A metabolic biosignature of early Lyme disease resulted in significantly greater diagnostic sensitivity and similar specificity, as compared with 2-tier serology, and correctly identified 77%–95% of early Lyme disease patients that were negative by 2-tier serology.
Keywords: Lyme disease, Borrelia burgdorferi, metabolomics, diagnostics, infectious diseases
Abstract
Background. Early Lyme disease patients often present to the clinic prior to developing a detectable antibody response to Borrelia burgdorferi, the etiologic agent. Thus, existing 2-tier serology-based assays yield low sensitivities (29%–40%) for early infection. The lack of an accurate laboratory test for early Lyme disease contributes to misconceptions about diagnosis and treatment, and underscores the need for new diagnostic approaches.
Methods. Retrospective serum samples from patients with early Lyme disease, other diseases, and healthy controls were analyzed for small molecule metabolites by liquid chromatography-mass spectrometry (LC-MS). A metabolomics data workflow was applied to select a biosignature for classifying early Lyme disease and non-Lyme disease patients. A statistical model of the biosignature was trained using the patients' LC-MS data, and subsequently applied as an experimental diagnostic tool with LC-MS data from additional patient sera. The accuracy of this method was compared with standard 2-tier serology.
Results. Metabolic biosignature development selected 95 molecular features that distinguished early Lyme disease patients from healthy controls. Statistical modeling reduced the biosignature to 44 molecular features, and correctly classified early Lyme disease patients and healthy controls with a sensitivity of 88% (84%–95%), and a specificity of 95% (90%–100%). Importantly, the metabolic biosignature correctly classified 77%–95% of the of serology negative Lyme disease patients.
Conclusions. The data provide proof-of-concept that metabolic profiling for early Lyme disease can achieve significantly greater (P < .0001) diagnostic sensitivity than current 2-tier serology, while retaining high specificity.
Lyme disease (LD), caused by Borrelia burgdorferi, is the most commonly reported tick-borne disease in the United States and Europe [1, 2]. Recent studies suggest that 300 000 cases of LD may occur in the United States each year [2]. Antibody-based diagnostics for LD are widely utilized in clinical practice [3], and the Centers for Disease Control and Prevention (CDC) recommends a 2-tier approach for serologic testing [4]. The detection of antibodies to B. burgdorferi is highly specific and sensitive in patients with late manifestations of LD; however, the sensitivity in patients with early LD is unsatisfactory (29%–40%) [3]. Direct diagnostic testing using culture or nucleic acid amplification on peripheral blood samples also has low sensitivity (≤50%) for early LD [5]. Thus, the diagnosis of early LD is usually based on recognition of the most common clinical manifestation, an erythema migrans (EM) skin lesion. Other skin lesions, however, such as tick-bite hypersensitivity reactions, STARI (southern tick associated rash illness), and certain cutaneous fungal infections, can be confused with EM [1, 6, 7].
Given the limitations of existing diagnostics for early LD, the feasibility of novel approaches that directly detect infecting spirochetes or the host's response to the pathogen should be evaluated. Modern “omic” technologies provide sensitive methods to investigate, discover, and validate individual molecules or panels of molecules as biomarkers or biosignatures of specific disease states [8, 9]. One such technology, metabolomics, allows for global analyses of low molecular mass (typically <1500 Da) biological molecules [9]. The metabolic activity of a biological system is strongly influenced by environmental factors, including infection. As a result, altered metabolic profiles may reflect a disease state and can be exploited for development of diagnostics [10]. Recently, metabolomics has resulted in the discovery of biosignatures for human infectious diseases, including diagnostic approaches for schistosomiasis and malaria [11, 12]. To test the feasibility of metabolic profiling as a diagnostic platform for LD, we evaluated a large retrospective cohort of sera from patients with early LD, other diseases and healthy controls. This resulted in a metabolic biosignature that yielded a sensitivity of 84%–95% for early LD detection while retaining high specificity (90%–100%), thus demonstrating the feasibility of a novel nonantibody test for improved laboratory diagnosis of early LD.
METHODS
Clinical Samples
Sera used for biosignature discovery and statistical modeling were procured from repositories at New York Medical College, the CDC [13], and Tufts University. Sera from early LD patients were collected pretreatment at the initial visit to the clinic. Healthy control serum donors were from endemic and nonendemic regions for LD. Other disease sera were from patients with infectious mononucleosis, fibromyalgia, severe periodontitis, or syphilis. Table 1 provides a detailed description of each patient population. All participating institutions obtained institutional review board approval.
Table 1.
Serum Samples Used in This Study
| Description of Samples | Sample Numbers | Sample Criteria for Inclusion | Sample Purpose | Sample Providera | Sample Set Abbreviation |
|---|---|---|---|---|---|
| Lyme Disease (n = 202) | |||||
| Early Lyme disease Age:16–72 Male (94), Female(46) |
140 | At least 1 EM present on initial visit to the clinic. Pretreatment samples collected at initial visit (all but 3 samples were collected within 30 d of onset). Positive culture and/or PCR test for B. burgdorferi. Patients lived in endemic area for Lyme disease. | Discovery and Test | New York Medical College (NYMC) | EL-NYMC |
| Early Lyme disease Age:21–80 Male (22), Female(18) |
40 | At least 1 EM present on initial visit to the clinic. Pretreatment samples collected at initial visit (collected within 10–35 d of onset). Positive culture and/or PCR test for B. burgdorferi in 65% of samples. Patients lived in endemic area for Lyme disease [13]. | Discovery and Test | CDC LSR | EL-CDC |
| C6-positive for Lyme disease Age: 9–83 Male (12), Female(10) |
22 | Clinically diagnosed with Lyme disease and positive by C6 EIA. Samples collected at initial visit to clinic, pretreatment, and within 20 d of onset. EM present in 6 patients, not present in 8 patients and EM status was unknown for 8 patients. Patients lived in endemic area for Lyme disease. | Test | Tufts University (TU) | EL-TU |
| Non-Lyme Disease Controls (n = 259) | |||||
| Healthy endemic Age:18–74 Male (26), Female(24) |
50 | No history of Lyme disease or tick-borne infection and individuals lived in an endemic area for Lyme disease for at least 5 years; no history of rheumatoid arthritis, multiple sclerosis, fibromyalgia, syphilis, or severe periodontitis was reported [13]. | Discovery and Test | CDC LSR | HEC-CDC |
| Healthy nonendemic Age:13–66 Male (39), Female(30) |
69 | No history of Lyme disease or tick-borne infection and had not lived in a Lyme disease endemic area within the previous 5 years; no history of rheumatoid arthritis, multiple sclerosis, fibromyalgia, syphilis, or severe periodontitis was reported [13]. | Discovery and Test | CDC LSR | HNC-CDC |
| Healthy endemic Age:27–49 Male (3), Female(4) |
7 | No history of Lyme disease, severe skin disease, diabetes, cancer, autoimmune disease, chronic hepatitis, HIV infection, or syphilis and lived in an endemic area for Lyme disease. | Test | Tufts University (TU) | HEC-TU |
| Healthy endemic Age:25–66 Male (1), Female(6) |
7 | No history of Lyme disease or tick-borne infection and individuals lived in an endemic area for Lyme disease for at least 5 years; no history of rheumatoid arthritis, multiple sclerosis, fibromyalgia, syphilis, or severe periodontitis was reported. | Test | New York Medical College (NYMC) | HEC-NYMC |
| Healthy nonendemic Age: Unknown Male (8), Female(17) |
25 | No history of tick-borne diseases in the past 12 mo and lived in a nonendemic area for Lyme disease. No history of an immunocompromising condition. | Test | CDC, Fort Collins CO. | HNC-CO |
| Diseases with overlapping clinical features Age:18–64b Male (53), Female(17)b |
101 | No history of Lyme disease; diagnosed with syphilis (n = 20), severe periodontitis (n = 20), infectious mononucleosis (n = 30), or fibromyalgia (n = 31) [13]. | Test | CDC LSR | LAD-CDC |
Abbreviations: CDC LSR, Centers for Disease Control and Prevention Lyme Serum Repository [13]; EIA, enzyme immunoassay; EL, early Lyme disease; EM, erythema migrans; HIV, human immunodeficiency virus; LAD, look-alike diseases; PCR, polymerase chain reaction.
a Sample handling varied among the laboratories that provided samples.
b Age and male/female ratio unknown for fibromyalgia patients.
Serologic Testing
Serologic testing was performed using the CDC recommended 2-tier testing algorithm [4]. The VIDAS Lyme immunoglobulin M (IgM) and immunoglobulin G (IgG) polyvalent assay (bioMérieux, Inc., Durham, North Carolina) was used as the first-tier enzyme immunoassay (EIA) and separate IgM and IgG immunoblots (MarDx Diagnostics, Inc., Carlsbad, California) were performed as second-tier tests. Serologic assays were performed according to the manufacturer's instructions, and the data were interpreted according to established CDC guidelines [4]. Duration of illness, however, was not considered in test interpretation. A C6 EIA (Immunetics, Boston, Massachusetts) was also performed as an alternative first- or second-tier test [14].
Sample Preparation and Liquid Chromatography-Mass Spectrometry (LC-MS)
Small molecule metabolites were extracted from aliquots (20 µL) of sera with 75% (final vol) HPLC grade methanol [15]. An aliquot equivalent to 5 µL of serum was analyzed by LC-MS (see Supplementary Material).
Data Analyses and Biosignature Selection
Sera and corresponding LC-MS data were randomly separated into discovery/training- and test-samples [16]. Figure 1A and Supplementary Material describes the metabolomics workflow for comparative analyses of early LD vs healthy control discovery-data, and the down-selection of molecular features (MFs, ie, metabolites defined by retention time and accurate mass). LC-MS data of the discovery-samples were processed with the Molecular Feature Extractor algorithm tool of the Agilent MassHunter Qualitative Analysis software. The Agilent Mass Profiler Pro software version B.12.01 was used to identify MFs that differed between the 2 groups. The abundances (area under the peak for the monoisotopic mass) of individual MF's were determined using the Agilent MassHunter Quantitative Analysis software version B.05.00.
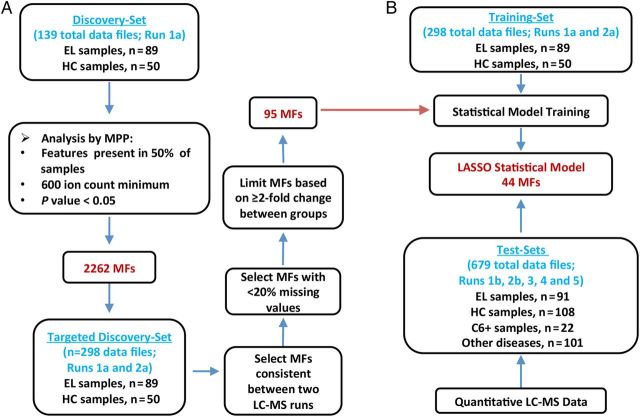
Figure 1.
Work flow for the discovery and testing of a serum biosignature that differentiates early Lyme disease (EL) from healthy controls (HC). A, Liquid chromatography-mass spectrometry (LC-MS) data from an initial discovery-set of samples (left) comprised of 89 EL patients and 50 HC (15 endemic and 35 nonendemic controls) (see Supplementary Material) were processed with the Molecular Feature Extractor algorithm tool of the Agilent MassHunter Qualitative Analysis software. The molecular features (MFs) were aligned between data files with a 0.25 minutes retention time window and 15 ppm mass tolerance. To reduce selection of MFs biased by uncontrolled variables (diet, other undisclosed illnesses, etc.), only those MFs present in greater than 50% of samples of at least one group and that differed between the groups with a significance of (P < .05) were selected. Agilent Mass Profiler Pro (MPP) software was used to identify MFs that differ between the 2 groups and this analysis resulted in 2262 MFs. A second LC-MS analysis of the same discovery-samples was performed. The abundance values for the 2262 MFs in both LC-MS data sets were combined to form the targeted discovery-sample data set. MFs were down-selected based on consistency between LC-MS runs and at least a 2-fold change in abundance from the median of the comparator group in replicate LC-MS analyses. This allowed for selection of an EL biosignature consisting of 95 MFs that were applied to statistical modeling. B, A training-data set along with the 95-MF biosignature list was used to train multiple statistical models [15]. The abundance values of targeted MFs used for model development were acquired with the Agilent MassHunter Quantitative Analysis software. Data from test-samples not included as samples for the training-data set were blindly tested against the statistical models. LASSO modeling selected 44 MFs for the refined biosignature and provided the most accurate classification of samples.
Statistical Analyses
For statistical modeling, classification analysis was accomplished using R, and model development was performed using targeted MFs. Figure 1B describes the workflow for model training and testing. The abundance values of targeted MFs used for model development were acquired with the Agilent MassHunter Quantitative Analysis software. Multiple classification approaches were applied: LDA [17]; classification tree (CT) analysis [18]; and LASSO logistic regression analysis [19]. Receiver operating characteristic (ROC) curves were created using the ROCR library [20].
Exact conditional logistic regression was used to compare sensitivities and specificities of sample classification based on LASSO modeling and serologic testing. The model response was scored as 1 if the test correctly classified the sample as early LD or non-LD, and 0 for an incorrect classification. The classification methodology (LASSO modeling or serology testing) was included as a predictor and each sample represented a stratum. Reported P-values are for the null hypothesis: the odds ratio of the 2 diagnostic methods correctly identifying a known case is 1. A linear, mixed-effects model [21] and LASSO model classification were employed to assess whether variables other than patient group affected MF abundance (Supplementary Material).
Metabolite Identification
The experimental accurate masses for individual MFs were used to predict chemical formulas [16], and searched against the publicly available Metlin compound database [22] and the Human Metabolome Database [23] for structural identifications.
RESULTS
Clinical Samples
Well-characterized retrospective serum samples selected based on defined criteria (Table 1) were randomly divided into discovery- and test-sample sets to allow development and testing of an early LD metabolic biosignature (Figure 1). Additionally, a small set of sera from patients clinically diagnosed with early LD and positive by the C6 EIA was included as test-samples (Table 1 and Figure 1B).
Biosignature Development
Although metabolomics studies performed by LC-MS yield abundance measurements of small molecule metabolites (ie, MFs), this technique when applied in a discovery phase is considered semiquantitative and can be influenced by run-to-run technical variances [15]. Thus, to generate a biosignature that differentiated early LD from healthy controls, duplicate LC-MS analyses were performed with 139 patient sera comprising the discovery-samples. A group comparison of the first dataset (Figure 1A) identified 2262 MFs that were present in at least 50% of either the LD or healthy control group samples, and that differed significantly in abundance between these 2 population groups (P < .05). The data of the second LC-MS analysis of the discovery-samples were used to down-select the 2262 MFs based on LC-MS run consistency and increased stringency (Figure 1A). This resulted in a biosignature of 95 MFs (Supplementary Figure 1 and Table 1) with 62 and 33 of the MFs increasing and decreasing in abundance in LD patient samples vs healthy controls, respectively.
Initial chemical identification for the 95 MFs resulted in 63 MFs with a predicted chemical formula and 49 MFs assigned a putative chemical structure (Supplementary Table 1). The putatively identified metabolites included: 11 polyunsaturated fatty acids (PUFAs) or lipids with PUFAs, and related to these, 6 products of prostaglandin metabolism; 8 structures of fatty acid or cholesterol metabolism; shingolipids; plasmalogens; products of tryptophan, purine, and heme metabolism; an endogenous alkaloid; and 7 peptides.
Biosignature Testing and Comparison With 2-Tier Serology
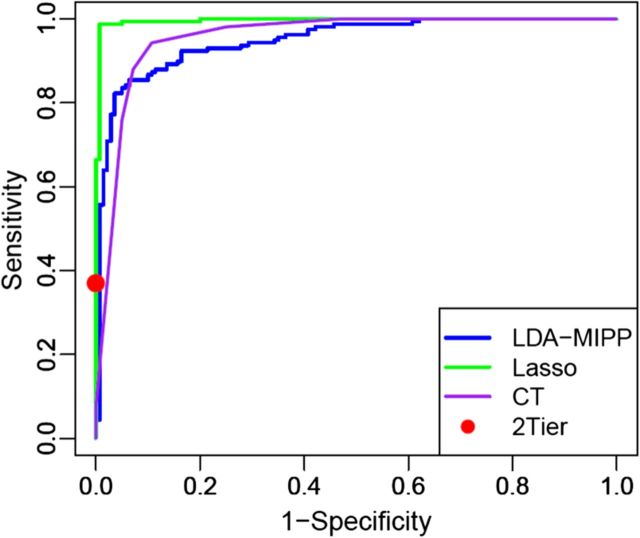
Statistical modeling was applied to assess whether metabolic profiling could accurately classify early LD patients vs healthy controls and other diseases. Several models (LDAmipp, CT, and LASSO) were trained against the 95-MF biosignature using data from the targeted discovery-samples (Figure 1B, Training-set). This training generated refined biosignatures (Supplementary Table 1), and a ROC curve was used to assess their relative performances. The LASSO model resulted in a refined biosignature of 44 MFs and was selected for further evaluation as it provided the most accurate prediction (Figure 2) with a 99% accuracy rate for both early LD and healthy controls. This accuracy was significantly (P < .0001) better than 2-tier testing with the same serum samples (Table 2). Further evaluation of the training-set based on leave-one-out cross-validation revealed an error rate of 7.4%.
Figure 2.
Receiver operating characteristic (ROC) curves to test model accuracies. ROC curves for the LDAmipp (blue), LASSO (green) and classification tree (CT) (purple) models were plotted and compared. The performance of the 2-tier testing algorithm (VIDAS/Marblot) (red dot) on the same sample set was included as a reference for the sensitivity and specificity of current laboratory-based Lyme disease diagnostics.
Table 2.
Sensitivity and Specificity Comparison Between 2-tier Serology and the Metabolomics LASSO Statistical Model
| No. of Samples Tested by Serologya | WCS EIA-VIDAS Results |
C6 EIA Results |
Immunoblot Resultsb (Marblot) % Pos. |
2-Tier Testing (VIDAS/Marblot)b |
2-Tier Testing (C6/Marblot)b |
Alternative 2-Tier Testing (VIDAS/C6)b |
Metabolomics LASSO Model |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % Pos. | % Pos. | IgM | IgG | IgM and IgG | No. Pos. Tests | Se. % | Sp. % | No. Pos. Tests | Se. % | Sp. % | No. Pos. Tests | Se. % | Sp. % | No. of Samples Testedc | No. Pos. Tests | Se. % | Sp. % | ||
| Training-Set | |||||||||||||||||||
| Subjects with early Lyme disease | |||||||||||||||||||
| Early Lyme | 89 | 58 | 52 | 30 | 3 | 9 | 33 | 37 | … | 31 | 35 | … | 37 | 42 | … | 158 | 156 | 99d | … |
| Non-Lyme disease controls | |||||||||||||||||||
| Healthy controls | 50 | 6 | 4 | 2 | 0 | 0 | 0 | … | 100 | 1 | … | 98 | 0 | … | 100 | 140 | 1 | … | 99e |
| Test-Set | |||||||||||||||||||
| Subjects with early Lyme disease | |||||||||||||||||||
| Early Lyme | 91 | 64 | 60 | 36 | 4 | 8 | 40 | 44 | … | 39 | 43 | … | 44 | 48 | … | 369 | 324 | 88d | … |
| C6-positive | 22 | 68 | 100 | 27 | 5 | 9 | 9 | 41 | … | 9 | 41 | … | 15 | 68 | … | 22 | 19 | 86f | … |
| Non-Lyme disease controls | |||||||||||||||||||
| Healthy controls | 108 | 10 | 0g | 4 | 0 | 0 | 0 | … | 100 | 0g | … | 100 | 0g | … | 100 | 187 | 10 | … | 95h |
| Other Diseases | 101 | 33 | 6 | 8 | 0 | 0 | 5 | … | 95 | 2 | … | 98 | 4 | … | 96 | 101 | 6 | … | 94i |
Abbreviations: CDC, Centers for Disease Control and Prevention; EIA, enzyme immunoassay; IgG, immunoglobulin G; IgM, immunoglobulin M; LC-MS, liquid chromatography-mass spectrometry; No., number; Pos., positive; Se., sensitivity; Sp., specificity; WCS, whole cell sonicate.
a Each sample was only tested one time.
b CDC 2-tier interpretation criteria were used [4]; however, all samples were tested by IgM immunoblots regardless of duration of illness.
c The serum samples tested included replicates due to multiple LC-MS runs.
d The sensitivity of LASSO modeling was significantly greater (P < .0001) than WCS EIA-VIDAS, C6 EIA, or 2-tier testing (VIDAS/Marblot). Statistical testing was not performed with the other 2-tier methods.
e The specificity of LASSO modeling was significantly greater (P < .003) than WCS EIA-VIDAS and not significantly different from C6 EIA (P = .06) or 2-tier testing (VIDAS/Marblot) (P = 1.00). Statistical testing was not performed with the other 2-tier methods.
f Sample size was not large enough to establish statistical significance for sensitivity.
g Healthy controls that were C6-positive were excluded from the test-set.
h The specificity of LASSO modeling did not differ significantly from WCS EIA-VIDAS (P = .14), C6 EIA (P = .08), or 2-tier testing (VIDAS/Marblot) (P = 1.00). Statistical testing was not performed with the other 2-tier methods.
i The specificity of LASSO modeling did not differ significantly from C6 EIA (P = 1.00) or 2-tier testing (VIDAS/Marblot) (P = .76), but was significantly better than the WCS EIA-VIDAS (P = .001). Statistical testing was not performed with the other 2-tier methods.
For more robust validation, LC-MS data of test-samples (ie, those not used for biosignature development or model training) were tested against the 44 MF LASSO model. The average accuracy achieved for classifying the early LD patients and healthy controls was 88% and 95%, respectively (Table 2). The relative abundance difference between each sample group for all 44 MFs (Supplementary Figure 2) allowed for the prediction accuracy of the LASSO model. As noted in the Supplementary Table 2, the data for the test-samples included 5 independent LC-MS runs with replicates of the samples. Across the 5 LC-MS runs sensitivity and specificity ranged from 84%–95% and 90%–100%, respectively. As expected, test-samples that were included in the same LC-MS runs as those for the training-set performed the best (93% sensitivity, 98% specificity). In comparison, the sensitivity of 2-tier testing for these early LD samples ranged from 43% to 48% with the highest sensitivity achieved with an alternative 2-tier testing algorithm consisting of 2 EIAs and no immunoblot (Table 2). Thus, LASSO modeling was significantly more sensitive than 2-tier testing (P < .0001). A significant difference was also observed when comparing LASSO modeling to the sensitivity of first-tier tests alone (VIDAS and C6 EIAs) (Table 2). Of importance, the metabolic profiling identified 77%–95% of the early LD samples that were negative by 2-tier testing (VIDAS/Marblot and C6/Marblot) (Table 3). This included 81%–96% and 83%–94% of those patients not diagnosed by the 2-tier IgM immunoblot assay and the 2-tier IgG immunoblot assay, respectively. As expected serological testing resulted in high specificity for healthy controls (100%). However, this specificity was not significantly better than that achieved by LASSO modeling (P = 1.0).
Table 3.
Comparison Between Positive and Negative Serology Tests and LASSO for Early Lyme Disease Test-samples
| LC-MS Run | 2-Tier Serologya vs LASSO |
IgM Immunoblot 2-Tier Serologya vs LASSO |
IgG Immunoblot 2-Tier Serologya vs LASSO |
IgM and IgG Immunoblot 2-Tier Serologya vs LASSO |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 2-Tier % | LASSO % Pos. | N | IgM % | LASSO % Pos. | N | IgG % | LASSO % Pos. | N | IgM/IgG % | LASSO % Pos. | |
| Subjects with Early Lyme Disease (n = 158) | ||||||||||||
| Run 1 (n = 20) | ||||||||||||
| Positive Serology | 7 | 35 | 100 | 4 | 20 | 100 | 2 | 10 | 100 | 1 | 5 | 100 |
| Negative Serology | 13 | 65 | 92 | 16 | 80 | 94 | 18 | 90 | 94 | 19 | 95 | 95 |
| Run 2 (n = 71) | ||||||||||||
| Positive Serology | 33 | 47 | 94 | 25 | 35 | 92 | 2 | 3 | 100 | 6 | 9 | 100 |
| Negative Serology | 38 | 53 | 95 | 46 | 65 | 96 | 69 | 97 | 94 | 65 | 91 | 94 |
| Run 3 (n = 140) | ||||||||||||
| Positive Serology | 66 | 47 | 83 | 50 | 36 | 84 | 4 | 3 | 100 | 12 | 9 | 75 |
| Negative Serology | 74 | 53 | 85 | 90 | 64 | 84 | 136 | 97 | 84 | 128 | 91 | 85 |
| Run 4 (n = 71) | ||||||||||||
| Positive Serology | 33 | 47 | 88 | 25 | 35 | 84 | 2 | 3 | 100 | 6 | 9 | 100 |
| Negative Serology | 38 | 53 | 92 | 46 | 65 | 94 | 69 | 97 | 90 | 65 | 91 | 89 |
| Run 5 (n = 67) | ||||||||||||
| Positive Serology | 32 | 48 | 91 | 24 | 36 | 88 | 2 | 3 | 100 | 6 | 9 | 100 |
| Negative Serology | 35 | 52 | 77 | 43 | 64 | 81 | 65 | 97 | 83 | 61 | 91 | 82 |
| C6-Positive Subjects (n = 22) | ||||||||||||
| Positive Serology | 9 | 41 | 100 | 6 | 27 | 100 | 1 | 5 | 100 | 2 | 9 | 100 |
| Negative Serology | 13 | 59 | 77 | 16 | 73 | 81 | 21 | 95 | 86 | 20 | 91 | 85 |
Abbreviations: CDC, Centers for Disease Control and Prevention; IgG, immunoglobulin G; IgM, immunoglobulin M; LC-MS, liquid chromatography-mass spectrometry; Pos., positive.
a CDC 2-tier interpretation criteria were used [4]; however, all samples were tested by IgM immunoblots regardless of duration of illness or first-tier test result; 2-tier serology was performed using VIDAS followed by Marblot immunoblots.
Two additional sample sets not included in LASSO model training were also tested: (1) sera from clinically diagnosed early LD patients that were C6 EIA positive, and (2) sera from patients with other diseases (Table 1). When challenged with the early LD samples collected based on clinical symptoms and C6 positivity, the LASSO model had a sensitivity of 86% (Table 2). These early LD C6-positive samples yielded 2-tier results (overall sensitivity of 41%) similar to the well-characterized early LD samples. Although these analyses demonstrated a large increase in sensitivity with LASSO modeling compared with 2-tier testing and corroborated the results obtained with the well-characterized early LD samples (Tables 2 and 3), the sample size was insufficient to assess statistical significance. When evaluating sera from other diseases the LASSO model yielded a specificity of 94% and did not differ significantly (P = .76) from the 95% specificity of 2-tier testing on these samples (Table 2).
Sample and LC-MS Variability
The range of sensitivity and specificity observed for sera analyzed in separate LC-MS runs reflected run-to-run variability [15]. However, the use of retrospective patient samples also introduced sample-handling variables. This included age of archived samples, heat-inactivation, and inter-lab differences in serum collection. Thus, we investigated the impact of run-to-run variability vs heat-inactivation of sera. Samples analyzed by LASSO modeling in 3 different LC-MS runs revealed inter-run variance of up to 10 percentage points based on classification accuracy (Supplementary Table 3). Analysis of inter-run variability of all 95 MFs in 3 replicate LC-MS runs with a linear, mixed effects model determined that the standard deviation for a given serum sample was 0.28 logs with a 95% confidence interval of .23–.34. This standard deviation did not vary substantially based on the MF being measured. In comparison, LC-MS analysis of 70 early LD sera that were heat-inactivated at 56°C for 30 min revealed four MFs that differed statistically (P < .05) in abundance from untreated samples. In spite of these four differences, the 44 MF LASSO model correctly classified the heat-inactivated and untreated samples with similar accuracies of 83% and 86%, respectively (Supplementary Table 3). Of the improperly classified samples, 6 were classified as non-LD in both the heat-inactivated and untreated groups.
DISCUSSION
In the natural course of LD, the human serves as a “dead-end” host for B. burgdorferi, thus early diagnosis is not a tool for disease control. Nevertheless, proper patient management can be influenced by early and accurate diagnosis. Multiple limitations exist for the diagnosis of early LD including: (1) poor sensitivity of current serological tests; (2) subjective interpretation of immunoblots; and (3) the subjectivity of clinical based diagnosis, even in the presence of an EM-like skin lesion [3, 6]. Thus, a significantly improved diagnostic test for early LD would enhance patient management, reduce over-testing [2] and help mitigate controversies associated with the diagnosis of LD [24].
The host inflammatory and immunological responses of LD are driven by B. burgdorferi infection and lead to the clinical symptoms of this disease [25, 26]. Thus, evaluation of metabolic biosignatures as a diagnostic platform of early disease is based on the hypothesis that the inflammatory responses of early LD distinguish it from healthy controls and diverge from those of other diseases with overlapping clinical features (eg, syphilis and fibromyalgia) [1, 3], serologic cross-reactivity (eg, infectious mononucleosis and syphilis) [13], and other spirochetal infections (eg, syphilis and severe periodontitis) [13, 27]. This study revealed a shift in the abundance of selected metabolites in patients with early LD as compared to healthy controls and patients diagnosed with other diseases. The refined early LD biosignature developed provides proof-of-concept for a novel diagnostic approach that has improvements over the currently recommended 2-tier serology algorithm. Most importantly, the early LD biosignature correctly diagnosed 77%–95% of 2-tier negative early LD patients, including 81%–96% of those patients not diagnosed by the 2-tier IgM assay, a test designed to detect early antibody responses [3]. Using well-characterized early LD samples, the refined metabolic biosignature yielded a greater sensitivity than the C6 EIA, another reported early marker of LD [28]. The specificity achieved with the metabolic biosignature was not significantly different from that of 2-tier serology for healthy controls or for patients with the other diseases assessed. Further optimization of the biosignature and assay must ensure judicious analysis of specificity vs sensitivity, to prevent false-positive test results in patient populations at risk for LD and to promote proper antibiotic stewardship. Overall, the current characteristics and performance of the metabolic biosignature revealed the potential for a novel diagnostic capable of detecting early LD prior to antibody responses.
The low sensitivity of serologic testing for early LD is a probable consequence of the time it takes to develop a humoral immune response [29–31]. In contrast, the shifts in metabolite profiles observed in this study likely reflect the innate immune response that emerges rapidly and underlies inflammation and pathology. C-reactive protein, a general marker of inflammation along with other protein markers or mediators of inflammation was shown to be elevated in LD patients and decrease with treatment [32, 33]. More recently, a multiplex-assay of inflammatory response associated proteins distinguished acute LD patients from healthy noninflammatory controls [34]. Consistent with these protein-based assays, several of the metabolites putatively identified in the reported biosignature are mediators or markers of inflammation. It is particularly interesting to note that the majority of metabolites putatively identified in the early LD biosignature are lipid or lipophilic structures. Thus, these initial efforts led to the hypothesis that B. burgdorferi infection elicits alterations in lipid mediators and markers of the inflammatory response. This hypothesis is supported by the recent report of altered eicosanoid production in an animal model of late LD [35].
Approximately 70%–80% of LD patients present with EM [6]; therefore, sera from clinically diagnosed early LD patients, with or without EM, but that were C6 EIA positive were included in our evaluations. These samples were correctly identified with an accuracy of 86%. Likewise the biosignature also performed well against other diseases (94% accuracy). It is noted that sera from the above patient groups were not included in LASSO model training; thus, they represented a more demanding evaluation of the LASSO model's ability to accurately classify patient samples. Continued development of a metabolomics based diagnostic test for early LD will require sera from patients with other clinical illnesses that might warrant consideration of a diagnosis of LD (eg, cellulitis, STARI, and cutaneous fungal infections among many possibilities), as well as patients with other tick-borne diseases present in LD endemic regions, such as anaplasmosis and babesiosis [1, 6, 36]. Assessment using sera from patients with other forms of LD including neurologic LD, Lyme carditis, and Lyme arthritis also will be required and may lead to additional or refined biosignatures that provide early recognition of these more severe disease manifestations.
For this study retrospective samples were used, and sample-handling variables that would not be associated with prospectively collected samples were a potential weakness. To account for these factors, large sample numbers collected from multiple laboratories were used to minimize or negate biases introduced from sample handling and storage. Moreover, stringent criteria were applied in biosignature selection to ensure that the most robust MFs were identified and used. The largest variability encountered in these studies was that which occurred among the LC-MS runs. Inter-run variability is an inherent issue with LC-MS based metabolomics studies targeted at discovery [15]. Such variability would be unacceptable for the clinical application of a diagnostic metabolic biosignature. Thus, along with evaluations of additional patient populations and prospective studies, an early LD diagnostic test that is deployable in a clinical setting will require refinement and standardization of LC-MS parameters, inclusion of internal standards for data normalization, establishment of system suitability protocols, and Food and Drug Administration (FDA) guidance [37–39]. It should be noted that LC-MS/MS based tests are currently used in clinical laboratories for the analyses of small molecule metabolites, such as for screening of inborn errors of metabolism. These tests are typically laboratory developed tests that fall under Clinical Laboratory Improvement Amendments guidelines; however, tests such as the Waters' NeoLynx Screening are FDA approved [40]. Thus, there is a developmental path and emerging infrastructure that would support a LC-MS based diagnostic platform for early LD.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online (http://cid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank Barbara J. Johnson (Centers for Disease Control and Prevention [CDC], Fort Collins) and Linden Hu (Tufts Medical Center, Boston) for kindly providing serum samples.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health [R21/R33 AI100228] and the CDC.
Potential conflicts of interest. All authors: No potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Stanek G, Wormser GP, Gray J, Strle F. Lyme borreliosis. Lancet 2012; 379:461–73. [DOI] [PubMed] [Google Scholar]
- 2.Hinckley AF, Connally NP, Meek JI, et al. Lyme disease testing by large commercial laboratories in the United. Clin Infect Dis 2014; 59:676–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguero-Rosenfeld ME, Wang G, Schwartz I, Wormser GP. Diagnosis of Lyme borreliosis. Clin Microbiol Rev 2005; 18:484–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Recommendations for test performance and interpretation from the Second National Conference on Serologic Diagnosis of Lyme Disease. Morb Mortal Wkly Rep 1995; 44:590–1. [PubMed] [Google Scholar]
- 5.Liveris D, Schwartz I, McKenna D, et al. Comparison of five diagnostic modalities for direct detection of Borrelia burgdorferi in patients with early Lyme disease. Diagn Microbiol Infect Dis 2012; 73:243–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wormser GP. Clinical practice. Early Lyme disease. N Engl J Med 2006; 354:2794–801. [DOI] [PubMed] [Google Scholar]
- 7.Dandache P, Nadelman RB. Erythema migrans. Infect Dis Clin North Am 2008; 22:235–60. [DOI] [PubMed] [Google Scholar]
- 8.Swan AL, Mobasheri A, Allaway D, Liddell S, Bacardit J. Application of machine learning to proteomics data: classification and biomarker identification in postgenomics biology. OMICS 2013; 17:595–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H, Hu H, Deng C, et al. Integrative system biology strategies for disease biomarker discovery. Comb Chem High Throughput Screen 2012; 15:286–98. [DOI] [PubMed] [Google Scholar]
- 10.Vinayavekhin N, Homan EA, Saghatelian A. Exploring disease through metabolomics. ACS Chem Biol 2010; 5:91–103. [DOI] [PubMed] [Google Scholar]
- 11.Balog CI, Meissner A, Göraler S, et al. Metabonomic investigation of human Schistosoma mansoni infection. Mol Biosyst 2011; 7:1473–80. [DOI] [PubMed] [Google Scholar]
- 12.Tritten L, Keiser J, Godejohann M, et al. Metabolic profiling framework for discovery of candidate diagnostic markers of malaria. Sci Rep 2013; 3:2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molins CR, Sexton C, Young JW, et al. Collection and characterization of samples for establishment of a serum repository for Lyme disease diagnostic test development and evaluation. J Clin Microbiol 2014; 52:3755–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Branda JA, Linskey K, Kim YA, Steere AC, Ferraro MJ. Two-tiered antibody testing for Lyme disease with use of 2 enzyme immunoassays, a whole-cell sonicate enzyme immunoassay followed by a VlsE C6 peptide enzyme immunoassay. Clin Infect Dis 2011; 53:541–7. [DOI] [PubMed] [Google Scholar]
- 15.Dunn WB, Broadhurst D, Begley P, et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc 2011; 6:1060–83. [DOI] [PubMed] [Google Scholar]
- 16.Mahapatra S, Hess AM, Johnson JL, et al. A metabolic biosignature of early response to anti-tuberculosis treatment. BMC Infect Dis 2014; 14:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soukup M, Cho H, Lee JK. Robust classification modeling on microarray data using misclassification penalized posterior. Bioinformatics 2005; 21:i423–30. [DOI] [PubMed] [Google Scholar]
- 18.Therneau T, Atkinson B, Ripley B. rpart: Recursive partitioning and regression trees. 2014; R package version 4.1–8.
- 19.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010; 33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 20.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics 2005; 21:3940–1. [DOI] [PubMed] [Google Scholar]
- 21.Dunnett CW. New tables for multiple comparisons with a control. Biometrics 1964; 20:482–91. [Google Scholar]
- 22.Smith CA, O'Maille G, Want EJ, et al. METLIN: a metabolite mass spectral database. Ther Drug Monit 2005; 27:747–51. [DOI] [PubMed] [Google Scholar]
- 23.Wishart DS, Knox C, Guo AC, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res 2009; 37(Database issue):D603–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King JK, Gerber NL, Ostroff SM, et al. Critical Needs and Gaps in Understanding Prevention, Amelioration, and Resolution of Lyme and Other Tick-Borne Diseases: The Short-Term and Long-Term Outcomes Workshop Report. Washington, DC: National Academies Press, 2011. [PubMed] [Google Scholar]
- 25.Oosting M, Buffen K, van der Meer JW, Netea MG, Joosten LA. Innate immunity networks during infection with Borrelia burgdorferi. Crit Rev Microbiol 2014; 25:1–12. [DOI] [PubMed] [Google Scholar]
- 26.Schröder NW, Eckert J, Stübs G, Schumann RR. Immune responses induced by spirochetal outer membrane lipoproteins and glycolipids. Immunobiology 2008; 213:329–40. [DOI] [PubMed] [Google Scholar]
- 27.Magnarelli LA, Miller JN, Anderson JF, Riviere GR. Cross-reactivity of nonspecific treponemal antibody in serologic tests for Lyme disease. J Clin Micro 1990; 28:1276–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wormser GP, Schriefer M, Aguero-Rosenfeld ME, et al. Single-tier testing with the C6 peptide ELISA kit compared with two-tier testing for Lyme disease. Diagn Microbiol Infect Dis 2013; 75:9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Craft JE, Fischer DK, Shimamoto GT, Steere AC. Antigens of Borrelia burgdorferi recognized during Lyme disease. Appearance of a new immunoglobulin M response and expansion of the immunoglobulin G response late in the illness. J Clin Invest 1986; 78:934–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dressler F, Whalen JA, Reinhardt BN, Steere AC. Western blotting in the serodiagnosis of Lyme disease. J Infect Dis 1993; 167:392–400. [DOI] [PubMed] [Google Scholar]
- 31.Nowalk AJ, Gilmore RD, Jr, Carroll JA. Serologic proteome analysis of Borrelia burgdorferi membrane-associated proteins. Infect Immun 2006; 74:3864–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan SS, Wong YC, Hodgkiss IJ. A preliminary study of C-reactive protein in the diagnosis and monitoring of Lyme disease. Biomed Environ Sci 1996; 9:424–9. [PubMed] [Google Scholar]
- 33.Pietruczuk A, Swierzbińska R, Pancewicz S, Pietruczuk M, Hermanowska-Szpakowicz T. Serum levels of interleukin-18 (IL-18), interleukin-1beta (IL-1beta), its soluble receptor sIL-1RII and C-reactive protein (CRP) in patients with Lyme arthritis. Infection 2006; 34:58–162. [DOI] [PubMed] [Google Scholar]
- 34.Soloski MJ, Crowder LA, Lahey LJ, Wagner CA, Robinson WH, Aucott JN. Serum inflammatory mediators as markers of human Lyme disease activity. PLoS One 2014; 9:e93243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pratt CL, Brown CR. The role of eicosanoids in experimental Lyme arthritis. Front Cell Infect Microbiol 2014; 4:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prusinski MA, Kokas JE, Hukey KT, Kogut SJ, Lee J, Backenson PB. Prevalence of Borrelia burgdorferi (Spirochaetales: Spirochaetaceae), Anaplasma phagocytophilum (Rickettsiales: Anaplasmataceae), and Babesia microti (Piroplasmida: Babesiidae) in Ixodes scapularis (Acari: Ixodidae) collected from recreational lands in the Hudson Valley Region, New York State. J Med Entomol 2014; 51:226–36. [DOI] [PubMed] [Google Scholar]
- 37.Chace DH, Kalas TA, Naylor EW. The application of tandem mass spectrometry to neonatal screening for inherited disorders of intermediary metabolism. Annu Rev Genomics Hum Genet 2002; 3:17–45. [DOI] [PubMed] [Google Scholar]
- 38.Abbatiello SE, Mani DR, Schilling B, et al. Design, implementation and multisite evaluation of a system suitability protocol for the quantitative assessment of instrument performance in liquid chromatography-multiple reaction monitoring-MS (LC-MRM-MS). Mol Cell Proteomics 2013; 12:2623–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sysi-Aho M, Katajamaa M, Yetukuri L, Oresic M. Normalization method for metabolomics data using optimal selection of multiple internal standards. BMC Bioinformatics 2007; 15:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu AH, French D. Implementation of liquid chromatography/mass spectrometry into the clinical laboratory. Clin Chim Acta 2013; 420:4–10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.