Abstract
Oestrogen-related receptor α (ERRα) is an orphan nuclear receptor which is important for adaptive metabolic responses under conditions of increased energy demand, such as cold, exercise and fasting. Importantly, metabolism under these conditions is usually accompanied by elevated production of organic acids, which may threaten the body acid–base status. Although ERRα is known to help regulate ion transport by the renal epithelia, its role in the transport of acid–base equivalents remains unknown. Here, we tested the hypothesis that ERRα is involved in acid–base regulation mechanisms by using zebrafish as the model to examine the effects of ERRα on transepithelial H+ secretion. ERRα is abundantly expressed in H+-pump-rich cells (HR cells), a group of ionocytes responsible for H+ secretion in the skin of developing embryos, and its expression is stimulated by acidic (pH 4) environments. Knockdown of ERRα impairs both basal and low pH-induced H+ secretion in the yolk-sac skin, which is accompanied by decreased expression of H+-secreting-related transporters. The effect of ERRα on H+ secretion is achieved through regulating both the total number of HR cells and the function of individual HR cells. These results demonstrate, for the first time, that ERRα is required for transepithelial H+ secretion for systemic acid–base homeostasis.
Keywords: oestrogen-related receptor α, H+ secretion, H+-ATPase-rich cell, zebrafish
1. Introduction
Oestrogen-related receptor α (ERRα) is an orphan nuclear receptor belonging to the oestrogen-related receptor family, a small subgroup of the nuclear receptor superfamily, which also includes ERRβ and ERRγ [1]. Although its natural cognate ligand has yet been identified, ERRα function has been linked to several physiological pathways. Initial studies on ERRα explored its role in modulating oestrogen signalling [2], bone formation [3] and breast cancer progression [4]. The identification of medium chain acyl-coenzyme A dehydrogenase, a rate-limiting enzyme in mitochondrial β-oxidation, as a target of ERRα [5] drew attention to the regulatory roles of ERRs in energy homeostasis. In its role as a transcription factor, ERRα controls various cellular metabolic activities through regulating the expression of energy metabolism genes, including the genes involved in carbohydrate metabolism, fatty acid oxidation, TCA cycle and oxidative phosphorylation [1,6,7]. Consistent with its regulatory role in these processes, ERRα is highly expressed in tissues with high metabolic needs, such as the heart, kidney, brown fat and skeletal muscle [1,7]. Moreover, ERRα expression is induced upon exposure to energy stresses such as cold [8], fasting [9] or exercise [10]. These findings reveal an important role of ERRα in the adaptive responses upon energetic stress stimuli. Physiological stress signals have been shown to elevate ERRα expression in a tissue-specific manner, and the increased ERRα during such challenges serves to produce the optimal responses that require a shift in energy production and utilization.
Maintaining internal pH homeostasis is critical for survival in all biological systems. When systemic acid–base status is disturbed by acid challenges, animals need to excrete excess acid equivalents to restore acid–base homeostasis. Vertebrates, including terrestrial and aquatic species, develop a specialized epithelium and organs to conduct acid/base transport. This process is mainly performed by kidneys in terrestrial mammals [11–13], whereas the gills account for approximately 90% of acid/base movement in aquatic teleost fish [14,15]. In mammals, the proximal tubule contributes to most transepithelial H+ secretion, a process which is primarily mediated by apical Na+/H+ exchanger 3 (NHE3). In addition, the collecting duct also contributes to H+ secretion through luminal V type H+-ATPase expressed in α-intercalated cells, thereby controlling final urinary acidification [11–13]. Similarly, NHE3 and H+-ATPase are also the apical major transporters involved in transepithelial H+ secretion in fish gills [15–17]. Transepithelial H+ secretion for body fluid acid–base homeostasis is tightly regulated by a variety of hormones and paracrine factors [11–13,18].
It is noteworthy that the aforementioned conditions of high metabolic demand (in which ERRα has been implicated) are usually accompanied by the threat of acid–base disturbance. Metabolism under these conditions would lead to elevated production of organic acids, such as lactic acid or ketone bodies [19–21]. These endogenous acids would result in an excessive acid load [22], and metabolism facilitated by ERRα may possibly exacerbate the situation. Preventing severe acid–base disturbance by diminishing excess acid through renal excretion appears to be critical for animals to cope with the physiological stimuli of energetic stress. The revelation that ERRα participates in the regulation of renal ion homeostasis [23], implicating it in the transepithelial transport process, has raised the interesting and important question of whether ERRα is involved in acid excretion; answering this question may help us understand the mechanisms underlying physiological adaptation to harsh environments. Studies using ERRα-knockout mice indicated that ERRα plays some roles in the renal regulation of blood pressure, via effects on ion homeostasis and the renin–angiotensin systems. ERRα null mice are hypotensive, with significant hypernatremia, hypokalaemia and slight hyperreninemia; further studies using a combination of genome-wide location analysis and expression profiling led to the proposal that ERRα regulates the expression of channels involved in renal Na+ and K+ handling (Scnn1a, Atp1a1, Atp1b1) [23]. However, no observations pertaining to ERRα regulation on renal acid–base transport have been reported so far.
To gain insights into the physiological role of ERRα in the adaptation to harsh environments, here we investigated the involvement of ERRα in body fluid acid–base homeostasis. Zebrafish has emerged as a useful in vivo model for the study of ionic and acid–base regulation in vertebrates [16,18,24]; ionocytes in the skin of developing zebrafish embryos play major roles in the transepithelial transport of ions and acid/base equivalents before the gills are fully developed [16,24,25]. Direct contact between the skin and the external environment makes zebrafish suitable for non-invasive measurements of ion and acid/base transport in vivo [26–28]. One subtype of ionocyte, H+-ATPase-rich (HR) cells—which are considered to be a functional analogue of α-intercalated cells in the mammalian kidney based on their molecular equipment, including apical V-ATPase [26,29] and basolateral anion exchanger 1 (AE1) [30]—have been identified as the cells specifically responsible for H+ secretion. Scanning ion-selective electrode technique (SIET) enables direct measurement of H+ current at the apical surface of a single HR cell in the yolk-sac epithelium of developing embryos, which revealed elevated H+ secretion from these cells upon low-pH stimulation [26,31]. In this study, zebrafish was used as a model to test the hypothesis that ERRα regulates body fluid acid–base homeostasis by controlling transepithelial H+ secretion. The effects of ERRα loss-of-function on transepithelial H+ secretion was determined, demonstrating that ERRα is an important regulator for acid–base homeostasis.
2. Material and methods
(a). Experimental animals
The AB strain of zebrafish was obtained from stocks held at the Institute of Cellular and Organismic Biology, Academia Sinica. Fish were kept in tanks with circulating freshwater (FW) (local tap water, pH 7.1–7.3) at 28.5°C under a 14 L : 10 D photoperiod. Embryos were collected within 30 min after fertilization and incubated in Petri dishes until they reached the required developmental stages.
(b). Acid acclimation
In accordance with previous studies, acidic FW (pH 4) was prepared by adding H2SO4 to local tap water. Adult zebrafish (males and females at the same ratio) were acclimated to acidic (pH 4 group) or control FW (local tap water, pH 7 group) for 7 days; abnormal behaviour and mortality were not observed during this period. A stable pH was maintained during the experiments by using an electrical pump to continually pump acidic FW into the bottom of the experimental tank. Fertilized zebrafish embryos were collected and immediately transferred to a Petri dish containing acidic or control FW for 3 or 7 days depending on the experiment design. Experimental water was changed twice per day to guarantee stable pH and optimal water quality.
(c). Whole-mount in situ hybridization
The esrra fragment was amplified by PCR with the following primer pair: forward, 5′-ATGTCTTCCAGAGAACGACGC-3′; reverse, 5′-CTAGGGTGAGTCCATCATGGC-3′. The resulting amplicon was then ligated into the pGEM-T Easy vector (Promega, Madison, WI, USA). The inserted fragment was amplified by PCR with the T7 and SP6 primers, and the products were used as templates for in vitro transcription with T7 and SP6 RNA polymerase (Roche, Penzberg, Germany) in the presence of digoxigenin (DIG)-UTP (Roche) to synthesize sense and antisense probes, respectively. Zebrafish embryos were anaesthetized and fixed with 4% paraformaldehyde in a PBS (1.4 mM NaCl, 0.2 mM KCl, 0.1 mM Na2HPO4 and 0.002 mM KH2PO4; pH 7.4) solution at 4°C overnight. Whole-mount in situ hybridization was performed according to previous reports [32,33]. Staining was conducted with the nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate system. For fluorescence in situ hybridization, staining was conducted with a commercial kit (TSA Plus Fluorescence Systems; Perkin-Elmer). Hybridization signals detected by the DIG-labelled RNA probes were amplified through fluorescein-tyramide signal amplification (TSA), while Alexa-488-tyramide substrate (Molecular Probes, Eugene, OR, USA) was used for the dinitrophenol-labelled probes. For subsequent double immunocytochemistry staining, the embryo samples were rehydrated in PBST and then incubated with 3% bovine serum albumin for 2 h to block non-specific binding. Samples were then incubated overnight at 4°C with a polyclonal antibody against the A subunit of zebrafish H+-ATPase. After being washed with PBST for 20 min, samples were further incubated in Alexa Fluor 488 goat anti-rabbit immunoglobulin G (IgG; Molecular Probes; diluted 1 : 200 with PBS) for 2 h at room temperature. Images were obtained with a confocal laser scanning microscope (TCS-SP5, Leica Lasertechnik, Heidelberg, Germany).
(d). Preparation of total RNA and reverse transcription
Whole zebrafish embryos or adult gills were homogenized in TRIzol reagent (Ambion, Woodward, TX, USA) and total RNA was purified following the manufacturer's protocol. To obtain sufficient RNA, 30 embryos were pooled to form one sample. As for sampling of adult gills, every single RNA sample was pooled from gills collected from one male and one female [34]. For complementary (c)DNA synthesis, 1–5 µg of total RNA was reverse-transcribed in a final volume of 20 µl containing 0.5 mM dNTPs, 2.5 µM oligo (dT)20, 250 ng random primers, 5 mM dithiothreitol, 40 units RNase inhibitor and 200 units Superscript RT (Invitrogen, Carlsbad, CA, USA) for 1 h at 50°C, followed by 15 min at 70°C.
(e). Real-time quantitative PCR
Expression levels of target gene mRNA were measured using Q-PCR with a LightCycler real-time PCR system (Roche). The reaction mixture consisted of 5 µl of 2× SYBR Green I Master Mix (Roche), primer pairs (300 nM) and 20–30 ng of cDNA, to a final volume of 10 µl. PCR products were subjected to a melting-curve analysis, and representative samples were electrophoresed to verify that only a single product was present. The standard curve of each gene was confirmed to be in a linear range, with ribosomal protein L13a used as an internal control. The primer sets used for Q-PCR are shown in electronic supplementary material, table S1.
(f). Translational knockdown with antisense morpholino oligonucleotides
Morpholino-modified antisense oligonucleotides were purchased from Gene Tools (Philomath, OR, USA). The sequence of the morpholino oligonucleotide (MO) against esrra (NM_ 212955.1) was 5′-CAGAGCGTCGTTCTCTGGAAGACAT-3′; this MO was prepared with 1× Danieau solution. Standard control MO (provided by Gene Tools) with a non-specific sequence (5′-CCTCTTACCTCAGTTACAATTTATA-3′) was injected in parallel as a control. Zebrafish embryos at the one- to two-cell stage were injected with MO solution containing 0.1% phenol red (as a visualizing indicator), using an IM-300 microinjection system (Narishige Scientific Instrument Laboratory, Tokyo, Japan). Various dosages (1, 2 and 4 ng per embryo) of MO were assessed; the 4 ng-injected group showed abnormal development (electronic supplementary material, figure S1a). Therefore, 2 ng per embryo was used in all subsequent experiments. The effectiveness of knockdown was verified by Western blot analysis (electronic supplementary material, figure S1).
(g). Measurement of surface pH of zebrafish embryos
Proton secretion in zebrafish embryos was determined by measuring the pH at the yolk surface. A non-invasive SIET was used to measure extracellular H+ activity (pH) at the surface of zebrafish embryos, as previously described [26]. Briefly, microelectrodes with a tip diameter of 3–4 µm were pulled from glass capillary tubes using a P-97 Flaming Brown pipette puller (Sutter Instruments, San Rafael, CA, USA), then baked at 200°C overnight, and vapour-silanized with dimethylchlorosilane (Fluka, Buchs, Switzerland) for 30 min. The microelectrodes were backfilled with a 1-cm column of 100 mM KCl/H2PO4 (pH 7.0), and then frontloaded with a 20- to 30-µm column of liquid ion exchanger cocktail (hydrogen ionophore I-cock-tail B; Fluka). The H+ microelectrode was positioned with a step-motor-driven three-dimensional positioner (Applicable Electronics, East Falmouth, MA, USA) via an Ag/AgCl wire electrode holder (World Precision Instruments, Sarasota, FL, USA), and the circuit was completed by placing a salt bridge. Data acquisition, preliminary processing and control of the three-dimensional electrode positioner were performed with ASET software (Science Wares, East Falmouth, MA, USA). The Nernstian properties of each microelectrode were measured by placing the microelectrode in a series of standard pH solutions (pH 6, 7 and 8).
To detect surface H+ activity of zebrafish embryos, SIET was performed at room temperature in a small plastic recording chamber filled with 1 ml of recording solution, which consisted of artificial media (0.5 mM NaCl, 0.2 mM CaSO4, 0.2 mM MgSO4, 0.05 mM KH2PO4 and 0.05 mM K2HPO4), 300 µM MOPS buffer (Sigma) and 0.1 mg l−1 Tricaine (3-aminobenzoic acid/ethyl ester; Sigma; pH 6.8). An anaesthetized embryo was positioned in the centre of the chamber, with its lateral side contacting the base of the chamber. The probe was then moved to a target position on the skin surface of the yolk sac and used to take recordings for 30 s; it was subsequently moved approximately 1 cm away from the embryo to record background levels in the media. The voltage outputs were converted to H+ concentrations according to the three-point calibration curve (described above), and Δ[H+] was used to represent the H+ gradients between the target point on the skin surface and the background.
(h). Measurement of H+ flux at HR cells
To record the local H+ flux at individual HR cells, SIET measurement was performed under a differential interference contrast microscope. The apical membrane of HR cells was identified in zebrafish skin, and the probe was then moved to a position 1–2 µm above the apical surface (membrane) of HR cells. The voltage difference in microvolts was measured by probing orthogonally to the surface at 10 μm intervals. At least 10 recordings were made at a single HR cell, and the median value was used for calculating the H+ flux of the cell. The measured ionocytes then were further confirmed as an HR cell by fluorescent Con-A, which is a specific and vital marker for HR cells. To calculate ionic flux, voltage differences were first converted into a concentration gradient ΔC (µmol l−1 cm−3), and ΔC was subsequently converted into ionic flux using Fick's law of diffusion in the following equation:
where J (pmol cm−2 s−1) is the net flux of the ion, D is the diffusion coefficient of the ion (9.4 × 10–5 cm2 s−1 for H+) and ΔX (cm) is the distance between the two points. The detailed operation for measurement and calculation of ionic flux were described in a previous report [26].
(i). Whole-mount immunocytochemistry and cell counting
Zebrafish embryos were fixed in 4% paraformaldehyde in PBST. Samples were then incubated with 3% bovine serum albumin for 2 h to block non-specific binding, before being incubated at 4°C overnight with one of the following: an α5 monoclonal antibody against the α-subunit of the avian Na+-K+-ATPase (Developmental Studies Hybridoma Bank, University of Iowa, Ames, IA, USA); a polyclonal antibody against the A subunit of zebrafish H+-ATPase (synthetic peptide: AEMPADSGYPAYLGARLA) [35]; a polyclonal antibody against the N-terminal domain of zebrafish Na+-Cl− cotransporter like 2 (synthetic peptide: IKKSRPSLDVLRNPPDD) [36]; or a monoclonal antibody against human P63 (Santa Cruz Biotechnology, Santa Cruz, CA, USA). After brief washing with PBST, samples were further incubated with Alexa Fluor 488 goat anti-rabbit immunoglobulin (IgG) (Molecular Probes; 1 : 200 dilution with PBS) or an Alexa Fluor 568 goat anti-mouse IgG antibody (Molecular Probes; 1 : 200 dilution with PBST) for 2 h at room temperature. Images were acquired with a Leica TCS-SP5 confocal laser scanning microscope (Leica Lasertechnik, Heidelberg, Germany). Cell number was quantified in 3 dpf control embryos and ERRa morphants using freehand selections made with an image processing program (ImageJ v. 1.45 s; Wayne Rasband, NIH), as previously described [37]. The total area of the yolk sac (one side for each embryo) was calculated by the image program based on the designated scale bar size of the image acquired from the microscope.
(j). Statistical analysis
Values are presented as means ± s.d. and were compared using Student's t-test or one-way analysis of variance (ANOVA).
3. Results
(a). Localization of ERRα in HR cells of zebrafish embryonic skin
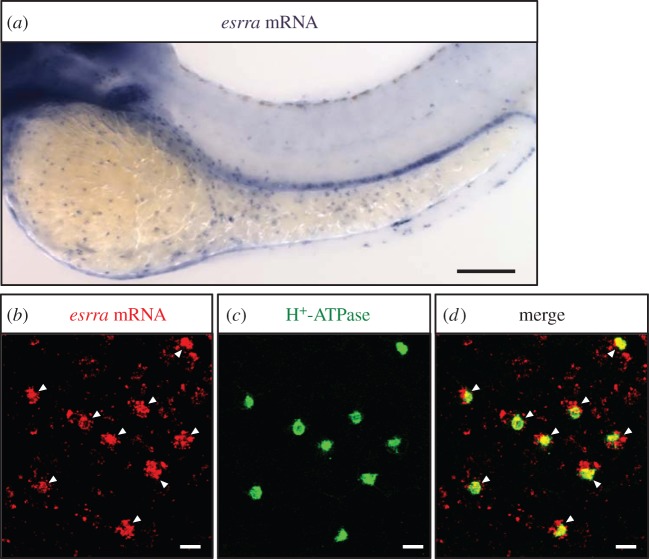
To determine whether ERRα is expressed in HR cells (a major ionocyte type responsible for transepithelial H+ secretion in zebrafish), we performed whole-mount in situ hybridization to detect esrra (encoding ERRα) mRNA expression in zebrafish embryos. As shown in figure 1a, the esrra signals were distributed at the surface of the yolk sac and yolk extension, and showed a ‘salt and pepper’ pattern (i.e. ionocyte pattern) in 3 dpf (days post fertilization) embryos. Subsequent double staining by in situ hybridization against esrra (figure 1b) and immunocytochemistry against H+-ATPase (an apical marker of HR cells) (figure 1c) showed that esrra mRNA signals colocalized with those of H+-ATPase in the same cells (figure 1d), indicating that ERRα is expressed in HR cells.
Figure 1.
Localization of esrra in HR cells in zebrafish embryos. (a) Whole-mount in situ hybridization against esrra mRNA in 3-dpf embryo. (b–d) Double labelling of esrra mRNA (by in situ hybridization, red signal) and H+-ATPase protein (by immunocytochemistry with anti-zebrafish H+-ATPase antibody, green signals) in yolk sac of 3 dpf embryos. (b) esrra mRNA; (c) H+-ATPase; (d) merged image of (b,c). Arrowheads indicate colocalization of esrra and H+-ATPase. Scale bars, (a) 100 µm; (b–d) 10 µm.
(b). Stimulation of ERRα by low pH environments
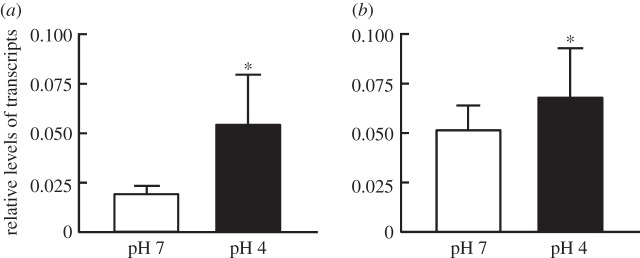
ERRα expression is induced by certain environmental cues. To examine whether ERRα expression is stimulated by low-pH environments in zebrafish, expression of esrra mRNA in zebrafish acclimated to different external pHs (for 7 days) was compared by Q-PCR. As shown in figure 2, the level of esrra mRNA was upregulated in the gills of adult zebrafish (figure 2a) as well as in whole embryos (figure 2b) exposed to acidic (pH 4) water, as compared with the control (pH 7), suggesting that ERRα is involved in the response to low pH challenge.
Figure 2.
Effects of acidic environment (pH 4) on esrra gene expression in (a) adult zebrafish gills and (b) zebrafish embryos. The levels of mRNA were analysed by Q-PCR, with rpl13a as an internal control. Asterisks denote significant difference from control (pH 7) group (p < 0.05, Student's t-test). Values were normalized to zrpl13a. Means ± s.d. (n = 6).
(c). Impaired H+ secretion of embryonic skin in ERRα-knockdown zebrafish
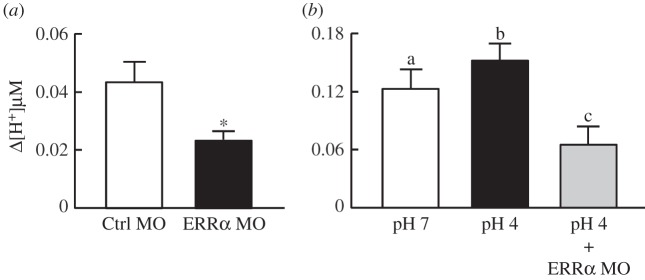
To investigate the contributions of ERRα to acid–base regulation in vivo, we injected MOs to perform loss-of-function experiments. Control MO or specific MO against ERRα was injected into embryos at the one- to two-cell stage. The effect of ERRα knockdown on H+ secretion at the yolk-sac skin of 3 dpf embryos was determined by SIET. As shown in figure 3a, ERRα-knockdown embryos (‘morphants') exhibited impaired H+ secretion at yolk-sac skin, indicating that ERRα is required for H+ secretion. In addition, the enhancement of H+ secretion by low pH, one of the adaptive responses upon acidic challenge [31], was prevented by ERRα knockdown, further supporting the importance of ERRα in the adaption to low-pH environments (figure 3b).
Figure 3.
Effects of ERRα knockdown on H+ secretion at the skin of zebrafish embryos. (a) ERRα knockdown (ERRα MO) decreased H+ secretion at the yolk sac of 3 dpf embryos. (b) ERRα knockdown prevented the increase in H+ secretion induced by low pH (pH 4). MOs were injected into embryos at the one- to two-cell stage. Embryos were transferred to pH 4 or pH 7 water immediately after MO injection, and H+ secretion was analysed by SIET at 3 dpf. Values are the mean ± s.d. (n = 10). Asterisk denotes significant difference from control (Ctrl MO) group (p < 0.05, Student's t-test). Different letters (a, b and c) indicate significant differences between treatments (one-way ANOVA).
(d). Decreased expression of H+-secreting-related transporters in ERRα morphants
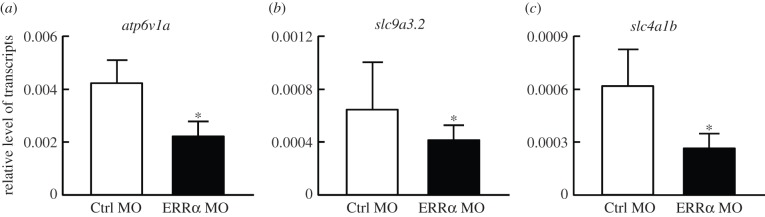
ERRα was previously reported to be involved in blood pressure and ion homeostasis through regulating the expression of renal ion transporters [23]. To examine whether ERRα regulates H+ secretion through affecting the expression of related transporters, we used Q-PCR to measure the expression of genes encoding transporters related to H+ secretion. While the majority of H+ secretion in HR cells is mediated by H+-ATPase [29], the sodium hydrogen exchanger 3b (NHE3b) and anion exchanger 1b (AE1b) are also involved, as evidenced by observations that inhibition or knockdown of these transporters impaired H+ secretion [27,30]. As compared with the control group, the expression levels of apt6v1a (a subunit of H+-ATPase) (figure 4a), slc9a3.2 (nhe3b) (figure 4b) and slc4a1b (ae1b) (figure 4c) were all significantly downregulated in ERRα morphants, suggesting the impairment of H+ secretion induced by ERRα knockdown may be due to defects in the transcriptional regulation of related transporters.
Figure 4.
Effects of ERRα knockdown on mRNA expression of H+ secretion-related transporters in 3 dpf zebrafish embryos. ERRα knockdown significantly downregulated the mRNA expression of (a) atp6v1a, (b) nhe3b and (c) ae1b. MOs were injected into embryos at the one- to two-cell stage. The levels of the indicated mRNAs were analysed by Q-PCR, with rpl13a as an internal control. Values are the mean ± s.d. (n = 6). Asterisks denote significant difference from the control (Ctrl MO) group (p < 0.05, Student's t-test).
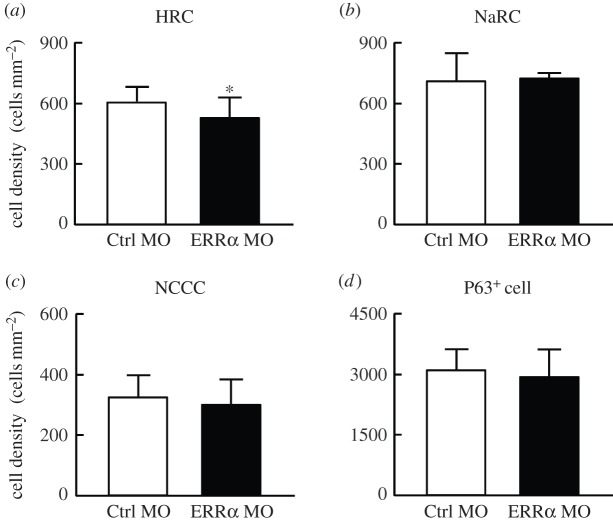
(e). Reduced HR cell number at yolk-sac skin in ERRα morphants
We next determined whether the impaired H+ secretion and reduced expression of related transporters arise from changes in HR cell number at the embryonic skin by using immunocytochemistry to compare HR cell numbers at the yolk-sac skin between ERRα morphants and control groups. The HR cells were labelled with anti-zebrafish H+-ATPase antibody, which demonstrated that the HR cell number at yolk-sac skin was slightly reduced in ERRα morphants (approx. 15%) compared with those in Ctrl MO-injected embryos (figure 5a). Zebrafish epidermal stem cells differentiate into multiple types of ionocytes [33,38]. ERRα knockdown did not affect the number of epidermal stem cells (with P63 as a marker; figure 5d), suggesting the decreased HR number of ERRα morphants did not arise from defects in the early events of ionocyte differentiation. On the other hand, other type of ionocytes, including Na+-K+-ATPase-rich cells (NaR cells; figure 5b) and Na+-Cl−-cotransporter cells (NCC cells; figure 5c), were not affected by ERRα knockdown at all, suggesting the effect of ERRα on ionocyte number is specific for HR cells.
Figure 5.
Effects of ERRα knockdown on numbers of ionocytes and epidermal stem cells in the yolk sac of 3 dpf embryos. ERRα knockdown slightly reduced the number of (a) HR cells, but did not affect those of (b) NaR cells, (c) NCC cells or (d) epidermal stem cells. MOs were injected into embryos at the one- to two-cell stage. HR cells, NaR cells and NCC cells were labelled with anti-zebrafish H+-ATPase antibody, α5 antibody and anti-zebrafish NCC antibody, respectively. Epidermal stem cells were labelled with an anti-human p63 antibody. Values are the mean ± s.d. (n = 8). Asterisk denotes significant difference from the control (Ctrl MO) group (p < 0.05, Student's t-test).
(f). Impaired H+ secretion of individual HR cells in ERRα morphants
To examine whether ERRα regulates H+ secretion through modulating the function of each HR cell, we determined the H+ secreting capacity of a single HR cell by SIET in 3 dpf embryos. As shown in figure 6, H+ secretion of HR cells was reduced by approximately 50% in ERRα morphants as compared with the control group, suggesting that ERRα regulates H+ secretion and transporter expression primarily through affecting the function of individual HR cells, rather than affecting the total HR cell number.
Figure 6.
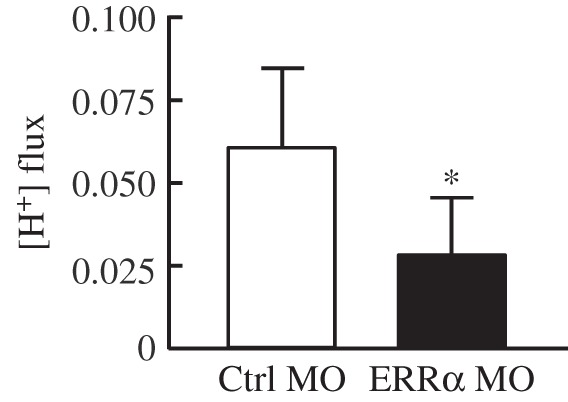
Effect of ERRα knockdown on H+ fluxes of individual HR cells in 3 dpf embryos. ERRα knockdown significantly reduced the H+ secretion ability of HR cells. MOs were injected into embryos at the one- to two-cell stage, and H+ flux was analysed by SIET at 3 dpf. Values are averages of 40 cells from six embryos (n = 6) in each group. Values are the mean ± s.d. Asterisk denotes significant difference from control (Ctrl MO) group (p < 0.05, Student's t-test).
4. Discussion
Systemic acid–base homeostasis must be maintained within a tight physiological range to sustain the essential biochemical and metabolic functions of all biological systems. Transepithelial transportation of acid/base equivalents is critical for acid–base regulation and is tightly controlled by various hormones, such as endothelins [39–41], angiotensin II [42,43] and corticosteroids [44,45]. By combining molecular, cellular and electrophysiological approaches in the zebrafish model, the present study identified ERRα as a novel regulator of body fluid acid–base homeostasis. ERRα is expressed in the acid-secreting HR cells and contributes to acid–base regulation by affecting transepithelial H+ secretion.
ERRα expression is abundant in high energy-consuming tissues, which are characterized by rich mitochondrial content [1]. Zebrafish HR cells are mitochondria-rich cells [28,46] that perform energy-consuming tasks, such as secretion of metabolic acid and sodium uptake, to maintain internal homeostasis under regular and harsh environments [27,28,31,46,47]. These features of HR cells are consistent with those of tissues in which ERRα is predominantly expressed. In addition, the expression pattern of ERRα in zebrafish embryonic skin is similar to that of HR cells (i.e. a spotty distribution at the surface of yolk sac and yolk extension, but not at the trunk; figure 1a [48]). These observations strongly link ERRα to the functions of HR cells. By employing in situ hybridization and immunocytochemistry approaches, this study demonstrated that ERRα is expressed in the HR cells of embryonic yolk skin (figure 1). Moreover, our acidic acclimation experiments revealed that low-pH water significantly stimulated the expression of ERRα in the adult gills as well as whole embryos (figure 2), suggesting that ERRα is involved in regulatory responses to cope with the challenges of acid–base disturbance. ERRα loss-of-function induced by a specific MO resulted in a significant decrease of H+ secretion at the yolk skin under both basal (figure 3a) and low-pH-stimulated (figure 3b) conditions, indicating that ERRα contributes to transepithelial H+ secretion. In agreement with the hypothesis that the zebrafish HR cell is analogous to the A type intercalated cell in mammalian kidney, our use of SIET to obtain non-invasive and real-time measurements of H+ secretion in an intact animal convincingly provides the first direct physiological evidence that ERRα is essential for transepithelial H+ secretion to achieve body fluid acid–base homeostasis in vertebrates.
Studies in the kidneys of ERRα-knockout mice indicated that, in addition to regulating energy metabolism, ERRα also directly controls expression of the transporters associated with transepithelial transport [23]. The present data suggest that the action of ERRα on H+ secretion also relies on these two processes. Knockdown of ERRα decreased the expression of metabolic genes (electronic supplementary material, figure S2) as well as the transporter genes related to H+ secretion (figure 4). Zebrafish HR cells express a set of transporters, including H+-ATPase [26], NHE3b [49] and AE1 [30], that are critical for H+ secretion of HR cells; knockdown or inhibition of these transporters has been demonstrated to impair H+ secretion in zebrafish embryos [27,29,30]. In this study, it is shown that the decrease in H+ secretion caused by ERRα knockdown (figure 3a) was accompanied by a significant reduction in gene expression of these three transporters (figure 4), suggesting the actions of ERRα on H+ secretion can be attributed to the impairment of transporter expression. In this context, one may further inquire as to whether the decreased expression of transporters arose from an alternation in the number of HR cells. To answer this question, we determined the effects of ERRα on HR cell number and H+ secretion at a single HR cell. While mRNA expression of the related transporters was decreased by approximately 50% (figure 4), the number of HR cells was reduced by about 15% in ERRα morphants (figure 5a), suggesting that not only HR cell number, but also transporters expressed in individual HR cells, were downregulated. Moreover, ERRα MO decreased the H+ secretion of a single HR cell (figure 6), further supporting the hypothesis that ERRα may regulate H+ secretion function at individual HR cells. Additionally, several predicted ERRα binding sites were identified (based on the human ERRα-response element, 5′-TNAAGGTCA-3′) [5] in the promoter of the zebrafish atp6v1a gene (e.g. −1189 to −1177 bp), which encodes a subunit of H+-ATPase critical for H+ secretion of HR cells, implying that ERRα directly affects the transcription of the transporter genes in HR cells. As such, while ERRα exerts some effects on HR cell number, it primarily regulates H+ secretion function at the cell level through modulating the expression of the related transporters in HR cells.
The observation that HR cell number was reduced by the knockdown of ERRα suggested that ERRα may be involved in the regulation of HR cell proliferation, differentiation and/or turnover, which are important mechanisms for coping with acidic challenge. According to the current model of zebrafish ionocyte differentiation [38], the embryonic ionocytes are derived from epidermal stem cells. Epidermal stem cells (expressing the proliferation marker p63) give rise to both keratinocytes and foxi3a-expressing ionocyte progenitor cells via a Delta-Notch fate determination mechanism [33]. These ionocyte progenitor cells then differentiate into different types of functional ionocytes. FOXI3a is the master regulator of ionocyte differentiation, and GCM2 is a transcription factor specifically controlling the specification of HR cells from the ionocyte progenitors [38]. In this study, ERRα knockdown did not affect the numbers of p63+ stem cells (figure 5b) or the other two types of ionocyte, NCC cells (figure 5c) and NaR cells (figure 5d), suggesting that regulation by ERRα may occur after the action of Foxi3a and may also be specific for HR cells. On the other hand, one cannot exclude the possibility that ERRα affects apoptosis to change the turnover rate of HR cells. The detailed mechanism behind the action of ERRα on controlling HR cell number requires further studies to clarify; however, the present findings provide a new direction from which to further explore the role of ERRα in the physiological adaption to pH challenge.
Revealing the regulatory role of ERRα in H+ secretion not only expands our understanding of the ERRα-mediated adaptive responses to environmental challenges, but may also provide new insights into how animals adapt to the harsh environments. Under stress conditions with increased metabolic needs, such as exposure to cold, hypoxia, fasting and exercise, metabolism is usually accompanied by the increased generation of organic acids, such as lactic acid or ketone bodies [19–21], and these endogenous acids (which are dissociated at physiological pH) may pose a substantial threat to acid–base homeostasis [22]. In fish, blood lactate level was demonstrated to be induced by exercise and hypoxia exposure [50,51]. The regulation of H+ secretion by ERRα may play a protective role to avoid body fluid acid–base disturbance, and thus ERRα regulation of metabolism continues to sustain the essential physiological functions. This mechanism would be of significance to fish because strenuous exercise is essential for survival (prey capture, predator avoidance or migration) and hypoxia is the common environmental condition in aquatic area [52]. On the other hand, lactic acid and ketone bodies are used as an energy source when glucose is not readily available under some conditions of high energy demand [22,53,54]. However, the increased production of lactic acid or keto acids would reduce systemic pH, which in turn inhibits the production of themselves as a negative feedback mechanism [22]. Therefore, the action of ERRα on acid–base regulation may also serve as a mechanism to ameliorate these feedback processes to ensure the sustained supply of fuel for tissues under these stressful conditions. Taken together, ERRα may possibly play an integrative role in modulating metabolism to provide energy for cell utilization and correcting the consequent acid–base disturbance, thus allowing the organism to achieve optimal adaptation under harsh environments. This notion about ERR's role on acid–base regulation awaits further studies in other animal species. Since the systemic acid–base status is relevant to various physiological processes [55] in which ERRα is implicated [56–60], it would be an interesting new direction from which to assess the physiologic role of ERRα in vivo.
The molecular physiology of how fish regulate their acid secretion to cope with to an acidic environment has been extensively elucidated in zebrafish [16,18]. In this study, a novel regulator, ERRα, for fish transepithelial H+ secretion mechanisms was identified. ERRα expression is abundant in HR cells (H+ secreting ionocytes) and is increased in response to low pH challenge in zebrafish. ERRα regulates H+ secreting capacity through modulating expressions of H+-secreting-related transporters. These findings reveal that ERRα is an important regulator of systemic acid–base homeostasis, and expand our understandings of the role of ERRα in the adaptation to environmental challenges.
Supplementary Material
Acknowledgements
We thank the ICOB Core Facility and the Taiwan Zebrafish Core Facility for technical support during the experiments. We thank Dr Li-Yih Lin (National Taiwan Normal University) and Ms Yi-Ling Chou (Academia Sinica) for their assistance in SIET analysis.
Ethics
All research described here adhered to local guidelines and all appropriate ethical approval and licences were obtained. Experimental protocols were approved by the Academia Sinica Institutional Animal Care and Utilization Committee (approval no.: RFIZOOHP220782).
Data accessibility
The datasets supporting this article have been uploaded as part of the electronic supplementary material.
Competing interests
The authors declare no conflict of interests.
Authors' contributions
Y.-J.G. and P.-P.H. conceived and designed the research. Y.-J.G. and C.-Y.Y. carried out the molecular laboratory work. Y.-J.G. and S.-T.L. performed SIET analysis. Y.-J.G. and P.-P.H. wrote the manuscript. P.-P.H. and C.-J.H. supervised the project. All authors approved the manuscript for publication.
Funding
This study was financially supported by grants to P.-P.H. from Academia Sinica and the Ministry of Science and Technology, Taiwan, Republic of China.
References
- 1.Giguere V. 2008. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr. Rev. 29, 677–696. (doi:10.1210/er.2008-0017) [DOI] [PubMed] [Google Scholar]
- 2.Giguere V. 2002. To ERR in the estrogen pathway. Trends Endocrinol. Metab. 13, 220–225. (doi:10.1016/S1043-2760(02)00592-1) [DOI] [PubMed] [Google Scholar]
- 3.Bonnelye E, Vanacker JM, Dittmar T, Begue A, Desbiens X, Denhardt DT, Aubin JE, Laudet V, Fournier B. 1997. The ERR-1 orphan receptor is a transcriptional activator expressed during bone development. Mol. Endocrinol. 11, 905–916. (doi:10.1210/Me.11.7.905) [DOI] [PubMed] [Google Scholar]
- 4.Deblois G, Giguere V. 2013. Oestrogen-related receptors in breast cancer: control of cellular metabolism and beyond. Nat. Rev. Cancer 13, 27–36. (doi:10.1038/nrc3396) [DOI] [PubMed] [Google Scholar]
- 5.Sladek R, Bader JA, Giguere V. 1997. The orphan nuclear receptor estrogen-related receptor alpha is a transcriptional regulator of the human medium-chain acyl coenzyme A dehydrogenase gene. Mol. Cell Biol. 17, 5400–5409. (doi:10.1128/MCB.17.9.5400) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ranhotra HS. 2010. The estrogen-related receptor alpha: the oldest, yet an energetic orphan with robust biological functions. J. Recept. Signal Transduct. Res. 30, 193–205. (doi:10.3109/10799893.2010.487493) [DOI] [PubMed] [Google Scholar]
- 7.Villena JA, Kralli A. 2008. ERRalpha: a metabolic function for the oldest orphan. Trends Endocrinol. Metab. 19, 269–276. (doi:10.1016/j.tem.2008.07.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Villena JA, Hock MB, Chang WY, Barcas JE, Giguere V, Kralli A. 2007. Orphan nuclear receptor estrogen-related receptor alpha is essential for adaptive thermogenesis. Proc. Natl Acad. Sci. USA 104, 1418–1423. (doi:10.1073/pnas.0607696104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ichida M, Nemoto S, Finkel T. 2002. Identification of a specific molecular repressor of the peroxisome proliferator-activated receptor γ coactivator-1 α (PGC-1 α). J. Biol. Chem. 277, 50 991–50 995. (doi:10.1074/jbc.M210262200) [DOI] [PubMed] [Google Scholar]
- 10.Cartoni R, et al. 2005. Mitofusins 1/2 and ERRalpha expression are increased in human skeletal muscle after physical exercise. J. Physiol. 567, 349–358. (doi:10.1113/jphysiol.2005.092031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koeppen BM. 2009. The kidney and acid–base regulation. Adv. Physiol. Educ. 33, 275–281. (doi:33/4/27510.1152/advan.00054.2009) [DOI] [PubMed] [Google Scholar]
- 12.Wagner CA, Devuyst O, Bourgeois S, Mohebbi N. 2009. Regulated acid-base transport in the collecting duct. Pflugers Arch. 458, 137–156. (doi:10.1007/s00424-009-0657-z) [DOI] [PubMed] [Google Scholar]
- 13.Wagner CA, Finberg KE, Breton S, Marshansky V, Brown D, Geibel JP. 2004. Renal vacuolar H+-ATPase. Physiol Rev. 84, 1263–1314. (doi:10.1152/physrev.00045.2003) [DOI] [PubMed] [Google Scholar]
- 14.Claiborne JB, Edwards SL, Morrison-Shetlar AI. 2002. Acid-base regulation in fishes: cellular and molecular mechanisms. J. Exp. Zool. 293, 302–319. (doi:10.1002/jez.10125) [DOI] [PubMed] [Google Scholar]
- 15.Evans DH, Piermarini PM, Choe KP. 2005. The multifunctional fish gill: dominant site of gas exchange, osmoregulation, acid-base regulation, and excretion of nitrogenous waste. Physiol. Rev. 85, 97–177. (doi:10.1152/physrev.00050.2003) [DOI] [PubMed] [Google Scholar]
- 16.Hwang PP, Chou MY. 2013. Zebrafish as an animal model to study ion homeostasis. Pflugers Arch. Eur. J. Physiol. 465, 1233–1247. (doi:10.1007/s00424-013-1269-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang PP, Lee TH. 2007. New insights into fish ion regulation and mitochondrion-rich cells. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 148, 479–497. (doi:10.1016/j.cbpa.2007.06.416) [DOI] [PubMed] [Google Scholar]
- 18.Guh YJ, Lin CH, Hwang PP. 2015. Osmoregulation in zebrafish: ion transport mechanisms and functional regulation. EXCLI J. 14, 627–659. (doi:10.17179/excli2015-246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phypers B, Pierce JT. 2006. Lactate physiology in health and disease. Contin. Educ. Anaesth. Crit. Care Pain 6, 128–132. (doi:10.1093/bjaceaccp/mkl018) [Google Scholar]
- 20.Goubern M, Cadot M. 1983. Ketone-bodies in cold-acclimated rats. Comp. Biochem. Phys. B 76, 741–744. (doi:10.1016/0305-0491(83)90387-5) [DOI] [PubMed] [Google Scholar]
- 21.Veech RL, Chance B, Kashiwaya Y, Lardy HA, Cahill GR. 2001. Ketone bodies, potential therapeutic uses. Iubmb Life 51, 241–247. (doi:10.1080/152165401753311780) [DOI] [PubMed] [Google Scholar]
- 22.Hood VL, Tannen RL. 1998. Protection of acid-base balance by pH regulation of acid production. New Engl. J. Med. 339, 819–826. (doi:10.1056/NEJM199809173391207) [DOI] [PubMed] [Google Scholar]
- 23.Tremblay AM, Dufour CR, Ghahremani M, Reudelhuber TL, Giguere V. 2010. Physiological genomics identifies estrogen-related receptor alpha as a regulator of renal sodium and potassium homeostasis and the renin-angiotensin pathway. Mol. Endocrinol. 24, 22–32. (doi:10.1210/me.2009-0254) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang PP, Perry SF. 2010. Ionic and acid–base regulation. In Fish physiology (eds Eekker M, Perry SF, Farrell AP, Brauner CJ), pp. 311–343. San Diego, CA: Elsevier Academia Press. [Google Scholar]
- 25.Hwang PP. 2009. Ion uptake and acid secretion in zebrafish (Danio rerio). J. Exp. Biol. 212, 1745–1752. (doi:10.1242/jeb.026054) [DOI] [PubMed] [Google Scholar]
- 26.Lin LY, Horng JL, Kunkel JG, Hwang PP. 2006. Proton pump-rich cell secretes acid in skin of zebrafish larvae. Am. J. Physiol. Cell Physiol. 290, C371–C378. (doi:10.1152/ajpcell.00281.2005) [DOI] [PubMed] [Google Scholar]
- 27.Shih TH, Horng JL, Liu ST, Hwang PP, Lin LY. 2012. Rhcg1 and NHE3b are involved in ammonium-dependent sodium uptake by zebrafish larvae acclimated to low-sodium water. Am. J. Physiol. Regul. Integr. Comp. Physiol. 302, R84–R93. (doi:10.1152/ajpregu.00318.2011) [DOI] [PubMed] [Google Scholar]
- 28.Esaki M, Hoshijima K, Kobayashi S, Fukuda H, Kawakami K, Hirose S. 2007. Visualization in zebrafish larvae of Na+ uptake in mitochondria-rich cells whose differentiation is dependent on foxi3a. Am. J. Physiol. Reg. Integr. Comp. Physiol. 292, R470–R480. (doi:10.1152/ajpregu.00200.2006) [DOI] [PubMed] [Google Scholar]
- 29.Horng JL, Lin LY, Huang CJ, Katoh F, Kaneko T, Hwang PP. 2007. Knockdown of V-ATPase subunit A (atp6v1a) impairs acid secretion and ion balance in zebrafish (Danio rerio). Am. J. Physiol. Regul. Integr. Comp. Physiol. 292, R2068–R2076. (doi:10.1152/ajpregu.00578.2006) [DOI] [PubMed] [Google Scholar]
- 30.Lee YC, Yan JJ, Cruz SA, Horng JL, Hwang PP. 2011. Anion exchanger 1b, but not sodium-bicarbonate cotransporter 1b, plays a role in transport functions of zebrafish H+-ATPase-rich cells. Am. J. Physiol. Cell Physiol. 300, C295–C307. (doi:10.1152/ajpcell.00263.2010) [DOI] [PubMed] [Google Scholar]
- 31.Horng JL, Lin LY, Hwang PP. 2009. Functional regulation of H+-ATPase-rich cells in zebrafish embryos acclimated to an acidic environment. Am. J. Physiol. Cell Physiol. 296, C682–C692. (doi:10.1152/ajpcell.00576.2008) [DOI] [PubMed] [Google Scholar]
- 32.Thisse C, Thisse B. 2008. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69. (doi:10.1038/nprot.2007.514) [DOI] [PubMed] [Google Scholar]
- 33.Hsiao CD, You MS, Guh YJ, Ma M, Jiang YJ, Hwang PP. 2007. A positive regulatory loop between foxi3a and foxi3b is essential for specification and differentiation of zebrafish epidermal ionocytes. PLoS ONE 2, e302 (doi:10.1371/journal.pone.0000302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chou MY, Hsiao CD, Chen SC, Chen IW, Liu ST, Hwang PP. 2008. Effects of hypothermia on gene expression in zebrafish gills: upregulation in differentiation and function of ionocytes as compensatory responses. J. Exp. Biol. 211, 3077–3084. (doi:10.1242/jeb.019950) [DOI] [PubMed] [Google Scholar]
- 35.Chang WJ, Wang YF, Hu HJ, Wang JH, Lee TH, Hwang PP. 2013. Compensatory regulation of Na+ absorption by Na+/H+ exchanger and Na+-Cl− cotransporter in zebrafish (Danio rerio). Front. Zool. 10, 46 (doi:10.1186/1742-9994-10-46) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang YF, Tseng YC, Yan JJ, Hiroi J, Hwang PP. 2009. Role of SLC12A10.2, a Na-Cl cotransporter-like protein, in a Cl uptake mechanism in zebrafish (Danio rerio). Am. J. Physiol. Regul. Integr. Comp. Physiol. 296, R1650–R1660. (doi:10.1152/ajpregu.00119.2009) [DOI] [PubMed] [Google Scholar]
- 37.Chou MY, Lin CH, Chao PL, Hung JC, Cruz SA, Hwang PP. 2015. Stanniocalcin-1 controls ion regulation functions of ion-transporting epithelium other than calcium balance. Int. J. Biol. Sci. 11, 122–132. (doi:10.7150/ijbs.10773) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang WJ, Hwang PP. 2011. Development of zebrafish epidermis. Birth Defects Res. C Embryo Today 93, 205–214. (doi:10.1002/bdrc.20215) [DOI] [PubMed] [Google Scholar]
- 39.Alpern RJ, Preisig PA. 2004. Dietary acid, endothelins, and sleep. Trans. Am. Clin. Climatol. Assoc. 115, 385–393; discussion 393–384. [PMC free article] [PubMed] [Google Scholar]
- 40.Wesson DE. 2011. Endothelins and kidney acidification. Contrib. Nephrol. 172, 84–93. (doi:10.1159/000328764) [DOI] [PubMed] [Google Scholar]
- 41.Guh YJ, Tseng YC, Yang CY, Hwang PP. 2014. Endothelin-1 regulates H+-ATPase-dependent transepithelial H+ secretion in zebrafish. Endocrinology 155, 1728–1737. (doi:10.1210/en.2013-1775) [DOI] [PubMed] [Google Scholar]
- 42.Wagner CA, Mohebbi N, Uhlig U, Giebisch GH, Breton S, Brown D, Geibel JP. 2011. Angiotensin II stimulates H+-ATPase activity in intercalated cells from isolated mouse connecting tubules and cortical collecting ducts. Cell Physiol. Biochem. 28, 513–520. (doi:10.1159/000335112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khadouri C, Marsy S, Barlet-Bas C, Doucet A. 1989. Short-term effect of aldosterone on NEM-sensitive ATPase in rat collecting tubule. Am. J. Physiol. 257, F177–F181. [DOI] [PubMed] [Google Scholar]
- 44.Ali R, Amlal H, Burnham CE, Soleimani M. 2000. Glucocorticoids enhance the expression of the basolateral Na+:HCO3- cotransporter in renal proximal tubules. Kidney Int. 57, 1063–1071. (doi:10.1046/j.1523-1755.2000.00933.x) [DOI] [PubMed] [Google Scholar]
- 45.Tojo A, Tisher CC, Madsen KM. 1994. Angiotensin II regulates H+-ATPase activity in rat cortical collecting duct. Am. J. Physiol. 267, F1045–F1051. [DOI] [PubMed] [Google Scholar]
- 46.Flynt AS, Thatcher EJ, Burkewitz K, Li N, Liu YZ, Patton JG. 2009. miR-8 microRNAs regulate the response to osmotic stress in zebrafish embryos. J. Cell Biol. 185, 115–127. (doi:10.1083/jcb.200807026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumai Y, Perry SF. 2011. Ammonia excretion via Rhcg1 facilitates Na+ uptake in larval zebrafish, Danio rerio, in acidic water. Am. J. Physiol. Regul. Integr. Comp. Physiol. 301, R1517–R1528. (doi:10.1152/ajpregu.00282.2011) [DOI] [PubMed] [Google Scholar]
- 48.Bertrand S, et al. 2007. Unexpected novel relational links uncovered by extensive developmental profiling of nuclear receptor expression. PLoS Genet. 3, 2085–2100. (doi:10.1371/journal.pgen.0030188) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan JJ, Chou MY, Kaneko T, Hwang PP. 2007. Gene expression of Na+/H+ exchanger in zebrafish H+ -ATPase-rich cells during acclimation to low-Na+ and acidic environments. Am. J. Physiol. Cell Physiol. 293, C1814–C1823. (doi:10.1152/ajpcell.00358.2007) [DOI] [PubMed] [Google Scholar]
- 50.Rees BB, Boily P, Williamson LAC. 2009. Exercise- and hypoxia-induced anaerobic metabolism and recovery: a student laboratory exercise using teleost fish. Adv. Physiol. Ed. 33, 72–77. (doi:10.1152/advan.90188.2008) [DOI] [PubMed] [Google Scholar]
- 51.Gleeson TT. 1996. Post-exercise lactate metabolism: a comparative review of sites, pathways, and regulation. Annu. Rev. Physiol. 58, 565–581. (doi:10.1146/annurev.physiol.58.1.565) [DOI] [PubMed] [Google Scholar]
- 52.Diaz RJ. 2001. Overview of hypoxia around the world. J. Environ. Qual. 30, 275–281. (doi:10.2134/jeq2001.302275x) [DOI] [PubMed] [Google Scholar]
- 53.Owen OE. 2005. Ketone bodies as a fuel for the brain during starvation. Biochem. Mol. Biol. Edu. 33, 246–251. (doi:10.1002/bmb.2005.49403304246) [Google Scholar]
- 54.Hood VL, Schubert C, Keller U, Muller S. 1988. Effect of systemic Ph on Phi and lactic-acid generation in exhaustive forearm exercise. Am. J. Physiol. 255, F479–F485. [DOI] [PubMed] [Google Scholar]
- 55.Kraut JA, Madias NE. 2010. Metabolic acidosis: pathophysiology, diagnosis and management. Nat. Rev. Nephrol. 6, 274–285. (doi:10.1038/nrneph.2010.33) [DOI] [PubMed] [Google Scholar]
- 56.Gallet M, Vanacker JM. 2010. ERR receptors as potential targets in osteoporosis. Trends Endocrin. Metab. 21, 637–641. (doi:10.1016/j.tem.2010.06.008) [DOI] [PubMed] [Google Scholar]
- 57.Murray J, Huss JM. 2011. Estrogen-related receptor alpha regulates skeletal myocyte differentiation via modulation of the ERK MAP kinase pathway. Am. J. Physiol. Cell Physiol. 301, C630–C645. (doi:10.1152/ajpcell.00033.2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Michalek RD, et al. 2011. Estrogen-related receptor-alpha is a metabolic regulator of effector T-cell activation and differentiation. Proc. Natl Acad. Sci. USA 108, 18 348–18 353. (doi:10.1073/pnas.1108856108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang T, McDonald C, Petrenko NB, Leblanc M, Wang T, Giguere V, Evans RM, Patel VV, Pei LM. 2015. Estrogen-related receptor α (ERR α) and ERRγ are essential coordinators of cardiac metabolism and function. Mol. Cell Biol. 35, 1281–1298. (doi:10.1128/Mcb.01156-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dufour CR, Wilson BJ, Huss JM, Kelly DP, Alaynick WA, Downes M, Evans RM, Blanchette M, Giguere V. 2007. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and gamma. Cell Metab. 5, 345–356. (doi:10.1016/j.cmet.2007.03.007) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article have been uploaded as part of the electronic supplementary material.