Abstract
This symposium summary, sponsored by the ASPET, was held at Experimental Biology 2015 on March 29, 2015, in Boston, Massachusetts. The symposium focused on: 1) the interactions of cytochrome P450s (P450s) with their redox partners; and 2) the role of the lipid membrane in their orientation and stabilization. Two presentations discussed the interactions of P450s with NADPH-P450 reductase (CPR) and cytochrome b5. First, solution nuclear magnetic resonance was used to compare the protein interactions that facilitated either the hydroxylase or lyase activities of CYP17A1. The lyase interaction was stimulated by the presence of b5 and 17α-hydroxypregnenolone, whereas the hydroxylase reaction was predominant in the absence of b5. The role of b5 was also shown in vivo by selective hepatic knockout of b5 from mice expressing CYP3A4 and CYP2D6; the lack of b5 caused a decrease in the clearance of several substrates. The role of the membrane on P450 orientation was examined using computational methods, showing that the proximal region of the P450 molecule faced the aqueous phase. The distal region, containing the substrate-access channel, was associated with the membrane. The interaction of NADPH-P450 reductase (CPR) with the membrane was also described, showing the ability of CPR to “helicopter” above the membrane. Finally, the endoplasmic reticulum (ER) was shown to be heterogeneous, having ordered membrane regions containing cholesterol and more disordered regions. Interestingly, two closely related P450s, CYP1A1 and CYP1A2, resided in different regions of the ER. The structural characteristics of their localization were examined. These studies emphasize the importance of P450 protein organization to their function.
Introduction
The cytochrome P450 (P450) system in eukaryotic organisms comprises numerous proteins that interact within the confines of a membrane environment. The microsomal P450s reside in the endoplasmic reticulum (ER) and catalyze numerous oxidative reactions of both exogenous and endogenous substrates (Omura and Sato, 1962; Omura et al., 1965; Rendic and Guengerich, 2015). Because the proteins are crowded in the ER, there are numerous “opportunities” for protein-protein interactions, leading to questions regarding how the P450 system proteins are organized in the membrane. The requirement for P450 enzymes to interact with their redox partners, NADPH-cytochrome P450 reductase and cytochrome b5 (b5), is well known (Hildebrandt and Estabrook, 1971); however, the mechanism that governs formation of these complexes remains unclear. Additionally, recent studies have shown that some P450 enzymes are capable of forming both homomeric and heteromeric P450•P450 complexes that affect monooxygenase function (Davydov, 2011; Reed and Backes, 2012). Consequently, P450 activities can be modulated by the specific substrates present, the relative amount of the catalytically pertinent P450 present, the relative amounts of the electron donors CPR and b5, whether the specific reaction is stimulated by the presence of b5, and whether a second P450 is present that can interact with the P450 responsible for substrate hydroxylation (Jansson and Schenkman, 1987; Im and Waskell, 2011).
The multiple interactions among P450 system proteins occur within a heterogeneous lipid environment of the ER membrane (Brignac-Huber et al., 2011). At one level, insertion of these proteins within the membrane restricts their ability to interact to two dimensions; however, owing to the flexibility of the proteins within the membrane, their movement around their membrane-binding segments results in a greater range of motion and increases their ability to form complexes (Baylon et al., 2013). The heterogeneity of the membrane can also affect P450 function by either concentrating or segregating proteins within membrane regions.
The goal of this symposium was to address issues related to how proteins of the P450 system are organized and how these proteins interact with the membrane. Dr. Emily Scott examined the interactions among CPR, b5, and CYP17A1 using solution NMR. Dr. Scott identified residues involved in the interaction of both CPR and b5 with CYP17A1, and showed that the b5•CYP17A1 complex was enhanced by the presence of 17α-hydroxypregnenolone, the substrate for the CYP17A1-mediated lyase reaction.
Dr. Roland Wolf examined the interactions of P450s with their redox partners in vivo by generating a series of mice having the murine P450 gene clusters deleted and then engineered to express the corresponding human P450 enzymes. These investigators then selectively knocked out hepatic b5 and showed the effect on the in vivo metabolism of CYP3A4- and CYP2D6-selective substrates. In these experiments, both CYP3A4 and CYP2D6 substrates that were known to require b5 were cleared from the hepatic b5 knockout mice.
Because the interactions among these proteins occur in a membrane environment, the role of the lipid membrane also serves as an integral component affecting P450 system function. Dr. Michal Otyepka used a computational approach to better understand the relationship among substrate, P450, and the membrane. He showed that the proximal side of the P450 molecule faced the aqueous environment, whereas the distal side was associated with the membrane-water interface. Substrate was modeled to approach the active site from the membrane through channels.
Using single-molecule total internal reflectance microscopy, Sara Humphreys reported on the interaction between CPR and the lipid bilayer and showed that this interaction was affected by the presence of detergent. In these studies, CPR was shown to move in and out of the membrane, a response exacerbated by the presence of detergent. Interestingly, this effect, referred to as helicoptering, is diminished by the presence of P450 proteins in the membrane.
The heterogeneity of the lipid components of the ER membrane were considered in the report by Dr. James (Rob) Reed, who showed that two closely related P450s, CYP1A1 and CYP1A2, reside in different regions of the ER. Whereas CYP1A2 exists in regions where the lipids are more ordered, CYP1A1 exists in the disordered regions. The investigators identified sequence motifs that governed their localization into the different membrane regions by generation of CYP1A1-CYP1A2 chimeric proteins.
Taken together, these studies show the interplay not only among of the protein components of the P450 system but also the role of the membrane on their organization. These protein-protein and protein-lipid interactions have a profound effect on the function of the P450 system.
Steroidogenic Cytochrome P450 17A1 Interactions with Catalytic Partners (D.F.E., J.S.L., and E.E.S.)
During the course of mono-oxygenation reactions, human P450 enzyme interaction with CPR is required to sequentially deliver the two electrons required for P450 catalysis, whereas interaction with b5 is not usually required but is variously reported to accelerate, inhibit, or have no effect on P450 catalysis, depending on the P450 and substrate (Akhtar et al., 2005; Zhang et al., 2008; Ortiz de Montellano, 2015). A biophysical and biochemical understanding of these critical protein-protein interactions has been limited by the absence of crystallographic structures for human P450 proteins interacting with these protein partners, although mutagenesis, crosslinking, in silico docking, and other techniques have provided substantial information (Bridges et al., 1998; Naffin-Olivos and Auchus, 2006; Im and Waskell, 2011; Zhao et al., 2012). The generation of X-ray structures of such P450-protein complexes is likely hindered by the transient nature of these P450-protein interactions and the fact that they are largely thought to be mediated by electrostatic interactions; however, defining and characterizing such P450-protein interactions at high-resolution are possible using solution NMR, which generates information on protein dynamics and conformations but is amenable to the lower affinities of P450-protein interactions. The generation of isotopically labeled, catalytically active, truncated forms of the human steroidogenic CYP17A1, CPR, and b5 proteins has permitted tracking the isotopically labeled resonances for individual amino acid residues as they broaden and/or shift upon experiencing changes in environment. In the set of experiments described herein, one of the three proteins is 15N-labeled and titrated with one or more of the other proteins that compose the P450 catalytic system. The human steroidogenic P450 used, CYP17A1, is saturated with pregnenolone, its substrate for an initial hydroxylase reaction; 17α-hydroxypregnenolone, the product of this hydroxylation reaction and substrate for a subsequent lyase reaction generating androgens; or the type II steroidal inhibitor abiraterone. CYP17A1 is an important system for evaluating P450-protein interactions because although CPR is required for both hydroxylase and lyase reactions, b5 results in a 10-fold increase in the lyase reaction without directly transferring electrons or substantially affecting the hydroxylation reaction (Auchus, et al., 1998). This selective b5 facilitation of the lyase reaction is physiologically important as adrenal increases in b5 levels drive androgen sex steroid production immediately before puberty in human development (Nakamura et al., 2009).
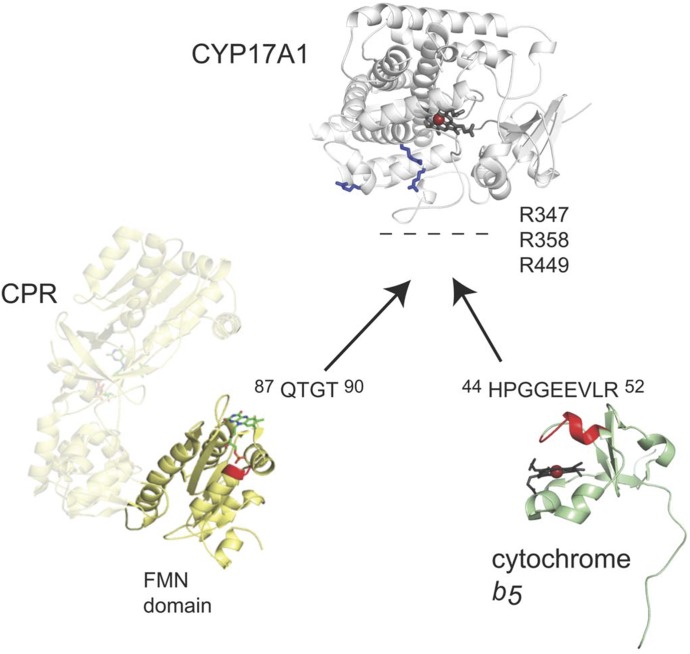
Previously published NMR studies (Estrada et al., 2013) have used 15N-b5 titrated with CYP17A1 to identify residues on b5 that are differentially line broadened (G47, E48, E49, V50, and surrounding residues) and compose the anionic b5 surface of the b5-CYP17A1 interface. Repeating such titration experiments with the R347H, R358Q, and R449L mutants of CYP17A1 reported to selectively depress the lyase activity (Geller et al., 1999) similarly prevented NMR changes indicative of b5-CYP17A1 physical complex formation. Thus, key residues of the CYP17A1-b5 interface were identified on the proximal surface of CYP17A1 and the α2 helix of b5 (Fig. 1). These interacting surfaces were the same regardless of the CYP17A1 ligand, but the strength of the b5-CYP17A1 interaction was modulated by the identity of the CYP17A1 substrate, suggesting communication between the b5 binding site on the proximal surface of CYP17A1 and the buried active site.
Fig. 1.
Surfaces of CYP17A1, cytochrome b5, and CPR involved in protein-protein interactions. The respective binding surfaces are shown as blue sticks for positively charged residues of CYP17A1, red for the negatively charged α2 helix of b5, and red for the positively charged loop 1 of the CPR FMN domain. Aggregated from data from Estrada et al., 2013, 2014, and 2015.
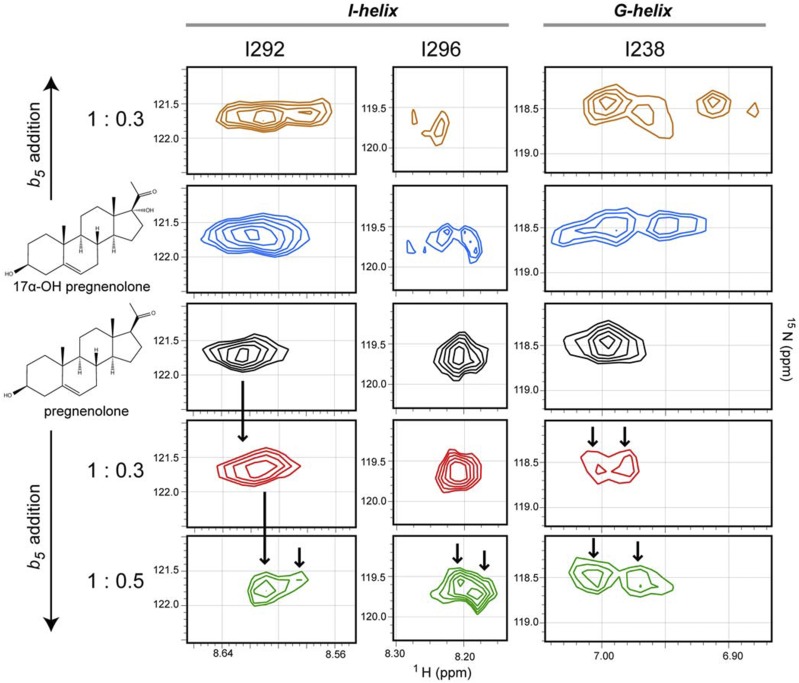
The reverse experiments titrating 15N-CYP17A1 with unlabeled b5 (Estrada et al., 2014) are much more complex owing to the number of resonances in CYP17A1 (494 amino acids) compared with b5 (114 amino acids) and, at present, can be only partially interpreted because of the availability of only partial resonance assignments; however b5 binding on the proximal surface of CYP17A1 clearly results in resonance splitting indicative of new backbone conformations for residues in the N terminus of the I helix (e.g., Fig. 2, I292 and I296) and the F and G helices (e.g., Fig. 2, I238) on the opposite, distal side of the protein from the b5 binding site—and does so in a substrate-dependent manner. Bending and straightening of the I helix and repositioning of the F and G secondary structure elements are often associated with ligand entry and exit, consistent with the idea that when the initial hydroxylase product 17α-hydroxypregnenolone occupies the CYP17A1 active site that b5 binding could promote a conformation that decreases ligand release, thereby promoting the second, lyase reaction to yield the androgen product. Differences in processivity have previously been observed for the two CYP17 enzymes in zebrafish, although in that case, processivity was not modulated by b5 (Pallan et al., 2015).
Fig. 2.
Effect of substrate and b5 on the conformational dynamics of CYP17A1. Distinct conformations are observed in the I-helix and G-helix residues of 15N-labeled CYP17A1when pregnenolone (black) versus 17α-hydroxypregnenolone (blue) is present in the active site. The addition of b5 to either substrate-bound sample further perturbs the conformations for these residues (top and bottom panels). When saturated with the hydroxylase substrate pregnenolone, serial titration of CYP17A1 with b5 [1:0.3 (red) and 1:0.5 (green)] induced a shift toward conformations more closely resembling those observed for CYP17A1 saturated with the lyase substrate 17α-hydroxypregnenolone state in the absence of b5. Vertical arrows emphasize differences between spectra. This research was originally published in Estrada et al. 2014.
The CPR interactions with CYP17A1 have been probed using solution NMR in a similar manner; however, instead of using the multidomain CPR protein in which the CPR FAD domain primarily interacts with its FMN domain and is only transiently available for interaction with CYP17A1, the isolated 15N-FMN domain of CPR was used. Titration of the CPR 15N-FMN domain with unlabeled CYP17A1 revealed that residues in a loop near the flavin (Q87-T90, Fig. 1) experienced the most significant line broadening, suggesting that this region is involved in the interaction between the FMN domain and CYP17A1; however, line broadening was much more broadly distributed and more severe for 15N-FMN domain binding to CYP17A1 than for 15N-b5 binding to CYP17A1, consistent with a higher-affinity interaction in the former case. Like the b5-CYP17A1 interactions, the affinity for the CYP17A1-FMN domain interaction was modulated by the identity of the CYP17A1 substrate.
When the experiment was inverted and 15N-CYP17A1 was titrated with the FMN domain of CPR, select residues in the B' helix, the F and G helices, and the N terminus of the I helix were line-broadened more than the average CYP17A1 residue. These resonances did not demonstrate peak splitting indicative of new backbone conformations, however, as they had upon addition of b5 (not shown). This suggests that the binding of the FMN domain alters the dynamics of CYP17A1, with specific effects on certain structural elements but does not alter the CYP17A1 conformation like b5 does. In other words, b5 has been shown to allosterically modulate CYP17A1 conformation, as suggested previously (Auchus et al., 1998).
Interactions between CYP17A1 and its catalytic partners can be further compared using solution NMR by evaluating the three proteins together. Experiments with 15N-b5 added to CYP17A1 demonstrate formation of the CYP17A1-b5 complex, which is then disrupted when full-length CPR (or the CPR FMN domain) is added, consistent with mutually exclusive binding and the partially overlapping binding sites for CPR and b5 on the proximal face of P450 enzymes as previously proposed (Zhang et al., 2007; Im and Waskell, 2011). Although single point mutations of R347H or R449L on the proximal surface of CYP17A1 were sufficient to prevent binding of b5, neither the R449L nor the R358Q mutation is individually sufficient to disrupt binding of the FMN domain of CPR. This result may indicate that these CYP17A1 residues are not involved in binding of the FMN domain or that the interaction with the CPR FMN domain is not easily disrupted by single point mutations, either of which is consistent with the functional observations (Geller et al., 1999).
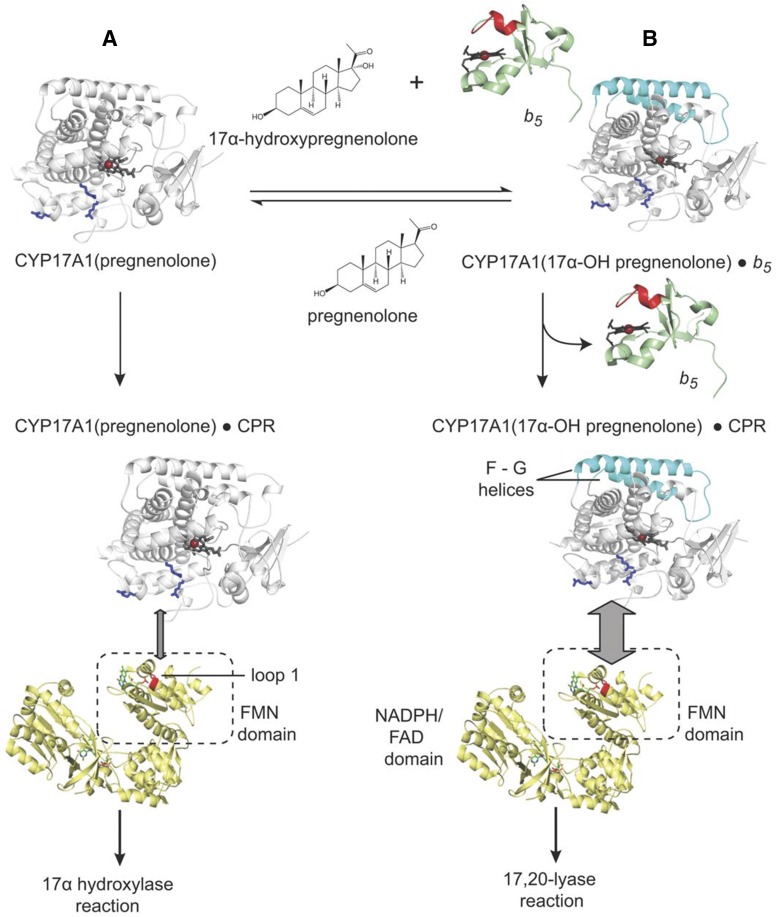
In aggregate, solution NMR studies, functional data, and structural data suggest a relatively simple comprehensive model for the observed CYP17A1 biochemistry, including selective facilitation of the lyase reaction by b5 (Fig. 3). NMR studies clearly demonstrate that in solution CYP17A1 exists in different conformational states with the hydroxylase substrate pregnenolone, the lyase substrate 17α-hydroxypregnenolone, and in the presence of b5. Binding of the hydroxylase substrate pregnenolone is likely to promote a conformational state that interacts with CPR and facilitates formation of the typical Fe(IV) oxo catalytic intermediate for the hydroxylation reaction. CYP17A1 can release the 17α-hydroxypregnenolone product or bind it de novo; however, it is likely that the 17α-hydroxy intermediate can also remain in the active site to undergo the second sequential reaction. Regardless, the presence of the lyase substrate in the CYP17A1 active site and b5 binding generate a distinct conformational state, which is likely to ultimately facilitate the subsequent lyase reaction. Because the presence of b5 facilitates the lyase reaction and because binding of b5 alters the backbone of elements implicated in substrate entry and exit, it is tempting to suggest that the binding of b5 to the CYP17A1/17α-hydroxypregnenolone complex promotes the processivity of the enzyme by promoting a closed conformational state. Regardless of how b5 does so, b5 and CPR binding are mutually exclusive, requiring b5 to dissociate for CPR to bind and deliver the two electrons necessary for catalysis. This is consistent with the NMR studies herein, which suggest that either CPR or the CPR FMN domain can outcompete b5. This model is internally consistent with the present state of knowledge regarding the interactions of CYP17A1, CPR, and b5 and significantly expands our knowledge of P450 dynamics and protein interactions beyond that previously understood for the human steroidogenic CYP17A1, with implications for other human P450 enzymes.
Fig. 3.
Model of CYP17A1 interaction with substrates, NADPH-cytochrome P450 reductase, and cytochrome b5 to perform either the 17α-hydroxylation (state A) or 17,20-lyase (state B) reaction consistent with current knowledge of the system. Interacting residues are shown as in Fig. 1. The CYP17A1 F-G helices that are altered by b5 binding are shown in cyan. Adapted from research originally published in Estrada et al. (2015).
Role of NADPH-Cytochrome P450 Reductase and Cytochrome b5 as Electron Donors to P450 (C.J.H., L.A.M., and C.R.W.)
Although the role of CPR in P450 function has been extensively studied and relatively well defined, that of b5 is less well understood, and, until recently, information on b5 had been essentially generated exclusively from in vitro experiments, yielding data that were not only difficult to interpret but often contradictory and not easily extrapolated to the in vivo situation. Within the last few years, however, we have generated mouse models with either a conditional (liver) or global deletion of b5 and used these, in conjunction with our previously described model of hepatic CPR deletion (hepatic reductase null mice) to investigate the role of both CPR and b5 in P450-mediated drug metabolism, disposition, and toxicity. Whereas CPR deletion was found, as expected, to have a significant effect on P450 activity both in vitro and in vivo, we also demonstrated, for the first time in vivo, that b5 deletion had a profound effect on P450 drug metabolism in a tissue- and substrate-dependent manner (Finn et al., 2008, 2011; McLaughlin et al., 2010). Furthermore, investigation of a residual P450 activity (∼10%) in vitro in HRN samples through the generation of a mouse line in which both CPR and b5 were deleted in the liver (hepatic b5 reductase null) led us to demonstrate that b5-Cyb5R can act as sole electron donors to the P450 system both in vitro and in vivo (Henderson et al., 2013).
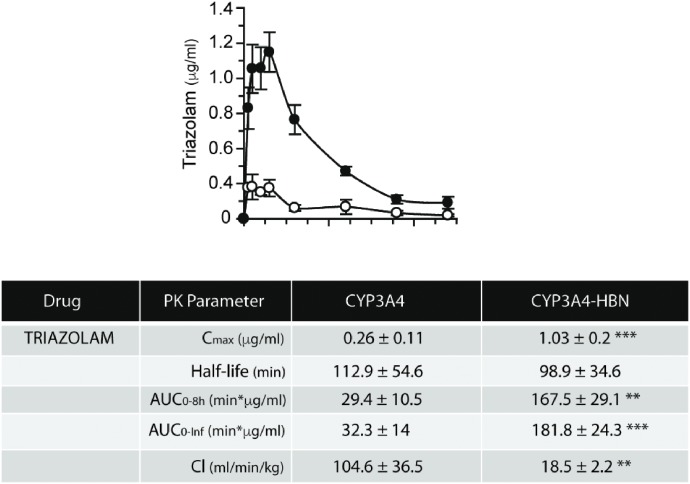
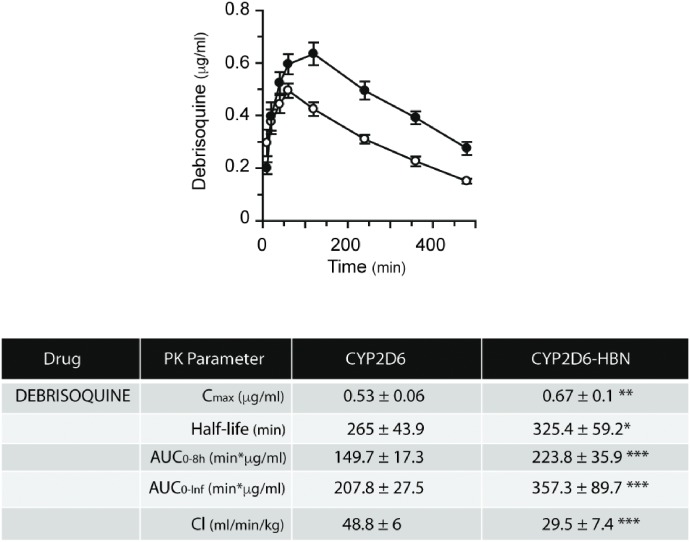
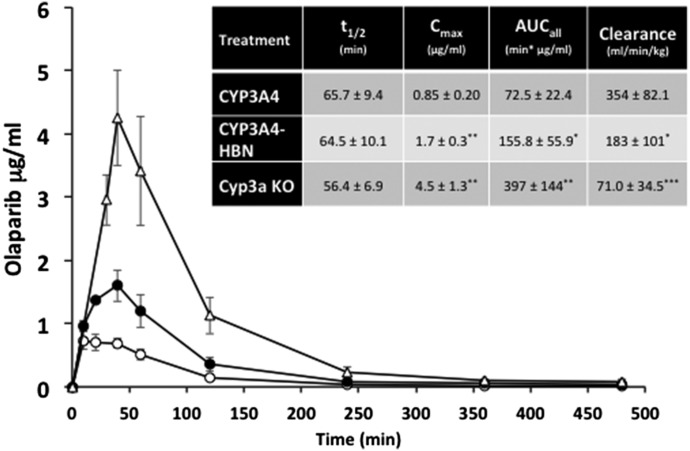
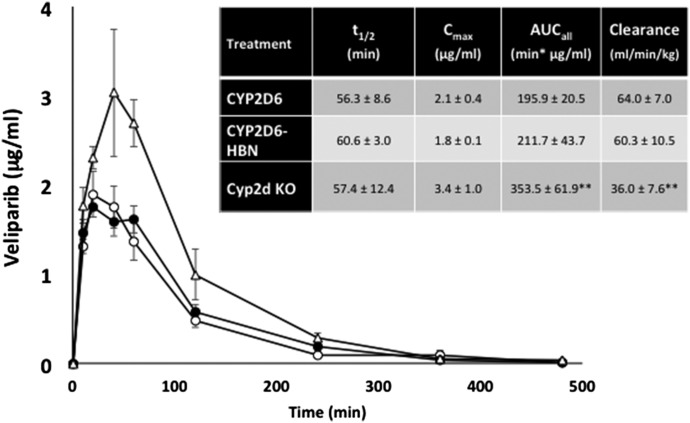
The importance of b5 in drug metabolism is underlined by the finding that expression of the enzyme varies significantly—by at least 5-fold—across the human population (Kurian et al., 2007; Henderson and Wolf, unpublished). Coupled with a recently discovered genetic variant in b5 (McKenna et al., 2014), there is clearly a need to better understand the role of b5 in relation to human P450 activity and to establish—in vivo—the extent to which human P450s exhibit a b5 dependency. To this end, we crossed the hepatic b5 null (HBN) (Finn et al., 2008) mouse with two mouse lines in which key human P450s—CYP2D6 or CYP3A4—are expressed on a Cyp2d or Cyp3a gene cluster null background (Hasegawa et al., 2011; Scheer et al., 2012). CYP2D6-HBN and CYP3A4-HBN mice (Henderson et al., 2015) were fertile and phenotypically normal and had only minor changes in hepatic lipids, perhaps surprising given the known involvement of b5 in lipid desaturation (Jeffcoat et al., 1977; Finn et al., 2011). Using substrates for human CYP3A4 (triazolam) and CYP2D6 (debrisoquine), it was shown in vitro in hepatic microsomes that the metabolism of these drugs was significantly reduced in the absence of b5. Moreover, in both cases, addition of exogenous b5 to reaction mixtures restored activity in a dose-dependent manner (Henderson et al., 2015). When triazolam (Fig. 4) and debrisoquine (Fig. 5) pharmacokinetics were determined in vivo in CYP3A4-HBN and CYP2D6-HBN mice, respectively, metabolism was again shown to be profoundly reduced in mice lacking hepatic b5, with significant increases in Cmax and AUC and decreased clearance relative to control animals. The minor perturbations in hepatic lipids reported in the CYP3A4-HBN and CYP2D6-HBN mice (Henderson et al., 2015) strongly support the view that the effects observed on drug disposition in vivo are not a result of changes to the lipid composition of the ER but rather reflect the consequences of a lack of b5. It is thus possible that the expression of b5, and its functionality, may make a significant contribution to the observed heterogeneity in plasma levels of many commonly prescribed drugs. To further illustrate this, the pharmacokinetics of the anticancer drugs and poly ADP ribose polymerase inhibitors olaparib (primarily metabolized by CYP3A4) and veliparib (primarily metabolized by CYP2D6) were determined in CYP3A4 and CYP2D6 mice, in the presence or absence of hepatic b5. For olaparib (Fig. 6), b5 significantly affected metabolism of this drug, with Cmax and area under the curve being increased and clearance decreased, in mice lacking b5 relative to CYP3A4 mice, whereas for veliparib (Fig. 7), b5 made no difference to metabolism. These data emphasize the importance of taking b5 expression into account for in vitro-in vivo extrapolation.
Fig. 4.
In vivo pharmacokinetic profiles of triazolam in CYP3A4 and CYP3A4-HBN mice. CYP3A4 (open circle) and CYP3A4-HBN (black circle) mice (female, n = 4) were treated with pregnenolone-16α-carbonitrile (PCN) at a dose of 10 mg/kg daily i.p. for 3 days. On day 4, mice were dosed p.o. with triazolam at 3 mg/kg body weight, and a pharmacokinetic study was carried out over 8 hours with blood sampling via the tail vein at the time points shown. Samples were analyzed for triazolam content by LC-MS/MS, and table shows pharmacokinetic parameters (mean ± S.E.M.). This figure was reproduced from Henderson et al. (2015).
Fig. 5.
In vivo pharmacokinetic profiles of debrisoquine in CYP2D6 and CYP2D6-HBN mice. CYP2D6 (open circle) and CYP2D6-HBN (black circle) mice (female, n = 4) were dosed p.o. with debrisoquine at 10 mg/kg of body weight, and a pharmacokinetic study was carried out over 8 hours with blood sampling via the tail vein at the time points shown. Samples were analyzed for debrisoquine content by LC-MS/MS, and table shows pharmacokinetic parameters (mean ± S.E.M.). This figure was reproduced from Henderson et al. (2015).
Fig. 6.
In vivo pharmacokinetic profiles of olaparib in CYP3A4, CYP3A4-HBN, and Cyp3a null mice. CYP3A4 (open circle), CYP3A4-HBN (black circle), and Cyp3a null (open triangle) mice (female; n = 4) were fed on a powdered diet (RM1) containing 250 mg/kg pregnenolone-16α-carbonitrile (PCN) for 3 days (equivalent to 50 mg/kg daily). On day 4, mice were dosed p.o. with olaparib at a final concentration of 25 mg/kg body weight, and a pharmacokinetic study carried out over 8 hours with blood sampling via the tail vein at the time points shown. Samples were analyzed for olaparib content by LC-MS/MS. Inset table shows pharmacokinetic parameters (mean ± S.D.). *P < 0.05, **P < 0.01 ***P < 0.001; CYP3A4-HBN or Cyp3a KO versus CYP3A4, t test.
Fig. 7.
In vivo pharmacokinetic profiles of veliparib in CYP2D6, CYP2D6-HBN, and Cyp2d KO mice. CYP2D6 (open circle), CYP2D6-HBN (black circle), and Cyp2d null (open triangle) mice (female; n = 4) were dosed p.o. with veliparib at a final concentration of 12.5 mg/kg body weight, and a pharmacokinetic study was carried out over 8 hours with blood sampling via the tail vein at the time points shown. Samples were analyzed for olaparib content by LC-MS/MS. Inset table shows pharmacokinetic parameters (mean ± S.D.). **P < 0.01; Cyp2d KO versus CYP2D6-HBN or CYP2D6, t test.
Positioning of Microsomal P450s and Drugs in Lipid Bilayers (M.O., M.P., V.N., K.B., P.A.)
Drugs enter cells via active and passive transport processes through lipid membranes and inside cells; both transport modes significantly contribute to the final drug disposition (Smith et al., 2014). Besides the mentioned processes, membrane partitioning, nonspecific protein binding, and biotransformation dominantly contribute to drug disposition in a cell (Anzenbacher and Anzenbacherova, 2001; Guengerich, 2006; Balaz, 2009; Seddon et al., 2009; Lucio et al., 2010; Endo et al., 2011; Nagar and Korzekwa, 2012). In humans, most marketed drugs undergo biotransformation processes catalyzed by P450 enzymes (Evans and Relling, 1999; Anzenbacher and Anzenbacherova, 2001; Zanger and Schwab, 2013), which are attached to membranes of the ER and mitochondria (Black, 1992). Experimental measurements suggested that P450 catalytic domains flow on the membrane surface being anchored to the lipid bilayer by an N-terminal α-helix; however the exact positioning of P450 in the membrane has not been yet determined by a direct experiment. This ignited an interest in modeling of P450s on the lipid bilayer. The early empirical models, which appeared soon after release of the crystal structures of mammalian P450s, were, however, not conclusive as they suggested very different membrane orientations of P450s (cf. Fig. 5 in Berka et al., 2011). Two independent models of membrane anchored CYP2C9 based on molecular dynamics simulations (Berka et al., 2011; Cojocaru et al., 2011) published in 2011 shared many similarities indicating that molecular dynamics might provide convergent and reliable membrane models of P450s. Since then, many independent models of membrane anchored P450s have been published, so we can discuss common features and differences among individual P450 forms (Denisov et al., 2012; Sgrignani and Magistrato, 2012; Baylon et al., 2013; Berka et al., 2013; Ghosh and Ray, 2013; Yu et al., 2015).
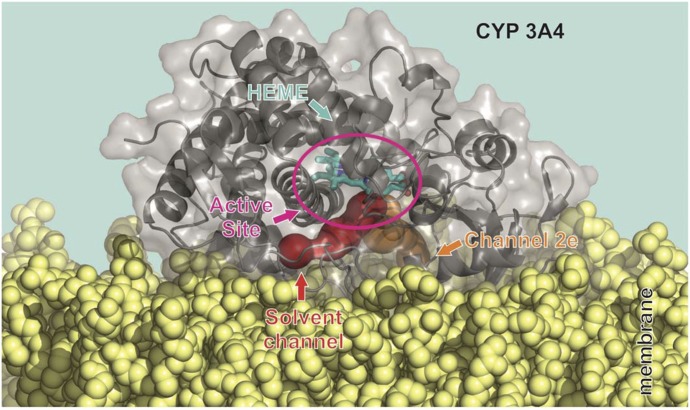
Common features of P450 membrane anchoring include the catalytic domain, which resides on the lipid bilayer being partially immersed to the membrane interior. The N-terminal part and F/G loop on the distal side of P450s are typically immersed to the nonpolar membrane interior (Fig. 8). The proximal side of P450 is exposed to the aqueous environment being available for interactions with redox partners. The active site, which is deeply buried in P450 structures (Otyepka et al., 2007; Johnson and Stout, 2013), is above the membrane surface and is accessible through a network of access channels (Cojocaru et al., 2007). Analysis of access channels by software tools CAVER (Petrek et al., 2006) and MOLE (Petrek et al., 2007) showed that openings of active-site access channels are positioned inside the membrane and the solvent channel exit (passing between F and I helices) faces the water/membrane interface (Fig. 9). The individual P450 forms (CYP1A2, 2A6, 2C9, 2D6, 2E1, and 3A4) show rather small variations in their membrane orientations, which affect positions of the channel openings with respect to the membrane (Berka et al., 2013).
Fig. 8.
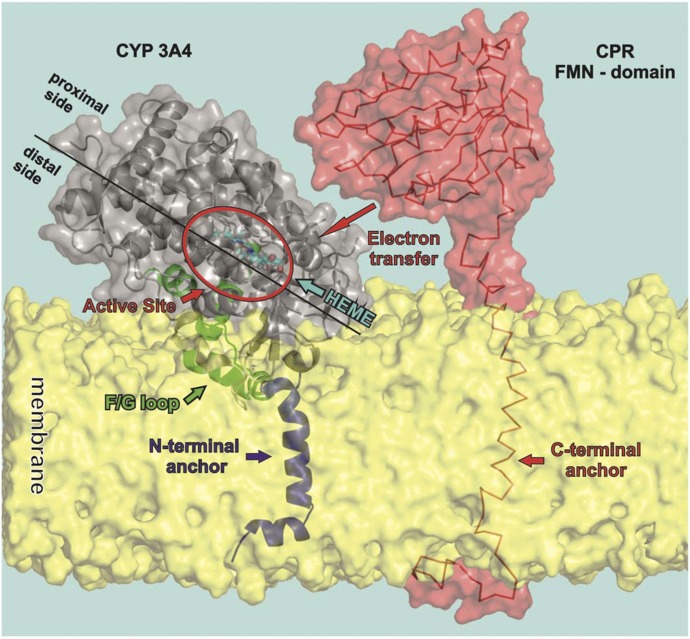
Model of the orientation of CYP3A4 and CPR with respect to the membrane. The figure shows a model of CYP3A4, taken from molecular dynamics simulations, floating on the membrane having a partially immersed catalytic domain and being anchored by one N-terminal α-helix crossing the membrane. The distal side faces to the membrane while the proximal side is oriented toward the cytosol and available for interaction with redox partners (here the FMN domain of CPR is shown).
Fig. 9.
Orientation of the CYP3A4 solvent channel and active site with respect to the membrane. The P450 catalytic domain (i.e., here CYP3A4 taken from molecular dynamics simulation) floats on the membrane and the access and exit channels (calculated by MOLE 2.0 software) to the deeply buried active site point toward the membrane opening above (for the solvent channel) and below (for channel no. 2e) the membrane surface.
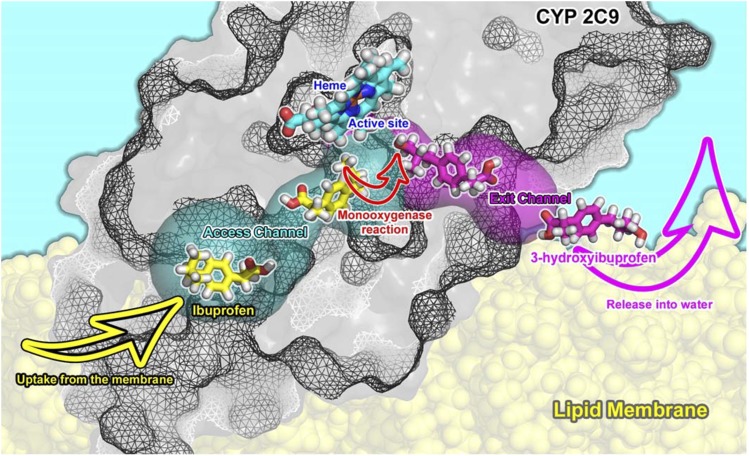
Most of the marketed drugs are amphiphilic compounds, and they have a large potential to accumulate in lipid bilayers (Seddon et al., 2009; Endo et al., 2011; Nagar and Korzekwa, 2012). It should be noted that the drug-metabolizing P450s catalyze the monooxygenase reaction as the most typical reaction. Formally, an oxygen atom is inserted onto a substrate molecule in the reaction, and one may expect that the amphiphilic substrate becomes more hydrophilic after the oxidation reaction. Comparison of affinities of drugs and their respective P450 metabolites to membranes shows that substrates have higher affinities for the membranes and are positioned deeper in the membrane structure than their respective metabolites (Paloncýová et al., 2013; Paloncýová et al., 2014). The drugs are typically localized inside the lipid bilayers just below the polar head group region, and the positions of P450 access active site channels openings are in the same membrane layer. Membrane positions of metabolites correspond to the solvent channel exit, which point toward the water-membrane interface (Fig. 10). Based on these findings, one may hypothesize that the drugs penetrate from the membrane to the P450 active sites. They are oxidized and released to the cytosol via the solvent channel.
Fig. 10.
Model of the substrate access channel of CYP2C9. Lipophilic and amphiphilic drugs can bind to CYP2C9 (taken from molecular dynamics simulation) active site from membrane, where they may accumulate. They pass via an access channel (calculated by MOLE 2.0) to the active site and there are transformed typically by a monooxygenase reaction and released via an exit channel to the water-membrane interface.
Super-resolution Protein-Lipid Interaction Mapping of P450 Reductase and Cytochrome P450 2C9 Interacting with Ternary Planar Lipid Bilayers (S.C.H., U.P.D., C.B., J.A.B., and J.P.J.)
Single-molecule techniques provide insight into protein-protein and protein-lipid interactions at a molecular level that is unresolvable by ensemble methods. Here, we investigated interactions between CPR, CYP2C9, and planar supported lipid bilayers using single molecule total internal reflectance microscopy (SM-TIRF) to generate super resolution protein-lipid interaction maps (SPLIMs). Specifically, we looked at how detergent concentration and time can affect protein-lipid interactions between CPR and planar lipid bilayers. We also examined how CYP2C9 perturbs this system.
Both CPR and CYP2C9 are localized to the cytoplasmic face of the hepatic ER in vivo via N-terminal α-helices (Williams and Kamin, 1962; Black, 1992).There is mounting evidence to suggest that under certain experimental conditions, CPR and P450s form higher-order complexes (Reed et al., 2010; Davydov, 2011; Davydov et al., 2015); however, the stoichiometry of these complexes and the dynamics governing their association and disassociation are not well understood. To understand how such complexes are formed, it is first important to investigate the simpler protein-lipid and protein-protein interactions essential to the system.
Detection and measurement of single ER proteins or protein complexes and their membrane counterparts are difficult in live cells owing to the complexity of the system and labeling shortcomings. Consequently, over decades, researchers have developed a range of cell membrane mimics to satisfy the finicky energetic requirements of membrane proteins for biphasic amphiphilic environments while removing much of the complexity of biologic membranes. Examples of P450 reconstituted systems include planar lipid bilayers, liposomes, and nanodiscs (Denisov and Sligar, 2011).
Although there are many advantages in using this type of reductionist approach, one distinct disadvantage is that reconstituted systems are prepared using detergents. Detergents are required to solubilize membrane proteins, maintain their stability, and facilitate their incorporation into the phospholipid bilayer (Helenius and Simons, 1975); however, detergents interfere with native protein-lipid and protein-protein interactions, and it is difficult to remove them from the system completely when they are no longer needed. The paradoxical beneficial and adverse roles of detergents in reconstituted systems can be explained by their chemical and thermodynamic properties. Commonly used detergents, such as (3-[3-cholamidopropyl)dimethylammonia]-1-propanesulfonate (CHAPS), structurally resemble cell membrane-associated compounds, including sterols and bile acids. In addition, akin to phospholipids, under certain conditions, detergents undergo organizational transformations to form lamellar micelles and bicelles.
Although attempts are often made to remove detergents using dialysis, hydrophobic adsorption, and gel filtration chromatography, based on our observations, we hypothesize that some unknown amount always remains. This could explain why incorporation of CPR and P450 into reconstituted systems is never complete (Reed et al., 2006). In support of this hypothesis, using SM-TIRF, we directly observed fluorescent-labeled proteins transiently interacting with lipid bilayers. This apparent dynamic binding and debinding phenomenon—helicoptering—was first described by (Ingelman-Sundberg and Glaumann, 1980) and is demonstrated in the (Supplemental Video S1). Furthermore, we examined how helicoptering is dependent on detergent concentration and time, as well on the strength of the protein-lipid interaction or protein complex-lipid interaction.
It is first important to provide a brief background on our CPR-CYP2C9 planar lipid bilayer system, SM-TIRF, and a data visualization technique we are calling super-resolution protein-lipid interaction mapping (SPLIM). We produce our reconstituted systems by laying down planar lipid bilayers on glass coverslips and allowing fluorescently labeled protein (Alexa 555 CPR and/or CYP2C9) to self-incorporate from above by incubation at 37°C. We prepare liposomes based on a method described by Guengerich and colleagues (Ingelman-Sundberg et al., 1996). From the liposomes, we prepare planar supported lipid bilayers using a method described by our group (Poudel et al., 2013).
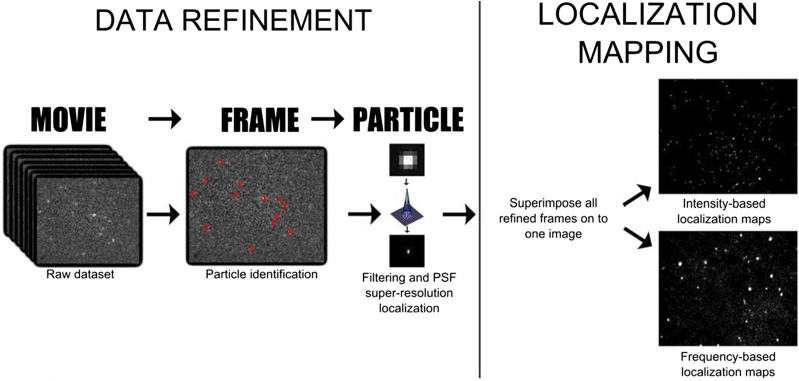
SM-TIRF involves focusing a laser beam through a high numerical aperture lens in such a way that it creates an evanescent field. This restricts the excitation of fluorophores to within ∼100 nm of the coverslip. Consequently, only fluorescently labeled proteins that are in or very close to the 5-nm-thick lipid bilayer on the surface of the coverslip are monitored. The light emitted from the excited fluorophores in a specific 2D region of the bilayer is captured by an electron multiplying charge coupled device camera in sequential 10-ms exposure frames (seen in S1). We use the ThunderSTORM plugin for ImageJ to process this movie data to achieve super-resolution localization of CPR interacting with the lipid bilayer (Fig. 11) (Ovesny et al., 2014). Briefly, each movie frame is filtered to improve the signal-to-noise ratio, and then fluorophores within each frame are identified according to a set of parameters (intensity, ellipticity, diameter, etc.). The center of each fluorophore’s point spread function is approximated to one pixel by fitting it with a two-dimensional (2D) Gaussian. Finally, the superlocalized positions of all fluorophores from all frames of the movie are superimposed to generate a 2D grayscale histogram that represents the spatial frequency distribution of all protein-lipid interactions within a precise region of the bilayer over a given period of time—hence, SPLIM.
Fig. 11.
Super-resolution protein-lipid interaction mapping (SPLIM) workflow. Single-molecule data collected by exciting individual Alexa-555-CPR proteins at the lipid bilayer in a TIRF evanescent field using a 532-nm laser. Emitted light was detected on an EM-CCD camera. Tens to hundreds of thousands of continuous 10-millisecond frames were collected from a single sample at a specific bilayer location. The ThunderSTORM plugin for ImageJ identifies individual proteins frame-by-frame. The super-resolution location of each protein was found by approximating the point spread function with a 2D Gaussian and finding the center. CPR-lipid interactions were ‘mapped’ by superimposing all super-resolution refined frames. Two different types of 2D grayscale maps can be generated: intensity-based maps (top left) and frequency-based maps (bottom left). Whereas cumulative molecular brightness (intensity) may be of interest in some applications, for example, in fixed cells, for SPLIM, we postulate that frequency-based localization better represents transient protein-lipid interactions, where ‘whiter’ regions are locations in the heterogeneous bilayer where the incidence of interaction is higher.
SPLIM differs from more conventional super-resolution image-reconstruction techniques because we plot frequency per pixel rather than cumulative intensity per pixel (Fig. 11, right). To explain, we use a grayscale 2D matrix to represent the region of the bilayer we collect data from; in the matrix, each element represents one pixel, and the pixel dimensions define the ultimate resolution of the image. Black is represented by zero, so before data collection begins, the matrix is populated with zeros. Shades of gray through to white are represented by integers greater than zero. Each time a protein “visits” a particular location on the bilayer, the value for that pixel in the matrix increases by one. The pixel that is “visited” the greatest number of times during data collection will be white and will have the highest corresponding integer value in the matrix. All other shades of gray will fall somewhere in between.
In conventional super-resolution imaging and reconstruction, the aim is to “map” fluorophore locations under the assumption that they are fixed. The blinking properties of fluorophores are exploited to attain sparsely populated images that are then superimposed to “fill in the gaps.” The cumulative intensity at each pixel location is used to populate the greyscale matrix. Cumulative intensity represents the total number of photons generated at a given pixel location rather than the number of times a protein is located there. A cumulative intensity image can differ significantly from a frequency plot because of dynamic changes in fluorophore quantum efficiencies with respect to their photophysical processes and local environment (i.e., in solution versus in the bilayer). We believe our SPLIM treatment better addresses the dynamic nature of our system, as we are most interested in the number of times a particle “visits” a certain location on the bilayer.
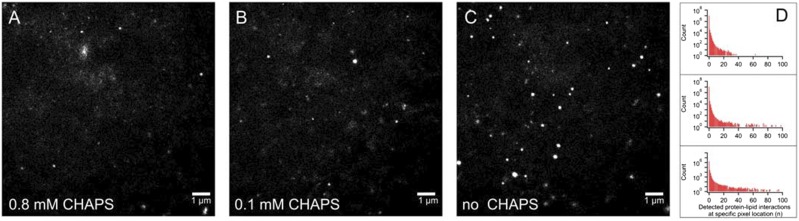
Using SM-TIRF and SPLIM, we investigated the role of detergent in CPR-bilayer interactions (Fig. 12). At high concentrations of detergent (CHAPS) (Fig. 12, A and B), CPR-bilayer interactions appear more spatially random, occupying more of the pixels in the field of view. In contrast, we observed that as the CHAPS concentration decreases, CPR appears to become less laterally mobile and is seen in the same pixel location more frequently (Fig. 12, C and D). It is important to note that based on these observations alone, we cannot distinguish between one particle that visits a particular region on the bilayer and is detected over many frames versus many particles visiting the same region.
Fig. 12.
SPLIM indicates diminishing detergent leads to changes in the frequency and distribution of protein-lipid interactions. Alexa 555-CPR (CPR purified according to Rock et al., (2001) and conjugated to Alexa 555 C2 maleimide according to the manufacturer (Thermofisher)] was added to a final concentration of 3.5 nM to ternary lipid bilayers (prepared in 200 mM K-HEPES (pH 7.4), 15 mM MgCl2, and 0.8 mM CHAPS). The degree of labeling of the final Alexa 555-CPR was 80%. After 30 minutes incubation at 37°C, the bilayer was rinsed in buffer containing 200 mM K-HEPES (pH 7.4), 15 mM MgCl2, and either 0.8 mM CHAPS (A), 0.1 mM CHAPS (B) or no CHAPS (C). These SPLIM images have been normalized for the number of particles instances (84,000 ± 600). A particle instance is defined as one spatially resolved CPR-lipid interaction in one 10-millisecond frame using ThunderSTORM. (A, B) were each generated by superimposing 20,000 frames, whereas (C) was generated from 12,200 frames Changes in the spatial frequency distribution demonstrated in these SPLIM images are further shown by one-dimensional histograms (D) where (A), (B), and (C) are the top, middle, and bottom, respectively. All SPLIM experiments were carried out at 37°C. Because SPLIM images are essentially 2D histograms that follow poissonian distributions, to enhance contrast, grayscale levels were adjusted. The same adjustments were applied to each SPLIM image. Thus, all pixels in all SPLIM images were treated identically.
One explanation for these observations is that CPR partitioning between the bilayer and solution is detergent-dependent. As detergent is removed, there are fewer solution-based amphiphilic molecules to interact with and stabilize CPRs’ hydrophobic N-terminal helix. This results in greater partitioning of CPR into the membrane, and the on/off rates of CPR-lipid helicoptering adjust accordingly. With less detergent, CPR is “on” the bilayer longer or permanently. This could account for the increased frequency of protein-lipid interactions at specific bilayer locations (i.e., we are detecting the same protein multiple times over multiple frames).
An alternative explanation is that at low detergent concentrations, helicoptering CPR could have higher affinity for specific regions within the heterogeneous bilayer [here, we used a ternary mix of DLPC, DOPC, DLPS (1:1:1 (w/w/w)], and what we are observing is it “visiting” these regions more often. It has been shown that CPR harbors a specific affinity for anionic phospholipids such as phosphatidylserine (Balvers et al., 1993). To investigate this question further, we looked at annexin V, a self-inserting transmembrane protein, interacting with bilayers under the high detergent conditions (0.8 mM CHAPS). Annexin V is known to interact specifically with phosphatidylserine (Tait and Gibson, 1992). We found that annexin V was localized to spatially distinct regions in a pattern similar to that seen in the “no CHAPS” sample, indicating that CPR does in fact localize to PS-rich domains (data not shown).
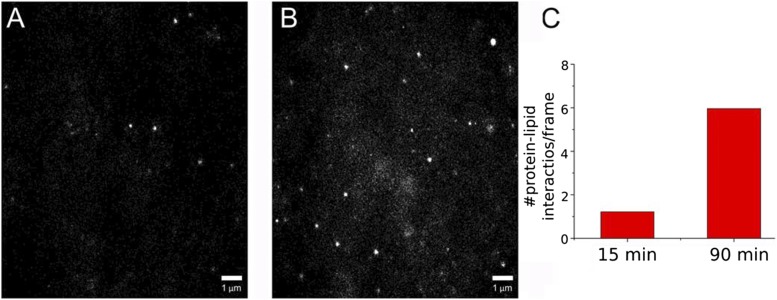
To ascertain whether CPR-lipid interactions displayed time-dependent variability, we collected SM-TIRF data from the same sample at two different time points after addition of CPR to the bilayer. From the SPLIM profiles (Fig. 13, A and B), we observed a change in the spatial distribution of protein-lipid interactions over time that was similar to what was seen with detergent removal. After a longer incubation, CPR appeared to frequent specific regions of the bilayer more often. In addition, the number of detected CPR-lipid interactions increased from 1.22 per frame to 5.97 per frame (Fig. 13C), confirming that we are observing a dynamic process that has not yet reached equilibrium. This finding corroborates the work of many groups who have described long protein-lipid incubation periods (hours to days) to achieve higher levels of protein incorporation into their reconstituted systems (Causey et al., 1990; Reed et al., 2006; Davydov et al., 2010). It is important to note, however, that we have been unable to find experimental evidence to suggest that protein incorporation is complete and irreversible, even after the removal of detergent.
Fig. 13.
Time-dependent CPR-lipid interactions using SPLIM. (A, B) are SPLIM images generated from 15,000 frames each (a total of 150 seconds). (A) Data were collected 15 minutes after CPR addition. (B) Data were collected 90 minutes after CPR addition. (C) Average number of particle instances detected per frame. Samples were prepared as in Fig. 12, using the 0.8 mM CHAPS-containing buffer. Contrast adjustments were performed as in Fig. 12.
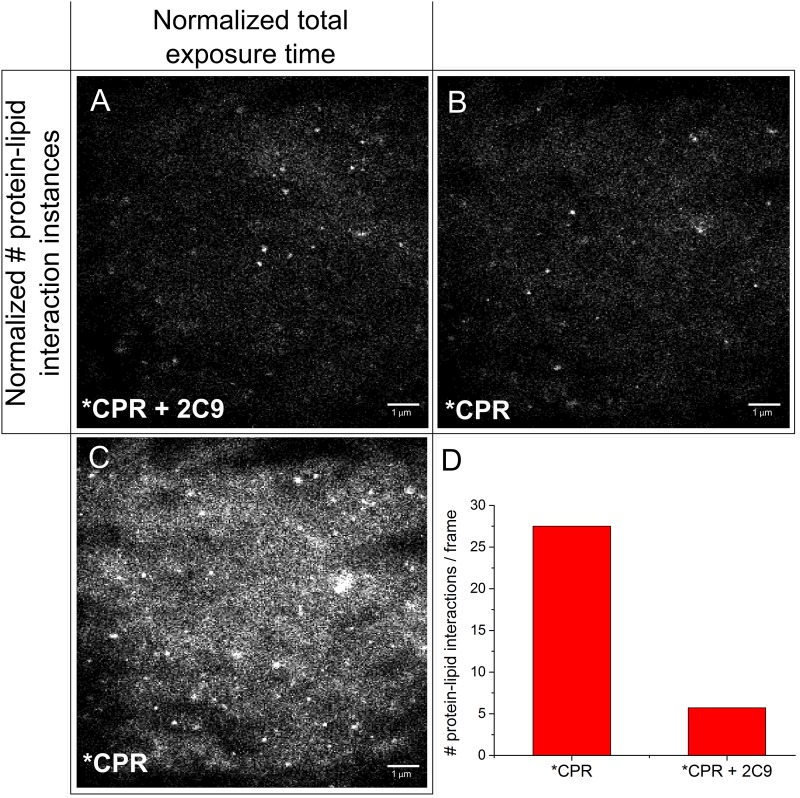
To investigate how CYP2C9 affects CPR-lipid interactions, we compared the SPLIM profiles of our CPR-lipid samples before and after the addition of CYP2C9 (Fig. 14). Once again, there were differences in the spatial distribution patterns (Fig. 14, A and B), as well as changes in the number of protein-lipid interaction instances over time (Fig. 14, A, C, and D). Most interestingly, there appeared to be a significant reduction (80%) in the number of recorded protein-lipid interaction instances when CYP2C9 was present. We believe this is could be due to Förster resonance energy transfer quenching of Alexa 555-CPR fluorescence by the P450 heme, a phenomenon that has been previously described in the literature (Isin and Guengerich, 2008). This would indicate that under these conditions, approximately 80% of all CPR molecules are within the Förster radius of a CYP2C9 molecule, signifying that CPR and CYP2C9 are forming heterodimers or oligomers. An alternative explanation could be that in the presence of CYP2C9, CPR “visits” the bilayer less frequently, although this is unlikely given that other groups have described the opposite to be true (Reed et al. 2006).
Fig. 14.
Using SPLIM to visually demonstrate perturbation of CPR-lipid interactions by CYP2C9. (A) A SPLIM image of a bilayer that has been incubated with Alexa 555-CPR and unlabeled CYP2C9 [3.5 nM each; CYP2C9 prepared according to Shimoji et al., (1998)]. It contains data from 19,801 frames and has 57,218 particle instances. (B, C) SPLIM images of the same Alexa-555 CPR sample (no CYP2C9), normalized according to (B) particle instances or (C) number of frames. (B) Generated from 4,508 frames. (C) Had 275,123 particle instances. (D) Bar graph showing the average number of particle instances per frame for each sample. Samples were prepared, and contrast adjustments were made as in Fig. 12.
In conclusion, we directly observed that CPR-lipid interactions are dependent on detergent concentration and time and that in the presence of CYP2C9, the CPR fluorophore is likely quenched via Förster resonance energy transfer to the P450 heme. Together, these findings highlight the unique information that can be extracted from reconstituted systems using SM-TIRF. In addition, we introduced SPLIM as a method to display protein-lipid interactions by localization frequency and spatial distribution. We postulate that by visualizing the data in this way, we can unveil information about the underlying structure of the lipid bilayer. In the future, SPLIM could be used to dynamically “map” lipid rafts and phase domains within membranes on cell or organelle surfaces. Cluster analysis such as the work described by Owen et al. (2010) may help to quantify SPLIM interactions further.
Elucidating the Macromolecular Determinants of Lipid Domain Localization for CYP1A1 and CYP1A2 in the ER (J.w.P., J.R.R., and W.L.B.)
Biologic membranes are heterogeneous with respect to both the composition and the ordering of the constituent lipids. These lipid bilayers are comprised primarily of a variety of phospholipids with varying lengths of fatty acyl chain moieties. Cholesterol is another component of biologic membranes that modulates the ordering of certain types of phospholipids in its proximity (Berkowitz, 2009). As a result, clusters of cholesterol and phospholipids containing a high proportion of saturated, straight-chained fatty acyl groups coalesce in the membranes as discrete zones that have been termed “lipid rafts” or “lipid microdomains” (Pike, 2009). The lipids in the microdomains are tightly packed which results in their lateral motion being highly restricted. These regions are described as being in the “liquid-ordered” state (Brown and London, 2000). The lipid microdomains are present in a surrounding medium containing phospholipids with a high degree of unsaturation in the fatty-acyl chains. This aspect of the lipids surrounding the microdomains prevents them from being tightly packed and results in the constituent lipids displaying an extreme state of random motion. As a result, this milieu has been referred to as being in the liquid-disordered phase (Brown and London, 2000).
One physical characteristic of the ordered domains that has allowed for their study is the fact that they are resistant to being solubilized by detergent treatment of the membrane (Ahmed et al., 1997; London and Brown, 2000). Thus, lipid microdomains have also been referred to as detergent-resistant membranes (DRMs). In studies with lipid microdomains, researchers have used detergent solubilization to identify the proteins in the membrane that predominantly localize to DRMs. In addition, researchers have commonly used a compound that tightly binds to cholesterol, methyl-β-cyclodextrin, to deplete cellular membranes of cholesterol. In addition to showing the dependence of lipid microdomains on the presence of cholesterol (Brignac-Huber et al., 2011), these studies have also elucidated some of the cellular functions of the microdomains. In the plasma membrane, DRMs have been shown to regulate diverse cellular processes, including signal transduction, endocytosis, cellular trafficking, and cytoskeletal tethering (reviewed in Head et al., 2014).
Lipid microdomains have recently been demonstrated to also exist in the ER (Browman et al., 2006), although their functions have not been well established. Our laboratory examined whether lipid microdomains influenced the distribution and activities of P450 in the ER membrane. Our initial studies determined that CYP1A2 predominantly resided in ordered membranes (Brignac-Huber et al., 2011). Furthermore, its localization in the membrane and its catalytic activity were greatly affected by cholesterol depletion. We subsequently used fluorescent lipid probes to demonstrate that ordered lipid domains were present in purified CYP1A2-reconstituted systems, with the lipid composition of total ER and that CYP1A2 resided in ordered domains as indicated by its resistance to detergent solubilization (Brignac-Huber et al., 2013). When comparing a homogeneous membrane reconstituted system prepared from bovine liver phosphatidylcholine and one prepared from a mixture of lipids that would favor the generation of ordered lipid microdomains, the presence of ordered domains was associated with a much higher apparent binding affinity between CYP1A2 and CPR (Brignac-Huber et al., 2011). Interestingly, the localization of CYP2B4 and CYP2E1 in the ER was distinct from that of CYP1A2 as the former distributed equally between ordered and disordered regions, whereas CYP2E1 was almost entirely within disordered regions (Park et al., 2014).
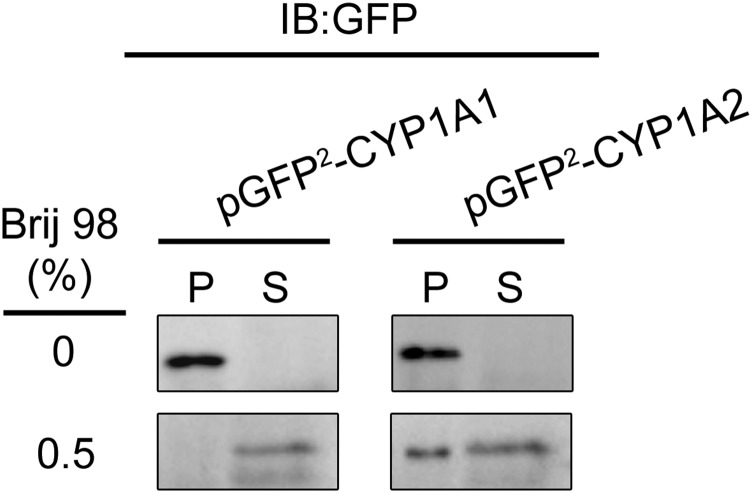
In the course of investigating the membrane distribution of CYP1A2, we noticed an intriguing distinction in the localization of CYP1A2 and CYP1A1 (Fig. 15). After the DRMs were isolated by high-speed centrifugation after treatment with 1% Brij 98, immune blotting was performed to monitor the P450 distribution by using a primary antibody reactive with both forms of CYP1A enzymes. As reported previously, CYP1A2 localized predominantly in ordered domains; however, CYP1A1 was found to reside almost entirely in disordered regions of the ER.
Fig. 15.
Localization of CYP1A2-GFP and CYP2B4-GFP fusion proteins in different lipid microdomains. The ordered and disordered lipid domains from HEK293T cells expressing either GFP-CYP1A1 or GFP-CYP1A2 were treated with 0.5% Brij 98 followed by high-speed centrifugation at 100,000g for 1 hour in a buffer containing 50 mM HEPES (pH 7.25), 150 mM NaCl, and 5 mM EDTA. S and P represent the supernatant (disordered) and P (ordered) fractions of the modified CYP1A proteins, showing that the fusion proteins reside in different membrane regions. (This research was originally reported in Park et al., unpublished.)
Because CYP1A1 and CYP1A2 share about 80% sequence identity, the protein regions responsible for the varying localization of the two P450s could be clearly determined by the generation of CYP1A1-CYP1A2 chimeras. Initially, it was important to ensure that we could accurately monitor the expression and localization of the two P450s in HEK293T cells. To easily monitor transient expression in the cells, we linked the cDNA of green fluorescent protein (GFP) to the C-terminal end of the CYP1A cDNAs and cloned into the pGFP2-N2 expression vector. After expression in the human embryonic kidney cell line HEK-293T cells, it was determined that the modified CYP1A proteins were expressed in the ER of the cells; after detergent solubilization with 0.5% Brij 98 of the postnuclear supernatant of the cells and high-speed centrifugation (100,000g for 1 hour), it was found that CYP1A1 and CYP1A2 were localized to disordered and ordered domains, respectively (data not shown). Thus, the cell system could be used with mutagenesis of the CYP1A sequences to determine the regions responsible for domain localization.
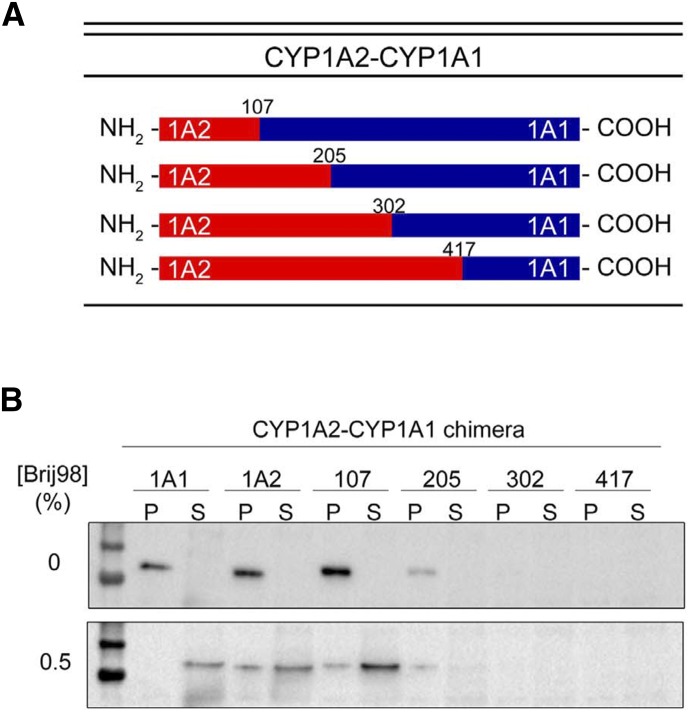
Chimeras were generated by overlap extension PCR (Ho et al., 1989). Four cut sites in the cDNA sequences of the P450s were selected based on their location in meander regions of the crystal structures (Sansen et al., 2007) and thus were not expected to interrupt key regions of secondary structure that were important for function. The cut sites were used to generate a combination of chimeras (shown in Fig. 16A). A complementary set of chimeras were also generated using the same cut sites by adding different lengths of the CYP1A1 N terminus to the C-terminal end of CYP1A2 (not shown). Substitution of the shortest N-terminal sequence in both chimera systems was sufficient to reverse the domain localization of the wild-type enzymes. More specifically, when the first 100 amino acids of CYP1A2 were combined with the C-terminal end of CYP1A1, the enzyme partially localized to the ordered domain (Fig. 16B), and with the opposite substitution, the chimera was found to reside entirely in the disordered region (not shown).
Fig. 16.
Membrane localization of CYP1A2-CYP1A1 chimeric proteins in HEK293T cells. Chimeric proteins were constructed that contained the N-terminal region of CYP1A2 fused to the C-terminal region of CYP1A1. (A) A diagram of the cut sites representing the junction of cDNAs for the chimeras. (B) The relative distribution of the expressed chimeras into ordered (P) and disordered (S) regions after cell lysis, isolation of the postnuclear supernatant, and high-speed centrifugation (100,000g for 1 hour). The 0% panel shows the immune blots resulting from no detergent treatment and the 0.5% panel shows the distribution after solubilization in 0.5% Brij 98. (This research was originally reported by Park et al., unpublished.)
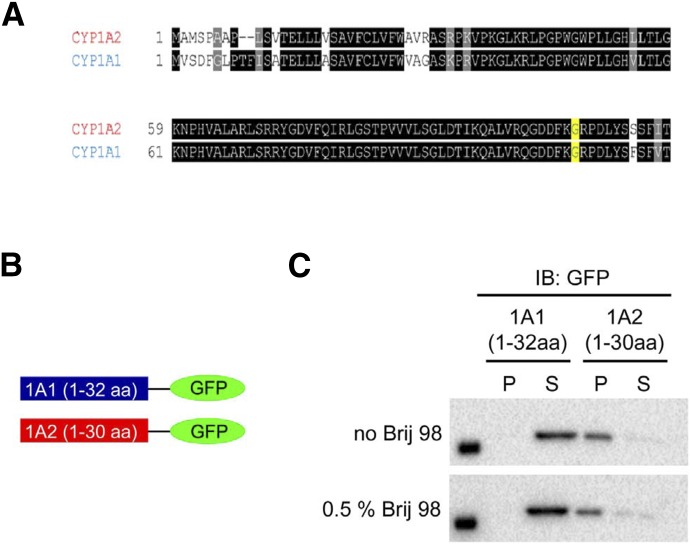
All the mammalian P450s involved in drug metabolism have an N-terminal sequence of approximately 30 amino acids in length that is hydrophobic and anchors the proteins to the membrane (Black and Coon, 1982). It is known that this segment functions to target the enzymes to the ER. Given the effect of substituting the N terminal ≈100 amino acids of CYP1A2 and CYP1A1 on domain localization, however, we suspected that the N-terminal hydrophobic tail of the P450 may also have a role in the lipid domain selectivity of the enzyme. Therefore, we generated expression plasmids with the N-terminal tails of CYP1A1 and CYP1A2 linked to the cDNA for GFP (Fig. 17B). These modified GFP proteins were then expressed in HEK 293T cells, and their membrane localization was determined by cell lysis and detergent solubilization of the postnuclear supernatant. After detergent treatment and high-speed centrifugation, it was found that the GFP with N-terminal CYP1A1 did not express in the membrane; however, the GFP with N-terminal CYP1A2 did express in the membrane and was predominantly localized in the ordered microdomains (Fig. 17C). Thus, the hydrophobic, N-terminal sequence (≈30 amino acids) of CYP1A2 appears to play a key role in targeting this enzyme to the ordered domains.
Fig. 17.
Membrane localization of CYP1A1-GFP and CYP1A2-GFP fusion proteins containing only their N-terminal regions. Construction of CYP1A-GFP chimeras derived by attaching the N-terminal regions of CYP1A1 (aa 1–32) and CYP1A2 (aa 1-30), respectively to the cDNA for GFP and their relative distribution into ordered and disordered lipid domains. (A) Comparison of the N-terminal regions of rabbit CYP1A1 and CYP1A2. (B) Illustration of the constructed fusion proteins. (C) Localization of the proteins before and after Brij 98 solubilization. CYP1A2 resides in the pellet before Brij 98 treatment and remains in the membrane after partial membrane solubilization. In contrast, the expressed fusion protein containing the CYP1A1 N-terminus does not reside in the membrane, even before to Brij 98. (This research was originally reported by Park et al., unpublished.)
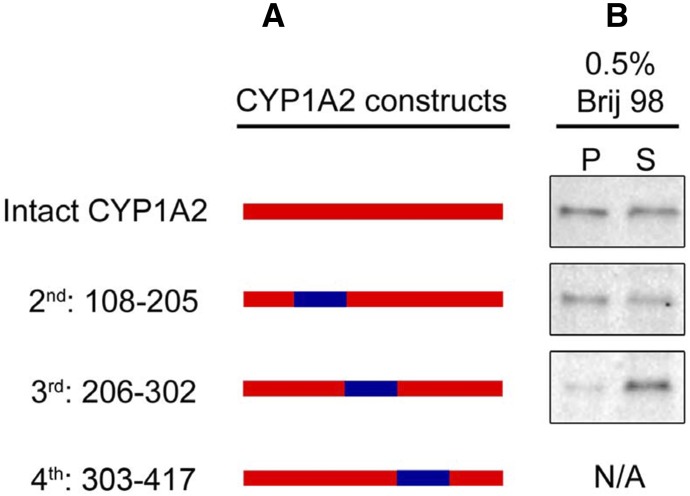
Examination of Fig. 16 shows that the N-terminal ≈100 amino acids of CYP1A2 resulted in only partial relocation of the CYP1A2-CYP1A1 chimera to ordered domains. As a result, we suspected that there might be an internal amino acid sequence of the P450s that also contributes to domain localization. To test this possibility, the same cleavage sites in the amino acid sequences of CYP1A1 and CYP1A2 were used to generate cDNAs for three chimeras in which one of the CYP1A1 internal sequences was substituted for that of CYP1A2 (Fig. 18). The chimera containing the CYP1A1 sequence from amino acid 303–417 did not express in the cells. The CYP1A1 substitution at amino acids 108 to 205 did not seem to alter the relative distribution into ordered and disordered domains; however, when amino acids 206–302 of CYP1A1 were substituted, a higher proportion of the P450 localized to disordered domains, suggesting this region also contributes to the domain selectivity of these enzymes. This region of the protein contains the F and G helices and F-G loop, which interact with the membrane, are involved in substrate binding from the membrane and constitute part of the ceiling of the P450 active site above the heme group (Williams et al., 2003; Yano et al., 2004; Pochapsky et al., 2010; Johnson et al., 2014). Our data show that both the hydrophobic N-terminal sequence and the F-G helices region confer the ordered microdomain selectivity of CYP1A enzymes. Because there is considerable sequence homology for CYP1A1 and CYP1A2 in these regions, future studies will focus on identifying the specific amino acids in these two sequences that are responsible for targeting the enzymes to disordered and ordered regions in the ER.
Fig. 18.
Role of the internal regions of CYP1A on membrane localization. Chimeric proteins where internal regions of CYP1A1 were inserted into the CYP1A2 sequence were generated. The constructs were then transfected into HEK293T cells and their localization determined. (A) Illustration of the chimeric constructs. (B) Localization of the chimeras after Brij 98 treatment of the membranes. This research was originally reported published by Park et al., unpublished.
P450s have been shown to associate with one another in lipid membranes. Both homomeric and heteromeric complexes of P450 have been demonstrated using a variety of techniques (reviewed in Davydov, 2011; Reed and Backes, 2012). Almost 40 years ago (Peterson et al., 1976), it was proposed and has been subsequently shown that the aggregation of P450s would allow for more efficient electron transfer by the CPR as electrons could be rapidly transferred to the aggregating P450s in a single interaction with the complex. The P450-P450 associations also directly influence the function of these enzymes because form-specific interactions have been shown to result in activation and/or inhibition of P450 catalytic activities (Hazai and Kupfer, 2005; Kelley et al., 2006; Subramanian et al., 2009, 2010; Reed et al., 2010). Thus, factors that influence the protein-protein associations of P450s in the ER can have important effects on P450 function. We are currently examining the effect of microdomain localization of different P450 enzymes on their ability to form P450-P450 complexes.
Summary
Although much is known about the factors controlling the expression and catalytic function of P450 system proteins, numerous questions remain regarding how these proteins are organized in the ER and how they interact with their protein partners and even with the membrane itself. These reports show that the interactions among these proteins are complex, and we must consider the specific proteins that are present as well as the membrane environment in which they reside.
Supplementary Material
Abbreviations
- b5
cytochrome b5
- CHAPS
(3-[3-cholamidopropyl)dimethylammonia]-1-propanesulfonate
- CPR
NADPH-cytochrome
- DRM
detergent-resistant membrane
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- HBN
hepatic b5 null
- NMR
nuclear magnetic resonance
- P450
cytochrome P450
- SM-TIRF
single-molecule total internal reflectance microscopy
- SPLIM
super-resolution protein-lipid interaction map
- 2D
two-dimensional
Authorship Contributions
Participated in research design: Park, Reed, Backes, Otyepka, Humphreys, Barnaba, Dahal, Brozik, Jones, Estrada, Laurence, Scott, Wolf, Henderson.
Conducted experiments: Park, Reed, Paloncýová, Navrátilová, Berka, Humphreys, Dahal, Estrada, McLaughlin.
Performed data analysis: Otyepka, Berka, Azenbacher, Park, Reed, Backes, Humphreys, Estrada, Laurence, Scott, McLaughlin.
Wrote or contributed to the writing of the manuscript: Otyepka, Park, Reed, Backes, Humphreys, Brozik, Jones, Estrada, Laurence, Scott, Henderson.
Footnotes
This work was supported in part by National Institutes of Health [R01 GM076343, GM102505] to E.E.S.; [F32 GM103069] to D.F.E.; [GM110790] to J.P.J.; and [R01 ES004344\ and [P42 ES013648] to W.L.B.; the Ministry of Education, Youth and Sports of the Czech Republic, project LO1305 to M.O. and the Grant Agency of the Czech Republic, [P303/12/G163] to P.A; and Cancer Research UK Programme Grant [C4639/A10822] to C.R.W.
This report is a summary of a session at Experimental Biology 2015 sponsored by the Drug Metabolism Division of ASPET.
Emily E. Scott, C. Roland Wolf, Michal Otyepka, Sara C. Humphreys, and James R. Reed were speakers at the symposium and are considered co-first authors.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Ahmed SN, Brown DA, London E. (1997) On the origin of sphingolipid/cholesterol-rich detergent-insoluble cell membranes: physiological concentrations of cholesterol and sphingolipid induce formation of a detergent-insoluble, liquid-ordered lipid phase in model membranes. Biochemistry 36:10944–10953. [DOI] [PubMed] [Google Scholar]
- Akhtar MK, Kelly SL, Kaderbhai MA. (2005) Cytochrome b(5) modulation of 17alpha hydroxylase and 17-20 lyase (CYP17) activities in steroidogenesis. J Endocrinol 187:267–274. [DOI] [PubMed] [Google Scholar]
- Anzenbacher P, Anzenbacherová E. (2001) Cytochromes P450 and metabolism of xenobiotics. Cell Mol Life Sci 58:737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auchus RJ, Lee TC, Miller WL. (1998) Cytochrome b5 augments the 17,20-lyase activity of human P450c17 without direct electron transfer. J Biol Chem 273:3158–3165. [DOI] [PubMed] [Google Scholar]
- Balaz S. (2009) Modeling kinetics of subcellular disposition of chemicals. Chem Rev 109:1793–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balvers WG, Boersma MG, Vervoort J, Ouwehand A, Rietjens IM. (1993) A specific interaction between NADPH-cytochrome reductase and phosphatidylserine and phosphatidylinositol. Eur J Biochem 218:1021–1029. [DOI] [PubMed] [Google Scholar]
- Baylon JL, Lenov IL, Sligar SG, Tajkhorshid E. (2013) Characterizing the membrane-bound state of cytochrome P450 3A4: structure, depth of insertion, and orientation. J Am Chem Soc 135:8542–8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berka K, Hendrychová T, Anzenbacher P, Otyepka M. (2011) Membrane position of ibuprofen agrees with suggested access path entrance to cytochrome P450 2C9 active site. J Phys Chem A 115:11248–11255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berka K, Paloncýová M, Anzenbacher P, Otyepka M. (2013) Behavior of human cytochromes P450 on lipid membranes. J Phys Chem B 117:11556–11564. [DOI] [PubMed] [Google Scholar]
- Berkowitz ML. (2009) Detailed molecular dynamics simulations of model biological membranes containing cholesterol. Biochim Biophys Acta 1788:86–96. [DOI] [PubMed] [Google Scholar]
- Black SD. (1992) Membrane topology of the mammalian P450 cytochromes. FASEB J 6:680–685. [DOI] [PubMed] [Google Scholar]
- Black SD, Coon MJ. (1982) Structural features of liver microsomal NADPH-cytochrome P-450 reductase: hydrophobic domain, hydrophilic domain, and connecting region. J Biol Chem 257:5929–5938. [PubMed] [Google Scholar]
- Bridges A, Gruenke L, Chang YT, Vakser IA, Loew G, Waskell L. (1998) Identification of the binding site on cytochrome P450 2B4 for cytochrome b5 and cytochrome P450 reductase. J Biol Chem 273:17036–17049. [DOI] [PubMed] [Google Scholar]
- Brignac-Huber L, Reed JR, Backes WL. (2011) Organization of NADPH-cytochrome P450 reductase and CYP1A2 in the endoplasmic reticulum--microdomain localization affects monooxygenase function. Mol Pharmacol 79:549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brignac-Huber LM, Reed JR, Eyer MK, Backes WL. (2013) Relationship between CYP1A2 localization and lipid microdomain formation as a function of lipid composition. Drug Metab Dispos 41:1896–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browman DT, Resek ME, Zajchowski LD, Robbins SM. (2006) Erlin-1 and erlin-2 are novel members of the prohibitin family of proteins that define lipid-raft-like domains of the ER. J Cell Sci 119:3149–3160. [DOI] [PubMed] [Google Scholar]
- Brown DA, London E. (2000) Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J Biol Chem 275:17221–17224. [DOI] [PubMed] [Google Scholar]
- Causey KM, Eyer CS, Backes WL. (1990) Dual role of phospholipid in the reconstitution of cytochrome P-450 LM2-dependent activities. Mol Pharmacol 38:134–142. [PubMed] [Google Scholar]
- Cojocaru V, Balali-Mood K, Sansom MSP, Wade RC. (2011) Structure and dynamics of the membrane-bound cytochrome P450 2C9. PLOS Comput Biol 7:e1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cojocaru V, Winn PJ, Wade RC. (2007) The ins and outs of cytochrome P450s. Biochim Biophys Acta 1770:390–401. [DOI] [PubMed] [Google Scholar]
- Davydov DR. (2011) Microsomal monooxygenase as a multienzyme system: the role of P450-P450 interactions. Expert Opin Drug Metab Toxicol 7:543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov DR, Davydova NY, Sineva EV, Halpert JR, Halpert JR. (2015) Interactions among cytochromes P450 in microsomal membranes: oligomerization of cytochromes P450 3A4, 3A5, and 2E1 and its functional consequences. J Biol Chem 290:3850–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov DR, Sineva EV, Sistla S, Davydova NY, Frank DJ, Sligar SG, Halpert JR. (2010) Electron transfer in the complex of membrane-bound human cytochrome P450 3A4 with the flavin domain of P450BM-3: the effect of oligomerization of the heme protein and intermittent modulation of the spin equilibrium. Biochim Biophys Acta 1797:378–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisov IG, Shih AY, Sligar SG. (2012) Structural differences between soluble and membrane bound cytochrome P450s. J Inorg Biochem 108:150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisov IG, Sligar SG. (2011) Cytochromes P450 in nanodiscs. Biochim Biophys Acta 1814:223–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo S, Escher BI, Goss KU. (2011) Capacities of membrane lipids to accumulate neutral organic chemicals. Environ Sci Technol 45:5912–5921. [DOI] [PubMed] [Google Scholar]
- Estrada DF, Laurence JS, Scott EE. (2013) Substrate-modulated cytochrome P450 17A1 and cytochrome b5 interactions revealed by NMR. J Biol Chem 288:17008–17018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada DF, Laurence JS, Scott EE. (2016) Cytochrome P450 17A1 interactions with the FMN domain of its reductase as characterized by NMR. J Biol ChemDOI: 291:3990–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada DF, Skinner AL, Laurence JS, Scott EE. (2014) Human cytochrome P450 17A1 conformational selection: modulation by ligand and cytochrome b5. J Biol Chem 289:14310–14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans WE, Relling MV. (1999) Pharmacogenomics: translating functional genomics into rational therapeutics. Science 286:487–491. [DOI] [PubMed] [Google Scholar]
- Finn RD, McLaughlin LA, Hughes C, Song C, Henderson CJ, Roland Wolf C. (2011) Cytochrome b5 null mouse: a new model for studying inherited skin disorders and the role of unsaturated fatty acids in normal homeostasis. Transgenic Res 20:491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, McLaughlin LA, Ronseaux S, Rosewell I, Houston JB, Henderson CJ, Wolf CR. (2008) Defining the in Vivo Role for cytochrome b5 in cytochrome P450 function through the conditional hepatic deletion of microsomal cytochrome b5. J Biol Chem 283:31385–31393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller DH, Auchus RJ, Miller WL. (1999) P450c17 mutations R347H and R358Q selectively disrupt 17,20-lyase activity by disrupting interactions with P450 oxidoreductase and cytochrome b5. Mol Endocrinol 13:167–175. [DOI] [PubMed] [Google Scholar]
- Ghosh MC, Ray AK. (2013) Membrane phospholipid augments cytochrome P4501a enzymatic activity by modulating structural conformation during detoxification of xenobiotics. PLoS One 8:e57919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich FP. (2006) Cytochrome P450s and other enzymes in drug metabolism and toxicity. AAPS J 8:E101–E111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Kapelyukh Y, Tahara H, Seibler J, Rode A, Krueger S, Lee DN, Wolf CR, Scheer N. (2011) Quantitative prediction of human pregnane X receptor and cytochrome P450 3A4 mediated drug-drug interaction in a novel multiple humanized mouse line. Mol Pharmacol 80:518–528. [DOI] [PubMed] [Google Scholar]
- Hazai E, Kupfer D. (2005) Interactions between CYP2C9 and CYP2C19 in reconstituted binary systems influence their catalytic activity: possible rationale for the inability of CYP2C19 to catalyze methoxychlor demethylation in human liver microsomes. Drug Metab Dispos 33:157–164. [DOI] [PubMed] [Google Scholar]
- Head BP, Patel HH, Insel PA. (2014) Interaction of membrane/lipid rafts with the cytoskeleton: impact on signaling and function: membrane/lipid rafts, mediators of cytoskeletal arrangement and cell signaling. Biochim Biophys Acta 1838:532–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A, Simons K. (1975) Solubilization of membranes by detergents. Biochim Biophys Acta 415:29–79. [DOI] [PubMed] [Google Scholar]
- Henderson CJ, McLaughlin LA, Scheer N, Stanley LA, Wolf CR. (2015) Cytochrome b5 is a major determinant of human cytochrome P450 CYP2D6 and CYP3A4 activity in vivo. Mol Pharmacol 87:733–739. [DOI] [PubMed] [Google Scholar]
- Henderson CJ, McLaughlin LA, Wolf CR. (2013) Evidence that cytochrome b5 and cytochrome b5 reductase can act as sole electron donors to the hepatic cytochrome P450 system. Mol Pharmacol 83:1209–1217. [DOI] [PubMed] [Google Scholar]
- Hildebrandt A, Estabrook RW. (1971) Evidence for the participation of cytochrome b 5 in hepatic microsomal mixed-function oxidation reactions. Arch Biochem Biophys 143:66–79. [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. [DOI] [PubMed] [Google Scholar]
- Im SC, Waskell L. (2011) The interaction of microsomal cytochrome P450 2B4 with its redox partners, cytochrome P450 reductase and cytochrome b(5). Arch Biochem Biophys 507:144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelman-Sundberg M, Glaumann H. (1980) Incorporation of purified components of the rabbit liver microsomal hydroxylase system into phospholipid vesicles. Biochim Biophys Acta 599:417–435. [DOI] [PubMed] [Google Scholar]
- Ingelman-Sundberg M, Hagbjörk AL, Ueng YF, Yamazaki H, Guengerich FP. (1996) High rates of substrate hydroxylation by human cytochrome P450 3A4 in reconstituted membranous vesicles: influence of membrane charge. Biochem Biophys Res Commun 221:318–322. [DOI] [PubMed] [Google Scholar]
- Isin EM, Guengerich FP. (2008) Substrate binding to cytochromes P450. Anal Bioanal Chem 392:1019–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson I, Schenkman JB. (1987) Influence of cytochrome b5 on the stoichiometry of the different oxidative reactions catalyzed by liver microsomal cytochrome P-450. Drug Metab Dispos 15:344–348. [PubMed] [Google Scholar]
- Jeffcoat R, Brawn PR, Safford R, James AT. (1977) Properties of rat liver microsomal stearoyl-coenzyme A desaturase. Biochem J 161:431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EF, Connick JP, Reed JR, Backes WL, Desai MC, Xu L, Estrada DF, Laurence JS, Scott EE. (2014) Correlating structure and function of drug-metabolizing enzymes: progress and ongoing challenges. Drug Metab Dispos 42:9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EF, Stout CD. (2013) Structural diversity of eukaryotic membrane cytochrome p450s. J Biol Chem 288:17082–17090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley RW, Cheng D, Backes WL. (2006) Heteromeric complex formation between CYP2E1 and CYP1A2: evidence for the involvement of electrostatic interactions. Biochemistry 45:15807–15816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian JR, Longlais BJ, Trepanier LA. (2007) Discovery and characterization of a cytochrome b5 variant in humans with impaired hydroxylamine reduction capacity. Pharmacogenet Genomics 17:597–603. [DOI] [PubMed] [Google Scholar]
- London E, Brown DA. (2000) Insolubility of lipids in triton X-100: physical origin and relationship to sphingolipid/cholesterol membrane domains (rafts). Biochim Biophys Acta 1508:182–195. [DOI] [PubMed] [Google Scholar]
- Lúcio M, Lima JLFC, Reis S. (2010) Drug-membrane interactions: significance for medicinal chemistry. Curr Med Chem 17:1795–1809. [DOI] [PubMed] [Google Scholar]
- McKenna JA, Sacco J, Son TT, Trepanier LA, Callan MB, Harvey JW, Arndt JW. (2014) Congenital methemoglobinemia in a dog with a promoter deletion and a nonsynonymous coding variant in the gene encoding cytochrome b₅. J Vet Intern Med 28:1626–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin LA, Ronseaux S, Finn RD, Henderson CJ, Roland Wolf C. (2010) Deletion of microsomal cytochrome b5 profoundly affects hepatic and extrahepatic drug metabolism. Mol Pharmacol 78:269–278. [DOI] [PubMed] [Google Scholar]
- Naffin-Olivos JL, Auchus RJ. (2006) Human cytochrome b5 requires residues E48 and E49 to stimulate the 17,20-lyase activity of cytochrome P450c17. Biochemistry 45:755–762. [DOI] [PubMed] [Google Scholar]
- Nagar S, Korzekwa K. (2012) Commentary: nonspecific protein binding versus membrane partitioning: it is not just semantics. Drug Metab Dispos 40:1649–1652. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Gang HX, Suzuki T, Sasano H, Rainey WE. (2009) Adrenal changes associated with adrenarche. Rev Endocr Metab Disord 10:19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura T, Sato R. (1962) A new cytochrome in liver microsomes. J Biol Chem 237:1375–1376. [PubMed] [Google Scholar]
- Omura T, Sato R, Cooper DY, Rosenthal O, Estabrook RW. (1965) Function of cytochrome P-450 of microsomes. Fed Proc 24:1181–1189. [PubMed] [Google Scholar]
- Ortiz de Montellano PR (2015) Cytochrome P450: Structure, Mechanism, and Biochemistry. Springer International Publishing, Switzerland. [Google Scholar]
- Otyepka M, Skopalík J, Anzenbacherová E, Anzenbacher P. (2007) What common structural features and variations of mammalian P450s are known to date? Biochim Biophys Acta 1770:376–389. [DOI] [PubMed] [Google Scholar]
- Ovesný M, Křížek P, Borkovec J, Svindrych Z, Hagen GM. (2014) ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics 30:2389–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen DM, Rentero C, Rossy J, Magenau A, Williamson D, Rodriguez M, Gaus K. (2010) PALM imaging and cluster analysis of protein heterogeneity at the cell surface. J Biophotonics 3:446–454. [DOI] [PubMed] [Google Scholar]
- Pallan PS, Nagy LD, Lei L, Gonzalez E, Kramlinger VM, Azumaya CM, Wawrzak Z, Waterman MR, Guengerich FP, Egli M. (2015) Structural and kinetic basis of steroid 17α,20-lyase activity in teleost fish cytochrome P450 17A1 and its absence in cytochrome P450 17A2. J Biol Chem 290:3248–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paloncýová M, Berka K, Otyepka M. (2013) Molecular insight into affinities of drugs and their metabolites to lipid bilayers. J Phys Chem B 117:2403–2410. [DOI] [PubMed] [Google Scholar]
- Paloncýová M, DeVane R, Murch B, Berka K, Otyepka M. (2014) Amphiphilic drug-like molecules accumulate in a membrane below the head group region. J Phys Chem B 118:1030–1039. [DOI] [PubMed] [Google Scholar]
- Park JW, Reed JR, Brignac-Huber LM, Backes WL. (2014) Cytochrome P450 system proteins reside in different regions of the endoplasmic reticulum. Biochem J 464:241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JW, Reed JR, Backes WL. (2015) The Localization of Cytochrome P450s CYP1A1 and CYP1A2 into Different Lipid Microdomains is Governed by their NH2-terminal and Internal Protein Regions. J Biol Chem 290:29449–29460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson JA, Ebel RE, O’Keeffe DH, Matsubara T, Estabrook RW. (1976) Temperature dependence of cytochrome P-450 reduction. A model for NADPH-cytochrome P-450 reductase:cytochrome P-450 interaction. J Biol Chem 251:4010–4016. [PubMed] [Google Scholar]
- Petrek M, Kosinová P, Koca J, Otyepka M. (2007) MOLE: a Voronoi diagram-based explorer of molecular channels, pores, and tunnels. Structure 15:1357–1363. [DOI] [PubMed] [Google Scholar]
- Petrek M, Otyepka M, Banás P, Kosinová P, Koca J, Damborský J. (2006) CAVER: a new tool to explore routes from protein clefts, pockets and cavities. BMC Bioinformatics 7:316–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike LJ. (2009) The challenge of lipid rafts. J Lipid Res 50 (Suppl):S323–S328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochapsky TC, Kazanis S, Dang M. (2010) Conformational plasticity and structure/function relationships in cytochromes P450. Antioxid Redox Signal 13:1273–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poudel KR, Jones JP, Brozik JA. (2013) A guide to tracking single transmembrane proteins in supported lipid bilayers. Methods Mol Biol 974:233–252. [DOI] [PubMed] [Google Scholar]
- Reed JR, Backes WL. (2012) Formation of P450 · P450 complexes and their effect on P450 function. Pharmacol Ther 133:299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JR, Eyer M, Backes WL. (2010) Functional interactions between cytochromes P450 1A2 and 2B4 require both enzymes to reside in the same phospholipid vesicle: evidence for physical complex formation. J Biol Chem 285:8942–8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JR, Kelley RW, Backes WL. (2006) An evaluation of methods for the reconstitution of cytochromes P450 and NADPH P450 reductase into lipid vesicles. Drug Metab Dispos 34:660–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendic SP, Guengerich FP. (2015) Survey of Human Oxidoreductases and Cytochrome P450 Enzymes Involved in the Metabolism of Chemicals. Chem Res Toxicol 28:38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock D, Rock D, Jones JP. (2001) Inexpensive purification of P450 reductase and other proteins using 2′,5′-adenosine diphosphate agarose affinity columns. Protein Expr Purif 22:82–83. [DOI] [PubMed] [Google Scholar]
- Sansen S, Yano JK, Reynald RL, Schoch GA, Griffin KJ, Stout CD, Johnson EF. (2007) Adaptations for the oxidation of polycyclic aromatic hydrocarbons exhibited by the structure of human P450 1A2. J Biol Chem 282:14348–14355. [DOI] [PubMed] [Google Scholar]
- Scheer N, Kapelyukh Y, McEwan J, Beuger V, Stanley LA, Rode A, Wolf CR. (2012) Modeling human cytochrome P450 2D6 metabolism and drug-drug interaction by a novel panel of knockout and humanized mouse lines. Mol Pharmacol 81:63–72. [DOI] [PubMed] [Google Scholar]
- Seddon AM, Casey D, Law RV, Gee A, Templer RH, Ces O. (2009) Drug interactions with lipid membranes. Chem Soc Rev 38:2509–2519. [DOI] [PubMed] [Google Scholar]
- Sgrignani J, Magistrato A. (2012) Influence of the membrane lipophilic environment on the structure and on the substrate access/egress routes of the human aromatase enzyme: a computational study. J Chem Inf Model 52:1595–1606. [DOI] [PubMed] [Google Scholar]
- Shimoji M, Yin H, Higgins L, Jones JP. (1998) Design of a novel P450: a functional bacterial-human cytochrome P450 chimera. Biochemistry 37:8848–8852. [DOI] [PubMed] [Google Scholar]
- Smith D, Artursson P, Avdeef A, Di L, Ecker GF, Faller B, Houston JB, Kansy M, Kerns EH, Krämer SD, et al. (2014) Passive lipoidal diffusion and carrier-mediated cell uptake are both important mechanisms of membrane permeation in drug disposition. Mol Pharm 11:1727–1738. [DOI] [PubMed] [Google Scholar]
- Subramanian M, Low M, Locuson CW, Tracy TS. (2009) CYP2D6-CYP2C9 protein-protein interactions and isoform-selective effects on substrate binding and catalysis. Drug Metab Dispos 37:1682–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian M, Tam HZhang H, Tracy TS.(2010) CYP2C9-CYP3A4 protein-protein interactions: role of the hydrophobic N terminus. Drug Metab Dispos 38:1003–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait JF, Gibson D. (1992) Phospholipid binding of annexin V: effects of calcium and membrane phosphatidylserine content. Arch Biochem Biophys 298:187–191. [DOI] [PubMed] [Google Scholar]
- Williams CH, Jr, Kamin H. (1962) Microsomal triphosphopyridine nucleotide-cytochrome c reductase of liver. J Biol Chem 237:587–595. [PubMed] [Google Scholar]
- Williams PA, Cosme J, Ward A, Angove HC, Matak Vinković D, Jhoti H. (2003) Crystal structure of human cytochrome P450 2C9 with bound warfarin. Nature 424:464–468. [DOI] [PubMed] [Google Scholar]
- Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF. (2004) The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-A resolution. J Biol Chem 279:38091–38094. [DOI] [PubMed] [Google Scholar]
- Yu X, Cojocaru V, Mustafa G, Salo-Ahen OMH, Lepesheva GI, Wade RC. (2015) Dynamics of CYP51: implications for function and inhibitor design. J Mol Recognit 28:59–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanger UM, Schwab M. (2013) Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 138:103–141. [DOI] [PubMed] [Google Scholar]
- Zhang H, Hamdane D, Im SC, Waskell L. (2008) Cytochrome b5 inhibits electron transfer from NADPH-cytochrome P450 reductase to ferric cytochrome P450 2B4. J Biol Chem 283:5217–5225. [DOI] [PubMed] [Google Scholar]
- Zhang H, Im SC, Waskell L. (2007) Cytochrome b5 increases the rate of product formation by cytochrome P450 2B4 and competes with cytochrome P450 reductase for a binding site on cytochrome P450 2B4. J Biol Chem 282:29766–29776. [DOI] [PubMed] [Google Scholar]
- Zhao C, Gao Q, Roberts AG, Shaffer SA, Doneanu CE, Xue S, Goodlett DR, Nelson SD, Atkins WM. (2012) Cross-linking mass spectrometry and mutagenesis confirm the functional importance of surface interactions between CYP3A4 and holo/apo cytochrome b(5). Biochemistry 51:9488–9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.