Abstract
Sacubitril was recently approved by the Food and Drug Administration for use in combination with valsartan for the treatment of patients with heart failure with reduced ejection fraction. As a prodrug, sacubitril must be metabolized (hydrolyzed) to its active metabolite sacubitrilat (LBQ657) to exert its intended therapeutic effects. Thus, understanding the determinants of sacubitril activation will lead to the improvement of sacubitril pharmacotherapy. The objective of this study was to identify the enzyme(s) responsible for the activation of sacubitril, and determine the impact of genetic variation on sacubitril activation. First, an incubation study of sacubitril with human plasma and the S9 fractions of human liver, intestine, and kidney was conducted. Sacubitril was found to be activated by human liver S9 fractions only. Moreover, sacubitril activation was significantly inhibited by the carboxylesterase 1 (CES1) inhibitor bis-(p-nitrophenyl) phosphate in human liver S9. Further incubation studies with recombinant human CES1 and carboxylesterase 2 confirmed that sacubitril is a selective CES1 substrate. The in vitro study of cell lines transfected with wild-type CES1 and the CES1 variant G143E (rs71647871) demonstrated that G143E is a loss-of-function variant for sacubitril activation. Importantly, sacubitril activation was significantly impaired in human livers carrying the G143E variant. In conclusion, sacubitril is selectively activated by CES1 in human liver. The CES1 genetic variant G143E can significantly impair sacubitril activation. Therefore, CES1 genetic variants appear to be an important contributing factor to interindividual variability in sacubitril activation, and have the potential to serve as biomarkers to optimize sacubitril pharmacotherapy.
Entresto, a combination of sacubitril, the first-in-class neprilysin inhibitor, and the angiotensin II receptor blocker valsartan, was approved by the Food and Drug Administration in July 2015 for the treatment of patients with heart failure with reduced ejection fraction (HFrEF). The randomized double-blind clinical trial PARADIGM-HF, which involved 8442 patients with class II–IV HFrEF, demonstrated that Entresto significantly reduced rates of mortality and morbidity compared with the angiotensin-converting enzyme inhibitor enalapril [e.g., first hospitalization for worsening heart failure (12.8 vs. 15.6%), death from cardiovascular causes (13.3 vs. 16.5%), and all-cause mortality (17.0 vs. 19.8%)] (McMurray et al., 2014). Due to its established efficacy, Entresto is expected to be widely used to treat patients with HFrEF, with annual sales estimated to reach as high as $8 billion by 2020. However, therapeutic outcomes varied significantly in patients treated with valsartan/sacubitril based upon previously published clinical studies (McMurray et al., 2014; Packer et al., 2015).
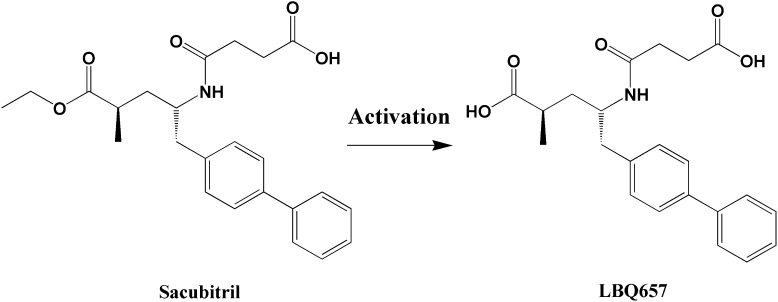
Sacubitril is an inactive ester prodrug that needs to be converted in vivo to its active metabolite sacubitrilat (LBQ657) (Fig. 1), a potent neprilysin inhibitor, to produce its intended pharmacological effects (Ksander et al., 1995). Thus, it is plausible to speculate that patients who cannot efficiently activate sacubitril may respond poorly to the treatment. To date, the enzyme(s) and organ(s) responsible for sacubitril activation have remained unknown. Significant interindividual variability in plasma concentrations of both sacubitril and its active metabolite LBQ657 was observed in clinical pharmacokinetic studies (Gu et al., 2010; Gan et al., 2016). However, the contributing factors to the variation in sacubitril activation and therapeutic outcomes have not been explored. The study of sacubitril-activating enzyme(s) could lead to a better understanding of the interindividual variability in sacubitril pharmacokinetics, and has the potential to identify patients who will or will not benefit from sacubitril therapy.
Fig. 1.
A scheme of the metabolism of sacubitril to its active metabolite LBQ657.
In the present study, we used several complementary in vitro experimental approaches to identify the enzyme(s) responsible for the activation of sacubitril and to determine the impact of genetic variation on sacubitril activation.
Materials and Methods
Materials.
Sacubitril was purchased from MedKoo Biosciences (Chapel Hill, NC). The hydrolytic active metabolite of sacubitril, LBQ657, was obtained in our laboratory following incubation of 100 μM sacubitril with 50 ng/μl recombinant human carboxylesterase 1 (CES1; R&D Systems Inc., Minneapolis, MN) at 37°C for 2 hours. Sacubitril was completely hydrolyzed to LBQ657 after incubation for 2 hours as determined by liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis. Ritalinic acid, enalapril, fluorescein diacetate, fluorescein, bis-(p-nitrophenyl) phosphate (BNPP), LC-MS–grade methanol, acetonitrile, and formic acid were all purchased from Sigma-Aldrich (St. Louis, MO). The enalapril hydrolytic metabolite enalaprilat dehydrate was purchased from Selleckchem (Houston, TX). Taq DNA polymerase with standard Taq buffer and deoxynucleotide (dNTP) solution mix were obtained from New England Biolabs (Ipswich, MA). Human intestine and kidney S9 fractions were purchased from XenoTech (Lenexa, KS). Recombinant human CES1 and carboxylesterase 2 (CES2) were obtained from R&D Systems Inc. All other chemicals and reagents were of analytical grade and commercially available.
A total of 53 individual normal human liver samples were obtained from XenoTech LLC, the Cooperative Human Tissue Network (Columbus, OH), and the Liver Tissue Cell Distribution System at the University of Minnesota (Minneapolis, MN). The demographic information of two samples was unknown. The other liver samples consisted of 25 males and 26 females with ages ranging from 1 to 81 years (57.6 ± 15.3 years). The donors included 49 Caucasians and two African-Americans.
Preparation of S9 Fractions from Individual Human Livers and CES1-Transfected Cells.
Individual human liver S9 fractions (HLS9) were prepared according to previous studies (Wang et al., 2015). Protein concentrations were determined using a Pierce BCA assay kit (Pierce, Rockford, IL). The Flp-In 293 cell lines stably expressing wild-type (WT) CES1 and the G143E variant (rs71647871) were developed in a study previously published by our laboratory (Zhu et al., 2008). Cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. After reaching approximately 95% confluence, cells were washed and harvested in phosphate-buffered saline (PBS) buffer. Afterward, cells were sonicated and then centrifuged at 9000g for 30 minutes at 4°C. The supernatant (S9 fractions) was collected and stored at –80°C until use.
Enzymatic Assays.
An in vitro incubation study was conducted to determine the activation of sacubitril in human plasma, liver, intestine, and kidney S9 fractions, as well as in recombinant CES1 and CES2 enzymes. The incubations were carried out in 1.5-ml Eppendorf tubes (Eppendorf, Hauppauge, NY) with a final volume of 40 μl. Sacubitril, tissue S9 fractions, plasma, and recombinant CES1 and CES2 were all prepared in PBS buffer. Hydrolytic reactions were initiated by mixing 20 μl of sacubitril solution with equal volumes of the S9 fractions, plasma, recombinant CES1, or recombinant CES2. The final concentrations of sacubitril, S9 fractions, and recombinant CES1 and CES2 were 200 µM, 50 µg/ml, 5 ng/µl, and 5 ng/µl, respectively. After incubation at 37°C for 10 minutes, the reactions were terminated by the addition of 120 μl of acetonitrile containing the internal standard (IS) ritalinic acid (33 μM). The mixture was vortexed for 5 minutes and centrifuged at 17,000g for 10 minutes to remove precipitated proteins. The supernatant was collected and quantified for LBQ657 concentrations by an LC-MS/MS assay, described later. The hydrolysis rate of sacubitril was calculated according to the formation of LBQ657. Incubations of sacubitril with PBS were carried out under the same experimental conditions as a negative control to determine the nonenzymatic hydrolysis of sacubitril.
For the kinetic study, recombinant CES1 was prepared in PBS buffer at a concentration of 5 ng/μl. The hydrolysis reaction was initiated by mixing 20 μl of recombinant CES1 and 20 μl of sacubitril with concentrations ranging from 31.25 to 2000 μM. The formation of LBQ657 was determined after incubation at 37°C for 10 minutes.
An in vitro study was conducted to evaluate the effect of CES1 inhibition on sacubitril activation. The formation of LBQ657 was determined after incubation of sacubitril (200 μM) with pooled HLS9 (0.05 mg/ml) at 37°C for 10 minutes in the presence of various concentrations of the CES1 inhibitor BNPP (0–100 μM).
The selective CES1 substrate enalapril and the selective CES2 substrate fluorescein diacetate were included in the in vitro incubation study as positive controls to measure CES1 and CES2 activity, respectively. The formation of enalaprilat, the hydrolytic metabolite of enalapril, was determined by an LC-MS/MS assay described previously (Wang et al., 2015). Fluorescein, the hydrolytic metabolite of fluorescein diacetate, was detected using a Synergy 2 Multi-Mode Microplate Reader (BioTek Inc., Winooski, VT) with an excitation wavelength of 485 nm and an emission wavelength of 525 nm, as reported by Wang et al. (2011). All incubation experiments were performed in triplicate, except for the study of individual HLS9 samples, for which the experiment was duplicated for each sample.
LC-MS/MS Analysis of Sacubitril and LBQ657.
The LC-MS/MS analysis was performed on a Shimadzu HPLC system (Shimadzu, Tokyo, Japan) coupled with an API 4000 triple quadrupole/linear ion trap (QTRAP) mass spectrometer (Applied Biosystems, Foster City, CA). Analytes were separated on a Shimadzu VP-ODS column (5 μm, 150 × 2.0 mm). The quantification of sacubitril and its metabolite LBQ657 was performed based on a previously reported method with slight modifications (Gu et al., 2010). The mobile phase consisted of water containing 0.1% formic acid (v/v; phase A) and acetonitrile containing 0.1% formic acid (v/v; phase B), and was delivered at a flow rate of 0.2 ml/min. A gradient elution was applied for the separation with the time program set as follows: phase B was increased from 35 to 90% during the time period of 0–7 minutes, maintained at 90% for 1 minute, then returned to 35% at 9 minutes, and maintained until the end of the run (12 minutes). MS was operated in positive ion mode using turbo electrospray ionization. The following transitions were monitored on a multiple reaction monitoring mode: sacubitril, m/z 412.7 > 266.7; LBQ657, m/z 384.7 > 266.7; IS, m/z 220.5 > 84.6.
Quantifications were performed based on the peak area ratios of analyte to IS. The regression coefficients of the calibration curves were greater than 0.99 for both sacubitril and LBQ657. Three quality controls representing low, medium, and high concentrations (100, 1000, and 5000 nM) of each analyte were included in every batch of analyzed samples. Interday and intraday relative standard deviations were less than 10% for both sacubitril and LBQ657.
CES1 Genotyping Study.
Genomic DNA was extracted from human liver samples using the PureLink Genomic DNA Mini Kit (Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. The extracted DNA was further purified using the PureLink Genomic DNA Mini Kit. The DNA concentrations were determined by the Qubit dsDNA High Sensitivity Assay (Life Technologies) using a Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA). The polymerase chain reaction (PCR) primers for the amplification of CES1 exon 4, where the CES1 variant G143E resides, were as follows: GACATCTTCTGAGTGCTCCCTG (forward); TGTGTCAACCTTCACCTGCTG (reverse). The PCR conditions involved initial denaturation at 95°C for 30 seconds; 30 cycles of 95°C for 30 seconds, 63°C for 45 seconds, and 68°C for 50 seconds; and a final extension for 5 minutes at 68°C. The PCR products were purified with the PureLink PCR Purification Kit (Life Technologies) and analyzed with agarose (2%) gel electrophoresis before being subjected to Sanger sequencing to determine the G143E genotypes.
Data Analysis.
Data are presented as the mean ± S.D). The Michaelis-Menten constant (Km) and maximal reaction rate (Vmax) of CES1-mediated sacubitril activation were calculated using the Michaelis-Menten equation [v = (Vmax × [S])/(Km + [S])] (v: enzymatic reaction velocity, [S]: substrate concentration). Apparent intrinsic clearance (CLint) was determined by Vmax/Km. The Mann-Whitney test was used to analyze the differences of CES1 expression and activity between the wild-type and the G143E genotype groups (GraphPad Prism software version 6.0; GraphPad Software, San Diego, CA). A P value less than 0.05 was considered statistically significant. The IC50 value in the inhibition study was calculated by nonlinear regression using the three-parameter Hill equation.
Results
Sacubitril Is a Selective CES1 Substrate, and Is Activated in the Liver.
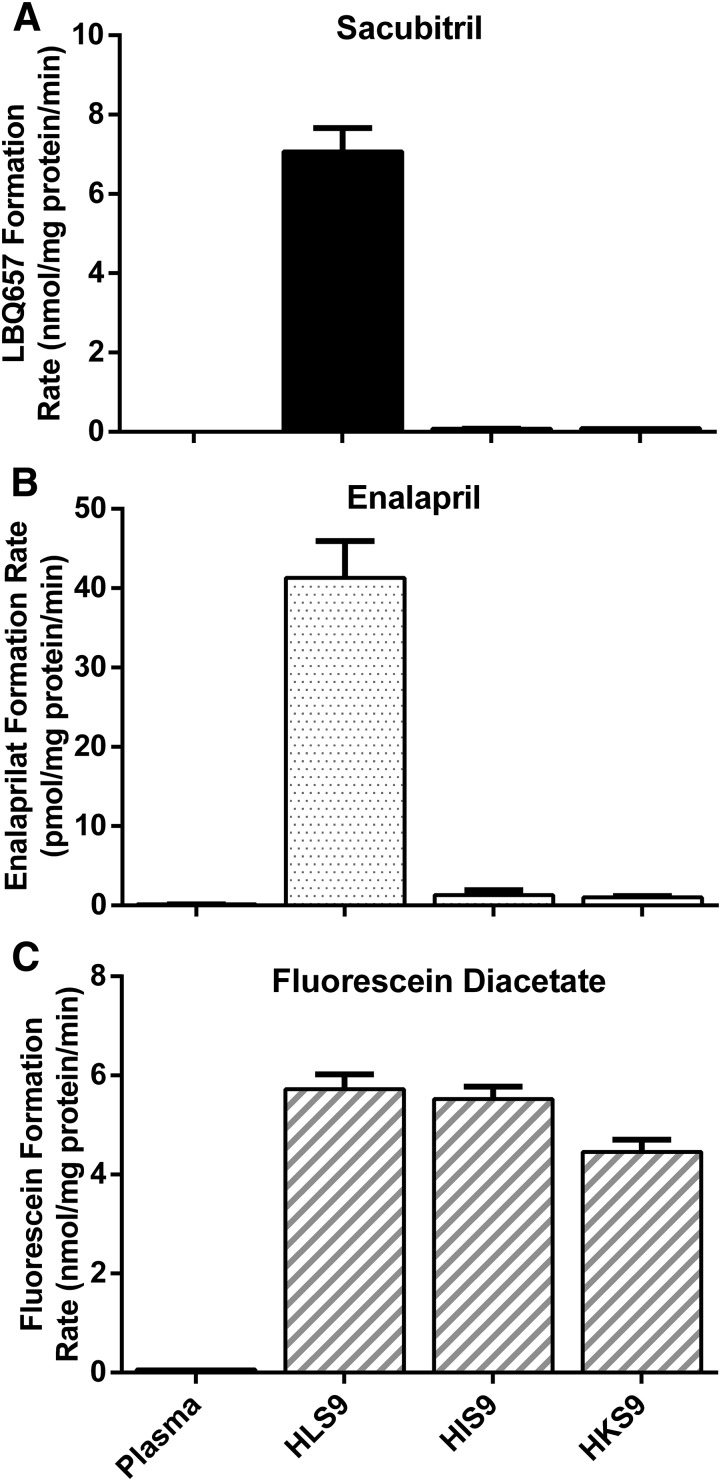
Sacubitril hydrolysis was determined after incubations with HLS9, human intestine S9 fractions (HIS9), and human kidney S9 fractions (HKS9), as well as with human plasma. As shown in Fig. 2, sacubitril was rapidly hydrolyzed to its active metabolite LBQ657 in HLS9 samples, whereas no appreciable hydrolysis was observed after incubation with human plasma, HIS9, or HKS9 samples.
Fig. 2.
Hydrolysis of sacubitril (A), enalapril (B), and fluorescein diacetate (C) by pooled human plasma, and the S9 fractions prepared from human liver (HLS9), intestine (HIS9), and kidney (HKS9). The hydrolytic products were determined after the substrates were incubated with the enzymes at 37°C for 10 minutes. The final protein concentrations of plasma, HLS9, HIS9, and HKS9 were 23.4, 0.05, 0.125, and 0.125 mg/ml, respectively. The concentrations of sacubitril, fluorescein diacetate, and enalapril were 200, 100, and 100 μM, respectively. Data are expressed as the formation rate of hydrolytic products (mean ± S.D., n = 3).
To evaluate hydrolytic activity of the plasma, HLS9, HIS9, and HKS9 samples, enalapril, the selective CES1 substrate, and fluorescein diacetate, the selective CES2 substrate, were included in the study as positive controls under the same experimental conditions as sacubitril (Fig. 2) (Wang et al., 2011, 2015). As expected, enalapril was efficiently hydrolyzed to its metabolite enalaprilat in HLS9 samples, but not in plasma, HIS9, or HKS9 samples. Fluorescein diacetate was readily hydrolyzed to its metabolite fluorescein in HLS9, HIS9, and HKS9 samples, where CES2 is present, but not in human plasma.
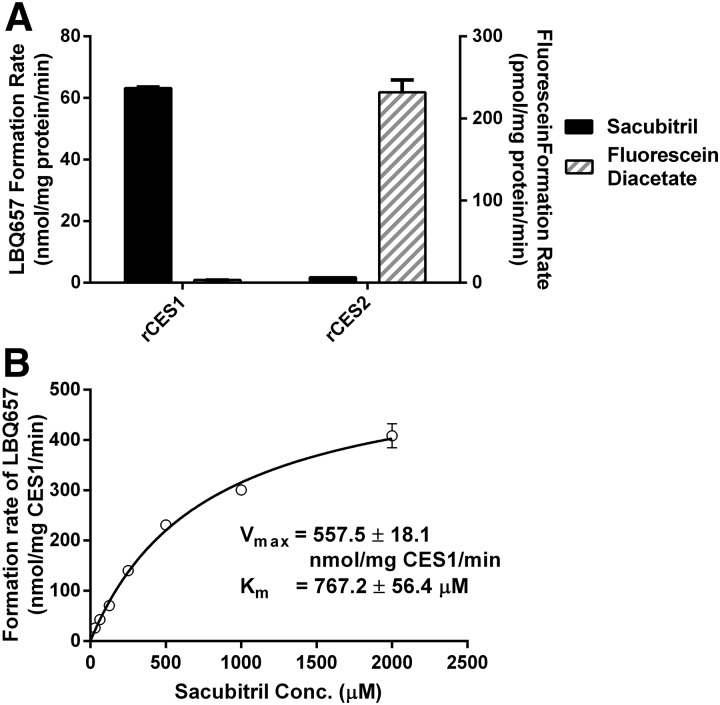
To further define the role of CESs in the hydrolysis of sacubitril, sacubitril and fluorescein diacetate were incubated with purified recombinant human CES1 and CES2. Sacubitril could only be hydrolyzed by CES1 (Fig. 3A). In contrast, fluorescein diacetate could only be metabolized by CES2.
Fig. 3.
Hydrolysis of sacubitril and fluorescein diacetate by recombinant human CES1 (rCES1) and recombinant human CES2 (rCES2) (A), and enzymatic kinetics of rCES1–catalyzed sacubitril hydrolysis (B). The hydrolytic reactions were carried out at 37°C for 10 minutes. The final concentration of recombinant CES1 and CES2 was 5 ng/μl. The concentrations of sacubitril and fluorescein diacetate were 200 and 100 μM, respectively (A), whereas the concentrations of sacubitril ranged from 31.25 to 2000 μM in the kinetic study (B). Data are presented as means ± S.D. (n = 3). Conc., concentration.
In addition, a kinetic study was carried out for sacubitril hydrolysis with recombinant human CES1. The data fit well according to Michaelis-Menten kinetics (Fig. 3B). Under our experimental conditions, the Vmax, Km, and CLint values were determined to be 557.5 ± 18.1 nmol/mg CES1/min, 767.2 ± 56.4 μM, and 726.9 ± 56.7 μl/mg CES1/min, respectively.
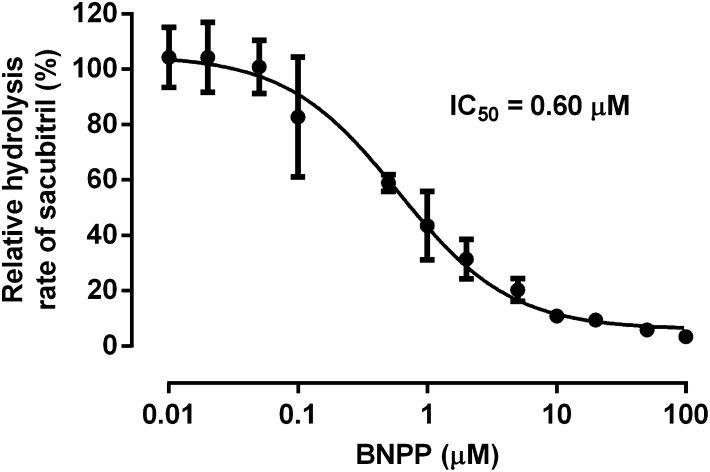
The in vitro inhibition study demonstrated that the CES1 inhibitor BNPP can inhibit sacubitril hydrolysis in HLS9 in a concentration-dependent manner (Fig. 4). Sacubitril activation was inhibited by approximately 90% when the concentration of BNPP reached 10 μM. The IC50 was determined to be 0.6 μM under the present experimental conditions.
Fig. 4.
Inhibition of sacubitril hydrolysis by the CES1 inhibitor BNPP in pooled HLS9. The formation of LBQ657 was determined after incubation of sacubitril (200 μM) with pooled HLS9 (0.05 mg/ml) at 37°C for 10 minutes in the presence of various concentrations of the CES1 inhibitor BNPP (0–100 μM). Hydrolytic activity was expressed as the percentage of the activity determined when BNPP is absent. Data are presented as means ± S.D. (n = 3).
The CES1 Polymorphism G143E Is a Loss-of-Function Variant for the Hydrolysis of Sacubitril, and Sacubitril Activation Is Impaired in Human Livers Carrying the 143E Allele.
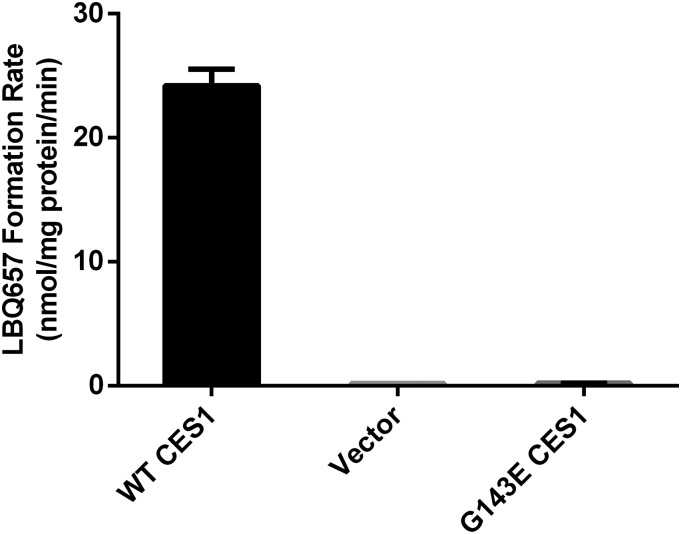
Sacubitril was incubated with the cell S9 fractions prepared from the Flp-In 293 cells stably expressing WT CES1 and the variant G143E. Similar to the HLS9 samples, the S9 fractions of WT CES1 cells efficiently hydrolyzed sacubitril to its active metabolite LBQ657. However, no appreciable hydrolytic metabolite LBQ657 was observed after incubation with the S9 samples from cells expressing the 143E allele (Fig. 5).
Fig. 5.
Activation of sacubitril in cell S9 fractions prepared from cells stably transfected with WT CES1 and the variant G143E. The formation of the active metabolite LBQ657 was determined after incubation of sacubitril with the S9 fractions at 37°C for 10 minutes. Final concentrations of sacubitril and cell S9 fractions were 200 μM and 0.05 mg/ml, respectively. Data are presented as means from three independent experiments with error bars representing S.D.
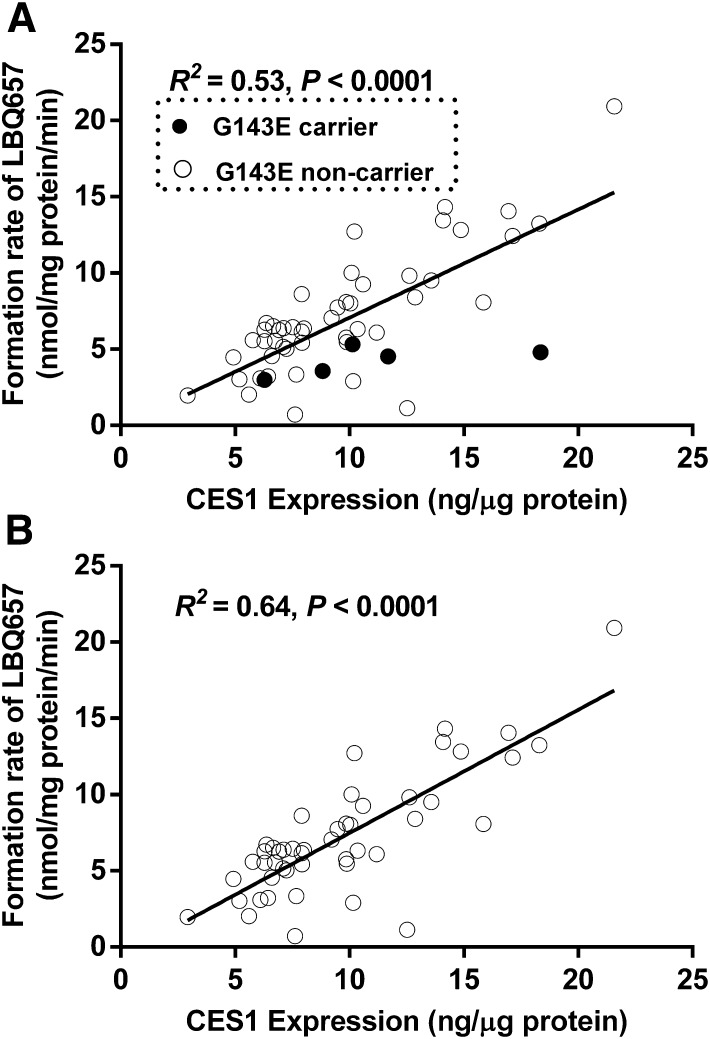
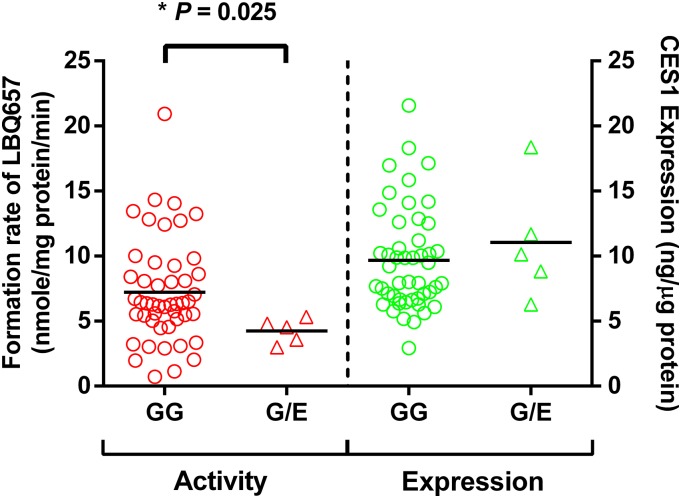
A total of 104 individual human liver samples were genotyped for the G143E variant. Five individuals were found to be the G143E heterozygotes, whereas the others were noncarriers (Wang et al., 2015). Sacubitril activation was studied in the five carriers and 48 randomly selected noncarriers. As shown in Fig. 6, both CES1 activity and expression varied markedly in the individual human liver samples, with 29- and 7-fold differences in sacubitril hydrolysis rate and CES1 protein expression, respectively. The CES1 activity on sacubitril activation was significantly correlated with CES1 expression in the tested individual human livers. The R2 values were 0.53 and 0.64 for the total 53 samples and the 48 non-G143E carriers, respectively. The activation rates of sacubitril were significantly lower in the carriers relative to that in the noncarriers (4.2 ± 1.0 vs. 7.2 ± 4.0 nmol/mg protein/min, P = 0.025; Fig. 7). CES1 expression in the carriers and noncarriers was determined in a previously published study (Wang et al., 2015), and no significant differences were found between the two groups (P = 0.47; Fig. 7).
Fig. 6.
Correlation analysis of the hydrolysis rate of sacubitril with CES1 expression in 53 HLS9 samples. The final concentrations of HLS9 and sacubitril in the reaction system were 0.05 mg/ml and 200 μM. The formation of LBQ657 was determined after incubation with individual HLS9 at 37°C for 10 minutes. Data are the means from two independent experiments. (A) All samples (n = 53). (B) G143E noncarriers only (n = 48).
Fig. 7.
Effect of the CES1 variant G143E on sacubitril activation (left) and CES1 protein expression (right) in human liver samples. The final concentrations of HLS9 and sacubitril in the reaction system were 0.05 mg/ml and 200 μM, respectively. LBQ657 formed from sacubitril hydrolysis was determined after incubation of the substrate with individual HLS9 at 37°C for 10 minutes. Data are mean values of two independent experiments. Horizontal bars represent mean values in each group. The Mann-Whitney test was used to test the differences of CES1 expression and activity between different G143E genotypes. A P value less than 0.05 was considered statistically significant.
Discussion
Although hydrolases are expressed in many human tissues, the liver, intestines, kidneys, and plasma are responsible for the majority of hydrolytic activities. Besides playing an important role in physiologic processes, such as lipid metabolism, these enzymes are critical for the metabolism of many therapeutic agents. Human hydrolases are distinct in substrate selectivity and specific expression patterns. For example, both CES1 and CES2 are expressed in human liver, but CES1 is the primary hepatic hydrolase, contributing to approximately 80–95% of hydrolytic activity in the liver (Imai, 2006). On the other hand, the intestines and kidneys have no CES1 expression, but instead express high levels of CES2 (Wang et al., 2015). In human plasma, the major hydrolases include paraoxonase and butyrylcholinesterase; neither CES1 nor CES2 is expressed (Bahar et al., 2012). Our in vitro incubation studies revealed that sacubitril is hydrolyzed by the liver, but not by the intestines, kidneys, or plasma, indicating that sacubitril is selectively activated by hepatic CES1. This conclusion is further supported by the observations that sacubitril was efficiently activated by purified recombinant CES1, but not CES2.
With regard to substrate specificity, in general, CES1 prefers substrates with a small alcohol group and large acyl group, such as oseltamivir and methylphenidate, whereas CES2 recognizes substrates with a large alcohol group and small acyl group, such as prasugrel and fluorescein diacetate (Sun et al., 2004; Shi et al., 2006; Hosokawa, 2008; Williams et al., 2008; Wang et al., 2011; Laizure et al., 2013). Sacubitril is an ester prodrug with a small alcohol group and a large acyl group (Fig. 1). Thus, the finding that sacubitril is metabolized by CES1 instead of CES2 is consistent with the substrate preferences of the two carboxylesterases.
The CLint value for CES1-mediated sacubitril hydrolysis was determined to be 726.9 μl/mg CES1/min, which is higher than that of the established CES1 substrates d-methylphenidate (35.6 μl/mg CES1/min) and dabigatran etexilate (27.2 μl/mg CES1/min), suggesting sacubitril is a highly efficient CES1 substrate (Sun et al., 2004; Laizure et al., 2014).
Significant interindividual variability in CES1 expression and activity has been consistently demonstrated by our research group and many others (Hosokawa et al., 1995; Shi et al., 2006; Yoshimura et al., 2008; Hagihara et al., 2009; Yang et al., 2009; Zhu et al., 2009; Ross et al., 2012). CES1 variants are considered an important contributing factor to CES1 variability (Zhu et al., 2008; Laizure et al., 2013; Rasmussen et al., 2015). We have found that hepatic CES1 expressions were significantly correlated with the activity on sacubitril metabolism, and the R2 was 0.53, indicating that approximately half of the variability in sacubitril hydrolysis could be explained by CES1 expression variation. In addition, we further analyzed the correlation after excluding samples carrying the G143E variant and observed an increase of correlation, with an R2 value of 0.64. Thus, the G143E variant appears to contribute to approximately 10% variation of sacubitril activation in our samples.
The CES1 genetic variant G143E is a loss-of-function variant that was originally discovered in a poor metabolizer of methylphenidate, a CES1 selective substrate, in a clinical pharmacokinetic study (Zhu et al., 2008). The minor allele frequency of G143E ranges from 2 to 4% among different populations except in Asians, in whom the variant is very rare (Zhu et al., 2008). Our incubation studies with transfected cell lines revealed that G143E is a loss-of-function variant for the activation of sacubitril. Consistent with the results from the transfected CES1 cells, sacubitril activation was significantly impaired in human livers with the 143G/E genotype relative to those with the 143G/G genotype. Several clinical studies have consistently demonstrated that this variant was associated with significantly altered pharmacokinetics and/or pharmacodynamics of several CES1 substrate drugs, including methylphenidate, clopidogrel, oseltamivir, and enalapril (Patrick et al., 2007; Zhu et al., 2008; Nemoda et al., 2009; Tarkiainen et al., 2012, 2015a,b; Lewis et al., 2013). For example, attention deficit hyperactivity disorder patients with the G143E genotype required significantly lower doses of methylphenidate for symptom reduction (0.41 ± 0.13 vs. 0.57 ± 0.15 mg/kg, P = 0.022) (Nemoda et al., 2009). In another study, the areas under the plasma concentration-time curves from 0 h to infinity of clopidogrel and its active metabolite were found to be 123% (P = 0.004) and 67% (P = 0.009) larger in the subjects carrying the G143E variant relative to the noncarriers. Consistent with the pharmacokinetic data, the carriers exhibited significantly greater inhibition of P2Y12-mediated platelet aggregation (Tarkiainen et al., 2015a).
It should be noted that marked interindividual variability in sacubitril activation exists even in human livers without the G143E variant (Fig. 7), indicating that other unknown genetic and/or nongenetic factors may affect sacubitril metabolism by CES1. The CES1 gene is highly polymorphic, with over 2000 genetic variants registered in the National Center for Biotechnology Information dbSNP database. However, very few of these variants have been studied for their potential effect on CES1 expression and/or activity. In addition to genetic variation, nongenetic factors, such as CES1 inhibitors, may affect CES1 function as well (Laizure et al., 2013). For example, sacubitril activation can be significantly inhibited by the model CES1 inhibitor BNPP (Fig. 4). Therefore, a wide range of genetic and nongenetic factors could affect CES1 activity and consequently alter the pharmacokinetics and the therapeutic outcomes of sacubitril.
In summary, our study has provided compelling evidence supporting the hypothesis that sacubitril is selectively activated by hepatic CES1 in humans, and that genetic variants of the CES1 enzyme can significantly affect sacubitril activation. Thus, patients who carry certain CES1 variants, such as G143E, may not have an adequate response to sacubitril due to the impairment of sacubitril activation. These patient groups may be unnecessarily exposed to a novel drug which may not successfully deliver the proposed therapeutic advantages of sacubitril therapy. Moreover, they may also find themselves with a considerable financial burden, as Entresto costs approximately $4000–$5,000/year, whereas enalapril costs approximately $50–$100/year. To improve its therapeutic outcomes, further basic and clinical research on CES1 pharmacogenetics is necessary to establish CES1 variants as valid biomarkers to optimize sacubitril pharmacotherapy.
Abbreviations
- BNPP
bis-(p-nitrophenyl) phosphate
- CES1
carboxylesterase 1
- CES2
carboxylesterase 2
- CLint
intrinsic clearance
- HFrEF
heart failure with reduced ejection fraction
- HIS9
human intestine S9 fractions
- HKS9
human kidney S9 fractions
- HLS9
human liver S9 fractions
- IS
internal standard
- Km
The Michaelis-Menten constant
- LBQ657
sacubitrilat
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- RCES
recombinant human CES
- Vmax
maximal reaction rate
- WT
wild type
Authorship Contributions
Participated in research design: Shi, Wang, Bleske, Zhu.
Conducted experiments: Shi, Wang, Nguyen.
Performed data analysis: Shi, Wang, Zhu.
Wrote or contributed to the writing of the manuscript: Shi, Wang, Nguyen, Wu, Bleske, Zhu.
Footnotes
Research reported in this publication was supported in part by the National Institutes of Health National Center for Advancing Translational Sciences [Grant 2UL1TR000433], National Institute on Aging [Grant R21AG048500], and American Association of Colleges of Pharmacy 2015 New Investigator Award.
References
- Bahar FG, Ohura K, Ogihara T, Imai T. (2012) Species difference of esterase expression and hydrolase activity in plasma. J Pharm Sci 101:3979–3988. [DOI] [PubMed] [Google Scholar]
- Gan L, Langenickel T, Petruck J, Kode K, Rajman I, Chandra P, Zhou W, Rebello S, Sunkara G. (2016) Effects of age and sex on the pharmacokinetics of LCZ696, an angiotensin receptor neprilysin inhibitor. J Clin Pharmacol 56:78–86. [DOI] [PubMed] [Google Scholar]
- Gu J, Noe A, Chandra P, Al-Fayoumi S, Ligueros-Saylan M, Sarangapani R, Maahs S, Ksander G, Rigel DF, Jeng AY, et al. (2010) Pharmacokinetics and pharmacodynamics of LCZ696, a novel dual-acting angiotensin receptor-neprilysin inhibitor (ARNi). J Clin Pharmacol 50:401–414. [DOI] [PubMed] [Google Scholar]
- Hagihara K, Kazui M, Kurihara A, Yoshiike M, Honda K, Okazaki O, Farid NA, Ikeda T. (2009) A possible mechanism for the differences in efficiency and variability of active metabolite formation from thienopyridine antiplatelet agents, prasugrel and clopidogrel. Drug Metab Dispos 37:2145–2152. [DOI] [PubMed] [Google Scholar]
- Hosokawa M. (2008) Structure and catalytic properties of carboxylesterase isozymes involved in metabolic activation of prodrugs. Molecules 13:412–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa M, Endo T, Fujisawa M, Hara S, Iwata N, Sato Y, Satoh T. (1995) Interindividual variation in carboxylesterase levels in human liver microsomes. Drug Metab Dispos 23:1022–1027. [PubMed] [Google Scholar]
- Imai T. (2006) Human carboxylesterase isozymes: catalytic properties and rational drug design. Drug Metab Pharmacokinet 21:173–185. [DOI] [PubMed] [Google Scholar]
- Ksander GM, Ghai RD, deJesus R, Diefenbacher CG, Yuan A, Berry C, Sakane Y, Trapani A. (1995) Dicarboxylic acid dipeptide neutral endopeptidase inhibitors. J Med Chem 38:1689–1700. [DOI] [PubMed] [Google Scholar]
- Laizure SC, Herring V, Hu Z, Witbrodt K, Parker RB. (2013) The role of human carboxylesterases in drug metabolism: have we overlooked their importance? Pharmacotherapy 33:210–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laizure SC, Parker RB, Herring VL, Hu Z-Y. (2014) Identification of carboxylesterase-dependent dabigatran etexilate hydrolysis. Drug Metab Dispos 42:201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JP, Horenstein RB, Ryan K, O’Connell JR, Gibson Q, Mitchell BD, Tanner K, Chai S, Bliden KP, Tantry US, et al. (2013) The functional G143E variant of carboxylesterase 1 is associated with increased clopidogrel active metabolite levels and greater clopidogrel response. Pharmacogenet Genomics 23:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray JJ, Packer M, Desai AS, Gong J, Lefkowitz MP, Rizkala AR, Rouleau JL, Shi VC, Solomon SD, Swedberg K, et al. PARADIGM-HF Investigators and Committees (2014) Angiotensin-neprilysin inhibition versus enalapril in heart failure. N Engl J Med 371:993–1004. [DOI] [PubMed] [Google Scholar]
- Nemoda Z, Angyal N, Tarnok Z, Gadoros J, Sasvari-Szekely M. (2009) Carboxylesterase 1 gene polymorphism and methylphenidate response in ADHD. Neuropharmacology 57:731–733. [DOI] [PubMed] [Google Scholar]
- Packer M, McMurray JJ, Desai AS, Gong J, Lefkowitz MP, Rizkala AR, Rouleau JL, Shi VC, Solomon SD, Swedberg K, et al. PARADIGM-HF Investigators and Coordinators (2015) Angiotensin receptor neprilysin inhibition compared with enalapril on the risk of clinical progression in surviving patients with heart failure. Circulation 131:54–61. [DOI] [PubMed] [Google Scholar]
- Patrick KS, Straughn AB, Minhinnett RR, Yeatts SD, Herrin AE, DeVane CL, Malcolm R, Janis GC, Markowitz JS. (2007) Influence of ethanol and gender on methylphenidate pharmacokinetics and pharmacodynamics. Clin Pharmacol Ther 81:346–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen HB, Bjerre D, Linnet K, Jürgens G, Dalhoff K, Stefansson H, Hankemeier T, Kaddurah-Daouk R, Taboureau O, Brunak S, et al. INDICES Consortium (2015) Individualization of treatments with drugs metabolized by CES1: combining genetics and metabolomics. Pharmacogenomics 16:649–665. [DOI] [PubMed] [Google Scholar]
- Ross MK, Borazjani A, Wang R, Crow JA, Xie S. (2012) Examination of the carboxylesterase phenotype in human liver. Arch Biochem Biophys 522:44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D, Yang J, Yang D, LeCluyse EL, Black C, You L, Akhlaghi F, Yan B. (2006) Anti-influenza prodrug oseltamivir is activated by carboxylesterase human carboxylesterase 1, and the activation is inhibited by antiplatelet agent clopidogrel. J Pharmacol Exp Ther 319:1477–1484. [DOI] [PubMed] [Google Scholar]
- Sun Z, Murry DJ, Sanghani SP, Davis WI, Kedishvili NY, Zou Q, Hurley TD, Bosron WF. (2004) Methylphenidate is stereoselectively hydrolyzed by human carboxylesterase CES1A1. J Pharmacol Exp Ther 310:469–476. [DOI] [PubMed] [Google Scholar]
- Tarkiainen EK, Backman JT, Neuvonen M, Neuvonen PJ, Schwab M, Niemi M. (2012) Carboxylesterase 1 polymorphism impairs oseltamivir bioactivation in humans. Clin Pharmacol Ther 92:68–71. [DOI] [PubMed] [Google Scholar]
- Tarkiainen EK, Holmberg MT, Tornio A, Neuvonen M, Neuvonen PJ, Backman JT, Niemi M. (2015a) Carboxylesterase 1 c.428G>A single nucleotide variation increases the antiplatelet effects of clopidogrel by reducing its hydrolysis in humans. Clin Pharmacol Ther 97:650–658. [DOI] [PubMed] [Google Scholar]
- Tarkiainen EK, Tornio A, Holmberg MT, Launiainen T, Neuvonen PJ, Backman JT, Niemi M. (2015b) Effect of carboxylesterase 1 c.428G > A single nucleotide variation on the pharmacokinetics of quinapril and enalapril. Br J Clin Pharmacol 80:1131–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Williams ET, Bourgea J, Wong YN, Patten CJ. (2011) Characterization of recombinant human carboxylesterases: fluorescein diacetate as a probe substrate for human carboxylesterase 2. Drug Metab Dispos 39:1329–1333. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang G, Shi J, Aa J, Comas R, Liang Y, Zhu HJ. (2015) CES1 genetic variation affects the activation of angiotensin-converting enzyme inhibitors. Pharmacogenomics J DOI: [published ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams ET, Jones KO, Ponsler GD, Lowery SM, Perkins EJ, Wrighton SA, Ruterbories KJ, Kazui M, Farid NA. (2008) The biotransformation of prasugrel, a new thienopyridine prodrug, by the human carboxylesterases 1 and 2. Drug Metab Dispos 36:1227–1232. [DOI] [PubMed] [Google Scholar]
- Yang D, Pearce RE, Wang X, Gaedigk R, Wan YJ, Yan B. (2009) Human carboxylesterases HCE1 and HCE2: ontogenic expression, inter-individual variability and differential hydrolysis of oseltamivir, aspirin, deltamethrin and permethrin. Biochem Pharmacol 77:238–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura M, Kimura T, Ishii M, Ishii K, Matsuura T, Geshi E, Hosokawa M, Muramatsu M. (2008) Functional polymorphisms in carboxylesterase1A2 (CES1A2) gene involves specific protein 1 (Sp1) binding sites. Biochem Biophys Res Commun 369:939–942. [DOI] [PubMed] [Google Scholar]
- Zhu HJ, Appel DI, Jiang Y, Markowitz JS. (2009) Age- and sex-related expression and activity of carboxylesterase 1 and 2 in mouse and human liver. Drug Metab Dispos 37:1819–1825. [DOI] [PubMed] [Google Scholar]
- Zhu HJ, Patrick KS, Yuan HJ, Wang JS, Donovan JL, DeVane CL, Malcolm R, Johnson JA, Youngblood GL, Sweet DH, et al. (2008) Two CES1 gene mutations lead to dysfunctional carboxylesterase 1 activity in man: clinical significance and molecular basis. Am J Hum Genet 82:1241–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]