Abstract
Resistance to β-lactam antibiotics in enteric Gram-negative bacilli may be difficult to detect using standard methods of either Kirby-Bauer disc diffusion (KBDD) or broth dilution for minimal inhibitory concentration (MIC). This difficulty is due to genetic differences in resistance determinants, differences in levels of gene expression, and variation in spectra of enzymatic activity against the substrate β-lactams used for susceptibility testing. We have examined 95 clinical isolates reportedly susceptible to ceftazidime and ceftriaxone, as originally determined by either KBDD or MIC methods. The organisms studied here were isolated in 2002 from two pediatric hospital centers (Seattle, USA and Shanghai, China). They belong to the inducible β-lactamase producing Gram-negative bacilli, such as Enterobacter spp., Citrobacter spp., Serratia spp., Morganella spp., Providencia spp., and Proteus vulgaris. A Kirby-Bauer disc approximation (KBDA) method identified inducible phenotypes of third-generation cephalosporin resistance in 76% of isolates, which would otherwise be considered susceptible by standard KBDD methods.
Introduction
Nosocomial infections due to antibiotic-resistant, enteric Gram-negative bacilli have increased at an alarming rate in intensive care facilities, and are frequently associated with immunocompromised hosts, for whom they may be particularly devastating [1-5]. Multidrug-resistant Escherichia coli and Klebsiella spp. (as well as other Enterobacteriaceae) carrying plasmid-borne extended-spectrum β-lactamases (ESBLs) have attracted much interest among clinical microbiologists, infectious disease specialists, and infection control practitioners [6-8]. In contrast, the carriage of chromosomally encoded AmpC β-lactamases by these organisms has been deemed a commensal trait of uncertain consequence, and thus the management of such infections has not benefited from the multidisciplinary approach taken with ESBL infections [9-12].
Standard Kirby-Bauer disc diffusion (KBDD) methods and automated minimal inhibitory concentration (MIC) instruments used in most clinical laboratories do not readily detect ampC-type of inducible resistance. Therapy based on such susceptible reports may result in selection of resistance in vivo [9-13]. A few instructive studies have reported that the observation of increased MICs to third-generation cephalosporins using high-density inocula (increasing sensitivity for detection of derepressed mutant subpopulations) may be used to predict clinical failures [14,15]. To address this gap, the National Committee for Clinical Laboratory Standards (NCCLS) has included warnings such as 'Enterobacter, Citrobacter, and Serratia spp. may develop resistance during prolonged therapy with third generation cephalosporins' in its interpretive guideline publications [16]. However, methods that can be used to detect such resistance have not been suggested.
In this study, we used the Kirby-Bauer disc approximation (KBDA) method [17-19] to detect and characterize several phenotypes of inducible β-lactamase production. The interpretation of antibiotic susceptibilities was based on a combination of inhibitary zone sizes and zone morphologies near a potent agent of β-lactamase induction.
Materials and methods
Bacterial isolates and testing conditions
The 95 clinical isolates of Enterobacter, Citrobacter, Serratia, Morganella, Providencia, and Proteus vulgaris examined in this study were considered etiologic agents in cases of meningitis, bacteremia, pneumonia, wound, and urinary tract infections. Of the 95 strains tested, 56 were recovered during the fourth quarter of 2002 at Children's Hospital and Regional Medical Center (CHRMC) in Seattle, WA, and 39 were recovered during 2002 at Shanghai Children's Medical Center (SCMC), a sister hospital in Shanghai, China. When initially tested by standard KBDD and/or Vitek MIC methods, these isolates were interpreted as susceptible to third-generation cephalosporins (i.e., ceftazidime and ceftriaxone), based on NCCLS criteria [16]. In addition, control strains of Escherichia coli ATCC 25922 and a cephalosporin-resistant E. cloacae strain A (as shown in fig. 1f) were included in the study.
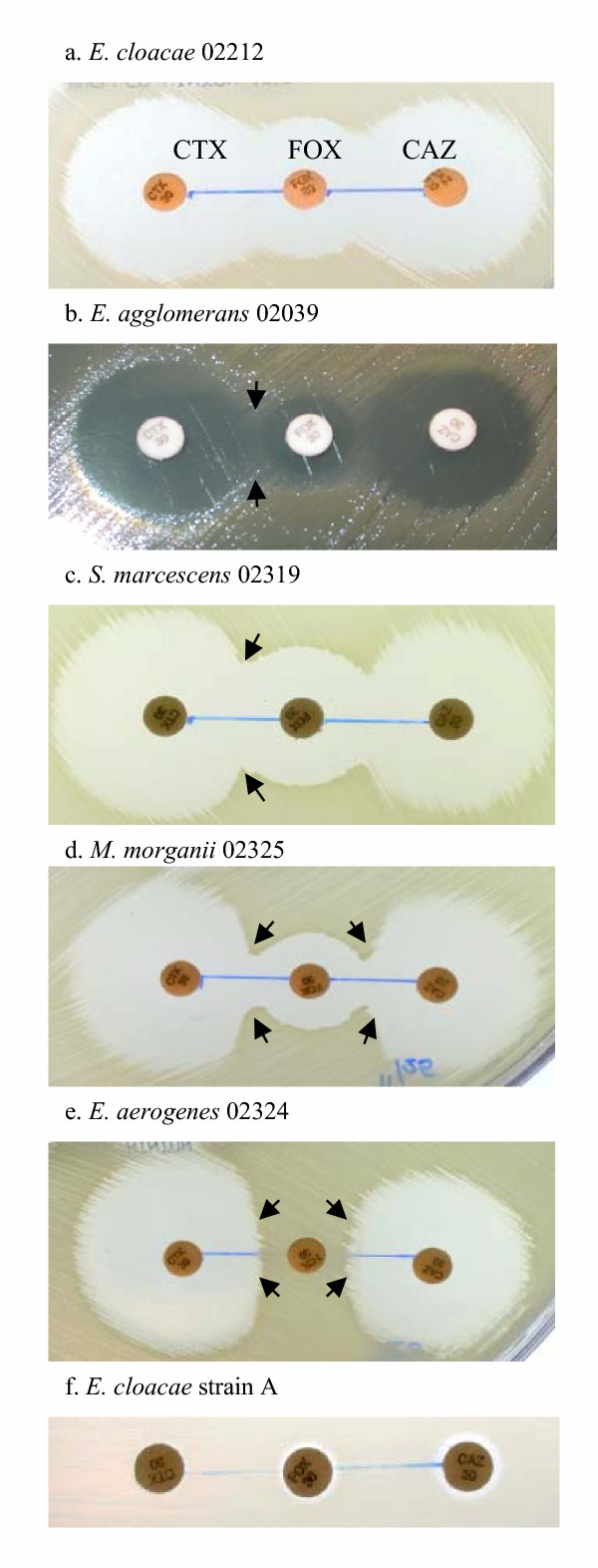
Figure 1.
Types of zones of inhibition determined by KBDA method. With FOX discs in the center, CTX discs are placed 15 mm to the left of FOX, while CAZ discs are 15 mm to the right of FOX as shown in fig 1a. (a) Susceptible zone: no growth between FOX and CTX/CAZ, E. cloacae 02212. (b) Sl zone: slight growth between FOX and CTX/CAZ, E. agglomerans 02039. (c) C zone: CTX zone distortion near FOX, S. marcescens 02319. (d) C zone: CTX zone distortion near FOX. M. morganii 02325. (e) D zone: CTX and CAZ zone truncation near FOX, E. aerogenes 02324. (f) Resistant zone: no zone of inhibition around any disc, E. cloacae strain A.
Testing conditions featured standard KB discs (Becton Dickinson Microbiology Systems, BBL, Sparks, MD, for CHRMC; Oxoid Limited, Basingstoke, Hampshire, England, for SCMC) and Mueller Hinton agar plates (BBL) with 35°C overnight incubation (~16 hours) in ambient air for KBDA analysis (NCCLS M2-A7) [20]. Standard 0.5 McFarland saline suspensions of bacteria were used to inoculate the Mueller Hinton agar media confluently with a cotton swab. The resultant zones of inhibition were measured with a caliper using transmitted light. We determined the diameter of circular zones of inhibition and the shortest radius of inducer-blunted zones.
KBDA for detection of third-generation cephalosporin resistance
The disc approximation technique was used as previously described [17] to detect inducible resistance to third-generation cephalosporins. We used cefoxitin (FOX) as a β-lactamase induction agent, and cefotaxime (CTX) and ceftazidime (CAZ) as the third-generation cephalosporin reporter agents [18,19,21]. Cefepime (FEP) was used for fourth-generation cephalosporin susceptibility testing. Based on preliminary results (described below), we selected a 15-mm edge-to-edge distance between the discs for β-lactamase induction testing of all isolates in the study. We also reviewed the initial susceptibility results of the 95 isolates to gentamicin (GM) and ciprofloxacin (CIP). To determine CAZ and CTX inhibitory activities upon induction, we doubled the radius measurements of zones of inhibition in the direction of the FOX discs, for direct comparison with the diameters of unaffected zones. Median zone measurement values were analyzed using the Wilcoxon signed-ranks test.
Results
Determination of disc distance for KBDA and measurements of antibiotic zones of inhibition
A pilot KBDA test was carried out with 13 strains of enteric Gram-negative bacilli, including E. coli ATCC 25922, C. freundii (n = 3), E. aerogenes (n = 1), E. agglomerans (also known as Pantoae agglomerans, n = 2), E. cloacae (n = 3), P. rettgeri (n = 1), and S. marcescens (n = 2). For each strain, we placed a FOX disc in the center and approximated CTX and CAZ discs on each side, comparing zones of inhibition for edge-to-edge distances of 15, 20, and 25 mm. FOX showed no effect on CTX nor CAZ susceptibilities in E. coli ATCC 25922, the E. agglomerans, and the P. rettgeri at any given disc distance. The most easily discernable patterns of FOX induced zone distortions were observed using the 15-mm disc distance in the remaining organisms.
We then tested each of 95 isolates against paired antibiotics using the 15-mm testing method. The test was repeated at least once with each organism. Averages of the zone measurements were used for final analysis. In the absence of FOX influence, the median zone sizes (and ranges) for CAZ and CTX were 27 mm (20–34 mm) and 29 mm (23–34 mm), respectively. With FOX induction, the radius from the center of the disc to the distorted zone edge near FOX ranged from 8 to 15 mm for CAZ and 8 to 16 mm for CTX, corresponding to median zone diameters of 23 mm and 24 mm for CAZ and CTX, respectively. For both CAZ and CTX, the FOX-induced zones were significantly smaller (p <0.0001) than the uninduced zones.
Interpretation of drug susceptibility patterns
Based on patterns of zone distortion, five types of susceptibility could be readily recognized among the test results. The five patterns (or zone morphologies) reflect incremental, visually discernable levels of bacterial resistance to CTX and CAZ, due to varying degrees of ampC induction (fig. 1a,1b,1c,1d,1e,1f). The two "extreme" patterns of drug susceptibility (fig. 1a and 1f) may be easily interpreted as drug-sensitive and drug-resistant, respectively. Resistant ('R') strains had no zones of inhibition around any of the three discs (fig. 1f, a CTX-resistant control E. cloacae strain A), while susceptible ('S') strains exhibited zones of inhibition above break points for CTX (≥ 23 mm) and CAZ (≥ 18 mm) and no growth between CTX-FOX or CAZ-FOX pairs (shown by E. cloacae 02212 in fig. 1a). Fifteen organisms were classified as susceptible to CTX and CAZ by the KBDA method: C. freundii (n = 1), C. koseri (n = 4), E. agglomerans (n = 2), E. amnigenus (n = 2), E. asburiae (n = 1), E. cloacae (n = 3), P. rettgeri (n = 1), and S. liquefaciens (n = 1).
However, not all patterns of inhibition could be judged solely by zone size measurement and thus interpreted using standard NCCLS guidelines. Hence, we distinguished three additional patterns, based on type of zone morphology around CTX (or CAZ) next to FOX and on bacterial growth between the disc pairs. 1) Slight growth ('Sl'-type zones) near FOX, but no identifiable zone distortion around CTX or CAZ (fig. 1b), was observed for 8 isolates: E. agglomerans (n = 5), E. sakazakii (n = 2), and P. vulgaris (n = 1). All exhibited zones of inhibition above the breakpoints for CTX and CAZ, and were thus interpreted as susceptible to these agents. 2) Zones of inhibition shaped like the letter 'C' ('C'-type zones), with bacterial growth pressing towards CTX or CAZ as a result of FOX induction (fig. 1c,1d), were observed (n = 23). Although the inducible growth was partially cleared by inhibitory action of FOX, producing letter 'C'-shaped zones, distortions of circular zones of inhibition around CTX were apparent. 3) Zones of inhibition resembling the letter 'D' ('D'-type zones), due to a flat edge of growth on both sides of FOX pressing against CTX and CAZ (fig. 1e), were also observed (n = 49). Because C- and D-types of growth around CTX and CAZ were clearly influenced by the close proximity of FOX, known for its potent effect on ampC induction, we have classified strains exhibiting such resistance patterns as 'Resistant (Inducible)'. Table 1 summarizes KBDA testing results for all 95 isolates, grouped into S-, Sl-, C- and D-types of zone categories. By classifying as 'susceptible' only those strains exhibiting S- and Sl-type growth patterns by the KBDA method, we have identified inducible resistance phenotypes in 76% (72/95) of the study isolates that were all reportedly susceptible to third-generation cephalosporins by standard KBDD or MIC methods. Inducible CTX resistance occurred in 80% (45/56) of CHRMC isolates and 69% (27/39) of SCMC isolates. The resistance patterns are not directly comparable due to differences in species distribution between the two groups. In figure 2, we summarize CTX susceptibility data generated by the KBDA method, by species. The same KBDA method was also employed to examine FEP susceptibility under the induction of FOX in all 95 isolates. FOX induction had minimal effect on the size or shapes of FEP zones of inhibition against this group of isolates (data not shown).
Table 1.
Types of zones of inhibition around CTX determined by KBDA.
| KBDA zone types around CTX | No. organisms (n = 95) | Percentage of the zone type | Susceptibility interpretation |
| S-type | 15 | 16% | Susceptible 24% |
| Sl-type | 8 | 8% | |
| C-type | 23 | 24% | Resistant (Inducible) 76% |
| D-type | 49 | 52% |
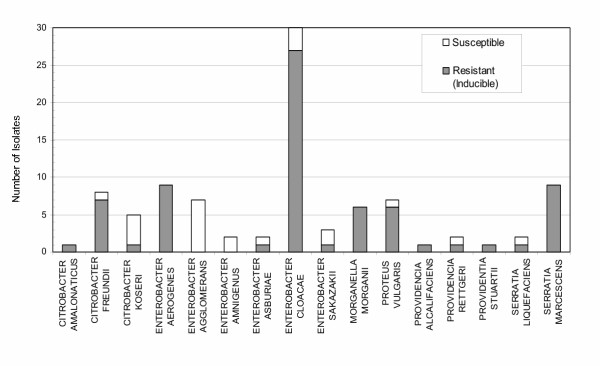
Figure 2.
Distribution of inducible resistance detected by KBDA among bacterial genera and species.
Discussion
Although in vitro susceptibility testing remains the cornerstone of clinical antibacterial therapy, not all naturally occurring mechanisms of resistance can be detected by standard laboratory methodologies [14]. Over 15 years ago, Sanders et al. [10,11] reported the emergence of bacterial resistance during cephalosporin treatment occurred in some 20–40% of systemic infections with these Gram-negative bacilli, accounting for treatment failure or relapse in at least 10% of such cases. Yet in the intervening years, individual hospital antibiograms have reported 75–90% of E. cloacae and S. marcescens isolates to be susceptible to third-generation cephalosporins. Thus, without accurate laboratory detection and informative reporting of such occult resistance phenotypes, the treatment of Gram-negative infections may remain suboptimal.
Of 95 isolates reported 'susceptible' to CTX and CAZ by conventional KBDD testing, 72 isolates (76%) exhibited C- and D-type inducible AmpC resistance. Moreover, the rates of resistance among this representative sample of common pathogens were rather alarming. Of the E. cloacae isolates, representing nearly 30% of the study strains, we found inducible resistance as high as 90% (fig. 2). Likewise, E. aerogenes, S. marcescens, and C. freundii, together accounting for 27% of study strains, exhibited a combined inducible resistance rate of 94%. This type of occult resistance phenotype may be missed by the standard testing methods and may have contributed to the frequency of treatment failures previously observed [9-13].
Distorted patterns of antibiotic zones of inhibition in the presence of β-lactamase induction agent FOX may provide clinical microbiologists with information not evident from conventional interpretations of zone size for this group of Gram-negative bacilli [18]. The statistical significance of the zone sizes between CTX and CTX-next-to-FOX strongly supports the notion of incorporating zone morphology into interpretation of antibiotic susceptibility results. We consider both 'C' and 'D' zone morphologies to be indicative of inducible resistance, and thus intend to categorize isolates exhibiting 'C' and 'D' zones as potentially resistant to third-generation cephalosporins. One potential strategy for communicating such data to clinicians would be to introduce a dual descriptor (such as 'S/R') to encompass results of both conventional and KBDA methods; a second level interpretation (such as 'Susceptibility testing indicates that resistance to this agent may emerge during prolonged therapy; this agent may still be useful for treatment of uncomplicated infections of the urinary tract, where high drug concentrations can be achieved') would accompany this descriptor [16,22,23].
The variety of zone phenotypes generated by KBDA may provide some insight into the regulation of these important genetic determinants. The distinctive zone morphologies observed among study strains may be indicative of the action of an auxiliary regulatory gene(s), such as ampR and ampD, on ampC expression rather than, for instance, point mutations in ampC promoters or regulators [24-27]. In contrast, the unmistakable no-zone type of AmpC resistance (a control strain shown in fig. 1f) is associated with permanent ampC derepression typically caused by promoter point mutations [28] or mutations in regulatory genes ampD or ampR [29]. Historically designated as de novo resistance, this phenotype would not be missed by conventional MIC and KBDD methods.
Consistent with previous findings, third-generation cephalosporin resistance conferred by ampC induction did not predict FEP resistance in this group of Gram-negative bacilli [30,31]. Many clinical isolates exhibiting inducible CTX and/or CAZ resistance in this collection remained susceptible to the fourth-generation cephalosporin FEP; those isolates resistant to FEP may have utilized a distinct mechanism, such as blockage of drug uptake [32,33], to escape its antibacterial activity. In the treatment of invasive infections caused by such pathogens, continued efficacy of alternative or combination therapy is supported by GM or CIP susceptibility at CHRMC [34]. Of interest, all 7 organisms resistant to these agents (5 strains resistant to GM and 2 resistant to CIP) were isolated from SCMC. No conclusive statements, however, can be made regarding the comparative rates of drug resistance between the two institutions due to differing bacterial genus and species make-up of the two collections.
The extent to which generalized treatment recommendations can be based solely on isolation of so-called SPICE-MP (Serratia, Proteus spp. – Indole positive,Citrobacter, Enterobacter, Morganella, and Providencia) organisms, the 'usual suspects' for ampC carriage, remains limited [22,23]. Those 23 (24%) isolates that did not exhibit recognizable zone distortion patterns were considered susceptible to CTX by KBDA, although some or all may contain a similar array of ampC determinants to the strains that exhibited resistant patterns. Of the AmpC family of β-lactamases, the spectrum of activity witnessed in our study isolates encompasses at least three Bush groups: group 1 for the majority, but group 2b for C. koseri and group 2e for P. vulgaris [35]. Thus, the genetics of both the structural ampC sequences and their regulatory pathways may still be highly polymorphic. In addition, organisms hosting both ESBL and ampC determinants have been found in many enteric Gram-negative bacilli [6,36], compounding the difficulties in recognition and detection. In short, accurate species identification does not allow simple assignment of the isolates to either ESBL- or AmpC-like β-lactam susceptibility patterns.
This study supports the use of a modified KBDD method, which would provide simple, visual information about bacterial resistance phenotypes. Further characterization of KBDA zone morphologies, considered jointly with zone size measurement, can be used to detect phenomena that even a refined quantitative system would be unable to measure. Laboratory tools need to be developed, protocols standardized and guidelines established to provide accurate detection and reporting practices that will enable more effective treatment strategies for these difficult infections.
Acknowledgments
Acknowledgements
This study was supported by Department of Laboratories and Pathology, Children's Hospital and Regional Medical Center, Seattle, USA. This work is also a collaborative effort between two pediatric hospitals (CHRMC and SCMC) through International Health Education, Project HOPE (Health Opportunities for People Everywhere). We thank Treva Tsosie for data organization and Excel presentation. We thank Drs. Carla R. Clausen, Marie B. Coyle, and Joe Rutledge for their critical reviews of the manuscript.
Contributor Information
Xuan Qin, Email: xuan.qin@seattlechildrens.org.
Scott J Weissman, Email: weissman@u.washington.edu.
Mary Frances Chesnut, Email: mary.f.chesnut@us.army.mil.
Bei Zhang, Email: bessiezh@yahoo.com.
Lisong Shen, Email: lshen@public1.sta.net.cn.
References
- Bujdakova H, Hanzen J, Jankovicova S, Klimackova J, Moravcikova M, Milosovic P, Michalkova-Papajova D, Kallova J, Jakab A, Kettner M. Occurrence and transferability of beta-lactam resistance in Enterobacteriaceae isolated in Children's University Hospital in Bratislava. Folia Microbiol (Praha) 2001;46:339–44. doi: 10.1007/BF02815624. [DOI] [PubMed] [Google Scholar]
- Fontana R, Lo Cascio G, Ligozzi M, Friscia O, Oldoni T. Antimicrobial susceptibility of respiratory isolates of Enterobacteriaceae and Staphylococcus aureus in Italy: incidence and trends over the period 1997–1999. Eur J Clin Microbiol Infect Dis. 2001;20:854–63. doi: 10.1007/s100960100628. [DOI] [PubMed] [Google Scholar]
- Hanberger H, Nilsson LE, Swedish Study Group High frequency of antibiotic resistance among Gram-negative isolates in intensive care units at 10 Swedish hospitals. Clin Microbiol Infect. 1997;3:208–215. doi: 10.1111/j.1469-0691.1997.tb00599.x. [DOI] [PubMed] [Google Scholar]
- Jones RN, Jenkins SG, Hoban DJ, Pfaller MA, Ramphal R. In vitro efficacy of six cephalosporins tested against Enterobacteriacea e isolated at 38 North American medical centres participating in the SENTRY Antimicrobial Surveillance Program, 1997–1998. Int J Antimicrob Agents. 2000;15:111–8. doi: 10.1016/S0924-8579(00)00152-7. [DOI] [PubMed] [Google Scholar]
- Pfaller MA, Jones RN. Antimicrobial susceptibility of inducible AmpC beta-lactamase-producing Enterobacteriaceae from the Meropenem Yearly Susceptibility Test Information Collection (MYSTIC) Programme, Europe 1997–2000. Int J Antimicrob Agents. 2002;19:383–8. doi: 10.1016/S0924-8579(02)00009-2. [DOI] [PubMed] [Google Scholar]
- Bradford PA. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001;14:933–51. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laboratory capacity to detect antimicrobial resistance, 1998. MMWR Morb Mortal Wkly Rep. pp. 1167–71. 2000, Jan 7. [PubMed]
- Steward CD, Rasheed JK, Hubert SK, Biddle JW, Raney PM, Anderson GJ, Williams PP, Brittain KL, Oliver A, McGowan JE, Jr, Tenover FC. Characterization of clinical isolates of Klebsiella pneumoniae from 19 laboratories using the National Committee for Clinical Laboratory Standards extended-spectrum beta-lactamase detection methods. J Clin Microbiol. 2001;39:2864–72. doi: 10.1128/JCM.39.8.2864-2872.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmidis J, Koratzanis G. Emergence of resistant bacterial strains during treatment of infections in the respiratory tract. Scand J Infect Dis Suppl. 1986;49:135–9. [PubMed] [Google Scholar]
- Sanders WE, Jr, Sanders CC. Inducible beta-lactamases: clinical and epidemiologic implications for use of newer cephalosporins. Rev Infect Dis. 1988;10:830–8. doi: 10.1093/clinids/10.4.830. [DOI] [PubMed] [Google Scholar]
- Sanders CC, Sanders WE., Jr beta-Lactam resistance in gram-negative bacteria: global trends and clinical impact. Clin Infect Dis. 1992;15:824–39. doi: 10.1093/clind/15.5.824. [DOI] [PubMed] [Google Scholar]
- Siebert JD, Thomson RB, Jr, Tan JS, Gerson LW. Emergence of antimicrobial resistance in gram-negative bacilli causing bacteremia during therapy. Am J Clin Pathol. 1993;100:47–51. doi: 10.1093/ajcp/100.1.47. [DOI] [PubMed] [Google Scholar]
- Fish DN, Piscitelli SC, Danziger LH. Development of resistance during antimicrobial therapy: a review of antibiotic classes and patient characteristics in 173 studies. Pharmacotherapy. 1995;15:279–91. [PubMed] [Google Scholar]
- Livermore DM. beta-Lactamases: quantity and resistance. Clin Microbiol Infect. 1997;3:S10–S19. [PubMed] [Google Scholar]
- Thomson KS, Moland ES. Cefepime, piperacillin-tazobactam, and the inoculum effect in tests with extended-spectrum beta-lactamase-producing Enterobacteriaceae. Antimicrob Agents Chemother. 2001;45:3548–54. doi: 10.1128/AAC.45.12.3548-3554.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Committee for Clinical Laboratory Standards. 1999, 2000, 2001, 2002, 2003. Performance Standards for Antimicrobial Susceptibility Testing. M100-S8, -S9, S10, S11, S-12, S13. National Committee for Clinical Laboratory Standards, Wayne, PA.
- Eliopoulos GM, Moellering RC., Jr . Antimicrobial combinations. In: Victor Lorian, editor. In Antibiotics in laboratory Medicine. 4. Williams & Wilkins; 1996. pp. 330–396. [Google Scholar]
- Livermore DM, Brown DF. Detection of beta-lactamase-mediated resistance. J Antimicrob Chemother. 2001;48:59–64. doi: 10.1093/jac/48.suppl_1.59. [DOI] [PubMed] [Google Scholar]
- Sanders CC, Sanders WE, Jr, Goering RV. In vitro antagonism of beta-lactam antibiotics by cefoxitin. Antimicrob Agents Chemother. 1982;21:968–75. doi: 10.1128/aac.21.6.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Committee for Clinical Laboratory Standards. Performance Standards for Antimicrobial Susceptibility Tests; Approved Standard – Seventh Edition M2-A7. National Committee for Clinical Laboratory Standards, Wayne, PA. 2000.
- Bauernfeind A, Schneider I, Jungwirth R, Sahly H, Ullmann U. A novel type of AmpC beta-lactamase, ACC-1, produced by a Klebsiella pneumoniae strain causing nosocomial pneumonia. Antimicrob Agents Chemother. 1999;43:1924–31. doi: 10.1128/aac.43.8.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore DM, Brown DF, Quinn JP, Carmeli Y, Paterson DL, Yu VL. Should third-generation cephalosporins be avoided against AmpC-inducible Enterobacteriaceae? Clin Microbiol Infect. 2004;10:84–5. doi: 10.1111/j.1469-0691.2004.00831.x. [DOI] [PubMed] [Google Scholar]
- Goldstein FW. Cephalosporinase induction and cephalosporin resistance: a longstanding misinterpretation. Clin Microbiol Infect. 2002;8:823–5. doi: 10.1046/j.1469-0691.2002.00492.x. [DOI] [PubMed] [Google Scholar]
- Bartowsky E, Normark S. Purification and mutant analysis of Citrobacter freundii AmpR, the regulator for chromosomal AmpC beta-lactamase. Mol Microbiol. 1991;5:1715–25. doi: 10.1111/j.1365-2958.1991.tb01920.x. [DOI] [PubMed] [Google Scholar]
- Lindquist S, Galleni M, Lindberg F, Normark S. Signalling proteins in enterobacterial AmpC beta-lactamase regulation. Mol Microbiol. 1989;3:1091–102. doi: 10.1111/j.1365-2958.1989.tb00259.x. [DOI] [PubMed] [Google Scholar]
- Mahlen SD, Morrow SS, Abdalhamid B, Hanson ND. Analyses of ampC gene expression in Serratia marcescens reveal new regulatory properties. J Antimicrob Chemother. 2003;51:791–802. doi: 10.1093/jac/dkg133. [DOI] [PubMed] [Google Scholar]
- Poirel L, Guibert M, Girlich D, Naas T, Nordmann P. Cloning, sequence analyses, expression, and distribution of ampC-ampR from Morganella morganii clinical isolates. Antimicrob Agents Chemother. 1999;43:769–76. doi: 10.1128/aac.43.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray BE. Antibiotic resistance. Adv Intern Med. 1997;42:339–67. [PubMed] [Google Scholar]
- Kuga A, Okamoto R, Inoue M. ampR gene mutations that greatly increase class C beta-lactamase activity in Enterobacter cloacae. Antimicrob Agents Chemother. 2000;44:561–7. doi: 10.1128/AAC.44.3.561-567.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller MA, Jones RN. MYSTIC (Meropenem Yearly Susceptibility Test Information Collection) results from the Americas: resistance implications in the treatment of serious infections. J Antimicrob Chemother. 2000;46:25–37. doi: 10.1093/jac/46.suppl_2.25. [DOI] [PubMed] [Google Scholar]
- Zhang YL, Li JT, Zhao MW. Detection of ampC in Enterobacter cloacae in China. Int J Antimicrob Agents. 2001;18:365–71. doi: 10.1016/S0924-8579(01)00414-9. [DOI] [PubMed] [Google Scholar]
- Charrel RN, Pages JM, De Micco P, Mallea M. Prevalence of outer membrane porin alteration in beta-lactam-antibiotic-resistant Enterobacter aerogenes. Antimicrob Agents Chemother. 1996;40:2854–8. doi: 10.1128/aac.40.12.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallea M, Chevalier J, Bornet C, Eyraud A, Davin-Regli A, Bollet C, Pages JM. Porin alteration and active efflux: two in vivo drug resistance strategies used by Enterobacter aerogenes. Microbiology. 1998;144:3003–9. doi: 10.1099/00221287-144-11-3003. [DOI] [PubMed] [Google Scholar]
- Chow JW, Yu VL. Combination antibiotic therapy versus monotherapy for gram-negative bacteremia: a commentary. Int J Antimicrob Agents. 1999;11:7–12. doi: 10.1016/S0924-8579(98)00060-0. [DOI] [PubMed] [Google Scholar]
- Bush K, Jacoby GA, Medeiros AA. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–33. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudron PE, Moland ES, Sanders CC. Occurrence and detection of extended-spectrum beta-lactamases in members of the family Enterobacteriaceae at a veteran's medical center: seek and you may find. J Clin Microbiol. 1997;35:2593–7. doi: 10.1128/jcm.35.10.2593-2597.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]