Abstract
Loading of water bodies with dissolved organic carbon (DOC) and dissolved total nitrogen (DTN) affects their integrity and functioning. Microbial interactions mitigate the negative effects of high nutrient loads in these ecosystems. Despite numerous studies on how biodiversity mediates ecosystem functions, whether and how diversity and complexity of microbial food webs (horizontal, vertical) and the underlying ecological mechanisms influence nutrient removal has barely been investigated. Using microbial microcosms accommodating systematic combinations of prey (bacteria) and predator (protists) species, we showed that increasing bacterial richness improved the extent and reliability of DOC and DTN removal. Bacterial diversity drove nutrient removal either due to species foraging physiology or functional redundancy, whereas protistan diversity affected nutrient removal through bacterial prey resource partitioning and changing nutrient balance in the system. Our results demonstrate that prey–predator diversity and trophic interactions interactively determine nutrient contents, thus implying the vital role of microbial trophic complexity as a biological buffer against DOC and DTN.
Keywords: biodiversity and ecosystem functioning, predator–prey interactions, nutrient removal, microbial model systems, trophic complexity and stability
1. Introduction
Excessive loading with dissolved organic carbon (DOC) and dissolved total nitrogen (DTN), for example, from agricultural run-off is deteriorating the receiving aquatic ecosystems as well as the terrestrial ecosystems from where these substances originate [1,2]. Whereas the role of bacteria as key consumers of DOC and DTN in water bodies is recognized [3–5], influences of bacterial species richness on nutrient removal have been experimentally addressed only in a few studies [6,7]. Ecological theory and studies show that species diversity acts as a buffer against environmental impacts [8] and often has positive effects on ecosystem functioning [9,10] and stability [11,12]. However, several studies indicate that the link between bacterial diversity and function cannot be generalized as it depends among others, on the microbial service, its environmental context and the ecosystem under study [6,13–15].
In natural ecosystems, in addition to bottom-up control by resource availability, bacterial abundance, activity and community composition are also top-down controlled by viruses and protistan predators [16–19]. Protists thus play a central role in energy and nutrient transfer to higher trophic levels and bioelement cycling in general. Trophic interactions between bacteria and their predators have received some attention for instance in the context of organic matter decomposition and pollutant degradation [20–22]. Despite theoretical considerations that diversity changes at linked trophic levels simultaneously affect resource removal, resource use efficiency and ecosystem stability [23,24], only few studies have analysed the combined effect of predator and prey diversity on ecosystem function [25–27].
Here, we performed a series of experiments using microbial microcosms to address the effect of bacterial species richness on DOC and DTN removal. Bacterial model communities were assembled with species richness from one to five in all possible combinations (n = 31). It was then investigated if grazing pressure by single or multiple protist predators with different feeding modes had an effect on DOC and DTN removal and its stability. In addition, we analysed whether biodiversity mechanisms such as complementarity and selection effects [9] on both trophic levels correlate with the extent of removal. We hypothesized (i) that communities of higher bacterial species richness reduce DOC and DTN concentrations more effectively and (ii) that the presence and identity of protist predators influence the relationship between bacterial richness and removal of DOC and DTN as well as the stability of the removal processes.
2. Material and methods
(a). Microbial prey (bacteria) and predator (protists) strains used in this study
We used five bacterial strains, i.e. the proteobacteria Agrobacterium sp. (B1) and Janthinobacterium sp. (B3), and the actinobacteria Micrococcus sp. (B2), Williamsia sp. (B4) and Rhodococcus sp. (B5), which form distinctly coloured colonies used for easy quantification [26]. All bacteria were isolated from soil, and none of these strains fixes molecular nitrogen. The bacterial strains were maintained in Brunner CR-2 medium [26,27]. Further, we used three predators which differ in their feeding modes: the amoeba Acanthamoeba sp. (a surface feeder), the flagellate Poterioochromonas sp. (a filter feeder) and the ciliate Tetrahymena sp. (a filter feeder). Filter feeders mostly prey upon suspended cells and can forage over larger areas, while surface feeders prey upon attached cells [28–30]. In natural ecosystems, these taxa are key representatives of bacterivorous protistan predators. Tetrahymena sp. and Poterioochromonas sp. were maintained axenically in NSY (3 g l−1), while Acanthamoeba sp. was maintained axenically in proteose peptone–yeast–glucose (PYG) medium at 25°C in an incubator without any shaking (see details in [26]).
(b). Experimental design
We selected a substitutive experimental design to test the impact of bacterial (prey) and protistan (predator) species richness on both carbon and nitrogen removal as described in [26,27]. Our experiment comprised three levels: (I) controls without predation, (II) predation by one out of three different predators and (III) multiple predation (all predators together). Bacterial pre-cultures were grown for 25 h in a closed Erlenmeyer flask on a closed rotating shaker at 25°C. The initial total density of bacteria (2.11 × 107 cells ml−1) in all combinations was kept the same across all richness levels in all predation and control experiments. The protists were grown in the respective media one week before starting the experiments. They were concentrated by centrifugation [26,27] and washed with Brunner CR-2 medium before using them in the predation experiments. The overall total initial density of protists in monocultures and mixtures was kept the same (5 × 104 cells ml−1). The total volume of microcosms was 1.2 ml (per well) of liquid in 24-well microtitre plates. The experiment consisted of 155 microcosms in triplicates (465 in total). These were incubated for 48 h at 25°C. After 48 h, samples of 100 µl were taken for bacterial plating (colony count ml−1), protistan cell counts per millilitre, and for analysis of DOC and DTN, separately. We conducted our experiment in Brunner CR-2 which is a modified form of the R2A complex medium [31] containing DOC and DTN sources and which is commonly used for cultivation of environmental bacteria. The medium contained (g l−1): Na2HPO4 (2.44), KH2PO4 (1.52), MgSO4 × 7 H2O (0.2), CaCl2 × 2 H2O (0.01), NaCl (1.0), proteose peptone (0.25), yeast extract (0.25), tryptone (0.25), casamino acids (0.25), starch (0.5), glucose (5.0) and (NH4)2 SO4 (0.5). The medium was supplemented with vitamin solution (5 ml l−1) and trace element solution SL 10 (1 ml l−1). The C : N ratio of the medium was 7.87. Further experimental details can be found in [26,27].
(c). Data measurement parameters
Two hundred microlitres of samples for nutrient analysis were taken from each well after 48 h of incubation and centrifuged at 15 000 r.p.m. for 30 min. The supernatant was analysed for total organic carbon (TOC that we defined as DOC) and total nitrogen (TN that we defined as DTN) on a TOC analyser (TOC-5000 Analyzer; Shimadzu, Germany).
(d). Statistical analysis
The concentrations of both DOC and DTN were determined in all samples. The difference in the concentrations of both inoculated and uninoculated blank controls (microbe-free) was used to determine the removed DOC and DTN.
ANOVA followed by a Tukey test was performed to determine significant differences of relative nutrient removal between bacterial species exposed to the predation treatments (figures 1 and electronic supplementary material, figure S2) and between different predation and no-predation experiments (electronic supplementary material, figures S3, S7–S10. The same analysis was performed to reveal significant differences of relative nutrient removal and transgressive nutrient removal (Dmax) between bacterial mixtures with and without species B3 (electronic supplementary material, figure S5), and between no-predation, single predation and multiple predation experiments (figure 3).
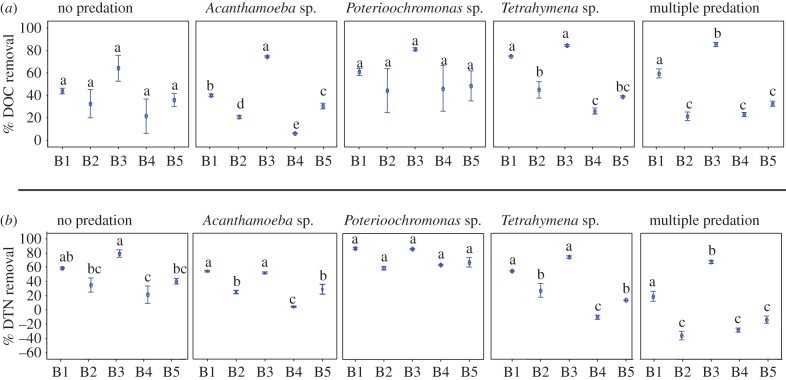
Figure 1.
Impact of predators on (a) relative carbon (DOC) removal and (b) on relative nitrogen (DTN) removal by different bacterial monocultures. The letters indicate statistically significant differences among treatments. The significance was determined using ANOVA test followed by Tukey's test. (Online version in colour.)
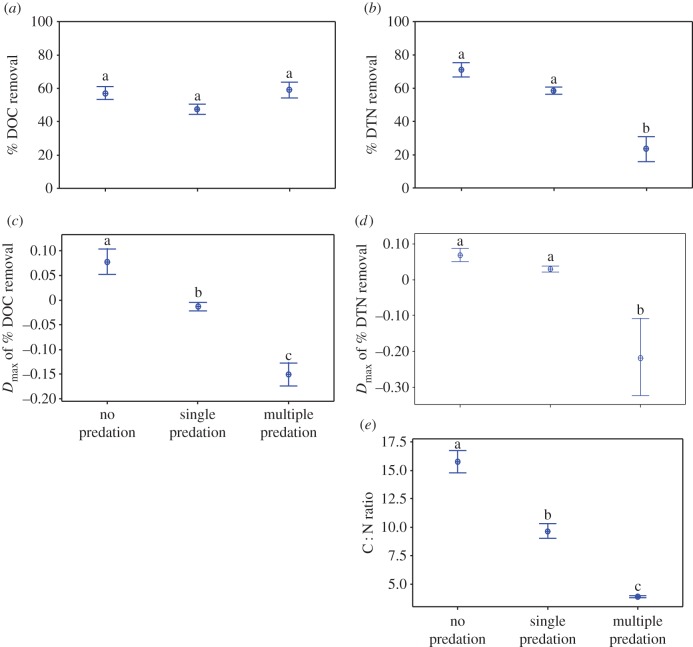
Figure 3.
Impact of predator richness on relative removal (a,b) and transgressive removal (Dmax) (c,d) of DOC and DTN and C : N ratio in the microcosms after 48 h of incubation (e) across all predation treatments. The results of the three single predator treatments (Tetrahymena sp., Poterioochromonas sp. and Acanthamoeba sp.) are averaged. Each point corresponds to the average of all replicates of each bacterial species combination across bacterial species richness. The bar represents standard error. The significance was determined using ANOVA test followed by Tukey's test. (Online version in colour.)
The transgressive nutrient removal (Dmax) was calculated for each bacterial mixture in all experiments by following [32] as given below.
| 2.10 |
The OT is the observed total removal of nutrients (DOC or DTN) by a mixture, and max (Mi) is maximum monoculture nutrient removal of the species found in that mixture. Dmax > 0 exhibits transgressive overyielding (i.e. nutrient removal) [32]. ANOVA with linear fitting of means was performed to assess the significance of bacterial species richness on nutrient removal, transgressive nutrient removal (Dmax) (figure 2) and stability (i.e. co-efficient of variation (C.V.)) (figure 4) in different predation experiments. Wilcoxon signed-rank test was performed to reveal the difference of Dmax from zero in all experiments (electronic supplementary material, figure S4). A general linear regression accompanied by ANOVA was done to show the relationship between predator production and DTN removal (electronic supplementary material, figure S6). Finally, we performed Spearman's rank order correlations analysis to determine the correlation of complementarity and selection effects as determined in [26,27] with nutrient removal determined in this study (electronic supplementary material, table S1).
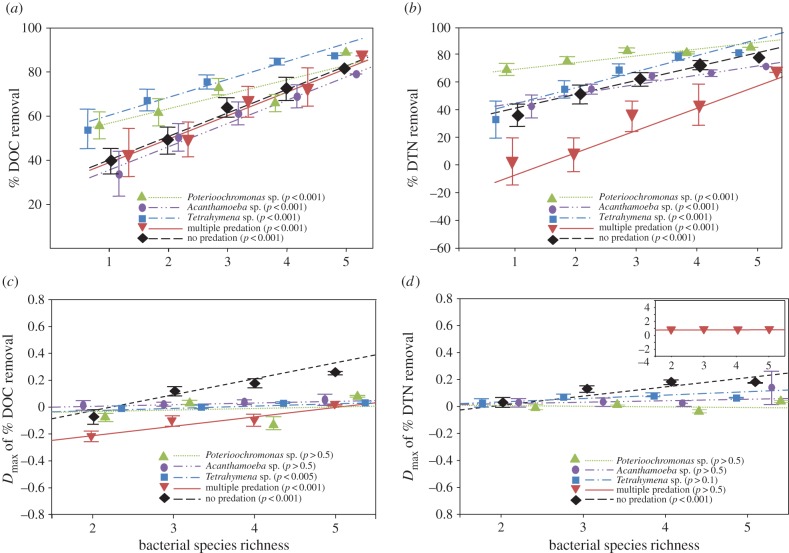
Figure 2.
Relative removal (a,b) and transgressive (Dmax) removal (c,d) of DOC and DTN as a function of bacterial species richness across all predation treatments. Each point corresponds to the average of all replicates of each bacterial species combination across all richness levels. The points referring to treatments at equal richness are slightly offset horizontally for clarity. The bar represents standard error. The significance was determined using ANOVA. (Online version in colour.)
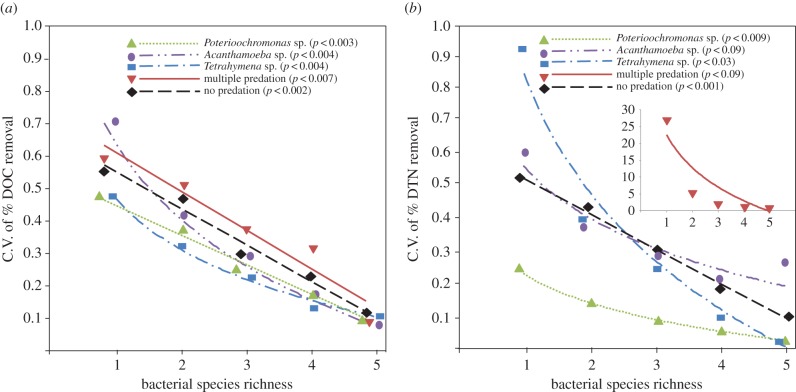
Figure 4.
C.V. of DOC (a) and DTN (b) removal as a function of bacterial species richness across all predation treatments and the predator-free control. The significance was determined using ANOVA. (Online version in colour.)
3. Results
(a). Nutrient removal by monocultures exposed to different predation pressures
In the absence of predators, DOC removal did not differ significantly between the individual monocultures (figure 1a), while DTN removal was relatively higher for the proteobacteria strains B1 (Agrobacterium sp.) and B3 (Janthinobacterium sp.) (figure 1b). Hence, the C : N ratios in the medium were higher after 48 h for these two strains than for the three actinobacteria strains B2, B4 and B5 (electronic supplementary material, figure S1).
Figure 1 and electronic supplementary material, figure S2, also illustrate the impact of any of the three predators or a mixture of all three predators on DOC and DTN removal by the monocultures: the proteobacterial strains B1 and B3 performed in almost all predation set-ups (except in the presence of Poterioochromonas) better than the other three bacterial monocultures (figure 1). Predator identity significantly affected DTN removal of individual bacterial species (electronic supplementary material, figure S2). For instance, predation by Poterioochromonas sp. tended to result in higher DTN removal by three out of five bacterial species; removal was even higher than in the microcosms without predators (electronic supplementary material, figure S2). Contrary, multiple predation resulted in the highest amount of remaining DTN in the system (electronic supplementary material, figures S2 and S3); in the case of B2, B4 and B5, DTN even reached higher concentrations than at the beginning of the experiment (electronic supplementary material, figure S2). A similar general effect by multiple predation was not observed for the removal of DOC (electronic supplementary material, figures S2 and S3).
(b). Impact of species richness on dissolved organic carbon and dissolved total nitrogen removal with and without predation pressure
Removal of DOC and DTN increased significantly with increasing bacterial species richness regardless of whether predators were present or not (figure 2a,b). Across the bacterial richness gradient, predator identity considerably affected DTN removal; microcosms exposed to Tetrahymena sp., Acathamoeba sp. and notably multiple predators resulted in significantly lower DTN removal and thus higher remaining DTN concentrations than in the control (electronic supplementary material, figure S3). In comparison, final DOC values in the different predation treatments varied much less (electronic supplementary material, figure S3).
(c). Relationship of biodiversity mechanisms with nutrient removal at different trophic levels
We further tested whether bacterial mixtures exhibited transgressive overyielding, i.e. higher nutrient removal of the mixture relative to the best monoculture in the corresponding mixture [32] and whether nutrient removal was correlated with mechanisms driving biodiversity effects, i.e. the complementarity and selection effects [9]. The latter analyses were based on bacterial (cfu ml−1) and protist (cells ml−1) yields of each species in monocultures and in mixtures as published in [26,27].
Transgressive nutrient removal (Dmax) tended to increase along the bacterial richness gradient. We observed a significant positive relationship between species richness and transgressive DOC removal (DmaxDOC) in predator-free communities, and in mixtures exposed to either Tetrahymena sp. or to multiple predators (figure 2c). Transgressive removal of DTN was positively correlated with bacterial richness only in microcosms without predators (figure 2d). On average, values of DmaxDTN were significantly higher than zero (Wilcoxon signed-rank test) in predator-free communities and in bacterial mixtures exposed either to Tetrahymena sp. or to Acanthamoeba sp. (electronic supplementary material, figure S4). Contrary, the Dmax of both DOC and DTN were significantly below zero in multiple predation experiments (electronic supplementary material, figure S4). As Dmax values close to zero indicate a positive selection effect [33], we compared the nutrient removal and corresponding transgressive nutrient removal between bacterial mixtures with and without the strain B3, which as a monoculture showed the highest removal of DOC and DTN (figure 1 and electronic supplementary material, figure S2). Bacterial mixtures with B3 removed more DOC and DTN than mixtures without this strain and showed transgressive overyielding in particular of DOC removal in the predation-free experiments and in mixtures with Tetrahymena sp. or multiple predators (electronic supplementary material, figure S5).
Contrary to the bacterial diversity gradient, average DTN removal and DmaxDTN significantly decreased at higher predator diversity (figure 3b,d). Though predator diversity did not affect DOC removal significantly, it significantly reduced the DmaxDOC of bacterial mixtures (figure 3a,c). Concomitantly, higher predator diversity resulted in a reduced C : N ratio of the microcosms (figure 3e).
Both bacterial complementarity and selection effects were not correlated with nutrient removal in predator-free experiments (electronic supplementary material, table S1). However, identity of the predators determined the direction of the relationship between these biodiversity mechanisms and nutrient removal. Bacterial microcosms exposed to multiple predators showed slightly significant correlation with nutrient removal across the bacterial richness gradient (electronic supplementary material, table S1).
The predators' complementarity and selection effects were negatively correlated with nutrient removal (electronic supplementary material, table S1). Increasing protist production in multiple predator assemblages negatively correlated with DTN removal (electronic supplementary material, figure S6). Eleven out of 31 microcosms exposed to those multiple predators were characterized by negative DTN removal indicating predation-driven recycling of N (electronic supplementary material, figure S7). Interestingly, none of these mixtures contained strain B3.
(d). Impact of species richness on stability of nutrient removal with and/or without predation pressure
The effect of increasing bacterial species richness on the stability of DOC and DTN removal was analysed by using the C.V., where higher values indicate lower stability. In all experiments, we observed that the variability in nutrient removal significantly decreased with increasing bacterial species richness independent of the trophic interactions present in the microcosms (figure 4a,b).
4. Discussion
Removal of DOC and DTN by bacterial monocultures varied under different predation pressures (figure 1 and electronic supplementary material, figure S2). Overall, nutrient removal by the proteobacteria B1 and B3 was relatively higher than that by the other bacteria. Though individual members from each phylum certainly vary in nutrient consumption, members of the proteobacteria have been suggested to be usually copiotrophic and fast growing due to their ability to more efficiently use labile nutrients [34]. Janthinobacteria have been shown to produce an anti-bacterial and anti-protistan pigment (violacein), which could be important for competing with other microbes [35,36]. However, we so far have not observed any inhibition of the other microorganism in our experiments with this species [26,27].
Depending on the species combinations, predation influenced DOC and DTN removal in different ways. To explain the counterintuitive stimulation of bacterial activity under predation pressure, various ecological mechanisms have been offered [37–40], which mainly fall into two categories. A non-consumptive effect is, for example, the promotion of actively growing prey by the selective elimination of rather inactive prey [41,42]. Non-consumptive effects may have thus provided additional niche space for the active bacteria leading to an improved nutrient removal. Consumptive effects [41,42] include the direct uptake or excretion of minerals and organic substances in the habitat colonized by both the prey and the predator; nutrient release may result in increased nutrient concentrations, and thus appear as decreased bacterial activity. Reduced DOC and DTN removal can also be a direct effect of predation eliminating active bacterial consumers (trophic cascade) and releasing bacterial carbon and nitrogen [42,43] into the system. The absence of predation effects might be explained by the compensation of the foregoing mechanisms [42,44].
Removal of DOC and DTN became more efficient (figure 2) and less variable (figure 4) with higher bacterial species richness. This richness effect was observed irrespective of bacterial identities and top-down control by predators. Functional stability of environmental processes is a fundamental ecosystem parameter [45,46], with high C.V. reflecting low ecosystem reliability [12,47]. Our results are consistent with earlier findings showing that species-rich ecosystems tend to use resources more efficiently [48] and that ecosystem processes stabilize with higher richness [49]. However, the relationship between bacterial richness and resource utilization may not be generalized. For instance, Langenheder et al. [50] showed that a combination of bacterial species richness and substrate richness enhanced substrate oxidation and that there was no direct interaction between only bacterial species richness and substrate utilization. Contrary to theoretical predictions that trophic interactions shift the diversity–functioning relationships in a nonlinear manner [23], we observed a linear relationship of bacterial richness versus DOC and DTN removal (figure 2).
There are several plausible interpretations with respect to the consumptive and non-consumptive effects of predator diversity on nutrient contents in the system, which however were not separated in our experimental set-up and would require isotope techniques: for instance, efficient prey exploitation by multiple predators as shown in [26,27] may possibly result in an imbalance between carbon leaving the system as CO2 as a result of intense predation and the development of excess nitrogen, which is not built into biomass because of the relative lack of carbon. Multiple predator production also showed a negative correlation with DTN removal (electronic supplementary material, figure S6), implying that higher predator production led to higher recycling of DTN into the system. The relatively high DOC removal (figure 3a), together with a potential recycling of nitrogen into the system, resulted in reduced nitrogen removal (figure 3b) and consequently narrowed down the C : N ratio (figure 3e) in the multiple predation experiment.
Overall, this work points at several aspects regarding nutrient stoichiometry and nutrient limitations, which are worthy of further investigation. First, in the case of a relatively higher C : N ratio as observed in the microcosms without predation and with Poterioochromonas sp. (electronic supplementary material, figure S8), carbon removal could be controlled by the rate at which bacteria can sequester nitrogen. Our data show that these two set-ups resulted in an overall higher nitrogen removal, both in the monocultures (electronic supplementary material, figure S9) and the mixtures (electronic supplementary material, figure S10). In contrast, the C : N ratio was relatively low in microcosms exposed to Acanthamoeba sp., Tetrahymena sp. and multiple predation (electronic supplementary material, figure S8), where nitrogen removal is very likely controlled by the carbon. Consequently, the observed low nitrogen might be also explained by carbon limitation (electronic supplementary material, figures S9 and S10). The larger predators (Acanthamoeba sp. and Tetrahymena sp.) might possibly recycle and thus excrete more nitrogen into the system than the smaller predator (Poterioochromonas sp.). However, active filter feeders such as Poterioochromonas sp. and Tetrahymena sp. may be more efficient predators than the relatively slow surface glider Acanthamoeba sp.
Interestingly, the assumed predator-mediated release of DTN was reduced when a broader spectrum of prey species was available (figure 2). This might be explained by the fact that more diverse prey assemblages build up biomass more efficiently [26], so that more N is immobilized [48]. Alternatively, increasing prey diversity may also result in a relatively reduced production of diverse predator assemblages [27].
Accordingly, high final DTN values were observed for poorly growing bacteria (e.g. strains B2 and B4 [26]), and in mixed cultures of low species richness containing these bacteria, especially when species B3 was not a part of these combinations (electronic supplementary material, figure S7). It is likely that the growth recovery or reproduction of these bacteria was not as fast as that of other species being controlled by multiple predators, thus leading to higher total DTN content in the system.
The additive partitioning approach by Loreau & Hector [9] allows mathematical calculation of potential mechanisms resulting in positive biodiversity effects: a complementarity effect may arise from either resource partitioning, niche differentiation or positive interactions among species, while the selection effect describes the increasing probability of including highly competitive species. Yet, there are different additional biological mechanisms, which need to be considered at the prey and the predator level. The observed Dmax of nutrients (electronic supplementary material, figure S4) could be explained by a more efficient nutrient foraging physiology of species B3 (i.e. a sort of nutrient selection effect). But in some cases, the bacterial mixtures containing B3 did not perform better than the best monoculture included (B3), thus pointing at a weak selection effect [33]. We recently showed that poorly performing (in terms of CFU production) bacterial species seemed to grow better under multiple predation pressure in this experimental set-up, resulting in more evenly distributed prey species and thus a species complementarity (i.e. apparent facilitation) [26]. However, these species alone could not exploit nutrient resources as readily as the mixture containing the best-performing species B3 (electronic supplementary material, figure S5a,b). These results suggest that a balanced community evenness caused by top-down control may not necessarily promote resource removal or partitioning by the species that inherently grow poorly. Generally, predation tended to reduce the performance (Dmax) of mixtures at lower species richness levels, and its effect on mixture-mediated nutrient removal decreased along the bacterial richness gradient (figure 2a,b). These results thus indicate stronger cascading effects of predators on less diverse bacterial communities.
The reduced Dmax of nutrients by bacterial mixtures exposed to predation (figure 3) is in line with a reduction of bacterial abundance across increasing predator richness [26,27]. This observation rejects the probability of strong selection or complementarity effects. Using the same experimental set-up, we recently showed that selection effects may be driving yield (calculated as cfu ml−1) in diverse bacterial assemblages without predation pressure, while the presence of predators resulted in increased complementarity effects [26,27]. However, we did not find any correlation of these mechanisms with nutrient removal in predation-free control experiments (electronic supplementary material, table S1). Overall, our results partly refute a role of species competition, synergistic interactions and niche breadth in nutrient processing [5,51]. Interpretation of bacterial processing of DOC and DTN may not be easy in the context of niche theory (i.e. resource partitioning) since many bacteria are generalists [51,52]. We used a complex medium with easy degradable carbon and nitrogen sources, which all bacterial strains were capable of using, indicating some degree of functional redundancy and/or generalism in our system. However, adding predation as a trophic factor tended to reshape the correlation between biodiversity mechanisms and nutrient removal, depending upon the identity of the predator. Both complementarity and selection effects showed different (positive, negative or absent) correlations with nutrient removal in single predation experiments, thus partly reflecting the divergent effect of protist predators on bacterial community composition [26], and consequently on nutrient removal. Similarly, a weak positive correlation was observed in bacterial microcosms exposed to multiple predators.
At the predator level, interestingly, both complementarity and selection effects showed negative correlation between DOC and DTN removal (electronic supplementary material, table S1). Regardless of whether protists exploit prey resources via complementarity or selection effects [27], there will be a negative impact on both prey productivity [27], and consequently on DOC and DTN removal. This might offer another explanation for the observed relatively low DTN removal in the multiple predation experiment. Consequently, predator complementarity and selection effects increase the consumptive and non-consumptive effects of predators on nutrient cycling in the system. Though predator influences on nutrient removal via a control of prey abundance and productivity have been described in the past [42], our results provide mechanistic information regarding predator diversity effects on DOC and DTN removal.
Bacterial species richness stabilized the process along the diversity gradient (figure 4), which is consistent with the theory that higher diversity stabilizes ecosystem-level processes such as biomass production [53]. To date, the few findings on the relationship between microbial richness and stability suggest that broad-scale ecosystem processes (such as resource use efficiency) may benefit from diverse microbial systems at least at larger scales [54]. However, for particular ecosystems, results are still contradictory. Decreasing diversity may in fact be correlated with higher predictability of ecosystem processes like microbial biomass production [55] and nitrogen cycling [56]. Eisenhauer & Schädler [57] observed an inconsistent impact of decomposer diversity on the stability of litter decomposition. Other studies highlight the importance of functional diversity (i.e. diversity of genes) to maintain and stabilize specific ecosystem functions, such as removal of pollutants [58]. The low variability in DOC and DTN removal in predation treatments further disprove predictions that systems with rather low species richness (like in this case) are more likely to display large differences and variability in their functioning [11,47]. Interestingly, the type of predators in general affected the variability of the microbial functions, while the presence of multiple predators tended to destabilize DTN removal mostly at lower species richness (figure 4). Such kind of variability in aggregated community functioning may arise in systems where species are distributed at different trophic levels, and prey–predator communities tend to dominate each other [11,47].
In conclusion, our results reveal that the trophic complexity across horizontal (bacterial prey) and vertical (protistan predators) axes was the primary mechanism explaining the effectiveness and stability of DOC and DTN removal. Our results therefore provide valuable aspects to be further tested with other organisms and more complex experimental set-ups regarding the impact of increased diversity and complexity of trophic networks on DOC and DTN removal from ecosystems.
Supplementary Material
Acknowledgements
We thank Jens Boenigk (University of Duisburg-Essen, Germany) and Sharon A. Huws (Aberystwyth University, UK) for kindly providing us the flagellate and the amoebae. We thank M. Brian Traw (University of Pittsburgh, USA) for his critical and useful comments on this manuscript. We are grateful to Verena Jaschik, Anett Heidtmann and Martina Kolbe for technical assistance and Roland Müller for support at the TOC-5000 Analyzer.
Data accessibility
Nitrogen and carbon data: Dryad: http://dx.doi.org/10.5061/dryad.q60vk.
Authors' contributions
All authors conceived the study framework and discussed the theoretical background. M.S., I.F. and A.C. planned the study. M.S. conducted the experiments and compiled the data; A.C. supervised the experiments. M.S., I.F. and A.C. contributed to the data analysis. M.S. wrote the first draft of the manuscript, and all authors contributed substantially to revisions. All authors gave final approval for publication.
Competing interests
We have no competing interest.
Funding
M.S. appreciates and acknowledges the financial support from a Helmholtz-DAAD PhD Fellowship, the Helmholtz Interdisciplinary GRADuate School for Environmental Research (HIGRADE) programme.
References
- 1.Vörösmarty CJ, et al. 2010. Global threats to human water security and river biodiversity. Nature 467, 555–561. ( 10.1038/nature09440) [DOI] [PubMed] [Google Scholar]
- 2.Woodward G, et al. 2012. Continental-scale effects of nutrient pollution on stream ecosystem functioning. Science 336, 1438–1440. ( 10.1126/science.1219534) [DOI] [PubMed] [Google Scholar]
- 3.Jetten MSM, et al. 2002. Improved nitrogen removal by application of new nitrogen-cycle bacteria. Rev. Environ. Sci. Biotechnol. 1, 51–63. ( 10.1023/A:1015191724542) [DOI] [Google Scholar]
- 4.Cotner JB, Biddanda BA. 2002. Small players, large role: microbial influence on biogeochemical processes in pelagic aquatic ecosystems. Ecosystems 5, 105–121. ( 10.1007/s10021-001-0059-3) [DOI] [Google Scholar]
- 5.McCarren J, Becker JW, Repeta DJ, Shi Y, Young CR, Malmstrom RR, Chisholm SW, DeLong EF. 2010. Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc. Natl Acad. Sci. USA 107, 16 420–16 427. ( 10.1073/pnas.1010732107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peter H, Beier S, Bertilsson S, Lindström ES, Langenheder S, Tranvik LJ. 2011. Function-specific response to depletion of microbial diversity. ISME J. 5, 351–361. ( 10.1038/ismej.2010.119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ylla I, Peter H, Romaní AM, Tranvik LJ. 2013. Different diversity–functioning relationship in lake and stream bacterial communities. FEMS Microbiol. Ecol. 85, 95–103. ( 10.1111/1574-6941.12101) [DOI] [PubMed] [Google Scholar]
- 8.Yachi S, Loreau M. 1999. Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc. Natl Acad. Sci. USA 96, 1463–1468. ( 10.1073/pnas.96.4.1463) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loreau M, Hector A. 2001. Partitioning selection and complementarity in biodiversity experiments. Nature 412, 72–76. ( 10.1038/35083573) [DOI] [PubMed] [Google Scholar]
- 10.Bell T, Newman JA, Silverman BW, Turner SL, Lilley AK. 2005. The contribution of species richness and composition to bacterial services. Nature 436, 1157–1160. ( 10.1038/nature03891) [DOI] [PubMed] [Google Scholar]
- 11.Naeem S, Li S. 1997. Biodiversity enhances ecosystem reliability. Nature 390, 507–509. ( 10.1038/37348) [DOI] [Google Scholar]
- 12.Haddad NM, Crutsinger GM, Gross K, Haarstad J, Tilman D. 2011. Plant diversity and the stability of foodwebs. Ecol. Lett. 14, 42–46. ( 10.1111/j.1461-0248.2010.01548.x) [DOI] [PubMed] [Google Scholar]
- 13.Cardinale BJ, Nelson K, Palmer MA. 2000. Linking species diversity to the functioning of ecosystems: on the importance of environmental context. Oikos 91, 175–183. ( 10.1034/j.1600-0706.2000.910117.x) [DOI] [Google Scholar]
- 14.Langenheder S, Lindström ES, Tranvik LJ. 2005. Weak coupling between community composition and functioning of aquatic bacteria. Limnol. Oceanogr. 50, 957–967. ( 10.4319/lo.2005.50.3.0957) [DOI] [Google Scholar]
- 15.Szabo KE, Itor POB, Bertilsson S, Tranvik L, Eiler A. 2007. Importance of rare and abundant populations for the structure and functional potential of freshwater bacterial communities. Aquat. Microb. Ecol. 47, 1–10. ( 10.3354/ame047001) [DOI] [Google Scholar]
- 16.Chan Y-F, Tsai A-Y, Chiang K-P, Hsieh C. 2009. Pigmented nanoflagellates grazing on Synechococcus: seasonal variations and effect of flagellate size in the coastal ecosystem of subtropical Western Pacific. Microb. Ecol. 58, 548–557. ( 10.1007/s00248-009-9569-x) [DOI] [PubMed] [Google Scholar]
- 17.Pernthaler J. 2005. Predation on prokaryotes in the water column and its ecological implications. Nat. Rev. Micro. 3, 537–546. ( 10.1038/nrmicro1180) [DOI] [PubMed] [Google Scholar]
- 18.Johnke J, Cohen Y, de Leeuw M, Kushmaro A, Jurkevitch E, Chatzinotas A. 2014. Multiple micro-predators controlling bacterial communities in the environment. Curr. Opin. Biotechnol. 27, 185–190. ( 10.1016/j.copbio.2014.02.003) [DOI] [PubMed] [Google Scholar]
- 19.Saleem M, Moe LA. 2014. Multitrophic microbial interactions for eco- and agro-biotechnological processes: theory and practice. Trends Biotechnol. 32, 529–537. ( 10.1016/j.tibtech.2014.08.002) [DOI] [PubMed] [Google Scholar]
- 20.Mattison RG, Harayama S. 2001. The predatory soil flagellate Heteromita globosa stimulates toluene biodegradation by a Pseudomonas sp. FEMS Microbiol. Lett. 194, 39–45. ( 10.1111/j.1574-6968.2001.tb09443.x) [DOI] [PubMed] [Google Scholar]
- 21.Cunningham JJ, Kinner NE, Lewis M. 2009. Protistan predation affects trichloroethene biodegradation in a bedrock aquifer. Appl. Environ. Microbiol. 75, 7588–7593. ( 10.1128/AEM.01820-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chatzinotas A, Schellenberger S, Glaser K, Kolb S. 2013. Assimilation of cellulose-derived carbon by microeukaryotes in oxic and anoxic slurries of an aerated soil. Appl. Environ. Microbiol. 79, 5777–5781. ( 10.1128/AEM.01598-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thébault E, Loreau M. 2003. Food-web constraints on biodiversity–ecosystem functioning relationships. Proc. Natl Acad. Sci. USA 100, 14 949–14 954. ( 10.1073/pnas.2434847100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loreau M, Thébault E. 2005. Food webs and the relationship between biodiversity and ecosystem functioning. In Dynamic food webs, volume 3: multispecies assemblages, ecosystem development and environmental change (eds de Ruiter PC, Wolters V, Moore GC), pp. 270–282. New York, NY: Academic Press. [Google Scholar]
- 25.Gamfeldt L, Hillebrand H, Jonsson PR. 2005. Species richness changes across two trophic levels simultaneously affect prey and consumer biomass. Ecol. Lett. 8, 696–703. ( 10.1111/j.1461-0248.2005.00765.x) [DOI] [Google Scholar]
- 26.Saleem M, Fetzer I, Dormann CF, Harms H, Chatzinotas A. 2012. Predator richness increases the effect of prey diversity on prey yield. Nat. Commun. 3, 1305 ( 10.1038/ncomms2287) [DOI] [PubMed] [Google Scholar]
- 27.Saleem M, Fetzer I, Harms H, Chatzinotas A. 2013. Diversity of protists and bacteria determines predation performance and stability. ISME J. 7, 1912–1921. ( 10.1038/ismej.2013.95) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Böhme A, Risse-Buhl U, Küsel K. 2009. Protists with different feeding modes change biofilm morphology. FEMS Microbiol. Ecol. 69, 158–169. ( 10.1111/j.1574-6941.2009.00710.x) [DOI] [PubMed] [Google Scholar]
- 29.Verni F, Gualtieri P. 1997. Feeding behaviour in ciliated protists. Micron 28, 487–504. ( 10.1016/S0968-4328(97)00028-0) [DOI] [Google Scholar]
- 30.Dopheide A, Lear G, Stott R, Lewis G. 2011. Preferential feeding by the ciliates Chilodonella and Tetrahymena spp. and effects of these protozoa on bacterial biofilm structure and composition. Appl. Environ. Microbiol. 77, 4564–4572. ( 10.1128/AEM.02421-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reasoner DJ, Geldreich EE. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loreau M. 1998. Separating sampling and other effects in biodiversity experiments. Oikos 82, 600–602. ( 10.2307/3546381) [DOI] [Google Scholar]
- 33.Jiang L. 2007. Negative selection effects suppress relationships between bacterial diversity and ecosystem functioning. Ecology 88, 1075–1085. ( 10.1890/06-1556) [DOI] [PubMed] [Google Scholar]
- 34.Fierer N, Lauber CL, Ramirez KS, Zaneveld J, Bradford MA, Knight R. 2012. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 6, 1007–1017. ( 10.1038/ismej.2011.159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pantanella F, Berlutti F, Passariello C, Sarli S, Morea C, Schippa S. 2007. Violacein and biofilm production in Janthinobacterium lividum. J. Appl. Microbiol. 102, 992–999. ( 10.1111/j.1365-2672.2006.03155.x) [DOI] [PubMed] [Google Scholar]
- 36.Matz C, Deines P, Boenigk J, Arndt H, Eberl L, Kjelleberg S, Jurgens K. 2004. Impact of violacein-producing bacteria on survival and feeding of bacteriovorans nanoflagellates. Appl. Environ. Microbiol. 70, 1593–1599. ( 10.1128/AEM.70.3.1593-1599.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verhagen FJM, Laanbroek HJ. 1992. Effects of grazing by flagellates on competition for ammonium between nitrifying and heterotrophic bacteria in chemostats. Appl. Environ. Microbiol. 58, 1962–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zweifell U, Blackburn N, Hagstrm A. 1996. Cycling of marine dissolved organic matter. I. An experimental system. Aquat. Microb. Ecol. 11, 65–77. ( 10.3354/ame011065) [DOI] [Google Scholar]
- 39.Hahn MW, Höfle MG. 2001. Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol. Ecol. 35, 113–121. ( 10.1111/j.1574-6941.2001.tb00794.x) [DOI] [PubMed] [Google Scholar]
- 40.Sherr EB, Sherr BF. 2002. Significance of predation by protists in aquatic microbial food webs. Antonie van Leeuwenhoek. 81, 293–308. ( 10.1023/A:1020591307260) [DOI] [PubMed] [Google Scholar]
- 41.Schmitz OJ. 2010. Resolving ecosystem complexity. Princeton, NJ: Princeton University Press. [Google Scholar]
- 42.Schmitz OJ, Hawlena D, Trussell GC. 2010. Predator control of ecosystem nutrient dynamics. Ecol. Lett. 13, 1199–1209. ( 10.1111/j.1461-0248.2010.01511.x) [DOI] [PubMed] [Google Scholar]
- 43.Strom SL, Benner R, Ziegler S, Dagg MJ. 1997. Planktonic grazers are a potentially important source of marine dissolved organic carbon. Limnol. Oceanogr. 42, 1364–1374. ( 10.4319/lo.1997.42.6.1364) [DOI] [Google Scholar]
- 44.Del Giorgio PA, Gasol JM, Vaque D, Mura P, Agusti S, Duarte CM. 1996. Bacterioplankton community structure: protists control net production and the proportion of active bacteria in a coastal marine community. Limnol. Oceanogr. 41, 1169–1179. ( 10.4319/lo.1996.41.6.1169) [DOI] [Google Scholar]
- 45.Pimm SL. 1984. The complexity and stability of ecosystems. Nature 307, 321–326. ( 10.1038/307321a0) [DOI] [Google Scholar]
- 46.McCann KS. 2000. The diversity–stability debate. Nature 405, 228–233. ( 10.1038/35012234) [DOI] [PubMed] [Google Scholar]
- 47.Leary DJ, Rip JMK, Petchey OL. 2011. The impact of environmental variability and species composition on the stability of experimental microbial populations and communities. Oikos 121, 327–336. ( 10.1111/j.1600-0706.2011.19523.x) [DOI] [Google Scholar]
- 48.Cardinale BJ. 2011. Biodiversity improves water quality through niche partitioning. Nature 472, 86–89. ( 10.1038/nature09904) [DOI] [PubMed] [Google Scholar]
- 49.Morin PJ, McGrady-Steed J. 2004. Biodiversity and ecosystem functioning in aquatic microbial systems: a new analysis of temporal variation and species richness-predictability relations. Oikos 104, 458–466. ( 10.1111/j.0030-1299.2004.13256.x) [DOI] [Google Scholar]
- 50.Langenheder S, Bulling MT, Solan M, Prosser JI. 2010. Bacterial biodiversity–ecosystem functioning relations are modified by environmental complexity. PLoS ONE 5, e10834 ( 10.1371/journal.pone.0010834) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N. 2009. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 3, 442–453. ( 10.1038/ismej.2008.127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mou X, Sun S, Edwards RA, Hodson RE, Moran MA. 2008. Bacterial carbon processing by generalist species in the coastal ocean. Nature 451, 708–711. ( 10.1038/nature06513) [DOI] [PubMed] [Google Scholar]
- 53.Balvanera P, Pfisterer AB, Buchmann N, He J, Nakashizuka T, Raffaelli D, Schmid B. 2006. Quantifying the evidence for biodiversity effects on ecosystem functioning and services. Ecol. Lett. 9, 1146–1156. ( 10.1111/j.1461-0248.2006.00963.x) [DOI] [PubMed] [Google Scholar]
- 54.Ptacnik R, Solimini AG, Andersen T, Tamminen T, Brettum P, Lepistö L, Willén E, Rekolainen S. 2008. Diversity predicts stability and resource use efficiency in natural phytoplankton communities. Proc. Natl Acad. Sci. USA 105, 5134–5138. ( 10.1073/pnas.0708328105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kokociński M, Soininen J. 2008. Temporal variation in phytoplankton in two lakes with contrasting disturbance regimes. Archiv. Hydrobiol. 171, 39–48. ( 10.1127/1863-9135/2008/0171-0039) [DOI] [Google Scholar]
- 56.Philippot L, Spor A, Hénault C, Bru D, Bizouard F, Jones CM, Sarr A, Maron P-A. 2013. Loss in microbial diversity affects nitrogen cycling in soil. ISME J. 7, 1609–1619. ( 10.1038/ismej.2013.34) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eisenhauer N, Schädler M. 2010. Inconsistent impacts of decomposer diversity on the stability of aboveground and belowground ecosystem functions. Oecologia 165, 403–415. ( 10.1007/s00442-010-1784-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cravo-Laureau C, Hernandez-Raquet G, Vitte I, Jézéquel R, Bellet V, Godon J-J, Caumette P, Balaguer P, Duran R. 2011. Role of environmental fluctuations and microbial diversity in degradation of hydrocarbons in contaminated sludge. Res. Microbiol. 162, 888–895. ( 10.1016/j.resmic.2011.04.011) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Nitrogen and carbon data: Dryad: http://dx.doi.org/10.5061/dryad.q60vk.