Abstract
Bone is a dynamic tissue that undergoes multiple changes throughout its lifetime. Its maintenance requires a tight regulation between the cells embedded within the bone matrix, and an imbalance among these cells may lead to bone diseases such as osteoporosis. Identifying cell populations and their proteins within bone is necessary for understanding bone biology. Immunolabeling is one approach used to visualize proteins in tissues. Efficient immunolabeling of bone samples often requires decalcification, which may lead to changes in the structural morphology of the bone. Recently, methyl-methacrylate embedding of non-decalcified tissue followed by heat-induced antigen retrieval has been used to process bone sections for immunolabeling. However, this technique is applicable for bone slices below 50-µm thickness while fixed on slides. Additionally, enhancing epitope exposure for immunolabeling is still a challenge. Moreover, imaging bone cells within the bone environment using standard confocal microscopy is difficult. Here we demonstrate for the first time an improved methodology for immunolabeling non-decalcified bone using a testicular hyaluronidase enzyme-based antigen retrieval technique followed by two-photon fluorescence laser microscopy (TPLM) imaging. This procedure allowed us to image key intracellular proteins in bone cells while preserving the structural morphology of the cells and the bone.
Keywords: confocal microscopy, testicular hyaluronidase, bone, two photon fluorescence laser microscopy, osteocytes, osteoblasts, immunolabeling
Introduction
Bone formation is a tightly regulated process that requires coordination between osteoclasts and osteoblasts in the bone. The interaction and communication among different cell types within the bone environment leads to a densely packed, rigid structure comprising inorganic minerals, and collagenous and non-collagenous matrices (Clarke 2008). The cells are embedded within the mineralized matrix, and this matrix is crucial for the proper functioning of bone (Clarke 2008). In progressive bone disorders like osteoporosis, bone structures are weakened because of an imbalance between bone formation and bone resorption, which leads to bone fractures. Studying the bone composition, especially the localization of cells within the bone matrix and their protein regulation, is important to better understand bone turnover. However, to achieve this goal, cells must be preserved in their native, calcified structures.
Immunohistochemistry (IHC) is an attractive tool to determine cell localization and protein expression within tissues. However, applying this technique to bone is limited by several drawbacks (Matos et al. 2010). Common practices of sample preparation for IHC involve paraffin embedding of the decalcified bone tissue (An 2003). However, decalcifying bone leads to the loss of trabecular integrity, and this causes changes in the overall morphology, making it difficult to maintain the same bone cell environment as compared to that of native mineralized bone. Alternatively, methyl methacrylate (MMA) embedding of non-decalcified bone can be used to preserve the bone structure with inorganic phosphates (Erben 1997). However, sectioning of these MMA-embedded bone samples is difficult. Following MMA embedding, conventional IHC is performed using heat-induced antigen retrieval, a complicated process that requires extreme precision in controlling the temperature to save the sample from being destroyed (Merchant et al. 2006). Microwave-based heat-induced retrieval is another approach (Blythe et al. 1997); however, once again, controlling the correct temperature is a very important aspect of this technique (Yang et al. 2003). Heat-induced antigen retrieval is commonly utilized for sections under 10-µm thickness for IHC. The samples are then mounted onto slides, and structural defects and changes in protein activity at the subcellular level are analyzed by confocal microscopy (Wittenburg et al. 2009). This technique is limited in the number of fluorophores that can be used and still requires other alternative steps to improve antigen availability (Yang et al. 2003). Additionally, the quality of the images obtained is limited as a result of the low fluorescence intensity of the samples. Finally, only small areas of around 0.18 mm2 can be analyzed as compared with the large area covered by the tile scan (approximately 1.62 mm2).
Here, we demonstrate for the first time the application of a testicular hyaluronidase-based antigen retrieval method on non-decalcified tissues followed by two-photon fluorescence laser microscopy (TPLM) imaging. Testicular hyaluronidase successfully exposes the antigen epitopes in multiple tissue types (Suetterlin et al. 2004; Jurukovski et al. 2005). However, it has not used to interrogate MMA-embedded bone samples until now. Our method used testicular hyaluronidase without heating of the sample for a milder antigen retrieval. This prevented morphing of the sample as well as the necessity of fixation on a slide. We used TPLM imaging to penetrate deeper into the bone; thus, we demonstrate here the imaging of a larger area and the processing of higher quality images. Using a combination of these techniques allowed us to identify intracellular proteins without altering the gross morphology of the bone, as well as clearly resolve the anatomical distinctions among different cell populations.
Materials & Methods
Mice
All C57BL/6J mice were obtained from The Jackson Laboratory (Bar Harbor, ME) and maintained under conventional conditions. The animal protocol was approved by the IUCAC at the University of Delaware and The Jackson Laboratory. At 8 weeks of age, female C57BL/6J mice (n=7 per group) were injected with 50 µl of PBS into the tail vein for 5 days. This protocol was a part of another previous study, as described elsewhere (Akkiraju et al. 2015). Mice were sacrificed 4 weeks after the last PBS injection, and femurs were isolated and fixed using 10% neutral-buffered formalin (NBF) for 48 hr at 4°C before processing. Three mice samples (n=3/7 of the total of group) were allocated to this study, with the remaining mice used for the other study (Akkiraju et al. 2015).
Histology
Bones fixed in 10% NBF were embedded using a modified protocol by O’Brien and colleagues (2000). Chemicals purchased for infiltration and embedding were methyl methacrylate (MMA) (Acros; Moris Plains, NJ), N-butyl pthalate, and benzoyl peroxide (wet) (Fisher Scientific, Fair Lawn, NJ) dried carefully. Fixed bones were washed with PBS thoroughly at room temperature. Subsequently, bones were dehydrated using an ethanol series of 70% (8 to 16 hr), 90% (8 to 16 hr), 95% (8 to 16 hr), 100% (8 to 16 hr), 100% (8 to 16 hr), two changes of 2-propanol (8 to 16 hr each rinse) and two changes of methyl salicylate clearing solution (4 hr each). After complete dehydration, samples were infiltrated first with MMA I (765 ml MMA + 140 ml N-butyl pthalate) solution at room temperature for 48 hr, next with MMA II (765 ml MMA + 140 ml N-butyl pthalate + 9.0 g dry benzoyl peroxide) solution at 4°C for 48 hr, and then in MMA III (765 ml MMA + 140 ml N-butyl pthalate + 17.75 g dry benzoyl peroxide) solution at 4°C for 48 hr. Thickened MMA III was polymerized in glass vials, where the infiltrated samples were then placed on the polymerized layer of plastic and allowed to polymerize for 7 days in a 40°C oven. Polymerized blocks were trimmed and sectioned at 200 µm (10.2 cm × 0.3 mm) using an IsoMet Low Speed Saw (Buehler; Lake Bluff, IL) with a diamond wafering blade (Buehler), and sanded down to even and thin the surface using CarbiMet Abrasive Discs, MicroCut PSA 12-inch 1200[P2500] (Buehler)-grade sand paper.
Morphological Staining
MMA-embedded samples were cut, sanded, and stained using Villanueva Osteochrome bone stain according to the manufacturer’s protocol (Polysciences, Inc.; Warrington, PA). Phase-contrast images of MMA sections were taken using a Nikon TMS (model TMS-F #211153; Nikon, Tokyo, Japan).
Testicular Hyaluronidase-induced Antigen Retrieval
MMA-embedded samples at 200-µm thickness were sanded and then incubated in xylene for 1 min to dissolve the top layer of the MMA section. These samples were then kept in pre-warmed testicular hyaluronidase solution: 47 ml of 0.1 M potassium phosphate and 3 ml of 0.1 M sodium phosphate with 0.025 g of testicular hyaluronidase (H3884; Sigma-Aldrich, St. Louis, MO) (Luna 1968; Sheehan and Hrapchak 1980; Bancroft and Stevens 1982). Samples were incubated in the pre-warmed solution at 37°C for 30 min. Post-incubation samples were washed with 1 × PBS three times for 5 min each to remove all remnants of the testicular hyaluronidase solution before immunostaining.
Heat-induced Antigen Retrieval
Cut and sanded samples were treated with xylene for 1 min and then placed in 400 ml of 0.01 M sodium citrate buffer (pH 6.0) that was brought to boiling temperature using an electric rice cooker. Sections were placed in a ceramic coverslip holder in pre-heated sodium citrate buffer for 30 min, and then taken out and allowed to cool on ice. Samples were then washed using 1 × PBS three times for 5 min each. This heat-induced antigen retrieval method was adopted and modified for use in an electric rice cooker instead of a microwave to better control the temperature of the solution (Blythe et al. 1997).
Immunostaining
Immunostaining of plastic embedded samples was performed by modification and optimization of the protocols described by O’Brien et al. (2000) and Su et al. (2010) using non-decalcified MMA-embedded bone sections. Sections labeled for Smad1/5/8, pERK1/2, osteocalcin, and alkaline phosphatase (ALP) were pretreated in testicular hyaluronidase for 30 min at 37°C followed by blocking with 3% BSA for 1 hr. Blocked samples were labeled as follows: (1) rabbit polyclonal Smad 1/5/8 IgG (Santa Cruz Biotechnology; Dallas, TX) diluted 1:500 in 3% BSA overnight at 4°C followed by a 1:500 dilution of Alexa Fluor 546 goat anti-rabbit (Invitrogen; Eugene, OR) for 1 hr at room temperature (RT); (2) mouse monoclonal p44/p42 MAPK (ERK1/2) antibody (Cell Signaling; Danvers, MA) pre-conjugated with Alexa Fluor 633 goat anti-mouse IgG (Invitrogen) diluted 1:500 in 3% BSA for 1 hr; (3) rabbit polyclonal osteocalcin (Santa Cruz Biotechnology) diluted 1:500 in 3% BSA overnight at 4°C followed by a 1:500 dilution of Alexa Fluor 488 donkey anti-rabbit (Invitrogen) for 1 hr at room temperature; or (4) goat polyclonal ALP (Santa Cruz Biotechnology) diluted 1:500 in 3% BSA overnight at 4°C followed by a 1:500 dilution of Alexa Fluor 568 donkey anti-goat (Invitrogen) for 1 hr at room temperature. The Hoechst (Bisbenzinamide 33342; Sigma-Aldrich) nuclear stain was applied for 10 min and washed using PBS.
Confocal Imaging
Samples (n=3 mice) were imaged using a Zeiss LSM 780 NLO with a 20× EC Plan Apochromat- 0.6 HD DIC M27 (Zeiss; Oberkochen, Germany). Two-photon excitation was used to penetrate deeper into the plastic sections. A Titanium:Sapphire (Ti:Al2O3) laser was tuned to 780 nm with a laser power of 3.5%, and a Helium Neon (HeNe2) laser set at an excitation of 633 nm at 7% laser power equipped with 20×/0.6 NA HD M27 air objective corrected for samples without coverslips. The Main Beam Splitter (MBS) of the confocal was set to 488/561/633, MBS 760+, with the filters set to measure Alexa Fluor 488, -546, -568, -633, and Hoechst (405 nm excitation). Using the LSM 780 as a conventional confocal, the main beam splitter was set to MBS 488/561/633 and MBS 405. Fluorophores were detected using an Argon laser tuned to 488 nm at 2% laser power; a diode pumped solid state (DPSS) laser tuned to 561 nm at 7% laser power; a HeNe laser tuned to 633 nm at 2% laser power; and a diode 405-30 tuned to 405 nm with 2% laser power, with the filters set to measure Alexa Fluor 488, Alexa Fluor 546, Alexa Fluor 568, Alexa Fluor 633, and Hoechst (405 nm excitation). Objective comparison was done between 20×/0.6NA HD M27 EC Epiplan Apochromat corrected for samples without coverslips and 20×/0.75NA Plan Apochromat corrected for a 0.17-mm coverslip. All images were processed using ImageJ (NIH, Bethesda, MD).
Results
MMA Embedding Preserved the Gross Morphology of Bone
The examination of bone histomorphometry is essential for measuring localized bone turnover and remodeling using histological sections of bone (Compston 2004). However, bone remodeling still remains a poorly understood process due to the many technical limitations in the fields of histology and imaging.
Conventional techniques used to identify signaling events in calcified tissue are limited, and require decalcification of the tissue for thin sections (An 2003). However, in this process, the gross morphology of the bone is lost. Likewise, bone cells, such as osteoblasts, which traverse the bone in its calcified regions, are lost. Therefore, we embedded non-decalcified bone into MMA. Conventional MMA embedding has been around for decades because of its simplicity (Schenk 1965; O’Brien et al. 2000). Utilizing this technique, we embedded the trabecular heads of femurs to study the trabecular architecture and its cellular activity. Following embedding, our samples were sectioned and sanded to thin even areas for further processing. To understand the base morphology of the trabecular bone makeup (Clarke 2008), we used Villanueva Osteochrome stain to mark the regions of new bone formation by osteoblasts and the calcified regions of the tissue (Villanueva and Lundin 1989), as shown in Fig. 1.
Figure 1.
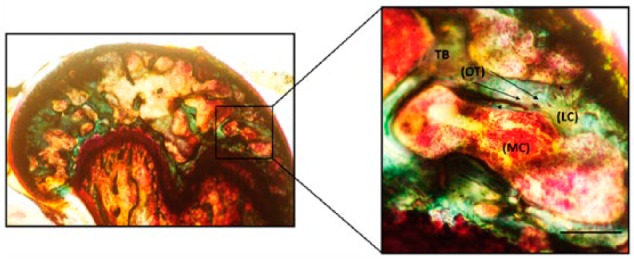
Trabecular morphology of the MMA-embedded femur sample. Femurs from mice were processed in MMA and stained by Villanueva Osteochrome. Phase-contrast images of the samples show osteocytes (OT) represented as dark brown dense osteoid seams of trabecular bone (TB; green) and osteoblasts or lining cells (LC) of the marrow cavity (MC; light red). Sections were imaged in the cancellous or TB of the mouse femur around the MC where the LC or the active osteoblasts reside alongside of OTs (indicated by arrows). Images were taken using 20× magnification. Scale, 50 µm. Imaging was conducted using a Nikon TMS (model TMS-F #211153).
Antigen Retrieval of Calcified Tissue by Testicular Hyaluronidase Digestion
In order to gain more insight into bone homeostasis, it is crucial to determine changes in cellular distribution and protein expression within bone. The use of imaging to examine cells and proteins in their native environment can provide an accurate evaluation of bone remodeling. Osteoblasts preserved in the calcified regions are responsible for overall bone formation. Smad1/5/8 and ERK1/2 signaling are required for osteoblast-mediated bone formation (Matsushita et al. 2009; Retting et al. 2009; Song et al. 2009). In addition, common biomarkers of osteoblasts, such as ALP and osteocalcin are used to determine the level of osteoblast activity during bone formation (Christenson 1997). Therefore, we chose these proteins for IHC of our bone sections.
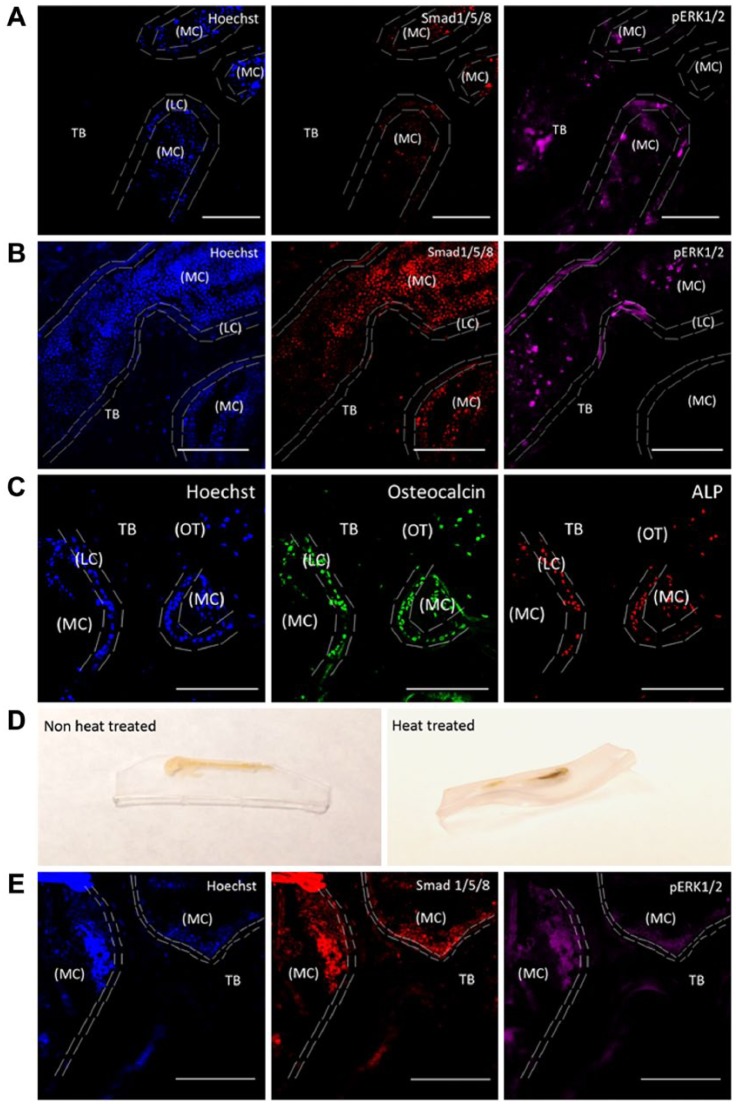
We used testicular hyaluronidase-based antigen retrieval to identify the cells from the tissue ECM, and this allowed us to immunostain for Smad, p-ERK, osteocalcin and ALP. Immunostaining was performed by pretreating bone sections with testicular hyaluronidase. Samples were then incubated with primary antibodies overnight followed by secondary antibodies at room temperature for 1 hr. As controls, we used heat-induced antigen retrieval, as described earlier (Jiao et al. 1999). We observed excellent antigen retrieval using our testicular hyaluronidase method as compared to the non-treated samples (Fig. 2A–2C). Heat-induced antigen retrieval led to the disfiguring of the sample making the imaging process an arduous task as it was hard to obtain a flat image surface (Fig. 2D). Additionally, the images obtained by this technique indicated low-quality staining (Fig. 2E).
Figure 2.
Testicular hyaluronidase-based antigen retrieval of MMA-embedded femur slices. MMA-embedded samples were immunostained for Smad 1/5/8, pERK1/2, osteocalcin and alkaline phosphatase (ALP). Hoechst staining was used to identify cells within the bone (blue). Heat-induced antigen retrieval was used as a comparison. Sections were imaged in the cancellous or the trabecular bone (TB) of the mouse femur around the marrow cavity (MC) where the lining cells (LC) or the active osteoblasts reside alongside osteocytes (OT). (A) MMA-embedded samples not treated with testicular hyaluronidase and imaged using TPLM. Tissue sections were stained with Hoechst (blue), regions of bone growth and proteins associated with bone cell activity were stained with Smad 1/5/8 (red) and pERK1/2 (magenta). (B) MMA samples treated with testicular hyaluronidase and stained as in (A) were imaged using TPLM. (C) MMA samples treated with testicular hyaluronidase and stained for osteocalcin (green), and ALP (red). (D) Heat-induced antigen retrieval caused morphing of the thick MMA-embedded bone sample if not mounted onto a permanent slide fixture. (E) MMA samples were heat treated and imaged using TPLM. Images were stained with Hoechst (blue), Smad 1/5/8 (red) and pERK1/2 (magenta). All images were taken using 20× magnification. Scale, 100 µM.
Two-photon Fluorescence Laser Microscopy (TPLM) Imaging of Thick MMA-embedded Sections
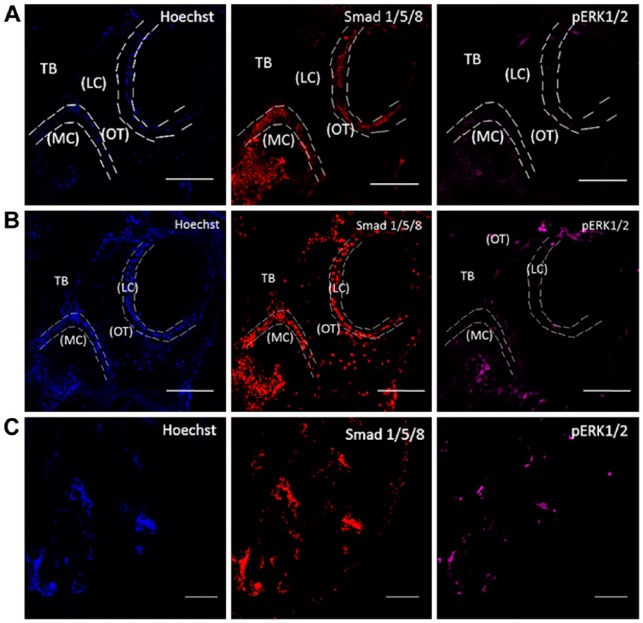
Conventional confocal microscopy is not the ideal source of imaging to view patterns of bone tissue on sections. This is because of its limited range of penetration through thicker bone tissue samples in the acquisition of images. To penetrate deeper into these thick samples, we used TPLM (Fig. 2) as the preferred technique, as it allows us to resolve microscopic structures with uneven ridges on an optically thick specimen (Piston 1999). To determine osteoblast protein expression levels in MMA-embedded samples, we immunolabeled sections for signaling factors (Smad1/5/8 and pERK1/2) and osteoblast biomarkers (ALP and osteocalcin). Sections were imaged using two-photon excitation microscopy. We compared the image quality obtained using TPLM versus conventional confocal microscopy. Using a tile scanning method, we stitched the images together to visualize the overall structure of the trabecular portion (Fig. 3). As can be seen, two-photon excitation microscopy surpassed the depth of penetration that could be achieved by conventional confocal microscopy, with a higher signal to noise ratio observed. This also allowed us to focus on protein expression at the cellular levels using high-resolution imaging to identify the cells expressing proteins of interest, as demonstrated in Fig. 4.
Figure 3.
Two-photon excitation laser microscopy imaging of MMA-embedded samples. Imaging of MMA-embedded samples stained with Hoechst (blue), Smad 1/5/8 (red) and pERK1/2 (magenta). (A) Conventional confocal setup. The collected images were of low-resolution and demonstrated high autofluorescence. (B) Two-photon excitation laser microscopy imaging. Images show nuclei and labeling for proteins. (C) A representative tile scan image that was used to examine the entire sample section. Sections were imaged in the cancellous or trabecular bone (TB) of the mouse femur around the marrow cavity (MC) where the lining cells (LC) or active osteoblasts reside alongside osteocytes (OT). Images were taken at 20× magnification. Scale (A, B) 100 µm; (C) 200 µm.
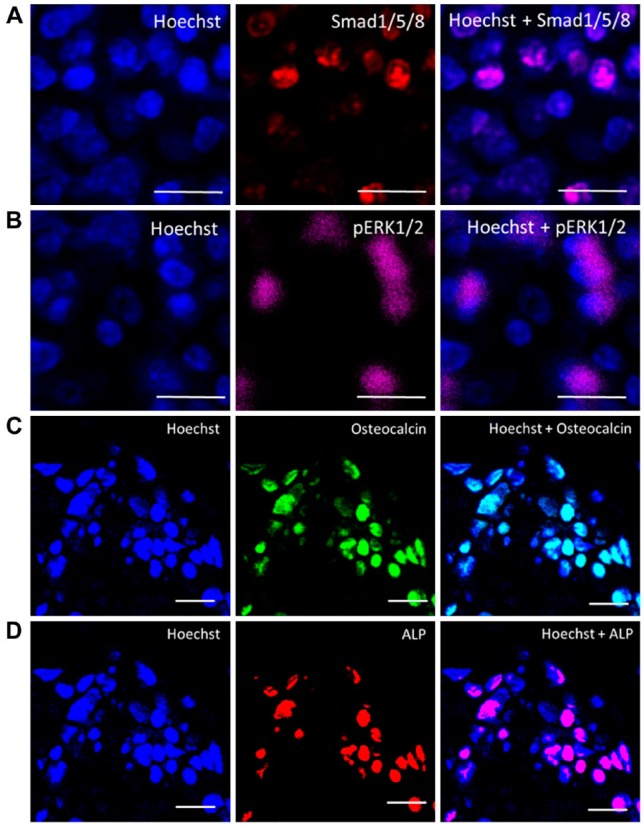
Figure 4.
High-resolution imaging of cells expressing Smad1/5/8, p-ERK, osteocalcin and alkaline phosphatase. Magnified regions of the images were taken using a 20× objective with 2× magnification at 4096 × 4096 pixels. (A) Smad1/5/8 (red), Hoechst (blue). (B) pERK1/2 (magenta). (C) Osteocalcin (green). (D) ALP (red) and Hoechst. The overlay images at high resolution clearly distinguish the different expressions of these proteins within the cells. Scale, 10 µm.
Selection of Objective Used for TLPM Imaging of Bone
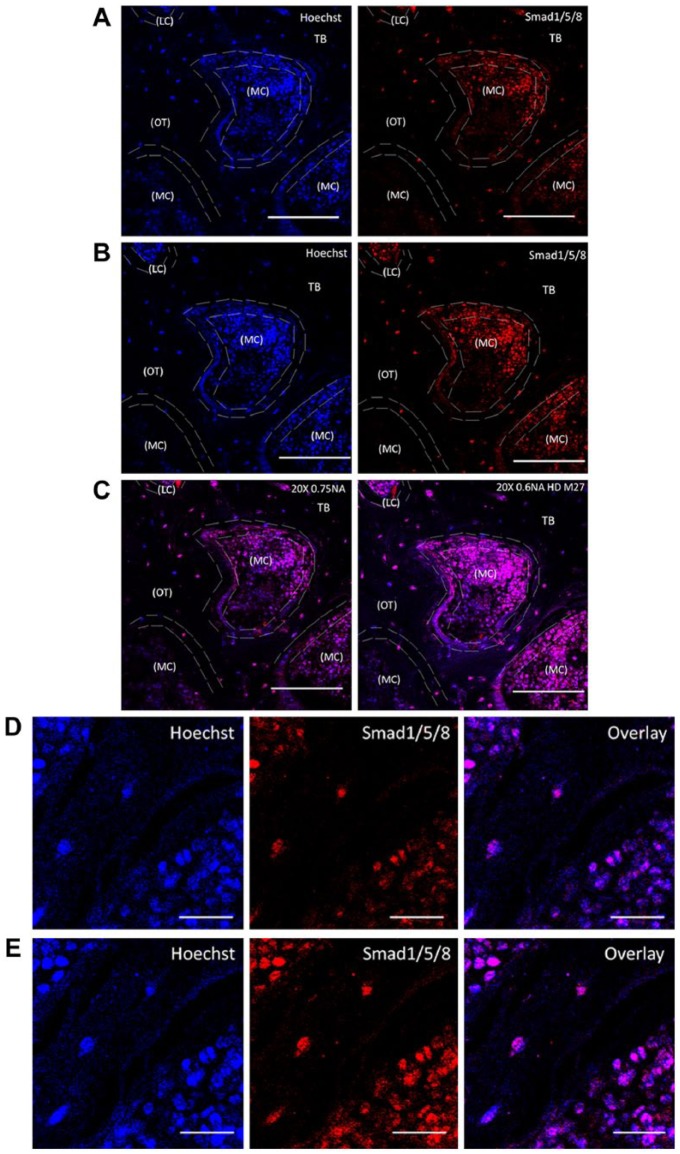
Thick bone samples were imaged without coverslips using a 20×/0.6NA HD M27 (EC Epiplan Apochromat) objective designed for samples without coverslips (Fig. 5A). For comparison, we also imaged the sample using 20×/0.75NA (Plan Apochromat) corrected for 0.17-mm coverslips (Fig. 5B). It is important to notice that the working distance of the objective effects the penetration of the laser beam into the sample (Piston 1999). The working distance of the 20×/0.6NA HD M27 objective is 1.7 mm, whereas the working distance for 20×/0.75NA is only 0.61 mm. This greater working distance allowed for the deeper penetration of the sample and compensated for the loss in NA to obtain a larger area of information (Fig. 5).
Figure 5.
Objective differences and their impact on sample brightness. MMA-embedded bone samples were TPLM-imaged on an inverted microscope Zeiss 780 using (A) 20×/NA 0.75 designed to be imaged with coverslips or a (B) 20×/NA 0.6 HD M27 for samples without coverslips. The minor differences in spatial resolution are compared based on the working distances of each objective. For the samples without coverslips, an objective of 20×/NA 0.6 HD M27 had a working distance of 1.7 mm, which provided the best imaging conditions for a better field of view, as shown in the overlay image in (C). Sections were imaged in the cancellous or trabecular bone (TB) of the mouse femur around the marrow cavity (MC) where the lining cells (LC) or the active osteoblasts reside alongside osteocytes (OT), labeled for Smad1/5/8 (red) and Hoechst (blue). bar representing 100 µm. High magnified images clearly distinguish objective differences in the staining of Smad (red) and Hoechst (blue) with overlay in images taken using (D) 20×/0.75NA, and (E) 20×/0.6NA HD M27. Images taken at 20× magnification. Scale (C) 100 µm; (D) 20 µm.
Troubleshooting
Understanding the proper parameters is important to obtain the best sample. Table 1 summarizes potential problems and their solutions during the process. Compiling this table allowed us to visualize the signaling proteins within the osteoblasts, the lining cells and the osteocytes.
Table 1.
Trouble Shooting of TPLM Imaging of Non-Decalcified Testicular Hyaluronidase Antigen Retrieved Bones Embedded in MMA.
| Problem | Possible Reasons | Solution |
|---|---|---|
| Casting of MMA with bubbles as a consequence. | Moisture retention in the MMA mixture. | No moisture should be introduced to the thickened MMA solution. |
| Improper penetration of MMA into the sample | Improper dehydration of sample | Adequate time for alcohol changes must be given |
| Uneven surface planes of cut sections | Uneven sanding of tissue samples. Usage of non-diamond-based wafered saws/ or without water | Use 1200 grade sand paper with water flow to sand the tissue sample. It is imperative to use even pressure throughout the sample for even sanding. |
| Softened tissue sample | Long xylene washes of samples | Start with 1 min to clear the surface plastic, taking care not to completely clear the sample of plastic. If needed, continue for 30 sec at a time to test the correct timing required for the plastic clearance. |
| Nonspecific staining of antibodies | Incorrect blocking buffer | Antibodies raised in the host tissue require prior blocking with either goat serum or horse serum to impede nonspecific binding. Pre-conjugation of the primary and secondary antibodies aid in staining without non-specific binding. |
| Improper resolution of sample | Usage of wrong objective | Using proper objectives designed for tissue sample imaging with or without coverslip along with matched NA for air will allow for the best resolution of the sample |
Discussion
Bone is a dynamic tissue that undergoes continuous formation and resorption, regulated by bone cells such as osteoblasts and osteoclasts, respectively. In progressive degenerative bone disorders like osteoporosis or osteomalacia, an imbalance between cellular activities is responsible for these bone defects. The majority of changes within bone occur within the cancellous bone or on the trabecular bone surface (Brandi 2009). Therefore, it is imperative to understand the cellular signaling activity that causes such major imbalances among bone cells.
We are able to better understand cellular activity in bone formation by identifying changes in osteoblast intracellular signaling activity. For example, previous work has demonstrated alterations to the activities of Smad1/5/8 and ERK1/2 during osteoblast differentiation and bone formation (Matsushita et al. 2009; Retting et al. 2009; Song et al. 2009). Changes in osteoblast activity can also be assessed using biomarkers, such as ALP and osteocalcin (Christenson 1997). However, direct labeling of these proteins in bone samples is challenging. Indeed, labeling proteins in general in cells within the bone environment is difficult, and the current practice of decalcifying bone may not be the best method to identify changes in these proteins and ascertain their dynamics in bone turnover or bone remodeling.
We demonstrate for the first time the imaging of intracellular signaling proteins in calcified bone tissue using testicular hyaluronidase-based antigen retrieval. We optimized the methodology for the immunostaining of multiple fluorophores and utilized two-photon microscopy for imaging sample specimens with maximal resolution. For decades, non-decalcified bone tissue has been commonly used, embedding the samples in a resin-type material for hard tissue sections. This included the cold processing of MMA for hard tissue embedding, similar to that achieved using the Technovit 9100, and this processing showed greater preservation of antigenicity (Erben 1997; An 2003; Willbold and Witte 2010). While the authors agree with its superiority in terms of preservation, we focused here only on the conventional MMA technique and remedial process for retrieving antigenicity to the tissue samples. We, therefore, utilized conventional MMA embedding for our experimental study. Moreover, due to chemical alterations during fixation and processing in conventional MMA embedding, the samples required harsh treatments for antigen retrieval. In general, there are two types of antigen retrieval methods: heat-induced epitope retrieval and enzyme-induced epitope retrieval (Shi et al. 1997; Mueller et al. 2000; Willbold and Witte 2010; Shi et al. 2011). However, only heat-induced antigen retrieval was successfully demonstrated in MMA-embedded samples.
Of notable concern is the trauma induced to the sample as a result of high temperature conditions and the disruption to tissue morphology (Merchant et al. 2006). Developing a process that uses a milder enzyme-based antigen retrieval could offer advantages over heat-induced antigen retrieval. This may allow for labeling of sections thicker than 10-µm thickness that are not mounted on slides. Furthermore, reducing the number of steps required for sample processing could ensure sample integrity and improve labeling of the intracellular proteins. A pragmatic approach utilizing enzyme-based antigen retrieval could also help preserve the tissue in its present form without adverse changes to the surrounding proteins or to the structure. As an enzyme-based antigen retrieval, testicular hyaluronidase is known to successfully expose the antigen epitopes in multiple tissue types (Suetterlin et al. 2004; Jurukovski et al. 2005). However, this was not demonstrated in MMA-embedded samples until now.
Advances in confocal microscopy avail us the opportunity to visualize the micro-anatomical components of bone. To image these immunostained samples, TPLM-based imaging is best suited for obtaining ideal resolution without background noise, which can be generated by the surrounding ECM and the uneven ridges present on the surface of calcified tissue. Moreover, TPLM allows for the penetration of thick specimens for imaging deeper depths of the tissue as compared with conventional confocal microscopy (Gerritsen and De Grauw 1999; Thériault et al. 2014). TPLM also permits the imaging of sensitive proteins with a reduced loss of fluorophore activity. Utilizing the microscope’s focal volume but not confocal pinhole allows for a higher signal. Moreover, the use of an emission pinhole in conventional confocal microscopy leads to inevitable scattering of fluorescent photons. TPLM, however, requires no pinhole; thus, this particular method can be exploited for exciting multiple fluorescent molecules by simultaneous absorption (Gerritsen and De Grauw 1999). Photo damage and photo bleaching of fluorophores are limited in TPLM as compared with conventional confocal microscopy (Rubart 2004). These advances allowed us high-resolution imaging of decalcified bone samples without the interference of background noise. Also, we used an objective that is corrected for samples without coverslips that covered a greater working distance for the enhancement of overall depth penetration and information acquired. It is important to ascertain the objective’s best working distance from the sample to obtain more information; this becomes a crucial aspect when imaging thick samples, as it allows for a deeper penetration into the tissue (Piston 1999).
The protocol presented here is straightforward; however, multiple problems can occur during the embedding process. We highlighted most of the common mistakes and their solution in Table 1.
The methodology described by us has multiple applications that are not only limited to bone diseases. It can be applied to identify multiple problems related to bone cancer metastasis by visualizing the metastatic nature of cancer cells (Hanna et al. 2012). Moreover, our findings may help to determine the protein expressions that may be involved in controlling the surrounding tissue. In conclusion, our improved methodology allows for the first time the identification of multiple protein targets within the intact bone environment.
Acknowledgments
The authors would like to thank Michael Moore of the Delaware Biotechnology Institute for aiding us with the image setup on the Zeiss 780. We would also like to thank Dr. Liyun Wang of University of Delaware for allowing us to utilize the Buehler Isomet saw for our experiments.
Footnotes
Author Contributions: Hemanth Akkiraju performed all of the experiments and wrote the manuscript. Jeremy Bonor extracted the animal tissue and helped in the embedding process. Anja Nohe analyzed the images and oversaw the writing of this manuscript.
Competing Interests: The authors declared no potential competing interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work has been supported by NIH grant R01AR064243.
References
- Akkiraju H, Bonor J, Olli K, Bowen C, Bragdon B, Coombs H, Donahue LR, Duncan R, Nohe A. (2015). Systemic injection of CK2.3, a novel peptide acting downstream of bone morphogenetic protein receptor BMPRIa, leads to increased trabecular bone mass. J Orthop Res 33:208-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An YH, Martin KL. (2003). Histological techniques for decalcified bone and cartilage Handbook of Histology Methods for Bone and Cartilage. New York: Humana Press. [Google Scholar]
- Bancroft JD, Stevens A. (1982). Theory and Practice of Histological Techniques, 2nd edition, New York: Churchill Livingstone. [Google Scholar]
- Blythe D, Hand NM, Jackson P, Barrans SL, Bradbury RD, Jack AS. (1997). Use of methyl methacrylate resin for embedding bone marrow trephine biopsy specimens. J Clin Pathol 50:45-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandi ML. (2009). Microarchitecture, the key to bone quality. Rheumatology (Oxford) 48 Suppl 4:iv3-8. [DOI] [PubMed] [Google Scholar]
- Christenson RH. (1997). Biochemical markers of bone metabolism: an overview. Clin Biochem 30:573-593. [DOI] [PubMed] [Google Scholar]
- Clarke B. (2008). Normal bone anatomy and physiology. Clin J Am Soc Nephrol 3 Suppl 3:S131-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compston J. (2004). Bone histomorphometry [mdash] the renaissance? IBMS BoneKEy 1:9-12. [Google Scholar]
- Erben RG. (1997). Embedding of bone samples in methylmethacrylate: an improved method suitable for bone histomorphometry, histochemistry, and immunohistochemistry. J Histochem Cytochem 45:307-313. [DOI] [PubMed] [Google Scholar]
- Gerritsen HC, De Grauw CJ. (1999). Imaging of optically thick specimen using two-photon excitation microscopy. Microsc Res Tech 47:206-209. [DOI] [PubMed] [Google Scholar]
- Hanna W, Barnes P, Berendt R, Chang M, Magliocco A, Mulligan AM, Rees H, Miller N, Elavathil L, Gilks B, Pettigrew N, Pilavdzic D, Sengupta S. (2012). Testing for her2 in breast cancer: current pathology challenges faced in Canada. Curr Oncol 19:315-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Sun Z, Lee T, Fusco FR, Kimble TD, Meade CA, Cuthbertson S, Reiner A. (1999). A simple and sensitive antigen retrieval method for free-floating and slide-mounted tissue sections. J Neurosci Methods 93:149-162. [DOI] [PubMed] [Google Scholar]
- Jurukovski V, Dabovic B, Todorovic V, Chen Y, Rifkin DB. (2005). Methods for measuring TGF-b using antibodies, cells, and mice. Methods Mol Med 117:161-175. [DOI] [PubMed] [Google Scholar]
- Luna LG. (1968). Manual of Histologic Staining Methods of the Armed Forces Institute of Pathology, 3rd ed. NY, Blackiston Division, McGraw-Hill. [Google Scholar]
- Matos LL, Trufelli DC, de Matos MG, da Silva Pinhal MA. (2010). Immunohistochemistry as an important tool in biomarkers detection and clinical practice. Biomark Insights 5:9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita T, Chan YY, Kawanami A, Balmes G, Landreth GE, Murakami S. (2009). Extracellular signal-regulated kinase 1 (ERK1) and ERK2 play essential roles in osteoblast differentiation and in supporting osteoclastogenesis. Mol Cell Biol 29:5843-5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SN, Burgess B, O’Malley J, Jones D, Adams JC. (2006). Polyester wax: a new embedding medium for the histopathologic study of human temporal bones. Laryngoscope 116:245-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller M, Wacker K, Hickey WF, Ringelstein EB, Kiefer R. (2000). Co-localization of multiple antigens and specific DNA. A novel method using methyl methacrylate-embedded semithin serial sections and catalyzed reporter deposition. Am J Pathol 157:1829-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien FJ, Taylor D, Dickson GR, Lee TC. (2000). Visualisation of three-dimensional microcracks in compact bone. J Anat 197 Pt 3:413-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piston DW. (1999). Imaging living cells and tissues by two-photon excitation microscopy. Trends Cell Biol 9:66-69. [DOI] [PubMed] [Google Scholar]
- Retting KN, Song B, Yoon BS, Lyons KM. (2009). BMP canonical Smad signaling through Smad1 and Smad5 is required for endochondral bone formation. Development 136:1093-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubart M. (2004). Two-photon microscopy of cells and tissue. Circ Res 95:1154-1166. [DOI] [PubMed] [Google Scholar]
- Schenk R. (1965). [On the histological processing of undecalcified bone]. Acta Anat (Basel) 60:3-19. [PubMed] [Google Scholar]
- Sheehan DC, Hrapchak BB. (1980). Theory and practice of Histotechnology, 2nd ed, Battelle Press. [Google Scholar]
- Shi SR, Cote RJ, Taylor CR. (1997). Antigen retrieval immunohistochemistry: past, present, and future. J Histochem Cytochem 45:327-343. [DOI] [PubMed] [Google Scholar]
- Shi SR, Shi Y, Taylor CR. (2011). Antigen retrieval immunohistochemistry: review and future prospects in research and diagnosis over two decades. J Histochem Cytochem 59:13-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B, Estrada KD, Lyons KM. (2009). Smad signaling in skeletal development and regeneration. Cytokine Growth Factor Rev 20:379-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su N, Sun Q, Li C, Lu X, Qi H, Chen S, Yang J, Du X, Zhao L, He Q, Jin M, Shen Y, Chen D, Chen L. (2010). Gain-of-function mutation in FGFR3 in mice leads to decreased bone mass by affecting both osteoblastogenesis and osteoclastogenesis. Hum Mol Genet 19:1199-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetterlin R, Baschong W, Laeng RH. (2004). Immunofluorescence and confocal laser scanning microscopy of chronic myeloproliferative disorders on archival formaldehyde-fixed bone marrow. J Histochem Cytochem 52:347-354. [DOI] [PubMed] [Google Scholar]
- Thériault G, Cottet M, Castonguay A, McCarthy N, De Koninck Y. (2014). Extended two-photon microscopy in live samples with Bessel beams: steadier focus, faster volume scans, and simpler stereoscopic imaging. Front Cell Neurosci 8:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva AR, Lundin KD. (1989). A versatile new mineralized bone stain for simultaneous assessment of tetracycline and osteoid seams. Stain Technol 64:129-138. [DOI] [PubMed] [Google Scholar]
- Willbold E, Witte F. (2010). Histology and research at the hard tissue-implant interface using Technovit 9100 New embedding technique. Acta Biomater 6:4447-4455. [DOI] [PubMed] [Google Scholar]
- Wittenburg G, Volkel C, Mai R, Lauer G. (2009). Immunohistochemical comparison of differentiation markers on paraffin and plastic embedded human bone samples. J Physiol Pharmacol 60 Suppl 8:43-49. [PubMed] [Google Scholar]
- Yang R, Davies CM, Archer CW, Richards RG. (2003). Immunohistochemistry of matrix markers in Technovit 9100 New-embedded undecalcified bone sections. Eur Cell Mater 6:57-71; discussion 71. [DOI] [PubMed] [Google Scholar]