Abstract
Selenium (Se) plays a critical role in testis, sperm, and reproduction, and testis Se levels are remarkably maintained in Se deficiency. In most other tissues, Se levels decrease dramatically as do levels of most selenoproteins and levels of a subset of Se-regulated selenoprotein mRNAs. Because of the recent identification of key molecules in the targeted trafficking of Se to the testis, we examined the hierarchy of Se regulation in testis by determining the dietary Se regulation of the full testis selenoproteome in rats fed graded levels of Se (0 to 0.8 μg Se/g) as Na2SeO3 for 28 d. Se status did not significantly affect testis weight or glutathione peroxidase-4 (Gpx4) activity (P > 0.05). qRT-PCR analysis of selenoprotein mRNA expression revealed that 21 of the 24 selenoprotein mRNAs and ApoER2 mRNA (the selenoprotein P (Sepp1) receptor) were also not regulated significantly by dietary Se status. In contrast, Gpx1 activity decreased to 28% of Se-adequate levels, and mRNA levels for Gpx1, Sepp1 and Sepw1 (selenoprotein W) decreased significantly in Se-deficient rats to 45, 46, and 55%, respectively, of Se-adequate plateau levels. Overlap of hyperbolic Gpx4 activity and Sepw1 mRNA response curves with testis Se concentration, all with minimum dietary Se requirements <0.016 μg Se/g, showed the priority for synthesis of Gpx4. Higher minimum dietary Se requirements of 0.04 μg Se/g for Gpx1 activity and Sepp1 mRNA, and the even higher minimum dietary Se requirement of 0.08 μg Se/g for Gpx1 mRNA, suggest that the hierarchy of these biomarkers reflects distinct, lower priority pools, cell-types and roles for Se within the testis.
Keywords: ApoER2, glutathione peroxidase, qRT-PCR, mRNA, selenoprotein P, selenoprotein W, selenoproteome
Introduction
Glutathione peroxidase-1 (Gpx14) activity, protein, and mRNA levels, as well as selenium (Se) concentrations, all fall exponentially, dramatically, and coordinately in liver to <10% of Se-adequate levels when rats are fed a Se-deficient diet (1-4). In contrast, liver Gpx4 activities only decrease to about 40% of Se-adequate levels, and liver Gpx4 mRNA levels do not decrease in the same animals (5). With the identification of all 24 rodent selenoproteins in the complete selenoprotein proteome (6), microarray analysis and qRT-PCR characterization of Se regulation of the complete selenoproteome in mouse liver and kidney found that mRNA levels for Selh and Sepw1 as well as Gpx1 were highly down-regulated by Se deficiency in mice (7). More recently, assessment of Se regulation of the complete selenoproteome in rats supplemented with graded levels of Se from deficient to super-nutritional (0 - 0.8 μg Se/g diet) found that 5 selenoprotein mRNAs in liver, 4 in kidney, and 2 in muscle decreased in Se deficiency to <41% of Se-adequate levels, but that the majority of selenoprotein mRNAs in each tissue were not significantly regulated by Se status (8). For these Se-regulated selenoprotein mRNAs, the minimum dietary Se levels necessary to raise mRNA levels to Se-adequate levels were all at 0.03-0.07 μg Se/g diet, and all were less than dietary Se levels necessary for plateau levels of liver and kidney Se (0.08-0.11 μg Se/g) (8).
Se plays a critical role in testis, sperm, and reproduction. Rodent testis Se concentrations are typically higher than for any other tissue except kidney (9). Early descriptions of the effects of Se deficiency reported that second-generation Se-deficient rats were sterile and lacked sperm or possessed immotile spermatozoa with separated heads and tails (10,11), and reported that Se-deficient rats labeled with 75Se preferentially retained 75Se in testis and concentrated this retained 75Se in the midpiece of sperm (12). Notably, total Se concentrations in rat testis, unlike other most other tissues except brain and endocrine tissues, generally do not decrease even with prolonged Se deficiency (13,14), showing that Se is preferentially maintained in the rodent testis (13,15). While Se in sperm was initially thought to be present associated with mitochondrial capsule protein, it is now clear that Gpx4 serves as a structural protein necessary for the integrity of the midpiece of mature sperm (16), and that the vast majority of the Se in testis is incorporated into Gpx4 (16-18). In humans as well, Gpx4 content of sperm is positively correlated with sperm viability, morphological integrity, and motility (19). Se-deficient rats rapidly incorporate injected carrier-free 75Se into testis such that 25% of the whole body 75Se is present as 75Se-labeled Gpx4 in the testis 24-72 hr post-injection (20), further illustrating the targeted delivery of Se to testis and to Gpx4.
Recent characterization in rodent models of the role of selenoprotein P (Sepp1) is rapidly unraveling the mechanism underlying the targeted trafficking of Se to testis. Deletion of the Sepp1 gene in mice results in marked decreases in testis Se even in mice supplemented with normally adequate or super-nutritional levels of Se (21,22), and results in sperm defects indistinguishable from those elicited by dietary Se deficiency (23,24). A Sepp1-specific receptor has recently been identified as the apolipoprotein E receptor (ApoER2) (25). ApoER2 is expressed in testis as well as brain, and deletion of ApoER2 in mice results in marked reduction in testis Se concentration and in sperm defects that are identical to those present in Sepp1-deletion mice or in Se-deficient mice and rats (25,26). Lastly, transgenic restoration of Sepp1 expression in just liver is fully sufficient to restore Se uptake by the testis, and to prevent the fragile sperm phenotype infertility (24). Thus we have a much improved molecular understanding of the targeted Se delivery to testis.
Because of this recent identification of Sepp1 and ApoER2 as key molecules in the targeted trafficking of Se to the testis, we decided to characterize the dietary Se regulation of the full selenoproteome in the testis using tissues we previously had harvested from rats in an earlier study (8). Our hypotheses were that the number of selenoproteome mRNAs that are regulated by Se status in testis would be substantially reduced relative to liver and kidney, and that the minimal dietary Se requirement of Se-regulated mRNAs would be substantially lowered due to the targeted maintenance of Se status in testis, but also that novel regulation patterns might emerge due to differential delivery of Se due to these targeting mechanisms.
Material and Methods
Reagents
Molecular biology reagents were purchased from Promega (Madison, WI), Invitrogen (Carlsbad, CA) or Sigma (St. Louis, MO). All other chemicals were of molecular biology or reagent grade.
Animals and diets
Sixty male weanling rats, 21 days old, were obtained from Holtzman (Madison, WI) and housed individually in hanging wire-mesh cages, following the care and treatment protocol approved by the Institutional Animal Care and Use Committee at the University of Wisconsin. Rats were fed the basal Se-deficient torula yeast-based diet (0.005 μg Se/g by analysis) supplemented with 100 mg/kg of all-rac-α-tocopherol acetate to ensure prevention of liver necrosis, and supplemented with 0.4% L-methionine to ensure adequate growth, as described previously (3,4). The rats were allocated randomly to treatment groups, and supplemented with graded levels of Se (0, 0.016, 0.04, 0.06, 0.08, 0.12, 0.16, 0.24, 0.4 or 0.8 μg Se/g) as Na2SeO3 for 28 d (6 rats/treatment). Body weight was measured bi-weekly. Animals had free access to feed and water.
Tissue analysis
These animals were part of a larger study (8) where the rats were anesthetized with ether, and the testis were removed and quick-frozen in liquid nitrogen for later analysis. Testis and dietary Se concentrations were determined by neutron activation analysis (27).
Enzyme assays
Testis supernatant was prepared by homogenization in 9 volumes (w/vol) of sucrose buffer (20 mM tris/HCl, pH 7.4, 0.25 M sucrose, 1.1 mM EDTA and 0.1% peroxide-free Triton X 100) and centrifuged (10,000 × g, 15 min, 4°C, model J2-21M, JA-21 rotor, Beckman Instruments, Palo Alto, CA), as described previously (4,5). Gpx4 activity was measured by the coupled assay procedure (5) using 78 μM phosphatidyl choline hydroperoxide (PCOOH), the specific substrate, and this activity is designated as Gpx4 activity. Substrate synthesis was modified from the described method (5) by using 30 mg L-α-phosphatidylcholine for a higher yield. In the same samples, Gpx1 activity was measured by the coupled assay procedure (28) using 120 μM H2O2, and corrected for the contribution due to Gpx4 (see below); this corrected activity is designated as Gpx1 activity. For both assays, 1 enzyme unit (EU) is the amount of enzyme that will oxidize 1 μmole GSH/min under these conditions. Protein concentrations were determined by the method of Lowry et al. (29).
Partial purification of testis Gpx4
Because rat testis have very high levels of Gpx4, and because H2O2 is also a good substrate for Gpx4 (30), the conventional assay for Gpx1 when used for rodent testis also includes substantial Gpx4-catalyzed activity. Anion exchange chromatography was used to purify rat testis Gpx4 away from Gpx1 (31,32). Briefly, testis from 3 adult Se-adequate Holtzman rats were homogenized in 3 vol of homogenization buffer (0.1 M Tris-HCl, 5 mM β-mercaptoethanol, pH 7.4) and centrifuged (10,000 × g, 30 min, 4°C, model J2-21M, JA-21 rotor, Beckman Instruments, followed by 100,000 × g, 60 min, 4°C, model L8-70M ultracentrifuge, rotor 70.1 TI, Beckman Instruments). The supernatant was dialyzed against equilibration buffer (10 mM potassium phosphate buffer, 5 mM β-mercaptoethanol, pH 7.5), applied to a DEAE-Sepharose 6B column (2.6 × 35 cm), and unbound proteins including Gpx1 were eluted with 5 column vol of equilibration buffer. Gpx4 was eluted with elution buffer (0.2 M potassium phosphate buffer, 0.5 mM β-mercaptoethanol, pH 7.0), and fractions assayed with both H2O2 and PCOOH following the standard assay conditions.
RNA isolation and analysis
Total RNA was isolated from rat testis (50-100 mg tissue, n=3/diet group) by homogenization in TRIzol Reagent (Catalog #15596-026, Invitrogen, Carlsbad, CA) followed by extraction with chloroform, microcentrifugation (12,100 x g, 15 min, 4°C) and precipitation with isopropanol following the manufacturer's protocol as previously described (8). The RNA pellet was dissolved in diethyl pyrocarbonate-treated water and quantitated spectrophotometrically by A260 (ND-1000 UV-Vis Spectrophotometer, NanoDrop Technologies, Wilmington, DE).
Quantitative real time polymerase chain reaction (qRT-PCR) analysis was used to measure relative mRNA abundance in testis. In a preliminary screen for dietary Se regulation in rat testis, transcript levels in total RNA for all 24 rat selenoproteins were determined by qRT-PCR for rats fed Se-deficient (0 μg Se/g), Se-adequate (0.16 μg Se/g) and super-nutritional Se (0.8 μg Se/g) diets using rat primers previously described (8). A complete analysis at all 10 levels of Se supplementation was conducted for the five selenoprotein mRNAs that were found to be down-regulated significantly by Se deficiency in this preliminary screen, as well as for mRNAs for Gpx4, Selh, Selk, Selv, and for the newly identified Sepp1 receptor, ApoER2 (forward primer 5′-TGTGCCTGTCCTGACACAAT-3′; reverse primer 5′-AGAGTCGTGGATGGTTGTCC-3′).
To determine relative mRNA abundance by qRT-PCR, RNA (1 μg) was reverse transcribed to cDNA using the RETROscript kit (AM1710, Ambion Inc., Austin, TX), following the manufacturer's instructions. Gene specific primers were designed to span a splice-junction and amplify ∼150 base segment (8). The final 25 μl real time reactions contained 10 ng reverse transcribed RNA, 0.2 mM gene specific forward and reverse primers, and 1X SybrGreen PCR Master Mix (#4309155, Applied Biosystems, Foster City, CA). Reactions were followed in an ABI Prism 7000 (Applied Biosystems) with initial stages of 50°C for 2 min and 95°C for 10 min, followed by 50 cycles consisting of 95°C for 15 sec and 60°C for 2 min. A dissociation curve was run for each plate to confirm the production of a single product. The amplification efficiency for each gene was determined using the DART-PCR program (33). The mRNA relative abundance was calculated according to Pfaffl (34), accounting for gene-specific efficiencies, normalized to the mean of β-Actin (Actb) and glyceraldehyde-3-phosphate dehydrogenase (Gapdh), and expressed as a percentage of Se-adequate levels (0.16 μg Se/g or plateau level).
Statistical Analysis
Data are presented as means ± SEM; n = 5 or 6/diet for testis enzyme activity analysis; n = 3/diet for mRNA analysis and Se concentrations. All data were analyzed by ANOVA using a fixed model testing the main effect of diet (SAS Inst. Inc., Cary, NC). When the main effect of diet was significant, differences between means were assessed by Duncan's multiple range analysis (P < 0.05), with Kramer's modification for unequal class sizes where necessary (35); for each parameter, variance of the means was tested by Bartlett's test (α = 0.05); no significant differences in variances were found for data presented here. For all tests, P < 0.05 was considered significant. The plateau breakpoint for each Se response curve, defined as the intersection of the line tangent to the point of steepest slope and the plateau, was calculated as described previously (3,4,36) using sigmoidal or hyperbolic regression analysis (Sigma Plot, Jandel Scientific) to estimate the minimum dietary Se necessary to obtain plateau responses.
Results
General experiment
This study on Se regulation of selenoprotein expression in testis was conducted on rats that were separately studied for Se regulation in liver, kidney and muscle (8). In this study, there was no significant effect of dietary Se on growth during the study (data not shown), nor on body weight at the end of the study (Table 1). As reported elsewhere, plasma Gpx3 activity, liver Se concentration, and liver Gpx1 activity in this study were decreased to 2, 3, and 2%, respectively, of levels in rats fed 0.24 μg Se/g diet, showing that the Se-deficient rats were Se deficient (8).
Table 1. Effect of dietary Se on final body weight and testis weight in ratsa.
| Diet | Body Weight | Testis Weight | Testis/Body Weight ratio |
|---|---|---|---|
|
| |||
| (μg Se/g) | (g) | (g) | % |
| 0 | 266 ± 11 | 2.40 ± 0.10 | 0.91 ± 0.06 |
| 0.016 | 274 ± 11 | 2.44 ± 0.10 | 0.90 ± 0.05 |
| 0.04 | 268 ± 8 | 2.20 ± 0.09 | 0.82 ± 0.04 |
| 0.06 | 275 ± 10 | 2.44 ± 0.15 | 0.90 ± 0.07 |
| 0.08 | 276 ± 7 | 2.38 ± 0.12 | 0.86 ± 0.04 |
| 0.12 | 269 ± 7 | 2.31 ± 0.17 | 0.86 ± 0.05 |
| 0.16 | 266 ± 9 | 2.33 ± 0.05 | 0.88 ± 0.04 |
| 0.24 | 278 ± 8 | 2.53 ± 0.04 | 0.91 ± 0.03 |
| 0.40 | 261 ± 8 | 2.20 ± 0.07 | 0.85 ± 0.04 |
| 0.80 | 267 ± 7 | 2.44 ± 0.09 | 0.91 ± 0.04 |
Values are expressed as means ± SEM. Each diet group contained n = 6 except the 0.16 μg Se/g group contained n=5. For body weight, testis weight, and testis/body weight ratio, ANOVA indicated P > 0.05.
Testis weight
The level of dietary Se did not affect testis weight (P > 0.05) (Table 1). Overall testis weight was 2.36 ± 0.03 g, and the ratio of testis weight to body weight was also not altered (P > 0.05) by dietary Se status.
Selenium in testis
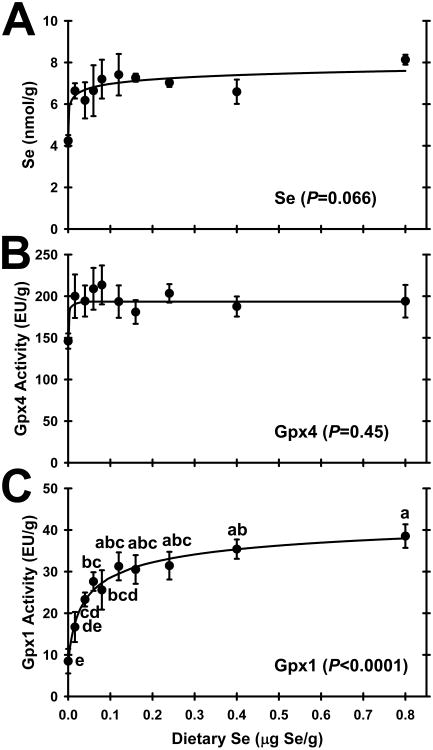
Testis Se concentrations in rats fed the Se-deficient diet were 58% of levels in rats fed the 0.16 μg Se/g diet (Fig. 1A). This difference was not significant by ANOVA (P = 0.066), but with increasing dietary Se supplementation, the Se response curve was hyperbolic (P <0.0025) with the plateau breakpoint at <0.016 μg/g diet (Table 2) such that rats fed diets containing 0.016 μg/g diet or higher were clearly on the plateau of the Se response curve.
Figure 1.
Effect of dietary Se on testis selenium concentration and enzyme activities. A. Testis Se concentration; B. Gpx4 activity; C. Gpx1 activity. Values are means ± SEM; n=3/diet for Se; n = 5 or 6/diet for Gpx4 and Gpx1. The level of significance by ANOVA is indicated in each panel; values with a common letter are not significantly different (P < 0.05). The testis Se response curve was significantly hyperbolic (P < 0.0025).
Table 2. Selenium requirement hierarchy in testis of growing rats.
| Biomarker | Minimum Requirementa | Extent of Regulationb |
|---|---|---|
|
| ||
| (μg Se/g) | ||
| Testis weight | <0.005 | Low (P = 0.40) |
| Gpx4 activity | <0.005 | Low (P = 0.45) |
| Selenium | <0.016 | Moderate (P = 0.066)c |
| Sepw1 mRNA | <0.016 | Moderate (P < 0.004) |
| Sepp1 mRNA | 0.04 | Moderate (P < 0.002) |
| Gpx1 activity | 0.04 | High (P < 0.0001) |
| Gpx1 mRNA | 0.08 | Moderate (P < 0.0001) |
Minimum dietary Se requirement for the growing rat as determined for each indicated biomarker. Requirements are the minimum dietary Se necessary for the indicated parameter to reach plateau levels when Se-adequate weanling rats are fed these diets from weaning, as determined by breakpoint analysis as described in the text.
Susceptibility to Se regulation of the indicated biomarker in Se-deficient rat tissue in Se-deficient versus Se-adequate rats: high = 11-40.9% of Se-adequate; moderate = 41-70% of Se-adequate; Low = >70% of Se-adequate. These ranges of regulation are the same as applied in previous studies (7,8). Significance of regulation (P-value for ANOVA) indicated in parentheses.
Testis Se response curve was significantly hyperbolic (P < 0.0025).
Enzyme activities
There was no significant effect (P = 0.45) of dietary Se level on testis Gpx4 activity (Fig. 1B). Because of the high abundance of Gpx4 in testis, Gpx activity in testis supernatants was measured using both the Gpx4-specific PCOOH substrate and assay, and the regular H2O2 substrate and assay. Under these conditions, the purified Gpx4 had a ratio of 0.26 ± 0.02 enzyme units assayed with H2O2 for every 1.0 enzyme unit assayed with PCOOH (data not shown), thus allowing calculation of Gpx1-specific enzyme activity in testis. In contrast to Gpx4 activity, testis Gpx1 activity was regulated significantly (P < 0.0001) by dietary Se (Fig. 1C). Gpx1 activity in Se-deficient rats was 28% of Se-adequate rats. The Se response curve for testis Gpx1 activity with increasing dietary Se was hyperbolic, with the steepest increase in activity between 0 and 0.016 μg Se/g diet, and with the plateau breakpoint at 0.04 μg Se/g diet. Super-nutritional levels of dietary Se (> 0.2 μg Se/g diet) did not further increase testis Gpx1 activity.
Selenoprotein mRNA transcript levels
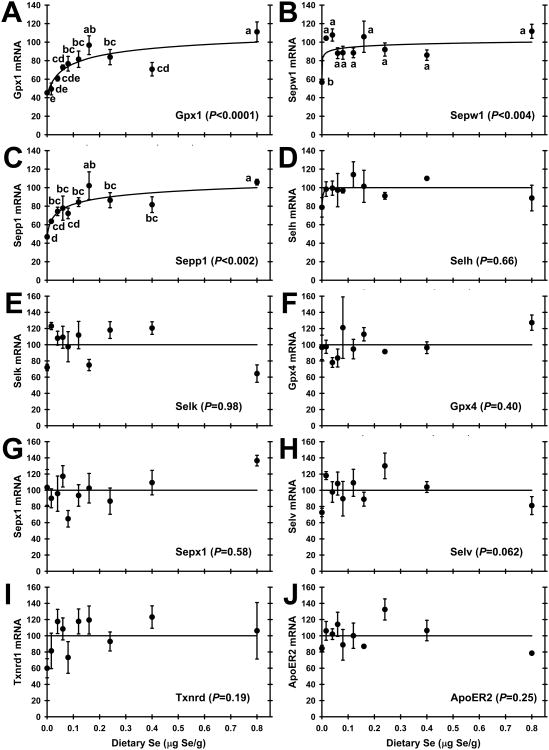
A preliminary screen using qRT-PCR for selenoprotein transcripts in total testis RNA from rats fed 0, 0.16 and 0.8 μg Se/g diet detected all 24 rodent selenoprotein mRNAs. Gpx4 mRNA, followed by Sepp1 and thioredoxin reductase-3 (Txnrd3) mRNA, appeared to be the most abundant based on relative cycle times. Seven selenoprotein mRNAs (Dio2, Dio3, Gpx2, Gpx3, Selm, Selo, and Sepn1), however, were present at less than 0.2% of apparent Gpx4 mRNA abundance (data not shown). There was no significant effect of Se status on levels of the control transcripts for Gapdh and Actb (data not shown). Quantitation of selenoprotein mRNA expression revealed that normalized levels of 19 of the 24 selenoprotein mRNAs in the initial screen were not regulated significantly by dietary Se (P > 0.05), whereas mRNAs for Gpx1, Sepx1, Sepp1, Sepw1, and Txnrd1 were found to be regulated significantly by Se (data not shown). Full qRT-PCR analyses of testis RNA from the 10 dietary treatments were conducted for 10 transcripts; the five significantly regulated mRNAs in the preliminary screen; two mRNAs (Selh, Selk) found to be regulated significantly by Se status in other tissues (8); one mRNA (Selv) reported to be expressed uniquely in testis; one mRNA (Gpx4) which is highly abundant in testis but not regulated by Se status in other tissues; ApoER2 mRNA which codes for the Sepp1 receptor found in testis. Testis Gpx1 mRNA expression (Fig. 2A) was regulated significantly by dietary Se (P < 0.0001). In Se-deficient rats, testis Gpx1 mRNA levels were reduced to 45% of plateau levels. There was a sigmoidal response in Gpx1 mRNA expression with increasing dietary Se supplementation; the steepest increase in Gpx1 mRNA levels occurred between 0.016 and 0.06 μg Se/g diet, with a plateau breakpoint at 0.08 μg Se/g diet, and super-nutritional levels of dietary Se above 0.24 μg Se/g diet did not significantly further increase Gpx1 mRNA levels. In testis, Sepw1 mRNA expression (Fig. 2B) was also moderately down-regulated in Se-deficiency to 55% of Se-adequate plateau levels (P < 0.004), with a plateau breakpoint at <0.016 μg Se/g diet. In testis, Sepp1 mRNA levels (Fig. 2C) decreased significantly in Se deficiency to 46% of Se-adequate plateau levels (P < 0.002), and increased with increasing dietary Se in a hyperbolic response with a plateau breakpoint at 0.04 μg Se/g diet.
Figure 2.
Effect of dietary Se concentration on selenoprotein transcript abundance. A. Gpx1; B. Sepw1; C. Sepp1; D. Selh; E. Selk; F. Gpx4; G. Sepx1; H. Selv; I. Txnrd1; J. ApoER2. Values are means ± SEM; n = 3/diet. Gpx1, Sepw1, and Sepp1 mRNA levels were significantly regulated by dietary Se level at the indicated level of significance; values with a common letter are not significantly different (P < 0.05). For transcripts that were not significantly regulated by dietary Se, a line in each panel indicated the mean of all normalized values.
Analysis of testis RNA from the ten dietary treatments, however, did not reveal significant decreases in mRNA for 6 additional selenoprotein mRNAs (Fig. 2D-2I). This included Selh, which was decreased significantly to <40% of Se-adequate levels in liver and kidney, and Selk which was decreased significantly to <40% of Se-adequate levels in liver (8). Gpx4 mRNA, the most abundant selenoprotein mRNA in testis, as well as mRNA levels for Sepx1, Selv, and Txnrd1 showed no decreases in Se deficiency, or significant increases with super-nutritional levels of Se supplementation.
Transcript levels for ApoER2, the Sepp1 receptor in testis, were also assayed. ApoER2 mRNA expression was detected in total RNA from rat testis, but was there no indication that dietary Se status regulated the expression of ApoER2 in rats fed Se-deficient or super-nutritional levels of Se (Fig. 2J).
Discussion
In this study, 21-day old male weanling rats were fed a Se-deficient diet for 28 days or supplemented with graded levels of Se from marginal to super-nutritional (0.016 to 0.8 μg Se/g diet), as reported elsewhere (8). Just as in a number of previous studies (3,5,36), regardless of level of Se supplementation, there was no significant effect of dietary Se on growth. Plasma Gpx3 activity, liver Se concentration, and liver Gpx1 activity in rats fed the Se-deficient basal diet, however, were decreased to 2, 3, and 2%, respectively, of levels in rats fed 0.24 μg Se/g diet, showing that these rats were Se deficient (8).
Reported here, there was no effect of Se deficiency or super-nutritional Se supplementation on testis weight. Like many previous studies, there was also no significant effect of Se deficiency (9,13) or super-nutritional Se supplementation (37) on testis Se concentrations, although rats fed the basal diet (0.005 μg Se/g) in this study had 58% of levels in Se-adequate rats; longer Se-deficiency studies have reported significant decreases in testis Se content (15,38). The plateau Se concentration of 7.0 ± 0.2 nmol Se/g (average for rats fed Se 0.016 μg Se/g and higher) in these 49-day old rats was intermediate between levels of 5.6 nmol Se/g reported in 45-day old rats and 9.0 nmol Se/g reported in 55-day old rats supplemented with 0.25 μg Se/g diet as selenite (39). Testis Gpx4 activity in this study was not significantly reduced in Se-deficient rats, and super-nutritional levels of dietary Se did not further increase Gpx4 activity. Gpx4 activities in liver and kidney but not muscle in this study (8) and in other rodent studies (40) typically fall to 30-70% of Se-adequate levels. In previous studies in our lab using a crystalline amino acid-based diet containing 0.002 μg Se/g, we also found a non-significant reduction in testis Gpx4 activity (5), whereas testis Gpx4 activity can fall significantly to 13% of Se-adequate levels in second-generation Se-deficient rats that grow at half the rate of Se-supplemented littermates (41). Thus this study affirmed the remarkable retention of Se and Gpx4 in testis in Se-deficient rats.
For this study, we purified testis Gpx4 away from Gpx1 and determined the relative activities both under our standard Gpx1 assay conditions using H2O2 and standard Gpx4 assay conditions using PCOOH, because the Gpx4 - H2O2 second-order rate constant is still a sizable 27% of the Gpx4 - PCOOH rate constant (30). We determined the ratio of 0.26 ± 0.02 H2O2/PCOOH enzyme units, which is virtually the same as the ratio of published rate constants, especially considering differences in assay buffers, detergents, and substrate concentrations. The result was that testis Gpx1 activity in Se-deficient rats decreased significantly to 28% of Se-adequate levels. Graded Se supplementation resulted in a hyperbolic response in testis Gpx1 activity, with a plateau breakpoint or minimum dietary Se requirement at 0.04 μg Se/g diet. Other studies have reported similar significant decreases in testis Gpx1 activity (5,37) using the conventional assay, and apparent Gpx1 activity in second-generation Se-deficient rats decreases to 8% of Se-adequate levels (41). The resulting minimum dietary Se requirement at 0.04 μg Se/g diet for Gpx1-specific activity, however, was unexpected because of the clear retention of Se by testis in this study.
In testis, mRNA levels for Gpx1, Sepp1 and Sepw1 decreased significantly in Se-deficient rats to 45, 46, and 55%, respectively, of Se-adequate plateau levels. Se supplementation resulted in hyperbolic responses in transcript levels for these three selenoproteins, with minimum dietary Se requirements at 0.08, 0.04, and <0.016 μg Se/g diet, respectively (Table 2). The Sepw1 mRNA Se response curve is similar to the Se concentration response curve, and thus as expected. The significant regulation of Sepp1 mRNA with the Se response curve breakpoint of 0.04 μg Se/g diet, however, was not expected in testis. In our previous studies, Sepp1 mRNA was not significantly down-regulated in Se-deficient rat liver (4,36,42). In the present experiment with additional lower levels of dietary Se, however, Sepp1 mRNA was down-regulated in Se-deficient liver and kidney to ∼55% of Se-adequate levels with minimum dietary Se requirements of 0.04 and 0.02 μg Se/g, respectively, and thus much lower than the minimum dietary Se required for plateau levels of tissue Se (8). Even more unexpected was that the Gpx1 mRNA Se response curve breakpoint (0.08 μg Se/g) is essentially the same as the Gpx1 mRNA breakpoint determined in liver (0.07 μg Se/g) in these same rats (8), and twice as high as the corresponding testis Gpx1 activity breakpoint.
This elevated minimum Se requirement for Gpx1 mRNA relative to the requirement for Gpx1 enzyme activity (Table 2) suggests unusual compartmentalization of Se in the testis. Until this study, we had always found for a given tissue that the minimum dietary Se levels (or breakpoints) needed to reach the plateau of the Sec response curves for selenoprotein enzyme activity or mRNA levels were the same or less than the minimum dietary Se requirements for plateau levels of Se (3-5,36,42). In testis, however, the minimum dietary Se necessary to reach the plateau for testis Se concentration was <0.016 μg Se/g, but testis Gpx1 activity, Gpx1 mRNA, and Sepp1 mRNA in this study required 0.04, 0.08 and 0.04 μg Se/g, respectively, for plateau levels. Especially puzzling is that the dietary Se level required to reach plateau levels of Gpx1 mRNA is higher than the level required for maximal Gpx1 activity. One possible explanation is that these young rats are just at the end of puberty. At weaning, rat testis has little Gpx4 activity but during puberty Gpx4 activity and testis Se increase dramatically (18,39). Thus with the rapid increases in Gpx4 synthesis, these levels may not have reached steadystate. Alternatively, the differences are more likely to arise due to the spacial distribution and roles of different cell types in the testis. ApoER2 receptors in the testis are located on the basal surface of the Sertoli cells and within the basal cytoplasm of the Sertoli cells (25). As the basal surfaces of Sertoli cells create the blood-testis barrier, this would allow delivery of blood-borne Se only as Sepp1 and only to Sertoli cells. From there, Se in an apparently different form is delivered to the developing spermatids where it provides Se for incorporation into Gpx4 (25). When Se supply is limited due to Se deficiency, incoming Se as Sepp1 would be delivered preferentially to the Sertoli cells, thereby maintaining testis Se concentrations. Overlap of testis Se concentration and testis Gpx4 activity response curves (Fig. 1A and 1B) clearly show the priority for Se is for synthesis of Gpx4, as both have minimum dietary Se requirements <0.016 μg Se/g. But under these Se-limited conditions, the higher minimum dietary Se requirement of 0.04 μg Se/g for Sepp1 mRNA, of 0.04 μg Se/g for Gpx1 activity, and even higher minimum dietary Se requirement of 0.08 μg Se/g for Gpx1 mRNA, suggest that these biomarkers reflect different pools of Se.
These pools are likely to be additional cell types that, in the trafficking of Se within the testis, are downstream of Sertoli cells and perhaps developing spermatids, or are outside of the blood-testis barrier. Riboprobes for ApoER2 mRNA detect ApoER2 transcripts only in Sertoli cells in the seminiferous epithelium, and ApoER2 transcripts are absent from developing spermatids (25). Similarly, Sepp1 mRNA is not detected in the seminiferous epithelium (43), and use of monoclonal antibodies localizes Sepp1 protein only within the Sertoli cells and not in spermatogenic cells or peritubular cells (25). In situ hybridization, however, indicates that Gpx4 mRNA is abundantly and preferentially expressed in spermatids (44), so imported Sepp1 appears to be degraded within the Sertoli cells and the released Se transferred to spermatogenic cells for Gpx4 synthesis. The coincidence of testes Se, Gpx4 activity, (and Sepw1 mRNA) response curves in this study places the preponderance of these components within the same hypothetical metabolic pool inside the blood-testis barrier. The synthesized Gpx4 in spermatids becomes oxidatively cross-linked and enzymatically inactive during sperm maturation (16), and thus Se originally in Gpx4 is lost from testis. Leydig cells, in contrast, reside outside of the blood-testis barrier, apparently do not express ApoER2 (25), do not contain appreciable Sepp1 protein (25), and thus obtain Se from forms other than Sepp1, perhaps as low molecular weight forms or plasma Gpx3. Leydig cells are the major cells expressing Sepp1 mRNA in testis (43,45), and thus could be part of the distinct pool of Se in testis distinguished by the higher minimal Se requirements for Sepp1 mRNA and Gpx1 activity. Sepp1 synthesis by these additional cell types in turn would facilitate recycling and retention within the testis (24). Lastly, the pool associated with Gpx1 mRNA with a minimum requirement of 0.08 μg Se/g diet could also be part of the Leydig cell pool, or possibly a third pool, as in situ hybridization for Gpx1 mRNA shows that interstitial Gpx1 mRNA is diffusely distributed in testis and appears to be largely associated with macrophages and not Leydig cells (44). In situ hybridizations, gene silencing, and perhaps identification of additional genes involved in Se trafficking will likely be needed to unravel the players in Se trafficking in testis.
In summary, 21 of 24 selenoprotein transcripts in total testis RNA, as well as transcript levels for the Sepp1 receptor, ApoER2, are not significantly regulated by Se status in the rapidly-growing Se-deficient rat, whereas transcript levels for Gpx1, Sepp1, and Sepw1 decrease to about 50% of Se-adequate levels in Se-deficient testis. The overlap of hyperbolic Gpx4 activity and Sepw1 mRNA response curves with testis Se concentration, all with minimum dietary Se requirements <0.016 μg Se/g, shows the priority for synthesis of Gpx4, and suggests that Sepw1 expression also originates within this same cell-type. Higher minimum dietary Se requirements of 0.04 μg Se/g for Gpx1 activity and Sepp1 mRNA, and the even higher minimum dietary Se requirement of 0.08 μg Se/g for Gpx1 mRNA, suggest that the hierarchy of these biomarkers reflects distinct, lower priority pools, cell-types, and roles for Se within the testis.
Acknowledgments
This Research was supported in part by the National Institutes of Health grant DK74184 and training grant T32-DK07665 (supporting K.M.B.), and by the University of Wisconsin Agricultural Experiment Station grant WIS04909.
Footnotes
Abbreviations used: Actb, β-actin; ApoER2, apolipoprotein ER2; Gpx1, glutathione peroxidase-1; Gpx3, plasma glutathione peroxidase; Gpx4, phospholipid hydroperoxide glutathione peroxidase; Gapdh, glyceraldehyde-3-phosphate dehydrogenase; PCOOH, phosphatidyl choline hydroperoxide; qRT-PCR, quantitative real time polymerase chain reaction; Se, selenium; Selh, selenoprotein H; Selk, selenoprotein K; Selv, selenoprotein V; Sepw1, selenoprotein W; Sepp1, selenoprotein P; Sepx1, methionine-R-sulfoxide reductase; Txnrd, thioredoxin reductase.
References
- 1.Hafeman DG, Sunde RA, Hoekstra WG. Effect of dietary selenium on erythrocyte and liver glutathione peroxidase in the rat. J Nutr. 1974;104:580–587. doi: 10.1093/jn/104.5.580. [DOI] [PubMed] [Google Scholar]
- 2.Knight SAB, Sunde RA. The effect of progressive selenium deficiency on anti-glutathione peroxidase antibody reactive protein in rat liver. J Nutr. 1987;117:732–738. doi: 10.1093/jn/117.4.732. [DOI] [PubMed] [Google Scholar]
- 3.Weiss SL, Evenson JK, Thompson KM, Sunde RA. The selenium requirement for glutathione peroxidase mRNA level is half of the selenium requirement for glutathione peroxidase activity in female rats. J Nutr. 1996;126:2260–2267. doi: 10.1093/jn/126.9.2260. [DOI] [PubMed] [Google Scholar]
- 4.Sunde RA, Evenson JK, Thompson KM, Sachdev SW. Dietary selenium requirements based on glutathione peroxidase-1 activity and mRNA levels and other selenium parameters are not increased by pregnancy and lactation in rats. J Nutr. 2005;135:2144–2150. doi: 10.1093/jn/135.9.2144. [DOI] [PubMed] [Google Scholar]
- 5.Lei XG, Evenson JK, Thompson KM, Sunde RA. Glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase are differentially regulated in rats by dietary selenium. J Nutr. 1995;125:1438–1446. doi: 10.1093/jn/125.6.1438. [DOI] [PubMed] [Google Scholar]
- 6.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Characterization of mammalian selenoproteins. Science (Washington, DC) 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 7.Sunde RA, Raines AM, Barnes KM, Evenson JK. Selenium status highly-regulates selenoprotein mRNA levels for only a subset of the selenoproteins in the selenoproteome. Biosci Rep. 2009;29 doi: 10.1042/BSR20080146. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnes KM, Evenson JK, Raines AM, Sunde RA. Transcript analysis of the selenoproteome indicates that dietary selenium requirements based on selenium-regulated selenoprotein mRNAs are uniformly less than requirements based on glutathione peroxidase activity in rats. J Nutr. 2009 Feb;139 doi: 10.3945/jn.108.098624. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Behne D, Höfer-Bosse T. Effects of a low selenium status on the distribution and retention of selenium in the rat. J Nutr. 1984;114:1289–1296. doi: 10.1093/jn/114.7.1289. [DOI] [PubMed] [Google Scholar]
- 10.McCoy KEM, Weswig PH. Some selenium responses in the rat not related to vitamin E. J Nutr. 1969;98:383–389. doi: 10.1093/jn/98.4.383. [DOI] [PubMed] [Google Scholar]
- 11.Wu SH, Oldfield JE, Whanger PD, Weswig PH. Effects of selenium, vitamin E, and antioxidants on testicular function in rats. Biol Reprod. 1973;8:625–629. doi: 10.1093/biolreprod/8.5.625. [DOI] [PubMed] [Google Scholar]
- 12.Brown DG, Burk RF. Selenium retention in tissues and sperm of rats fed a torula yeast diet. J Nutr. 1973;103:102–108. doi: 10.1093/jn/103.1.102. [DOI] [PubMed] [Google Scholar]
- 13.Behne D, Höfer T, Berswordt-Wallrabe R, Elger W. Selenium in the testis of the rat: studies on its regulation and its importance for the organism. J Nutr. 1982;112:1682–1687. doi: 10.1093/jn/112.9.1682. [DOI] [PubMed] [Google Scholar]
- 14.Wu ASH, Oldfield JE, Shull LR, Cheeke PR. Specific effect of selenium deficiency on rat sperm. Biol Reprod. 1979;20:793–798. doi: 10.1095/biolreprod20.4.793. [DOI] [PubMed] [Google Scholar]
- 15.Behne D, Hilmert H, Scheid S, Gessner H, Elger W. Evidence for specific selenium target tissues and new biologically important selenoproteins. Biochim Biophys Acta. 1988;966:12–21. doi: 10.1016/0304-4165(88)90123-7. [DOI] [PubMed] [Google Scholar]
- 16.Ursini F, Heim S, Kiess M, Majorino M, Roveri A, Wissing J, Flohé L. Dual function of the selenoprotein PHGPx During Sperm Maturation. Science (Washington, DC) 1999;285:1393–1396. doi: 10.1126/science.285.5432.1393. [DOI] [PubMed] [Google Scholar]
- 17.Behne D, Wolters W. Distribution of selenium and glutathione peroxidase in the rat. J Nutr. 1983;113:456–461. doi: 10.1093/jn/113.2.456. [DOI] [PubMed] [Google Scholar]
- 18.Roveri A, Casasco A, Maiorino M, Dalan P, Calligaro A, Ursini F. Phospholipid hydroperoxide glutathione peroxidase of rat testis. Gonadotropin dependence and immunocytochemical identification. J Biol Chem. 1992;267:6142–6146. [PubMed] [Google Scholar]
- 19.Foresta C, Flohé L, Garolla A, Roveri A, Ursini F, Maiorino M. Male fertility is linked to the selenoprotein phospholipid hydroperoxide glutathione peroxidase. Biol Reprod. 2002;67:967–971. doi: 10.1095/biolreprod.102.003822. [DOI] [PubMed] [Google Scholar]
- 20.Evenson JK, Sunde RA. Selenium incorporation into selenoproteins in the Se-adequate and Se-deficient rat. Proc Soc Exp Biol Med. 1988;187:169–180. doi: 10.3181/00379727-187-42651. [DOI] [PubMed] [Google Scholar]
- 21.Hill KE, Zhou J, McMahan WJ, Motley AK, Atkins JF, Gesteland RF, Burk RF. Deletion of selenoprotein P alters distribution of selenium in the mouse. J Biol Chem. 2003;278:13640–13646. doi: 10.1074/jbc.M300755200. [DOI] [PubMed] [Google Scholar]
- 22.Schomburg L, Schweizer U, Holtmann B, Flohé L, Sendtner M, Köhrle J. Gene disruption discloses role of selenoprotein P in selenium delivery to target tissues. Biochem J. 2003;370:397–402. doi: 10.1042/BJ20021853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olson GE, Winfrey VP, Nagdas SK, Hill KE, Burk RF. Selenoprotein P is required for mouse sperm development. Biol Reprod. 2005;73:201–211. doi: 10.1095/biolreprod.105.040360. [DOI] [PubMed] [Google Scholar]
- 24.Renko K, Werner M, Renner-Muller I, Cooper TG, Yeung CH, Hollenbach B, Scharpf M, Köhrle J, Schomburg L, Schweizer U. Hepatic selenoprotein P (SePP) expression restores selenium transport and prevents infertility and motor-incoordination in Sepp-knockout mice. Biochem J. 2008;409:741–749. doi: 10.1042/BJ20071172. [DOI] [PubMed] [Google Scholar]
- 25.Olson GE, Winfrey VP, Nagdas SK, Hill KE, Burk RF. Apolipoprotein E receptor-2 (ApoER2) mediates selenium uptake from selenoprotein P by the mouse testis. J Biol Chem. 2007;282:12290–12297. doi: 10.1074/jbc.M611403200. [DOI] [PubMed] [Google Scholar]
- 26.Olson GE, Winfrey VP, Hill KE, Burk RF. Sequential development of flagellar defects in spermatids and epididymal spermatozoa of selenium-deficient rats. Reproduction. 2004;127:335–342. doi: 10.1530/rep.1.00103. [DOI] [PubMed] [Google Scholar]
- 27.McKown DM, Morris JS. Rapid measurement of selenium in biological samples using instrumental neutron activation analysis. J Radioanal Nucl Chem. 1978;43:411–420. [Google Scholar]
- 28.Lawrence RA, Sunde RA, Schwartz GL, Hoekstra WG. Glutathione peroxidase activity in rat lens and other tissues in relation to dietary selenium intake. Exp Eye Res. 1974;18:563–569. doi: 10.1016/0014-4835(74)90062-1. [DOI] [PubMed] [Google Scholar]
- 29.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 30.Maiorino M, Gregolin C, Ursini F. Phospholipid hydroperoxide glutathione peroxidase. Meth Enzymol. 1990;186:448–457. doi: 10.1016/0076-6879(90)86139-m. [DOI] [PubMed] [Google Scholar]
- 31.Sunde RA, Hoekstra WG. Incorporation of selenium into liver glutathione peroxidase in the Se-adequate and Se-deficient rat. Proc Soc Exp Biol Med. 1980;165:291–297. doi: 10.3181/00379727-165-40973. [DOI] [PubMed] [Google Scholar]
- 32.Sunde RA, Evenson JK. Serine incorporation into the selenocysteine moiety of glutathione peroxidase. J Biol Chem. 1987;262:933–937. [PubMed] [Google Scholar]
- 33.Peirson SN, Butler JN, Foster RG. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 2003;31:e73. doi: 10.1093/nar/gng073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steel RGD, Torrie JH. Principles and Procedures of Statistics. New York, NY: McGraw-Hill Book; 1960. [Google Scholar]
- 36.Weiss SL, Evenson JK, Thompson KM, Sunde RA. Dietary selenium regulation of glutathione peroxidase mRNA and other selenium-dependent parameters in male rats. J Nutr Biochem. 1997;8:85–91. doi: 10.1016/S0955-2863(96)00178-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whanger PD, Butler JA. Effects of various dietary levels of selenium as selenite or selenomethionine on tissue selenium levels and glutathione peroxidase activity in rats. J Nutr. 1988;118:846–852. doi: 10.1093/jn/118.7.846. [DOI] [PubMed] [Google Scholar]
- 38.Deagen JT, Butler JA, Beilstein MA, Whanger PD. Effects of dietary selenite, selenocystine and selenomethionine on selenocysteine lyase and glutathione peroxidase activities and on selenium levels in rat tissues. J Nutr. 1987;117:91–98. doi: 10.1093/jn/117.1.91. [DOI] [PubMed] [Google Scholar]
- 39.Behne D, Duk M, Elger W. Selenium content and glutathione peroxidase activity in the testis of the maturing rat. J Nutr. 1986;116:1442–1447. doi: 10.1093/jn/116.8.1442. [DOI] [PubMed] [Google Scholar]
- 40.Weitzel F, Ursini F, Wendel A. Phospholipid hydroperoxide glutathione peroxidase in various mouse organs during selenium deficiency and repletion. Biochim Biophys Acta. 1990;1036:88–94. doi: 10.1016/0304-4165(90)90018-r. [DOI] [PubMed] [Google Scholar]
- 41.Thompson KM, Haibach H, Evenson JK, Sunde RA. Liver selenium and testis phospholipid hydroperoxide glutathione peroxidase are associated with growth during selenium repletion of second-generation Se-deficient male rats. J Nutr. 1998;128:1289–1295. doi: 10.1093/jn/128.8.1289. [DOI] [PubMed] [Google Scholar]
- 42.Weiss Sachdev S, Sunde RA. Selenium regulation of transcript abundance and relative translational efficiency of glutathione peroxidase 1 and 4 in rat liver. Biochem J. 2001;357:851–858. doi: 10.1042/0264-6021:3570851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koga M, Tanaka H, Yomogida K, Tsuchida J, Uchida K, Kitamura M, Sakoda S, Matsumiya K, Okuyama A, Nishimune Y. Expression of selenoprotein-P messenger ribonucleic acid in the rat testis. Biol Reprod. 1998;58:261–265. doi: 10.1095/biolreprod58.1.261. [DOI] [PubMed] [Google Scholar]
- 44.Maiorino M, Wissing JB, Brigelius-Flohé R, Calabrese F, Roveri A, Steinert P, Ursini F, Flohé L. Testosterone mediates expression of the selenoprotein PHGPx by induction of spermatogenesis and not by direct transcriptional gene activation. FASEB J. 1998;12:1359–1370. doi: 10.1096/fasebj.12.13.1359. [DOI] [PubMed] [Google Scholar]
- 45.Steinert P, Bachner D, Flohé L. Analysis of the mouse selenoprotein P gene. Biol Chem. 1998;379:683–691. doi: 10.1515/bchm.1998.379.6.683. [DOI] [PubMed] [Google Scholar]