ABSTRACT
Alphaviruses represent a diverse set of arboviruses, many of which are important pathogens. Chikungunya virus (CHIKV), an arthritis-inducing alphavirus, is the cause of a massive ongoing outbreak in the Caribbean and South America. In contrast to CHIKV, other related alphaviruses, such as Venezuelan equine encephalitis virus (VEEV) and Semliki Forest virus (SFV), can cause encephalitic disease. E2, the receptor binding protein, has been implicated as a determinant in cell tropism, host range, pathogenicity, and immunogenicity. Previous reports also have demonstrated that E2 contains residues important for host range expansions and monoclonal antibody binding; however, little is known about what role each protein domain (e.g., A, B, and C) of E2 plays on these factors. Therefore, we constructed chimeric cDNA clones between CHIKV and VEEV or SFV to probe the effect of each domain on pathogenicity in vitro and in vivo. CHIKV chimeras containing each of the domains of the E2 (ΔDomA, ΔDomB, and ΔDomC) from SFV, but not VEEV, were successfully rescued. Interestingly, while all chimeric viruses were attenuated compared to CHIKV in mice, ΔDomB virus showed similar rates of infection and dissemination in Aedes aegypti mosquitoes, suggesting differing roles for the E2 protein in different hosts. In contrast to CHIKV; ΔDomB, and to a lesser extent ΔDomA, caused neuron degeneration and demyelination in mice infected intracranially, suggesting a shift toward a phenotype similar to SFV. Thus, chimeric CHIKV/SFV provide insights on the role the alphavirus E2 protein plays on pathogenesis.
IMPORTANCE Chikungunya virus (CHIKV) has caused large outbreaks of acute and chronic arthritis throughout Africa and Southeast Asia and has now become a massive public health threat in the Americas, causing an estimated 1.2 million human cases in just over a year. No approved vaccines or antivirals exist for human use against CHIKV or any other alphavirus. Despite the threat, little is known about the role the receptor binding protein (E2) plays on disease outcome in an infected host. To study this, our laboratory generated chimeric CHIKV containing corresponding regions of the Semliki Forest virus (SFV) E2 (domains A, B, and C) substituted into the CHIKV genome. Our results demonstrate that each domain of E2 likely plays a critical, but dissimilar role in the viral life cycle. Our experiments show that manipulation of E2 domains can be useful for studies on viral pathogenesis and potentially the production of vaccines and/or antivirals.
INTRODUCTION
The alphaviruses represent a diverse family of arthropod-borne viruses (arboviruses), many of which are important veterinary or human pathogens. Their transmission cycles involve both an arthropod vector and vertebrate host, resulting in unique evolutionary restrictions. The genus Alphavirus (family Togaviridae) currently includes 29 species that are grouped into 10 complexes based on antigenic, genetic, and/or geographic similarities (1). The aquatic alphaviruses infect marine mammals and have been isolated in lice, although their role as a vector remains unknown (2). The New World alphaviruses include important veterinary and human pathogens, such as the equine encephalitis viruses, Venezuelan (VEEV), eastern (EEEV), and western (WEEV), all of which cause fatal encephalitic disease in both humans and animals such as horses and birds (3). The Old World alphaviruses, present mostly in Africa and Southeast Asia, are commonly associated with arthritic disease, fever, and rash.
Chikungunya virus (CHIKV) is the most medically relevant Old World alphavirus and is the current cause of an outbreak of arthritic disease in the Americas, with more than 1 million cases in at least 44 countries, including the United States (4). CHIKV, which reemerged in 2004, has previously caused explosive epidemics of acute and chronic arthritis in humans in Africa, India, and southern Europe (5). Clinically, it resembles several other arboviral diseases, but it is often associated with high fever and severe, incapacitating joint pain, particularly in the small joints, that can last for months or years (6). Semliki Forest virus (SFV), the type species of the SFV complex of Old World alphaviruses (of which CHIKV is a member), is one of its most studied members. Despite its laboratory importance, SFV is not considered to be a medically important arbovirus, although it was the cause of a fatal case of laboratory acquired meningoencephalitis (7). SFV has caused small outbreaks of disease in the Central African Republic (CAR) in 1987, with symptoms characterized by fever and extremely severe, persistent headaches (8). In addition, serosurveys in Kenya (9), Uganda (10), Nigeria (11), and CAR (12), among other African nations, have detected up to 25% prevalence of SFV specific antibody, indicating that many human infections do occur.
Alphaviruses are small, enveloped, positive-sense, single-stranded RNA viruses with a worldwide distribution. The roughly 12-kb alphavirus genome is capped and polyadenylated and has two open reading frames, encoding nonstructural (nsPs) and structural (sPs) polyproteins, respectively (13). Translation of the nsPs produces a polyprotein that is cleaved into nsP1, nsp2, nsp3, and nsp4 proteins, which function to replicate the genome, modulate cellular functions, and cleave the polyprotein, among other functions (14). The sPs are translated from a subgenomic RNA that is produced by the replicase recognizing an internal promoter in the minus-strand replicative intermediate RNA (15). The sPs consist of the capsid, E3, E2, 6K/TF, and E1 proteins. The E1 protein is responsible for membrane fusion and is thought to play a role in immunogenicity and host range (16). E2, the receptor binding protein, contains residues critical for immunogenicity, host range, and tissue/cell tropism (reviewed in reference 17). The E2 protein consists of three domains (A, B, and C), of which A and B have been found to contain the majority of residues that affect cell attachment and or tissue/cell tropism (17). While individual mutations have been shown to have various effects on these factors, the role of each of E2 domain on replication, pathogenesis, host range, and tropism is not well understood.
Previous efforts to understand alphavirus proteins and protein domains have led to a deeper appreciation of the complexity of the alphavirus-host cell interplay and demonstrated the utility of using chimeric viruses to understand basic cellular and virological processes. For example, envelope protein (E3-E2-6K-E1) chimeras between Sindbis virus (SINV) and SFV provided insight on alphavirus protein interactions (18). It was determined that SFV/SIN but not SIN/SFV was viable, likely due to abrogated capsid-spike interaction. Furthermore, chimeric SINV/Ross River virus constructs have been used to identify important residues in budding, nucleocapsid/envelope-protein interactions, and virus assembly (19–22). Chimeric alphaviruses also have been developed as vaccine candidates, usually swapping the entire sPs from one virus into another for attenuation (23, 24). Others have used chimeras to probe the role of viral proteins on important viral factors such as host tropism or infectivity. For example, the use of CHIKV/O'nyong nyong virus (ONNV) chimeras demonstrated that ONNV requires the entire sPs for infection of anopheline mosquitoes (25). Further, more comprehensive work looked at individual genes with chimeric viruses and determined that nsp3 is the critical determinant allowing for ONNV's rare ability to infect anopheline mosquitoes (26).
To date, chimeric viruses between Old and New World alphaviruses have been restricted to swapping the complete nsP or sP polyprotein for use as vaccine candidates (24, 27, 28). Although useful as vaccines, and potentially for studying the interaction between nsP and sP proteins, these types of chimeric viruses do not provide sufficient specificity to examine the role of individual proteins. Few reports exist of attempting to swap individual genes or domains of genes of alphaviruses, and we are not aware of any that look specifically at E2 or its domains. Accordingly, we constructed and characterized chimeras between CHIKV and VEEV/SFV to understand the role of E2 protein domains on alphavirus pathogenicity. We have previously described these chimeras and the role of the E2 domains in protective immunity and antibody neutralization (29). In this report, using these chimeras, we were able to identify domains of the E2 protein which are important for replication and pathogenicity of CHIKV.
MATERIALS AND METHODS
Ethics statement.
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The IACUC protocol (protocol V01380) was approved by the Institutional Animal Care and Use Committee of the University of Wisconsin.
Cells and viruses.
BHK-21 (ATCC CCL-10, hamster kidney fibroblast), MRC-5 (ATCC CCL-171, human lung fibroblast), BV-2 (murine microglia; courtesy of Grace Sun, University of Missouri), SHSY5Y (ATCC CRL-2266, human neuroblastoma) were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), nonessential amino acids, sodium pyruvate, 10 mM HEPES, and penicillin-streptomycin at 37°C under 5% CO2. HMEC-1 (ATCC CRL-3243, human endothelial) cells were maintained in DMEM with 10 ng of epidermal growth factor (EGF)/ml, 1 ng of hydrocortisone/ml, 10 mM l-glutamine, and 10% FBS. CG-4 cells (rat oligodendrocyte precursors; courtesy of Ian Duncan, University of Wisconsin) were maintained and differentiated as previously described (30). Briefly, CG-4 precursors were grown in DMEM with 30% conditioned medium from B104 (B104-CM) (rat neuroblastoma cells), N1 supplement (50 μg of transferrin/ml, 5 μg of insulin/ml, 100 mM putrescine, 20 nM progesterone, and 30 nM selenium), and 10 ng of biotin/ml. Differentiation to mature oligodendrocytes was achieved by incubating CG-4 cells with DMEM-N1-biotin lacking B104-CM for 48 h, followed by the addition of 20% FBS for an additional 4 days. CG-4 cells were differentiated into type II astrocytes in the same manner, except that 20% FBS was immediately added. The cells were then allowed to differentiate for 6 days before infection. Differentiation was confirmed by staining with oligodendrocyte- or astrocyte-specific antibodies (CNPase and GFAP, respectively [data not shown]). C6/36 (Aedes albopictus) cells were maintained at 28°C in MEM with 10% FBS. Chikungunya virus SL-CK1 and VEEV vaccine strain TC-83 were obtained from Scott Weaver (University of Texas Medical Branch). The neurovirulent Semliki Forest virus strain L10 was obtained from John Fazakerley (The Pirbright Institute).
Construction of chimeric viruses.
A Sindbis virus infectious clone under the control of a cytomegalovirus (CMV) promoter was used as the backbone for construction of all cDNA clones (courtesy of Brian Geiss, Colorado State University) (31). The CMV promoter allowed for direct transfection or electroporation in mammalian cells without production of RNA. Cloning was performed using overlap extension PCR (OE-PCR) (32), replacing the 5′ untranslated region (5′UTR)/nonstructural proteins and the structural proteins/3′UTR in two cloning steps. Each of the viruses was engineered to express green fluorescent protein (GFP) under a second subgenomic promoter 5′ to the structural polyprotein (33). Chimeric viruses were constructed using OE-PCR to replace the different domains of the CHIKV E2 with those corresponding to VEEV or SFV. Primer and clone sequences are available upon request.
Sequencing.
For all viruses generated, the entire genome was sequenced from the cDNA clone. In addition, the entire structural polyprotein was sequenced for all virus stocks recovered from transfections, which maintained 100% match with cDNA sequences. Sequencing was performed by first extracting viral RNA using a ZR Viral RNA kit (Zymo Research, Irvine, CA). Reverse transcription (RT) was then performed to produce cDNA using the Superscript III reverse transcriptase first-strand synthesis system (Invitrogen, Carlsbad, CA). cDNA then was used as a template for PCR with virus-specific primers using Q5 high-fidelity polymerase (New England BioLabs, Ipswich, MA). Amplicons were sequenced at the University of Wisconsin Biotechnology Center DNA sequencing facility according to established protocols. Sequences were assembled and aligned using Vector NTI (version 11.5; Invitrogen).
Electroporation and virus production.
Infectious virus was rescued by electroporation of 2 μg of Maxi-Prep purified plasmid (Zymo Research, Orange, CA) into an 80 to 90% confluent T175 flask of BHK-21 cells using Cytomix electroporation buffer at infinite resistance, 300 V, and 960-μF capacitance in a 2-mm cuvette (34). Supernatant was harvested 1 to 3 days (when ∼50 to 75% cytopathic effect [CPE] was evident) after electroporation, and cellular debris was removed by centrifugation at 2,000 × g for 20 min at 4°C. Virus was then pelleted by centrifugation at 13,500 × g overnight at 4°C. The viral pellet was resuspended in TEN buffer and stored in small volume single-use aliquots at −80°C. The virus titer was determined by plaque assay on BHK-21 cells as previously described (35). Briefly, 10-fold serial dilutions of virus were inoculated onto six-well plates of confluent BHK-21 cells. After 1 h for virus adsorption, the virus inoculum was removed, and the cells were washed extensively with phosphate-buffered saline (PBS). Then, 1.5% carboxymethyl cellulose (CMC) in DMEM with 10% FBS was added as a viral overlay. After 36 h of incubation, the CMC was discarded, and the cells were fixed in 4% paraformaldehyde (PFA), followed by staining with crystal violet (CV). Plaques were then counted by hand and titers are reported as PFU/ml. Rescued viruses were sequenced and subjected to RT-PCR to confirm the chimeric genotype. To determine plaque size after electroporation, plasmids were electroporated as described, except that the cells were subjected to serial 10-fold dilutions into uninfected cells and then seeded into six-well plates. After a 3-h attachment period, a CMC overlay was added to allow plaque formation. Then, 36 h later, the plaques were stained with CV and imaged, and photos taken using an Evos FL microscope. Plaque size was then measured for 21 individual plaques for each virus using ImageJ software and expressed as squared pixels.
Molecular modeling.
CHIKV, SFV, and VEEV structural data were obtained from the Protein Data Bank (PDB codes 2XFC, 3J0C, 3J2W, and 3N42). CHIKV E2 protein with SFV and VEEV domain sequence substitutions were threaded onto the native CHIKV envelope structure (2XFC) (17) using iTasser (36). Predicted structures were then visualized with Pymol (Schrödinger, LLC), and specific amino acid interactions were manually visualized and compared.
Growth curves.
Cells for growth curves were seeded into 24-well plates the night before infection. The following day, 80 to 90% confluent cells were infected at a multiplicity of infection (MOI) of 0.1 (multistep) or 10 (one-step) PFU/cell in duplicate in 24-well plates to assess viral growth kinetics in vitro. After 1 h of absorption, monolayers were washed three times with PBS to remove unbound virus, and prewarmed medium was added. This was considered time point zero. At the time points indicated after infection, 200 μl of supernatant was removed and replaced with fresh prewarmed medium. The infectious virus titer was assessed using a 50% tissue culture infective dose (TCID50) assay on BHK-21 cells and are reported as the TCID50/ml (37). Each growth curve was repeated twice or more, with consistent results.
Genome/PFU ratios.
Quantitative real-time PCR (qRT-PCR) was used to determine the number of CHIKV or chimeric virus genomes/ml in three different cell lines (BHK-21, C6/36, and CG-4 oligodendrocytes). The approach we used was very similar to the method previously reported by Silva et al. (38), including the same primer and probe set. Briefly, BHK-21, C6/36, or CG-4 oligodendrocytes were infected with each virus at an MOI of 0.1 in triplicate. After 24 h of incubation, the supernatant was harvested, and the cell debris was removed via centrifugation. Then, 50 μl of supernatant was treated with RNase A (Fermentas) to remove nonencapsulated viral RNA, and RNA was extracted using a Direct-Zol RNA miniprep kit (Zymo Research) according to the manufacturer's instructions. An iTaq Universal probe supermix (Bio-Rad) was used for quantification, with final concentrations of 450 nM forward primer, 900 nM reverse primer, and 200 nM probe (IDT) and 5 μl of RNA. The cycling conditions were as follows: 10 min at 50°C, 3 min at 95°C, and then 40 cycles of 15 s at 95°C and 30 s at 60°C, with data acquisition using FAM reporter during the latter step. In vitro-transcribed RNA from a CHIKV SL-CK1 infectious clone under the control of an SP6 promoter was used to generate a standard curve, using serial 10-fold dilutions. Threshold cycle (CT) values from virus samples were plotted against the standard curve to obtain genome equivalents. The mean of two technical replicates for each of the three biological replicates was then expressed as genomes/ml. This value was then divided by the mean number of PFU/ml for three biological replicates and is expressed as genome equivalents (GE)/PFU.
Heparin competition assays.
Assays to assess the effect of heparin on virus infection were performed as previously described (39). Virus was diluted to an MOI of 2.5 in Opti-MEM (Invitrogen) and then mixed with either heparin or bovine serum albumin (BSA; final concentration, 200 μg/ml). The virus-heparin or virus-BSA mixtures were allowed to incubate at 4°C for 30 min and then used to infect either Vero or BHK-21 cells in black 96-well plates (Corning). After 2 h of incubation at 37°C for virus adsorption, the virus inoculum was removed and replaced with complete medium containing 20 mM NH4Cl. Subsequently, after 18 h of incubation at 37°C, the cells were fixed with fresh 4% PFA in PBS and then stained with Hoechst, followed by five washes with PBS. Enhanced GFP (eGFP) and Hoechst levels were measured using a BioTek Cytation 3 plate reader with specific parameters (eGFP, excitation at 485 nm [20-nm bandwidth] and emission at 515 nm [20-nm bandwidth]; Hoechst, excitation at 360 nm [35-nm bandwidth] and emission at 465 nm [20-nm bandwidth]), using the bottom reading mode and covering the entire well. eGFP was normalized to the amount of cells in each well (with Hoechst) and was used as a proxy for virus replication. Data from each virus were normalized to infection when incubated with only BSA.
Mosquito infections.
The vector competence of the Aedes aegypti black-eyed Liverpool (LVP) strain was evaluated for chimeric alphaviruses. The LVP used in these studies was maintained at the University of Wisconsin—Madison as previously described (40). Infection, dissemination, and transmission rates were determined according to long-established procedures (41, 42), with one exception. After homogenization and inoculation on BHK cell culture, the cells were monitored for GFP expression to confirm virus-positive samples. The cells fixed with PFA were examined for GFP expression using phase-contrast optics on an EVOS fl inverted fluorescence microscope at ×10 magnification (Life Technologies). Briefly, mosquitoes were exposed to virus-infected blood meals via water-jacketed membrane feeders maintained at 36.5°C (43). Blood meals consisted of defibrinated sheep blood (HemoStat Laboratories) and chimeric virus from frozen stock, yielding an infectious blood meal concentration of 7.0 log10 PFU/ml for each virus. Five- to seven-day-old female mosquitoes were sucrose starved for 14 to 16 h prior to blood feeding. Mosquitoes that fed to repletion were separated into cartons and maintained on 0.3 M sucrose in an environmental chamber at 26.5°C ± 1°C and 75 ± 5% relative humidity and with a 16-h photoperiod within the Department of Pathobiological Sciences BSL3 Insectary Facility at the University of Wisconsin—Madison. Infection was determined by virus-positive bodies, whereas dissemination was indicated by virus-positive legs. Transmission was defined as the release of infectious virus with salivary secretions, i.e., the potential ability to infect another host, and was indicated by virus-positive salivary secretions.
Mouse experiments.
Ifnar1 −/− mice in a 129/Sv background (A129) were obtained from B&K Universal, Limited (Hull, England), and bred onsite at the University of Wisconsin—Madison. C57BL/6 mice were purchased from Jackson Laboratories (Bar Harbor, ME) and were maintained at the University of Wisconsin. Six- to nine-week-old mixed sex mice were used for all experiments involving A129 mice. Six-week-old male mice were used for all experiments in C57BL/6 mice. Virus was diluted in RPMI 1640 containing 1% BSA, 10 mM HEPES, and penicillin-streptomycin for all mouse experiments. Mice were infected in the left hind footpad with either 102 or 104.5 PFU for A129 mice or 105 PFU for C57BL/6 mice. For intracranial (i.c.) infections, mice were placed under heavy anesthesia with isoflurane and then infected with 20 μl of virus using a 27-gauge needle. Then, 102 and 103 PFU of virus were used for the i.c. infection of A129 and C57BL/6 mice, respectively. After infection, mice were monitored twice daily for the duration of the study. Mice that were moribund or that lost greater than 20% of starting weight were humanely euthanized using CO2. Submandibular blood draws were performed using a 4-mm lancet and serum was collected by centrifugation at 3,000 × g for 15 min.
Histopathology and fluorescent microscopy.
Brains were harvested from mice and fixed for 16 to 24 h in 2% PFA. Tissues for paraffin embedding were submitted to the Histology Laboratory at the School of Veterinary Medicine at the University of Wisconsin—Madison, where they were processed and sectioned before staining with hematoxylin and eosin. For frozen sections, fixed brains were cryoprotected with 30% sucrose overnight in PBS, embedded in OCT medium, snap-frozen on metal blocks partially submerged in liquid nitrogen, and stored at −80°C until sectioning. Sectioning was performed at the University of Wisconsin—Madison by the UWCCC Experimental Pathology Laboratory. Slides containing tissues were stored at −80°C until use. To assess GFP expression, frozen slides were warmed to room temperature, followed by acetone treatment at −20°C for 10 min. Autofluorescence was then blocked with 300 mM glycine in PBS (pH 7.4) for 1 h. Sections were then mounted in ProLong Gold Antifade Reagent with DAPI (4′,6′-diamidino-2-phenylindole; Cell Signaling Technology, Danvers, MA) and coverslipped. An EVOS fl inverted fluorescence microscope was used for imaging equipped with ×10 and ×20 objective lenses. Tissues from multiple experiments and depth of brain tissue were examined, and representative images are presented from each group.
Data analysis.
Statistical analyses were run using GraphPad Prism (version 6; GraphPad, San Diego, CA). Replication and viral load data were analyzed by using the Mann-Whitney test. One-way and two-way analysis of variance (ANOVA) was used to assess plaque sizes and genome equivalent/PFU ratios. Mosquito infection rates were compared by using the Fisher exact test. Survival analyses were performed using Kaplan-Meier curves with the log-rank test. An alpha of 0.05 was used for all studies as the threshold for significance. Variances were compared by using the F test. All experiments other than mosquito infections were repeated twice or more with consistent results.
RESULTS
CHIKV/SFV, but not CHIKV/VEEV E2 chimeras can be rescued and replicate in vitro.
Using OE-PCR (32), a panel of chimeric CHIKV/VEEV and CHIKV/SFV cDNA clones were constructed to replace different portions of the CHIKV E2 with those corresponding to the same region in the VEEV or SFV genome. When CHIKV/VEEV clones were electroporated into BHK-21 cells, they were uniformly nonviable, since no CPE was observed, nor was infectious virus detected by plaque assay (data not shown). In contrast, many of the CHIKV/SFV chimeras were viable and replicated well in vitro (Table 1). The inability to rescue CHIKV/VEEV, but not CHIKV/SFV chimeras is likely due to lower amino acid sequence similarity, which likely resulted in disrupted protein interactions. To test this, parental and chimeric CHIKV/VEEV and CHIKV/SFV E2 proteins were mapped onto the three-dimensional structure of the CHIKV envelope glycoprotein complex (PDB 2XFC) (17) (Fig. 1). Table 2 summarizes the structural effect that variances in key wild-type (WT) amino acids in both CHIKV/VEEV and CHIKV/SFV have on interactions between E1 and E2 in these chimeras. The loss of several hydrogen binding sites and charge interactions, accompanied by steric disturbances, likely result in the inability to rescue any of the CHIKV/VEEV constructs. No significant disruptions in key amino acid interactions between E3 and E2 were observed for either set of chimeric viruses (data not shown).
TABLE 1.
Chimeric CHIKV/SFV viruses constructed and their viabilities
| Gene or domain swapped | Viability |
|---|---|
| Full E2 | No |
| E2 (without cytoplasmic domain) | No |
| E3-E2 (without cytoplasmic domain) | No |
| E2 domain A | Yes |
| E2 domain B | Yes |
| E2 domain C | Yes |
FIG 1.
CHIKV/SFV E2 chimeras have several differences in interacting residues. (A) Top view of the published structure 2XFC in a cartoon tube. CHIKV E1 subunits are shown in blue, red, and orange. CHIKV E2 subunits are shown in green, yellow, and purple. E3 (from PDB 3N42) is shown as the lime green surface. Subunit backbone colorations are the same in subsequent panels. (B) Expanded view of the CHIKV E2 subunit. Domain A is yellow, domain B is orange, and domain C is blue. SFV residues which are substituted in the three different chimeras are shown in cyan, magenta, and yellow sticks for domains A, B, and C, respectively. (C to E) Wild-type (WT) CHIKV residues are shown as sticks. Residues in E1 are shown as sticks colored to match the backbone. Green sticks represent WT E2 residues which differ in the chimeras, and orange sticks represent WT residues that do not change in the chimeras. (C) Two intermolecular hydrogen bonds and one intramolecular salt bridge are lost, and one intermolecular salt bridge is weakened when SFV residues replace CHIKV residues in domain A. (D) One hydrogen bond is lost with a backbone carbonyl of E1 when K200 in CHIKV is substituted with N200 of SFV domain B. E3 does not make contact with substituted residues. (E) CHIKV E2 interacts with two E1 subunits at domain C. Three substitutions disrupt a hydrophobic pocket for I387 of one E1 subunit, and two hydrogen bonds and one stacking/hydrophobic interaction are lost in the other E1 when SFV substitutions are made. For panels C to E, see Table 2 for specific residues and interactions. (F) The amino acid sequences of CHIKV, VEEV, and SFV E2 were aligned using ClustalW and are displayed using ESPript. Colored bars above the alignment represent each of the domains, which correspond to the color shown in panels A to E.
TABLE 2.
Interactions disrupted by SFV (strain L10) and VEEV (strain TC-83) domain swapping in CHIKV (strain SL-CK1) E2 protein
| Swapped domain | Originating virus | Total no. of residue changes | Critical interacting residue substitutions | Specific disruption |
|---|---|---|---|---|
| Domain A | SFV | 39 | R104T | Inter-E2 salt bridge with D43 |
| H18Y | Inter-E2 H-bond with T228 | |||
| R36A | Salt bridge with E112 of E1 (R38 may partially reduce this loss) | |||
| Domain B | SFV | 22 | K200N | Weakened H-bonding w/backbone carbonyl of E1 (C62) |
| Domain C | SFV | 34 | P340S | Hydrophobic interaction/side chain orientation |
| Y278H, Y338L | Hydrophobic interaction w/I387 of E1; also loss of intramolecular stacking interaction | |||
| K314A | H-bond with backbone of K241 in E1 | |||
| Y288H | Stacking/hydrophobic interaction w/P237 of E1 | |||
| T277I | H-bond with R196 of E1 | |||
| Domain A | VEEV | 77 + 2 deletions | E24V | H-bond (T156/158) or possibly salt bridge (R168) |
| D21R | Inter-E2 salt bridge with R144 or H-bond with S143 | |||
| R36A, R38K | Salt bridge with E112 of E1 | |||
| H26− | Possible H-bond loss | |||
| H18R | Steric clash | |||
| Domain B | VEEV | 43 + 3 insertions | K200E | H-bond loss |
| H299R | Steric clash | |||
| A231Y | Steric clash | |||
| N203E | (–) charge close to D97 of E1 | |||
| K208+ | Addition closes off hydrophobic pocket for F95 | |||
| R178S | Intramolecular salt bridge w/E223 | |||
| Domain C | VEEV | 48 | L345E | (–) charge close to TM domain/membrane (difficult to say w/o other data) |
| K314A | H-bond to backbone K241 of E1 | |||
| Y288H | Stacking/hydrophobic interaction w/P237 of E1 | |||
| R272P | H-bond w/Q235 of E1 | |||
| T275M | H-bond w/Q218 of E1 | |||
| K270L | H-bond w/Q222 of E1 |
CHIKV/SFV chimeras had fewer amino acid differences, particularly those with strong structural effects, as expected given their genetic relatedness (Table 3). Three CHIKV/SFV chimeras (here called ΔDomA, ΔDomB, and ΔDomC) were rescued successfully and selected for further study (Fig. 2). Each represents the corresponding domain from the SFV E2 replaced into the backbone of CHIKV. GFP also was included in the genome of all viruses to allow visualization of virus infection both in vitro and in vivo. Alphaviruses expressing recombinant proteins through a second subgenomic promoter have been well characterized and have been shown to be useful for tracking viral replication and spread (33, 44–46). RT-PCR and DNA sequencing of RT-PCR products confirmed that rescued viruses maintained their chimeric composition (data not shown).
TABLE 3.
Amino acid differences between select CHIKV (strain SL-CK1) proteins (and domains) and VEEV (strain TC-83) and SFV (strain L10)
| Alphavirus protein or domain | Length (no. of amino acids) | % identity with CHIKV |
|
|---|---|---|---|
| VEEV | SFV | ||
| Full E2 | 423 | 35.7 | 57.4 |
| E2 domain A | 132 | 35.8 | 59.8 |
| E2 domain B | 59 | 25.8 | 62.7 |
| E2 domain C | 73 | 34.2 | 53.4 |
FIG 2.
Genome organization of chimeric CHIKV/SFV viruses. Genome organization of chimeric CHIKV/SFV viruses. The different domains of E2 from SFV were inserted into the CHIKV genome in the corresponding position in individual constructs using a PCR-based cloning approach. Each virus expressed the GFP protein under the control of a second subgenomic promoter. Red portions of the E2 represent genetic sequences of SFV, whereas CHIKV is shown in blue.
Chimeric CHIKV/SFV have different levels of attenuation in vitro.
In an effort to determine whether chimeric viruses had altered replication kinetics, BHK-21 and C6/36 cells were inoculated at MOIs of 0.1 and 10 PFU/cell, and comparisons between chimeric viruses and wild-type viruses (CHIKV and SFV) were made over the course of 24 h (one-step) or 36 h (multistep) (Fig. 3). No significant differences in replication were observed between CHIKV and any of the chimeras in one-step growth curves on BHK-21 cells (Fig. 3A). In contrast, ΔDomC was significantly attenuated in multi- and one-step growth curves on C6/36 and in multistep on BHK-21 cells (Fig. 3B to D). CHIKV, ΔDomA and ΔDomB viruses had no significant differences in growth at any of the time points tested. SFV, as expected, replicated to significantly higher titers on BHK-21 cells in both one- and multistep growth curves. The role of nsPs can be inferred from studies on alphavirus nsP/sP chimeras, which show replication kinetics similar to the parental virus of its nsPs (47). In contrast, on C6/36 cells, CHIKV and SFV replication kinetics were similar in one-step but not multistep growth curves, with the latter producing significantly less virus at 10 and 13 h postinfection (Fig. 3B and D).
FIG 3.
In vitro characterization of CHIKV/SFV chimeras. (A to D) Growth curves of each virus in BHK-21 (A and C) and C6/36 (B and D). Cells were exposed to either high (MOI = 10)- or low (MOI = 0.1)-MOI infection in one-step (A and B) and multistep (C and D) growth curves, respectively. The supernatant was removed at the indicated time points postinfection, and then infectious virus titers were measured using a TCID50 assay.
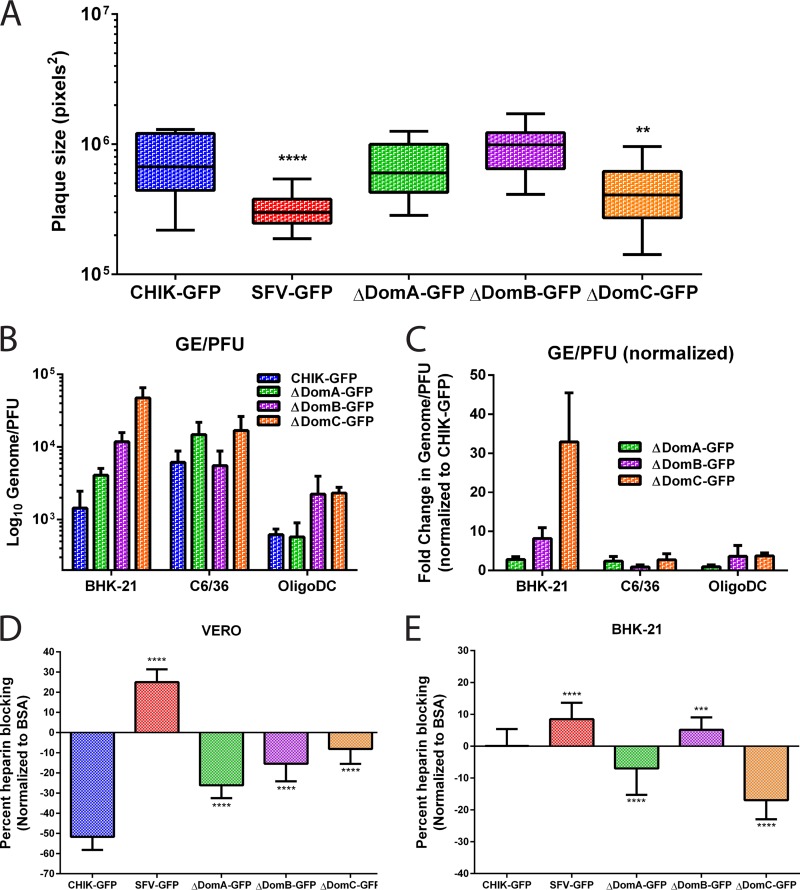
To examine plaque size and phenotypic homogeneity following viral rescue, cells electroporated with virus from each construct were diluted with uninfected cells and then allowed to form plaques under a semisolid overlay. There were no differences in plaque size between CHIKV or the ΔDomA or ΔDomB viruses (Fig. 4A). Plaque sizes following electroporation were significantly smaller for ΔDomC or SFV compared to CHIKV (both P < 0.01 using one-way ANOVA with Tukey's multiple-comparison test). There was no difference between the variance of plaque sizes in ΔDomA or ΔDomB compared to CHIKV. However, ΔDomC had a significantly different variance for plaque sizes (P = 0.02), suggesting more heterogeneity in plaque size, which may be suggestive of virus adaptation.
FIG 4.
Plaque size, specific infectivity, and effects of soluble heparin on infectivity of chimeric viruses. (A) Plaque formation after electroporation of parental and chimeric CHIKV/SFV. Infectious clones were electroporated into BHK-21 cells and then mixed with uninfected cells. After attachment, the cells were overlaid with 1.5% CMC media and incubated for 48 h. After incubation, the CMC was discarded, and the cells were fixed with 4% paraformaldehyde in PBS and subsequently stained with crystal violet to visualize plaques. Twenty-one plaques for each virus were imaged using an Evos FL microscope, and plaque sizes were measured using ImageJ software. (B and C) Specific infectivity (GE/PFU) of CHIK-GFP and chimeric viruses (B) or GE/PFU values for the chimeric viruses normalized to CHIK-GFP (C). GE/PFU ratios were evaluated in three cell types; BHK-21, C6/36, and OligoDC. (D and E) Effect of soluble heparin on infection of parental or chimeric viruses in Vero (D) or BHK-21 (E) cells. Virus was mixed with heparin or BSA (200 μg/ml) at 4C for 30 min and then used to infect cells. Virus infection was measured using GFP expression as a proxy for virus replication and normalized to both the amount of cells (using Hoechst) and to virus infection with BSA instead of heparin.
To assess viral fitness, genome/PFU ratios were determined for three cell types (BHK-21, C6/36, and CG-4 oligodendrocytes). Parental CHIKV had mean genome/PFU ratios of 1,435, 6,116, and 617, respectively, in the three cell lines tested; the value in BHK-21 cells is consistent with a previous report with a similar strain without GFP (38) (Fig. 4B). Although mean genome/PFU ratios were mostly higher for the chimeric viruses than CHIKV, the only significant difference observed was between CHIKV and ΔDomC in BHK-21 cells (P = 0.0002). When the chimeric viruses were compared to one another by normalizing the genome/PFU ratio to CHIKV, ΔDomC had a significantly higher ratio than either ΔDomA or ΔDomB in BHK-21 cells (P < 0.01 for both) (Fig. 4C).
The effect of soluble heparin in infectivity varies for each virus and between cell types.
Previous studies have shown that glycosoaminoglycans (GAGs), such as heparin sulfate (HS) have a major influence on cell binding of CHIKV and infectivity (38, 39). To investigate the effect that chimerization may have had on GAG binding, we incubated the viruses with the HS analog, heparin, and measured the effect on infectivity in both Vero and BHK-21 cells. In Vero cells, a significant enhancement in infectivity (as measured by GFP prior to the virus completing a round of replication) was observed in parental CHIK-GFP and all of the chimeras compared both to BSA or when normalized and compared to SFV-GFP (P < 0.01 for all comparisons by One-way ANOVA) (Fig. 4D). SFV-GFP produced significantly less GFP when incubated with heparin, a finding suggestive of reduced infectivity (P < 0.001). Interestingly, the chimeric viruses all had significantly increased GFP expression in the presence of heparin, though at intermediate levels between CHIK-GFP and SFV-GFP, likely reflecting HS binding sites within the E2 protein in each of the domains (all P < 0.01). In contrast, in BHK-21 cells, similar to data previously reported (38), no effect was observed on infectivity for parental CHIKV (Fig. 4E). However, there was a significant reduction in the amount of GFP produced by SFV after incubation with heparin, as in Vero cells (P < 0.001). The ΔDomA and ΔDomC viruses showed significantly increased infectivity, similar to Vero cells but in contrast to parental CHIK-GFP (P < 0.001). In contrast to this, ΔDomB showed a reduction in GFP production similar to that of SFV-GFP, indicating a similar effect of heparin on infectivity in this cell line.
Infection and dissemination in Aedes aegypti mosquitoes.
To assess the effect that E2 domains have on mosquito vector competence, groups of female A. aegypti were exposed to infectious blood meals containing each virus. Fully engorged mosquitoes were placed in new containers and subsequently were assayed for infection, dissemination, and transmission potential at 7 and 12 days post-blood feeding (PF) (Table 4). As expected, infection and dissemination rates were high for A. aegypti exposed to blood containing CHIK-GFP at both 7 and 12 days PF (33). In contrast, there was a significant reduction (Fisher exact test, P < 0.05) in infection, dissemination, and transmission rates for A. aegypti exposed to blood containing SFV-GFP or ΔDomC. SFV was originally isolated from Aedes abnormalis mosquitos (48) and in previous epidemiological studies was rarely isolated from A. aegypti (8). ΔDomC had the lowest rates of infection, dissemination, and transmission compared to CHIK-GFP on both days 7 and 12 PF. However, no significant differences were observed in the vector competence of A. aegypti for ΔDomC and SFV-GFP, i.e., this mosquito species displayed poor peroral vector competence for both viruses. Although significant differences were observed in the dissemination and transmission rates between CHIK-GFP and ΔDomA on day 7, the latter virus had similar rates by day 12, a finding consistent with the slower replication rates observed in vitro. Interestingly, no differences were observed between CHIK-GFP and ΔDomB, suggesting that this domain plays a limited role in vector competence of A. aegypti for CHIKV.
TABLE 4.
Rates of infection, dissemination, and transmission potential in Aedes aegypti mosquitoesa
| Virus | Infection rate (%) |
|||||
|---|---|---|---|---|---|---|
| Day 7 postfeeding |
Day 12 postfeeding |
|||||
| Infection | Dissemination | Transmission | Infection | Dissemination | Transmission | |
| CHIK-GFP | 40/40 (100) | 37/40 (92.5) | 14/40 (35) | 39/40 (97.5) | 35/39 (90) | 10/39 (26) |
| ΔDomA-GFP | 38/39 (97.4) | 11/38 (29) | 2/38 (5) | 37/40 (92.5) | 32/37 (86) | 17/37 (46) |
| ΔDomB-GFP | 40/40 (100) | 37/40 (92.5) | 17/40 (42.5) | 40/40 (100) | 38/40 (95) | 18/40 (45) |
| ΔDomC-GFP | 8/40 (20) | 2/8 (25) | 1/8 (13) | 11/40 (27.5) | 4/11 (36) | 3/11 (27) |
| SFV-GFP | 17/40 (42.5) | 5/17 (29) | 0/40 (0) | 13/40 (32.5) | 1/13 (8) | 0/13 (0) |
Values in boldface indicate a statistically significant difference compared to CHIKV, as determined by the Fisher exact test (P < 0.05). Dissemination, dissemination of infected mosquitoes; transmission, transmission of infected mosquitoes.
Effect of E2 domains on infection of mice.
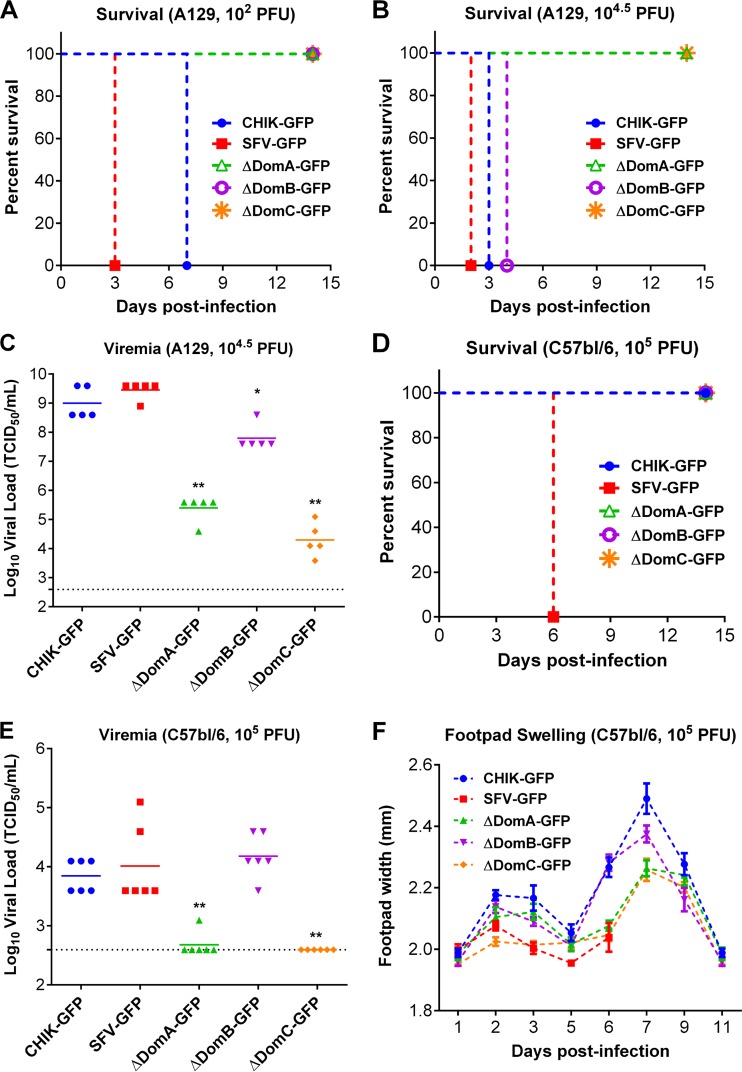
To determine the role of different E2 domains in mice, we performed infections of both Ifnar1−/− mice in the 129/Sv background (A129) and wild-type C57BL/6 mice. Ifnar1−/− mice have previously been shown to be a useful vaccine and pathogenicity model for CHIKV, producing similar pathology (with similar cell tropism) and high viremia, which are characteristic of CHIKV disease in humans (49, 50). When given either 102 PFU or 104.5 PFU in the footpad, CHIK-GFP and SFV-GFP were uniformly lethal in this model (Fig. 5A and B). In contrast, none of the mice infected at 102 PFU with any of the chimeric viruses succumbed to infection (Fig. 5A). However, when the infectious dose was raised to 104.5 PFU, all mice infected with ΔDomB succumbed to infection, while ΔDomA and ΔDomC-challenged mice retained 100% survival (Fig. 5B). Viremia, measured at 2 days postinfection, revealed a similar trend, since all chimeric viruses produced viral loads significantly lower than either CHIKV or SFV (ΔDomB, P < 0.05; ΔDomA and ΔDomC, P < 0.01) (Fig. 5C).
FIG 5.
Infection of A129 and wild-type mice with CHIKV/SFV chimeras. (A and B) Survival analysis of 6- to 9-week-old A129 mice infected with either 102 (A) or 104.5 (B) PFU of each chimeric virus in the left hind footpad. (C) Viremia in A129 mice infected with 104.5 PFU of each virus. *, Statistically significant difference in viral load (*, P < 0.05; **, P < 0.01). (D) Survival analysis of C57BL/6 mice infected with 105 PFU of each virus in the left hind footpad. (E and F) Viremia (E) and footpad swelling (F) in infected C57BL/6 mice. Viremia (C and E) was measured via TCID50 assay in BHK-21 cells. (F) Footpad swelling was assessed by measuring the width of the footpad over the course of the infection using a digital caliper.
Immunocompetent C57BL/6 mice have been used as a model of CHIKV arthritis and SFV neuropathology (51, 52) and were thus selected for further characterization of CHIKV/SFV chimeras. When infected with a high dose of each virus (105 PFU) subcutaneously, all mice infected with SFV-GFP succumbed to infection, while all other mice remained healthy throughout the duration of the study, with the exception of footpad swelling (Fig. 5D and F). In contrast to A129 mice, C57BL/6 mice infected with CHIK-GFP, SFV-GFP, or ΔDomB all had similar viremia levels on day 2 postinfection (Fig. 5E). ΔDomA and ΔDomC produced a significantly lower serum viral load than CHIK-GFP (P < 0.01), with only one mouse in the former group producing detectable viremia at this time point. For additional markers of morbidity, the width of the injected footpad was used as an indicator of arthritis (Fig. 5F). All mice, except for those challenged with SFV-GFP, experienced significant footpad swelling after infection, with CHIK-GFP and ΔDomB producing similar peak levels of inflammation that were significantly higher than in mice infected with ΔDomA or ΔDomC, a finding consistent with the viremia data. These data suggest that modification of domain A or C of CHIKV E2 is highly detrimental in vivo, significantly reducing replication in mosquitoes, as well as significantly reducing replication and lethality in immunocompromised and immunocompetent mice. Altering E2 domain B, in contrast, did not result in significant attenuation except when delivered in a low dose to A129 mice.
Intracranial delivery of CHIKV/SFV chimeras results in altered disease phenotype.
Previous studies have shown that SFV infects brain cells, including neurons and oligodendrocytes in mice (53). In contrast, field strains of CHIKV are not known to infect either of these cell types but have previously been shown to infect leptomeningeal and ependymal cells, but not glial cells, in the mouse brain (49). Since previous studies have shown that alphavirus E2 domains A and B are important for cell and tissue tropism (reviewed in reference 17), we hypothesized that mice infected with these chimeras may show different pathology. To investigate this, A129 and C57BL/6 mice were infected i.c. with 102 and 103 PFU, respectively. Mice were euthanized 3 or 7 days postinfection for A129 or C57BL/6, respectively (except for mice challenged with SFV-GFP, which required euthanasia on day 5 postinfection). Brains were harvested and stored at −80°C for virus titration or fixed in 2% PFA for tissue sectioning. Although A129 mice infected with CHIK-GFP, SFV-GFP, and ΔDomB rapidly showed clinical signs and were moribund at 3 days postinfection, the other two groups, along with mock-infected animals, did not succumb. SFV-GFP was uniformly lethal in C57BL/6 mice, whereas all other mice survived following infection (data not shown).
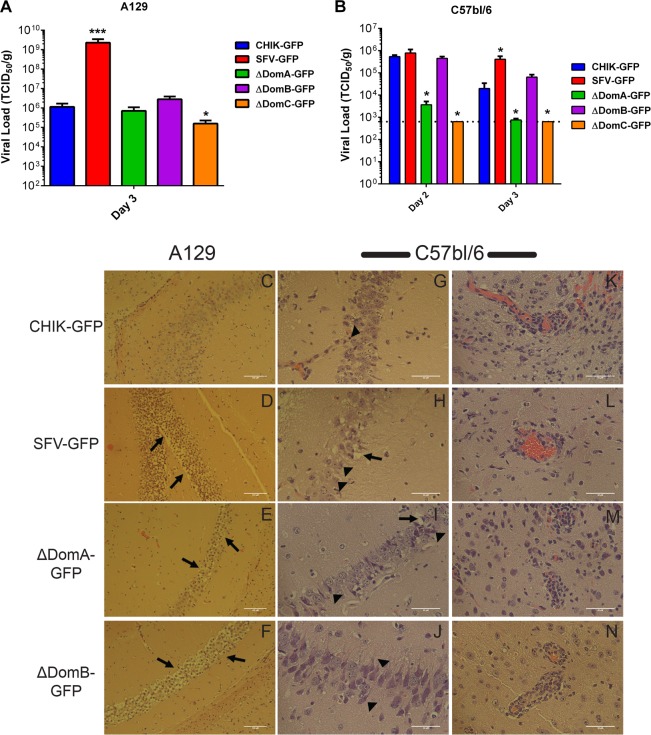
To assess viral replication in the brains of infected mice, the brains were homogenized in a 10% (wt/vol) solution in RPMI 1640–1% BSA, and virus titration was performed in BHK-21 cells. All viruses replicated well in the brains of A129 mice, with only ΔDomC showing significantly reduced titers on day 3 postinfection (Fig. 6A). In C57BL/6 mice, ΔDomA and ΔDomC produced significantly less virus on both days 2 and 3 postinfection, while no significant differences were observed with ΔDomB (Fig. 6B). Interestingly, titers observed for all viruses besides SFV-GFP were significantly lower on day 3 compared to day 2, suggesting viral clearance.
FIG 6.
Comparative histological imaging of mouse brains after infection with chimeric viruses. Intracranial (i.c.) infection of different mouse strains with chimeric viruses resulted in altered pathology and viral loads. (A) TCID50/g viral titers in A129 mice brains. (B) TCID50/g viral titers in C57BL/6 mice brains. (C) Neuronal cells in the hippocampus remained mostly intact in A129 mice following i.c. infection with CHIK-GFP. (E to F) In contrast, SFV-GFP-infected A129 mice revealed massive demyelination and neuron degeneration in the hippocampus (D), as did ΔDomA-infected (E) and ΔDomB-infected (F) A129 mice. A similar pattern was observed with C57BL/6 mice infected with the chimeric viruses: representative images of hippocampal neurons from infected C57BL/6 mice (G to J) and of inflammation and perivascular cuffing in the white matter of C57bl/6 mice (K to N) are shown. Neuron degeneration is indicated by arrowheads, while arrows represent demyelination. *, Significant increase in viral load in the brain (*, P < 0.05; **, P < 0.01). Scale bars: C to F, 100 μm; G to N, 50 μm.
Histological analyses of infected brains confirmed pathology associated with infection. Previous studies with SFV identified the hippocampus as a major target of SFV infection (53); therefore, we undertook a comparative histological analysis of the hippocampus from SFV-, CHIKV-, and CHIKV/SFV chimera-infected mice, specifically surveying for obvious morphological changes associated with the different virus infections. Examination of hematoxylin-eosin-stained sections revealed that neuronal cells in the hippocampus remained mostly intact in A129 mice following i.c. infection with CHIK-GFP (Fig. 6C), despite these mice becoming sick. In contrast, SFV-GFP-infected mice revealed massive demyelination and neuron degeneration in the hippocampus (Fig. 6D). Interestingly, mice infected with ΔDomB, and to a lesser extent ΔDomA, showed demyelination with neuron degeneration that appeared similar to SFV-GFP (Fig. 6E and F). No lesions in the hippocampus of mice infected with ΔDomC were observed and, as a result, images are not shown to conserve space. A similar disease phenotype was observed in C57BL/6 mice, with CHIK-GFP-infected mice maintaining healthy hippocampal neurons (Fig. 6G). Demyelination and neuronal cell death were observed in SFV-GFP-infected brains (Fig. 6H). ΔDomB-infected mice presented with moderate neuron degeneration but little demyelination, whereas ΔDomA-infected mice showed mild neuron degeneration (Fig. 6I and J). Massive inflammation and perivascular cuffing were observed in CHIK-GFP mice (Fig. 6K) despite the lack of neuron damage. Infiltration of mononuclear cells and perivascular cuffing was observed in the brains of mice infected with SFV-GFP, ΔDomA, and ΔDomB viruses (Fig. 6L and N). It is interesting that ΔDomB induced significant hippocampal neuron degeneration despite appearing to produce less overall inflammation of the brain (Fig. 6N). These data indicate that E2 domains A and B play a role in pathology produced by infection in both immunodeficient and immunocompetent mice.
In vitro infection of brain cells to assess cell tropism changes of CHIKV/SFV chimeras.
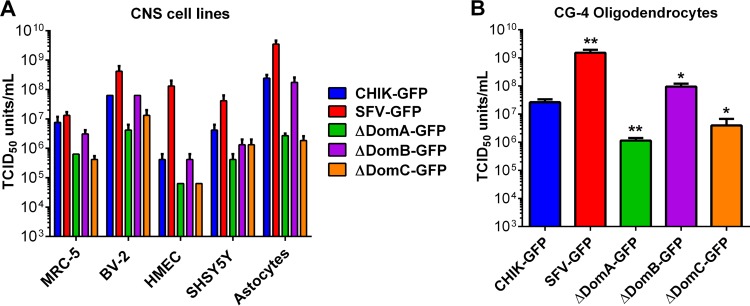
To determine the role of cell tropism in the altered brain pathology observed with CHIKV/SFV chimeras, we infected various brain cell cultures at an MOI of 0.1 PFU/cell and measured the titer of infectious virus at 24 h postinfection. MRC-5 (human lung fibroblast) cells were used as a positive control, since they are highly susceptible to CHIKV (40). There were no significant differences in the replication of CHIK-GFP compared to ΔDomB on microglial (BV-2), endothelial (HMEC), neuroblastoma (SHSY5Y), and astrocyte cells (Fig. 7A), whereas ΔDomA and ΔDomC were uniformly attenuated on all cell lines tested (the latter in SHSY5Y the only exception). ΔDomA and ΔDomC were similarly attenuated for virus replication in oligodendrocytes, while ΔDomB and SFV-GFP produced significantly higher infectious virus titers compared to CHIK-GFP at 24 h after infection (P < 0.05) (Fig. 7B).
FIG 7.
Growth of CHIK/SFV chimeras in different CNS cell types. The replication of parental and chimeric viruses in brain cells was examined. (A) MRC-5 (fibroblasts), BV-2 (microglia), HMEC (endothelial), SHSY5Y (neuroblastoma), CG-4 (astrocytes), and (B) CG-4 (oligodendrocytes) were infected at an MOI of 0.01 PFU/cell, and the infectious virus released 24 h postinfection was measured by TCID50 assay in BHK-21 cells. CG-4 oligodendrocyte progenitors were differentiated into either type II astrocytes (A) or oligodendrocytes (B) according to well-established methods. *, Statistically significant differences in viral titer (*, P < 0.05; ** P < 0.01).
Chimeric viruses present altered GFP distribution in the mouse brain.
To assess the tropism of chimeric viruses in the brains of mice, frozen sections from i.c.-infected A129 mice were analyzed for GFP expression via fluorescence microscopy using an Evos FL inverted microscope. CHIK-GFP tropism was very similar to previously reported results, with GFP expression primarily found in the choroid plexus and ependymal wall (49) outside the hippocampus (Fig. 8A and B). In contrast, SFV-GFP was found both in the ependymal cells and inside the hippocampus, primarily infecting the cells which appear to be pyramidal neurons (Fig. 8C), a finding consistent with previous reports (53). In addition, SFV-GFP infection was observed in cells in white matter tracts which formed chains characteristic of oligodendrocytes (Fig. 8D), also consistent with previous reports (53, 54). Interestingly, ΔDomA and ΔDomB viruses appeared to spread further into the hippocampus, with the latter appearing to infect cells surrounding the hippocampal pyramidal neurons (Fig. 8E to H), albeit with a different distribution than SFV-GFP. GFP expression in white matter tracts of chain-like cells characteristic of oligodendrocytes was observed with both ΔDomA- and ΔDomB-infected mice (Fig. 8F and H), which was not detected in CHIK-GFP-infected mice (Fig. 8B). ΔDomC virus replication was highly attenuated in the brain, and little GFP expression was observed in infected mice. Replication of ΔDomC appeared to be restricted to cells with distribution characteristic of ependymal cells (Fig. 8I and J).
FIG 8.
Distribution of CHIKV/SFV chimeras in the mouse brain. GFP expression was used to monitor tropism of CHIKV/SFV chimeras in the brains of A129 mice. Cryosectioned tissues were cut using a cryostat to 10 μm and placed onto positively charged slides. GFP expression was monitored using an Evos FL microscope with either a ×10 (A, C, E, G, and I) or ×20 (B, D, F, H, and J) objective lens. Scale bars represent 50 μm (A, C, E, G, and I) and 20 μm (B, D, F, H, and J). Blue color represents cellular nuclei stained with DAPI. White arrows represent chains of cells that have an oligodendrocyte-like morphology.
DISCUSSION
Although the roles of individual amino acid substitutions in the E2 have been established for many alphaviruses (55–57), the importance of these changes in other alphaviruses, or the larger function of protein domains of E2, remains poorly understood. We hypothesized that constructing chimeric viruses between CHIKV and either VEEV or SFV would facilitate studies elucidating the importance of these domains in terms of replication, structure, and pathogenicity, among other factors. However, CHIK/VEEV chimeras were nonviable, likely due to large sequence variations between the two highly divergent viruses which resulted in disruption of amino acid interactions between E1 and E2 (Tables 2 and 3). CHIKV/SFV chimeras, in contrast, could be rescued and grown to high titers for further study.
In the studies described here, using in vitro and in vivo (mosquitoes and mice) approaches, we established the importance of alphavirus E2 domains. Through replication kinetics, mosquito vector competence, infection of immunocompetent and interferon signaling-deficient mice, we determined that the role of E2 domain C is crucial to the complete virus transmission cycle. Although residues associated with tissue tropism or host range have not been shown to map to domain C, it appears likely that its structural role is indispensable for proper viral function. The recent publication of the crystal structure of CHIKV reveals the close association between domain C and E1 (17), and it is likely that this interaction is disrupted in the ΔDomC chimera. Structural analysis of the chimeric envelope proteins showed that many amino acid interactions were disrupted in the ΔDomC virus, which is likely responsible for its attenuation. Domain A, like domain C, interacts strongly with E1, and possibly due to this ΔDomA was also attenuated, albeit to a lesser degree than ΔDomC. This is supported by the increased amino acid similarity between CHIKV and SFV in ΔDomA, compared to ΔDomC, and also the number of important residues for protein interaction disrupted. In contrast to domain C, which is positioned between two E1 molecules, domain A has residues internal and at the surface of the envelope spikes. The interaction of the internal residues with E1 was likely the cause of attenuation of ΔDomA, while the residues on the surface may have contributed to the altered pathology seen in mice infected i.c. As suggested by the attenuation of the viruses, ΔDomC had lower viral fitness, as measured by genome/PFU ratios. Surprisingly, no differences were observed between ΔDomA and ΔDomB in genome/PFU ratio, despite the differences in attenuation levels. The reason for this is unclear but may be the result of measuring levels in cell culture. It is quite possible that ΔDomA and ΔDomB would show greater differences in vivo. In future studies it may be prudent to examine domain A broken into different “subdomains,” since this would likely result in less attenuation, making it easier to observe differences in infection and pathology.
ΔDomB, in contrast to ΔDomA and ΔDomC, showed similar replication kinetics in vitro and in vivo compared to CHIK-GFP, except for the low-dose infection of A129 mice. Similarly, ΔDomB had fewer amino acid differences between CHIKV and SFV than the other two domains and also had only minor structural differences due to chimerization. Likely as a result, ΔDomB was less attenuated than the other two chimeras and showed only slight differences to parental CHIKV. Interestingly, ΔDomB replicated to significantly higher peak titers in cultured oligodendrocytes, a finding suggestive but not conclusive of a cell tropism shift conferred by the SFV domain B. Furthermore, viral infection of cells characteristic of oligodendrocytes was observed in both SFV-GFP, ΔDomA- and ΔDomB-infected mice. Infection of these cells was not observed with CHIK-GFP, indicating that these domains may be playing a role in cell tropism in these cells. This may have been due to interactions with GAGs, since ΔDomB virus showed a similar reduction in infectivity in BHK-21 cells in the presence of heparin as SFV. It is well established that SFV can infect oligodendrocytes and is even used as a model for experimental autoimmune encephalomyelitis (58). The data presented here suggest that a widespread virus like CHIKV could cause demyelinating disease following infection if E2 mutations are selected for which result in increased oligodendrocyte tropism. CNS involvement has become increasingly more common (59, 60), and cases of demyelinating disease following CHIKV infection have been reported (61–64), warranting further research.
Future studies focusing on infection rates instead of replication may determine a precise role for both domains A and B in cell tropism. Using i.c. infection of different mouse strains, we showed drastically altered pathology, which more closely mimicked SFV-GFP than CHIK-GFP for ΔDomB and, to a lesser degree, ΔDomA. Although ΔDomC did not show an altered infection phenotype, it is possible that this could be related to its attenuation and therefore may play an important role in infection. Demyelination and neuron degeneration observed in infected mice may have been due to these viruses' increased ability to infect oligodendrocytes or neurons. Previous studies attempting to identify the correlates for SFV's neurotropism have proven that it likely involves a combination of viral factors, since many residues or cis-acting RNA elements have been implicated (65–71). Therefore, while domain B may not be the only factor involved in infection of these cells, it is likely that it is playing a role in receptor mediated entry (reviewed in reference 17). The position of domain B at the surface of the E2/E1 spike, with minimal contact with E1, offers an explanation for the limited attenuation observed with this chimera. In addition to E1, interactions between E2 and E3 have been shown to play an important role in virus replication and assembly (74). This interaction was shown to be clade specific, which is in agreement with our data that no important interactions between E3 and E2 were disrupted in the CHIKV/SFV chimeric viruses presented within the manuscript, since both are in the SFV complex of Old World alphaviruses. The overwhelming number of amino acid differences between CHIKV and VEEV E2 make it difficult to determine what role the E2-E3 interaction played in the inability to rescue CHIKV/VEEV chimeric viruses.
Our studies demonstrate that each of the domains of the alphavirus E2 plays a critical role in viral infection. We showed that changing domains A or C resulted in viruses that are highly attenuated both in vitro and in vivo, highlighting the importance of the E1-E2 interaction which is critical for viral replication. These viruses can be useful to study this interaction, since serial passaging in cell culture or in vivo (mosquitoes or mice) should allow for identification of additional residues important for the E1-E2 interaction. In addition to studying virus protein interaction, the greatly reduced replication rates and mortality of the ΔDomA and ΔDomC chimeras suggest that they may have utility as vaccines. This attenuation, which is likely mediated by differences in many amino acid residues compared to the wild type, suggests that these viruses could be highly safe CHIKV vaccines or recombinant vaccine vectors producing a foreign antigen in the same manner GFP is currently expressed. Our studies also highlight the importance of E2 domain B, implicating it in pathology and potentially cell tropism. Since E2 domain B is where many neutralizing antibodies map (72, 73), it should be explored as a vaccine antigen that could be cross-reactive for other alphaviruses. In addition, the relative lack of attenuation of ΔDomB, despite only sharing ∼60% amino acid sequence similarity between CHIKV and SFV, suggests that this region could be utilized to express foreign antigens to alter cellular tropism or possibly target specific cells.
In summary, these studies highlight the importance of studying the role of individual domains of viral proteins. It was determined that all three E2 domains are important for viral replication and pathogenesis in vitro and in vivo (mosquitoes and mice). In addition, we established the importance of assessing infection and replication in both the vertebrate and invertebrate host for arboviruses, since differences are likely to be observed. The chimeric viruses described here may be useful for future studies to understand the role of alphavirus E2 domains in assembly, immunogenicity (including vaccines), and further infection/pathogenicity studies.
ACKNOWLEDGMENT
This research was supported by National Institutes of Health grant R01 AI093491-01.
REFERENCES
- 1.Forrester NL, Palacios G, Tesh RB, Savji N, Guzman H, Sherman M, Weaver SC, Lipkin WI. 2012. Genome-scale phylogeny of the alphavirus genus suggests a marine origin. J Virol 86:2729–2738. doi: 10.1128/JVI.05591-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.La Linn M, Gardner J, Warrilow D, Darnell GA, McMahon CR, Field I, Hyatt AD, Slade RW, Suhrbier A. 2001. Arbovirus of marine mammals: a new alphavirus isolated from the elephant seal louse, Lepidophthirus macrorhini. J Virol 75:4103–4109. doi: 10.1128/JVI.75.9.4103-4109.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weaver SC, Reisen WK. 2010. Present and future arboviral threats. Antiviral Res 85:328–345. doi: 10.1016/j.antiviral.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2015. Chikungunya: geographic distribution. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/chikungunya/geo/index.html. [Google Scholar]
- 5.Morrison TE. 2014. Reemergence of Chikungunya virus. J Virol 88:11644–11647. doi: 10.1128/JVI.01432-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Powers AM. 2010. Chikungunya. Clin Lab Med 30:209–219. doi: 10.1016/j.cll.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Willems WR, Kaluza G, Boschek CB, Bauer H, Hager H, Schutz HJ, Feistner H. 1979. Semliki Forest virus: cause of a fatal case of human encephalitis. Science 203:1127–1129. doi: 10.1126/science.424742. [DOI] [PubMed] [Google Scholar]
- 8.Mathiot CC, Grimaud G, Garry P, Bouquety JC, Mada A, Daguisy AM, Georges AJ. 1990. An outbreak of human Semliki Forest virus infections in Central African Republic. Am J Trop Med Hyg 42:386–393. [DOI] [PubMed] [Google Scholar]
- 9.Tigoi C, Lwande O, Orindi B, Irura Z, Ongus J, Sang R. 2015. Seroepidemiology of selected arboviruses in febrile patients visiting selected health facilities in the lake/river basin areas of Lake Baringo, Lake Naivasha, and Tana River, Kenya. Vector Borne Zoonotic Dis 15:124–132. doi: 10.1089/vbz.2014.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodhain F, Gonzalez JP, Mercier E, Helynck B, Larouze B, Hannoun C. 1989. Arbovirus infections and viral haemorrhagic fevers in Uganda: a serological survey in Karamoja district, 1984. Trans R Soc Trop Med Hyg 83:851–854. doi: 10.1016/0035-9203(89)90352-0. [DOI] [PubMed] [Google Scholar]
- 11.Adekolu-John EO, Fagbami AH. 1983. Arthropod-borne virus antibodies in sera of residents of Kainji Lake Basin, Nigeria 1980. Trans R Soc Trop Med Hyg 77:149–151. doi: 10.1016/0035-9203(83)90053-6. [DOI] [PubMed] [Google Scholar]
- 12.Sureau P, Jaeger G, Pinerd G, Palisson MJ, Bedaya-N′Garo S. 1977. Sero-epidemiological survey of arbovirus diseases in the Bi-Aka pygmies of Lobaye, Central African Republic. Bull Soc Pathol Exot Filiales 70:131–137. (In Spanish.) [PubMed] [Google Scholar]
- 13.Strauss JH, Strauss EG. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev 58:491–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garmashova N, Gorchakov R, Volkova E, Paessler S, Frolova E, Frolov I. 2007. The Old World and New World alphaviruses use different virus-specific proteins for induction of transcriptional shutoff. J Virol 81:2472–2484. doi: 10.1128/JVI.02073-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fata CL, Sawicki SG, Sawicki DL. 2002. Alphavirus minus-strand RNA synthesis: identification of a role for Arg183 of the nsP4 polymerase. J Virol 76:8632–8640. doi: 10.1128/JVI.76.17.8632-8640.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S. 2007. A single mutation in Chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog 3:e201. doi: 10.1371/journal.ppat.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voss JE, Vaney MC, Duquerroy S, Vonrhein C, Girard-Blanc C, Crublet E, Thompson A, Bricogne G, Rey FA. 2010. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 468:709–712. doi: 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]
- 18.Smyth J, Suomalainen M, Garoff H. 1997. Efficient multiplication of a Semliki Forest virus chimera containing Sindbis virus spikes. J Virol 71:818–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhn RJ, Niesters HG, Hong Z, Strauss JH. 1991. Infectious RNA transcripts from Ross River virus cDNA clones and the construction and characterization of defined chimeras with Sindbis virus. Virology 182:430–441. doi: 10.1016/0042-6822(91)90584-X. [DOI] [PubMed] [Google Scholar]
- 20.Lopez S, Yao JS, Kuhn RJ, Strauss EG, Strauss JH. 1994. Nucleocapsid-glycoprotein interactions required for assembly of alphaviruses. J Virol 68:1316–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn RJ, Griffin DE, Owen KE, Niesters HG, Strauss JH. 1996. Chimeric Sindbis-Ross River viruses to study interactions between alphavirus nonstructural and structural regions. J Virol 70:7900–7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao JS, Strauss EG, Strauss JH. 1996. Interactions between PE2, E1, and 6K required for assembly of alphaviruses studied with chimeric viruses. J Virol 70:7910–7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roy CJ, Adams AP, Wang E, Leal G, Seymour RL, Sivasubramani SK, Mega W, Frolov I, Didier PJ, Weaver SC. 2013. A chimeric Sindbis-based vaccine protects cynomolgus macaques against a lethal aerosol challenge of eastern equine encephalitis virus. Vaccine 31:1464–1470. doi: 10.1016/j.vaccine.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang E, Volkova E, Adams AP, Forrester N, Xiao SY, Frolov I, Weaver SC. 2008. Chimeric alphavirus vaccine candidates for Chikungunya. Vaccine 26:5030–5039. doi: 10.1016/j.vaccine.2008.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanlandingham DL, Tsetsarkin K, Klingler KA, Hong C, McElroy KL, Lehane MJ, Higgs S. 2006. Determinants of vector specificity of O'nyong nyong and Chikungunya viruses in Anopheles and Aedes mosquitoes. Am J Trop Med Hyg 74:663–669. [PubMed] [Google Scholar]
- 26.Saxton-Shaw KD, Ledermann JP, Borland EM, Stovall JL, Mossel EC, Singh AJ, Wilusz J, Powers AM. 2013. O'nyong nyong virus molecular determinants of unique vector specificity reside in non-structural protein 3. PLoS Negl Trop Dis 7:e1931. doi: 10.1371/journal.pntd.0001931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang E, Kim DY, Weaver SC, Frolov I. 2011. Chimeric Chikungunya viruses are nonpathogenic in highly sensitive mouse models but efficiently induce a protective immune response. J Virol 85:9249–9252. doi: 10.1128/JVI.00844-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darwin JR, Kenney JL, Weaver SC. 2011. Transmission potential of two chimeric Chikungunya vaccine candidates in the urban mosquito vectors, Aedes aegypti and Aealbopictus. Am J Trop Med Hyg 84:1012–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weger-Lucarelli J, Aliota MT, Kamlangdee A, Osorio JE. 2015. Identifying the role of E2 domains on alphavirus neutralization and protective immune responses. PLoS Negl Trop Dis 9:e0004163. doi: 10.1371/journal.pntd.0004163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Louis JC, Magal E, Muir D, Manthorpe M, Varon S. 1992. CG-4, a new bipotential glial cell line from rat brain, is capable of differentiating in vitro into either mature oligodendrocytes or type-2 astrocytes. J Neurosci Res 31:193–204. doi: 10.1002/jnr.490310125. [DOI] [PubMed] [Google Scholar]
- 31.Steel JJ, Henderson BR, Lama SB, Olson KE, Geiss BJ. 2011. Infectious alphavirus production from a simple plasmid transfection. Virol J 8:356. doi: 10.1186/1743-422X-8-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bryksin AV, Matsumura I. 2010. Overlap extension PCR cloning: a simple and reliable way to create recombinant plasmids. Biotechniques 48:463–465. doi: 10.2144/000113418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsetsarkin K, Higgs S, McGee CE, De Lamballerie X, Charrel RN, Vanlandingham DL. 2006. Infectious clones of Chikungunya virus (La Reunion isolate) for vector competence studies. Vector Borne Zoonotic Dis 6:325–337. doi: 10.1089/vbz.2006.6.325. [DOI] [PubMed] [Google Scholar]
- 34.van den Hoff MJ, Moorman AF, Lamers WH. 1992. Electroporation in ‘intracellular’ buffer increases cell survival. Nucleic Acids Res 20:2902. doi: 10.1093/nar/20.11.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hernandez R, Sinodis C, Brown DT. 2010. Sindbis virus: propagation, quantification, and storage. Curr Protoc Microbiol Chapter 15:Unit15B 11. [DOI] [PubMed] [Google Scholar]
- 36.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2015. The I-TASSER Suite: protein structure and function prediction. Nat Methods 12:7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Hyg 27 493–497. [Google Scholar]
- 38.Silva LA, Khomandiak S, Ashbrook AW, Weller R, Heise MT, Morrison TE, Dermody TS. 2014. A single-amino-acid polymorphism in Chikungunya virus E2 glycoprotein influences glycosaminoglycan utilization. J Virol 88:2385–2397. doi: 10.1128/JVI.03116-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gardner CL, Hritz J, Sun C, Vanlandingham DL, Song TY, Ghedin E, Higgs S, Klimstra WB, Ryman KD. 2014. Deliberate attenuation of chikungunya virus by adaptation to heparan sulfate-dependent infectivity: a model for rational arboviral vaccine design. PLoS Negl Trop Dis 8:e2719. doi: 10.1371/journal.pntd.0002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Christensen BM, Sutherland DR. 1984. Brugia-Pahangi: exsheathment and midgut penetration in Aedes aegypti. Trans Am Microscopical Soc 103:423–433. doi: 10.2307/3226478. [DOI] [Google Scholar]
- 41.Ebel GD, Carricaburu J, Young D, Bernard KA, Kramer LD. 2004. Genetic and phenotypic variation of West Nile virus in New York, 2000-2003. Am J Trop Med Hyg 71:493–500. [PubMed] [Google Scholar]
- 42.Aliota MT, Jones SA, Dupuis AP, Ciota AT, Hubalek Z, Kramer LD. 2012. Characterization of Rabensburg virus, a flavivirus closely related to West Nile Virus of the Japanese encephalitis antigenic group. PLoS One 7:e39387. doi: 10.1371/journal.pone.0039387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rutledge LC, Ward RA, Gould DJ. 1964. Studies on the feeding response of mosquitoes to nutritive solutions in a new membrane feeder. Mosquito News 24:407–419. [Google Scholar]
- 44.Brault AC, Foy BD, Myles KM, Kelly CL, Higgs S, Weaver SC, Olson KE, Miller BR, Powers AM. 2004. Infection patterns of o'nyong nyong virus in the malaria-transmitting mosquito, Anopheles gambiae. Insect Mol Biol 13:625–635. doi: 10.1111/j.0962-1075.2004.00521.x. [DOI] [PubMed] [Google Scholar]
- 45.Reynaud JM, Kim DY, Atasheva S, Rasalouskaya A, White JP, Diamond MS, Weaver SC, Frolova EI, Frolov I. 2015. IFIT1 differentially interferes with translation and replication of alphavirus genomes and promotes induction of type I interferon. PLoS Pathog 11:e1004863. doi: 10.1371/journal.ppat.1004863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heise MT, Simpson DA, Johnston RE. 2000. Sindbis-group alphavirus replication in periosteum and endosteum of long bones in adult mice. J Virol 74:9294–9299. doi: 10.1128/JVI.74.19.9294-9299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paessler S, Fayzulin RZ, Anishchenko M, Greene IP, Weaver SC, Frolov I. 2003. Recombinant Sindbis/Venezuelan equine encephalitis virus is highly attenuated and immunogenic. J Virol 77:9278–9286. doi: 10.1128/JVI.77.17.9278-9286.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smithburn KC, Haddow AJ. 1944. Semliki Forest virus. J Immunol 49:141–157. [Google Scholar]
- 49.Couderc T, Chretien F, Schilte C, Disson O, Brigitte M, Guivel-Benhassine F, Touret Y, Barau G, Cayet N, Schuffenecker I, Despres P, Arenzana-Seisdedos F, Michault A, Albert ML, Lecuit M. 2008. A mouse model for Chikungunya: young age and inefficient type-I interferon signaling are risk factors for severe disease. PLoS Pathog 4:e29. doi: 10.1371/journal.ppat.0040029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weger-Lucarelli J, Chu H, Aliota MT, Partidos CD, Osorio JE. 2014. A novel MVA vectored Chikungunya virus vaccine elicits protective immunity in mice. PLoS Negl Trop Dis 8:e2970. doi: 10.1371/journal.pntd.0002970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gardner J, Anraku I, Le TT, Larcher T, Major L, Roques P, Schroder WA, Higgs S, Suhrbier A. 2010. Chikungunya virus arthritis in adult wild-type mice. J Virol 84:8021–8032. doi: 10.1128/JVI.02603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mokhtarian F, Swoveland P. 1987. Predisposition to EAE induction in resistant mice by prior infection with Semliki Forest virus. J Immunol 138:3264–3268. [PubMed] [Google Scholar]
- 53.Fragkoudis R, Tamberg N, Siu R, Kiiver K, Kohl A, Merits A, Fazakerley JK. 2009. Neurons and oligodendrocytes in the mouse brain differ in their ability to replicate Semliki Forest virus. J Neurovirol 15:57–70. doi: 10.1080/13550280802482583. [DOI] [PubMed] [Google Scholar]
- 54.Fazakerley JK, Boyd A, Mikkola ML, Kaariainen L. 2002. A single amino acid change in the nuclear localization sequence of the nsP2 protein affects the neurovirulence of Semliki Forest virus. J Virol 76:392–396. doi: 10.1128/JVI.76.1.392-396.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsetsarkin KA, McGee CE, Volk SM, Vanlandingham DL, Weaver SC, Higgs S. 2009. Epistatic roles of E2 glycoprotein mutations in adaption of chikungunya virus to Aedes albopictus and Ae. aegypti mosquitoes. PLoS One 4:e6835. doi: 10.1371/journal.pone.0006835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brault AC, Powers AM, Weaver SC. 2002. Vector infection determinants of Venezuelan equine encephalitis virus reside within the E2 envelope glycoprotein. J Virol 76:6387–6392. doi: 10.1128/JVI.76.12.6387-6392.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brault AC, Powers AM, Holmes EC, Woelk CH, Weaver SC. 2002. Positively charged amino acid substitutions in the e2 envelope glycoprotein are associated with the emergence of Venezuelan equine encephalitis virus. J Virol 76:1718–1730. doi: 10.1128/JVI.76.4.1718-1730.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jerusalmi A, Morris-Downes MM, Sheahan BJ, Atkins GJ. 2003. Effect of intranasal administration of Semliki Forest virus recombinant particles expressing reporter and cytokine genes on the progression of experimental autoimmune encephalomyelitis. Mol Ther 8:886–894. doi: 10.1016/j.ymthe.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 59.Das T, Jaffar-Bandjee MC, Hoarau JJ, Krejbich Trotot P, Denizot M, Lee-Pat-Yuen G, Sahoo R, Guiraud P, Ramful D, Robin S, Alessandri JL, Gauzere BA, Gasque P. 2010. Chikungunya fever: CNS infection and pathologies of a re-emerging arbovirus. Prog Neurobiol 91:121–129. doi: 10.1016/j.pneurobio.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 60.Kalita J, Kumar P, Misra UK. 2013. Stimulus-sensitive myoclonus and cerebellar ataxia following Chikungunya meningoencephalitis. Infection 41:727–729. doi: 10.1007/s15010-013-0406-2. [DOI] [PubMed] [Google Scholar]
- 61.Maity P, Roy P, Basu A, Das B, Ghosh US. 2014. A case of ADEM following Chikungunya fever. J Assoc Physicians India 62:441–442. [PubMed] [Google Scholar]
- 62.Bass JW, Chan DS, Creamer KM, Thompson MW, Malone FJ, Becker TM, Marks SN. 1997. Comparison of oral cephalexin, topical mupirocin and topical bacitracin for treatment of impetigo. Pediatr Infect Dis J 16:708–710. doi: 10.1097/00006454-199707000-00013. [DOI] [PubMed] [Google Scholar]
- 63.Musthafa AK, Abdurahiman P, Jose J. 2008. Case of ADEM following Chikungunya fever. J Assoc Physicians India 56:473. [PubMed] [Google Scholar]
- 64.Wielanek AC, Monredon JD, Amrani ME, Roger JC, Serveaux JP. 2007. Guillain-Barre syndrome complicating a Chikungunya virus infection. Neurology 69:2105–2107. doi: 10.1212/01.wnl.0000277267.07220.88. [DOI] [PubMed] [Google Scholar]
- 65.Atkins GJ. 1983. The avirulent A7 Strain of Semliki Forest virus has reduced cytopathogenicity for neuroblastoma cells compared to the virulent L10 strain. J Gen Virol 64(Pt 6):1401–1404. doi: 10.1099/0022-1317-64-6-1401. [DOI] [PubMed] [Google Scholar]
- 66.Sheahan BJ, Gates MC, Caffrey JF, Atkins GJ. 1983. Oligodendrocyte infection and demyelination produced in mice by the M9 mutant of Semliki Forest virus. Acta Neuropathol 60:257–265. doi: 10.1007/BF00691874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Glasgow GM, Killen HM, Liljestrom P, Sheahan BJ, Atkins GJ. 1994. A single amino acid change in the E2 spike protein of a virulent strain of Semliki Forest virus attenuates pathogenicity. J Gen Virol 75(Pt 3):663–668. doi: 10.1099/0022-1317-75-3-663. [DOI] [PubMed] [Google Scholar]
- 68.Glasgow GM, Sheahan BJ, Atkins GJ, Wahlberg JM, Salminen A, Liljestrom P. 1991. Two mutations in the envelope glycoprotein E2 of Semliki Forest virus affecting the maturation and entry patterns of the virus alter pathogenicity for mice. Virology 185:741–748. doi: 10.1016/0042-6822(91)90545-M. [DOI] [PubMed] [Google Scholar]
- 69.Logue CH, Sheahan BJ, Atkins GJ. 2008. The 5′ untranslated region as a pathogenicity determinant of Semliki Forest virus in mice. Virus Genes 36:313–321. doi: 10.1007/s11262-008-0209-1. [DOI] [PubMed] [Google Scholar]
- 70.Tuittila M, Hinkkanen AE. 2003. Amino acid mutations in the replicase protein nsP3 of Semliki Forest virus cumulatively affect neurovirulence. J Gen Virol 84:1525–1533. doi: 10.1099/vir.0.18936-0. [DOI] [PubMed] [Google Scholar]
- 71.Tuittila MT, Santagati MG, Roytta M, Maatta JA, Hinkkanen AE. 2000. Replicase complex genes of Semliki Forest virus confer lethal neurovirulence. J Virol 74:4579–4589. doi: 10.1128/JVI.74.10.4579-4589.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Strauss EG, Stec DS, Schmaljohn AL, Strauss JH. 1991. Identification of antigenically important domains in the glycoproteins of Sindbis virus by analysis of antibody escape variants. J Virol 65:4654–4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stec DS, Waddell A, Schmaljohn CS, Cole GA, Schmaljohn AL. 1986. Antibody-selected variation and reversion in Sindbis virus neutralization epitopes. J Virol 57:715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Snyder AJ, Mukhopadhyay S. 2012. The alphavirus E3 glycoprotein functions in a clade-specific manner. J Virol 86:13609–13620. doi: 10.1128/JVI.01805-12. [DOI] [PMC free article] [PubMed] [Google Scholar]