ABSTRACT
The mammalian host responds to viral infections by inducing expression of hundreds of interferon-stimulated genes (ISGs). While the functional significance of many ISGs has yet to be determined, their cell type and temporal nature of expression suggest unique activities against specific pathogens. Using a combination of ectopic expression and gene silencing approaches in cell culture, we previously identified Ifi27l2a as a candidate antiviral ISG within neuronal subsets of the central nervous system (CNS) that restricts infection by West Nile virus (WNV), an encephalitic flavivirus of global concern. To investigate the physiological relevance of Ifi27l2a in the context of viral infection, we generated Ifi27l2a−/− mice. Although adult mice lacking Ifi27l2a were more vulnerable to lethal WNV infection, the viral burden was greater only within the CNS, particularly in the brain stem, cerebellum, and spinal cord. Within neurons of the cerebellum and brain stem, in the context of WNV infection, a deficiency of Ifi27l2a was associated with less cell death, which likely contributed to sustained viral replication and higher titers in these regions. Infection studies in a primary cell culture revealed that Ifi27l2a−/− cerebellar granule cell neurons and macrophages but not cerebral cortical neurons, embryonic fibroblasts, or dendritic cells sustained higher levels of WNV infection than wild-type cells and that this difference was greater under conditions of beta interferon (IFN-β) pretreatment. Collectively, these findings suggest that Ifi27l2a has an antiviral phenotype in subsets of cells and that at least some ISGs have specific inhibitory functions in restricted tissues.
IMPORTANCE The interferon-stimulated Ifi27l2a gene is expressed differentially within the central nervous system upon interferon stimulation or viral infection. Prior studies in cell culture suggested an antiviral role for Ifi27l2a during infection by West Nile virus (WNV). To characterize its antiviral activity in vivo, we generated mice with a targeted gene deletion of Ifi27l2a. Based on extensive virological analyses, we determined that Ifi27l2a protects mice from WNV-induced mortality by contributing to the control of infection of the hindbrain and spinal cord, possibly by regulating cell death of neurons. This antiviral activity was validated in granule cell neurons derived from the cerebellum and in macrophages but was not observed in other cell types. Collectively, these data suggest that Ifi27l2a contributes to innate immune restriction of WNV in a cell-type- and tissue-specific manner.
INTRODUCTION
West Nile virus (WNV) is a positive-stranded, enveloped RNA virus that belongs to the Flavivirus genus of the Flaviviridae family. WNV and related flaviviruses typically are transmitted by arthropod vectors and include members that cause encephalitis (e.g., Japanese encephalitis virus [JEV], Saint Louis encephalitis virus [SLEV], and tick-borne encephalitis virus [TBEV]) or systemic and/or visceral disease (e.g., dengue virus [DENV] and yellow fever virus [YFV]). WNV transmission occurs between Culex species mosquitoes and selected avian hosts, with incidental, dead-end infection of horses, humans, and other vertebrate animals. Humans can develop severe disease following WNV infection, as the virus can invade the central nervous system (CNS) and cause flaccid paralysis, meningitis, or encephalitis, often leading to long-term neurological sequelae or death (1). In the CNS, WNV replicates principally in neurons, and infection may lead to focal lesions, cell injury, and cell death within the brain and spinal cord (2–4). Factors governing WNV entry into and replication within the CNS are complex and include the age of the host, the genetic background (5–8), the quality of the immune response, and the integrity of the blood-brain barrier (for reviews, see references 9–12).
In response to viral infections, most mammalian cells secrete type I interferon (IFN), which promotes an antiviral state in an autocrine and paracrine manner by inducing expression of hundreds of interferon-stimulated genes (ISGs). The gene signature and inhibitory activity promoted by type I IFNs vary depending on the cell type, specific viral pathogen, and possible pathogen-induced immune evasion mechanisms. Within the CNS, the innate immune response must balance the need to restrict virus infection while simultaneously protecting nonrenewable neurons. Indeed, selected regions of the brain and CNS have evolved distinct antiviral programs and mechanisms to restrict infection by different RNA and DNA viruses (13–18). Neurons derived from the cerebral cortex are more permissive of infection by multiple viruses, with IFN-β pretreatment reducing infection of several viruses only minimally (14). In comparison, granule cell neurons (GCN) derived from the cerebellum are less permissive of viral infection at the baseline state and produce a stronger antiviral response following IFN-β pretreatment. Microarray analysis revealed differences in the basal and induced expression of ISGs in GCN compared to cortical neurons (CN) (14). As an example, Ifi27l2a is an ISG expressed at higher levels in GCN than in CN under basal conditions, after IFN-β pretreatment, or following WNV infection. Ectopic expression of Ifi27l2a in CN suppressed infection of a neurotropic flavivirus (WNV) and coronavirus (murine hepatitis virus [MHV]) but not an alphavirus (Venezuelan equine encephalitis virus [VEEV]). Reciprocally, gene silencing of Ifi27l2a in GCN resulted in enhanced WNV infection (14).
Ifi27l2a (also termed ISG12b) is a 7.9-kDa protein belonging to a larger family of genes, including related Ifi27/IFI27 genes and the human IFI6-16 gene (19), which are distinguished by an “ISG12” motif of unknown function (20). Family members are small and highly hydrophobic and may be localized to either mitochondrial membranes (21, 22) or nuclear membranes (23, 24), although the exact localization has not been fully elucidated. Several Ifi27 genes are IFN inducible (19), and yet others are not, and among the family members, some orthologs are not conserved across species. As an example, IFI6-16 is an IFI27 human gene family member that inhibits infection of YFV, WNV, and hepatitis C virus (HCV) (25–28) but that does not have an apparent ortholog in mice. Although Ifi27l2a is induced broadly in peripheral organs after IFN stimulation, it is expressed in selected regions in the brain during development in an age-dependent manner (29), with high levels within the hippocampus (30). Cell culture studies have suggested that some Ifi27 gene orthologs (e.g., ISG12a) promote apoptosis and cell death (21, 31, 32).
Ifi27l2a and its closest orthologs have been evaluated as candidate antiviral genes. Despite the strong upregulation of Ifi27l2a mRNA in lung tissues, largely as a result of infiltration of immune cells, Ifi27l2a−/− mice were not more susceptible to influenza A virus (IAV) infection than wild-type (WT) mice (33). In contrast, ectopic expression of human IFI27 inhibited HCV and Newcastle disease virus (NDV) infection in hepatocellular carcinoma cells; reciprocally, gene silencing of IFI27 resulted in increased HCV and NDV infection (28, 34). Apart from their possible antiviral activity, Ifi27 genes may regulate inflammation, as mice lacking Ifi27l2a sustained less vascular injury (24) and exhibited less septic shock after administration of endotoxin (35).
To characterize further the antiviral effects of Ifi27l2a, we generated Ifi27l2a−/− mice directly in a C57BL/6 background. Ifi27l2a−/− GCN from the cerebellum and bone marrow-derived macrophages (Mϕ) supported higher levels of WNV infection. Following in vivo infection with WNV, Ifi27l2a−/− mice exhibited increased mortality and higher viral burden in the cerebellum, brain stem, and spinal cord. The enhanced viral burden in the cerebellum and brain stem of Ifi27l2a−/− mice was associated with lower levels of neuron death at early stages of CNS infection. Our findings suggest that Ifi27l2a contributes to an antiviral state against WNV within the CNS and protects subsets of cell types and regions of the brain against infection.
MATERIALS AND METHODS
Virus.
A WNV-New York (WNV-NY) stock was generated in C6/36 Aedes albopictus cells (ATCC) from a single passage of strain 3000.0259 isolated from a mosquito in New York in 2000 (36). The WNV Madagascar strain (WNV-MAD) stock was generated by passaging virus in Vero or C6/36 cells as described previously (37). WNV titers were assessed by plaque assay performed on BHK21-15 cells (38, 39). All virus stocks were stored at −80°C.
Mice.
WT C57BL/6 mice were obtained from Jackson Laboratory (Bar Harbor, ME). Ifi27l2a−/− (ISG12b1; 76933) mice were generated at Washington University after receiving heterozygous sperm from C57BL/6 mice containing a deleted gene [Ifi27l2atm1(KOMP)Vlcg] from the Knockout Mouse Project Repository (KOMP; University of California, Davis). Sperm was used for in vitro fertilization of eggs from C57BL/6 recipient female mice. Heterozygous Ifi27l2a+/− mice were backcrossed to establish the Ifi27l2a−/− colony. Ifi27l2a−/− mice produced normal litter sizes of expected Mendelian ratios, with all progeny appearing healthy. All animals were maintained in the pathogen-free animal facility of Washington University School of Medicine.
Mouse infection experiments.
The experimental protocols were approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine (Assurance Number A3381-01). Studies were performed on sex- and age-matched mice between 11 and 12 weeks of age. Peripheral infection was performed by subcutaneous inoculation into the footpad with 102 PFU of virus diluted in 50 μl Hanks' balanced salt solution (HBSS) with 1% heat-inactivated fetal bovine serum (HI-FBS). Survival was followed for 30 days. For viral burden studies after subcutaneous infection, mice were sacrificed at days 2, 4, 6, 8, 10, and 14 and peripheral organ and CNS tissues were collected following extensive perfusion with phosphate-buffered saline (PBS) and stored at −80°C. Serum was collected after cardiac puncture according to standard procedures. Intracranial infection was performed by injecting 101 PFU of WNV-NY or WNV-MAD in a solution of 10 μl HBSS supplemented with 1% HI-FBS. For analysis of viral burden after intracranial infection, brain and spinal cord tissues were collected at 3 and 5 days and processed as described for tissues from peripheral infection. Brains were divided by dissection into brain stem, cerebellum, olfactory bulb, gray matter (cerebral cortex), and subcortex (corpus callosum, hippocampus, thalamus, and hypothalamus). Plaque assays were performed with Vero cells as previously described (38). Levels of WNV RNA in serum were measured by quantitative reverse transcription-PCR (qRT-PCR) as previously described (38, 40).
Generation and infection of primary cell cultures.
All primary cell culture preparation and virus infection studies were performed as described below. In some experiments, cells were pretreated with indicated doses of mouse IFN-β prior to infection.
(i) MEFs.
Murine embryonic fibroblasts (MEFs) were generated from embryonic day 14 WT and Ifi27l2a−/− mice. Embryos were decapitated, livers were removed, and the remaining minced tissue was digested in 0.25% (wt/vol) trypsin for 10 min at 37°C with periodic, gentle agitation and mechanical disassociation. Following trypsin neutralization with FBS, cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 20% HI-FBS, 1% HEPES, 1% GlutaMAX (Life Technologies), 100 U/ml penicillin and streptomycin (Gibco), and 1% nonessential amino acids (Gibco). Cells were infected at a multiplicity of infection (MOI) of 0.01. A subset of cells was pretreated with 5 U/ml of mouse IFN-β (PBL Assay Science) (derived from Escherichia coli) for 12 h prior to infection with WNV.
(ii) Macrophages and dendritic cells.
Mϕ and dendritic cells (DCs) were generated as previously described (41) from bone marrow of WT or Ifi27l2a−/− mice. Mϕ and DCs were stimulated in culture for 8 days with 40 ng/ml recombinant murine granulocyte-macrophage colony-stimulating factor (M-CSF) (Peprotech) and 20 ng/ml recombinant murine IL-4 and recombinant murine granulocyte-macrophage colony-stimulating factor (GM-CSF) (Peprotech), respectively. Mϕ and DCs were infected with WNV-NY at an MOI of 0.01 and an MOI of 0.001, respectively.
(iii) Cortical neurons.
CN were prepared from the cerebral cortex of embryonic day 15 WT and Ifi27l2a−/− mice as described previously (42). Tissues were dissociated at 37°C in 1 ml 0.25% (wt/vol) trypsin–0.25 mg DNase I (Sigma)–HBSS for 20 min. Trypsin was neutralized with 10% HI-FBS–DMEM, and cells were filtered through a 70-μm-pore-size filter and seeded at 5 × 105 cells/well on poly-d-lysine and laminin (10 μg/ml)-coated 24-well cell-culture-treated plates. CN cells were cultured in Neurobasal medium (Life Technologies) supplemented with 2% B-27 (Gibco), 1% GlutaMAX, and 100 U/ml penicillin and streptomycin.
(iv) Granule cell neurons.
GCN were prepared from the cerebellum of 7-day-old pups and dissociated using the same protocol as for CN. These cells were cultured in the same medium as the CN with the addition of 40 mM KCl. Medium changes (50% of starting volume) were performed every 2 to 3 days, and neurons were maintained for 21 days in culture. In some experiments, neurons were pretreated for 24 h with 100 U/ml of IFN-β. Neurons were infected for 1.5 h at 37°C, rinsed with HBSS twice, and cultured in their respective complete neuronal media. GCN were infected with WNV-NY or WNV-MAD at an MOI of 0.01 or 0.1, respectively. In some experiments, GCN were pretreated with IFN-β (150 or 100 IU/ml for WNV-NY or WNV-MAD, respectively) for 24 h. Viral titers were determined by focus-forming (FF) assay, as previously described (43).
The purity of cultured neuron populations was defined by immunofluorescence microscopy analysis after incubating with antibodies to S100-β (Abcam 52642) (1:200 dilution), NeuN (Milipore MAB377B) (1:100 dilution), and Iba1 (WAKO 019-19741) (1:500 dilution) to identify astrocytes, neurons, and microglia, respectively. Secondary Alexa Fluor 488 or 555-conjugated dyes (Invitrogen) (1:400 dilution) were used for detection. Samples were imaged with a Nuance FX multiplex biomarker imaging system (PerkinElmer). Using this analysis, our GCN cultures were shown to be comprised of 85% neurons and 12% astrocytes.
Cytokine and chemokine profiling.
Cytokines and chemokines were profiled from serum at days 4 and 6 after peripheral WNV-NY infection. Protein levels were assayed with the Bio-Plex Pro cytokine assay per the manufacturer's protocol.
Immune cell analysis.
Splenocytes and peripheral blood mononuclear cells were harvested from WT and Ifi27l2a−/− mice at day 8 after subcutaneous infection with WNV. Cells were stained for the following surface antigens following a 10-min preincubation with Fc-block (eBioscience) (1:25 dilution): CD3 (BD Horizon; 500A2) (1:25 dilution), CD4 (BioLegend; RM4[5) (1:100 dilution), CD8α (Biolegend; 53-6.7) (1:100 dilution), and CD19 (BioLegend; 6D5) (1:100 dilution). Dead cells were excluded from analysis using eFluor viability dye (eBioscience) (1:300 dilution). Cells were washed, fixed, permeabilized, and stained for granzyme B (Invitrogen; GB11) (1:50 dilution) and the allophycocyanin (APC)-conjugated Db-restricted NS4B peptide (SSVWNATTAI) tetramer (NIH Tetramer Facility [Atlanta, GA]) (1:300 dilution). Blood monocytes were detected after staining with Gr-1 (BioLegend; RB6-8C5) (1:100 dilution), CD115 (eBioscience; AFS98) (1:100 dilution), CD8α (1:100 dilution), and F4/80 (Serotec; CI:A3-1) (1:100 dilution) antibodies. CD8− CD115+ F4/80+ cells were designated monocytes after extensive gating analysis (44). The monocytes in blood included circulating CD115+ F4/80+ Gr-1lo monocytes that likely do not become resident within tissues. Circulating CD115+ F4/80+ Gr-1hi monocytes may enter tissue during inflammation (“inflammatory monocytes”) and differentiate into macrophages (44–47). All samples were processed on a LSR Fortessa cell analyzer, and data were analyzed by FlowJo software (Tree-Star).
CNS leukocytes were isolated according to a published method (48). Briefly, 8 days following subcutaneous WNV infection, mice were perfused extensively with PBS. Brain tissue was minced and digested in HBSS supplemented with 0.05% collagenase D (Sigma), 0.1 μg/ml trypsin inhibitor TLCK (N-α-p-tosyl-l-lysine chloromethyl ketone), 10 μg/ml DNase I (Sigma), and 10 mM HEPES (pH 7.3) for 30 min. CNS cells were strained with a 70-μm-pore-size filter and subjected to Percoll gradient (30% [vol/vol]) purification (1,200 × g, 30 min). Cells were washed, incubated with Fc-block, and stained for CD8α (1:100 dilution), CD11b (1:100 dilution), CD19 (1:100 dilution), and CD45 (1:100 dilution) with eFluor viability dye (1:300 dilution) for 1 h at 4°C and then rinsed and incubated with commercial cell fixative (eBioscience). Samples were processed on an LSR Fortessa flow cytometer and analyzed with FlowJo commercial software (Tree-Star). Neutrophils (CD11bhigh CD45high) were identified by their unique high-side-scatter profile. Mϕ (CD11bhigh CD45high) and microglial (CD11bhigh CD45low) populations were identified by their relative levels of expression of CD11b and CD45 and their low-to-medium-side-scatter profiles.
The TFH and germinal center B cell responses were measured in the draining lymph node (DLN) 8 days postinfection with WNV-NY. Cells were stained as previously described with fluorochrome or biotin-conjugated antibodies purchased from BD Biosciences, Biolegend, and eBioscience as follows: CD3ε (145-2C11), CD4 (RM4-5), CD19 (1D3), PD1 (29F.1A12), Fas (Jo2), and GL7 and CXCR5 (2G8) (49).
Serum antibody analysis.
WNV-specific IgG and IgM dilution endpoint titers were determined by enzyme-linked immunosorbent assay (ELISA) against purified WNV E protein, as previously described (50). Assays using the 50% focus reduction neutralization test (FRNT50) were performed on Vero cells following serial dilution of serum with 100 focus-forming units (FFU) of WNV as previously described (39, 49).
TUNEL staining.
Brain tissue was harvested from mice 9 days after subcutaneous WNV infection. Mice were perfused with 30 ml PBS, and half of the brain was retained for viral titer analysis, while the other brain half was fixed in 4% paraformaldehyde (PFA)–PBS overnight. This was followed by 6 h of incubation of brains in 20% sucrose solution and overnight incubation in 30% sucrose solution, all at 4°C. Selected brains (from WT or Ifi27l2a−/− mice) with equivalent viral titers (1 to 8 × 105 PFU/g) were embedded in Optimal Cutting Temperature medium (OTC; Tissue-Tek), frozen at −80°C, and sectioned in 10-μm-thick slices on a Microm HM505N cryostat on positively charged slides (Globe Scientific; 1358W). Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) staining was performed with a Roche in situ cell death detection kit using TMR red per the manufacturer's instructions. Neurons were costained with anti-NeuN (Millipore; A60) (1:100 dilution) and secondary Alexa Fluor 488 (1:400 dilution), and nuclei were visualized with DAPI (4′,6-diamidino-2-phenylindole). Slides were mounted with Prolong Gold Diamond anti-fade mounting media (Invitrogen). Tissues were imaged on a Zeiss LSM880 confocal microscope with a Plan-Apochromat 40×/1.4 oil differential inference contrast (DIC) M27 objective at the Washington University Microscopy Core Facility. DAPI, Alexa Fluor 488, and TMR red were detected with the respected wavelength channels: 415 to 470 nm, 491 to 553 nm, and 553 to 624 nm. The image, approximately 637.64 μm by 637.64 μm, was composed by automated tiling of 9 panels (Zeiss Zen), with the central panel being selected for TUNEL-positive cells in the same region of the brain tissue in each animal. Four mice were imaged per genotype with two tissue slices per mouse and two images per tissue slice for both brain stem and cerebellum. TUNEL-positive events were counted within the 9-tiled composite image.
qRT-PCR assays.
WNV, Oas1a, and Ifit1 mRNA was analyzed using RNA extracted from GCN following treatment with IFN-β (100 U/ml), poly(I·C) (InvivoGen) (50 μg/ml), WNV-NY (MOI, 5), or WNV-MAD (MOI, 5) for 8 h prior to collection in lysis buffer. qRT-PCR was performed as previously described for WNV, Oas1a, and Ifit1, and gene expression was normalized to the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene expression (51). Commercially available Ifi27l2a primer-probe assay was purchased from IDT. A TaqMan RNA-to-CT 1-Step kit was used for qRT-PCR.
Blood-brain barrier permeability.
Blood-brain barrier (BBB) studies were performed as previously described (52). Briefly, 4 days after subcutaneous infection with WNV-NY, WT and Ifi27l2a−/− mice were injected via the intraperitoneal route with 100 μl of 100 mg/ml fluorescein (Sigma)–PBS. Dye was allowed to circulate for 45 min, serum was collected as a normalization control, mice were perfused with 20 ml PBS, and brain regions were collected for analysis as previously described (52).
Statistical analyses.
All data were analyzed with Prism software (GraphPad Prism, San Diego, CA). qRT-PCR data from comparisons between three or more groups were analyzed by one-way analysis of variance (ANOVA) with Tukey's honest significant difference (HSD) post hoc analysis. qRT-PCR data from comparisons between two groups were analyzed by Student's t test, with corrections for multiple comparisons performed using the Holm-Sidak method. Serum cytokine levels were analyzed by Student's t test, with corrections for multiple comparisons performed using the Holm-Sidak method. Kaplan-Meier survival curves were analyzed by the Mantel-Cox log-rank test. Viral burden in tissues was analyzed by the Mann-Whitney test. Serum antibody titers were analyzed by Student's t test. For analyses of viral growth kinetics in cell culture, the log-transformed viral titer was analyzed by Student's t test. Results of flow cytometry-based assays, where the total cell count or percent total cell count was measured, were also analyzed by Student's t test.
RESULTS
A deficiency of Ifi27l2a increases susceptibility to WNV infection.
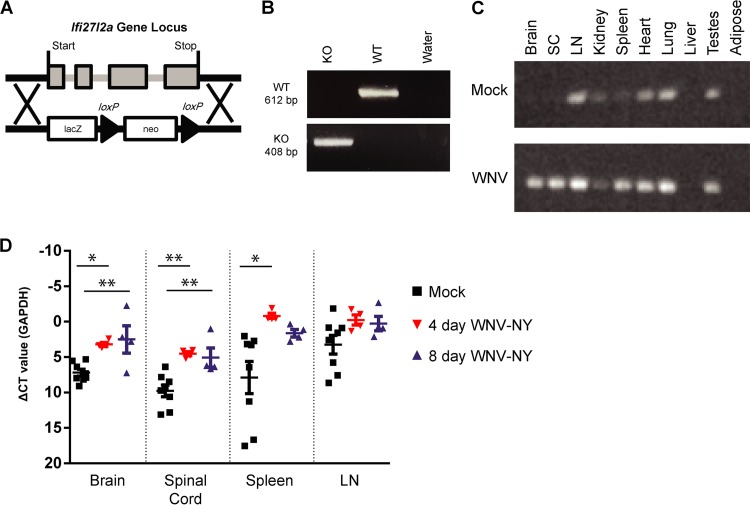
The Ifi27l2a ISG is differentially upregulated in selected neurons of the brain after WNV infection, and ectopic expression of Ifi27l2a in cultured cortical neurons inhibited infection by WNV (14). To explore an antiviral role for this relatively poorly characterized ISG in vivo, we generated Ifi27l2a−/− mice using a targeted gene deletion strategy (Fig. 1A); deletion of Ifi27l2a was validated by PCR (Fig. 1B). In WT mice, basal Ifi27l2a mRNA expression was observed in lymph node, heart, lung, and testes and, to a lesser extent, in the kidney and spleen. Following WNV infection, Ifi27l2a mRNA expression was induced in the brain and spinal cord (Fig. 1C). To quantify Ifi27l2a mRNA expression in response to viral infection, we analyzed selected tissues at successive time points following peripheral inoculation (Fig. 1D). At 4 and 8 days postinfection, Ifi27l2a mRNA expression was enhanced in the brain (2.0-fold and 3.3-fold, respectively; P < 0.005) and the spinal cord (2.1-fold and 2.0-fold, respectively; P < 0.005). Within the spleen, 12-fold-higher levels of Ifi27l2a mRNA were observed 4 days after infection (P < 0.05). We also analyzed Ifi27l2a−/− and wild-type (WT) mice for possible differences in immune cell subsets in the spleen and blood. Although the numbers of CD4+ and CD8+ T cells and CD19+ B cells were similar, Ifi27l2a−/− mice had slightly greater numbers of splenic NK cells (NK1.1+) than WT mice (Table 1); while noteworthy, this phenotype may be less important in the context of WNV infection, as NK cell depletion does not impact WNV pathogenesis in mice (12, 53). Within the peripheral blood leukocyte compartment, we observed similar numbers of myeloid cells, monocytes, and subsets of granulocytes (Table 2).
FIG 1.
Generation of Ifi27l2a−/− mice and tissue expression of Ifi27l2a. (A) Scheme of Ifi27l2a locus with targeting cassette. Exons are noted in gray, and the target location is noted with a black arrow. Ifi27l2a gene deletion was verified by PCR. Genotyping was verified with a positive-control plasmid containing wild-type Ifi27l2a and negative controls with a null plasmid control. (B) Ifi27l2a deletion was confirmed by the presence of a 408-bp band (KO, knockout), whereas WT Ifi27l2a manifested as a 612-bp band. (C) The Ifi27l2a RT-PCR product was screened for in brain, spinal cord (SC), lymph node (LN), spleen, kidney, lung, liver, white fat (adipose), and testes. Selected mice were infected subcutaneously with WNV-NY, and tissues were collected 4 days after infection and compared to those of mock-infected animals. The results are representative of 2 to 3 mice per treatment group. (D) Following peripheral infection by WNV-NY, selected tissues were collected at 4 and 8 days after infection and expression of Ifi27l2a mRNA was compared to the results seen with mock-infected animals. Ifi27l2a mRNA was measured in brain, spinal cord, spleen, and lymph node and was normalized to the GAPDH gene by qRT-PCR. Means of the results determined for the mock-infected and infected groups were compared using one-way ANOVA followed by Tukey's HSD post hoc analysis (*, P < 0.05; **, P < 0.005). Bars represent means ± standard errors of the means (SEM). CT, threshold cycle.
TABLE 1.
Immunophenotyping of lymphocytes in the spleen of naive WT and Ifi27l2a−/− micea
| Cell type | Cell type subset | WT |
Ifi27l2a−/− |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Absolute no. |
Percent |
Absolute no. |
Percent |
||||||
| Avg | ±SD | Avg | ±SD | Avg | ±SD | Avg | ±SD | ||
| CD4+ | 2.84E + 06 | 4.74E + 05 | 14.20 | 1.78 | 2.95E + 06 | 2.72E + 05 | 14.13 | 1.08 | |
| CD44high CD62llow | 3.05E + 05 | 2.26E + 04 | 10.88 | 1.08 | 3.52E + 05 | 8.57E + 04 | 11.93 | 2.85 | |
| CD44low CD62lhigh | 2.10E + 06 | 4.22E + 05 | 73.60 | 2.68 | 2.14E + 06 | 2.56E + 05 | 72.39 | 5.09 | |
| CD8+ | 2.01E + 06 | 3.23E + 05 | 10.04 | 1.06 | 2.05E + 06 | 2.67E + 05 | 9.79 | 0.62 | |
| CD44high CD62llow | 3.41E + 04 | 6.47E + 03 | 1.75 | 0.47 | 4.69E + 04 | 2.19E + 04 | 2.31 | 1.13 | |
| CD44low CD62lhigh | 1.93E + 06 | 3.19E + 05 | 96.20 | 0.94 | 1.95E + 06 | 2.79E + 05 | 94.93 | 2.77 | |
| CD19+ | 1.39E + 07 | 2.43E + 06 | 69.26 | 2.65 | 1.43E + 07 | 2.01E + 06 | 67.86 | 2.63 | |
| IgMhigh | 1.36E + 07 | 2.34E + 06 | 98.14 | 0.31 | 1.40E + 07 | 1.91E + 06 | 98.02 | 0.29 | |
| NK | 4.80E + 05* | 8.13E + 04 | 2.41* | 0.30 | 7.31E + 05* | 1.13E + 05 | 3.47* | 0.21 | |
Data represent means (Avg) ± standard deviations (SD) for cells from WT and Ifi27l2a−/− mice (n = 5 mice each; *, P < 0.05). The percentages of the indicated populations were calculated as proportions of the total parent populations.
TABLE 2.
Immunophenotyping of myeloid cells in peripheral blood of naive WT and Ifi27l2a−/− micea
| Cell type | Cell type subset | WT |
Ifi27l2a−/− |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Absolute no. |
Percent |
Absolute no. |
Percent |
||||||
| Avg | ±SD | Avg | ±SD | Avg | ±SD | Avg | ±SD | ||
| Monocyte | 2.31E + 05 | 2.90E + 04 | 7.54 | 0.52 | 2.72E + 05 | 1.03E + 05 | 8.98 | 2.50 | |
| Gr-1high | 1.68E + 05 | 2.60E + 04 | 72.62 | 3.68 | 1.96E + 05 | 9.22E + 04 | 70.58 | 5.44 | |
| Gr-1low | 6.17E + 04 | 8.17E + 03 | 26.90 | 3.73 | 7.49E + 04 | 1.58E + 04 | 29.04 | 5.48 | |
| Neutrophil | 3.53E + 05 | 1.30E + 05 | 11.20 | 2.44 | 3.16E + 05 | 9.53E + 04 | 10.48 | 2.02 | |
| Eosinophil | 3.07E + 04 | 1.84E + 04 | 0.96 | 0.41 | 3.02E + 04 | 1.78E + 04 | 0.99 | 0.47 | |
Data represent numbers of cells per milliliter of blood (Avg) ± standard deviation (SD) from WT and Ifi27l2a−/− (n = 5) mice. The percentages of the indicated populations were calculated as proportions of the total parent populations.
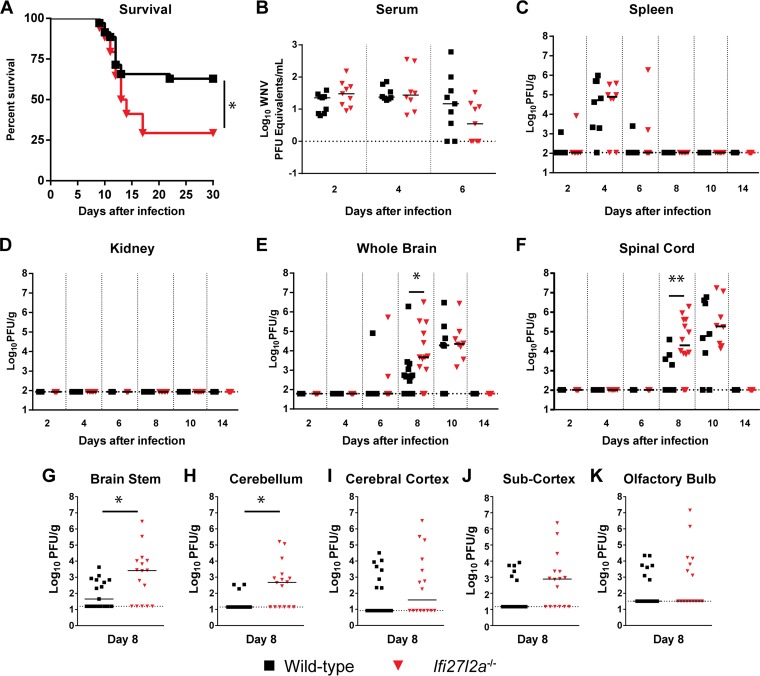
We next infected WT and Ifi27l2a−/− congenic mice with a pathogenic isolate of WNV (strain 3000.0259; WNV-NY). After subcutaneous infection with 102 PFU of WNV-NY, Ifi27l2a−/− mice exhibited a decreased survival rate compared to WT animals (29% versus 63%, P < 0.05), although the mean times to death in the two groups were similar (Fig. 2A).
FIG 2.
Survival and viral burden analysis for WT and Ifi27l2a−/− mice infected with WNV. (A) Survival analysis of 11-to-12-week-old WT or Ifi27l2a−/− mice after inoculation with 102 PFU of WNV-NY by subcutaneous injection in the footpad. In the experiments represented in panel A, WT (n = 39) and Ifi27l2a−/− (n = 34) mice were used for survival curve determinations. Asterisks indicate differences that were statistically significant compared to WT mouse results (Mantel-Cox log rank test analysis; P < 0.05). (B to F) Viral burden after WNV-NY infection of WT or Ifi27l2a−/− mice was measured by qRT-PCR (B) or plaque assay (C to G) in samples from serum (B), spleen (C), kidney (D), brain (E), and spinal cord (F) at the indicated time points after infection. (G to K) Selected brain regions were assayed for viral burden at 8 days post-subcutaneous infection with WNV-NY. Data points represent individual mice. Bars indicate medians of values obtained with 16 to 17 mice per tissue. Asterisks indicate statistical significance as determined by the Mann-Whitney test (*, P < 0.05; **, P < 0.005). The dotted line indicates the limit of detection for each tissue. Data are pooled from the results of at least three independent experiments.
WNV burden in the CNS of Ifi27l2a−/− mice.
To understand why an absence of Ifi27l2a resulted in enhanced pathogenicity of WNV-NY, viral burden was examined at different days (2, 4, 6, 8, 10, or 14) in serum, peripheral organs (spleen and kidney), and CNS tissues (brain and spinal cord). Levels of WNV viremia in WT and Ifi27l2a−/− mice at days 2, 4, and 6 were similar (Fig. 2B). At all time points tested, levels of viral burden in the spleen of WT and Ifi27l2a−/− mice were also similar (Fig. 2C). Moreover, a deficiency in Ifi27l2a did not result in productive infection of the kidney (Fig. 2D), an organ that is typically resistant to WNV-NY infection in WT mice and yet is permissive in animals with defects in type I IFN induction, signaling, or effector functions (54–58). However, within the CNS at day 8 after infection, the WNV-NY burden increased in the brain (2.4-fold increase, P < 0.05) and spinal cord (170-fold increase, P < 0.005) of Ifi27l2a−/− mice (Fig. 2E and F). This difference in viral titer was not apparent at later time points, and by day 14, infectious virus was not detectable within the CNS or peripheral tissues in surviving animals of both genotypes, suggesting that Ifi27l2a−/− animals did not have a defect in the clearance phase of WNV, which requires CD8+ effector T cells (36).
To corroborate these findings, we performed plaque assays on tissue homogenates isolated from specific regions of the CNS. WT and Ifi27l2a−/− mice were infected with WNV-NY via the subcutaneous route, and viral burden in the brain stem, cerebellum, cerebral cortex, subcortex (defined in Materials and Methods), and olfactory bulb was measured at day 8 after infection (Fig. 2G to K). Although we observed no differences in WNV titers in the cerebral cortex, subcortex, or olfactory bulb, higher levels of infection were observed in the cerebellum and brain stem (590-fold [P < 0.05] and 5,200-fold [P < 0.05], respectively) of Ifi27l2a−/− mice. These data suggest that Ifi27l2a has a protective, antiviral role in selected regions of the CNS.
WNV infection after direct intracranial inoculation.
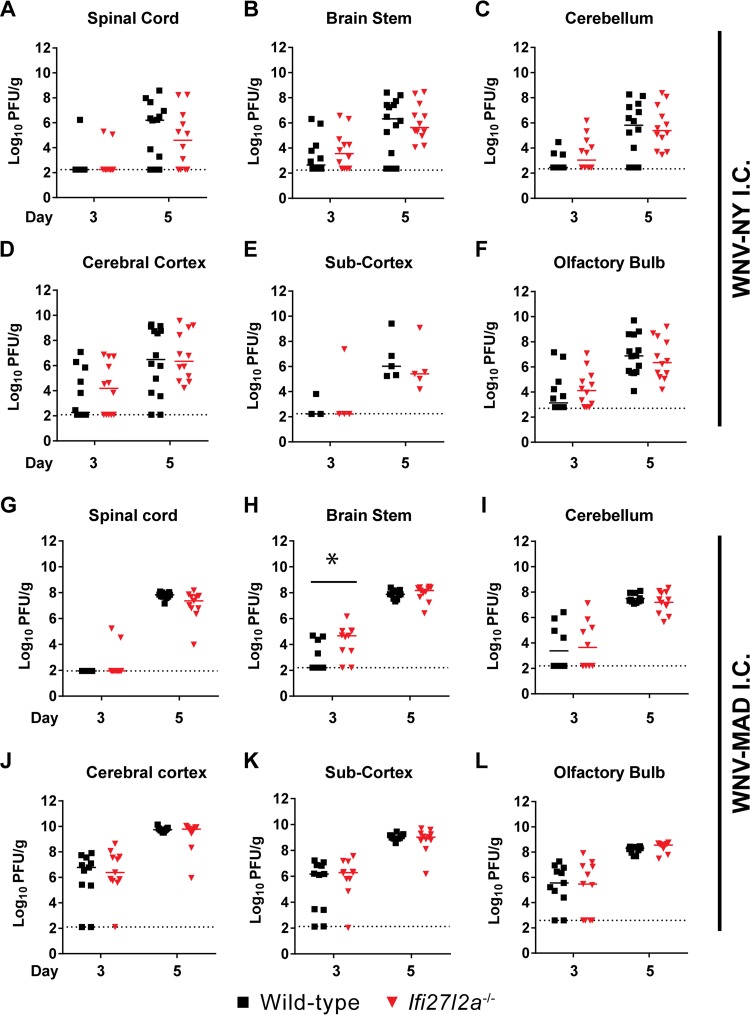
As Ifi27l2a−/− mice exhibited higher viral titers in the brain stem and cerebellum, we postulated that Ifi27l2a might protect specific regions when virus was administered directly into the CNS. Unexpectedly, following intracranial inoculation of WNV-NY into the cerebral cortex, we observed no differences in viral titers within different regions of the CNS at either 3 or 5 days after infection (Fig. 3A to F). Because the WNV-NY strain is highly virulent, we repeated intracranial infection studies with the attenuated WNV Madagascar strain (WNV-MAD), which is more sensitive to the antiviral effects of type I IFN (14, 37, 59). We observed a modest (17-fold, P < 0.05) and yet statistically significant phenotype, with greater WNV-MAD infection at day 3 after infection in the brain stem of Ifi27l2a−/− mice (Fig. 3G to L). Thus, Ifi27l2a had an antiviral effect when virus was introduced directly into the CNS, although its magnitude was limited and apparent only with an attenuated, more IFN-sensitive strain.
FIG 3.
Viral titers in the brain after intracranial infection of WT and Ifi27l2a−/− mice. Mice were infected with 101 PFU of either WNV-NY (A to F) or WNV-MAD (G to L) via an intracranial (I.C.) route, selected CNS regions were harvested, and viral burden was determined by plaque assay. Data points represent individual mice. Bars indicate median values obtained with 4 to 10 mice per time point per tissue. Dotted lines represent the limit of detection of the assay. Asterisks indicate statistical significance as determined by the Mann-Whitney test (*, P < 0.05).
Ifi27l2a does not alter adaptive cellular or humoral immune responses during acute WNV infection.
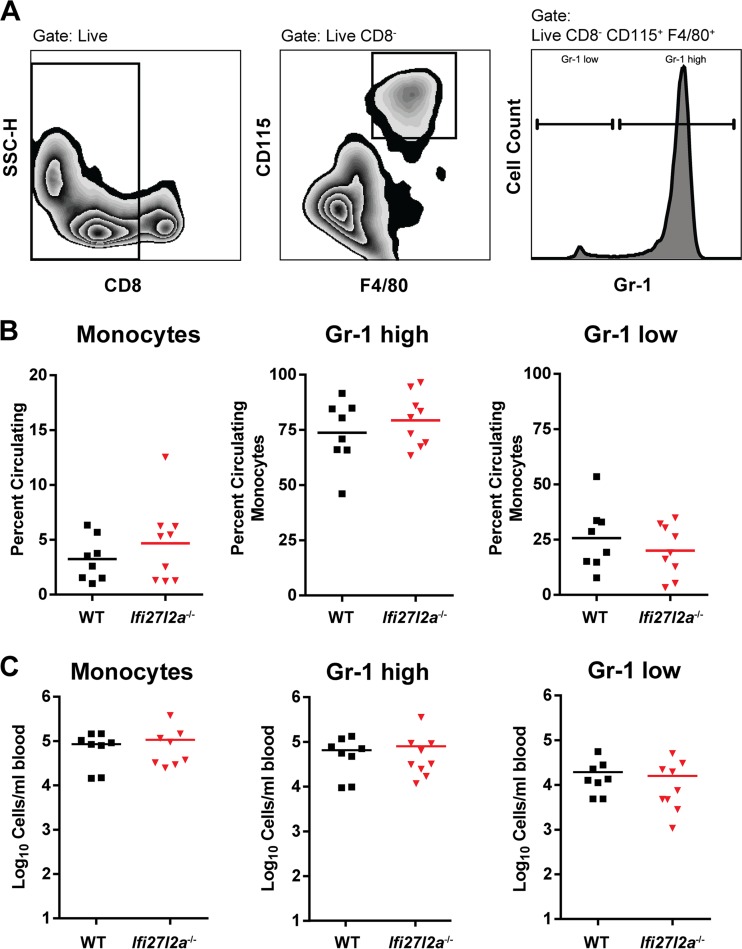
As depressed antiviral CD8+ T cell or antibody responses can facilitate dissemination to and replication of WNV within the CNS (reviewed in reference 12), we investigated whether an absence of Ifi27l2a influenced the development of adaptive immunity during WNV-NY infection. Initially, we examined the effects of Ifi27l2a on lymphocyte numbers in the peripheral tissues. At baseline, normal numbers and percentages of B cells, CD4+ T cells, and CD8+ T cells were present in the blood and spleen. Because a previous study suggested that Ifi27l2a modulates inflammation, possibly through regulation of Mϕ differentiation (24, 35), we profiled monocytes in blood during WNV infection (Fig. 4A). At day 8 after infection, Ifi27l2a−/− and WT mice had similar percentages and numbers of circulating and inflammatory blood monocytes as determined on the basis of differential levels of expression of the surface markers F4/80, CD115, and Gr-1 (Ly6C and Ly6G) (Fig. 4B and C and Materials and Methods).
FIG 4.
Circulating monocytes isolated from the blood of WT and Ifi27l2a−/− mice. (A) Circulating blood monocytes were gated as CD8− F4/80+ CD115+ and analyzed for expression of additional surface markers, including Gr-1 (Ly6C and Ly6G). (B and C) WT and Ifi27l2a−/− monocytes were present at similar levels at 8 days postinfection in the blood. Specific monocyte populations of Gr-1high and Gr-1low cells were phenotyped according to prior studies (45, 71), and the results are presented as either percentages (B) or total numbers (C) of cells per milliliter of blood from WT and Ifi27l2a−/− mice. For each group, Student's t test was used to compare cells from WT mice to cells from Ifi27l2a−/− mice. Bars indicate mean values of the results from three independent experiments for 8 to 9 mice for each genotype.
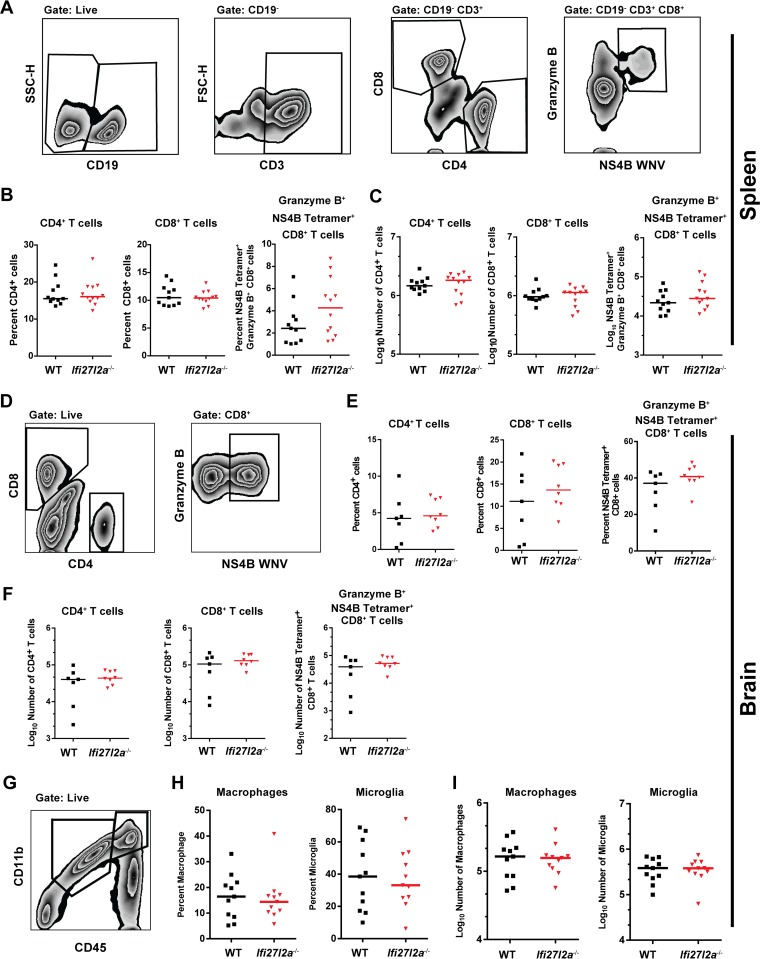
We next evaluated T cell responses in peripheral organs by characterizing the relative levels of CD4+ and CD8+ T cells (Fig. 5A). At day 8 after WNV-NY infection, equivalent percentages and numbers of CD4+ and CD8+ cells were observed in the spleen (Fig. 5B and C). Furthermore, no difference in the levels of granzyme B+ NS4B tetramer+ antigen-specific CD8+ T cells was observed in the spleens of WT and Ifi27l2a−/− mice. We assessed whether leukocyte accumulation in the CNS was altered, which could independently affect disease outcome. Leukocytes were isolated from brains of WT and Ifi27l2a−/− mice at day 8 and analyzed by flow cytometry (Fig. 5D). We observed similar percentages and numbers of CD4+ and CD8+ T cells or granzyme B+ NS4B tetramer+ CD8+ T cells within the brain (Fig. 5E and F). Microglia and infiltrating Mϕ were characterized by CD45 and CD11b surface expression (Fig. 5G). We also observed no differences in the percentage or numbers of activated microglia (CD11bhigh CD45low) or Mϕ (CD11bhigh CD45high) (Fig. 5H and I) in the brains of WT and Ifi27l2a−/− mice after WNV-NY infection. Thus, a deficiency of Ifi27l2a did not affect priming, recruitment, or activation of antigen-specific or innate immune cells in the CNS of WNV-infected mice.
FIG 5.
Splenic T cell and brain-specific immune response to WNV infection in WT and Ifi27l2a−/− mice. (A) T cells were identified by inclusion of CD19− and CD3+ cells and were analyzed by CD4 and CD8. CD8+ cells were additionally analyzed as granzyme B+- and WNV-specific NS4B tetramer+ cells. FSC-H, forward scatter height; SSC-H, side scatter height. (B and C) At 8 days after subcutaneous infection with WNV-NY, splenocytes were harvested. Similar percentages (B) and similar absolute cell numbers (C) were observed for CD4+ T cells, CD8+ T cells, and WNV-specific granzyme B+ NS4B tetramer+ CD8+ T cells (n = 11). For each group, Student's t test was used to compare values from WT mice to values from Ifi27l2a−/− mice (P < 0.05). Bars indicate mean values. Brain cells were purified by Percoll gradient centrifugation from brains of mice at 8 days postinfection. (D) Cells were gated as CD8+ positive, and WNV-specific CD8+ T cells were identified by costaining for granzyme B and with WNV-specific NS4B tetramer. (E) The percentages of CD4+ and CD8+ T cells, as well as of NS4B-specific cells, from WT and Ifi27l2a−/− mice were similar. (F) No difference in absolute numbers of infiltrating CD4+ and CD8+ T cells or in the WNV specificities of CD8+ T cells was observed. (G) Brains were analyzed for numbers of macrophages (CD45high CD11b+) and microglia (CD45low and CD11b+). (H and I) The percentages and numbers of macrophages and microglia from WT and Ifi27l2a−/− mice were similar. Percentages and absolute numbers of results from WT and Ifi27l2a−/− samples determined in three independent experiments were compared with Student's t test (n = 7 to 11 mice; *, P < 0.05). Bars indicate mean values. y-axis data are cell type dependent.
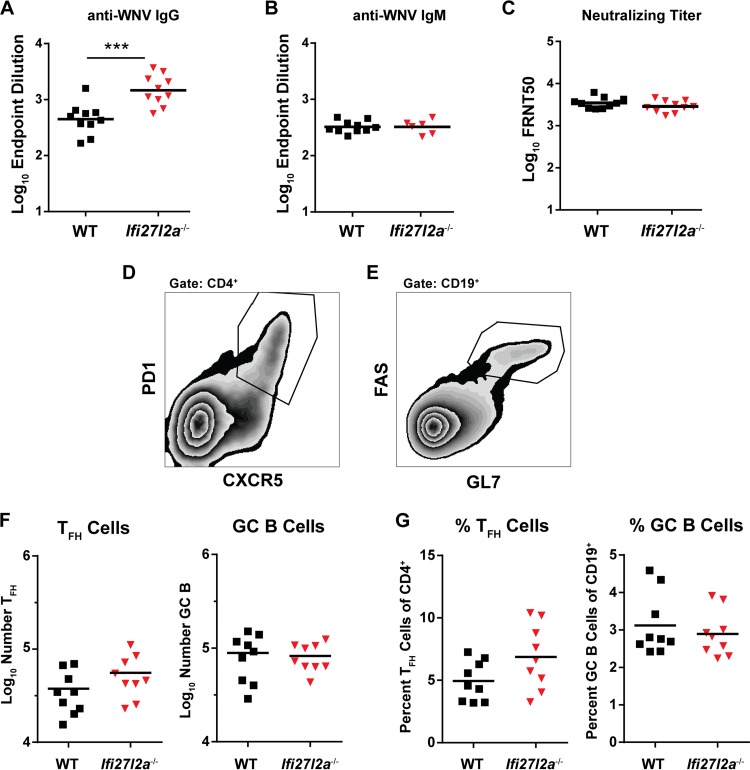
To assess the effect of Ifi27l2a on WNV-specific antibody responses, we analyzed serum from Ifi27l2a−/− and WT mice on day 8 after infection for binding to the WNV E protein. We observed IgG titers that were elevated (3.2-fold, P < 0.0005) in Ifi27l2a−/− mice compared to WT mice (Fig. 6A) but no difference in IgM titers (Fig. 6B). However, neutralization assays detected no difference in the abilities of serum-derived antibody from WT and Ifi27l2a−/− mice to neutralize WNV-NY infection (Fig. 6C).
FIG 6.
Antibody responses in WT and Ifi27l2a−/− mice after WNV infection. (A and B) Serum was obtained from WNV-infected WT and Ifi27l2a−/− mice, and IgG levels (A) or IgM levels (B) were determined at 8 days after infection and measured by ELISA for reactivity with WNV E protein. (C) Neutralizing antibody titers were determined by a focus-reduction assay from serum at day 8. Results are shown as a scatter plot and represent samples from 7 to 10 mice per group. Data are plotted as the log10 endpoint neutralization titer or log10 50% focus reduction neutralization test (FRNT50) values. Student's t test was used to compare data from WT and Ifi27l2a−/− mice (***, P < 0.0005). (D and E) Populations of TFH cells (PD1+ and CXCR5+) (D) and GC B cells (Fas+ and GL7+) (E) were identified in the DLN at 8 days post-WNV-NY infection. (F and G) Total numbers of TFH cells and GC B cells (F) and percentages of TFH cells of total CD4+ cells and percentages of GC B cells of total CD19+ cells (G) of WT and Ifi27l2a−/− mice were similar. Bars indicate mean values.
Because of the increased IgG titers in Ifi27l2a−/− mice, we next characterized whether there were differences in the T cell-dependent germinal center response in DLN of WNV-NY infected mice at 8 days postinfection. T follicular helper cells were characterized as CD4+, PD1+, and CXCR5+ (TFH; Fig. 6D), and germinal center B cells were classified as CD19+, Fas+, and GL7+ (GC B; Fig. 6E). As the total numbers and percentages of TFH and GC B cells were similar in WNV-infected WT and Ifi27l2a−/− mice, a deficiency in Ifi27l2a did not appear to alter the germinal center response within the DLN.
Cytokine and chemokine expression profiles in serum of WNV-infected Ifi27l2a−/− mice.
Because specific vasoactive cytokines (e.g., tumor necrosis factor alpha [TNF-α], IL-1β, and IL-6) can alter the blood-brain barrier (BBB) and affect transit of WNV into the brain parenchyma and early replication (reviewed in references 10 and 11), we measured whether a deficiency of Ifi27l2a affected systemic production of cytokines and chemokines at 4 or 6 days after WNV-NY infection. In WNV-infected mice, we observed greater expression of IL-1β and eotaxin in WT mice than in Ifi27l2a−/− mice at 4 days after infection (3.0-fold [P < 0.05] and 1.2-fold [P < 0.05]) but not of other cytokines and chemokines (Table 3). To assess whether this small variation in cytokine expression levels in serum impacted BBB permeability and, possibly, virus entry into the CNS, we injected the small molecule sodium fluorescein via an intraperitoneal route into WT and Ifi27l2a−/− mice at 4 days after WNV infection and then measured extravasation into different regions of the CNS. Notably, similar levels of sodium fluorescein accumulated in the cerebral cortex, cerebellum, brain stem, and spinal cord (data not shown). Thus, the small increases in serum cytokine levels in the absence of Ifi27l2a did not substantively impact BBB permeability.
TABLE 3.
Serum cytokine levels at days 4 and 6 after subcutaneous inoculation of WNVa
| Cytokine | LOD (U/ml) | Level (U/ml) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 4 dpi |
6 dpi |
||||||||
| WT |
Ifi27l2a−/− |
WT |
Ifi27l2a−/− |
||||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | ||
| IL-1α | 4.66 | 6.4 | 4.7 | 4.6 | 0.8 | 5.4 | 1.3 | 4.7 | 0.0 |
| IL-1β | 32.95 | 182.2* | 88.2 | 60.1* | 44.0 | 53.2 | 43.6 | 78.9 | 145.8 |
| IL-2 | 2.92 | 19.4 | 8.6 | 27.0 | 10.4 | 19.1 | 14.2 | 16.3 | 10.6 |
| IL-3 | 1.32 | 4.7 | 2.6 | 2.8 | 1.4 | 2.9 | 1.0 | 1.9 | 1.2 |
| IL-4 | 6.11 | 7.2 | 2.0 | 5.7 | 1.2 | 5.3 | 1.6 | 5.7 | 4.6 |
| IL-5 | 1.76 | 91.8 | 190.4 | 23.1 | 6.1 | 29.9 | 20.8 | 22.1 | 12.0 |
| IL-6 | 0.86 | 26.1 | 55.4 | 5.5 | 0.9 | 10.9 | 16.2 | 5.2 | 2.5 |
| IL-9 | 26.47 | 26.5 | 0.0 | 26.5 | 0.0 | 26.5 | 0.0 | 26.5 | 0.0 |
| IL-10 | 6.32 | 62.3 | 40.9 | 47.6 | 20.3 | 38.1 | 19.2 | 24.0 | 14.5 |
| IL-12(p40) | 1.7 | 229.5 | 114.9 | 179.2 | 54.0 | 151.5 | 46.4 | 134.2 | 27.2 |
| IL-12(p70) | 0.99 | 34.1 | 16.3 | 24.5 | 9.3 | 19.5 | 14.4 | 8.4 | 8.2 |
| IL-13 | 63.51 | 212.9 | 54.2 | 162.3 | 57.9 | 178.0 | 57.6 | 144.8 | 18.7 |
| IL-17 | 2.94 | 29.7 | 16.4 | 28.6 | 7.9 | 25.2 | 10.9 | 26.7 | 13.4 |
| Eotaxin | 38.95 | 454.6* | 90.9 | 370.2* | 157.6 | 371.8 | 125.7 | 379.5 | 138.4 |
| G-CSF | 4.11 | 39.5 | 13.6 | 26.4 | 8.7 | 36.7 | 11.9 | 16.8 | 6.8 |
| GM-CSF | 51.12 | 116.2 | 29.4 | 78.0 | 35.9 | 96.1 | 24.5 | 61.0 | 32.8 |
| IFN-γ | 1.55 | 5.7 | 1.6 | 4.6 | 1.5 | 3.9 | 1.9 | 3.3 | 1.3 |
| KC | 1.43 | 97.1 | 48.0 | 120.6 | 44.4 | 66.2 | 35.9 | 67.1 | 35.3 |
| MCP-1 | 15.39 | 156.3 | 54.0 | 147.5 | 20.1 | 130.7 | 46.6 | 110.7 | 39.3 |
| MIP-1α | 1.76 | 11.2 | 2.6 | 10.9 | 1.3 | 9.8 | 2.0 | 8.6 | 2.4 |
| MIP-1β | 6.81 | 41.1 | 38.8 | 24.4 | 8.7 | 15.5 | 10.6 | 17.1 | 14.7 |
| RANTES | 0.81 | 34.6 | 8.1 | 32.5 | 12.6 | 33.8 | 10.9 | 37.4 | 15.4 |
| TNF-α | 3.01 | 148.2 | 110.4 | 97.7 | 27.5 | 98.6 | 33.5 | 82.9 | 21.5 |
Cytokine levels were assayed via Bioplex Pro to determine differences between WT and Ifi27l2a−/− mice (n = 8 to 9 mice per genotype per time point) at 4 and 6 days postinfection (dpi) for 102 FFU administered by the subcutaneous route. Data represent means ± standard deviations (SD). Levels were compared for WT and Ifi27l2a−/− cells by Student's t test (*, P < 0.05), with correction for multiple comparisons performed by the Holm-Sidak method. LOD, limit of detection for each cytokine determined on the basis of the manufacturer's protocol for establishing standard curves for each parameter measured; G-CSF, granulocyte colony-stimulating factor; GM-CSF, granulocyte-macrophage colony-stimulating factor; KC, keratinocyte chemoattractant; MCP-1, monocyte chemoattractant protein-1; MIP-1, macrophage inflammatory protein-1.
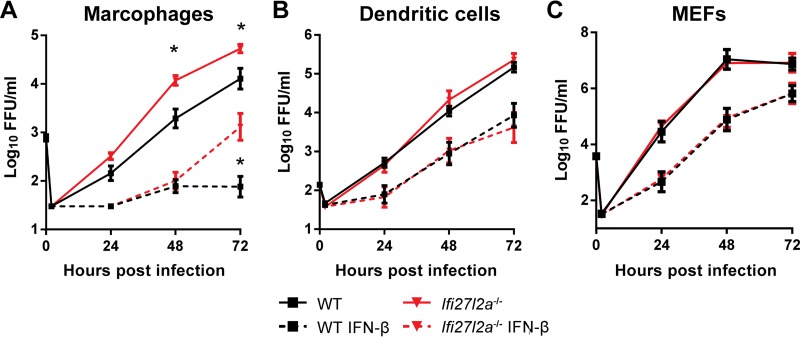
Ifi27l2a exhibits antiviral effects on WNV in Mϕ but not DCs or MEFs.
Although Ifi27l2a is expressed after WNV infection in primary and secondary lymphoid tissues, we did not observe greater viral burden in peripheral organs. We speculated that the antiviral effect of Ifi27l2a against WNV infection might not occur in nonneuronal cell types. To evaluate this hypothesis, we generated bone marrow-derived Mϕ and DCs and primary MEFs from WT and Ifi27l2a−/− mice. Cells were either pretreated with IFN-β or not treated and were then subsequently infected at a low MOI with WNV-NY. We observed increased WNV-NY replication in untreated or IFN-β-treated Ifi27l2a−/− Mϕ at later time points (P < 0.05) (Fig. 7A). However, we observed no difference in levels of viral infection at any time point in DC or MEF cultures generated from WT and Ifi27l2a−/− mice (Fig. 7B and C).
FIG 7.
Ifi27l2a restricts WNV replication in Mϕ cells but not in DCs or MEFs. (A) Bone marrow-derived Mϕ were infected with WNV (MOI, 0.01), and viral replication kinetics were followed for 72 h. A subset of Mϕ cells was pretreated with IFN-β (1 U/ml for 6 h) prior to infection. (B) Bone marrow-derived DCs were infected with WNV (MOI, 0.001), and viral kinetics were followed for 72 h. A subset of DCs was pretreated with IFN-β (10 U/ml for 6 h) prior to infection. (C) MEFs were infected with WNV (MOI, 0.01), and viral kinetics were followed for 72 h. A subset of MEFs was pretreated with IFN-β (10 U/ml for 6 h). The data were analyzed by a Student's t test for each time point for comparisons between treatment groups and are expressed as the log10 median titers ± SEM as reflects pooled data from 3 to 4 independent experiments performed with three technical replicates per independent experiment for each cell type. Ranges of y-axis data are cell type dependent.
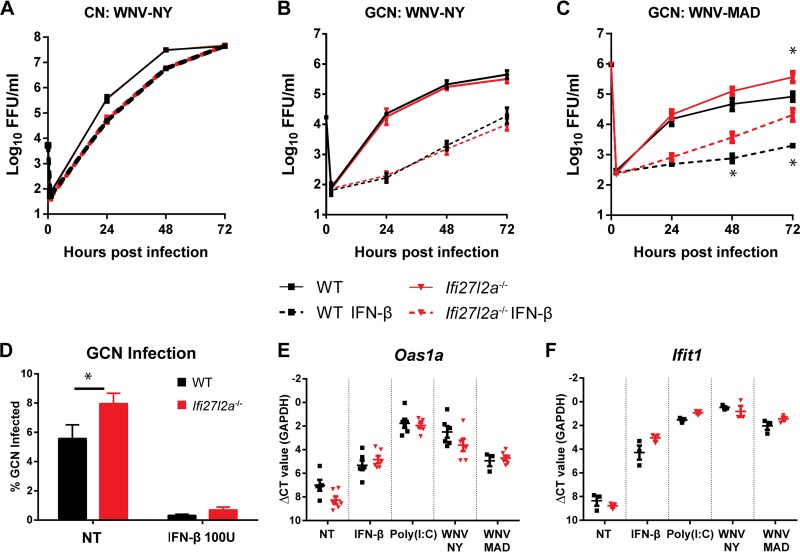
Subsets of primary neurons from Ifi27l2a−/− mice exhibit enhanced WNV infection.
Given that the virologic phenotype occurred in selected brain regions of Ifi27l2a−/− mice, we investigated whether Ifi27l2a had differential antiviral activity in different neuron populations. We prepared primary neurons from the cerebral cortex (CN) and cerebellum (GCN) of WT and Ifi27l2a−/− mice, pretreated cells with IFN-β, and measured viral growth kinetics after infection with WNV-NY. We detected no differences in replication kinetics in CN, with only mild suppression of infection with IFN-β pretreatment (Fig. 8A), as seen previously (14). Somewhat unexpectedly, we observed similar viral growth kinetics in GCN from WT and Ifi27l2a−/− mice for WNV-NY, with replication being suppressed to a greater degree following IFN-β pretreatment (Fig. 8B), also as reported previously (14). We reassessed viral growth kinetics with the more IFN-sensitive strain, WNV-MAD. Ifi27l2a−/− GCN supported higher levels of WNV-MAD infection; this effect was more pronounced when cells were pretreated with IFN-β (18-fold, P < 0.05) (Fig. 8C), and a difference in viral replication (55-fold) was present in non-IFN-β-treated cells. Consistent with these data, by 72 h after infection, a greater proportion of WNV-MAD-infected GCN was observed in Ifi27l2a−/− than in WT GCN (Fig. 8D). Because we observed differences in WNV restriction in WT and Ifi27l2a−/− GCN, we considered whether a deficiency of Ifi27l2a altered the general ISG response in GCN. We treated cells with known ISG inducers [IFN-β, poly(I·C), WNV-NY, and WNV-MAD] and assessed expression of Oas1a and Ifit1 mRNA. As Oas1a and Ifit1 induction levels were similar in WT and Ifi27l2a−/− GCN (Fig. 8E and F), a deficiency of Ifi27l2a did not broadly impact expression of other antiviral ISGs.
FIG 8.
Viral infection of WT and Ifi27l2a−/− primary neurons. Primary neuron cultures were generated from WT and Ifi27l2a−/− mice and infected with WNV-NY or WNV-MAD. Cell supernatants were harvested at the indicated time points and titrated in a focus-forming assay. (A to C) CN (A) and GCN (B and C) were infected at the following MOIs: WNV-NY, 0.01; WNV-MAD, 0.1. In some experiments, GCN and CN were pretreated with IFN-β for 24 h (for CN, 150 U/ml for WNV-NY; for GCN, 150 U/ml for WNV-NY or 100 U/ml for WNV-MAD). (D) WT and Ifi27l2a−/− GCN were infected with WNV-MAD for 72 h, and cells of infected neurons (microtubule-associated protein 2 positive [MAP2+]) were counted by automated high-throughput imaging. (E and F) WT and Ifi27l2a−/− GCN were analyzed by qRT-PCR for expression of Oas1a (E) and Ifit1 (F). Viral replication data were analyzed at each time point and for each treatment by Student's t test for each treatment group (*, P < 0.05; n = 3 for independent replicates). GCN infection assay results were analyzed by Student's t test (two experimental replicates; three to four sample wells per replicate) (*, P < 0.05). For high-throughput imaging, three wells per treatment group were analyzed for each biological replicate (n = 3). For each well, 60 computer-randomized images were collected and analyzed by the use of a GE IN Cell 2000 imager and IN Cell software. qRT-PCR data were analyzed by Student's t test with correction for multiple comparisons by the Holm-Sidak method (*, P < 0.05). NT, not treated.
Ifi27l2a−/− mice exhibit less neuronal death in the cerebellum and brain stem after WNV infection.
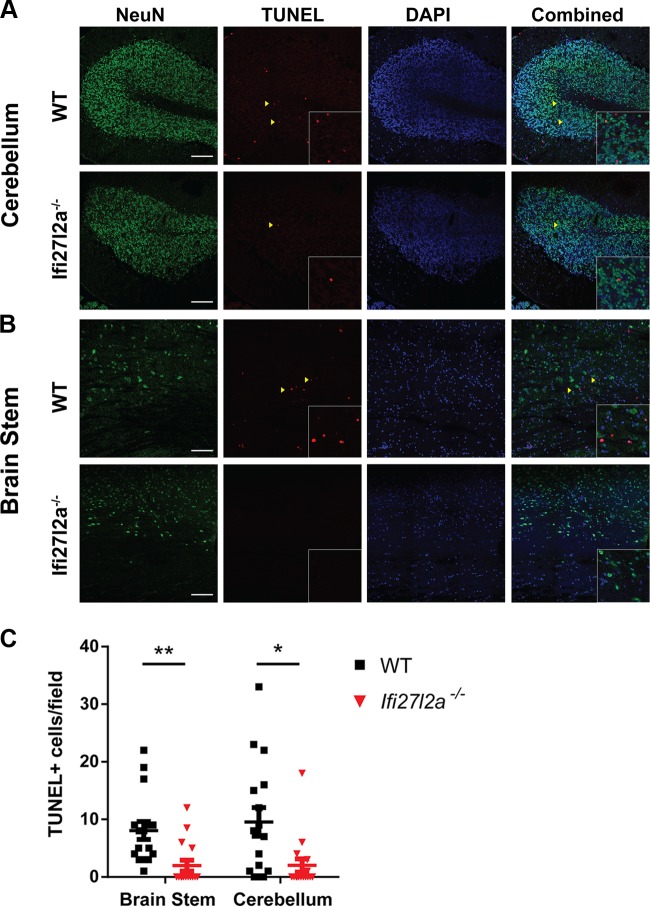
To provide a mechanistic link between our in vitro and in vivo phenotypes with Ifi27l2a−/− cells and mice, we prepared brain tissue sections for histological and immunohistochemical analyses. Although we have historically detected WNV antigen staining in neurons of different brain regions at day 9 after infection in younger (e.g., 5-and-8-week-old) mice (60, 61), despite multiple attempts, the results of viral antigen staining in 11-to-12-week-old WT or Ifi27l2a−/− mice were inconclusive. The levels of viral antigen in the CNS were at the threshold of detection, with only sporadic staining of infected neurons in different brain regions of a subset of the mice (data not shown). Because of this, and given prior reports suggesting that some Ifi27 family members (e,g., ISG12a) are required for IFN-induced cellular apoptosis (21, 31, 32), we evaluated neuronal cell death in WT and Ifi27l2a−/− mice that had equivalent WNV titers in the brain at 9 days after infection. We noted significantly more cell death in the cerebellum (5-fold, P < 0.05) (Fig. 9A and C) and brain stem (4-fold, P < 0.005) (Fig. 9B and C) of WT mice, whereas TUNEL-positive neurons were largely absent in the hindbrain regions of Ifi27l2a−/− mice.
FIG 9.
Neuronal death within the cerebellum and brain stem of WNV-infected mice. Nine days following subcutaneous infection with WNV-NY, brains of selected mice with similar levels of virus were sectioned and stained for neurons (NeuN; green), cell death (as determined by TUNEL staining; red), and nuclei (DAPI; blue). Cerebellum (A) and brain stem (B) tissues were analyzed by confocal microscopy, and fields of view containing TUNEL staining were quantitated for numbers of events per field of view (C) (n = 4 mice per genotype, 2 sections per mouse, 2 fields of view per section). The relative numbers of TUNEL+ cells in WT and Ifi27l2a−/− mice were analyzed by Student's t test, with bars indicating the means (*, P < 0.05; **, P < 0.005). Scale bars = 100 μm. Yellow arrowheads indicate examples of TUNEL+ nuclei. Inset boxes are higher-magnification images of areas denoted with yellow arrowheads.
DISCUSSION
Viral replication and the subsequent immune response within the CNS can result in significant morbidity and mortality. Because neurons are largely nonrenewable, it is imperative that the host clears viral infection while protecting cells from direct or collateral immune-mediated damage. We previously identified Ifi27l2a as a putative inhibitory ISG that was expressed preferentially within neurons of the cerebellum compared to those from the cerebral cortex (14). Here, we established a protective antiviral role for Ifi27l2a in vivo against WNV. Ifi27l2a−/− mice were more susceptible to WNV-induced mortality and sustained higher viral titers in the cerebellum, brain stem, and spinal cord. Remarkably, at day 9 after infection, Ifi27l2a−/− mice had less cell death in neurons of the cerebellum and brain stem. Ifi27l2a−/− mice showed no apparent defects in their ability to generate a peripheral or CNS cellular immune response, and levels of WNV replication were equivalent in several other cell types that lacked or expressed Ifi27l2a.
Several members of the Ifi27 family have been studied in the context of viral infections. Many viruses, including influenza A virus, Sindbis virus, WNV, JEV, and human immunodeficiency virus type 1, induce expression of Ifi27 family members (14, 29, 62–64). Our prior study reported that ectopic expression of Ifi27l2a in CN reduced WNV infection whereas small interfering RNA (siRNA)-mediated gene silencing in GCN resulted in enhanced viral replication (14). Work by others has shown that ectopic expression of human IFI27 (ISG12A) inhibited replication of HCV in Huh-7.5 cells and that, reciprocally, siRNA-mediated silencing of human IFI27 enhanced HCV replication (28). High levels of expression of human IFI27 also inhibited Newcastle disease virus (NDV) replication and oncolytic activity in Huh7 cells (34).
Ifi27l2a−/− mice have been investigated in other contexts. Although Ifi27l2a was identified as an upregulated ISG in lung tissue following IAV infection (63), Ifi272la−/− C57BL/6 mice did not sustain higher levels of viral burden or altered pathology in the lungs of infected animals (33). In another Ifi27l2a−/− mouse of mixed genetic background, gene-deficient animals were protected against cecal ligation-induced sepsis, lipopolysaccharide (LPS) endotoxemia, and vascular injury after arterial ligation (24, 35). In contrast, our Ifi27l2a−/− C57BL/6 mice succumbed to LPS administration at a rate similar to that seen with WT mice (T. M. Lucas and M. S. Diamond, unpublished observations). Although Ifi27l2a has a postulated role in regulating inflammation, at least in the context of WNV infection, we failed to observe hypercytokinemia, changes in the infiltrating immune cells, or altered adaptive immunity in Ifi27l2a−/− mice.
The unique cell type expression, subcellular localization, and induction patterns of Ifi27 family genes suggest possible modular functions. Humans have four IFI27 members, of which only IFI27 and IFI6-16 (IFI6) are IFN inducible. Mice have three gene paralogs, Ifi27 (Ifi27l1), Ifi27l2a, and Ifi27l2b, all of which are IFN inducible, with Ifi27l2a exhibiting the greatest induction after type I IFN treatment (19). IFI27 family members have been suggested to localize to the mitochondria (21, 22) or to the nuclear membrane (23, 24); in the latter case, IFI27 interacts with and sequesters the NR4A1 nuclear receptor, which regulates expression of anti-inflammatory genes (24). IFI27 family members also appear to have proapoptotic effects (21, 32, 34, 65, 66), possibly as a result of stabilization of the mitochondrial membrane and regulation of caspase activity (21, 65). Perhaps because of these proposed pleotropic activities, IFI27 family members have been associated with overexpression in certain cancers (66, 67), promotion of skin keratinocyte replication (68), and DNA damage-induced apoptosis and cytochrome c release (21). IFI6-16 (IFI6) is an ISG12 motif-containing family member that may share some functions with Ifi27l2a. In the context of DENV infection, a deficiency of IFI6-16 was associated with increased caspase levels and decreased Bcl-2 expression and mitochondrial membrane stabilization (65). Additionally, ectopic expression of IFI6-16 has been shown to suppress infection of YFV (69). Our in vivo data are most consistent with a model in which Ifi27l2a expression is induced by type I IFN in response to WNV infection and (for reasons that remain uncertain) promotes cell death in a cell-type-specific manner. In its absence (in Ifi27l2a−/− mice), subsets of virally infected neurons (e.g., in the cerebellum, brain stem, and possibly spinal cord) live longer, which allows greater yields of virus to accumulate, at least during the early phase of CNS infection. We speculate that the increased rate of death of WNV-infected Ifi27l2a−/− mice is ultimately caused by virus-induced injury of neurons in regions of the CNS that control key or autonomic function. The results of our study, along with the work of others, suggest multiple possible functions for different Ifi27 family members, with some of the antiviral properties being linked to cell death phenotypes in infected cells.
The predominant effect of Ifi27l2a in the CNS suggests a specialization of the host antiviral immune response. Analogously, preferential antiviral roles in the CNS for other ISGs (Ifit2 and Rsad2 [viperin]) have been reported. In Ifit2−/− mice, higher WNV and VSV titers were observed in the olfactory bulb, brain stem, and cerebellum (13, 70). In Rsad2−/− mice, an increase in WNV infection was observed in the cerebral cortex, white matter, and spinal cord (54). We observed differences in the regional restriction of WNV in the CNS mostly in the context of peripheral viral but not intracranial infection, with the exception of the brain stem. Viral infection of peripheral immune tissues (e.g., lymph node or spleen) may induce systemic accumulation of type I IFN that primes the antiviral response in the brain either earlier or prior to viral entry into the CNS, whereas direct administration of virus to the CNS may permit rapid replication of WNV in neurons in the context of a less robust type I IFN response. Although future studies are needed to determine why region-specific antiviral effects of individual ISGs occur, we speculate that these genes do not function in isolation, and partner proteins that are expressed differentially may regulate their activity. While our findings suggest an antiviral function within select neurons and Mϕ, the precise cellular mechanism of action of Ifi27l2a warrants further investigation.
In summary, we have identified a protective role for Ifi27l2a in the CNS following WNV infection. Ifi27l2a-mediated restriction of WNV was greatest in the cerebellum, brain stem, and spinal cord and correlated with an antiviral and cell survival effect in subsets of neurons and myeloid cells. These findings suggest that Ifi27l2a may have a discrete antiviral activity within selected regions of the CNS.
ACKNOWLEDGMENTS
We thank A. Pinto and J. Mine for help with the serum cytokine and BBB experiments, J. Williams and G. Randolph for advice on monocyte gating and analysis, M. Ilagan at the Washington University High Throughput Screening Core for assistance with the high-content imaging and analyses, and W. Beatty and B. Anthony at the Washington University Microscopy Core for assistance with confocal imaging.
REFERENCES
- 1.Carson PJ, Konewko P, Wold KS, Mariani P, Goli S, Bergloff P, Crosby RD. 2006. Long-term clinical and neuropsychological outcomes of West Nile virus infection. Clin Infect Dis 43:723–730. doi: 10.1086/506939. [DOI] [PubMed] [Google Scholar]
- 2.Chan CK, Limstrom SA, Tarasewicz DG, Lin SG. 2006. Ocular features of West Nile virus infection in North America: a study of 14 eyes. Ophthalmology 113:1539–1546. doi: 10.1016/j.ophtha.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 3.Leis AA, Fratkin J, Stokic DS, Harrington T, Webb RM, Slavinski SA. 2003. West Nile poliomyelitis. Lancet Infect Dis 3:9–10. doi: 10.1016/S1473-3099(03)00478-X. [DOI] [PubMed] [Google Scholar]
- 4.Hainline ML, Kincaid JC, Carpenter DL, Golomb MR. 2008. West Nile poliomyelitis in a 7-year-old child. Pediatr Neurol 39:350–354. doi: 10.1016/j.pediatrneurol.2008.07.027. [DOI] [PubMed] [Google Scholar]
- 5.Bigham AW, Buckingham KJ, Husain S, Emond MJ, Bofferding KM, Gildersleeve H, Rutherford A, Astakhova NM, Perelygin AA, Busch MP, Murray KO, Sejvar JJ, Green S, Kriesel J, Brinton MA, Bamshad M. 2011. Host genetic risk factors for West Nile virus infection and disease progression. PLoS One 6:e24745. doi: 10.1371/journal.pone.0024745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glass WG, McDermott DH, Lim JK, Lekhong S, Yu SF, Frank WA, Pape J, Cheshier RC, Murphy PM. 2006. CCR5 deficiency increases risk of symptomatic West Nile virus infection. J Exp Med 203:35–40. doi: 10.1084/jem.20051970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim JK, McDermott DH, Lisco A, Foster GA, Krysztof D, Follmann D, Stramer SL, Murphy PM. 2010. CCR5 deficiency is a risk factor for early clinical manifestations of West Nile virus infection but not for viral transmission. J Infect Dis 201:178–185. doi: 10.1086/649426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim JK, Lisco A, McDermott DH, Huynh L, Ward JM, Johnson B, Johnson H, Pape J, Foster GA, Krysztof D, Follmann D, Stramer SL, Margolis LB, Murphy PM. 2009. Genetic variation in OAS1 is a risk factor for initial infection with West Nile virus in man. PLoS Pathog 5:e1000321. doi: 10.1371/journal.ppat.1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho H, Diamond MS. 2012. Immune responses to West Nile virus infection in the central nervous system. Viruses 4:3812–3830. doi: 10.3390/v4123812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suen W, Prow N, Hall R, Bielefeldt-Ohmann H. 2014. Mechanism of West Nile virus neuroinvasion: a critical appraisal. Viruses 6:2796–2825. doi: 10.3390/v6072796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neal JW. 2014. Flaviviruses are neurotropic, but how do they invade the CNS? J Infect 69:203–215. doi: 10.1016/j.jinf.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 12.Suthar MS, Diamond MS, Gale M Jr. 2013. West Nile virus infection and immunity. Nat Rev Microbiol 11:115–128. doi: 10.1038/nrmicro2950. [DOI] [PubMed] [Google Scholar]
- 13.Cho H, Shrestha B, Sen GC, Diamond MS. 2013. A role for Ifit2 in restricting West Nile virus infection in the brain. J Virol 87:8363–8371. doi: 10.1128/JVI.01097-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho H, Proll SC, Szretter KJ, Katze MG, Gale M, Diamond MS. 2013. Differential innate immune response programs in neuronal subtypes determine susceptibility to infection in the brain by positive-stranded RNA viruses. Nat Med 19:458–464. doi: 10.1038/nm.3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nair S, Michaelsen-Preusse K, Finsterbusch K, Stegemann-Koniszewski S, Bruder D, Grashoff M, Korte M, Köster M, Kalinke U, Hauser H, Kröger A. 2014. Interferon regulatory factor-1 protects from fatal neurotropic infection with vesicular stomatitis virus by specific inhibition of viral replication in neurons. PLoS Pathog 10:e1003999. doi: 10.1371/journal.ppat.1003999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farmer JR, Altschaefl KM, O'Shea KS, Miller DJ. 2013. Activation of the type I interferon pathway is enhanced in response to human neuronal differentiation. PLoS One 8:e58813. doi: 10.1371/journal.pone.0058813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosato PC, Leib DA. 2014. Intrinsic innate immunity fails to control herpes simplex virus and vesicular stomatitis virus replication in sensory neurons and fibroblasts. J Virol 88:9991–10001. doi: 10.1128/JVI.01462-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yordy B, Iijima N, Huttner A, Leib D, Iwasaki A. 2012. A neuron-specific role for autophagy in antiviral defense against herpes simplex virus. Cell Host Microbe 12:334–345. doi: 10.1016/j.chom.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parker N, Porter A. 2004. Identification of a novel gene family that includes the interferon-inducible human genes 6-16 and ISG12. BMC Genomics 5:8. doi: 10.1186/1471-2164-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheriyath V, Leaman DW, Borden EC. 2011. Emerging roles of FAM14 family members (G1P3/ISG 6-16 and ISG12/IFI27) in innate immunity and cancer. J Interferon Cytokine Res 31:173–181. doi: 10.1089/jir.2010.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosebeck S, Leaman DW. 2008. Mitochondrial localization and pro-apoptotic effects of the interferon-inducible protein ISG12a. Apoptosis 13:562–572. doi: 10.1007/s10495-008-0190-0. [DOI] [PubMed] [Google Scholar]
- 22.Li B, Shin J, Lee K. 2009. Interferon-stimulated gene ISG12b1 inhibits adipogenic differentiation and mitochondrial biogenesis in 3T3-L1 cells. Endocrinology 150:1217–1224. doi: 10.1210/en.2008-0727. [DOI] [PubMed] [Google Scholar]
- 23.Martensen PM, Søgaard TMM, Gjermandsen IM, Buttenschøn HN, Rossing AB, Bonnevie-Nielsen V, Rosada C, Simonsen JL, Justesen J. 2001. The interferon alpha induced protein ISG12 is localized to the nuclear membrane. Eur J Biochem 268:5947–5954. doi: 10.1046/j.0014-2956.2001.02545.x. [DOI] [PubMed] [Google Scholar]
- 24.Papac-Milicevic N, Breuss JM, Zaujec J, Ryban L, Plyushch T, Wagner GA, Fenzl S, Dremsek P, Cabaravdic M, Steiner M, Glass CK, Binder CJ, Uhrin P, Binder BR. 2012. The interferon stimulated gene 12 inactivates vasculoprotective functions of NR4A nuclear receptors. Circ Res 110:e50–e63. doi: 10.1161/CIRCRESAHA.111.258814. [DOI] [PubMed] [Google Scholar]
- 25.Schoggins JW, Rice CM. 2011. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol 1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, Dabelic R, Manicassamy B, Aitchison JD, Aderem A, Elliott RM, Garcia-Sastre A, Racaniello V, Snijder EJ, Yokoyama WM, Diamond MS, Virgin HW, Rice CM. 2014. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505:691–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Ding SC, Cho H, Chung BC, Gale M, Chanda SK, Diamond MS. 2013. A short hairpin RNA screen of interferon-stimulated genes identifies a novel negative regulator of the cellular antiviral response. mBio 4:e00385-13. doi: 10.1128/mBio.00385-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Itsui Y, Sakamoto N, Kakinuma S, Nakagawa M, Sekine-Osajima Y, Tasaka-Fujita M, Nishimura-Sakurai Y, Suda G, Karakama Y, Mishima K, Yamamoto M, Watanabe T, Ueyama M, Funaoka Y, Azuma S, Watanabe M. 2009. Antiviral effects of the interferon-induced protein guanylate binding protein 1 and its interaction with the hepatitis C virus NS5B protein. Hepatology 50:1727–1737. doi: 10.1002/hep.23195. [DOI] [PubMed] [Google Scholar]
- 29.Labrada L, Liang X, Zheng W, Johnston C, Levine B. 2002. Age-dependent resistance to lethal alphavirus encephalitis in mice: analysis of gene expression in the central nervous system and identification of a novel interferon-inducible protective gene, mouse ISG12. J Virol 76:11688–11703. doi: 10.1128/JVI.76.22.11688-11703.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mody M, Cao Y, Cui Z, Tay K-Y, Shyong A, Shimizu E, Pham K, Schultz P, Welsh D, Tsien JZ. 2001. Genome-wide gene expression profiles of the developing mouse hippocampus. Proc Natl Acad Sci U S A 98:8862–8867. doi: 10.1073/pnas.141244998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu N, Zuo C, Wang X, Chen T, Yang D, Wang J, Zhu H. 2014. miR-942 decreases TRAIL-induced apoptosis through ISG12a downregulation and is regulated by AKT. Oncotarget 5:4959–4971. doi: 10.18632/oncotarget.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang D, Meng X, Xue B, Liu N, Wang X, Zhu H. 2014. MiR-942 mediates hepatitis C virus-induced apoptosis via regulation of ISG12a. PLoS One 9:e94501. doi: 10.1371/journal.pone.0094501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tantawy MA, Hatesuer B, Wilk E, Dengler L, Kasnitz N, Weiß S, Schughart K. 2014. The interferon-induced gene Ifi27l2a is active in lung macrophages and lymphocytes after influenza A infection but deletion of Ifi27l2a in mice does not increase susceptibility to infection. PLoS One 9:e106392. doi: 10.1371/journal.pone.0106392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu N, Long Y, Liu B, Yang D, Li C, Chen T, Wang X, Liu C, Zhu H. 2014. ISG12a mediates cell response to Newcastle disease viral infection. Virology 462–463:283–294. [DOI] [PubMed] [Google Scholar]
- 35.Uhrin P, Perkmann T, Binder B, Schabbauer G. 2013. ISG12 is a critical modulator of innate immune responses in murine models of sepsis. Immunobiology 218:1207–1216. doi: 10.1016/j.imbio.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shrestha B, Diamond MS. 2004. Role of CD8+ T cells in control of West Nile virus infection. J Virol 78:8312–8321. doi: 10.1128/JVI.78.15.8312-8321.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keller BC, Fredericksen BL, Samuel MA, Mock RE, Mason PW, Diamond MS, Gale M. 2006. Resistance to alpha/beta interferon is a determinant of West Nile virus replication fitness and virulence. J Virol 80:9424–9434. doi: 10.1128/JVI.00768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diamond MS, Shrestha B, Marri A, Mahan D, Engle M. 2003. B cells and antibody play critical roles in the immediate defense of disseminated infection by West Nile encephalitis virus. J Virol 77:2578–2586. doi: 10.1128/JVI.77.4.2578-2586.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brien JD, Lazear HM, Diamond MS. 2013. Propagation, quantification, detection, and storage of West Nile virus. Curr Protoc Microbiol 31:15D.3.1–15D.3.18. doi: 10.1002/9780471729259.mc15d03s31. [DOI] [PubMed] [Google Scholar]
- 40.Lanciotti RS, Kerst AJ, Nasci RS, Godsey MS, Mitchell CJ, Savage HM, Komar N, Panella NA, Allen BC, Volpe KE, Davis BS, Roehrig JT. 2000. Rapid detection of West Nile virus from human clinical specimens, field-collected mosquitoes, and avian samples by a TaqMan reverse transcriptase-PCR assay. J Clin Microbiol 38:4066–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daffis S, Samuel MA, Keller BC, Gale M Jr, Diamond MS. 2007. Cell-specific IRF-3 responses protect against West Nile virus infection by interferon-dependent and -independent mechanisms. PLoS Pathog 3:e106. doi: 10.1371/journal.ppat.0030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klein RS, Lin E, Zhang B, Luster AD, Tollett J, Samuel MA, Engle M, Diamond MS. 2005. Neuronal CXCL10 directs CD8+ T-cell recruitment and control of West Nile virus encephalitis. J Virol 79:11457–11466. doi: 10.1128/JVI.79.17.11457-11466.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fuchs A, Pinto AK, Schwaeble WJ, Diamond MS. 2011. The lectin pathway of complement activation contributes to protection from West Nile virus infection. Virology 412:101–109. doi: 10.1016/j.virol.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tacke F, Ginhoux F, Jakubzick C, van Rooijen N, Merad M, Randolph GJ. 2006. Immature monocytes acquire antigens from other cells in the bone marrow and present them to T cells after maturing in the periphery. J Exp Med 203:583–597. doi: 10.1084/jem.20052119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. 2010. Development of monocytes, macrophages, and dendritic cells. Science 327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geissmann F, Auffray C, Palframan R, Wirrig C, Ciocca A, Campisi L, Narni-Mancinelli E, Lauvau G. 2008. Blood monocytes: distinct subsets, how they relate to dendritic cells, and their possible roles in the regulation of T-cell responses. Immunol Cell Biol 86:398–408. doi: 10.1038/icb.2008.19. [DOI] [PubMed] [Google Scholar]
- 47.Auffray C, Fogg DK, Narni-Mancinelli E, Senechal B, Trouillet C, Saederup N, Leemput J, Bigot K, Campisi L, Abitbol M, Molina T, Charo I, Hume DA, Cumano A, Lauvau G, Geissmann F. 2009. CX3CR1+ CD115+ CD135+ common macrophage/DC precursors and the role of CX3CR1 in their response to inflammation. J Exp Med 206:595–606. doi: 10.1084/jem.20081385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shrestha B, Pinto AK, Green S, Bosch I, Diamond MS. 2012. CD8+ T cells use TRAIL to restrict West Nile virus pathogenesis by controlling infection in neurons. J Virol 86:8937–8948. doi: 10.1128/JVI.00673-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richner JM, Gmyrek GB, Govero J, Tu Y, van der Windt GJW, Metcalf TU, Haddad EK, Textor J, Miller MJ, Diamond MS. 2015. Age-dependent cell trafficking defects in draining lymph nodes impair adaptive immunity and control of West Nile virus infection. PLoS Pathog 11:e1005027. doi: 10.1371/journal.ppat.1005027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mehlhop E, Diamond MS. 2006. Protective immune responses against West Nile virus are primed by distinct complement activation pathways. J Exp Med 203:1371–1381. doi: 10.1084/jem.20052388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lazear HM, Lancaster A, Wilkins C, Suthar MS, Huang A, Vick SC, Clepper L, Thackray L, Brassil MM, Virgin HW, Nikolich-Zugich J, Moses AV, Gale M Jr, Früh K, Diamond MS. 2013. IRF-3, IRF-5, and IRF-7 coordinately regulate the type I IFN response in myeloid dendritic cells downstream of MAVS signaling. PLoS Pathog 9:e1003118. doi: 10.1371/journal.ppat.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lazear HM, Daniels BP, Pinto AK, Huang AC, Vick SC, Doyle SE, Gale M, Klein RS, Diamond MS. 2015. Interferon-λ restricts West Nile virus neuroinvasion by tightening the blood-brain barrier. Sci Transl Med 7:284ra59. doi: 10.1126/scitranslmed.aaa4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shrestha B, Samuel MA, Diamond MS. 2006. CD8+ T cells require perforin to clear West Nile virus from infected neurons. J Virol 80:119–129. doi: 10.1128/JVI.80.1.119-129.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szretter KJ, Brien JD, Thackray LB, Virgin HW, Cresswell P, Diamond MS. 2011. The interferon-inducible gene viperin restricts West Nile virus pathogenesis. J Virol 85:11557–11566. doi: 10.1128/JVI.05519-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suthar MS, Ma DY, Thomas S, Lund JM, Zhang N, Daffis S, Rudensky AY, Bevan MJ, Clark EA, Kaja M-K, Diamond MS, Gale M Jr. 2010. IPS-1 is essential for the control of West Nile virus infection and immunity. PLoS Pathog 6:e1000757. doi: 10.1371/journal.ppat.1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Daffis S, Suthar MS, Szretter KJ, Gale M Jr, Diamond MS. 2009. Induction of IFN-β and the innate antiviral response in myeloid cells occurs through an IPS-1-dependent signal that does not require IRF-3 and IRF-7. PLoS Pathog 5:e1000607. doi: 10.1371/journal.ppat.1000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Samuel MA, Diamond MS. 2005. Alpha/beta interferon protects against lethal West Nile virus infection by restricting cellular tropism and enhancing neuronal survival. J Virol 79:13350–13361. doi: 10.1128/JVI.79.21.13350-13361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lazear HM, Pinto AK, Vogt MR, Gale M, Diamond MS. 2011. Beta interferon controls West Nile virus infection and pathogenesis in mice. J Virol 85:7186–7194. doi: 10.1128/JVI.00396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suthar MS, Brassil MM, Blahnik G, Gale M. 2012. Infectious clones of novel lineage 1 and lineage 2 West Nile virus strains WNV-TX02 and WNV-Madagascar. J Virol 86:7704–7709. doi: 10.1128/JVI.00401-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shrestha B, Gottlieb DI, Diamond MS. 2003. Infection and injury of neurons by West Nile encephalitis virus. J Virol 77:13203–13213. doi: 10.1128/JVI.77.24.13203-13213.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Samuel MA, Morrey JD, Diamond MS. 2007. Caspase 3-dependent cell death of neurons contributes to the pathogenesis of West Nile virus encephalitis. J Virol 81:2614–2623. doi: 10.1128/JVI.02311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wie S-H, Du P, Luong TQ, Rought SE, Beliakova-Bethell N, Lozach J, Corbeil J, Kornbluth RS, Richman DD, Woelk CH. 2013. HIV downregulates interferon-stimulated genes in primary macrophages. J Interferon Cytokine Res 33:90–95. doi: 10.1089/jir.2012.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pommerenke C, Wilk E, Srivastava B, Schulze A, Novoselova N, Geffers R, Schughart K. 2012. Global transcriptome analysis in influenza-infected mouse lungs reveals the kinetics of innate and adaptive host immune responses. PLoS One 7:e41169. doi: 10.1371/journal.pone.0041169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clarke P, Leser JS, Bowen RA, Tyler KL. 2014. Virus-induced transcriptional changes in the brain include the differential expression of genes associated with interferon, apoptosis, interleukin 17 receptor A, and glutamate signaling as well as flavivirus-specific upregulation of tRNA synthetases. mBio 5:e00902-14. doi: 10.1128/mBio.00902-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qi Y, Li Y, Zhang Y, Zhang L, Wang Z, Zhang X, Gui L, Huang J. 2015. IFI6 inhibits apoptosis via mitochondrial-dependent pathway in dengue virus 2 infected vascular endothelial cells. PLoS One 10:e0132743. doi: 10.1371/journal.pone.0132743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Makovitzki-Avraham E, Daniel-Carmi V, Alteber Z, Farago M, Tzehoval E, Eisenbach L. 2013. The human ISG12a gene is a novel caspase dependent and p53 independent pro-apoptotic gene, that is overexpressed in breast cancer. Cell Biol Int Rep 20:37–46. doi: 10.1002/cbi3.10009. [DOI] [Google Scholar]
- 67.Li S, Xie Y, Zhang W, Gao J, Wang M, Zheng G, Yin X, Xia H, Tao X. 2015. Interferon alpha-inducible protein 27 promotes epithelial-mesenchymal transition and induces ovarian tumorigenicity and stemness. J Surg Res 193:255–264. [DOI] [PubMed] [Google Scholar]
- 68.Hsieh WL, Huang YH, Wang TM, Ming YC, Tsai CN, Pang JHS. 2015. IFI27, a novel epidermal growth factor-stabilized protein, is functionally involved in proliferation and cell cycling of human epidermal keratinocytes. Cell Prolif 48:187–197. doi: 10.1111/cpr.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fensterl V, Wetzel JL, Ramachandran S, Ogino T, Stohlman SA, Bergmann CC, Diamond MS, Virgin HW, Sen GC. 2012. Interferon-induced Ifit2/ISG54 protects mice from lethal VSV neuropathogenesis. PLoS Pathog 8:e1002712. doi: 10.1371/journal.ppat.1002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garcia MR, Ledgerwood L, Yang Y, Xu J, Lal G, Burrell B, Ma G, Hashimoto D, Li Y, Boros P, Grisotto M, van Rooijen N, Matesanz R, Tacke F, Ginhoux F, Ding Y, Chen SH, Randolph G, Merad M, Bromberg JS, Ochando JC. 2010. Monocytic suppressive cells mediate cardiovascular transplantation tolerance in mice. J Clin Invest 120:2486–2496. doi: 10.1172/JCI41628. [DOI] [PMC free article] [PubMed] [Google Scholar]