Abstract
Digital image analysis of histology sections provides reliable, high-throughput methods for neuropathological studies but data is scant in frontotemporal lobar degeneration (FTLD), which has an added challenge of study due to morphologically diverse pathologies. Here, we describe a novel method of semi-automated digital image analysis in FTLD subtypes including: Pick’s disease (PiD, n=11) with tau-positive intracellular inclusions and neuropil threads, and TDP-43 pathology type C (FTLD-TDPC, n=10), defined by TDP-43-positive aggregates predominantly in large dystrophic neurites. To do this, we examined three FTLD-associated cortical regions: mid-frontal gyrus (MFG), superior temporal gyrus (STG) and anterior cingulate gyrus (ACG) by immunohistochemistry. We used a color deconvolution process to isolate signal from the chromogen and applied both object detection and intensity thresholding algorithms to quantify pathological burden. We found object-detection algorithms had good agreement with gold-standard manual quantification of tau- and TDP-43-positive inclusions. Our sampling method was reliable across three separate investigators and we obtained similar results in a pilot analysis using open-source software. Regional comparisons using these algorithms finds differences in regional anatomic disease burden between PiD and FTLD-TDP not detected using traditional ordinal scale data, suggesting digital image analysis is a powerful tool for clinicopathological studies in morphologically diverse FTLD syndromes.
Keywords: FTLD, TDP-43, Tau, digital image analysis, Pick’s disease
Introduction
Traditional neuropathological assessments of neurodegenerative disease are based on semi-quantitative ratings of disease burden on an ordinal scale (i.e. 0 = none/rare, 1 = mild, 2 = moderate, 3 = severe) (Montine et al. 2012). These ratings are based on an examiner’s subjective impressions of the burden of neurodegenerative pathology based on visual inspection. However, there may be inter-individual variability even among experts (Armstrong 2003), and ordinal scales provide limited statistical power for empirical studies (Neltner et al. 2012). This is of particular importance in the context of less common and neuropathologically heterogeneous conditions like frontotemporal lobar degeneration (FTLD). For example, roughly 50% of behavioral variant frontotemporal dementia (bvFTD) clinical syndrome is associated with either tau inclusions (FTLD-Tau) or TDP-43 (FTLD-TDP) inclusions (Forman et al. 2006; Irwin et al. 2015), and quantitative comparative studies of FTLD pathological subtypes within a particular clinical phenotype are crucial for efforts to identify endophenotypes that reliably predict neuropathology during life (Irwin et al. 2015). Thus, a reliable and high-throughput method that provides a continuous measure of histopathology is urgently needed for clinicopathological research studies of FTLD and related neurodegenerative diseases.
Stereology (West et al. 1991) and manual quantification (Armstrong et al. 1999a; Armstrong and Cairns 2012) can provide a continuous measure of pathological burden but these time-intensive methods are prohibitive for large-scale comparative studies. Emerging methods for automated digital image analysis of histology may provide a high-throughput means of quantification. Indeed, digital image analysis methods for quantifying Alzheimer’s disease (AD) neuropathology have been previously described (Byrne et al. 2009; Neltner et al. 2012; Samaroo et al. 2012), but there are minimal data on the neuropathology of FTLD. Further, FTLD spectrum pathology has a range of intracellular inclusions such as Pick bodies (PBs) as well as large dystrophic neurites (axonal or proximal dendritic inclusions; i.e., DNs; Fig. 1J–1O) and small diffuse neuritic threads (distal dendritic inclusions; i.e., NTs; Fig. 1F, yellow overlay), which require different image analysis methods for quantification. Thus, the focus of this study was on developing a reliable method for sampling cortical tissue and detection of FTLD neuropathology in Pick’s disease (PiD) and FTLD-TDP subtype C (FTLD-TDPC). PiD and FTLD-TDPC were chosen as representative groups for the two main subtypes of FTLD (i.e., FTLD-Tau and FTLD-TDP), as PiD has predominantly cytoplasmic inclusions (CI) and diffuse NTs (Dickson 2004; Dickson et al. 2011; Zhukareva et al. 2002) whereas FTLD-TDPC has mainly large DNs with minimal CI or diffuse NTs (Mackenzie et al. 2011; Neumann et al. 2006; Tan et al. 2013). We demonstrate that digital analysis methods are reliable among users of varying experience for these diverse pathologies and we observed regional differences in pathological burden not detected by traditional ordinal scores.
Figure 1.
Color deconvolution and detection algorithms. Photomicrographs depict sample images of mid-frontal gyrus (MFG) from a Pick’s disease case (PiD; A–I) and frontotemporal lobar degeneration (FTLD) with TDP-43 inclusions (FTLD-TDPC) case (J–R) with digital image analysis outputs. Background reactivity in negative controls stained in the absence of primary antibody are shown for PiD (i.e., PID NC) (G–I) and FTLD-TDPC (i.e. FTLD-TDPC NC) (P–R). Raw images (A, D, G, H, J, M, P) are deconvoluted into chromogen signals (B, E, H, K, N, Q) for both the inclusion detection (i.e., Tau CI/mm2 or TDP DN/mm2) and %AO algorithms. Final markup images show detected tau CI (C) and TDP-43 DN (L) in red overlay and total %AO of stain in PiD (F) and FTLD-TDP-C (O) in yellow/orange/red overlay. In contrast, negligible %AO stain is seen in negative control tissue (I, R), conferring specificity of algorithms for pathology. DN, dystrophic neurites. Scale, 100 µm.
Materials & Methods
Patients
Patients were seen at the University of Pennsylvania (Penn) Perelman School of Medicine Frontotemporal Degeneration Center or Alzheimer’s Disease Center, and autopsies were performed at the Penn Center for Neurodegenerative Disease Research. All procedures were performed with informed consent in accordance with Penn Institutional Review Board guidelines. Neuropathological assessment was performed as reported previously (Toledo et al. 2013) using established criteria (Mackenzie et al. 2011; Mackenzie et al. 2010; Montine et al. 2012). Patient demographics (Table 1) and semi-quantitative pathology rating scores from expert pathologists (EBL, JQT) were obtained from the Penn Integrated Neurodegenerative Disease Database (INDD) (Xie et al. 2011). We selected cases with a clinical diagnosis of bvFTD and a primary neuropathological diagnosis of PiD (n=11) or FTLD-TDPC (n=10) and normal controls with a neuropathological diagnosis of unremarkable adult brain (n=5) (Table 1). Genetic screening for C9orf72, GRN, and MAPT mutations, as reported elsewhere (Irwin et al. 2013a; Irwin et al. 2013b), revealed that one PiD case had a pathogenic mutation in MAPT gene (p.Leu266Val) that was associated with PBs composed of 3R isoforms of tau with the same morphology as in sporadic disease (Hogg et al. 2003). Further, one additional case harbored a C9orf72 repeat expansion (i.e. >30 repeats) with typical FTLD-TDP type C pathology, as reported previously (Irwin et al. 2013b).
Table 1.
Patient Demographics.
| Primary Diagnosis | Gender (M/F) | Brain Weight (g) | PMI (hr) | Age at Onset (years) | Age at Death (years) | Disease Duration (years) | Braak Stage (n per stage) | CERAD Score (n per stage) |
|---|---|---|---|---|---|---|---|---|
| Pick’s Disease n=11 | 7/4 | 1011.5 (141.9) | 12.7 (6.2) | 54.4 (12.8) | 64.3 (14.2) | 9.9 (3.9) | 0 = 9 | 0 = 9 |
| I-II = 2 | A = 1 | |||||||
| III-IV = 0 | B = 1 | |||||||
| V-VI = 0 | C = 0 | |||||||
| FTD-TDPC n=10 | 7/3 | 1114.5 (119.9) | 9.2 (6.1)* | 61.7 (10.7) | 71.3 (10.8) | 9.6 (4.6) | 0 = 6 | 0 = 5 |
| I-II = 4 | A = 1 | |||||||
| III-IV = 0 | B = 2 | |||||||
| V-VI = 0 | C = 2 | |||||||
| Normal n=5 | 2/3 | 1279.6 (98.8) | 15.5 (7.1) | NA | 60.6 (14.3) | NA | 0 =3 | 0 = 5 |
| I-II = 2 | A = 0 | |||||||
| III-IV = 0 | B = 0 | |||||||
| V-VI = 0 | C = 0 |
Data missing for one case. PMI, post mortem interval; NA, not applicable. Continuous variables reported as mean (standard deviation). CERAD = Consortium to Establish a Registry for Alzheimer’s Disease Plaque Score; Braak = Modified Tau Braak Stage (please see Montine et al, 2012 for details on these scales).
Immunohistochemistry
Fresh tissue samples obtained at autopsy were fixed overnight in 70% ethanol with 150 mM sodium chloride (referred to simply as EtOH) or 10% neutral-buffered formalin (NBF). Tissue samples were trimmed, placed into cassettes and processed through a series of alcohol, xylene and Surgipath EM-400 paraffin embedding media (Leica Microsystems; Buffalo Grove, IL) with incubations overnight (70% ethanol × 2 hr, 80% ethanol × 1 hr, 95% ethanol × 1 hr, 95% ethanol × 2 hr, 100% ethanol × 2 hr, twice, xylene × 30 min, xylene × 1 hr, xylene × 1.5 hr, and paraffin × 1 hr, three times) in a Shandon tissue processor (Thermo Scientific; Waltham, MA). All incubations were done under vacuum and at ambient temperature except paraffin (62°C). Tissue was then embedded into paraffin blocks and 6-µm-thick sections were cut for analysis. All tissue was processed in an identical manner. Regions sampled included the mid-frontal gyrus (MFG; i.e., Brodman area BA46), superior and mid-temporal gyri (STG and MTG; i.e., BAs 22,21) and the anterior cingulate gyrus (ACG; i.e., BA32) according to a standard anatomic atlas (Toledo et al. 2013). Quantitative analysis was focused on MFG, STG and ACG. Immunohistochemistry (IHC) was performed using well-characterized antibodies for phosphorylated tau (i.e., mouse monoclonal pS396/404 PHF-1; a gift from Peter Davies) (Otvos et al. 1994) in PiD, and TDP-43 (i.e., rat monoclonal TAR5P-1D3 pS409/410 TDP-43; Ascenion, Munich, Germany) (Neumann et al. 2009) in FTLD-TDPC. IHC was performed as previously described (Forman et al. 2006) using 3,3’-diaminobenzidine as the chromogen and hematoxylin as the counter stain. Each staining run included a cortical section of a typical case of AD and FTLD-TDP as a positive control. As a negative control, we stained five PiD (i.e., PiD NC) and five FTLD-TDPC (i.e., FTLD-TDPC NC) MFG sections in the absence of primary antibodies for analysis to detect background endogenous peroxidase activity.
Digital Image Acquisition
Digital images of histology slides at 20× magnification were obtained using a Lamina (Perkin Elmer, Waltham, MA) slide scanning system with a pixel size of 6.5 µm2 (i.e., pixel resolution of 0.325 µm), camera resolution of 2560 × 2160, and a bit depth of 16 (.mrxs image file). Each slide was autocorrected through capture of 10 empty fields of view to create a compensation image used to create evenly illuminated composite images.
Automated Inclusion Detection Algorithms
Halo digital image software v1.90 (Indica Labs; Albuquerque, NM) was utilized to develop detection algorithms to quantify tau-positive CI (CI-i.e. PBs and glial inclusions) in PiD and TDP-43-positive DNs in FTLD-TDP using the “object co-localization tool” v1.1. This tool used parameters of red, green, blue (RGB) optical density (OD) for color deconvolution to isolate signals from the chromogen and counterstain (Fig. 1). Initial parameters of RGB OD (scale range 0–2.4) were developed through averaged values obtained from characteristic inclusions in five different random slides. The minimum/maximum object size was determined from measuring the smallest/largest inclusions found in these slides. Next, various parameters—blur radius (BR; scale range 0-30) for smoothing of adjacent pixels to prevent over-segmentation of single inclusions; contrast threshold (CT; scale range 0-1) to differentiate IHC chromogen signal from tissue; and minimum stain OD for stain detection sensitivity—were initially developed through systematic variation and compared with digital images using visual inspection and the “real-time tuning” window of the software. Other image detection parameters were unchanged during the development process, including “connect length” (i.e., minimum distance to connect neighboring object fragments), “minimal tissue OD” (i.e., minimum OD of non-stained tissue to eliminate non-tissue “glass” regions from analysis), “tissue edge thickness” (i.e., thickness of tissue edge to exclude from analysis), “object contrast radius” (i.e., smallest radius of space to differentiate inclusions from background) and “fill holes” (i.e., filling a non-stained hole in a concentric inclusion), as these parameters reflect constant features of images. Connect length was set to the minimum object size to prevent over-counting. Algorithms were developed on slides stained simultaneously in the same batch and we also performed a secondary analysis evaluating adjacent or near-adjacent slides that were stained in a separate batch (Supplementary Table 1).
Gold-Standard Manual Object Count and Algorithm Validation
Initial algorithms were further optimized by incremental variation of BR, CT and stain OD and tested for accuracy against manually detected counts from a preliminary survey of 10 randomly selected 500 µm tiles (area = 249,801 µm2) in each slide (Supplementary Table 1) using the “Manual Click Counter” tool at 20× magnification. Tiles were generated using the “tile partitioning tool” from manually segmented grey matter using the “pen” and “magnetic pen” annotation tools. Manual counts were performed using morphological criteria (intracellular tau PBs or glial inclusions with a visible nucleus or cell membrane in PiD and large, coarse dystrophic neurites in longitudinal or cross-sectional orientation in FTLD-TDPC). Linear regression was used to compare manual counts to digital counts from each preliminary algorithm and the optimum algorithm was selected with the highest R2 value in a randomly divided training set (PiD, n=6; FTLD-TDPC, n=5) and test set (n=5) (Supplemental Table 1). A Bland-Altman method was used to test the agreement between the optimal algorithm digital counts and the gold-standard manual counts from new manual count data obtained in the 175-µm tiles used in our random sampling scheme (see below). The mean difference between the gold-standard manual counts and the digital counts of pathology was plotted against the mean value across these measures (jitter was added to reveal identical data points). The limits of agreement were calculated as 1.96 multiplied by the standard deviation of the difference in values, and the mean bias was calculated as the mean difference in values between measurements across all tiles. Manual and digital counts were not normally distributed, as determined using a Shapiro-Wilk analysis (p<0.001). As such, a Wilcoxon signed-rank test was used to test the significance of the bias: the null-hypothesis is that the median difference between the manual and digital counts was equal to zero (i.e., median bias is not significantly different from 0).
Sampling Method Validation
Since FTLD neuropathology can often preferentially involve specific cortical layers (Armstrong et al. 1999a; Armstrong et al. 1999b; Mackenzie et al. 2011; Tan et al. 2013), it was necessary to sample the parallel-oriented cortex, as curved or tangentially cut areas of tissue may over- or under-represent cortical layers and influence “whole-slide” random plot sampling methods (Armstrong 2003; Neltner et al. 2012). As such, we selected the longest area of intact grey matter parallel to the pia matter (i.e., vertical transect sampling (Armstrong 2003) of the entire width of grey matter) for the region of interest (ROI) for measurement. A standardized operating procedure for this method was developed independently and implemented by three separate investigators (DJI, MDB, FC). Briefly, intact and straight regions of a cortical ribbon (i.e., exclusion of sulcal depths or gyral peaks) were traced at the grey–white and grey–pial interfaces using the “magnetic pen” and “pen” tools. The “layer thickness” tool was used to measure the thickness of the cortical ribbon at 0.25-mm intervals. The smoothing function was adjusted to create the maximum number of parallel measurements per region. The “ruler drawing tool” was used to measure the longest contiguous area of parallel cortex defined by an overall deviation in cortical thickness of 0.2 mm per region (~5%–10% of cortical thickness). In slides with multiple regions of parallel cortex, the longest region was selected for analysis using a random sampling scheme (Fig. 2). We used the “tile partitioning tool” to create 175 µm tiles (i.e. area= 3049.6 µm2) for analysis. We chose this size because it maximized the number of tiles for random sampling (i.e. majority of tissue samples had >50 tiles) and the upper limit of inclusion counts per tile to prevent manual count fatigue (Neltner et al. 2012). To determine the optimum number of random tiles to sample in our random sampling scheme, we performed a permutation analysis (Supplemental Fig. 1). This analysis method evaluates 10,000 estimates of a random subset of tiles (noise) relative to the true measure of pathology (signal). An optimum number of random tiles would then be a quantity that reliably falls within a 99% confidence interval of the true signal. In this context, we consider a reliable estimate as one that includes less than 10% of random sampling falling outside the 99% confidence interval of the true signal (i.e., mean value from entire ROI). ROIs were reviewed to confirm accurate delineation of the grey–white junction by less-experienced investigators and data from digital algorithms in random samples from these ROIs were compared for reproducibility (Supplemental Table 3) using a Wilcoxon sign-rank test to test the significance of the bias between measurements by investigators. Data from an experienced investigator (DJI) is reported in Table 2.
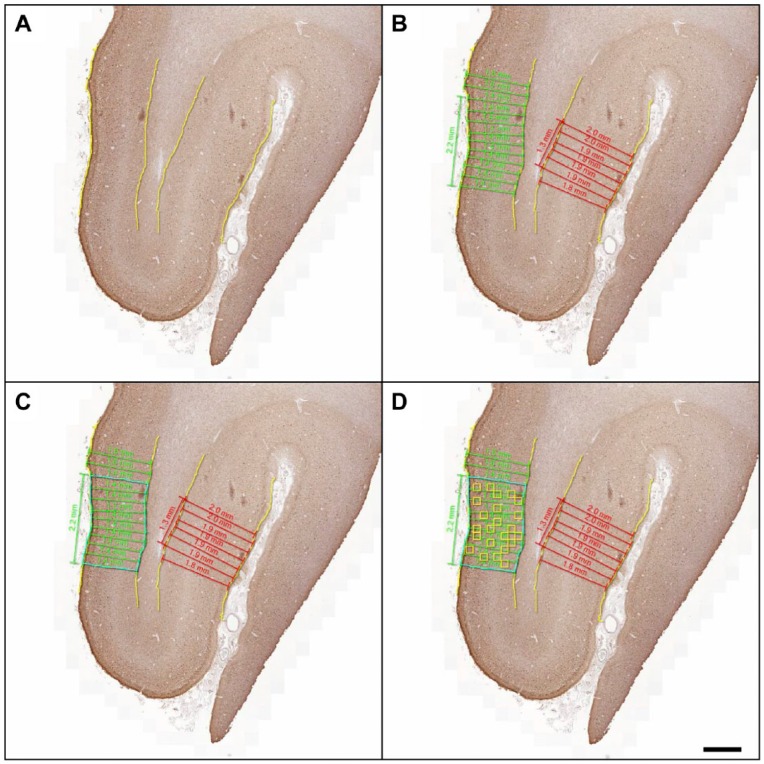
Figure 2.
Sampling method for representative cortex. Photomicrographs depict low-power images showing all available cortex for mid-frontal gyrus (MFG) in a representative case of Pick’s disease (PiD) and steps for selection of analysis region. (A) Grey matter was manually segmented (yellow) and (B) measured for cortical thickness (green and red measurements) in all intact parallel-oriented cortices (i.e., not on gyral peaks or sulci). In this example, two regions met the criteria for measurement; the longest intact area with parallel measurements of <0.2-mm deviation (2.2 mm of cortex = green) (C) was manually segmented (blue). (D) 24 random 175 µm tiles were created for the final analysis. Scale, 1mm.
Table 2.
Regional Neuropathological Measures in PiD and FTLD-TDPC.
| PiD | FTLD-TDPC | P value | ||
|---|---|---|---|---|
| MFG | Inclusions/mm2 | 92.8 (57.9) | 148.0 (143.3)‡ | - |
| %AO | 17.9 (11.4)% | 0.37 (0.4)%‡ | - | |
| Ordinal Score | 3 (3,3) | 2 (1-3) ‡‡ | >0.1 | |
| %Total | 36.9% (14.1%) | 18.6% (11.9%) | <0.01 | |
| STG | Inclusions/mm2 | 46.0 (47.5)*,** | 248.9 (174.8) | - |
| %AO | 7.4 (5.4)%* | 0.61 (0.4)% | - | |
| Ordinal Score | 3 (1,3) | 3 (2.75-3) | >0.1 | |
| %Total | 19.1% (17.9%) | 33.7% (13.6%) | 0.05 | |
| ACG | Inclusions/mm2 | 104.6 (61.7) | 336.5 (201.8) | - |
| %AO | 18.7 (15.6)% | 0.91 (0.5)% | - | |
| Ordinal Score | 3 (2,3) | 2.75 (3-3) | >0.1 | |
| %Total | 44.0% (17.7%) | 47.7% (8.2%) | >0.1 |
p<0.04 STG vs MFG; **p<0.01 STG vs ACG; ‡p<0.03 MFG vs ACG; ‡‡ p=0.05 MFG vs ACG, STG. All variables reported as mean (standard deviation) with the exception of ordinal scores reported as median (inter-quartile range). %AO (% area occupied) is %AOnt (%AO by neuropil threads) for PiD and %AOt (total %AO) for FTLD-TDPC. PiD, Pick’s disease; FTLD, frontotemporal lobular degeneration; TDPC, TDP-43 pathology type C; MFG, mid-frontal gyrus; STG, superior temporal gyrus; ACG, anterior cingulate gyrus.
%AO of IHC Immunoreactivity
Algorithms for quantification of the total percentage area occupied (i.e. %AOt) by tau (Fig. 1F) and TDP-43 (Fig. 1O) immunoreactivity (Supplemental Table 2) were developed using the “area quantification” v1.0 tool. These algorithms were based on stain RGB OD for color deconvolution to identify chromogen signal and thresholding using the minimum OD of positive stain to differentiate from the background signal. Algorithms were developed using the stain RGB values established from the inclusion detection process above and verified for accuracy using visual inspection. The program allows for three levels of OD thresholding (i.e., yellow is weak, orange is moderate, red is strong); the algorithms were tuned so to ensure minimal reactivity with non-pathological background (Fig. 1) and reported as %AOt (i.e., sum of yellow, orange and red intensity levels). Further validation was performed through comparison with traditional ordinal scale scores for each slide (Fig. 4) and analysis of background signal in negative and normal control tissue (Fig. 1G–1I, 1P–1R; Supplemental Table 4). Since PiD has significant diffuse NT pathology that cannot be manually quantified, we calculated the %AO of diffuse NTs (i.e., %AOnt) by subtracting the %AO of intracellular inclusions (i.e., %AOi; obtained from the inclusion detection algorithm output) from the total %AOt. Since FTLD-TDPC has minimal additional diffuse threads (Fig. 1J–1O), we examined the %AOt of TDP-43 immunoreactivity instead.
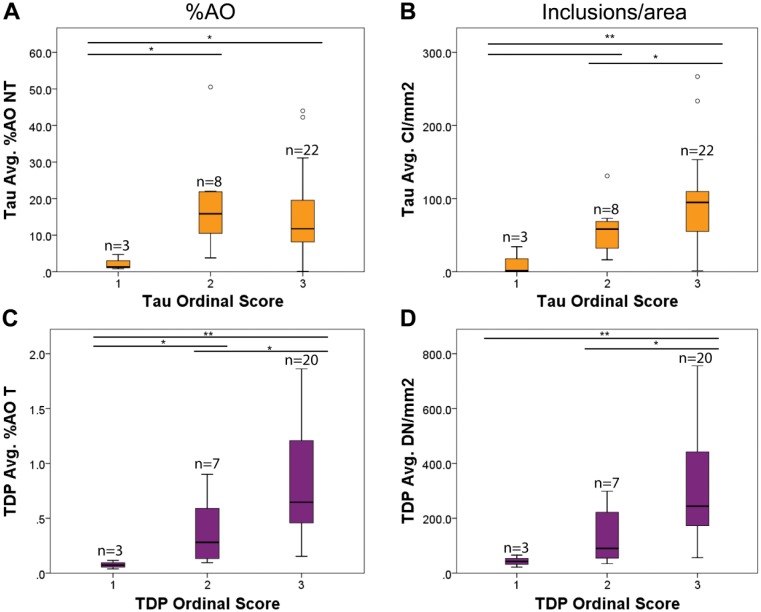
Figure 4.
Comparison of %AO and inclusion/area algorithm data with traditional ordinal scale scores. Box plots represent %AO (A, C) and inclusion/area (B, D) data from each slide (i.e., MFG, STG and ACG from 11 PiD and 10 FTLD-TDPC cases) plotted for each original ordinal scale score rating (i.e., 1, 2, 3). Inclusion/area (B) was more effective in detecting differences between all score categories compared with %AOnt (A) for PiD, whereas FTLD-TDPC was similarly differentiated using %AOt (C) and inclusions/area (D). *p≤0.05, **p<0.01 between score groups. Bar without asterisk denotes p=0.07. PiD, Pick’s disease; FTLD, frontotemporal lobular degeneration; TDPC, TDP-43 pathology type C; MFG, mid-frontal gyrus; STG, superior temporal gyrus; ACG, anterior cingulate gyrus.
Comparison with Other Digital Image Analysis Platform
To test the generalizability of our method, we examined the accuracy of our digital image analysis algorithms in a separate freely available software (Image J v1.49; http://imagej.nih.gov/ij/). Since Image J is not compatible with the .mrxs files used by HALO, we needed to convert images into full resolution non-compressed .tiff format. We used Pannoramic Viewer software v.1.15.4 (3D Histech; Budapest, Hungary) to select and capture identical 175 µm × 100 µm regions for analysis (i.e., one copy in full resolution .tiff format for ImageJ and one copy in .mrxs format for analysis in HALO) from five random PiD and FTLD-TDPC images. We chose this size because larger sized areas required prohibitively long processing times due to the extremely large size of uncompressed .tiff files. These images were then uploaded into their respective analysis programs with identical analysis being conducted on both images. For ImageJ analysis, images were first converted to a single channel using the “Image-Type-RGB Color” command. ImageJ and HALO analyze color differently: HALO uses absorbance (RGB OD on 0–2.4 scale) whereas ImageJ uses emittance (Intensity on 0–255 scale). We used the Color Deconvolution plugin (Ruifrok and Johnston 2001) in ImageJ to convert HALO RGB OD values to emittance with the formula: Intensity = 10.0^-OD x WhiteLevel. We then used the deconvoluted image representing the IHC chromogen signal for analysis of TDP %AOt using converted OD thresholds from our HALO algorithm (Supplemental Table 2) for intensity thresholding in ImageJ. To test tau CI quantification in PiD, we converted images to binary and converted the size parameters from our tau CI algorithm (Supplemental Table 2) from µm2 to pixel2 for ImageJ based on image resolution of pixel size of 6.5 µm2. Since ImageJ does not have an identical parameter to BR, we chose to use the “Erode” function at a level of 3.0 to approximate BR in our HALO algorithm. The within-subject %CV between measurements was calculated (%CV = standard deviation/mean of measurements between platforms) for each image and reported as an average from the five cases (Supplemental Table 5).
Regional Neuroanatomic Analyses and Demographics Statistical Analyses
Continuous measures of inclusion counts/%AO were assessed for normal distribution using a Shapiro-Wilk test and were found to be non-normally distributed (p≤0.02). Comparisons between regions and correlations were performed using non-parametric Mann-Whitney U and Spearman correlation analyses, respectively. To directly compare the relative anatomic burden between PiD and FTLD-TDPC, we calculated the percentage of the total pathology burden for each region (e.g., MFG/ MFG+STG+ACG) and compared these between PiD and FTLD-TDPC using an independent t-test (this variable was normally distributed; Shaprio-Wilk, p>0.2). Comparison of frequencies of categorical variables between groups was performed using a Chi-squared analysis. All analyses were two-sided with α = 0.05. Statistical analyses were performed with SPSS v21.0 (SPSS Inc.; Chicago, IL), R v3.2.1 (www.r-project.org) or STATA v12.1 (StataCorp; College Station, TX).
Results
Patients
PiD and TDP-43 patient groups showed similar ages of onset, death, and disease duration and post-mortem interval (PMI), with no significant differences between the disease groups (Table 1). All cases had minimal age-associated or AD-like neurofibrillary tangle tau pathology limited to the medial temporal lobe (i.e., Braak stage 0-II).
Automated Inclusion Detection Algorithm Validation
To test the agreement between our optimal digital image analysis algorithms for tau CI and TDP DN quantification with gold-standard manual counts, we used a Bland-Altman method to determine the mean bias (i.e., mean difference between measures) and limits of agreement (Fig. 3). We found that the majority of samples fell within the limits of agreement and that the median bias (tau CI = 0.01, TDP DN = -0.47) did not significantly differ from 0 (p>0.1). Thus, there is adequate agreement between our digital counts and gold-standard manual counts. Post-hoc analyses confirmed that the fixation method did not affect correspondence between manual and digital counts for FTLD-TDPC (NBF, n=18 slides; EtOH, n=12 slides), as the median bias was not significantly different than zero (p>0.1) for both fixation methods. We were not able to assess fixative methods for PiD because all cases were fixed in NBF.
Figure 3.
Bland-Altman analysis of digital algorithms for inclusion counts compared with gold-standard manual counts. Graphs depict test–re-test agreement of digital image analysis inclusion detection algorithms for (A) Tau cytoplasmic inclusions (CI) and (B) TDP dendritic inclusions (DN). Solid red lines represent the limits of agreement, and the dotted green line represents the mean bias.
Sampling Method Validation
Both groups had similar mean lengths of intact, parallel-oriented, cortical ribbon segmented in ROIs for random sampling (PiD mean = 1.75 µm, SD = 0.7; FTLD-TDP mean 1.83 µm, SD = 0.8). This method was chosen to reduce bias from overrepresentation of cortical layers in gyral crests or sulcal depths of cortical tissue (Armstrong 2003). To perform a user-independent random sampling scheme, we subdivided these ROIs into 175 µm2 tiles. To determine the optimum number of random tiles to sample, we performed permutation analyses to compare the frequency of estimations from the average of increasing numbers of tiles sampled to the 99% confidence interval of the “true mean” of the total ROI. First, due to the predominance of FTLD pathology in differing cortical layers (Armstrong et al. 1999b; Tan et al. 2013), we chose to compare random sampling from the entire ROI, independent of the cortical layers. We found that 24 random tiles from the entire ROI had a low frequency of estimations (i.e., <5%) that were outside of the 99% confidence interval of the mean for the entire ROI sampled (Fig. 3). Second, to evaluate whether preferential burden of a particular cortical layer (i.e., layer I-II, III-IV and V-VI) may bias random sampling estimates, we repeated our permutation procedure by sampling an equal number of ROIs from each layer. In this analysis, we observed that 8 tiles per layer (24 tiles total) yielded very similar results (<10% outside 99% confidence interval) as the whole ROI procedure; thus our ROIs had an equal representation of cortical laminae. Therefore, all subsequent analyses used the average output from 24 random tiles independent of cortical layers. In slides with insufficient tissue for 24 total tiles (n=10), we report the average for all available tiles.
To further validate the sampling procedures, three investigators independently segmented cortical ribbon into ROIs using a standardized operating procedure and applied the random sampling scheme above for analysis. Digital image analysis data from each investigator (Supplemental Table 3) were similar and median bias between investigators was not significant (p>0.1); thus, the sampling method was reproducible.
Comparison of Inclusion Counts and %AO by IHC Staining
We compared digital tau CI counts and %AOnt to traditional ordinal scale sores for further validation of our methods. Tau CI/mm2 counts correlated with traditional ordinal scores (rho = 0.5, p<0.01) but %AOnt did not correlate with ordinal scores (rho = 0.1, p>0.05); Indeed, comparisons of ordinal scores revealed a significant difference in %AOnt between “low” and “moderate” or “severe” sections but “moderate” and “severe” cases had similar levels of %AOnt (Fig. 4A). In contrast, tau CI/mm2 more readily distinguished all three ordinal categories (Fig. 4B). In FTLD-TDPC, the quantitative data on large DNs were highly correlated with %AOt (rho = 0.9, p<0.001) and both correlated with ordinal TDP-43 scores (both rho = 0.6, p<0.001). Further, both DN/mm2 and %AOt could distinguish “moderate” and “severe” sections, but %AOt had better discrimination of “low” pathology sections (Fig. 4C, 4D). Analysis of negative control tissue for endogenous peroxidase background stain finds a very low average %AOt and inclusion count for both tau and TDP-43 algorithms (tau CI/mm2 = 0, tau %AOt = 0.001; TDP DN/mm2 = 1.2, TDP %AOt = 0.003; Supplemental Table 4). Further, analysis of non-neurodegenerative control tissue found similar low average levels of the signal detected (tau CI/mm2 = 0, tau %AOt = 0.01; TDP DN/mm2 = 0.9, TDP %AOt = 0.002; Supplemental Table 4). These control values are between two to four orders of magnitude lower than those observed in patient groups (Table 2), suggesting the chromogen signal quantified is specific for pathology and has negligible influence from background signal.
Comparison with Other Digital Image Analysis Platform
To test the generalizability of our method, we applied our optimal tau CI object detection and TDP %AO algorithms (Supplemental Table 2) in randomly sampled areas from a subset of cases (PiD, n=5; FTLD-TDPC, n=5) in an open-source image analysis software (ImageJ). Due to differences in the accepted file format and units of measure, it was necessary to convert image files and algorithm parameters into equivalent units (see Materials & Methods for details). We found, overall, that the algorithms detected similar levels of pathology (Supplemental Fig. 2) with reasonable accuracy for HALO derived quantification (i.e., %CV<25, Supplemental Table 5).
Regional Neuroanatomic Analyses
We evaluated regional pathological burden in the cases studied here and observed lower tau CI/mm2 count in STG compared with ACG and MFG and with lower %AOnt in STG compared with MFG (p<0.04; Table 2, Supplemental Fig. 1). Examination of regional pathology in FTLD-TDPC cases showed a lower DN/mm2 count and %AOt TDP in MFG that reached significance compared with ACG (p<0.02; Table 2, Supplemental Fig. 1). Direct comparison of the relative anatomic burden between PiD and FTLD-TDPC revealed that PiD cases had a trend for lower mean percent of total pathology in STG (19%) compared with FTLD-TDPC (34%; p=0.05), whereas FTLD-TDPC cases had a lower mean percent of total pathology in MFG (19%) compared with PiD (37%; p<0.01, Table 2). Trends for these associations were recognized using traditional ordinal scores but they did not reach significance (p≥0.05; Table 2).
Discussion
Here we describe a novel method of semi-automated digital image analysis and non-biased sampling method for cortical sections in bvFTD patients with underlying PiD or FTLD-TDPC. The automated detection algorithms showed a high level of agreement relative to gold-standard manually detected pathology (i.e., tau CI, TDP DN), at a level comparable with previous reports for AD (Neltner et al. 2012). Further, examination of the sampling methods revealed consistent results among the examiners and reasonable accuracy between software platforms. Finally, the neuroanatomic analyses from this pilot series demonstrated feasibility and applicability of these novel methods with findings not detected by traditional ordinal rating scales.
To our knowledge, the only reported digital image analysis of FTLD neuropathology to date has been performed for %AO of co-morbid TDP-43 inclusions in clinical/pathological AD (Josephs et al. 2014b). This study only examined the hippocampal dentate gyrus with digital image analysis but it did not examine FTLD-TDP cases. Further, continuous measures of TDP-43 pathology as %AO were re-categorized into an ordinal scale for analysis. %AO has been used to quantify AD neuropathology (Mitchell et al. 2002; Robinson et al. 2011), as it can provide useful information on the total burden of tau and Aβ deposits; %AO represents the fraction of total pixels containing IHC chromogen signal (i.e., coverage) (Armstrong 2003), but it does not allow for separation of individual pathological features stained with the same antibody. Using a similar approach to NT in AD (Mitchell et al. 2002; Mitchell et al. 2000), we were able to calculate the %AO of NT (%AOnt) separately in our analyses of PiD and we observed that %AOnt was able to distinguish cases with low levels of pathology, but could not easily distinguish between cases with moderate to severe pathology (Fig. 4a). This suggests that diffuse NT in PiD may reach a ceiling of maximum detectable intensity in moderate to severe disease, or, alternatively, that small diffuse NTs are less influential than intracellular inclusions in rater designation of ordinal scale scores (Armstrong 2003). Our object detection algorithms allow for measurement of specific morphological features, such as CI and DN, and can be validated through comparison with manually detected objects. Indeed, comparison of both measures finds additional sensitivity for higher severity cases using tau CI/mm2 in PiD. In contrast, since total FTLD-TDPC pathology is largely DN and there are negligible NTs and intracellular TDP-43 inclusions in FTLD-TDPC, DN/mm2 counts and total %AO measures were very similar. Thus, analysis of both measures is useful, and indeed we found similar results in our comparisons of regional pathologies (Table 2). Other features of PiD or FTLD-TDP, such as ballooned neurons, neuron loss and gliosis were not included in the current work and will be of importance in future studies. Co-morbid neuropathology is common in neurodegenerative conditions. Indeed, our sample included a range of low amyloid plaque burden but there was minimal AD-associated neurofibrillary tangle tau pathology (i.e., Braak 0-II; Table 1) to confound our quantification of PiD tau pathology. It will also be important to consider and measure the effect of multiple pathologies in future clinicopathological studies.
To demonstrate the feasibility of this approach, we performed preliminary neuroanatomic analyses in our pilot sample of FTLD neuropathology. We restricted analyses to cases with a clinical phenotype of bvFTD to limit variability on regional pathological burden and because this clinical syndrome is associated with equal frequencies of FTLD-Tau and FTLD-TDP (Forman et al. 2006; Irwin et al. 2015). Further comparative studies of bvFTD with FTLD-Tau or FTLD-TDP neuropathology are lacking (Rohrer 2011). Indeed, non-random hierarchical patterns of TDP-43 pathology have been described in bvFTD (Brettschneider et al. 2014) and AD (Josephs et al. 2014a)—suggesting sequential neuron-to-neuron spread of protein inclusions—but it is unclear how FTLD-Tau subtypes may progress through frontal and temporal brain regions in bvFTD. These stages are based on neuroanatomic regions relatively free of pathology, but qualitative accumulation of pathology within a region across stages can be difficult to document with a limited ordinal scale. Staging efforts have not been performed in FTLD-Tau, and digital image analysis could provide a complimentary role to staging efforts for FTLD. Indeed, our data finds a lower level of tau inclusions in the STG for PiD compared with frontal neocortical (MFG) and limbic (ACG) regions in group-wise comparisons; although some individual cases did not have a lower tau CI/mm2 in STG. Indeed, previous reports of PiD (Uchihara and Tsuchiya 2008) and FTLD-Tau with a MAPT mutation (Spina et al. 2008) described a relative sparing of the STG. In contrast, our data in FTLD-TDPC finds similar disease burden in STG and ACG compared with a lower amount of TDP-43 pathology in MFG. However, we previously found MFG, STG and ACG affected in the same early phase of pathology through examination of 70-µm-thick sections (Brettschneider et al. 2014); that study included other FTLD-TDP subtypes (i.e., A, B) and ordinal ratings of both STG and MTG combined, in comparison to our present focused study of STG, which may explain this partial discrepancy. Further study is needed to determine the exact nature of associations between staging and quantitative measures of pathological burden in FTLD. A direct comparison of the percent of total pathological burden finds a double-dissociation, where PiD has a lower percent of total pathology in STG whereas FTLD-TDPC has a lower total percent in MFG (Table 2). These preliminary associations in our pilot study suggest that FTLD neuropathological subtypes may have subtle differences in the pattern of disease in bvFTD, which could have potential clinical implications for improved diagnostics of underlying neuropathology (i.e., development of endophenotypes) (Irwin et al. 2015). Future studies in larger samples of bvFTD, including other forms of FTLD-Tau and FTLD-TDP, will help to confirm these observations. Importantly, these correlations were not detected by traditional ordinal scale data from our brain bank, which highlights the usefulness of digital image analysis for continuous measures of disease burden.
There are several limitations to the current study. We did not examine all forms of FTLD-Tau and FTLD-TDP but, instead, focused our analyses on PiD and FTLD-TDPC to capture the spectrum of FTLD morphological features for quantification (i.e., PBs and glial CI and diffuse NTs in PiD and large DNs in FTLD-TDPC). Future work will examine these methods in other FTLD subtypes. Tissue processing methods could potentially influence staining results. Phospho-tau epitopes in neurodegenerative disease are stable for prolonged PMIs (Lee et al. 1991; Matsuo et al. 1994) and insoluble TDP-43 aggregations are reliably identified in post-mortem FTLD-TDP brains (Neumann et al. 2006). Further, the PMI was ≤24 hr for all cases; thus, variations in the low PMI in this study should not affect the sensitivity of the antibodies used in this study. We also examined the effects of fixatives and found a similar level of agreement between digital and manual counts in the subset of ethanol-fixed sections as compared with NBF sections, but future digital image analysis studies should continue to examine pre-analytic factors of tissue preparation carefully. It is important to note that this method of digital image analysis does not represent a quantitative biochemical measure of pathological insoluble tau and TDP-43 proteins but, instead, represents a reliable, high-throughput method for obtaining continuous measures of disease burden in FTLD neuropathologies with vastly different morphological features. Indeed, the dynamic range of IHC is small, prohibiting a true analytic measurement of protein concentration from tissue (Rimm 2006). Further, analysis of cortical tissue, as compared with specific brain nuclei, is challenging due to the variable volume and orientation of cortical tissue in a given section (Neltner et al. 2012). The vertical transection sampling method presented here identifies “representative cortex” to evaluate disease burden through random sampling to minimize sampling bias (Armstrong 2003), similar to methods used in manual quantification studies of neocortical FTLD neuropathology (Armstrong et al. 1999a; Armstrong and Cairns 2012); but this approach does not replicate stereology (i.e., estimating 3-dimensional volume and cell/inclusion density from 2-dimensional images) (West et al. 1991). Thus, readers must use caution in interpretation of the meaning of our quantitative measures of tau CI/mm2 or TDP DN/mm2, as these are not absolute quantification of inclusions as in stereology, but rather represent a relative index of pathological burden that provides increased granularity for statistical associations. Future work using novel multiplexed immunofluorescence (Gerdes et al. 2013), image registration to map serial sections into a three-dimensional space (Adler et al. 2014) or tissue clearing technologies (Ando et al. 2014) may provide new avenues to explore these limitations further. Finally, our methods were developed using a proprietary software with specific algorithm building tools, as in reports of digital image analysis in AD (Byrne et al. 2009; Josephs et al. 2014b; Neltner et al. 2012; Samaroo et al. 2012) but we also show that our algorithms can be applied to a freely available software package using conversion factors with similar results (Supplemental Fig. 2, Supplemental Table 5). Further validation is necessary prior to combining data obtained from different software packages in a single study, but these results demonstrate the generalizability of our methods.
In summary, digital image analysis is a useful tool to provide continuous measures of FTLD neuropathologies both with largely intracellular (PiD) and neuritic inclusions (FTLD-TDPC), and there may be subtle differences between regional pathological burden between bvFTD with FTLD-Tau and FTLD-TDP neuropathology. Future studies using these methods in larger samples with other forms of FTLD neuropathology will help confirm these observations.
Supplementary Material
Acknowledgments
We thank Manuela Neumann and Elisabeth Kremmer for providing the phosphorylation specific TDP-43 antibody 1D3 and Dr. Peter Davies for his generous contribution of PHF-1 antibody. We would also like to thank the patients and their families for their meaningful contributions to research, for without this work would not be possible.
Footnotes
Author Contribution: DJI designed the experiments, performed digital image analysis experiments, performed statistical analysis, drafted the manuscript and provided funding. MDB and FC performed the immunohistochemistry and digital image analysis experiments. CTM performed statistical analyses. SXX supervised the statistical analyses and provided critical revisions of the manuscript. SEA, EBL, VVD, MG and JQT provided patient data. MG, SEA, CTM, VMYL and JQT provided funding, guidance for the experimental design and implementation and provided critical revisions of the manuscript. All authors have read and approved the final manuscript.
Competing Interests: The authors declared no potential competing interests with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study is supported by NIH grants NS044266, AG038490, AG015116, AG010124, P01NS053488, P01AG032953, P01AG017586, AG043503, NS088341 and the Wyncote Foundation.
References
- Adler DH, Pluta J, Kadivar S, Craige C, Gee JC, Avants BB, Yushkevich PA. (2014). Histology-derived volumetric annotation of the human hippocampal subfields in postmortem MRI. Neuroimage 84:505-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando K, Laborde Q, Lazar A, Godefroy D, Youssef I, Amar M, Pooler A, Potier MC, Delatour B, Duyckaerts C. (2014). Inside Alzheimer brain with CLARITY: senile plaques, neurofibrillary tangles and axons in 3-D. Acta Neuropathol 128:457-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong R, Cairns N, Lantos P. (1999a). The spatial patterns of Pick bodies, Pick cells and Alzheimer’s disease pathology in Pick’s disease. Neuropathology 19:64-70. [DOI] [PubMed] [Google Scholar]
- Armstrong RA. (2003). Quantifying the pathology of neurodegenerative disorders: quantitative measurements, sampling strategies and data analysis. Histopathology 42:521-529. [DOI] [PubMed] [Google Scholar]
- Armstrong RA, Cairns NJ. (2012). Different molecular pathologies result in similar spatial patterns of cellular inclusions in neurodegenerative disease: a comparative study of eight disorders. J Neural Transm 119:1551-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong RA, Cairns NJ, Lantos PL. (1999b). Laminar distribution of pick bodies, pick cells and Alzheimer disease pathology in the frontal and temporal cortex in Pick’s disease. Neuropathol Appl Neurobiol 25:266-271. [DOI] [PubMed] [Google Scholar]
- Brettschneider J, Del Tredici K, Irwin DJ, Grossman M, Robinson JL, Toledo JB, Fang L, Van Deerlin VM, Ludolph AC, Lee VM, Braak H, Trojanowski JQ. (2014). Sequential distribution of pTDP-43 pathology in behavioral variant frontotemporal dementia (bvFTD). Acta Neuropathol 127:423-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne UT, Ross JM, Faull RL, Dragunow M. (2009). High-throughput quantification of Alzheimer’s disease pathological markers in the post-mortem human brain. J Neurosci Methods 176:298-309. [DOI] [PubMed] [Google Scholar]
- Dickson D. (2004). ‘Sporadic Tauopaties: Pick’s disease, corticobasal degeneration, progressive supranuclear palsy and argyrophilic grain disease’ in Esiri M LVM-Y, Trojanowski JQ. (eds.) The Neuropathology of Dementia, NY:Cambridge University Press. [Google Scholar]
- Dickson DW, Kouri N, Murray ME, Josephs KA. (2011). Neuropathology of frontotemporal lobar degeneration-tau (FTLD-tau). J Mol Neurosci 45:384-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman MS, Farmer J, Johnson JK, Clark CM, Arnold SE, Coslett HB, Chatterjee A, Hurtig HI, Karlawish JH, Rosen HJ, Van Deerlin V, Lee VM, Miller BL, Trojanowski JQ, Grossman M. (2006). Frontotemporal dementia: clinicopathological correlations. Ann Neurol 59:952-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes MJ, Sevinsky CJ, Sood A, Adak S, Bello MO, Bordwell A, Can A, Corwin A, Dinn S, Filkins RJ, Hollman D, Kamath V, Kaanumalle S, Kenny K, Larsen M, Lazare M, Li Q, Lowes C, McCulloch CC, McDonough E, Montalto MC, Pang Z, Rittscher J, Santamaria-Pang A, Sarachan BD, Seel ML, Seppo A, Shaikh K, Sui Y, Zhang J, Ginty F. (2013). Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc Natl Acad Sci U S A 110:11982-11987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg M, Grujic ZM, Baker M, Demirci S, Guillozet AL, Sweet AP, Herzog LL, Weintraub S, Mesulam MM, LaPointe NE, Gamblin TC, Berry RW, Binder LI, de Silva R, Lees A, Espinoza M, Davies P, Grover A, Sahara N, Ishizawa T, Dickson D, Yen SH, Hutton M, Bigio EH. (2003). The L266V tau mutation is associated with frontotemporal dementia and Pick-like 3R and 4R tauopathy. Acta Neuropathol 106:323-336. [DOI] [PubMed] [Google Scholar]
- Irwin DJ, Cairns NJ, Grossman M, McMillan CT, Lee EB, Van Deerlin VM, Lee VM, Trojanowski JQ. (2015). Frontotemporal lobar degeneration: defining phenotypic diversity through personalized medicine. Acta Neuropathol 129:469-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin DJ, Cohen TJ, Grossman M, Arnold SE, McCarty-Wood E, Van Deerlin VM, Lee VM, Trojanowski JQ. (2013a). Acetylated tau neuropathology in sporadic and hereditary tauopathies. Am J Pathol 183:344-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin DJ, McMillan CT, Brettschneider J, Libon DJ, Powers J, Rascovsky K, Toledo JB, Boller A, Bekisz J, Chandrasekaran K, Wood EM, Shaw LM, Woo JH, Cook PA, Wolk DA, Arnold SE, Van Deerlin VM, McCluskey LF, Elman L, Lee VM, Trojanowski JQ, Grossman M. (2013b). Cognitive decline and reduced survival in C9orf72 expansion frontotemporal degeneration and amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry 84:163-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephs KA, Murray ME, Whitwell JL, Parisi JE, Petrucelli L, Jack CR, Petersen RC, Dickson DW. (2014a). Staging TDP-43 pathology in Alzheimer’s disease. Acta Neuropathol 127:441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephs KA, Whitwell JL, Weigand SD, Murray ME, Tosakulwong N, Liesinger AM, Petrucelli L, Senjem ML, Knopman DS, Boeve BF, Ivnik RJ, Smith GE, Jack CR, Jr., Parisi JE, Petersen RC, Dickson DW. (2014b). TDP-43 is a key player in the clinical features associated with Alzheimer’s disease. Acta Neuropathol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee VM, Balin BJ, Otvos L, Jr., Trojanowski JQ. (1991). A68: a major subunit of paired helical filaments and derivatized forms of normal Tau. Science 251:675-678. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, Neumann M, Baborie A, Sampathu DM, Du Plessis D, Jaros E, Perry RH, Trojanowski JQ, Mann DM, Lee VM. (2011). A harmonized classification system for FTLD-TDP pathology. Acta Neuropathol 122:111-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Neumann M, Bigio EH, Cairns NJ, Alafuzoff I, Kril J, Kovacs GG, Ghetti B, Halliday G, Holm IE, Ince PG, Kamphorst W, Revesz T, Rozemuller AJ, Kumar-Singh S, Akiyama H, Baborie A, Spina S, Dickson DW, Trojanowski JQ, Mann DM. (2010). Nomenclature and nosology for neuropathologic subtypes of frontotemporal lobar degeneration: an update. Acta Neuropathol 119:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo ES, Shin RW, Billingsley ML, Van deVoorde A, O’Connor M, Trojanowski JQ, Lee VM. (1994). Biopsy-derived adult human brain tau is phosphorylated at many of the same sites as Alzheimer’s disease paired helical filament tau. Neuron 13:989-1002. [DOI] [PubMed] [Google Scholar]
- Mitchell TW, Mufson EJ, Schneider JA, Cochran EJ, Nissanov J, Han LY, Bienias JL, Lee VM, Trojanowski JQ, Bennett DA, Arnold SE. (2002). Parahippocampal tau pathology in healthy aging, mild cognitive impairment, and early Alzheimer’s disease. Ann Neurol 51:182-189. [DOI] [PubMed] [Google Scholar]
- Mitchell TW, Nissanov J, Han LY, Mufson EJ, Schneider JA, Cochran EJ, Bennett DA, Lee VM, Trojanowski JQ, Arnold SE. (2000). Novel method to quantify neuropil threads in brains from elders with or without cognitive impairment. J Histochem Cytochem 48:1627-1638. [DOI] [PubMed] [Google Scholar]
- Montine TJ, Phelps CH, Beach TG, Bigio EH, Cairns NJ, Dickson DW, Duyckaerts C, Frosch MP, Masliah E, Mirra SS, Nelson PT, Schneider JA, Thal DR, Trojanowski JQ, Vinters HV, Hyman BT, National Institute on A, Alzheimer’s A (2012). National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease: a practical approach. Acta Neuropathol 123:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neltner JH, Abner EL, Schmitt FA, Denison SK, Anderson S, Patel E, Nelson PT. (2012). Digital pathology and image analysis for robust high-throughput quantitative assessment of Alzheimer disease neuropathologic changes. J Neuropathol Exp Neurol 71:1075-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Kwong LK, Lee EB, Kremmer E, Flatley A, Xu Y, Forman MS, Troost D, Kretzschmar HA, Trojanowski JQ, Lee VM. (2009). Phosphorylation of S409/410 of TDP-43 is a consistent feature in all sporadic and familial forms of TDP-43 proteinopathies. Acta Neuropathol 117:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. (2006). Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314:130-133. [DOI] [PubMed] [Google Scholar]
- Otvos L, Jr., Feiner L, Lang E, Szendrei GI, Goedert M, Lee VM. (1994). Monoclonal antibody PHF-1 recognizes tau protein phosphorylated at serine residues 396 and 404. J Neurosci Res 39:669-673. [DOI] [PubMed] [Google Scholar]
- Rimm DL. (2006). What brown cannot do for you. Nat Biotechnol 24:914-916. [DOI] [PubMed] [Google Scholar]
- Robinson JL, Geser F, Corrada MM, Berlau DJ, Arnold SE, Lee VM, Kawas CH, Trojanowski JQ. (2011). Neocortical and hippocampal amyloid-beta and tau measures associate with dementia in the oldest-old. Brain 134:3708-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer JD. (2011). Behavioural variant frontotemporal dementia–defining genetic and pathological subtypes. J Mol Neurosci 45:583-588. [DOI] [PubMed] [Google Scholar]
- Ruifrok AC, Johnston DA. (2001). Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol 23:291-299. [PubMed] [Google Scholar]
- Samaroo HD, Opsahl AC, Schreiber J, O’Neill SM, Marconi M, Qian J, Carvajal-Gonzalez S, Tate B, Milici AJ, Bales KR, Stephenson DT. (2012). High throughput object-based image analysis of beta-amyloid plaques in human and transgenic mouse brain. J Neurosci Methods 204:179-188. [DOI] [PubMed] [Google Scholar]
- Spina S, Farlow MR, Unverzagt FW, Kareken DA, Murrell JR, Fraser G, Epperson F, Crowther RA, Spillantini MG, Goedert M, Ghetti B. (2008). The tauopathy associated with mutation +3 in intron 10 of Tau: characterization of the MSTD family. Brain 131:72-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan RH, Shepherd CE, Kril JJ, McCann H, McGeachie A, McGinley C, Affleck A, Halliday GM. (2013). Classification of FTLD-TDP cases into pathological subtypes using antibodies against phosphorylated and non-phosphorylated TDP43. Acta Neuropathol Commun 1:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo JB, Van Deerlin VM, Lee EB, Suh E, Baek Y, Robinson JL, Xie SX, McBride J, Wood EM, Schuck T, Irwin DJ, Gross RG, Hurtig H, McCluskey L, Elman L, Karlawish J, Schellenberg G, Chen-Plotkin A, Wolk D, Grossman M, Arnold SE, Shaw LM, Lee VM, Trojanowski JQ. (2013). A platform for discovery: The University of Pennsylvania Integrated Neurodegenerative Disease Biobank. Alzheimers Dement 10:477-484.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchihara T, Tsuchiya K. (2008). Neuropathology of Pick body disease. Handb Clin Neurol 89:415-430. [DOI] [PubMed] [Google Scholar]
- West MJ, Slomianka L, Gundersen HJ. (1991). Unbiased stereological estimation of the total number of neurons in thesubdivisions of the rat hippocampus using the optical fractionator. Anat Rec 231:482-497. [DOI] [PubMed] [Google Scholar]
- Xie SX, Baek Y, Grossman M, Arnold SE, Karlawish J, Siderowf A, Hurtig H, Elman L, McCluskey L, Van Deerlin V, Lee VM, Trojanowski JQ. (2011). Building an integrated neurodegenerative disease database at an academic health center. Alzheimers Dement 7:e84-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhukareva V, Mann D, Pickering-Brown S, Uryu K, Shuck T, Shah K, Grossman M, Miller BL, Hulette CM, Feinstein SC, Trojanowski JQ, Lee VM. (2002). Sporadic Pick’s disease: a tauopathy characterized by a spectrum of pathological tau isoforms in gray and white matter. Ann Neurol 51:730-739. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.