Abstract
The diversity and structure of ecosystems has been found to depend both on trophic interactions in food webs and on other species interactions such as habitat modification and mutualism that form non-trophic interaction networks. However, quantification of the dependencies between these two main interaction networks has remained elusive. In this study, we assessed how habitat-modifying organisms affect basic food web properties by conducting in-depth empirical investigations of two ecosystems: North American temperate fringing marshes and West African tropical seagrass meadows. Results reveal that habitat-modifying species, through non-trophic facilitation rather than their trophic role, enhance species richness across multiple trophic levels, increase the number of interactions per species (link density), but decrease the realized fraction of all possible links within the food web (connectance). Compared to the trophic role of the most highly connected species, we found this non-trophic effects to be more important for species richness and of more or similar importance for link density and connectance. Our findings demonstrate that food webs can be fundamentally shaped by interactions outside the trophic network, yet intrinsic to the species participating in it. Better integration of non-trophic interactions in food web analyses may therefore strongly contribute to their explanatory and predictive capacity.
Keywords: consumer–resource interactions, non-trophic interactions, facilitation, ecological networks, ecosystem engineering, foundation species
1. Introduction
One of the great challenges in ecology is to elucidate how different types of species interactions drive the structure and dynamics of communities and ecosystems. Ever since Darwin [1] coined the term ‘web of life’ [1], food webs have been intensively studied as paradigmatic examples of natural complex systems [2–4]. To date, analyses investigating the stability and structure of species interaction networks have primarily focused on the properties of the network formed by feeding interactions between species [5–12]. Studies typically investigate the topology of trophic interactions (e.g. links per species, connectance) [5,9], variation in interaction strength (e.g. across trophic levels) and the nature of trophic interactions (e.g. predator–prey, plant–herbivore) [8,11,13,14].
However, species do not only interact through feeding interactions. Non-trophic interactions, such as mutualism and habitat modification, are pervasive in ecosystems and, through their impact on species abundance and the strength of individual trophic links, may transform the topology and dynamics of the overall network [15,16]. Despite urgent calls from recent studies to integrate non-trophic interactions [4,17–20], food webs are still typically studied without considering species interactions outside the trophic network, and quantification of the impacts of non-trophic effects on food web structure has thus far remained elusive. Therefore, even after 150 years, the question remains: are food webs mostly ‘self-shaped’ by trophic interactions alone or are they fundamentally contingent on non-trophic interactions?
Here, we empirically test the hypothesis that, in ecosystems dominated by organisms that strongly modify their abiotic environment (hereafter called ‘habitat modifiers’), overall food web complexity is enhanced by these modifications, beyond previously documented single-species facilitation effects. Habitat modifiers, also described as ‘ecosystem engineers’ or ‘foundation species’, are increasingly recognized as important drivers of ecosystem functions [16,21–28]. Although habitat modifiers are part of the food web like any other species (e.g. as prey or predator), they also have non-trophic effects on associated species by creating new habitat, altering resource availability and modifying physical environmental conditions. In theory, these non-trophic effects can be positive for some species (facilitation) [16], and negative for others, meaning the overall impact of non-trophic interactions on food web structure may be positive, negative or neutral [19]. Despite their ubiquity and pronounced, well-documented direct effects on specific species and individual trophic interactions, it remains unclear (i) how habitat modifiers affect the overall food web, (ii) how important non-trophic interactions by habitat modifiers are compared to their own trophic interactions, and (iii) how these non-trophic effects compare in importance to those species with the highest number of trophic links in the food web (hereafter called ‘most highly connected species’) [20].
To investigate whether key food web properties are indeed contingent on non-trophic facilitation by habitat modifiers as hypothesized, we carried out detailed field-based studies in two ecosystems: (i) temperate fringing salt marshes on the cobble beaches of New England (USA, North America) and (ii) tropical seagrass meadows on the intertidal flats of the Banc d'Arguin (Mauritania, Africa). Both salt marsh and seagrass ecosystems are essential components of coastal zones worldwide, serving as vital habitats for many species, functioning as carbon and nutrient sinks and playing an important role in coastal protection [29,30]. Using the natural dynamics and heterogeneity in each ecosystem, we defined three distinct stages of habitat modification. Within each stage, we intensively sampled all species across trophic levels and reconstructed the food web structure and non-trophic facilitation linkages using stable isotope analyses, mixing models and literature surveys. This allowed the separation of trophic and non-trophic effects of habitat modifiers on overall food web structure, and the comparison of these effects to those of the most highly connected species in the food web.
2. Material and methods
(a). Study sites
The intertidal zone of New England (North America) is typically composed of a top layer of unconsolidated cobbles (5- to 15-cm diameter) deposited by receding glaciers with a coarse, sandy sediment underneath (median grain size: 386 ± 5 µm (mean ± s.e.); electronic supplementary material, figure S1a). Heat is an important stressor causing mortality in summer as cobbles on these beaches can heat up to over 40°C [31]. Additionally, cobble movement during storms can crush any organism present [23]. Bare cobble habitat can become colonized by patches of cordgrass (Spartina alterniflora) that stabilize the cobbles between their shoot/root system and shade the substrate with their canopy in summer (electronic supplementary material, figure S1b) [23]. As these colonizing cordgrass patches mature and become more established over time, this habitat modification allows ribbed mussels (Geukensia demissa) to form dense aggregations (electronic supplementary material, figure S1c). These aggregations further modify conditions by providing hard and stable substrate and crevice space for attachment (e.g. algae, barnacles) and by cooling the surface through active evapotranspiration [23].
Whereas cordgrass and mussels occur on relatively narrow fringes (less than 25 m wide) of intertidal cobble beach and modify their habitat at scales of tens of centimetres, habitat modification by seagrasses and crabs occurs at scales of tens of metres within the much larger (more than 100 ha) intertidal flats of the Banc d'Arguin. Bare habitat is typified by coarse sandy substrate with many dead shells of the bivalve Senilia senilis (median grain size: 175 ± 9 µm; electronic supplementary material, figure S1d). This habitat can become colonized by patches of seagrass (Zostera noltii) that trap and accumulate fine suspended sediment from the water layer between their roots [32,33]. In the first few years (i.e. less than approx. 5 years old) seagrass habitat typically consists of a mosaic of seagrass patches alternating with bare sediment (electronic supplementary material, figure S1e). Due to sediment trapping, the sandy substrate within seagrass patches becomes covered by a approximately 5-cm (measured by a gauge rod) thick silt layer (approx. 71% less than 63 µm silt fraction, approximately 8% organic matter). As seagrass habitat ages, seagrass cover and the thickness of the silt layer gradually increase over time. In long-term established meadows (more than 40 years old; electronic supplementary material, figure S1f), seagrass cover increases to around 90% and the silt layer reaches a height of approximately 90 cm. This thick silt layer allows large numbers of swimming crabs (approx. 3300 ha−1), through their intensive burrowing activities, to create large permanently water-filled pools (size up to approx. 75 m2) in the silt layer that cover approximately 30% of these areas (see electronic supplementary material, text S1 and figure S2).
Occasionally, storms and/or ice scour (in New England) and excessive sediment accumulation after, for example, major dust storms (causing overexposure at low tide) followed by erosion (in Banc d'Arguin) reset cordgrass and seagrass habitats to bare cobbles and sand, respectively [32,34], yielding mosaics of different ecosystem development stages. The ‘natural experiments’ formed by the resulting habitat mosaics of different stages of ecosystem development provide an excellent opportunity to study how habitat modifiers affect food webs. Based on if and how long an area had been colonized by cordgrass or seagrass (see ‘Habitat selection’), we defined three distinct stages of habitat modification in both ecosystems: (i) bare areas not yet affected by habitat modifiers, (ii) colonizing (1- to 4-year-old) areas characterized by primary habitat modifiers (cordgrass/seagrass), but unaffected by secondary habitat modification, and (iii) established cordgrass (more than 10-years old) and seagrass (more than 40-years old) areas that were also affected by secondary habitat modifiers (mussels/crabs).
(b). Habitat selection
For the fringing marshes of Narragansett Bay, Rhode Island, New England (41°35′ N; 71°20′ W), we combined Google Earth images taken at low tide in 2002, 2004, 2006, 2007, 2008 and 2010 with ground truthing in 2013 to select four paired replicate sampling stations of three habitat types: (i) cobble habitat that had been bare from 2002 onward, (ii) 1- to 2-year-old colonizing habitat growing adjacently at the same tidal elevation, and (iii) neighbouring established cordgrass habitat that was established before 2002. Colonizing habitat sampling stations were selected onsite from expanding edges of established cordgrass patches. The age of these stations was estimated by measuring the distance to the outer edge and calculating the time of colonization by assuming a growth rate of 0.25–0.80 cm d−1 and a six-month growing season [35].
At Banc d'Arguin (19°53′ N; 16°18′ W), we used the normalized differences vegetation index calculated from Landsat 5 and 7 images (U.S. Geological Survey) taken at low tide in 1973, 1985, 1994, 1999–2003, 2007, 2009 and 2010 combined with ground truthing in 2011 to select four replicates for three types of habitat: (i) bare habitat that had been bare from at least 1973 onwards, (ii) 2- to 4-year-old colonizing seagrass habitat that had become vegetated after 2007, and (iii) at least 40-years old established seagrass meadows that were continuously vegetated since at least 1973. As habitat modification effects occur at much larger scales (see ‘Study sites’) in these seagrass meadows compared with the New England salt marshes, it was not possible to use a paired design here. Instead, to avoid spatial auto-correlation due to environmental gradients, all sampling stations were selected based on a random spatial distribution, with similar elevation, distance to the gully, maximum fetch length and Exposure Index—an integrative measure of wave exposure [36] (electronic supplementary material, figure S2 and table S1).
(c). Food web sampling
For each sampling station, we collected and identified all dominant resident species (representing more than 95% of the biomass in each trophic group and not migrating with the tides) and planktonic sources (see below and electronic supplementary material, table S2), measured nitrogen and carbon stable isotope values per species, and constructed the trophic interaction matrices based on literature, databases, abundance and isotope data, and mixing models.
(i). Fringing marshes, New England
At each sampling station (approx. 10 × 25 m), we randomly sampled six replicate plots using a 25 × 25-cm quadrat in which we determined the number, abundance and size of resident species. Next, we manually collected all epibenthic organisms by hand-picking within the quadrat and took a 5-cm2 5-cm deep sediment sample using a PVC corer for isotopic analysis of sediment particulate organic matter (sPOM). Finally, we sampled for endobenthic species to a depth of 20 cm using a 38-cm2 steel corer after which the samples were sieved over a 1-mm mesh. All fauna was identified to species level in the laboratory. Additionally, we collected benthic microbial mats by scraping from rocks at each station. Water column particulate organic matter (wPOM) samples were sampled into 5-l containers, filtered over a 200-µm zooplankton mesh and finally precipitated onto pre-combusted Whatman GF/F glass fibre filters. Zooplankton was concentrated using a zooplankton net, and subsequently filtered onto pre-combusted Whatman GF/F filters.
(ii). Seagrass meadows, Banc d'Arguin
To standardize sampling at the much larger intertidal mudflats, we established a 50-m diameter circle at each sampling station during low tide. Within this circle, we selected four replicate areas for sediment and (endo)benthos samples. Sediment samples were taken with a 5-cm deep, 12.5-cm2 PVC corer for isotopic analysis of sPOM. Benthos samples were taken with a 179-cm2 stainless steel corer to a depth of 20 cm, after which the samples were sieved over a 1-mm mesh. To determine crustacean densities, we took four 5-m-long hauls with a 40-cm-wide shrimp net at each station. At the established stations, the water column of four intertidal pools was separately sampled. Crustaceans were sampled by taking one haul with a shrimp net from the edge to the centre of a pool, while fish were sampled by pulling a beach seine net through each pool. All fauna was identified to species level in the laboratory. Additionally, benthic diatoms were scraped from the sediment surface at each station. After migration through an 80-µm mesh into combusted sand, they were collected in filtered seawater and precipitated onto pre-combusted Whatman GF/F filters. wPOM were sampled into 5-l containers, filtered over a 200-µm zooplankton mesh and precipitated onto pre-combusted Whatman GF/F filters. Zooplankton was concentrated using a zooplankton net (mesh size: 200 µm), and subsequently precipitated onto pre-combusted Whatman GF/F filters.
(d). Stable isotope measurements (δ13C and δ15N)
We took muscle tissue samples from fish and soft tissue from invertebrates wherever possible, but used the whole animal for smaller samples. All samples (including primary producers) were rinsed with demineralized water, oven-dried at 50°C for 48 h and ground. We took sub-samples for separate carbon and nitrogen analyses when samples contained inorganic calcified structures. Samples for carbon analysis were decalcified prior to analysis by addition of 3 M HCl. Stable isotope ratios were measured using an elemental analyser coupled to an IRMS (Thermo Scientific).
(e). Food web analyses
Based on abundance data, we excluded rare observations to include only ecologically relevant species (representing more than 95% of the biomass in each trophic group). Next, we used published literature, the WoRMS (World Register of Marine Species) database, FishBase and connected online databases, to determine all potential trophic relations for each species and constructed a theoretical, maximized dichotomous interaction matrix for each sampling station that included all potential trophic links. In other words, we first linked each species to all its potential resources. Next, we used size data (i.e. of some species we only found juveniles with a different diet and suite of consumers than adults), stable isotope biplots (δ15N versus δ13C) and Bayesian mixing models (R-package SIAR) to estimate the percentage contribution of each potential resource to a consumer's diet at each station [37,38]. Biplots and mixing models were constructed for each consumer at each sampling station using δ13C and δ15N stable isotope data with at least two replicate measurements per species. Based on these analyses, we constrained the theoretical, maximized matrices by removing trophic links where a resource contributed less than 5% to the diet of the consumer. As such, we only include regular, empirically important consumer–resource interactions, and omit incidental interactions. Finally, we used the constrained trophic interaction matrix to calculate six commonly used measures of food web structure. We use species richness (number of species or food web nodes; S) as an indicator of diversity, link density (number of links per species; L/S) and connectance (C; realized fraction of all possible links; L/S2) as metrics of topological complexity of the food web, and the percentages of top (species without consumers), basal (species without resources) and intermediate species as trophic distribution metrics [5,7,11]. Food web images presented in figures 1 and 2 were constructed using the software Network3D [39].
Figure 1.
Conceptual representation of performed removal procedures. (a,b) To test the hypothesis that observed differences in food web structure between habitats resulted from non-trophic facilitation by a habitat modifier, we first removed its non-trophic effect by removing species that depend obligatorily on its non-trophic facilitation (e.g. as attachment substrate). (a–c) Second, to test whether food webs differed due to the trophic effects of the habitat modifier, we deleted it and species exclusively feeding on it from the food web. (a–d) Third, we compared the trophic and non-trophic effects by habitat modifiers to those of the most highly connected species.
Figure 2.
Salt marsh and seagrass system food webs in the absence and presence of primary (cordgrass/seagrass) and secondary (mussels/crabs) habitat modifiers. Bare sites are typified by relatively simple food webs (a,d). Food webs have higher species richness and link density in colonizing habitats with primary habitat modifiers (b,e) and these effects are further enhanced by secondary habitat modifiers in established habitats (c,f). Node colour changes from red (basal species) to yellow with increasing trophic level.
To investigate whether observed differences in food web structure between bare, colonizing and established habitat indeed resulted from non-trophic facilitation as hypothesized, we first compared food web structure between these three habitats. Next, we examined the abiotic habitat requirements of each species in literature and databases (see deposited data). Based on those requirements, we constructed a second dichotomous interaction matrix for each sampling station that included obligatory non-trophic dependencies for each species. Next, we removed non-trophic links and, consequently, the species depending on these links. In other words, we removed those species from the trophic interaction matrix (both rows and columns) that are obligately dependent on non-trophic facilitation by a habitat modifier and compared habitats again using the resulting new food web matrices (figure 1a,b). Specifically, we first focused on secondary habitat modifiers (mussels/crabs) and removed species from the matrix that depend on attachment to mussels in the established salt marsh habitat and on the intertidal pools formed by crabs in the established seagrass meadows and recalculated all food web metrics. Second, we removed species from the original matrix that depend on primary habitat modifiers (cordgrass/seagrass) in both established and colonizing habitat. In colonizing habitat, these are the species that directly depend on shading or substrate stabilization by cordgrass or on aboveground structure or silt accumulation by seagrass. In established habitat, however, this procedure also resulted in the removal of species that indirectly depend on primary habitat modification through their direct dependence on secondary modification, because primary modification is a prequisite for secondary modification (e.g. pool formation by crabs is impossible without a silt layer accumulated by seagrass).
To test whether food web differences between habitat types could result from trophic rather than non-trophic effects by habitat-modifying species, we removed primary or secondary habitat modifiers themselves from the original matrix and, as a result, also species that fed exclusively on the removed habitat modifier (figure 1a,c). Similar to the previous procedure, we then compared the three habitats again using the resulting new matrices.
Finally, to test the importance of non-trophic facilitation by habitat modifiers for food web structure relative to species with a well-documented key trophic position, we compared the trophic role of the most highly connected species in the food web with the non-trophic effects of habitat modifiers [9,40]. To this end, we removed the species with the highest number of trophic links from the original interaction matrices in colonizing and established habitat, respectively, as well as species solely connected to this node (figure 1a,d). Next, we calculated the relative contribution of this species to each food web metric (i.e. 1—value calculated for modified matrix/value calculated for original matrix). We then did the same calculations for the non-trophic effects of habitat modifiers.
(f). Statistical analyses
To compare between habitats, we used one-way ANOVA for the seagrass meadows for all metrics. For the salt marshes, and to compare relative contributions of the most highly connected species with those of non-trophic effects of habitat modifiers to food web metrics, we applied generalized linear mixed models with a Gaussian distribution and sampling station as a random factor with Satterthwaite approximation of the degrees of freedom. All model residuals were checked for normality using Shapiro–Wilk tests (p = 0.05). We applied Tukey HSD post hoc tests to detect significant effects between bare, colonizing and established habitat.
3. Results
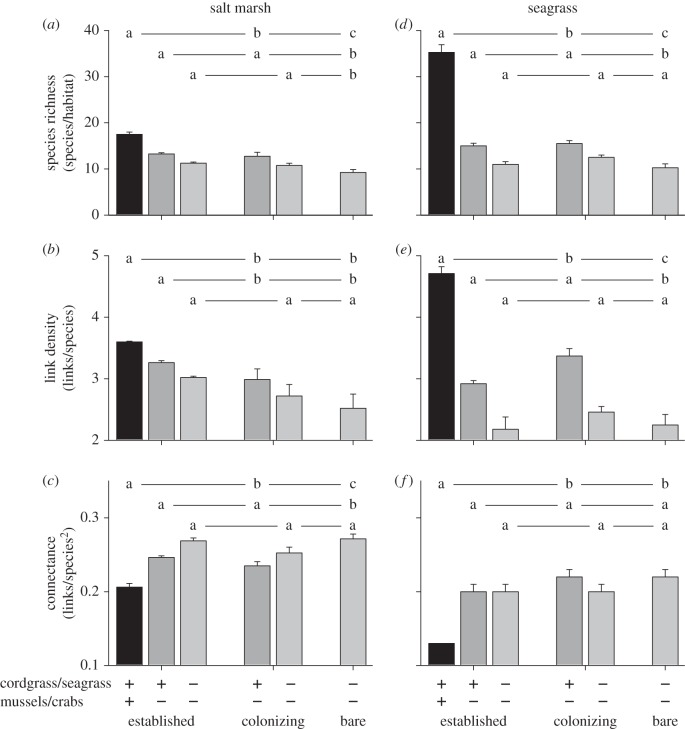
Despite obvious differences between the two ecosystems in terms of non-trophic habitat modification and trophic structure, the results showed that food web properties in the salt marsh and seagrass systems had pronounced and remarkably similar responses to the presence of habitat modifiers (figure 2). Both species richness and link density increased as cordgrass and seagrass beds matured (figure 3; electronic supplementary material, tables S3 and S4). Species richness and link density in the salt marsh increased 1.4 and 1.2 times from bare to colonizing cordgrass, respectively, and were another 1.4 and 1.2 times enhanced in established habitats. The seagrass system demonstrated similar and even stronger trends with both species richness and link density increasing by around 1.5 times from bare to colonizing and another 2.5 (species richness) and 1.5 (link density) times from colonizing to established habitats. Connectance followed an opposite trend, decreasing by 0.8 (salt marsh) to 0.6 (seagrass) times from bare to established, probably because the number of links needed to hold the network together relative to all possible links strongly increases as networks become very small. Trophic distribution metrics were not significantly affected despite the increase in species richness, indicating that habitat modifiers affected species similarly across multiple trophic levels.
Figure 3.
Species richness (a,d), link density (b,e) and connectance (c,f) as conditional on primary and secondary habitat modifiers. For each habitat, the most left bar indicates the natural situation, and subsequent bars depict the outcomes of the removal of species dependent on habitat modification (i.e. removal of non-trophic effects). Letters indicate post hoc grouping; error bars represent s.e.m. Species richness and link density are significantly higher in modified habitat, whereas connectance is generally lower. Removal of species dependent on habitat modification increased similarity to unmodified, bare habitat.
The removal procedure of the non-trophic effects of habitat modifiers revealed that 11% of all species depended on primary habitat modification in both systems, and that another 24% and 64% depended on the combined effects of primary and secondary habitat modification in the salt marsh and seagrass system, respectively. When non-trophic effects of secondary habitat modifier (mussels/crabs) were excluded, three food web metrics (species richness, link density and connectance) in established habitat became statistically indistinguishable from colonizing habitat, but continued to deviate from bare habitat (figure 3; electronic supplementary material tables S3 and S4). Moreover, removal of the non-trophic effects of primary habitat modifiers (cordgrass/seagrass) caused these food web metrics in both established and colonizing habitats to converge towards the simplified characteristics of bare, unmodified habitat in both ecosystems, although species richness remained somewhat enhanced in the salt marsh. In contrast, removal of the trophic effect of primary or secondary habitat modifiers from the food web caused relatively minor changes in diversity and complexity metrics, demonstrating that the trophic role of habitat modifiers could not explain the observed changes in food web structure from bare to established habitat (electronic supplementary material, tables S3 and S4). Trophic distribution metrics (percentage of top, intermediate and basal species) did not change consistently in response to the removal of non-trophic interactions or trophic interactions from the network.
The comparison of the relative contribution to food web metrics by non-trophic habitat modification versus the most highly connected species demonstrated that habitat modification was much more important for diversity and typically of more, or similar, importance for food web complexity. In both the salt marsh and seagrass system, species richness was more sensitive to the removal of habitat modifiers than to the removal of the most highly connected species (figure 4; electronic supplementary material, table S5). Link density and connectance in established salt marsh and seagrass habitat were also more affected by primary habitat modification than by the most highly connected species, although effects on link density in the smaller salt marsh food were similar. Because species that are facilitated by habitat modifiers exhibit relatively low levels of trophic connectance compared with the most highly connected species, habitat modifiers and the most highly connected species had opposite effects on connectance in established habitats. Whereas non-trophic interactions by habitat modifiers reduced connectance, this metric was enhanced by the most highly connected species. In colonizing habitat, link density and connectance were more affected by the most highly connected species, whereas in the seagrass system, effects on connectance were similar and link density was more affected by habitat modification. Finally, the effects on trophic distribution metrics varied depending on both the ecosystem (salt marsh or seagrass) and habitat type (established or colonizing). In general, however, we found these metrics to be more affected by habitat modifiers in the seagrass system, whereas the importance of habitat modifiers and the most highly connected species were similar in the salt marsh (electronic supplementary material, table S5).
Figure 4.
The relative effect of secondary and primary habitat modifiers, and the most highly connected species on species richness (a,d), link density (b,e), and conductance (c,f). Relative effects are calculated as (1 − value calculated for each modified food web matrix) / (value calculated for the original food web matrix). Letters indicate statistical grouping; error bars represent s.e.m. The analyses show that habitat modification was more important for diversity and of more or similar importance for both link density and conductance measures of complexity.
4. Discussion
Overall, our findings demonstrate that habitat modification strongly changes food web structure—not only by facilitating species and thus enhancing diversity, but also by increasing the number of trophic interactions that species have with other species in the food web. We found these non-trophic, indirect effects of habitat modifiers to be much more pronounced than their trophic roles. Furthermore, depending on the metric and habitat type, we found habitat modification of more or similar importance compared to the trophic role of the most highly connected species in the food web. This is important because the most highly connected node has been repeatedly documented to have a key structuring role in food webs as well in networks in general [9,40], thus emphasizing the importance of habitat modification for food web structure in our study systems. Finally, although trophic distribution metrics were affected by habitat modifiers, we did not identify consistent shifts in the percentage of top, intermediate and basal species as we removed the non-trophic effects of habitat modifiers. This implies that their modification of environmental conditions alters food web structure across multiple trophic levels, for example by affecting the outcome of trophic and competitive interactions between species.
Studies from a wide range of ecosystems have shown that amelioration of physical stress by habitat-modifying organisms can profoundly impact the associated community by facilitating other species [16], and recent work demonstrated that this effect may not only be local [41]. On cobble beaches, for instance, many studies (including this one) have revealed non-trophic facilitation of the local community [23,42] and that at a scale of metres to tens of metres, cordgrass patches function as wave breaks to facilitate wave-sensitive forb species [43]. Similarly, intertidal mussel beds in the Wadden Sea were found to facilitate the community at a scale exceeding 100 m by baffling waves and changing sediment characteristics [24,44]. Although we focused on habitat modifier-effects within a single habitat type, the contrasting spatial scales at which non-trophic facilitation impacted food webs in salt marshes (less than 1 m) and seagrass meadows (more than 10 m), respectively, suggest that habitat-modifying species can affect not only a few species, but also whole food web dynamics beyond the habitat in which they grow. Furthermore, the investigated habitat types reflect distinct stages of ecosystem succession and changes in food webs structure can largely be explained by habitat modification (figure 3). This illustrates that non-trophic interactions can play a key role in moderating community assembly as well as in maintaining complex food webs. In addition to habitat provision and alleviation of physical stress, habitat modification may also indirectly stimulate the development of complex food webs by increasing productivity and mediating how energy and nutrients move through ecosystems. Habitat modification thus affects important factors that control the stability of assembling food webs during succession [12,45].
Until now, food web structure and stability have been typically analysed as a function of the properties of the trophic network itself, like the number of species and links, connectance or the strength of loops [7–12]. Our findings indicate that, in ecosystems dominated by habitat-modifying organisms, those properties themselves can be highly contingent on interactions outside the trophic network, yet intrinsic to the species participating in it. Most likely, this is not only the case in ecosystems where these organisms drive succession as investigated here, but also in systems where habitat modifiers create alternative stable states by facilitating their own growth or survival [46]. We therefore suggest that integrating non-trophic interactions in food web analyses is important in increasing their explanatory and predictive capacity, and consider our approach as a vital first step in that direction. We propose that future work should focus on the development of quantitative hybrid networks of multiple interaction types, firmly grounded in empirical data. One potential problem in this regard is the large diversity of non-trophic interactions observed in real ecosystems. A first conceptual approach to overcome this issue may be to construct dynamic models, in which trophic interactions are modelled as energy flows or consumer–resource interactions, and non-trophic interactions are integrated using functional classes defined by how trophic interactions are modified [4,19].
Increased predictive capacity is urgently needed by society because ecosystems dominated by habitat modifiers like seagrass meadows, salt marshes, coral reefs, peatlands and rainforests are now degrading at alarming rates worldwide due to anthropogenic disturbances with relatively low success of restoration efforts [29,30,47]. Our findings suggest that the development of the typically complex species interaction networks of these systems can take decades due to long-term cumulative effects of multiple habitat modifiers. This implies that ecosystem conservation and restoration efforts should not focus on trophic interactions alone (e.g. only on recovery of top predators), but like future approaches to network analyses, should consider various interaction types and potential synergistic or antagonistic effects between them.
Supplementary Material
Acknowledgements
We thank J. Eygensteyn and K. Donkers for technical assistance; and A. J. P. Smolders, S. Kosten and L. P. M. Lamers for their comments on the manuscript.
Data accessibility
Data are available from the Dryad Digital Repository: http://doi.org/10.5061/dryad.22p7r.
Authors' contributions
E.v.d.Z., T.v.d.H., C.A., H.O., A.H.A. and B.R.S. generated hypotheses and designed field sampling. E.v.d.Z., T.v.d.H., C.A., L.L.G., K.J.v.d.R., J.A.v.G., M.J.A.C. and M.v.d.G. collected field data. E.v.d.Z., T.v.d.H. and C.A. conducted analyses and wrote the first draft of the manuscript. All authors contributed substantially to revisions.
Competing interests
The authors declare no competing interests.
Funding
E.v.d.Z. and H.O. were financially supported by the ‘Waddenfonds’ programme; C.A. by NSF GRFP DGE-0802270 and University of Florida Graduate Alumni Fellowship awards; B.R.S. by NSF Career award 1056980; M.v.d.G. and T.P. by the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO)–WOTRO Integrated Programme grant W.01.65.221.00 awarded to T.P.; J.v.G. by NWO–VIDI grant 864.09.002; T.v.d.H. by NWO-VENI grant 863.12.003.
References
- 1.Darwin C. 1859. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle of life. London, UK: John Murray. [PMC free article] [PubMed] [Google Scholar]
- 2.Dunne JA, Williams RJ, Martinez ND. 2002. Food-web structure and network theory: the role of connectance and size. Proc. Natl Acad. Sci. USA 99, 12 917–12 922. ( 10.1073/pnas.192407699) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allesina S, Alonso D, Pascual M. 2008. A general model for food web structure. Science 320, 658–661. ( 10.1126/science.1156269) [DOI] [PubMed] [Google Scholar]
- 4.Kéfi S, et al. 2012. More than a meal … integrating non-feeding interactions into food webs. Ecol. Lett. 15, 291–300. ( 10.1111/j.1461-0248.2011.01732.x) [DOI] [PubMed] [Google Scholar]
- 5.May RM. 1972. Will a large complex system be stable? Nature 238, 413–414. ( 10.1038/238413a0) [DOI] [PubMed] [Google Scholar]
- 6.Pimm SL, Lawton JH, Cohen JE. 1991. Food web patterns and their consequences. Nature 350, 669–674. ( 10.1038/350669a0) [DOI] [Google Scholar]
- 7.Williams RJ, Martinez ND. 2000. Simple rules yield complex food webs. Nature 404, 180–183. ( 10.1038/35004572) [DOI] [PubMed] [Google Scholar]
- 8.Neutel AM, Heesterbeek JAP, de Ruiter PC. 2002. Stability in real food webs: weak links in long loops. Science 296, 1120–1123. ( 10.1126/science.1068326) [DOI] [PubMed] [Google Scholar]
- 9.Dunne JA, Williams RJ, Martinez ND. 2002. Network structure and biodiversity loss in food webs: robustness increases with connectance. Ecol. Lett. 5, 558–567. ( 10.1046/j.1461-0248.2002.00354.x) [DOI] [Google Scholar]
- 10.Bascompte J, Melian CJ, Sala E. 2005. Interaction strength combinations and the overfishing of a marine food web. Proc. Natl Acad. Sci. USA 102, 5443–5447. ( 10.1073/pnas.0501562102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gross T, Rudolf L, Levin SA, Dieckmann U. 2009. Generalized models reveal stabilizing factors in food webs. Science 325, 747–750. ( 10.1126/science.1173536) [DOI] [PubMed] [Google Scholar]
- 12.Neutel A-M, Heesterbeek JAP, van de Koppel J, Hoenderboom G, Vos A, Kaldeway C, Berendse F, de Ruiter PC. 2007. Reconciling complexity with stability in naturally assembling food webs. Nature 449, 599–602. ( 10.1038/nature06154) [DOI] [PubMed] [Google Scholar]
- 13.Stouffer DB, Bascompte J. 2011. Compartmentalization increases food-web persistence. Proc. Natl Acad. Sci. USA 108, 3648–3652. ( 10.1073/pnas.1014353108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunne JA, et al. 2013. Parasites affect food web structure primarily through increased diversity and complexity. PLoS Biol. 11, e1001579 ( 10.1371/journal.pbio.1001579) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stachowicz JJ. 2001. Mutualism, facilitation, and the structure of ecological communities. Bioscience 51, 235–246. ( 10.1641/0006-3568(2001)051%5B0235:MFATSO%5D2.0.CO;2) [DOI] [Google Scholar]
- 16.Bruno JF, Stachowicz JJ, Bertness MD. 2003. Inclusion of facilitation into ecological theory. Trends Ecol. Evol. 18, 119–125. ( 10.1016/s0169-5347(02)00045-9) [DOI] [Google Scholar]
- 17.Olff H, Alonso D, Berg MP, Eriksson BK, Loreau M, Piersma T, Rooney N. 2009. Parallel ecological networks in ecosystems. Phil. Trans. R. Soc. B 364, 1755–1779. ( 10.1098/rstb.2008.0222) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baiser B, Whitaker N, Ellison AM. 2013. Modeling foundation species in food webs. Ecosphere 4, 146. ( 10.1890/ES13-00265.1) [DOI] [Google Scholar]
- 19.Sanders D, Jones CG, Thébault E, Bouma T, van der Heide T, van Belzen J, Barot S. 2014. Integrating ecosystem engineering and food webs. Oikos 123, 513–524. ( 10.1111/j.1600-0706.2013.01011.x) [DOI] [Google Scholar]
- 20.Kéfi S, Berlow EL, Wieters EA, Joppa LN, Wood SA, Brose U, Navarrete SA. 2015. Network structure beyond food webs: mapping non-trophic and trophic interactions on Chilean rocky shores. Ecology 96, 291–303. ( 10.1890/13-1424.1) [DOI] [PubMed] [Google Scholar]
- 21.Dayton P. 1972. Toward an understanding of community resilience and the potential effects of enrichments to the benthos at McMurdo Sound, Antarctica. In Proceedings of the colloquium on conservation problems in Antarctica (ed. Parker BC.), pp. 81–95. Lawrence, KS: Allen Press. [Google Scholar]
- 22.Jones CG, Lawton JH, Shachak M. 1994. Organisms as ecosystem engineers. Oikos 69, 373–386. ( 10.2307/3545850) [DOI] [Google Scholar]
- 23.Altieri AH, Silliman BR, Bertness MD. 2007. Hierarchical organization via a facilitation cascade in intertidal cordgrass bed communities. Am. Nat. 169, 195–206. ( 10.1086/510603) [DOI] [PubMed] [Google Scholar]
- 24.Van der Zee EM, van der Heide T, Donadi S, Eklof JS, Eriksson BK, Olff H, van der Veer HW, Piersma T. 2012. Spatially extended habitat modification by intertidal reef-building bivalves has implications for consumer–resource interactions. Ecosystems 15, 664–673. ( 10.1007/s10021-012-9538-y) [DOI] [Google Scholar]
- 25.Van der Heide T, Eklof JS, van Nes EH, van der Zee EM, Donadi S, Weerman EJ, Olff H, Eriksson BK. 2012. Ecosystem engineering by seagrasses interacts with grazing to shape an intertidal landscape. PLoS ONE 7, e42060 ( 10.1371/journal.pone.0042060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Angelini C, Silliman BR. 2013. Secondary foundation species as drivers of trophic and functional diversity: evidence from a tree–epiphyte system. Ecology 95, 185–196. ( 10.1890/13-0496.1) [DOI] [PubMed] [Google Scholar]
- 27.Van der Heide T, Tielens E, van der Zee EM, Weerman EJ, Holthuijsen S, Eriksson BK, Piersma T, van de Koppel J, Olff H. 2014. Predation and habitat modification synergistically interact to control bivalve recruitment on intertidal mudflats. Biol. Conserv. 172, 163–169. ( 10.1016/j.biocon.2014.02.036) [DOI] [Google Scholar]
- 28.Van der Zee EM, Tielens E, Holthuijsen S, Donadi S, Eriksson BK, van der Veer HW, Piersma T, Olff H, van der Heide T. 2015. Habitat modification drives benthic trophic diversity in an intertidal soft-bottom ecosystem. J. Exp. Mar. Biol. Ecol. 465, 41–48. ( 10.1016/j.jembe.2015.01.001) [DOI] [Google Scholar]
- 29.Waycott M, et al. 2009. Accelerating loss of seagrasses across the globe threatens coastal ecosystems. Proc. Natl Acad. Sci. USA 106, 12 377–12 381. ( 10.1073/pnas.0905620106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gedan KB, Silliman BR, Bertness MD. 2009. Centuries of human-driven change in salt marsh ecosystems. Annu. Rev. Mar. Sci. 1, 117–141. ( 10.1146/annurev.marine.010908.163930) [DOI] [PubMed] [Google Scholar]
- 31.Bertness MD. 1989. Intraspecific competition and faciliation in a northern acron barnacle population. Ecology 70, 257–268. ( 10.2307/1938431) [DOI] [Google Scholar]
- 32.Folmer EO, van der Geest M, Jansen E, Olff H, Anderson TM, Piersma T, van Gils JA. 2012. Seagrass-sediment feedback: an exploration using a non-recursive structural equation model. Ecosystems 15, 1380–1393. ( 10.1007/s10021-012-9591-6) [DOI] [Google Scholar]
- 33.Van der Heide T, et al. 2012. A three-stage symbiosis forms the foundation of seagrass ecosystems. Science 336, 1432–1434. ( 10.1126/science.1219973) [DOI] [PubMed] [Google Scholar]
- 34.Ewanchuk PJ, Bertness MD. 2003. Recovery of a northern New England salt marsh plant community from winter icing. Oecologia 136, 616–626. ( 10.1007/s00442-003-1303-7) [DOI] [PubMed] [Google Scholar]
- 35.Newton C, Thornber C. 2013. Ecological impacts of macroalgal blooms on salt marsh communities. Estuaries Coasts 36, 365–376. ( 10.1007/s12237-012-9565-0) [DOI] [Google Scholar]
- 36.Howes D, Morris M, Zacharia M. 1999. British Columbia estuary mapping system. Victoria, BC, Canada: Coastal Task Force Resource Inventory Committee Secretariat. [Google Scholar]
- 37.Parnell AC, Inger R, Bearhop S, Jackson AL. 2010. Source partitioning using stable isotopes: coping with too much variation. PLoS ONE 5, e9672 ( 10.1371/journal.pone.0009672) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parnell A, Jackson A. 2011. SIAR: stable isotope analysis in R. R package v. 4.1. 3. See https://cran.r-project.org/web/packages/siar/index.html. [Google Scholar]
- 39.Williams RJ. 2010. Network3D [Software]. Cambridge, UK: Microsoft Research. [Google Scholar]
- 40.Jordan F. 2009. Keystone species and food webs. Phil. Trans. R. Soc. B 364, 1733–1741. ( 10.1098/rstb.2008.0335) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van de Koppel J, van der Heide T, Altieri AH, Eriksson BK, Bouma TJ, Olff H, Silliman BR. 2015. Long-distance interactions regulate the structure and resilience of coastal ecosystems. Annu. Rev. Mar. Sci. 7, 139–158. ( 10.1146/annurev-marine-010814-015805) [DOI] [PubMed] [Google Scholar]
- 42.Angelini C, van der Heide T, Griffin JN, Morton JP, Derksen-Hooijberg M, Lamers LPM, Smolders AJP, Silliman BR. 2015. Foundation species’ overlap enhances biodiversity and multifunctionality from the patch to landscape scale in southeastern United States salt marshes. Proc. R. Soc. B 282, 20150421. ( 10.1098/rspb.2015.0421) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van de Koppel J, Altieri AH, Silliman BR, Bruno JF, Bertness MD. 2006. Scale-dependent interactions and community structure on cobble beaches. Ecol. Lett. 9, 45–50. ( 10.1111/j.1461-0248.2005.00843.x) [DOI] [PubMed] [Google Scholar]
- 44.Donadi S, van der Heide T, van der Zee EM, Eklof JS, van de Koppel J, Weerman EJ, Piersma T, Olff H, Eriksson BK. 2013. Cross-habitat interactions among bivalve species control community structure on intertidal flats. Ecology 94, 489–498. ( 10.1890/12-0048.1) [DOI] [PubMed] [Google Scholar]
- 45.Moore JC, Deruiter PC, Hunt HW. 1993. Influence of productivity on the stability of real and model ecosystems. Science 261, 906–908. ( 10.1126/science.261.5123.906) [DOI] [PubMed] [Google Scholar]
- 46.Scheffer M, Carpenter S, Foley JA, Folke C, Walker B. 2001. Catastrophic shifts in ecosystems. Nature 413, 591–596. ( 10.1038/35098000) [DOI] [PubMed] [Google Scholar]
- 47.Rands MRW, et al. 2010. Biodiversity conservation: challenges beyond 2010. Science 329, 1298–1303. ( 10.1126/science.1189138) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available from the Dryad Digital Repository: http://doi.org/10.5061/dryad.22p7r.