ABSTRACT
Gammaherpesviruses are important human and animal pathogens. Despite the fact that they display the classical architecture of herpesviruses, the function of most of their structural proteins is still poorly defined. This is especially true for tegument proteins. Interestingly, a potential role in immune evasion has recently been proposed for the tegument protein encoded by Kaposi's sarcoma-associated herpesvirus open reading frame 63 (ORF63). To gain insight about the roles of ORF63 in the life cycle of a gammaherpesvirus, we generated null mutations in the ORF63 gene of murid herpesvirus 4 (MuHV-4). We showed that disruption of ORF63 was associated with a severe MuHV-4 growth deficit both in vitro and in vivo. The latter deficit was mainly associated with a defect of replication in the lung but did not affect the establishment of latency in the spleen. From a functional point of view, inhibition of caspase-1 or the inflammasome did not restore the growth of the ORF63-deficient mutant, suggesting that the observed deficit was not associated with the immune evasion mechanism identified previously. Moreover, this growth deficit was also not associated with a defect in virion egress from the infected cells. In contrast, it appeared that MuHV-4 ORF63-deficient mutants failed to address most of their capsids to the nucleus during entry into the host cell, suggesting that ORF63 plays a role in capsid movement. In the future, ORF63 could therefore be considered a target to block gammaherpesvirus infection at a very early stage of the infection.
IMPORTANCE The important diseases caused by gammaherpesviruses in human and animal populations justify a better understanding of their life cycle. In particular, the role of most of their tegument proteins is still largely unknown. In this study, we used murid herpesvirus 4, a gammaherpesvirus infecting mice, to decipher the role of the protein encoded by the viral ORF63 gene. We showed that the absence of this protein is associated with a severe growth deficit both in vitro and in vivo that was mainly due to impaired migration of viral capsids toward the nucleus during entry. Together, our results provide new insights about the life cycle of gammaherpesviruses and could allow the development of new antiviral strategies aimed at blocking gammaherpesvirus infection at the very early stages.
INTRODUCTION
Gammaherpesviruses (γHVs) are widespread viruses that cause lifelong infections in many mammalian species (1) and represent a significant cause of diseases (2, 3). Epstein-Barr virus (EBV; genus Lymphocryptovirus) and Kaposi's sarcoma-associated herpesvirus (KSHV; genus Rhadinovirus genus) are highly prevalent in human populations (3, 4) and are associated with several cancers (5). Despite the burden associated with these infections in some regions of the world, there is still no standard treatment (6). A better understanding of their biological cycle is therefore needed to develop new prophylactic or therapeutic strategies. As EBV and KSHV replicate poorly in vitro and have no established in vivo infection models, animal γHVs, such as murid herpesvirus 4 (MuHV-4), have emerged as efficient and relevant models to study γHV biology.
γHVs display a morphological organization which is typical of all herpesviruses (7). Briefly, infectious virions contain a double-stranded DNA genome which is incorporated in a large icosahedral nucleocapsid. This capsid is surrounded by a thick protein layer called tegument which is enclosed in a lipid bilayer envelope spiked with glycoproteins. Until now, knowledge about the organization and function of tegument proteins has largely derived from studies on alphaherpesviruses, including herpes simplex virus 1 (HSV-1) and pseudorabies virus (PrV). Tegument proteins represent approximately one-third of the volume of the virion and form one of the more complex and diverse structures of the herpesvirus particle. Thus, we recently estimated that 13 of the 31 structural proteins of MuHV-4 extracellular virions are located in the tegument (8). As for the other herpesviruses, the γHV tegument proteins have a duality of functions due to the roles that they play during the early steps (incoming of the virus into the host cells) and/or during the late phase of the infection (egress of progeny virions from the infected cells) (9–13). However, the function of most of the γHV tegument proteins remains largely unknown.
MuHV-4 open reading frame 63 (ORF63) encodes a 938-amino-acid protein which has orthologs in all γHVs. However, the function of this ancestral gene is still poorly characterized. On the one hand, a recent study on KSHV revealed that KSHV ORF63 interacts with different members of the NLR (nucleotide binding and oligomerization, leucine-rich repeat) family of proteins, including NLRP1, NLRP3, and NOD2. This inhibits NLR-mediated innate immunity against KSHV, including caspase-1 activation and processing of interleukin 1β (IL-1β) and IL-18 (14), thereby revealing a role of KSHV ORF63 in immune evasion of innate immunity. On the other hand, MuHV-4 ORF63 belongs to one of the seven core gene blocks encoded by members of the Herpesviridae, and its product is supposed to be homologous to proteins encoded by the alphaherpesvirus HSV-1 and PrV UL37. In these viruses, UL37 has been shown to be required for both entry and egress. Thus, although UL37 is not required for the transport of incoming capsids to the nuclear pores (15), its absence delays this process (16). During egress, the absence of UL37 leads to the accumulation of unenveloped capsids in the cytosol and the release of infectious virions is either strongly impaired (17, 18) or completely blocked (17, 19). These observations support a role for alphaherpesvirus UL37 in capsid transport and in secondary envelopment. It is totally unknown whether proteins encoded by γHVs ORF63 share similar functions.
To gain insight about the roles of ORF63 in the biology of γHV infection, we constructed MuHV-4 strains in which the ORF63 coding sequence was disrupted by the insertion of STOP codons, and we analyzed the growth of these strains both in vitro and in vivo.
MATERIALS AND METHODS
Cells and viruses.
BHK-21 and NIH 3T3 cells were propagated in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 2 mM glutamine, 100 U of penicillin ml−1, 100 mg of streptomycin ml−1, and 10% fetal calf serum (FCS). We used the wild-type (WT) MHV-68 strain of MuHV-4 (20). The MuHV-4 strain expressing luciferase under the control of the M3 promoter was described previously (21).
Alignment and three-dimensional structure.
Sequences were aligned by MUSCLE v3.8.31 (22), while the secondary-structure prediction was performed via the 1D Protein Structure Prediction Server (http://biomine.ece.ualberta.ca/1D/1D.html) (23–25). All the subsequent information was imported and integrated in CLC Main Workbench.
Production of the MuHV-4 ORF63 mutant strains.
The recombinant strains ORF63 STOP and ORF63 STOP Luc were derived from bacterial artificial chromosomes (BACs) (21, 26). We disrupted the MuHV-4 ORF63 coding sequence (GenBank accession no. U97553.1, genomic coordinates 83751 to 86567) by introducing STOP codons into the coding sequence of ORF63. MuHV-4 ORF63 STOP was produced using two-step galactokinase (galK) positive and negative selection in bacteria (27). The first recombination process (galK positive selection) consisted of introducing the galK gene in ORF63 (genomic coordinate 84218). Recombination was achieved using the ORF63 galK cassette. It consisted of the galK gene flanked by 50-bp sequences corresponding to ORF63 regions (coordinates 84168 to 84218 and 84219 to 84269 of the MuHV-4 WUMS strain genome). This cassette was produced by PCR using pgalK vector (27) as the template and ORF63-fwd-galK (5′-GGCGCTGACAACGACTCTATTATATCAAATTCTAACATCCTAAAGGATTGCCCTGTTGACAATTAATCATCGGCA-3′) and ORF63-rev-galK (5′-AGAAGTTGACATCTGTTCAGTTTCTAATTTTGTGAGGTACCAAGACAGCACTGTCCTGCTCCTT-3′) as forward and reverse primers, respectively. The second recombination process (galK negative selection) consisted of replacing the galK sequence with an ORF63 STOP cassette. This cassette consisted of a synthetic double-stranded DNA (Eurogentec) corresponding to genomic coordinates 84168 to 84269 with the introduction of 36 nucleotides coding for in-frame STOP codons and restriction sites after genomic position 84218. These 36 nucleotides do not insert STOP codons in any of the 5 other frames of the genome. The MuHV-4 ORF63 Rev plasmid was produced similarly from MuHV-4 ORF63 STOP plasmid. The first recombination process (galK positive selection) was identical to the one described above. The second recombination process (galK negative selection) consisted of restoring ORF63 to generate a revertant BAC plasmid. This cassette was produced by PCR using the MuHV-4 genome as the template and ORF63-zone-rec-sens (5′-AAATCCTCCAAGCAGACCTC-3′) and ORF63-zone-rec-rev (5′-AGATACTTTCTATTAGGTGTCC-3′) as forward and reverse primers, respectively. The same strategy was followed to produce the MuHV-4 ORF63 STOP Luc and ORF63 Rev Luc plasmids. Reconstitution of infectious virus from BAC plasmids was achieved by transfection in BHK cells (BAC+ strains). For in vivo experiments, the loxP-flanked viral BAC-eGFP cassette was removed by passage through NIH 3T3-CRE cells (28) (BAC− strains). Virus stocks were grown in BHK-21 cells.
Southern blotting.
Southern blot analysis of BAC DNA digested with BamHI was performed with a probe corresponding to nucleotides 83819 to 84693 of the MuHV-4 WUMS strain genome (coding for ORF63).
Virus purification.
Virions were purified as previously described (8). Briefly, after removal of the cell debris by low-speed centrifugation (1,000 × g for 30 min at 4°C), virions present in the infected cell supernatant were harvested by ultracentrifugation (100,000 × g for 2 h at 4°C) through a 30% (wt/vol) sucrose cushion. Virions were then banded by isopycnic gradient ultracentrifugation in a 20 to 50% (wt/vol) potassium tartrate gradient in phosphate-buffered saline (PBS) (100,000 × g, 2 h at 4°C). The band containing virions was collected (∼3 ml), diluted 10-fold in PBS, and pelleted by ultracentrifugation (100,000 × g for 2 h at 4°C). The pellet was finally resuspended in PBS, and virus-enriched preparations were stored at −80°C.
Genome sequencing.
Viral genomic DNA was extracted from purified virions as described previously (29). For full-length genome sequencing, viral DNA was prepared using the Nextera XT DNA library preparation kit by following the manufacturer's recommendation (Illumina, San Diego, CA). The samples were analyzed using MiSeq DNA sequencer running v2 chemistry (Illumina). Approximately 3 million 250-nucleotide paired-end reads were obtained per sample. The reads were prepared for assembly using Trim Galore v. 0.4.0 (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore). Sequences were assembled de novo for the different strains using ABySS (v1.5.2) (30). The resulting contigs were ordered against the MUHV-4 genome sequence (GenBank accession no. U97553.2), stitched into scaffold with gap filling, and subsequently annotated by using the Post-Assembly Genome Improvement Toolkit (PAGIT, v1.01) (31). Meanwhile, GapFiller v1-10 (32), Gapcloser (33), and custom Perl scripts were employed as well for gap closing. To create sequence logos, the reads falling in ORF63 region were extracted from Bam files, and only the reads covering the insertion site of STOP codons and its immediate flanking regions (12 bp on both sides) were retained for further analysis. Afterwards, the sequence logos were generated by using seqLogo implemented in R (https://www.bioconductor.org/packages/release/bioc/html/seqLogo.html).
Gene expression analysis.
Subconfluent monolayers of BHK cells were infected at a multiplicity of infection (MOI) of 1 PFU/cell. At various times postinfection (p.i.), RNA was isolated by using an RNeasy minikit (Qiagen). Contaminating DNA was removed by DNase treatment. cDNA was produced by using a First Strand cDNA synthesis kit (Roche Applied Science) with a poly(dT) primer. The cDNA products were amplified by PCR with Taq polymerase (New England BioLabs) and specific primer pairs. ORF25fwd (5′-ATGGTATAGCCGCCTTTGTG-3′) and ORF25rev (5′-ACAAGTGGATGAAGGGTTGC-3′) were used to amplify ORF25. ORF50fwd (5′-GCCTGGAGGCGCTTAGG-3′) and ORF50rev (5′-AACACATTGCGCCCAAATG-3′) were used to amplify ORF50 (RTA). ORF62fwd (5′-GAGAGAAACCTGCATGAGGGACTTG-3′) and ORF62rev (5′-GGCCGTGGGAAAGTGACTCATT-3′) were used to amplify ORF62. ORF63fwd (5′-AAATCCTCCAAGCAGACCTC-3′) and ORF63rev (5′-AGATACTTTCTATTAGGTGTCC-3′) were used to amplify ORF63. ORF64fwd (5′-CTACCTTGCTCACAGTCATCAACGC-3′) and ORF64rev (5′-GCTCGGGCCATGTCTCATCAATA-3′) were used to amplify ORF64.
Growth curve.
The growth kinetics of mutant and revertant viruses were compared to that of the WT. Cell cultures were infected at an MOI of 0.01 (multistep assay) or at an MOI of 1 (single-step assay). After 2 h of adsorption, the cells were washed then overlaid with DMEM containing 10% FCS. Supernatants of infected cultures and infected cells were harvested at successive intervals, and the amount of infectious virus was determined by plaque assay on BHK-21 cells (34). When stated, growth curves were performed in the presence of the pan-caspase inhibitor Z-VAD-FMK (carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone, 20 μM; Promega) or of the NLRP3 inflammasome inhibitor glyburide (25 μg/ml; Sigma). In order to test if supernatants from ORF63 STOP-infected cells could contain antiviral factors, growth curves were also performed in the presence of culture supernatant from BHK-21 cells previously infected with the WT, ORF63 STOP, or ORF63 Rev strains. These supernatants had previously been collected 48 h p.i., ultracentrifuged (100,000 × g for 2 h at 4°C), and filtered through a 0.1-μm filters to remove virions. The absence of virions was checked by titration on BHK-21 cells. Then, naive cells were incubated with these supernatants (final concentration, 50%) for 3 h before starting the growth assay.
Plaque size.
Plaque sizes were measured as previously described (35). Briefly, BHK-21 cells grown on coverslips were infected with the different BAC+ MuHV-4 strains and then overlaid with DMEM containing 10% FCS and 0.6% (wt/vol) medium-viscosity carboxymethylcellulose (CMC; Sigma) in order to obtain isolated plaques. At successive intervals after infection, plaques were fixed, mounted, and imaged based on enhanced green fluorescent protein (eGFP) fluorescence (the BAC cassette encodes an eGFP reporter gene). Images were captured with a Leica DM2000 light-emitting diode (LED) microscope and a charge-coupled-device (CCD) camera system (DFC450 C), and plaque areas were determined with Leica Application Suite software. For each virus, 20 plaques were measured per time point.
Animals.
Females BALB/c mice were purchased from Harlan Laboratories. All the animals were housed at the University of Liège, Department of Infectious Diseases. The animals were infected with MuHV-4 when 6 to 12 weeks old. Anesthetized animals were infected intranasally (30 μl per animal). For luciferase imaging, animals were anesthetized with isoflurane, injected intraperitoneally with luciferin (150 mg/kg), and then scanned with an IVIS Spectrum system (Caliper Life Sciences) 10 min after luciferin injection. For quantitative comparisons, we used Living Image software (Caliper Life Sciences) to obtain the maximum radiance (photons per second per square centimeter per steradian) over the whole body of the animal. All experiments conformed to guidelines from the local animal ethics committee of the University of Liège.
Viral infectivity assays.
Virus stocks were titrated by plaque assay on BHK-21 cells (34). Cell monolayers were incubated with virus (2 h at 37°C), overlaid with 0.6% carboxymethylcellulose, and 4 days later fixed and stained for plaque counting. Infectious virus in lungs was measured by homogenizing them in 6 ml of complete medium prior to plaque assay. Latent virus in ex vivo tissues was measured by infectious-center assay (34): spleen suspensions were cocultured with BHK-21 cells and then fixed and stained for plaque counting after 5 days. Preformed infectious virus titers in lymphoid tissue, as measured by plaque assay of freeze-thawed cells, were always <1% of the infectious-center assay titers, so the latter essentially measured reactivable latent virus.
Viral genome quantification.
Viral genome loads were measured by real-time PCR. DNA from organs (100 ng) was used to amplify MuHV-4 genomic coordinates 43015 to 43,138 (iCycler, Bio-Rad) (gene ORF25; ORF25fwd, 5′-ATGGTATAGCCGCCTTTGTG-3′, and ORF25rev, 5′-ACAAGTGGATGAAGGGTTGC-3′). The PCR products were quantified by hybridization with a TaqMan probe (genomic coordinates 43117 to 43027; 5′-6-carboxyfluorescein [FAM]-CAACCACTGGATCAGCATAAAACTTATGAA-black hole quencher [BHQ1]-3′) and converted to genome copies by comparison with a standard curve of cloned plasmid template serially diluted in control spleen DNA and amplified in parallel. Cellular DNA was quantified by amplifying part of the interstitial retinoid binding protein (IRBP) gene (forward primer, 5′-ATCCCTATGTCATCTCCTACYTG-3′, and reverse primer, 5′-CCRCTGCCTTCCCATGTYTG-3′). The PCR products were quantified with Sybr green (Invitrogen). The copy number was calculated by comparison with standard curves of cloned Mus musculus IRBP template amplified in parallel. Amplified products were distinguished from paired primers by melting-curve analysis and the correct sizes of the amplified products confirmed by electrophoresis and staining with ethidium bromide.
Quantification of anti-MuHV-4 antibodies by ELISA.
Nunc Maxisorp enzyme-linked immunosorbent assay (ELISA) plates (Nalgene Nunc) were coated for 18 h at 37°C with 0.1% Tween 20-disrupted MuHV-4 virions (2 × 106 PFU/well), blocked in PBS–0.1% Tween 20–3% bovine serum albumin (BSA), and incubated with mouse sera (diluted 1/300 in PBS–0.1% Tween 20–3% BSA). Bound antibodies were detected with alkaline phosphatase-conjugated goat anti-mouse Ig polyclonal antibody (pAb) (Sigma). Washing was performed with PBS–0.1% Tween 20–3% BSA. p-Nitrophenylphosphate (Sigma) was used as the substrate, and absorbance was read at 405 nm using a Benchmark ELISA plate reader (Thermo).
Lung histology.
Portions of lungs were fixed in buffered formol saline, processed routinely to 5-mm paraffin wax-embedded sections, stained with hematoxylin and eosin, and examined by light microscopy.
Apoptosis assay.
For annexin-propidium iodide (PI) analysis, cells were washed in 1× annexin-V binding buffer (BD Bioscience), resuspended in 0.5 μg/ml of PI and 5 μl of annexin V-allophycocyanin (APC) per 100 μl (BD Bioscience), incubated for 15 min, washed in binding buffer, and, finally, analyzed by flow cytometry on a BD FACS ARIA flow cytometer.
Transmission electron microscopy (TEM).
Samples were prepared as previously described (35). Briefly, cells were washed with PBS and fixed directly in the dish with cacodylate buffer containing 2.5% glutaraldehyde and 2% paraformaldehyde. The cells were then scraped off and prepared for electron microscopy. Epon blocks and sections were prepared as described elsewhere (36). Sections were analyzed using a Technai Spirit transmission electron microscope (FEI, Eindhoven, The Netherlands), and electron micrographs were taken using a bottom-mounted 4-by-4 K Eagle camera (FEI).
Western blotting.
Purified virions were lysed and denatured by heating (95°C for 5 min) in SDS-PAGE sample buffer (31.25 mM Tris-HCl [pH 6.8], 1% [wt/vol] SDS, 12.5% [wt/vol] glycerol, 0.005% [wt/vol] bromophenol blue, 2.5% [vol/vol] 2-mercaptoethanol). Proteins were resolved by electrophoresis on Mini-PROTEAN TGX (Tris-glycine extended) precast 7.5% resolving gels (Bio-Rad) in SDS-PAGE running buffer (25 mM Tris base, 192 mM glycine, 0.1% [wt/vol] SDS) and transferred to polyvinylidene difluoride membranes (Immobilon-P transfer membrane, 0.45-μm pore size; Millipore). The membranes were blocked with 3% nonfat milk in PBS–0.1% Tween 20 and then incubated with anti-MuHV-4 rabbit polyserum in the same buffer. Bound antibodies were detected with horseradish peroxidase-conjugated goat anti-rabbit IgG polyclonal Ab (Dako Corporation), followed by washing in PBS–0.1% Tween 20, development with ECL substrate (GE Healthcare), and exposure to X-ray film.
Antibodies.
MuHV-4-specific monoclonal antibodies (MAbs) were derived from MuHV-4-infected BALB/c mice. The MAbs used in this study were as follows: BN-8C3 (ORF75c; IgG1) (37), 3F7 (gN; IgG2a) (38), and 12B8 (ORF65; IgG2a) (39). The rat IgG2a MAb against alpha-tubulin was from Thermo Scientific (clone YL1/2).
Immunofluorescence.
BHK-21 cells were seeded overnight onto glass coverslips. MuHV-4 virions (particles equivalent to 30 WT PFU/cell) were bound to the cells (2 h at 4°C). The cells were then washed 3 times with ice-cold PBS to remove unbound virions and then fixed either immediately or after an incubation period at 37°C in complete medium. After one wash in ice-cold PBS, fixation was achieved by adding ice-cold 4% formaldehyde in PBS and leaving at room temperature (RT) for 30 min followed by 3 washes in PBS. The cells were then permeabilized with 0.1% Triton X-100 (30 min at RT), blocked with 10% FCS–1% bovine serum albumin–0.3 M glycine (1 h at 37°C), stained with primary MAbs (1 h at RT), washed 3 times in PBS–0.1% Tween 20, stained with secondary antibodies (1 h at RT), washed 3 times in PBS–0.1% Tween 20 and once in H2O, and mounted in ProLong Gold Antifade Mountant with 4′,6-diamidino-2-phenylindole (DAPI; Life Technologies). Secondary antibodies (goat anti-mouse IgG1 and IgG2a and goat anti-rat IgG labeled with Alexa Fluor 488, 568, or 633) were all from Life Technologies. Images were acquired on a Leica TCS SP5 confocal laser scanning microscope and analyzed with ImageJ.
Statistics.
Statistical analysis was performed using Prism software (GraphPad).
RESULTS
Sequence variations and structure analysis of pORF63 homologs in various herpesviruses.
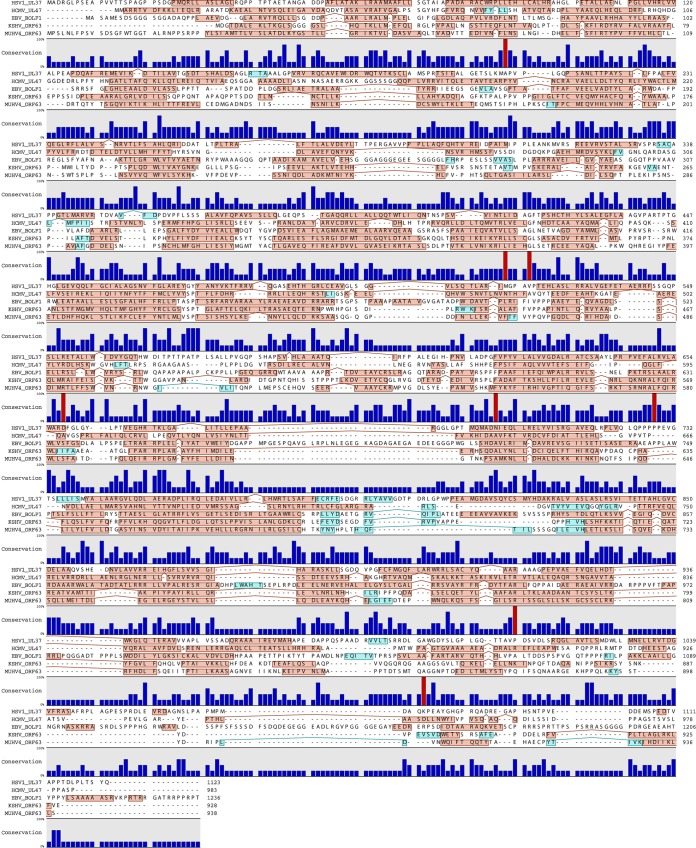
Sequence comparison of pORF63 with its positional homologs in HSV-1 (pUL37; GI 692148201), human cytomegalovirus (HCMV) (pUL47; GI 44903273), EBV (BOLF1; GI 764007616), and KSHV (pORF63; GI 139472855) revealed a very poor conservation of the primary amino acid sequence. Indeed, when comparing two by two, we observed, on average, only 13% amino acid similarities between these sequences (with minimum and maximum similarity scores of 9.66% and 20.63%, respectively). In particular, our alignment identified only 8 amino acids that were conserved across the different species (Fig. 1). Moreover, the region of KSHV ORF63 (around amino acids 210 to 360 of KSHV pORF63) that has been previously shown to interact with human NLRP1 and inhibit the inflammasome (14) also displayed poor similarities with homologs in other herpesviruses. In contrast, when we looked at the secondary structure, we observed a very similar pattern made of successive α-helices (Fig. 1) suggesting a possible common function despite their extremely low sequence conservation.
FIG 1.
Comparison of MuHV-4 ORF63 with its homologs in other herpesviruses. Shown is a multiple-sequence alignment of the protein encoded by MuHV-4 ORF63 with its homologs in different herpesviruses: HSV-1 UL37 (GI 692148201), HCMV UL47 (GI 44903273), EBV BOLF1 (GI 764007616), and KSHV ORF63 (GI 139472855). Αlpha-helices are highlighted in red boxes, while beta-sheets are highlighted in blue. The conservation at individual amino acids positions is shown below the sequences. Conservation in all the strains is highlighted in red.
Production of ORF63-deficient MuHV-4 strains.
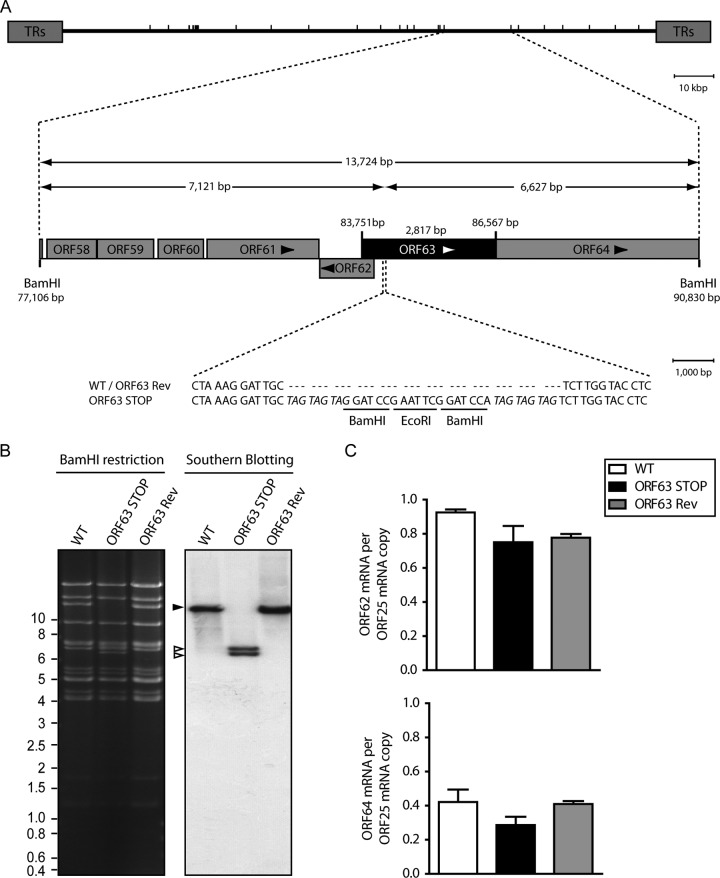
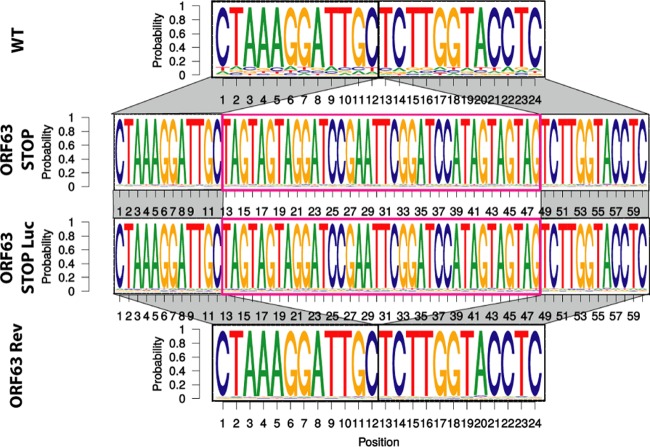
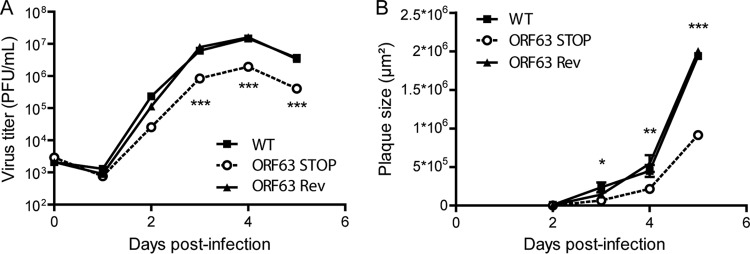
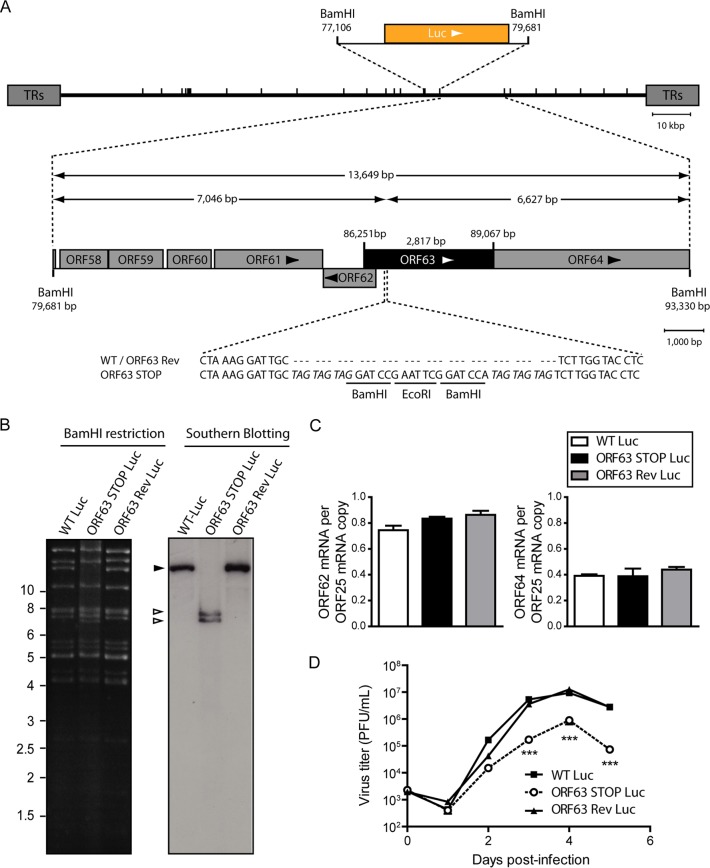
We therefore tested the functional importance of ORF63 for MuHV-4 replication by disrupting its sequence. In the MuHV-4 WUMS strain, ORF63 is predicted to be located from genomic coordinates 83751 to 86567. It encodes a predicted protein of 938 amino acids. As a second in-frame ATG is present at genomic coordinates 84165 to 84167, we disrupted the ORF63 coding sequence by inserting STOP codons at genomic coordinate 84168 to generate the ORF63 STOP strain as described in Materials and Methods (Fig. 2A). A revertant strain, called ORF63 Rev, was constructed to validate the ORF63 STOP mutant. The predicted molecular structures of the recombinant strains were confirmed by BamHI restriction mapping and Southern blotting (Fig. 2B) and further by DNA sequencing of genomic DNA from purified virions (Fig. 3). The ORF63 STOP mutant produced infectious virus after BAC DNA transfection into BHK-21 cells, although a delay in plaque formation was observable for the ORF63 STOP mutant in comparison with the WT and the ORF63 Rev strains. As there is no intergenic region between ORF62, ORF63, and ORF64, we investigated if the introduction of the STOP codons into ORF63 could have a polar effect on the expression of ORF62 and/or ORF64. Quantitative reverse transcription-PCR (qRT-PCR) analysis revealed that despite slight variations, the expressions of these two genes were not significantly different in the WT, ORF63 STOP, and ORF63 Rev strains (Fig. 2C). As the relatively slow spread of the viral infection after transfection of the ORF63 STOP BAC DNA into BHK-21 cells suggested that ORF63 is important for MuHV-4 replication in vitro, we addressed the role of ORF63 in MuHV-4 lytic infection in vitro by multistep growth and plaque assays with BHK-21 cells (Fig. 4). The results obtained for the multistep growth assay showed an important ORF63-dependent growth deficit (Fig. 4A). Accordingly, the ORF63 STOP plaques were significantly smaller than those produced by the WT and the ORF63 Rev strains (Fig. 4B). In parallel, a MuHV-4 strain deficient for ORF63 expression and expressing luciferase was generated (Fig. 5A to C) based on the MuHV-4 Luc strain that had been previously described (21, 26). A similar ORF63-dependent growth deficit was observed in this background (Fig. 5D). Together, these results showed that MuHV-4 ORF63 is nonessential but that its deletion is associated with a severe in vitro growth deficit.
FIG 2.
Generation of a ORF63-deficient MuHV-4 mutant. (A) Schematic representation of the strategy followed to produce the recombinant MuHV-4 strains. The ORF63-deficient MuHV-4 mutant was derived from a cloned MuHV-4 BAC by a galK counterselection method. The ORF63 coding sequence was disrupted by inserting stop codons (ORF63 STOP). The mutation incorporated new BamHI restriction sites. This virus was reverted by homologous recombination with a WT genomic segment (ORF63 Rev). TRs, terminal repeats. (B) Verification of the molecular structure. BAC DNA was digested with BamHI, resolved by agarose gel electrophoresis, and hybridized with a 32P-labeled probe, corresponding to nucleotides 83819 to 84693 of the MuHV-4 WUMS strain genome. The black arrowhead shows the WT ORF63 fragment (13,724 bp). Open arrowheads show the restriction fragments that contain ORF63 STOP (7,121 bp and 6,627 bp, respectively, for the left and the right fragments). Sizes in kilobase pairs are indicated on the left. (C) Viral transcription in ORF63+ and ORF63− viruses. BHK-21 cells were infected with WT, ORF63 STOP, and ORF63 Rev MuHV-4 strains (0.5 PFU/cell), and 24 h later, RNA was extracted, reverse transcribed, and assayed for viral transcripts by qRT-PCR amplification of part of each gene. ORF62 and ORF64 flank the ORF63 gene of MuHV-4. ORF25 is a control viral gene. The data are averages from triplicate measurements ± SEMs and were analyzed by 1-way analysis of variance (ANOVA) and Bonferroni posttests.
FIG 3.
Verification of the molecular structures of the viral genomes after in vitro growth. Shown is a schematic representation as a sequence logo of the aligned sequences of the region encompassing the insertion point of the stop codons for the WT, ORF63 STOP, ORF63 STOP Luc, and ORF63 Rev strains. These sequencing reactions have been performed on viral DNA from purified virions. All the genomes displayed the expected molecular structures.
FIG 4.
In vitro effect of the ORF63 deficiency on growth of MuHV-4. (A) Effect of ORF63 deficiency on MuHV-4 growth in vitro. BHK-21 cells were infected with WT, ORF63 STOP, and ORF63 Rev MuHV-4 strains in 6-well cluster dishes at an MOI of 0.01 PFU per cell. Supernatant and infected cells were harvested at different times after infection, and the amount of infectious virus was determined by plaque assay on BHK-21 cells. The data are averages from triplicate measurements ± SEMs and were analyzed by 2-way ANOVA and Bonferroni posttests. ***, P < 0.001. At time zero p.i., the inocula were retitrated to ensure that similar amounts of virus were put on the cells. (B) Effect of the absence of pORF63 on MuHV-4 plaque size. BHK-21 cells grown on coverslips were infected with MuHV-4 WT, ORF63 STOP, and ORF63 Rev strains and then overlaid with DMEM containing CMC as described in Materials and Methods. At successive intervals after infection, plaques were fixed and measured. Each datum point is the average ± SEM for the measurement of 20 plaques per time point. The data were analyzed by 2-way ANOVA and Bonferroni posttests. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
FIG 5.
Generation of an ORF63-deficient MuHV-4 mutant expressing luciferase. (A) Schematic representation of the strategy followed to produce the recombinant MuHV-4 Luc strains. The ORF63 STOP Luc strain was derived from a cloned MuHV-4 BAC-Luc strain by a galK counterselection method. The ORF63 coding sequence was disrupted by inserting stop codons (ORF63 STOP). The mutation incorporated new BamHI restriction sites. This virus was reverted by homologous recombination with an unmutated genomic segment (ORF63 Rev). (B) Verification of the molecular structure. BAC DNA was digested with BamHI, resolved by agarose gel electrophoresis, and hybridized with a 32P-labeled probe corresponding to nucleotides 83819 to 84693 of the MuHV-4 WUMS strain genome. The black arrowhead shows WT ORF63 fragment (13,649 bp). Open arrowheads show the restriction fragments that contain ORF63 STOP (7,046 bp and 6,627 bp, respectively, for the left and the right fragments). Sizes in kilobase pairs are indicated on the left. (C) Viral transcription in ORF63+ and ORF63− Luc viruses. BHK-21 cells were infected with WT Luc, ORF63 STOP Luc, and ORF63 Rev Luc MuHV-4 strains (0.5 PFU/cell), and 24 h later RNA was extracted, reverse transcribed, and assayed for viral transcripts by qRT-PCR amplification of part of each gene. ORF62 and ORF64 flank the ORF63 gene of MuHV-4. ORF25 is a control viral gene. The data are averages from triplicate measurements ± SEMs and were analyzed by 1-way ANOVA and Bonferroni posttests. (D) Effect of ORF63 deficiency on in vitro growth of MuHV-4 strains expressing Luc. BHK-21 cells were infected with WT Luc, ORF63 STOP Luc, and ORF63 Rev Luc MuHV-4 strains in 6-well cluster dishes at an MOI of 0.01 PFU per cell. Supernatant and infected cells were harvested at different times after infection, and the amount of infectious virus was determined by plaque assay on BHK-21 cells. The data are averages from triplicate measurements ± SEMs and were analyzed by 2-way ANOVA and Bonferroni posttests; ***, P < 0.001. At time zero p.i., the inocula were retitrated to ensure that similar amounts of virus were put on the cells.
In vivo infection of BALB/c mice by MuHV-4 ORF63 STOP strains.
In order to have a global picture of the in vivo infection without having to rely on new virion production as a readout, we followed infection with the strains encoding Luc as a reporter gene. It has to be noted that luciferase expression is under the dependence of the early lytic M3 promoter and is therefore not expressed during the latent phase of the infection (21). We infected anesthetized animals intranasally with 104 PFU of the luciferase-positive MuHV-4 strains and tracked the infection over time by d-luciferin injection and charge-coupled-device (CCD) camera scanning. Based on previous analysis (21), we considered thoracic signals to come from the lungs, abdominal signals from the spleen, and neck signals from the superficial cervical lymph nodes (SCLNs). A strong signal was visible in the lungs of WT Luc-infected mice at the peak of lytic replication (5 to 7 days p.i.) (Fig. 6). In contrast, we observed a severe reduction of the luciferase signal originating from the lungs of ORF63 STOP Luc-infected mice and a delay of the peak of lytic replication (that occurred ∼10 days p.i.). Moreover, while we observed signal originating from the spleen and SCLNs corresponding to latency amplification (which is associated with serial replication in lymphoid and myeloid cells [40]) in WT Luc-infected mice, signals in the corresponding sites were weak or undetectable in ORF63 STOP-infected mice (Fig. 6). Finally, we did not observe genital excretion in the ORF63 STOP Luc-infected group, while it was readily detectable in the WT Luc group (day 17 p.i.). Together, these results suggest a severe impairment of ORF63 STOP lytic replication in vivo and justify further investigation.
FIG 6.
In vivo infection by luciferase-expressing MuHV-4 strains. Female mice were infected intranasally (1 × 104 PFU) with MuHV-4 WT Luc (top row) or ORF63 STOP Luc (bottom rpw) strains under general anesthesia. The mice were then injected with luciferin and imaged at the indicated time points. Images show a representative mouse (from a group of 5 mice) at days 3, 5, 7, 10, 12, 14, 17, and 19 p.i. The scale bar shows the color scheme for signal intensity.
As explained, luciferase expression by the luciferase-positive strains reflects predominantly lytic gene expression (21). Therefore, to also explore the establishment of latency, we infected mice intranasally with 104 PFU of the WT, ORF63 STOP, or ORF63 Rev strain. Consistent with the bioluminescence imaging results, lytic replication as measured by plaque assay was greatly reduced in ORF63 STOP-infected mice compared to the levels in mice of the WT and ORF63 Rev groups (Fig. 7A). At day 7 p.i., plaque assay titers were nearly 1,000 times lower in the ORF63 STOP group than in the 2 other groups (no infectious virus was detected in the lungs of most animals at later time points). Accordingly, a lower antibody response against MuHV-4 was also observed in the ORF63 STOP group at day 21 p.i. (Fig. 7B), confirming the lower virus replication and antigen load in this group. However, despite these differences in amounts of infectious viruses, focal lymphocyte infiltrations with potential lymphatic follicle formation, previously described as inducible bronchus-associated lymphoid tissue (iBALT) (41, 42), were visible in all the groups (Fig. 7C).
FIG 7.
Effect of ORF63 deficiency on replication of MuHV-4 in vivo. (A) BALB/c mice were infected intranasally with WT, ORF63 STOP, and ORF63 Rev MuHV-4 strains (1 × 104 PFU). At the indicated times p.i., the infectious virus titers in lungs were determined by plaque assay. (B) Individual sera collected at the different time points were analyzed for MuHV-4-specific IgG by ELISA. Pooled naive sera provided the negative control. The data are averages from 5 mice ± SEMs and were analyzed by 2-way ANOVA and Bonferroni posttests. ***, P < 0.001; **, P < 0.01. (C) Lung histology. Seven days after infection with the different strains of MuHV-4, lungs were removed and fixed in formaldehyde before hematoxylin-eosin staining. Rectangles identify regions that are highlighted in higher-magnification pictures. Arrows indicate perivascular and peribronchial lymphocyte accumulation. The images are representative of data from at least 5 animals. (D) Spleens from the same mice were analyzed individually by infectious-center assay. (E) DNA was extracted from individual spleens. The viral genome copy number was then determined by real-time PCR.
The colonization of spleens by latent virus was then determined by infectious-center assay. In the case of WT and ORF63 Rev-infected mice, the recovery of replication-competent virus from spleens reached a peak at 14 days p.i. (Fig. 7D), consistent with published data (42). Virus recovery from the spleens of mice infected by the MuHV-4 ORF63 STOP virus was much lower, especially at day 14 p.i. However, at 21 days p.i., MuHV-4 ORF63 STOP latent infection in spleens reached levels similar to the ones observed for the MuHV-4 WT and ORF63 Rev strains. This result suggests therefore that latency amplification occurs for the MuHV-4 ORF63 STOP strain but is delayed, as also suggested by the fact that no infectious centers were detected for this strain at day 7 p.i., in contrast to the case with the WT and ORF63 Rev strains. Lower virus recovery by infectious-center assays could reflect a lower capacity of virus reactivation from MuHV-4 ORF63 STOP-infected spleen cells explanted onto BHK-21 monolayers. In order to evaluate latent loads in spleens further, we quantified viral genomes by quantitative PCR. We detected a trend similar to the one observed for infectious-center assays (Fig. 7E). Thus, latent loads of MuHV-4 WT and ORF63 Rev viruses were maximal at 14 days p.i. (Fig. 7E). In contrast, latent loads of MuHV-4 ORF63 STOP virus were lower, especially at day 14 p.i., but were not statistically significantly different at day 21 p.i. These results suggest, therefore, again that latency amplification occurs for the MuHV-4 ORF63 STOP virus but is delayed in comparison with that in WT and revertant strains. Moreover, comparisons of Fig. 7D and E do not highlight a decreased efficiency of reactivation for the ORF63 STOP mutant. Indeed, in Fig. 7E (viral genome copies) at day 14 postinfection, we observe differences of 7,649- and 2,904-fold between the ORF63 STOP and the WT and ORF63 Rev samples, respectively. In Fig. 7D (infectious-center assay), these differences are 788- and 631-fold between the ORF63 STOP and the WT and ORF63 Rev samples, respectively. We therefore do not observe a decreased efficiency of reactivation. Together, these experiments confirm that ORF63 deficiency in MuHV-4 is mainly associated with a replication deficit both in vitro and in vivo.
Apoptosis and release of antiviral factors.
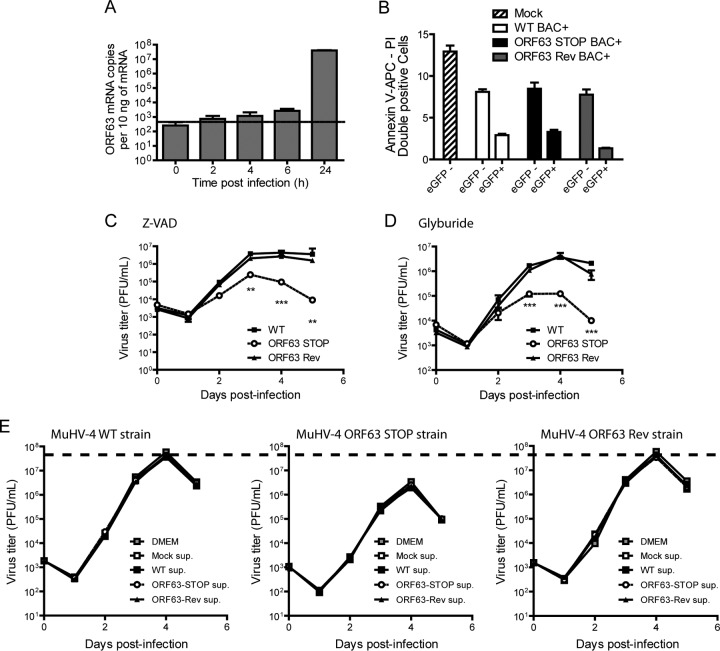
The MuHV-4 growth deficit associated with the absence of pORF63 could be the consequence of increased cell death consecutive, for example, to inflammasome activation and release of some antiviral factors. Indeed, Gregory et al. previously showed that KSHV pORF63 is an NLR homolog that blocks inflammasome activation (14). First, we compared cell death after infection by the different strains at 24 h p.i., a time at which ORF63 is expressed (Fig. 8A). Briefly, BHK-21 cells were mock infected or infected by the MuHV-4 WT, ORF63 STOP, or ORF63 Rev strain. In order to distinguish infected from noninfected cells, we used BAC+ strains expressing eGFP. Twenty-four hours p.i., cells were harvested and cell viability was analyzed in eGFP+ (infected) and eGFP− (noninfected) populations. No increase in cell mortality was observed in the ORF63 STOP BAC+ strain-infected cells (Fig. 8B). Second, the MuHV-4 growth deficit associated with ORF63 deficiency was still observed when the cells were cultured in the presence of Z-VAD, a well-known inhibitor of caspase-1 (Fig. 8C), or of glyburide, a potent inhibitor of NLRP3 inflammasome (43) (Fig. 8D). Finally, in order to test if the reduced growth of the ORF63 STOP strain could be associated with the release of some antiviral factor in the culture medium, the WT, ORF63 STOP, and ORF63 Rev strains were cultured in normal medium or in supernatants from mock-, WT-, ORF63 STOP-, and ORF63 Rev-infected cell cultures. The growth of the different strains was not affected by any of these different conditions (Fig. 8E), suggesting that the ORF63 STOP growth deficit is not associated with the release of antiviral factors in the culture medium.
FIG 8.
The growth deficit of the MuHV-4 ORF63 STOP mutant strain is not associated with an increased cell death or with the activation of the inflammasome. (A) Kinetic of ORF63 expression. BHK-21 cells were infected with MuHV-4 (MOI of 0.5 PFU/cell). At the indicated time postinfection, expression of ORF63 was studied by a Sybr green qRT-PCR approach as described in Materials and Methods. Time zero represents uninfected cells. The data are averages from triplicate measurements ± SEMs and were analyzed by 1-way ANOVA and Bonferroni posttests. ***, P < 0.001. (B) BHK-21 cells were mock infected or infected with WT BAC+, ORF63 STOP BAC+, and ORF63 Rev BAC+ MuHV-4 strains at an MOI of 0.5 PFU/cell. Twenty-four hours after infection, cell viability was assessed by annexin V-APC and propidium iodide labeling and flow cytometry analysis. Percentages of doubly positive cells were measured in eGFP− and eGFP+ populations. The data are averages from triplicates ± SEMs and were analyzed by 2-way ANOVA and Bonferroni posttests. (C and D) Effect of Z-VAD (C) or glyburide (D) on growth of MuHV-4 in vitro. BHK-21 cells were infected with WT, ORF63 STOP, and ORF63 Rev MuHV-4 strains in 6-well cluster dishes at an MOI of 0.01 PFU per cell in the presence of the pan-caspase inhibitor Z-VAD (20 μM) or the NLRP3 inflammasome inhibitor glyburide (25 μg/ml). Supernatant and infected cells were harvested at different times after infection, and the amounts of infectious virus were determined by plaque assay on BHK-21 cells. The data are averages from triplicate measurements ± SEMs and were analyzed by 2-way ANOVA and Bonferroni posttests. **, P < 0.01; ***, P < 0.001. At time zero p.i., the inocula were retitrated to ensure that similar amounts of virus were put on the cells. (E) Effect of infection supernatant on the growth of MuHV-4 in vitro. BHK-21 cells were infected with WT, ORF63 STOP, and ORF63 Rev MuHV-4 strains in 24-well cluster dishes at an MOI of 0.01 PFU per cell in the presence of supernatant of BHK-21 cells previously infected with WT, ORF63 STOP, and ORF63 Rev MuHV-4 strains (500 μl/well; 50% final concentration). Supernatant and infected cells were harvested at different times after infection and the amounts of infectious virus were determined by plaque assay on BHK-21 cells. The data are averages from triplicate measurements ± SEMs and were analyzed by 2-way ANOVA and Bonferroni posttests. **, P < 0.01; ***, P < 0.001. At time zero p.i., the inocula were retitrated to ensure that similar amounts of virus were put on the cells. To allow comparisons between graphs, a dashed line has been added across the graphs at the mean maximal value measured for WT and ORF63 Rev strains.
Effect of ORF63 disruption on virion morphogenesis.
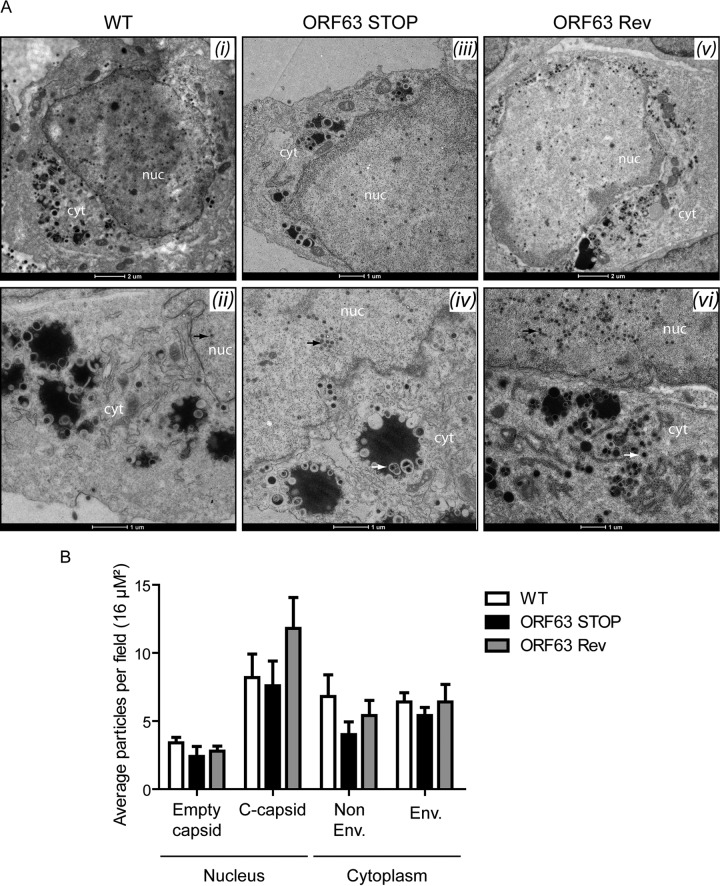
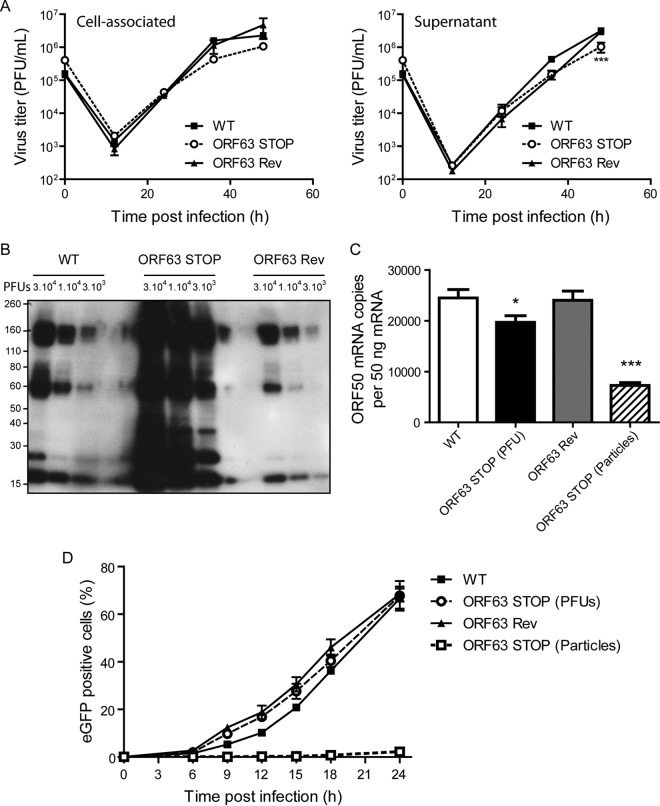
Analysis of the role of the MuHV-4 pORF63 homologs in alphaherpesviruses suggested a role in virion morphogenesis (18, 19). To investigate such a role in MuHV-4, TEM was performed on WT-, ORF63 STOP-, and ORF63 Rev-infected BHK-21 cells. No difference was observed between the different strains (Fig. 9). Moreover, in order to quantify some possible differences between the strains, we have categorized the different types of nuclear and cytoplasmic particles and provided additional statistical analysis (Fig. 9B). No statistical difference has been observed between the WT, the ORF63 STOP, and the ORF63 Rev strains. Especially, in contrast with PrV (18) and HSV-1 (19) strains deficient for UL37, we did not observe large accumulations of intracytoplasmic capsids that could suggest a defect in secondary envelopment. Accordingly, while PrV and HSV-1 strains deficient for UL37 exhibited a marked reduction of production of infectious virions in single-step growth assays, the absence of ORF63 only barely affected the growth of MuHV-4 in a high-MOI growth curve assay (Fig. 10A). Indeed, we observed only a slight reduction of ORF63 STOP infectious-particle production in the supernatant 48 h p.i. in comparison with WT and ORF63 Rev strains. We then determined the ratio between physical particles and PFU for the WT, ORF63 STOP, and ORF63 Rev strains. Interestingly, for the same number of PFU, the viral protein content was more than 10 times higher in the ORF63 STOP stocks of purified virions (Fig. 10B). Together, these results suggested that ORF63 is not involved in the morphogenesis and/or egress of MuHV-4 virions. In contrast, the higher particle/PFU ratio observed for the ORF63 STOP strain in purified virus stocks suggested that ORF63 deficiency could be associated with a defect in virus entry. Accordingly, for a similar amount of viral particles (determined by Western blotting), we observed a decreased transcription of the immediate early (IE) ORF50 encoding the viral immediate early transactivator protein RTA at 5 h postinfection in ORF63 STOP-infected cells in comparison with that in WT- and ORF63 Rev-infected cells (Fig. 10C). This was confirmed by analyzing eGFP expression at different times postinfection, as the eGFP is under the control of the HCMV IE promoter in the BAC cassette (26). For a similar amount of viral particles (determined by Western blotting), we observed a dramatically reduced percentage of eGFP-positive cells in the ORF63 STOP-infected samples in comparison with WT- and ORF63 Rev-infected cells (Fig. 10D).
FIG 9.
ORF63 deficiency is not associated with a morphogenesis defect or an egress deficit but is associated with an increased particle/PFU ratio. (A) Transmission electron microscopic (TEM) analysis for morphogenesis of the virions. BHK-21 cells were infected with WT (i and ii), ORF63 STOP (iii and iv), and ORF63 Rev (v and vi) MuHV-4 strains (1 PFU/cell, 48 h), washed with PBS, and fixed in TEM fixation buffer for TEM. The scale bars are shown below the images. nuc, nucleus; cyt, cytoplasm. No difference was observed in the assembly of nucleocapsids and their transport from the nucleus (black arrows; approximate diameter, 100 nm) among the infected cells by different viral strains. The enveloped viruses are shown with the white arrows in the cytoplasms of the infected cells. No difference in virus egress was found among the samples. (B) Distribution of different virus capsids and particle types in TEM micrographs of BHK-21 cells that were infected with the WT, ORF63 STOP, or ORF63 Rev strains for 48 h at an MOI of 0.5 PFU/cell. Average numbers of particles from 6 to 9 different micrographs spanning at least 3 different infected cells for each sample were identified based on their established characteristics (61) and enumerated before statistical analysis was performed (GraphPad software).
FIG 10.
ORF63 deficiency is associated with an increased particle/PFU ratio and a deficit in entry. (A) High-MOI growth curve. BHK-21 cells were infected with WT, ORF63 STOP, and ORF63 Rev MuHV-4 strains in 6-well cluster dishes at an MOI of 1 PFU per cell. Supernatant and infected cells were harvested independently at different times after infection, and the amounts of infectious virus were determined for both kinds of samples by plaque assay on BHK-21 cells. The data are the averages from triplicate measurements ± SEMs and were analyzed by 2-way ANOVA and Bonferroni posttests. ***, P < 0.001. At time zero p.i., the inocula were retitrated to ensure that similar amounts of virus were put on the cells. (B) Comparison of the structural proteins content for different amounts of PFU (3 × 104, 1 × 104, and 3 × 103) between the different strains. MuHV-4 WT, ORF63 STOP, and ORF63 Rev stocks were compared for viral protein content by immunoblotting with anti-MuHV-4 rabbit polyserum. (C) A total of 106 BHK-21 cells were infected with WT, ORF63 STOP, and ORF63 Rev MuHV-4 strains at an MOI of 0.5 PFU/cell. For the ORF63 STOP strain, an additional sample of cells infected by a number of particles equivalent to the WT and ORF63 Rev strains was added. Six hours later, RNA was extracted, reverse transcribed, and assayed for ORF50 expression by qPCR amplification. The data are averages from triplicate measurements ± SEMs and were analyzed by 1-way ANOVA and Bonferroni posttests. (D) BHK-21 cells were exposed to eGFP expressing (BAC+) WT, ORF63 STOP, and ORF63 Rev strains (0.5 PFU/cell). For the ORF63 STOP strain, an additional sample of cells infected by a number of particles equivalent to those of the WT and ORF63 Rev strains (determined by Western blotting) was added. After binding for the times indicated, the cells were washed with PBS and assayed by flow cytometry for eGFP expression. The data are the averages ± SEMs from triplicate measurements. The data were analyzed by 2-way ANOVA and Bonferroni posttests. ***, P < 0.001.
Role of ORF63 in MuHV-4 entry.
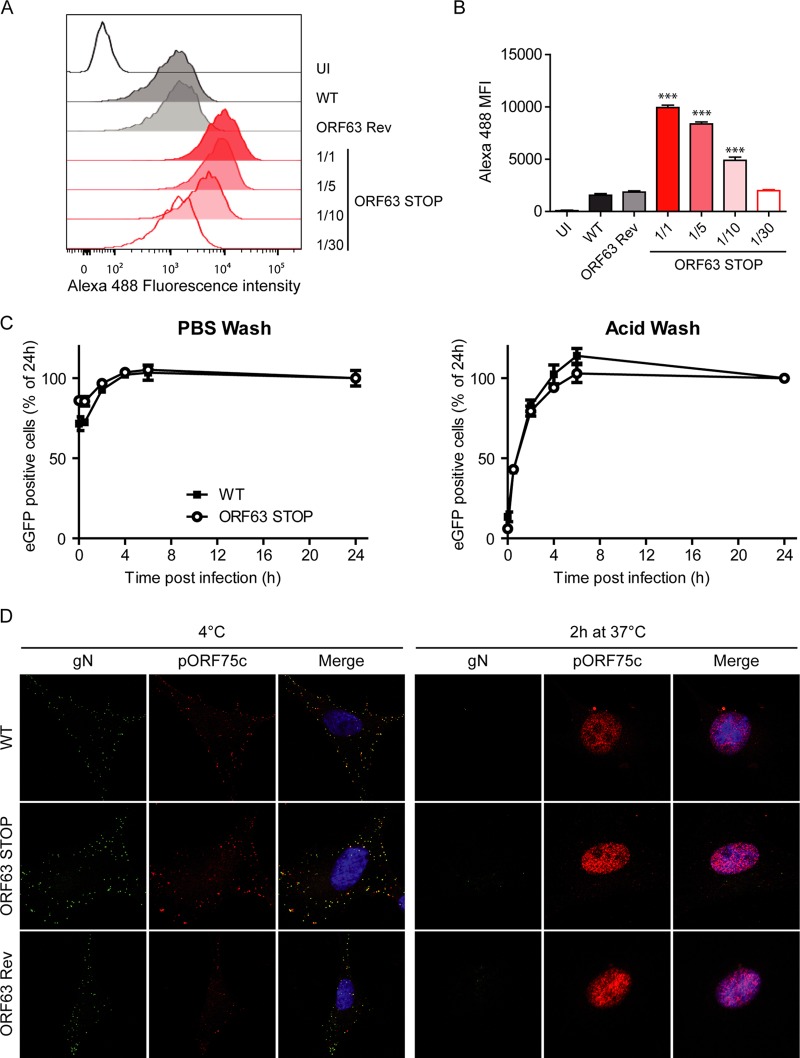
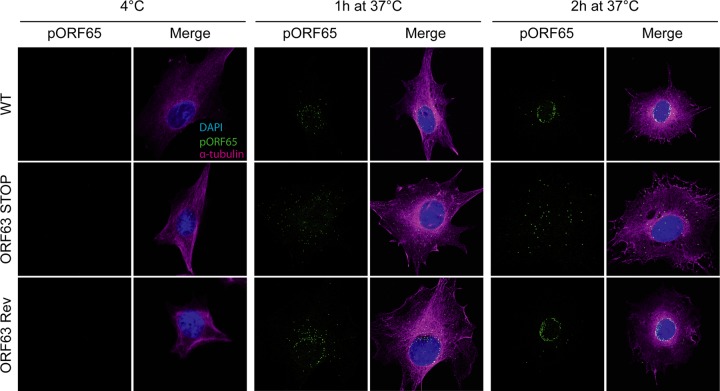
We investigated successively the different steps of herpesvirus entry in order to unravel a putative role for MuHV-4 ORF63. First, we did not observe any binding deficit associated with the absence of pORF63 (Fig. 11A and B). Second, we showed that WT and ORF63 STOP virions were equally sensitive to postbinding acid wash treatment (34, 44), indicating that the ORF63 STOP growth deficit was not associated with a default of penetration (Fig. 11C). Finally, we examined further virus entry by immunofluorescence using release of the abundant tegument component encoded by ORF75c (13) as a marker of virion membrane fusion. Indeed, pORF75c is rapidly transported to the cell nucleus after release in the cytoplasm and so provides a useful marker of fusion. ORF75c staining also shows a marked increase in intensity after fusion, presumably because the protein confined within the virion tegument is poorly accessible to antibody (45). Interestingly, the pORF75c of virions from all strains was rapidly transported to the cell nucleus (Fig. 11D), indicating that the growth deficit of the ORF63 STOP strain is not associated with a defect in fusion or tegument protein release. Finally, we tracked the release of capsids and their migration to the perinuclear margin. We used MAb MG-12B8, which recognizes an ORF65 capsid epitope that is inaccessible in virions unless they have uncoated (39, 46). There was no MG-12B8 staining of WT, ORF63 STOP, or ORF63 Rev virions after binding to BHK-21 cells at 4°C (Fig. 12). After incubation at 37°C, the capsids of WT and ORF63 Rev virions had become visible and reached progressively the nuclear margin. After 2 h at 37°C, most of the capsids were located around the nuclei of infected cells. In contrast, although ORF63 STOP virion capsids were accessible to MAb MG-12B8, confirming that fusion had occurred, the majority remained scattered throughout the cytoplasm (Fig. 12). Since capsids are normally released from late endosomes (46, 47), these results suggest that the ORF63 STOP growth deficit is associated with a default of transport of released capsids to the nuclear periphery.
FIG 11.
Binding, endocytosis, and fusion of ORF63 STOP virions. (A and B) WT, ORF63 STOP, and ORF63 Rev virions were bound to BHK-21 cells (3 h, 4°C) with either the same numbers of PFU for the three strains or dilutions of this amount (1/5, 1/10, 1/30) for the ORF63 STOP strain. Cell surface-bound virions were detected by washing, fixing, and staining them for gN with MAb 3F7 (38). Secondary detection was with Alexa 488-conjugated goat anti-mouse IgG pAb. The cells were then analyzed by flow cytometry (A). This experiment was performed in triplicates (B), and the differences in gN detection (mean fluorescence intensity [MFI]) between samples were analyzed by 1-way ANOVA and Bonferroni posttests. ***, P < 0.001 compared to the value for the WT sample. (C) BHK-21 cells were exposed to WT, ORF63 STOP, and ORF63 Rev BAC+ (expressing eGFP) strains (0.5 PFU/cell) for the times indicated and then washed either with PBS (pH 7.4) or with isotonic buffer (pH 3; acid wash). Viral infection was assayed by measuring eGFP expression 24 h p.i. by flow cytometry. The data are average ± SEMs for triplicate measurements. The data were analyzed by 2-way ANOVA and Bonferroni posttests. (D) MuHV-4 WT, ORF63 STOP, and ORF63 Rev virions were bound to BHK-21 cells (particles equivalent to 30 WT PFU/cell, 3 h, 4°C). The cells were then washed with PBS and either fixed immediately or first further incubated (2 h, 37°C) to allow virion endocytosis and membrane fusion. The cells were then stained for the gN envelope glycoprotein (IgG2a [green]) and for the ORF75c virion tegument protein with MAb BN-8C3 (IgG1 [red]), and with DAPI (blue). Red and green colocalization appears as yellow. Equivalent data were obtained in a repeat experiment. The data are fully representative of at least 100 cells examined. The confocal settings were the same for the corresponding images at 4°C and after 2 h at 37°C.
FIG 12.
Effect of ORF63 deficiency on capsid localization during entry. MuHV-4 WT, ORF63 STOP, and ORF63 Rev virions were bound to BHK-21 cells (particles equivalent to 30 WT PFU/cell, 3 h, 4°C). The cells were then washed with PBS and either fixed immediately or first further incubated (1 or 2 h, 37°C) to allow virion endocytosis, membrane fusion, and capsid release into the cytoplasm. The cells were then stained for the ORF65 capsid protein with MAb 12B8 (IgG2a [green]; epitope only revealed after fusion) and for the alpha-tubulin (rat IgG [magenta]), and with DAPI (blue). Equivalent data were obtained in a repeat experiment. The data shown are fully representative of those obtained for at least 100 cells examined. The confocal settings were the same for the corresponding images at 4°C and after 1 or 2 h at 37°C.
DISCUSSION
Despite being considered one of the core proteins of herpesviruses, pORF63 exhibits minimal conservations among members of the Herpesviridae family. Accordingly, MuHV-4 pORF63 shares minimal sequence conservation with its positional homologs encoded by γHVs or more generally by alpha- and betaherpesviruses (Fig. 1). The function of ORF63 homologs in the biological cycle of γHVs therefore remained elusive. In this study, we showed that ORF63 deficiency in MuHV-4 was associated with a major growth deficit both in vitro and in vivo (Fig. 2 to 7). This deficiency was not the consequence of an increased death of the infected cells (Fig. 8) or of a deficit in egress of progeny virions (Fig. 9) but reflected a problem during entry (Fig. 10) and, more precisely, during the trafficking of incoming capsids to the nucleus (Fig. 12).
The transport of intracellular components is of particular importance to all forms of eukaryotic organisms (48). However, the ability to undergo directional transport in the host cytoplasm is also crucial for many viruses (49). Indeed, during entry, viruses have to deliver their genome to the replication compartment, while during egress, progeny virions have to find their way out of the infected host cell. As herpesvirus DNA replication occurs in the nucleus, incoming particles have to cross the cytoplasm following entry into the host cell. In most of the cases, γHVs enter the cells via an endocytic mechanism (37). Then, capsids are released in the cytoplasm and migrate to nuclear pores to release the viral genome.
Our results show that ORF63 STOP MuHV-4 virions display a deficit in the trafficking of incoming capsids. The release of pORF75c (Fig. 11) and epitope accessibility to MG-12B8 MAb (Fig. 12) revealed that it was not related to a defect of viral envelope fusion with cellular membrane. In contrast, once released into the cytoplasm, capsids of MuHV-4 ORF63 STOP virions appeared to not reach the nuclear periphery. Indeed, we never observed accumulation of capsids around the nucleus for the MuHV-4 ORF63 STOP virions as has been observed for WT and ORF63 Rev virions (Fig. 12). Moreover, the appearance of the MG-12B8-positive capsids and their blockage in ORF63 STOP virions reveal that fusion does not occur at the nuclear margin but that capsids are released in the cytoplasm and have to be actively transported to the nuclear periphery. MuHV-4 pORF63 appears, therefore, to be a central player in the control of capsid trafficking in the very early stages of the infection. As the transport of γHV incoming capsids appears to mainly use the microtubule (MT) network (50, 51), we hypothesize that after fusion, pORF63 directly recruits specific MT motors to uncoated capsids and allows their directional transport to the nucleus. This hypothesis will have to be tested in the future.
Deletion of the ORF63 positional homolog UL37 also appeared to delay the entry of PrV virions without affecting penetration (16). Interestingly, HSV-1 pUL37 has recently been shown to interact with the MT-binding protein dystonin/BPAG1 to promote capsid transport on microtubules (52). Moreover, the same group showed that dystonin silencing during entry impaired HSV-1 capsid accumulation as a ring around the nuclear periphery (53), a phenotype very similar to the one observed for the MuHV-4 ORF63 STOP mutant (Fig. 12). These authors explained this phenotype based on the organization of the cellular MT network (53). Indeed, the MT network is typically organized around one or more MT-organizing centers (MTOC), with MT minus ends anchored at the MTOC while the plus ends radiate outwards. Therefore, herpesviruses coming along MT have to switch polarity during their journey from the plasma membrane to the nucleus. Thus, they first have to travel to the MTOC by minus-end-directed transport and then must travel from the MTOC to the nucleus by plus-end-directed transport (48). In the HSV-1 context, it is postulated that dystonin has an important role in the plus-end-directed transport of capsids from the centrosome to the nucleus and that pUL37 mediates the interaction between capsids and dystonin (53). Dystonin silencing would therefore impair HSV-1 migration from the centrosome to the nucleus. Interestingly, a similar mechanism could explain the phenotype observed for the ORF63 STOP MuHV-4 virions. However, this is a hypothesis and, in the future, interaction of pORF63 and MuHV-4 capsids with MT, MT motors, and MT-binding proteins such as dystonin will have to be tested. Also, the position of incoming capsids with regard to the MTOC will have to be determined.
Despite the existence of similarities, important differences exist between the phenotypes of the MuHV-4 ORF63 STOP strain and UL37-deficient alphaherpesviruses. Indeed, one of the most striking consequences of the absence of pUL37 in PrV or HSV-1 is that virion formation is severely impaired or totally abolished, respectively (17–19). In both cases, nonenveloped capsids accumulated in clusters in the cytoplasm, revealing a defect in virion morphogenesis (17–19). This was associated with a defect of progeny capsids from a UL37-deficient strain to be transported to the trans-Golgi network, where secondary envelopment occurs (54). Moreover, in HSV-1, the first observed consequence of dystonin depletion was a strong inhibition of capsid movement in the cytoplasm during egress (52). In contrast, similar observations were not made for the ORF63 STOP strain of MuHV-4 and production of extracellular virions was not affected (Fig. 10), indicating that MuHV-4 pORF63 is not involved in virion morphogenesis or egress. This indicates that, at least in MuHV-4, different mechanisms are involved in capsid trafficking during entry and egress.
One of the features of the MuHV-4 ORF63 STOP mutant was a strong reduction of the size of the plaques (Fig. 4B). Interestingly, this had also been observed in a PrV UL37-deficient strain (18). Moreover, the recent resolution of the crystal structure of the PrV UL37 N terminus (49) identified structural similarities between this protein and cellular multisubunit tethering complexes (MTCs) which control cellular vesicular trafficking by tethering transport vesicles to their destination membranes. This could therefore allow pUL37 to control virus trafficking to cell junctions for cell-cell spread. Interestingly, point mutations in the R2 region of UL37 of PrV were associated with a reduction of plaque size without any defect in virion morphogenesis or production in the supernatant (49). Similarly, the MuHV-4 ORF63 STOP strain displayed a reduction in plaque size (Fig. 4B) without any deficit in extracellular virion production (Fig. 10A and B). MuHV-4 pORF63 could therefore be involved in cell-cell spread similarly to PrV pUL37. However, due to very poor sequence conservation, structural similarities between the PrV pUL37 R2 region and MuHV-4 pORF63 were very difficult to decipher (data not shown) and will require further analysis to determine if MuHV-4 pORF63 also share structural similarities with MTCs.
The phenotype of the MuHV-4 ORF63 STOP mutant could be the direct consequence of the absence of pORF63 or could reflect differences in the incorporation or in the conformation of other tegument proteins. Especially, pUL37 of alphaherpesviruses have been shown to interact with pUL36 (55–58). The positional homolog of alphaherpesvirus UL36 is ORF64 in rhadinoviruses. Therefore, in the future, interaction of MuHV-4 pORF63 with pORF64 will have to be tested and comparison of the phenotype of ORF63 and ORF64 mutants will have to be performed. More generally, the consequence of the absence of pORF63 on the incorporation of other MuHV-4 tegument proteins will have to be determined.
Finally, KSHV pORF63 has recently been shown to be a viral NLR homolog that inhibits the inflammasome (14). Based on our results, we cannot determine whether MuHV-4 pORF63 shares the same property. Based on sequence analysis, MuHV-4 pORF63 does not display the critical residues identified in KSHV to mediate interaction with inflammasome. However, subsequent in silico analysis of the KSHV pORF63 suggested that the capacity of KSHV pORF63 to inhibit the inflammasome could rely more on its predominant alpha-helical structure, which would allow this protein to wrap around some target proteins and consequently inhibit conformational changes or recruitment of interacting partners (59). As shown in this study, despite poor sequence conservation, MuHV-4 pORF63 shares an almost entirely alpha-helical structure with its positional homologs (Fig. 1). Similarly to KSHV pORF63, it could therefore inhibit innate immunity in some particular settings. In order to address this question, specific experimental conditions will have to be set up. However, even if this last hypothesis is possible, our results show that in BHK-21 cells (which are likely deficient in interferon production [60]), the main deficiency associated with a lack of pORF63 in MuHV-4 is a reduction of virus particle infectivity due to a defect of incoming capsid migration to the nucleus.
In summary, this study shows that an MuHV-4 strain deficient for pORF63 displays a major growth deficit both in vitro and in vivo. This deficit was associated with neither an increased death of infected cells, a morphogenesis deficit, nor a default of egress. Instead, the migration of incoming capsids appeared to be severely impaired after membrane fusion in the absence of pORF63. This shows therefore that despite extremely poor sequence conservation, γHV pORF63 shares some functional properties with its positional homolog of alphaherpesviruses. However, important differences also exist. In the future, detailed analysis of these similarities and differences could allow us to gain insight into the life cycle of γHVs in particular but also of herpesviruses in general. Ultimately, this could allow us to identify conserved mechanisms that could be targeted for the development of new drugs against herpesvirus infections.
ACKNOWLEDGMENTS
B.M. is a postdoctoral researcher of the Fonds de la Recherche Scientifique–Fonds National Belge de la Recherche Scientifique (FRS-FNRS).
We thank M. Ledecq, C. Delforge, J. Javaux, E. Deglaire, L. Dams, and C. Espert for excellent technical and secretarial assistance.
REFERENCES
- 1.Ackermann M. 2006. Pathogenesis of gammaherpesvirus infections. Vet Microbiol 113:211–222. doi: 10.1016/j.vetmic.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 2.Thorley-Lawson DA, Gross A. 2004. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N Engl J Med 350:1328–1337. doi: 10.1056/NEJMra032015. [DOI] [PubMed] [Google Scholar]
- 3.Verma SC, Robertson ES. 2003. Molecular biology and pathogenesis of Kaposi sarcoma-associated herpesvirus. FEMS Microbiol Lett 222:155–163. doi: 10.1016/S0378-1097(03)00261-1. [DOI] [PubMed] [Google Scholar]
- 4.Henle G, Henle W, Clifford P, Diehl V, Kafuko GW, Kirya BG, Klein G, Morrow RH, Munube GM, Pike P, Tukei PM, Ziegler JL. 1969. Antibodies to Epstein-Barr virus in Burkitt's lymphoma and control groups. J Natl Cancer Inst 43:1147–1157. [PubMed] [Google Scholar]
- 5.Damania B. 2004. Oncogenic gamma-herpesviruses: comparison of viral proteins involved in tumorigenesis. Nat Rev Microbiol 2:656–668. doi: 10.1038/nrmicro958. [DOI] [PubMed] [Google Scholar]
- 6.Coen N, Duraffour S, Snoeck R, Andrei G. 2014. KSHV targeted therapy: an update on inhibitors of viral lytic replication. Viruses 6:4731–4759. doi: 10.3390/v6114731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai W, Jia Q, Bortz E, Shah S, Liu J, Atanasov I, Li X, Taylor KA, Sun R, Zhou ZH. 2008. Unique structures in a tumor herpesvirus revealed by cryo-electron tomography and microscopy. J Struct Biol 161:428–438. doi: 10.1016/j.jsb.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vidick S, Leroy B, Palmeira L, Machiels B, Mast J, Francois S, Wattiez R, Vanderplasschen A, Gillet L. 2013. Proteomic characterization of murid herpesvirus 4 extracellular virions. PLoS One 8:e83842. doi: 10.1371/journal.pone.0083842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bortz E, Wang L, Jia Q, Wu TT, Whitelegge JP, Deng H, Zhou ZH, Sun R. 2007. Murine gammaherpesvirus 68 ORF52 encodes a tegument protein required for virion morphogenesis in the cytoplasm. J Virol 81:10137–10150. doi: 10.1128/JVI.01233-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo H, Wang L, Peng L, Zhou ZH, Deng H. 2009. Open reading frame 33 of a gammaherpesvirus encodes a tegument protein essential for virion morphogenesis and egress. J Virol 83:10582–10595. doi: 10.1128/JVI.00497-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia Q, Chernishof V, Bortz E, McHardy I, Wu TT, Liao HI, Sun R. 2005. Murine gammaherpesvirus 68 open reading frame 45 plays an essential role during the immediate-early phase of viral replication. J Virol 79:5129–5141. doi: 10.1128/JVI.79.8.5129-5141.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen S, Guo H, Deng H. 2014. Murine gammaherpesvirus-68 ORF38 encodes a tegument protein and is packaged into virions during secondary envelopment. Protein Cell 5:141–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaspar M, Gill MB, Losing JB, May JS, Stevenson PG. 2008. Multiple functions for ORF75c in murid herpesvirus-4 infection. PLoS One 3:e2781. doi: 10.1371/journal.pone.0002781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregory SM, Davis BK, West JA, Taxman DJ, Matsuzawa S, Reed JC, Ting JP, Damania B. 2011. Discovery of a viral NLR homolog that inhibits the inflammasome. Science 331:330–334. doi: 10.1126/science.1199478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts AP, Abaitua F, O'Hare P, McNab D, Rixon FJ, Pasdeloup D. 2009. Differing roles of inner tegument proteins pUL36 and pUL37 during entry of herpes simplex virus type 1. J Virol 83:105–116. doi: 10.1128/JVI.01032-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krautwald M, Fuchs W, Klupp BG, Mettenleiter TC. 2009. Translocation of incoming pseudorabies virus capsids to the cell nucleus is delayed in the absence of tegument protein pUL37. J Virol 83:3389–3396. doi: 10.1128/JVI.02090-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leege T, Granzow H, Fuchs W, Klupp BG, Mettenleiter TC. 2009. Phenotypic similarities and differences between UL37-deleted pseudorabies virus and herpes simplex virus type 1. J Gen Virol 90:1560–1568. doi: 10.1099/vir.0.010322-0. [DOI] [PubMed] [Google Scholar]
- 18.Klupp BG, Granzow H, Mundt E, Mettenleiter TC. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J Virol 75:8927–8936. doi: 10.1128/JVI.75.19.8927-8936.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desai P, Sexton GL, McCaffery JM, Person S. 2001. A null mutation in the gene encoding the herpes simplex virus type 1 UL37 polypeptide abrogates virus maturation. J Virol 75:10259–10271. doi: 10.1128/JVI.75.21.10259-10271.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blaskovic D, Stancekova M, Svobodova J, Mistrikova J. 1980. Isolation of five strains of herpesviruses from two species of free living small rodents. Acta Virol 24:468. [PubMed] [Google Scholar]
- 21.Milho R, Smith CM, Marques S, Alenquer M, May JS, Gillet L, Gaspar M, Efstathiou S, Simas JP, Stevenson PG. 2009. In vivo imaging of murid herpesvirus-4 infection. J Gen Virol 90:21–32. doi: 10.1099/vir.0.006569-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen K, Kurgan L. 2007. PFRES: protein fold classification by using evolutionary information and predicted secondary structure. Bioinformatics 23:2843–2850. doi: 10.1093/bioinformatics/btm475. [DOI] [PubMed] [Google Scholar]
- 24.Chen K, Kurgan LA, Ruan J. 2008. Prediction of protein structural class using novel evolutionary collocation-based sequence representation. J Comput Chem 29:1596–1604. doi: 10.1002/jcc.20918. [DOI] [PubMed] [Google Scholar]
- 25.Homaeian L, Kurgan LA, Ruan J, Cios KJ, Chen K. 2007. Prediction of protein secondary structure content for the twilight zone sequences. Proteins 69:486–498. doi: 10.1002/prot.21527. [DOI] [PubMed] [Google Scholar]
- 26.Adler H, Messerle M, Wagner M, Koszinowski UH. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J Virol 74:6964–6974. doi: 10.1128/JVI.74.15.6964-6974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gillet L, Stevenson PG. 2007. Antibody evasion by the N terminus of murid herpesvirus-4 glycoprotein B. EMBO J 26:5131–5142. doi: 10.1038/sj.emboj.7601925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thiry J, Widen F, Gregoire F, Linden A, Belak S, Thiry E. 2007. Isolation and characterisation of a ruminant alphaherpesvirus closely related to bovine herpesvirus 1 in a free-ranging red deer. BMC Vet Res 3:26. doi: 10.1186/1746-6148-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res 19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swain MT, Tsai IJ, Assefa SA, Newbold C, Berriman M, Otto TD. 2012. A post-assembly genome-improvement toolkit (PAGIT) to obtain annotated genomes from contigs. Nat Protoc 7:1260–1284. doi: 10.1038/nprot.2012.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boetzer M, Pirovano W. 2012. Toward almost closed genomes with GapFiller. Genome Biol 13:R56. doi: 10.1186/gb-2012-13-6-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu SM, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam TW, Wang J. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gillet L, May JS, Colaco S, Stevenson PG. 2007. Glycoprotein L disruption reveals two functional forms of the murine gammaherpesvirus 68 glycoprotein H. J Virol 81:280–291. doi: 10.1128/JVI.01616-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Machiels B, Lete C, de Fays K, Mast J, Dewals B, Stevenson PG, Vanderplasschen A, Gillet L. 2011. The bovine herpesvirus 4 Bo10 gene encodes a nonessential viral envelope protein that regulates viral tropism through both positive and negative effects. J Virol 85:1011–1024. doi: 10.1128/JVI.01092-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mast J, Nanbru C, van den Berg T, Meulemans G. 2005. Ultrastructural changes of the tracheal epithelium after vaccination of day-old chickens with the La Sota strain of Newcastle disease virus. Vet Pathol 42:559–565. doi: 10.1354/vp.42-5-559. [DOI] [PubMed] [Google Scholar]
- 37.Glauser DL, Gillet L, Stevenson PG. 2012. Virion endocytosis is a major target for murid herpesvirus-4 neutralization. J Gen Virol 93:1316–1327. doi: 10.1099/vir.0.040790-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.May JS, Colaco S, Stevenson PG. 2005. Glycoprotein M is an essential lytic replication protein of the murine gammaherpesvirus 68. J Virol 79:3459–3467. doi: 10.1128/JVI.79.6.3459-3467.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gillet L, Gill MB, Colaco S, Smith CM, Stevenson PG. 2006. Murine gammaherpesvirus-68 glycoprotein B presents a difficult neutralization target to monoclonal antibodies derived from infected mice. J Gen Virol 87:3515–3527. doi: 10.1099/vir.0.82313-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frederico B, Milho R, May JS, Gillet L, Stevenson PG. 2012. Myeloid infection links epithelial and B cell tropisms of murid herpesvirus-4. PLoS Pathog 8:e1002935. doi: 10.1371/journal.ppat.1002935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hughes DJ, Kipar A, Sample JT, Stewart JP. 2010. Pathogenesis of a model gammaherpesvirus in a natural host. J Virol 84:3949–3961. doi: 10.1128/JVI.02085-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.François S, Vidick S, Sarlet M, Michaux J, Koteja P, Desmecht D, Stevenson PG, Vanderplasschen A, Gillet L. 2010. Comparative study of murid gammaherpesvirus 4 infection in mice and in a natural host, bank voles. J Gen Virol 91:2553–2563. doi: 10.1099/vir.0.023481-0. [DOI] [PubMed] [Google Scholar]
- 43.Lamkanfi M, Mueller JL, Vitari AC, Misaghi S, Fedorova A, Deshayes K, Lee WP, Hoffman HM, Dixit VM. 2009. Glyburide inhibits the cryopyrin/Nalp3 inflammasome. J Cell Biol 187:61–70. doi: 10.1083/jcb.200903124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lété C, Machiels B, Stevenson PG, Vanderplasschen A, Gillet L. 2012. Bovine herpesvirus type 4 glycoprotein L is nonessential for infectivity but triggers virion endocytosis during entry. J Virol 86:2653–2664. doi: 10.1128/JVI.06238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Glauser DL, Kratz AS, Gillet L, Stevenson PG. 2011. A mechanistic basis for potent, glycoprotein B-directed gammaherpesvirus neutralization. J Gen Virol 92:2020–2033. doi: 10.1099/vir.0.032177-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gillet L, Colaco S, Stevenson PG. 2008. Glycoprotein B switches conformation during murid herpesvirus 4 entry. J Gen Virol 89:1352–1363. doi: 10.1099/vir.0.83519-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gillet L, Colaco S, Stevenson PG. 2008. The murid herpesvirus-4 gL regulates an entry-associated conformation change in gH. PLoS One 3:e2811. doi: 10.1371/journal.pone.0002811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dodding MP, Way M. 2011. Coupling viruses to dynein and kinesin-1. EMBO J 30:3527–3539. doi: 10.1038/emboj.2011.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pitts JD, Klabis J, Richards AL, Smith GA, Heldwein EE. 2014. Crystal structure of the herpesvirus inner tegument protein UL37 supports its essential role in control of viral trafficking. J Virol 88:5462–5473. doi: 10.1128/JVI.00163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang W, Greene W, Gao SJ. 2012. Microtubule- and dynein-dependent nuclear trafficking of rhesus rhadinovirus in rhesus fibroblasts. J Virol 86:599–604. doi: 10.1128/JVI.06129-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naranatt PP, Krishnan HH, Smith MS, Chandran B. 2005. Kaposi's sarcoma-associated herpesvirus modulates microtubule dynamics via RhoA-GTP-diaphanous 2 signaling and utilizes the dynein motors to deliver its DNA to the nucleus. J Virol 79:1191–1206. doi: 10.1128/JVI.79.2.1191-1206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pasdeloup D, McElwee M, Beilstein F, Labetoulle M, Rixon FJ. 2013. Herpesvirus tegument protein pUL37 interacts with dystonin/BPAG1 to promote capsid transport on microtubules during egress. J Virol 87:2857–2867. doi: 10.1128/JVI.02676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McElwee M, Beilstein F, Labetoulle M, Rixon FJ, Pasdeloup D. 2013. Dystonin/BPAG1 promotes plus-end-directed transport of herpes simplex virus 1 capsids on microtubules during entry. J Virol 87:11008–11018. doi: 10.1128/JVI.01633-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pasdeloup D, Beilstein F, Roberts AP, McElwee M, McNab D, Rixon FJ. 2010. Inner tegument protein pUL37 of herpes simplex virus type 1 is involved in directing capsids to the trans-Golgi network for envelopment. J Gen Virol 91:2145–2151. doi: 10.1099/vir.0.022053-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klupp BG, Fuchs W, Granzow H, Nixdorf R, Mettenleiter TC. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J Virol 76:3065–3071. doi: 10.1128/JVI.76.6.3065-3071.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mijatov B, Cunningham AL, Diefenbach RJ. 2007. Residues F593 and E596 of HSV-1 tegument protein pUL36 (VP1/2) mediate binding of tegument protein pUL37. Virology 368:26–31. doi: 10.1016/j.virol.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 57.Vittone V, Diefenbach E, Triffett D, Douglas MW, Cunningham AL, Diefenbach RJ. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J Virol 79:9566–9571. doi: 10.1128/JVI.79.15.9566-9571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Desai P, Sexton GL, Huang E, Person S. 2008. Localization of herpes simplex virus type 1 UL37 in the Golgi complex requires UL36 but not capsid structures. J Virol 82:11354–11361. doi: 10.1128/JVI.00956-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boyle JP, Monie TP. 2012. Computational analysis predicts the Kaposi's sarcoma-associated herpesvirus tegument protein ORF63 to be alpha helical. Proteins 80:2063–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karki S, Li MM, Schoggins JW, Tian S, Rice CM, MacDonald MR. 2012. Multiple interferon stimulated genes synergize with the zinc finger antiviral protein to mediate anti-alphavirus activity. PLoS One 7:e37398. doi: 10.1371/journal.pone.0037398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tandon R, Mocarski ES, Conway JF. 2015. The A, B, Cs of herpesvirus capsids. Viruses 7:899–914. doi: 10.3390/v7030899. [DOI] [PMC free article] [PubMed] [Google Scholar]