/ml), 8 (MAb SM874-CM355, raised against 2006 Saga GII.4 P domain; 1.0 mg/ml), and 9 (PAb, raised against 2012 NSW VLPs; 2.0 mg/ml) (24). All experiments were performed in triplicate (error bars shown), and the cutoff was at an optical density at 490 nm (OD490) of >0.15 (dashed line; i.e., 3 times the value of ∼0.05 for the average blank) (23).
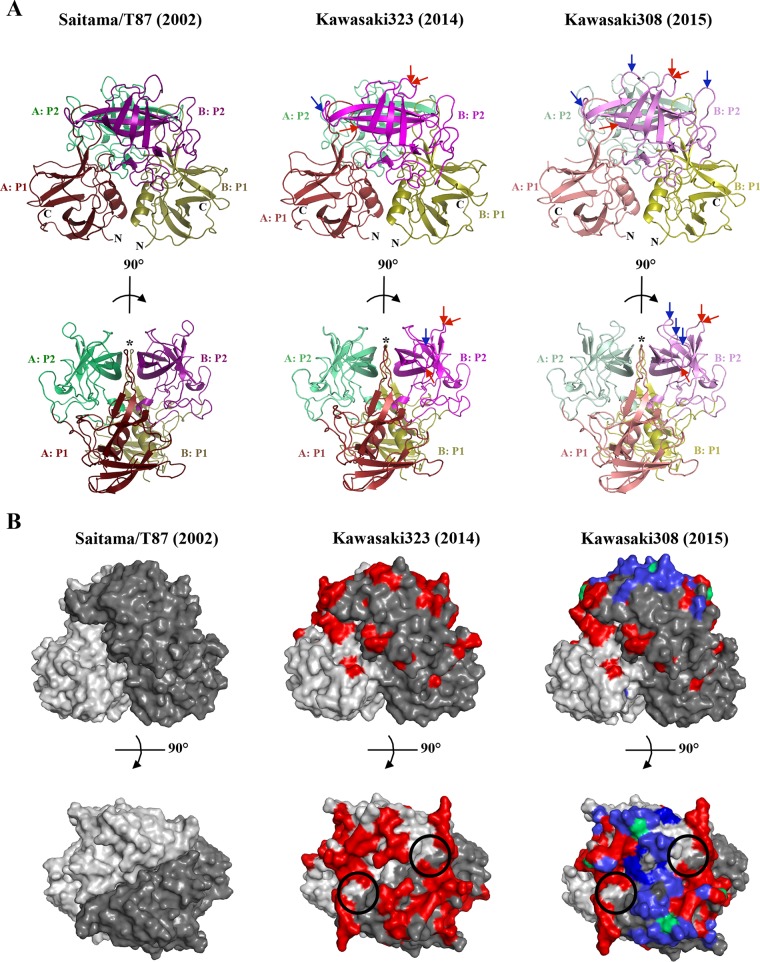
To characterize the structural changes among the three GII.17 strains, we determined the X-ray crystal structures of the P domains. All three P domains formed rectangular plate-like crystals by a hanging-drop diffusion method in a crystallization solution containing 0.2 M MgCl2, 20% (wt/vol) polyethylene glycol (PEG 8000), and 0.1 M Tris-HCl (pH 8.5). Diffraction data were processed as previously described (24). Briefly, diffraction data were processed and scaled using XDS (25). Molecular replacement was performed in PHASER (26). The search model used for molecular replacement was a homology model of the GII.17 P domain generated by homology modeling server SWISS-MODEL (27) using norovirus GII.10 (PDB accession code 3ONU), which shared 70% sequence identity. The structure was refined in PHENIX (28) with multiple rounds of manual model building in COOT (29) and validated with Molprobity (30) and PDBePISA (31).
The P domain of Saitama/T87 crystallized as a monomer per asymmetric unit in space group C2221, whereas Kawasaki323 and Kawasaki308 crystallized as a dimer in space groups P43212 and P212121, respectively. All three GII.17 P domains had well-defined electron density. Data collection and refinement statistics are provided in Table 1. The overall secondary structure of the GII.17 P domains was equivalent to those of other human norovirus P domains (Fig. 2A). For example, the P1 subdomain comprised β-sheets and one α-helix, while the P2 subdomain contained six antiparallel β-strands that formed a barrel-like structure. The P1-interface loop, which interacts with HBGAs (24), was also identical in size and orientation to those of the epidemic GII.4 strains.
TABLE 1.
Data collection and refinement statistics of GII.17 P domainsa
| Parameter | Result(s)b |
||
|---|---|---|---|
| Saitama/T87 (5F4J) | Kawasaki323 (5F4M) | Kawasaki308 (5F4O) | |
| Data collection | |||
| Space group | C2221 | P43212 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 73.30, 100.75, 83.37 | 121.51, 121.51, 156.6 | 75.72, 86.97, 97.35 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution range (Å) | 48.31–1.93 (2.00–1.93) | 19.61–2.24 (2.32–2.24) | 43.48–1.58 (1.64–1.58) |
| Rmerge | 9.73 (77.49) | 16.55 (86.52) | 6.70 (68.69) |
| I/σI | 11.32 (1.92) | 11.05 (2.50) | 11.75 (1.71) |
| Completeness (%) | 99.18 (96.80) | 99.17 (91.61) | 96.57 (96.33) |
| Redundancy | 4.6 (4.6) | 8.8 (8.1) | 3.9 (3.7) |
| Refinement | |||
| Resolution range (Å) | 48.31–1.93 | 19.61–2.27 | 43.48–1.59 |
| No. of reflections | 23,273 | 56,183 | 85,678 |
| Rwork/Rfree | 16.62/19.75 | 15.94/18.64 | 15.14/17.40 |
| No. of atoms | |||
| Total | 2,528 | 5,145 | 5,414 |
| Protein | 2,327 | 4,793 | 4,813 |
| Ligand/ion | 8 | 8 | 38 |
| Water | 193 | 344 | 563 |
| Avg B factors (Å2) | |||
| Protein | 32.60 | 38.90 | 21.20 |
| Ligand/ion | 33.80 | 35.80 | 29.10 |
| Water | 34.10 | 38.30 | 28.30 |
| RMSD | |||
| Bond length (Å) | 0.006 | 0.007 | 0.008 |
| Bond angle (°) | 0.99 | 1.08 | 1.19 |
The data set was collected from single crystal. RMSD, root mean square deviation.
Values in parentheses are for highest-resolution shell.
FIG 2.
X-ray crystal structures of GII.17 P domains. (A) Cartoon representation of the GII.17 P dimers. The 2002 Saitama/T87 P domain contained one monomer per asymmetric unit (dimer shown) and was subdivided into P1 (chain A, dark ruby; chain B, dark olive) and P2 (chain A, dark lime green; chain B, dark magenta); the 2014 Kawasaki323 P domain apo structure was subdivided into P1 (chain A, ruby; chain B, olive) and P2 (chain A, lime green; chain B, magenta); and the 2015 Kawasaki308 P domain apo structure was subdivided into P1 (chain A, light ruby; chain B, light olive) and P2 (chain A, light lime green; chain B, pink). Insertions (blue arrows) and deletions (red arrows) were located in the uppermost part of the P dimer. The P1 interface loop (marked with a five-pointed star) was identical in size to those of the GII.4 P domains. (B) Amino acid substitutions are highlighted on GII.17 P dimers (side view [top panel] and top view [bottom panel]), showing changes from 2002 to 2014 (red); changes from 2014 to 2015 (blue); and conformations from 2002 that reappeared (green). The circles represent the equivalent GII.4 HBGA binding pocket (24) on the GII.17 P domains.
Structural evolution of the three GII.17 P domains from 2002 to 2014 and 2015 indicated that most amino acid substitutions occurred in P2 subdomain, while the P1 subdomain remained relatively conserved (Fig. 2B). Furthermore, all insertions and deletions were located in the uppermost part of the P2 subdomain (Fig. 2A). Of significance, an insertion in the GII.4 P2 subdomain after 1998 likely resulted in the emergence of current epidemic GII.4 variant strains (1–3). Interestingly, in the case of these GII.17 P domains, there were three deletions and one insertion from 2002 to 2014 and two insertions from 2014 to 2015. These results suggested that the GII.17 P domains might have evolved from a nonprevalent 2002 strain into a prevalent 2014-2015 GII.17 variant strain. Furthermore, a recent study estimated that the number GII.17 capsid nucleotide substitutions per site per year was 1 order of magnitude higher than the GII.4 rate, possibly allowing their rapid emergence (8).
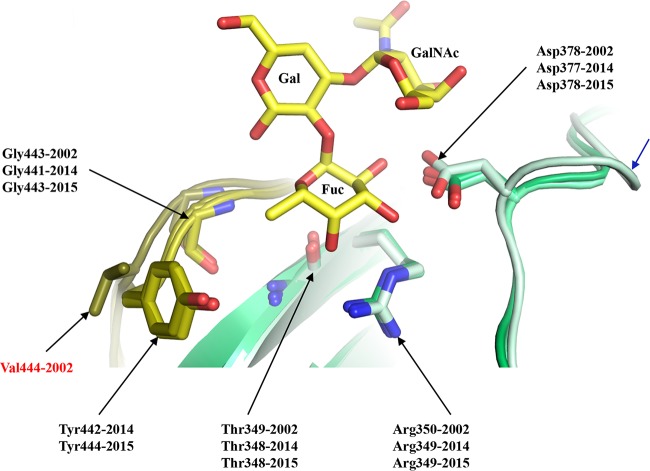
Four of five GII.4 residues that commonly interact with the fucose moiety of the HBGAs, i.e., Gly443/441/443, Thr349/348/348, Arg350/349/349, and Asp378/377/378, in Saitama/T87, Kawasaki323, and Kawasaki308, respectively (24), were highly conserved in these GII.17 P domains. Superposition of an HBGA A-trisaccharide onto the GII.17 P domain structures indicated that these four residues were well positioned to interact with the fucose (Fig. 3). Interestingly, Kawasaki308 had an insertion on the loop hosting Asp378, and yet the orientation of the Asp378 side chain was maintained as seen in the other GII.17 P domains (Fig. 3). The one exception to the common set of residues interacting with fucose was Val444 in Saitama/T87, which was Tyr442 in Kawasaki323, Tyr444 in Kawasaki308, and Tyr444 in GII.4. Our previous study showed that Tyr444 provided a hydrophobic interaction to the fucose methyl group in GII.4 and GII.10 (Tyr452) (22, 24). Mutagenesis of the Tyr to Ala was shown to block HBGA binding (32). These results suggested that Tyr444 in Kawasaki323 and Kawasaki308 might also be critical for HBGA binding. Two reports have suggested that, prior to the emergence of the 2014-2015 GII.17 strains, the earlier GII.17 strains may not have interacted HBGAs (4, 8). Therefore, a larger portion of the population might be more susceptible to 2014-2015 GII.17 strains (4, 8). Unfortunately, our cocrystallization and soaking experiments with GII.17 P domains and HBGAs were unsuccessful and the exact HBGA binding site remains unknown.
FIG 3.
Model of the GII.17 HBGA binding pocket. The figure presents a superposition of an A-trisaccharide (A-tri; yellow sticks) from the GII.10 P domain A-trisaccharide complex structure (3PA1) onto the GII.17 P domains (colored as described in the Fig. 2 legend), Saitama/T87 (dark olive and dark lime green), Kawasaki323 (olive and lime green), and Kawasaki308 (light olive and light lime green), showing the common set of residues (side chain and main chain) interacting with the fucose (Fuc) moiety of HBGAs. The 2002 Saitama/T87 Val444 residue (labeled in red) was substituted to Tyr442/Tyr444 in 2014 and 2015 (Kawasaki323 and Kawasaki303, respectively). The 2015 Kawasaki308 P domain had an insertion (blue arrow) on the loop hosting Asp378. The A-tri is an α-l-fucose(Fuc)-(1-2)-α-d-galactose(Gal)-(3-1)-N-acetyl-α-d-galactosamine (GalNAc).
In summary, our structural evolutionary data showed that the GII.17 noroviruses have undergone a large number of surface-exposed substitutions from 2002 to 2014 and 2015. A possible explanation for the increase in prevalence might be related to a change in the herd immunity, comparable to the evolving GII.4 strains (33). Also, the HBGA binding profile of GII.17 may have changed with the emergence of the 2014-2015 GII.17 variants. Currently, many of the norovirus vaccines in clinical trials are directed against GII.4 strains (20, 21). Therefore, antigenicity and structural information provided in this study could be critical for future norovirus vaccine development, considering that the GII.17 strains showed little or no cross-reactivity to GII.4 antisera.
Protein structure accession numbers.
Atomic coordinates and the structure factor of GII.17 P domains were deposited in the Protein Databank under PDB accession codes 5F4J, 5F4M, and 5F4O.
ACKNOWLEDGMENTS
We acknowledge the European Synchrotron Radiation Facility (ID23-1 and BM30A) for provision of synchrotron radiation facilities and the Protein Crystallization Platform, CellNetworks, Heidelberg, Germany, for assistance with protein crystallization. We also thank Doug McAllister (ViroStat, USA) for providing the MAbs.
REFERENCES
- 1.Siebenga JJ, Lemey P, Kosakovsky Pond SL, Rambaut A, Vennema H, Koopmans M. 2010. Phylodynamic reconstruction reveals norovirus GII.4 epidemic expansions and their molecular determinants. PLoS Pathog 6:e1000884. doi: 10.1371/journal.ppat.1000884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siebenga JJ, Vennema H, Renckens B, de Bruin E, van der Veer B, Siezen RJ, Koopmans M. 3 July 2007. Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006. J Virol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noel JS, Fankhauser RL, Ando T, Monroe SS, Glass RI. 1999. Identification of a distinct common strain of “Norwalk-like viruses” having a global distribution. J Infect Dis 179:1334–1344. doi: 10.1086/314783. [DOI] [PubMed] [Google Scholar]
- 4.Zhang XF, Huang Q, Long Y, Jiang X, Zhang T, Tan M, Zhang QL, Huang ZY, Li YH, Ding YQ, Hu GF, Tang S, Dai YC. 2015. An outbreak caused by GII.17 norovirus with a wide spectrum of HBGA-associated susceptibility. Sci Rep 5:17687. doi: 10.1038/srep17687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsushima Y, Ishikawa M, Shimizu T, Komane A, Kasuo S, Shinohara M, Nagasawa K, Kimura H, Ryo A, Okabe N, Haga K, Doan YH, Katayama K, Shimizu H. 2015. Genetic analyses of GII.17 norovirus strains in diarrheal disease outbreaks from December 2014 to March 2015 in Japan reveal a novel polymerase sequence and amino acid substitutions in the capsid region. Euro Surveill 20:pii=21173. doi: 10.2807/1560-7917.ES2015.20.26.21173. [DOI] [PubMed] [Google Scholar]
- 6.Lee CC, Feng Y, Chen SY, Tsai CN, Lai MW, Chiu CH. 1 December 2015, posting date. Emerging norovirus GII.17 in Taiwan. Clin Infect Dis doi: 10.1093/cid/civ647. [DOI] [PubMed] [Google Scholar]
- 7.Fu J, Ai J, Jin M, Jiang C, Zhang J, Shi C, Lin Q, Yuan Z, Qi X, Bao C, Tang F, Zhu Y. 2015. Emergence of a new GII.17 norovirus variant in patients with acute gastroenteritis in Jiangsu, China, September 2014 to March 2015. Euro Surveill 20:pii=21157. doi: 10.2807/1560-7917.ES2015.20.24.21157. [DOI] [PubMed] [Google Scholar]
- 8.Chan MC, Lee N, Hung TN, Kwok K, Cheung K, Tin EK, Lai RW, Nelson EA, Leung TF, Chan PK. 2015. Rapid emergence and predominance of a broadly recognizing and fast-evolving norovirus GII.17 variant in late 2014. Nat Commun 6:10061. doi: 10.1038/ncomms10061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rackoff LA, Bok K, Green KY, Kapikian AZ. 2013. Epidemiology and evolution of rotaviruses and noroviruses from an archival WHO Global Study in Children (1976–79) with implications for vaccine design. PLoS One 8:e59394. doi: 10.1371/journal.pone.0059394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nenonen NP, Hannoun C, Svensson L, Toren K, Andersson LM, Westin J, Bergstrom T. 2014. Norovirus GII.4 detection in environmental samples from patient rooms during nosocomial outbreaks. J Clin Microbiol 52:2352–2358. doi: 10.1128/JCM.00266-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xi JN, Graham DY, Wang KN, Estes MK. 1990. Norwalk virus genome cloning and characterization. Science 250:1580–1583. doi: 10.1126/science.2177224. [DOI] [PubMed] [Google Scholar]
- 12.Prasad BV, Hardy ME, Dokland T, Bella J, Rossmann MG, Estes MK. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287–290. doi: 10.1126/science.286.5438.287. [DOI] [PubMed] [Google Scholar]
- 13.Huang P, Farkas T, Marionneau S, Zhong W, Ruvoen-Clouet N, Morrow AL, Altaye M, Pickering LK, Newburg DS, LePendu J, Jiang X. 2003. Noroviruses bind to human ABO, Lewis, and secretor histo-blood group antigens: identification of 4 distinct strain-specific patterns. J Infect Dis 188:19–31. doi: 10.1086/375742. [DOI] [PubMed] [Google Scholar]
- 14.Huang P, Farkas T, Zhong W, Tan M, Thornton S, Morrow AL, Jiang X. 2005. Norovirus and histo-blood group antigens: demonstration of a wide spectrum of strain specificities and classification of two major binding groups among multiple binding patterns. J Virol 79:6714–6722. doi: 10.1128/JVI.79.11.6714-6722.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrington PR, Lindesmith L, Yount B, Moe CL, Baric RS. 2002. Binding of Norwalk virus-like particles to ABH histo-blood group antigens is blocked by antisera from infected human volunteers or experimentally vaccinated mice. J Virol 76:12335–12343. doi: 10.1128/JVI.76.23.12335-12343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rockx BH, Vennema H, Hoebe CJ, Duizer E, Koopmans MP. 2005. Association of histo-blood group antigens and susceptibility to norovirus infections. J Infect Dis 191:749–754. doi: 10.1086/427779. [DOI] [PubMed] [Google Scholar]
- 17.Lindesmith LC, Donaldson EF, Baric RS. 2011. Norovirus GII.4 strain antigenic variation. J Virol 85:231–242. doi: 10.1128/JVI.01364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Debbink K, Lindesmith LC, Donaldson EF, Baric RS. 2012. Norovirus immunity and the great escape. PLoS Pathog 8:e1002921. doi: 10.1371/journal.ppat.1002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atmar RL, Bernstein DI, Harro CD, Al-Ibrahim MS, Chen WH, Ferreira J, Estes MK, Graham DY, Opekun AR, Richardson C, Mendelman PM. 2011. Norovirus vaccine against experimental human Norwalk virus illness. N Engl J Med 365:2178–2187. doi: 10.1056/NEJMoa1101245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan M, Huang P, Xia M, Fang PA, Zhong W, McNeal M, Wei C, Jiang W, Jiang X. 2011. Norovirus P particle, a novel platform for vaccine development and antibody production. J Virol 85:753–764. doi: 10.1128/JVI.01835-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atmar RL, Estes MK. 2012. Norovirus vaccine development: next steps. Expert Rev Vaccines 11:1023–1025. doi: 10.1586/erv.12.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansman GS, Biertumpfel C, Georgiev I, McLellan JS, Chen L, Zhou T, Katayama K, Kwong PD. 2011. Crystal structures of GII.10 and GII.12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability. J Virol 85:6687–6701. doi: 10.1128/JVI.00246-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansman GS, Natori K, Shirato-Horikoshi H, Ogawa S, Oka T, Katayama K, Tanaka T, Miyoshi T, Sakae K, Kobayashi S, Shinohara M, Uchida K, Sakurai N, Shinozaki K, Okada M, Seto Y, Kamata K, Nagata N, Tanaka K, Miyamura T, Takeda N. 2006. Genetic and antigenic diversity among noroviruses. J Gen Virol 87:909–919. doi: 10.1099/vir.0.81532-0. [DOI] [PubMed] [Google Scholar]
- 24.Singh BK, Leuthold MM, Hansman GS. 2015. Human noroviruses' fondness for histo-blood group antigens. J Virol 89:2024–2040. doi: 10.1128/JVI.02968-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kabsch W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 26:795–800. doi: 10.1107/S0021889893005588. [DOI] [Google Scholar]
- 26.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 32.Tan M, Xia M, Cao S, Huang P, Farkas T, Meller J, Hegde RS, Li X, Rao Z, Jiang X. 2008. Elucidation of strain-specific interaction of a GII-4 norovirus with HBGA receptors by site-directed mutagenesis study. Virology 379:324–334. doi: 10.1016/j.virol.2008.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Debbink K, Lindesmith LC, Donaldson EF, Costantini V, Beltramello M, Corti D, Swanstrom J, Lanzavecchia A, Vinje J, Baric RS. 1 August 2013, posting date. Emergence of new pandemic GII.4 Sydney norovirus strain correlates with escape from herd immunity. J Infect Dis doi: 10.1093/infdis/jit370. [DOI] [PMC free article] [PubMed] [Google Scholar]