Abstract
Species distribution models (SDMs) assume species exist in isolation and do not influence one another's distributions, thus potentially limiting their ability to predict biodiversity patterns. Community-level models (CLMs) capitalize on species co-occurrences to fit shared environmental responses of species and communities, and therefore may result in more robust and transferable models. Here, we conduct a controlled comparison of five paired SDMs and CLMs across changing climates, using palaeoclimatic simulations and fossil-pollen records of eastern North America for the past 21 000 years. Both SDMs and CLMs performed poorly when projected to time periods that are temporally distant and climatically dissimilar from those in which they were fit; however, CLMs generally outperformed SDMs in these instances, especially when models were fit with sparse calibration datasets. Additionally, CLMs did not over-fit training data, unlike SDMs. The expected emergence of novel climates presents a major forecasting challenge for all models, but CLMs may better rise to this challenge by borrowing information from co-occurring taxa.
Keywords: ecological niche modelling, co-occurrence, no-analogue, pollen, Quaternary, palaeoecology
1. Introduction
Species distribution models (SDMs) remain among the most widely used methods for forecasting regional- to global-scale changes in species distributions, species assemblages and patterns of biodiversity in response to climate change [1]. When modelling entities or attributes above the species level using SDMs, a ‘predict first, assemble later’ approach [2] is typically employed, wherein individual models are fitted and projected for each species, the mapped predictions of which are then aggregated or ‘stacked’ to infer potential changes in community-level patterns such as species richness (e.g. [3]). An alternative, but relatively rarely used, approach involves combining data from multiple species to simultaneously analyse and map patterns of biodiversity at the community level. This ‘assemble and predict together’ strategy [2] underlies community-level models (CLMs) that fit a species co-occurrence matrix to environmental variables to predict both community structure and the distributions of individual species [2]. CLMs implicitly capture any process driving co-occurrence patterns, including shared climatic requirements of species, responses to unmeasured environmental variables and possibly biotic interactions [4]. SDMs implicitly also capture these processes (by setting the boundaries of a species' realized niche), but do so in a manner that is isolated from the broader community context in which species occur (a Gleasonian approach).
By simultaneously modelling all observed species within a region of interest and incorporating information on co-occurrence, CLMs may have the potential to predict species distributions and changes in community composition better than SDMs [2], especially for large climatic shifts and novel climate regimes where individual taxon–climate relationships may break down [5], but broader biodiversity–climate relationships may remain comparatively stable [6]. The potentially greater transferability of CLMs to novel climates may be beneficial given the predicted emergence of no-analogue climates in the near future [7,8]. However, these ideas remain untested, and the relative ability of SDMs and CLMs to simulate the past emergence of no-analogue communities is unknown. Ferrier & Guisan [2] suggested that CLMs may effectively balance the assumptions underlying predictions of community types (that species move together as fixed community types in response to climate change—a Clementsian approach) versus the assumptions underlying predictions of individual species distributions (that the environmental variables shaping distributions may differ markedly between species) by modelling multiple species in relation to a shared set of environmental variables and by implicitly accounting for other drivers of co-occurrence [9]. The palaeo-record offers evidence that neither of these assumptions is fully valid [10,11]. CLMs may implicitly account for any factors that drive patterns of co-occurrence (including biotic interactions) and, therefore, could be advantageous given the well-documented role of biotic interactions in mediating responses of species and communities to climate change [12] and associated reshuffling of communities during current and past environmental changes [13,14]. Another major advantage of CLMs is an ability to model very large numbers of species, including rare (or poorly sampled) species [15,16]. By detecting shared environmental responses across species, CLMs can ‘borrow strength’ from more common (or better sampled) species and potentially produce better predictions than possible from SDMs [2,17–19]. Thus, for predicting species distributions and assemblages, CLMs allow fuller consideration of species that might otherwise be excluded from SDM-based analyses owing to an insufficiency of observations.
The extent to which the potential benefits of CLMs translate into improved predictions remains unknown. Most CLMs are multiresponse (i.e. multispecies) extensions of commonly used SDMs (e.g. constrained quadratic ordination (CQO) is a multiresponse version of generalized linear models (GLM)), while other CLMs have no direct SDM counterpart (e.g. generalized dissimilarity modelling). Previous studies that compared the performance of SDMs and CLMs when predicting species distributions, community composition and/or species richness have had mixed results, with some studies finding that CLMs tend to outperform SDMs [20–22], and others finding the opposite [4,15,23,24]. Only one of these analyses directly compared CLM and SDM predictions against observed changes in species assemblages [15], and this analysis only compared one CLM with one SDM. The analysis presented here is the first to compare model predictions with observed species assemblages using head-to-head evaluations of multiple SDM algorithms and their direct CLM counterparts. In addition, previous analyses have not tested the transferability of CLMs to predict species distributions and community composition across longer time periods (more than 25 years), across periods of climate change similar in magnitude to that expected for this century, and in no-analogue climates.
Here, we used fossil pollen from sediment cores in eastern North America spanning the past 21 000 years (figure 1) to perform the first comprehensive SDM and CLM model comparison across the large and rapid climate changes of the Late Quaternary in order to tease apart the differences in SDMs and CLMs and to evaluate how these models may perform in predicting species distributions and assemblages under climate change. The palaeontological record, in general, and the fossil-pollen record of the Late Quaternary, in particular, provides a unique opportunity to test and compare ecological forecasting models across periods of climatic and biotic change [8,27]. We tested the ability of five SDM algorithms and their direct CLM counterparts to predict observed changes in fossil-pollen distributions and assemblages through time and test whether simultaneously modelling co-occurring taxa in a region increased model performance compared with SDMs. By comparing SDMs with their direct CLM counterparts, we controlled for differences in predictive performance among different classes of algorithms. We assessed how model performance changes as a function of the climatic and compositional novelty between time periods, in order to estimate the limits of model predictability and thereby provide guidelines for future projections. Lastly, we tested the hypothesis that CLMs have a higher predictive skill for rare taxa.
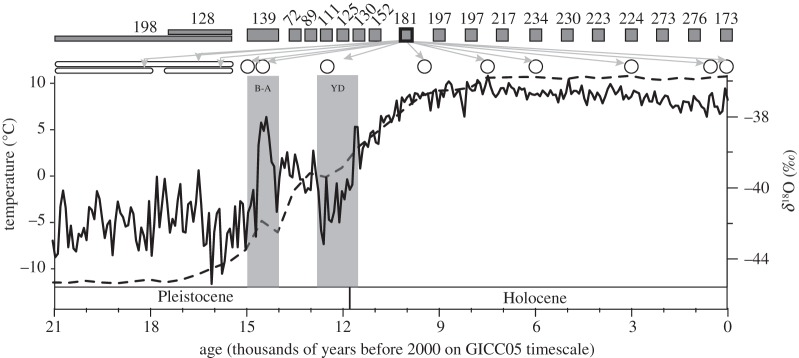
Figure 1.
Temperature change and model comparison scheme. Solid line shows oxygen isotope data from 21 000 years ago (kyr BP) to the present from the North Greenland Ice Core Project (GICC05) [25] with the 2005 Greenland Ice Core Chronology age model [26]. Dashed line is mean maximum temperature from climate simulations used in this analysis. The Bølling–Allerød (B-A) and Younger Dryas (YD) Chronozones are highlighted in grey. The time periods used to fit SDM and CLM models are indicated with shaded boxes/rectangles, while projected time periods are indicated with open ellipses. This is illustrated for the 10 kyr BP time period (shaded box with thickened outline and grey arrows indicating projected time periods). Data were pooled for 15 to 14 kyr BP, 17.5 to 15.5 kyr BP and 21 to 15.5 kyr BP when building models (shaded rectangles) and for 17.5 to 15.5 kyr BP, 21 to 15.5 and 21 to 18 kyr BP when projecting models (open rounded rectangles). The number of localities used to fit models is indicated above the grey box representing the model fitting time period.
2. Material and methods
(a). Setting
Since the Last Glacial Maximum (LGM—21 kyr BP (thousands of years before present)), global mean temperature increased from 3°C to 5°C, with periods of rapid (decadal- to centennial-scale) warming and cooling (e.g. the Bølling–Allerød and Younger Dryas onset and terminations; figure 1). Plant distributions [13] and realized niches [11,28] shifted in response to these climate changes, resulting in a reshuffling of taxon associations [13–14,29,30]. These climatic and ecological changes in the past were characterized by periods of climatic novelty and no-analogue communities as compared to contemporary climates and communities. The last deglaciation (21 kyr BP to present) is not an exact analogue for contemporary climate change given lower CO2 concentrations, different trends in insolation and temperature seasonality, and a smaller human footprint; however, the two time periods share similarities such as high rates of climate change, regions and times with highly novel climates, species extinctions and increased human pressure.
(b). Taxonomic occurrences
Taxon occurrence data were from the eastern North America fossil-pollen relative abundance dataset from Blois et al. [30], with the following modifications: (i) we removed sites that had marine/estuarine or unknown depositional environments, (ii) pollen abundances were expressed as the pollen sum for a particular taxon divided by the total sum for all genus-level taxa, rather than divided by the total upland sum for the site (which includes both genera as well as families and other higher level taxa), and (iii) pollen relative abundances were interpolated to 500-year time slices from 21 000 to present, rather than 1000-year time slices. Pollen data quality was calculated as in Blois et al. [30], and for each time period, only sites with a weighted quality value above 0.75 were included. If multiple sites fell within the same grid cell, their pollen abundances were averaged. Taxa co-occurrences were within a grid cell, with the majority of grid cells containing co-occurrences from within a single lake sediment core. Following Nieto-Lugilde et al. [31], the relative abundance pollen matrix was converted to a presence/absence matrix after applying a threshold scaled to 5% of the maximum abundance (see electronic supplementary material, table S1 and figure S1). Because the majority of fossil-pollen types considered here can be consistently identified, absences are considered true absences. We chose the 19 most abundant-through-time taxa at the generic level (see electronic supplementary material), the lowest taxonomic level in which most fossil pollen can be identified—except for Ostrya and Carpinus (Ostrya/Carpinus), which are palynologically indistinguishable, and Ambrosia-type, which can include Iva. Even though interspecific differences in climatic and ecological tolerances may differ from genus-level tolerances, the same problem exists, although to a lesser extent, for populations within species (e.g. [32]). In addition, pollen records best match forest inventory data at the genus level [31]; thus, analyses at the genus level are as appropriate as analyses at the species level, especially for pollen data, but may introduce another source of uncertainty. See Blois et al. [30] for a complete list of pollen types assigned to each genus.
(c). Environmental variables
We used palaeoclimate simulations from the Community Climate System Model Version 3 (CCSM3) SynTrace transient simulation with seasonally averaged model outputs saved at a decadal time step from 21 kyr BP to the present [33]. Climate variables were de-biased and downscaled to a 0.5° × 0.5° grid (approx. 50 × 50 km) and processed to create yearly, quarterly and monthly variables for every 500 years since 21 kyr BP. All climate variables are means across 200 years, centred on the 500 year time slices (e.g. the 1.5 kyr BP climate variables are averaged across the years 1.6–1.4 kyr BP). From the original 27 variables (nine yearly, quarterly and monthly), we chose six uncorrelated variables (maximum temperature of the warmest quarter, minimum precipitation of the driest quarter, maximum precipitation of the wettest quarter, mean yearly potential evapotranspiration, mean yearly actual evapotranspiration and mean yearly water deficit index) using a Pearson correlation coefficient cut-off of 0.75 (see electronic supplementary material, figure S2).
(d). Model implementation
To enable direct comparison between the SDM and CLM versions of each model-class, we compared five SDMs and their direct CLM counterparts, covering a wide range of model-class types: (i) GLM [34] and CQO [35]; (ii) generalized additive models (GAM [34]) and constrained additive ordination (CAO [36]); (iii) single and multiresponse multivariate adaptive regression splines (MARS and MMARS, respectively, [37,38]); (iv) single classification and regression trees and multivariate regression trees (CARTs and MRTs, respectively, [2,19,39–41]) and (v) single and multiresponse artificial neural networks (ANN and MANN, respectively [42]). A description of each model-class type is provided in the electronic supplementary material, table S2. Models were fitted for 20 selected time periods and then projected to 12 time periods representing distinct climates through the Late Quaternary (figure 1). For fitting models, we pooled taxon occurrence and climate data for 15–14, 17.5–15.5 and 20–15.5 kyr BP to overcome low sample sizes (number of localities) while covering as many time periods as possible during the end of the last glacial period. Similarly, for model projections, we pooled 17.5–15.5, 21–18 and 21–15.5 kyr BP. Data were split into 10 random partitions of training (70%) and testing (30%) sets and models were fit and evaluated using each random set. All models were fitted with the same six climatic variables (i.e. we did not perform variable selection) with the exception of MARS/MMARS and CARTs/MRTs because these algorithms build variable selection into their model fitting. SDM and CLM pairs were tuned so that parameter complexity was equal between the SDM and its CLM counterpart (see electronic supplementary material, table S3). For example, with artificial neural networks, the number of nodes in the hidden layer was set to 20 for each taxon in the SDM (ANN) as well as for the CLM (MANN). To tune parameter values, models were fit with the 0 kyr BP occurrence and climate data and then the chosen parameters of the best model were propagated to the other time periods, thereby controlling model complexity across time. We also created step functions for variable selection and training functions to tune models for the 0 kyr BP time period and then projected these models to the selected 12 time periods, in order to examine if variable selection and parameter tuning influenced SDM and CLM performance (see the electronic supplementary material). All models and analyses were run in R v. 3.1.1 [43]. All R code, including the full list of R packages used and citations, are available on GitHub (https://github.com/fitzLab-AL/SDM-CLMcomp).
(e). Model evaluation
We evaluated the ability of models to predict taxa distributions in terms of both discrimination and reliability (also known as ‘calibration’) [44], which are useful metrics when comparing continuous model outputs to binary observations of the presences/absences. Reliability assesses the overall agreement between observations and predictions, and in particular, whether the model correctly predicts the probability that sites will be occupied or unoccupied. Reliability was measured using the Brier score [45]. The Brier score is equivalent to mean squared error but is applicable when comparing continuous probabilities to mutually exclusive discrete outcomes, in this case the presence or absence. The Brier score ranges from 0 to 1, in which 0 indicates complete agreement between observed and predicted and 1 indicates complete disagreement. Discrimination quantifies the ability of the model to correctly distinguish between occupied and unoccupied sites (i.e. the extent to which predicted probabilities for occupied sites are higher than those for unoccupied sites), regardless of the correctness of the predicted probability of occupation. We measured discrimination using the area under the receiver operating characteristic curve (AUC). AUC ranges from 0 to 1, with a value of 1 indicating perfect agreement and 0.5 indicating model performance no better than random. We also evaluated the ability of the models to predict community composition by comparing predicted and observed community composition at each locality and examining their similarity using 1 − Bray–Curtis dissimilarity index. Bray–Curtis dissimilarity was calculated between the predicted probabilities of occurrence and observed presence/absence (in other words, observed absence is represented by a probability of 0 and observed presence is represented by a probability of 1). We assessed the statistical significance of differences in model performance between each SDM/CLM comparison and overall SDM and CLM performance using Wilcoxon tests on all 10 iterations for the 19 taxa for each metric.
(f). Model performance versus climatic and compositional novelty
Model performance was evaluated against climatic and compositional (biotic) novelty to examine the extent to which models can be reliably projected to new climatic regimes and no-analogue communities. Climatic novelty for each combination of projected and fitted time periods was calculated by averaging the minimum Euclidean distances between an occupied grid cell for the time period in which the model was fitted and all occupied grid cells in the time period to which the model was projected [7,14], based on the first and second principal component axes of a PCA analysis on the six climate variables from all occupied grid cells from all time periods, scaled to have unit variance. Similarly, compositional novelty for each combination of fitted and projected time periods was calculated as the mean of the minimum dissimilarities in relative abundance between each site in the fitted time period compared to all sites in the projected-to time period using Bray–Curtis dissimilarity [14]. To determine whether SDM and CLM performance was influenced by climatic novelty, composition novelty or by both factors, we built linear models between model performance and each of the factors. We then evaluated the linear models with the Akaike information criteria (AIC) and calculated the AIC weights using the akaike.weights function in the qpcR package.
(g). Model performance and number of occurrences
One of the proposed strengths of CLMs is that by pooling data from all taxa they may better detect shared environmental responses across taxa with fewer occurrences, leading to improved predictions of their distributions. The number of occurrences (or the presence records) available for modelling a given taxon is a function of the number of available localities (sample size) and the prevalence of the taxon (the proportion of these sites at which the taxon of interest is present). The number of localities in the fitted time periods varied between 72 and 276 (including pooled time periods), potentially contributing to differences in model performance between SDMs and CLMs. To examine the influence of this on differences in performance between the SDMs and CLMs, we subsampled the number of localities from the 1 kyr BP (276 localities) and 9 kyr BP (197 localities) time periods at 50, 100, 150 and 200 (1 kyr BP only) localities and reran the analyses. We also examined the effect of prevalence (taxon rarity) on model performance to determine if CLMs model rare taxa better than SDMs.
3. Results
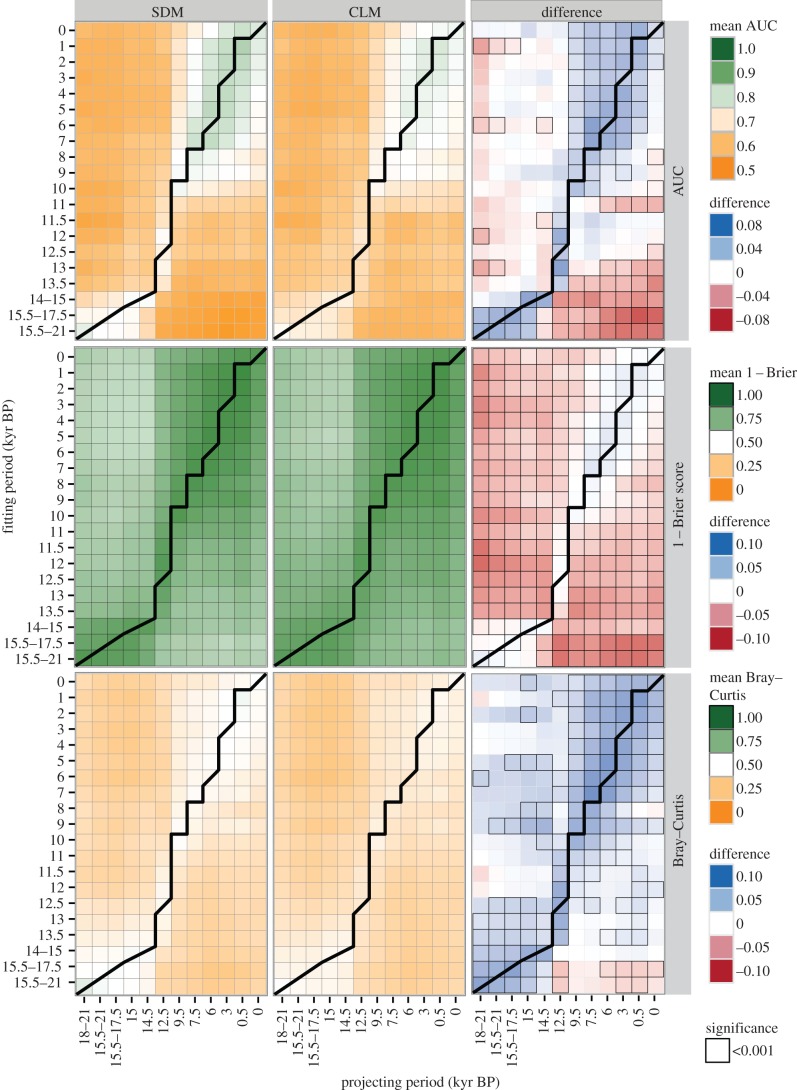
When models were projected to climatically similar time periods, both SDMs and CLMs had good ability to discriminate between occupied and unoccupied sites for individual species (AUC scores more than 0.7), with SDMs tending to outperform CLMs under these circumstances (figure 2a and figure 3, top row). However, the opposite was true when models were projected to climatically dissimilar time periods: both SDMs and CLMs performed poorly and their relative performance switched, with CLMs tending to outperform SDMs (figure 2a), especially when forecasting from the data-sparse LGM to more recent time periods (figure 3, top row). This pattern is especially evident in the GLM–CQO and GAM–CAO comparisons (electronic supplementary material, figure S4a), in which GLM and GAM performed particularly poorly relative to their CLM counterpart when forecasting. MRTs and MMARS also had higher AUC scores than their SDM counterparts, while ANN and MANN performed similarly through time (electronic supplementary material, figure S4).
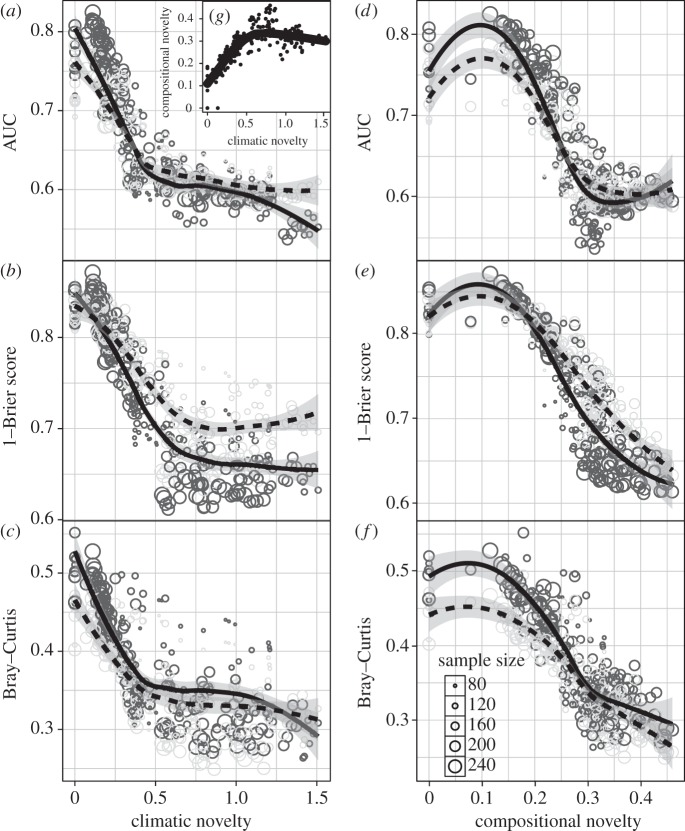
Figure 2.
Model performance as a function of climatic and compositional novelty. AUC scores (a,d), 1−Brier scores (b,e) and Bray–Curtis similarity (1 – Bray–Curtis dissimilarity) (c,f) as a function of climatic novelty (a–c) and compositional novelty (d–f), fitted with a local polynomial regression (LOESS) line. Black circles and solid black lines represent SDM values. Grey circles and dashed lines represent CLM values. Circle size reflects sample size. (g) (inset in (a)) Compositional novelty as a function of climatic novelty.
Figure 3.
Mean SDM and CLM performance. Mean AUC scores (top row), 1−Brier scores (middle row) and Bray–Curtis similarity (bottom row) of SDMs (left column) and CLMs (middle column) for each combination of fit in and projected to periods. Each square represents the average of all five modelling algorithms for all genera. Green shading indicates high performance and orange shading indicates low performance. The right column shows the differences between SDM and CLM scores across all five modelling algorithms and all genera. Blue shading indicates superior SDM performance while red shading indicates better CLM performance. Cells with black outlines indicate statistically significant differences between SDMs and CLMs based on a Wilcoxon test on the mean values for each metric. The thick solid black lines indicate models that were fit and projected to the same time period and thus divide hindcasted models (above line) from forecasted models (below line).
In terms of model reliability (Brier score), SDMs and CLMs predicted species' probability of occurrence similarly when projected to climatically similar time periods (figures 2b, and 3, middle row); however, when models were hindcasted or forecasted to climatically novel periods, CLMs outperformed SDMs. This pattern was observed in three of the five paired model-class comparisons, except for GLM–CQO and GAM–CAO (electronic supplementary material, figure S4b). For these two model-classes, SDMs outperformed CLMs when models were fitted in, and projected to, climatically similar and temporally close time periods; however, they were similar to the other model-classes in that the CLM consistently outperformed the SDM in climatically novel and temporally distant time periods.
As with AUC and Brier scores, CLMs and stacked SDMs predicted community composition better when models were fitted in, and projected to, climatically similar time periods, with stacked SDMs tending to outperform CLMs (figures 2c, and 3, bottom row). When models were fitted and projected to climatically novel time periods, the models exhibited equal performance, with SDMs hindcasting slightly better and CLMs forecasting slightly better (figure 3, bottom row). When individual model-classes were examined, there were no significant differences between the SDM and CLM (electronic supplementary material, figure S4).
The performance of SDMs and CLMs tracked one another as compositional novelty increased between the fitted and projected time periods (figure 2d–f), with subtle differences: CLMs slightly outperformed SDMs at higher levels of compositional novelty according to the Brier Score, while SDMs outperformed CLMs according to Bray–Curtis similarity (1 − Bray–Curtis dissimilarity) at high and low values of compositional novelty. The inclusion of both climatic and compositional novelty as predictors of model performance is statistically supported over either variable alone (electronic supplementary material, table S4), suggesting that climatic novelty and compositional novelty each have partially independent effects on model performance. When each variable was examined individually, compositional novelty was statistically more supported than climatic novelty according to all three metrics, the only exception to this being that climatic novelty is more supported for SDM discrimination (AUC).
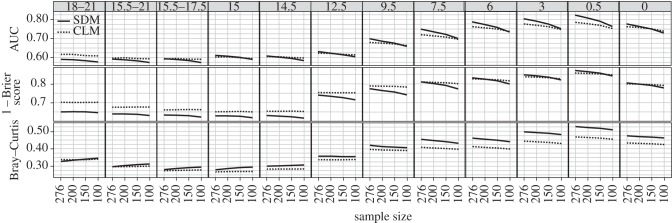
The number of localities included in the model affected the performance (AUC) of SDMs more than CLMs (figure 4). As the number of localities decreased in the fitted time period (1 kyr BP), the performance of CLMs projected to other time periods did not change substantially according to all three metrics examined, whereas performance of SDMs tended to decrease (figure 4). We also observed this pattern in sample-size sensitivity tests using the 9 kyr BP time period (electronic supplementary material, figure S5). We found no evidence that the predictive skill of CLMs was consistently superior to SDMs for species with low prevalence (electronic supplementary material, figure S6). While CLMs performed better than SDMs at low prevalence for some taxa, other taxa showed the opposite or no trend, indicating that CLMs did not model rarer taxa better than SDMs overall.
Figure 4.
Number of localities and performance. Performance of models based on AUC scores (top row), 1 − Brier scores (middle row) and Bray–Curtis similarity (bottom row) when the number of localities used to fit the model in 1 kyr BP is the original number (n = 276) and subsampled to 100, 150 and 200 localities and then projected to chosen time periods. Solid line represents the mean value of each metric across all SDMs and the dashed line represents the mean across all CLMs.
4. Discussion
A major challenge in predicting future patterns of biodiversity is accommodating the complex and shifting relationships between the environment, species distributions and communities as the environment changes and communities reshuffle in response to those changes [8]. For example, realized niches can change as climates change [11], owing to changing biotic interactions [46,47] and emergence of novel climates [10]. Here, we found that overall model transferability was low (AUC < 0.65; 1 − Brier score < 0.75; Bray–Curtis similarity < 0.35) in the periods of greatest climatic and compositional novelty, irrespective of whether SDMs or CLMs were used. By simultaneously modelling all co-occurring taxa in a region, CLMs outperformed SDMs for the most novel climates and when forecasting from the data-sparse periods of the Pleistocene. This is most likely because CLMs (i) identify the primary climatic gradients driving biodiversity patterns and thereby buffer against idiosyncratic changes in individual species–environment relationships that can reduce the performance of SDMs and (ii) are more robust to small sample sizes (fewer localities or total number of presences and absences, but not necessarily taxon prevalence).
(a). Model performance and transferability
Overall differences between the combined averages (i.e. across all iterations, taxa and model types) of SDMs and CLMs were small (AUC < 0.1; Brier score < 0.15, Bray–Curtis similarity < 0.15). Small differences were to be expected, given that the algorithm for each SDM/CLM pair is essentially the same and differed only in whether the response was univariate or multivariate. Additionally, all models were fitted with the same climatic variables and each SDM/CLM pair was fitted with the same parameter complexity. However, average differences between SDMs and CLMs remained small even when models were permitted to choose the environmental variables and complexity of parameters (AUC < 0.1; Brier score < 0.15, Bray–Curtis similarity < 0.15). Neural networks (ANN and MANN) were a notable exception to this pattern as ANN outperformed MANN more frequently when variable selection and parameter complexity were chosen using a step function (electronic supplementary material, figure S3). The more revealing aspect of this analysis is the inability of models to predict patterns of biodiversity in novel climates with different community compositions. The transferability of a model between time periods is affected by the extent to which biological patterns are controlled by climate, the stability of these relationships between the two time periods of interest and the adequacy of the data. Modelled individual taxon–climate relationships (SDMs) and composition–climate relationships (CLMs) will break down as climates become increasingly novel relative to our reference window and new taxon associations emerge in response to these climates. Thus, model performance is expected to decline as climates become increasingly distant from modern analogues, for which taxon–climate and community–climate relationships are poorly understood [7,10,30]. Our results clearly demonstrate this decline (figure 2a–c). SDMs exhibited greater declines in their ability to predict taxon occurrences than CLMs under increasingly novel climates (figure 2a,b), suggesting that CLMs may be able to extrapolate taxon occurrences beyond known climatic regimes better than SDMs, though perhaps only marginally so. The higher performance of CLMs in predicting taxon occurrences in the most novel communities (figure 2e) is somewhat surprising given that SDMs allow individualistic responses while CLMs are somewhat more constrained [2,15]. Our analysis suggests that enough information was maintained about taxa associations along climate gradients that CLMs could borrow strength and make more informed predictions using low sample sizes. This result did not hold up for discrimination (figure 2d); however, discrimination capacity in SDMs depends on the representativeness of the environmental domain and therefore is a context-dependent property [48] and may not be a good measure of model transferability in this case. SDMs consistently over-fit the training data in this study (electronic supplementary material, figure S7) and others [4] (but see [15]), whereas CLMs showed less over-fitting, perhaps because they were constrained by multi-taxa inputs. This may explain why SDMs tended to have higher predictive skill for temporally adjacent time periods, whereas CLMs predicted better in novel climatic regimes and no-analogue communities than their SDM counterparts.
The relative discrimination capacity of SDMs and their CLM counterparts varied, with some SDMs generally outperforming their CLM counterpart (i.e. GLM, similar to Baselga & Araújo [4] and Bonthoux et al. [15], and GAM), while some CLMs generally outperformed their SDM counterpart (i.e. MMARS, similar to Leathwick et al. [38], and MRTs). ANN and MANN performed similarly, possibly given their use of a machine-learning algorithm designed for both univariate and multivariate analyses [42]. Model reliability (Brier score), on the other hand, clearly shows the superiority of all CLM algorithms to accurately predict the presence/absence, as discussed above.
Although others found CLMs better predicted community composition than SDMs [21,49], our analysis shows the opposite (figure 2c,f, and 3, bottom row; also see [23]). The one exception was forecasting from the LGM, where CLMs predicted community composition better than SDMs, likely because there were fewer localities during the LGM compared with more recent time periods, and CLMs were better than SDMs at predicting taxon occurrences with few sampling localities (figure 4). Neither stacked SDMs nor CLMs predicted community composition into novel climates particularly well (figure 2c). As compositional novelty increased, both SDMs and CLMs predicted community composition poorly, but on average, SDMs better predicted community composition (figure 2f). This mean result was largely driven by the superiority of GLM and GAM to predict community composition (electronic supplementary material, figure S4c). The SDM and CLM versions of the other three model-classes predicted community composition equally. Overall, results suggest that CLMs may not be able to capture emergent community properties in extreme instances of climatic and compositional novelty any better than stacked SDMs owing to individualistic taxon responses [15].
The differences in model performance and transferability between SDMs and CLMs is not owing just to the emergence of novel climates or the emergence of novel communities; rather, both are necessary to fully explain variation in model performance (electronic supplementary material, table S4). Climatic and compositional novelties were positively correlated until compositional novelty reached a plateau (figure 2g). This pattern was recovered for the 19 taxa in this analysis as well as all 109 taxa available in the original dataset and has been observed both temporally (this analysis, but see [14]) and spatially [50]. At the point at which compositional novelty plateaued in this analysis (compositional novelty approx. 0.3 and climatic novelty approx. 0.5) there was a shift in model performance across all metrics, after which model performance stabilized (figure 2a–f). After this threshold, model performance was mostly influenced by climatic novelty, when CLMs predicted distributions better than SDMs, further suggesting the superiority of CLMs when projecting to novel climates.
(b). Model performance and number of occurrences
The number of occurrences available to model a taxon's distribution, which itself results from the interplay between the number of available localities and taxon prevalence, drove differences between model performance. When fitted with few localities, CLMs may have outperformed SDMs because of their multiresponse nature in which they can ‘borrow strength’ across taxa. On the other hand, when more localities were available, parameter estimation from SDMs was as good as or better than CLMs. Interestingly in our study, when sample size effect was removed, taxon prevalence was inconsistently correlated with model performance (electronic supplementary material), nor was there any consistent difference in the effect of prevalence on the predictive skill of SDMs and CLMs. It has been hypothesized [2,51] and demonstrated [17–19] that CLMs improve predicted distributions of rare taxa by incorporating information on co-occurrences with other taxa that have similar environmental tolerances (but see [15,19]). Whether or not CLMs ‘borrow strength’ to support rare taxa is complex—Bonthoux et al. [15] found that CLMs improved the understanding of rare species patterns but did not predict their distributions better than SDMs. Here we suggest that the superior predictive ability of CLMs may not be just the result of their ability to model rare taxa but also because they are more robust to low numbers of localities.
Overall, the difference in predictive performance of SDMs versus CLMs was modest even when variable selection and parameter complexity were introduced [19]. Similar to this study, several previous analyses examining the model performance of CLMs against SDMs reported slight differences or mixed results [4,15,22,24], indicating that model performance may be case-dependent and related to individual taxon–environment relationships [4]. Model performance might also be influenced by inaccurate palaeoclimatic simulations as well as uncertainties in the pollen data and age models, or missing important covariates such as CO2 concentrations [6]. However, SDMs and CLMs should be equally affected by these uncertainties and therefore are unlikely to affect the main conclusions and comparative analyses presented here.
5. Conclusion
Both SDMs and CLMs performed poorly when projected into novel climates and assemblages. This finding calls into question the dependability and utility of using empirical models to project future distributions and communities. Overall, however, our results both reveal a path towards better predictions of future ecological assemblages given the large magnitude of climatic change expected for the future and highlight the need for better approaches given the expected increase in compositional novelty [7,52]. We confirm that CLMs are a similar or modestly superior approach to SDMs for modelling the responses of species diversity and distributions to climate change. Although CLMs are no better at predicting community composition than stacked SDMs [23] (but see [21]), they generally have higher predictive skill (with respect to discrimination and reliability) under conditions of novel climates and low taxon occurrences. For these reasons, CLMs deserve greater attention, application and examination than they have received to date.
Supplementary Material
Acknowledgements
The authors thank Bette Otto-Bliesner, Zhengyu Liu and Feng He for the original climate simulations. We thank David Harris for helpful discussions and Janet Franklin and three anonymous reviewers for comments that greatly improved the manuscript. This is UMCES- Appalachian Laboratory Scientific contribution no. 5172.
Data accessibility
The presence/absence matrices of pollen taxa at each site per time slice and a list of final sites included in the analysis are available at Dryad data repository (http://dx.doi.org/10.5061/dryad.hk400).
Authors' contributions
J.L.B., M.C.F., D.N.L., K.C.M., J.W.W. designed the study; J.L.B. compiled the pollen data; D.J.L. downscaled and prepared the climate data; D.N.L. wrote the R code and scripts; D.N.L. and K.C.M. performed the analyses; J.L.B., M.C.F., S.F., D.N.L., K.C.M., J.W.W. analysed the output data; K.C.M. wrote the manuscript; all the authors contributed substantially to revisions, and gave final approval for publication.
Competing interests
The authors declare that they have no competing interests.
Funding
This work was supported by NSF (DEB-1257033 to J.L.B., DEB-1257164 to M.C.F. and DEB-1257508 to J.W.W.)
References
- 1.Urban MC. 2015. Accelerating extinction risk from climate change. Science 348, 571–573. ( 10.1126/science.aaa4984) [DOI] [PubMed] [Google Scholar]
- 2.Ferrier S, Guisan A. 2006. Spatial modelling of biodiversity at the community level. J. Appl. Ecol. 43, 393–404. ( 10.1111/j.1365-2664.2006.01149.x) [DOI] [Google Scholar]
- 3.Fitzpatrick MC, Gove AD, Sanders NJ, Dunn RR. 2008. Climate change, plant migration, and range collapse in a global biodiversity hotspot: the Banksia (Proteaceae) of Western Australia. Glob. Change Biol. 14, 1337–1352. ( 10.1111/j.1365-2486.2008.01559.x) [DOI] [Google Scholar]
- 4.Baselga A, Araújo MB. 2009. Individualistic vs community modelling of species distributions under climate change. Ecography 32, 55–65. ( 10.1111/j.1600-0587.2009.05856.x) [DOI] [Google Scholar]
- 5.Elith J, Leathwick J. 2009. Species distribution models: ecological explanation and prediction across space and time. Annu. Rev. Ecol. Evol. Syst. 40, 677–697. ( 10.1146/annurev.ecolsys.110308.120159) [DOI] [Google Scholar]
- 6.Blois JL, Williams JW, Fitzpatrick MC, Jackson ST, Ferrier S. 2013. Space can substitute for time in predicting climate-change effects on biodiversity. Proc. Natl Acad. Sci. USA 110, 93 734–99 379. ( 10.1073/pnas.1220228110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams JW, Jackson ST, Kutzbach JE. 2007. Projected distributions of novel and disappearing climates by 2100 AD. Proc. Natl Acad. Sci. USA 104, 5738–5742. ( 10.1073/pnas.0606292104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maguire KC, Nieto-Lugilde D, Fitzpatrick MC, Williams JW, Blois JL. 2015. Modeling species and community responses to past, present, and future episodes of climatic and ecological change. Annu. Rev. Ecol. Evol. Syst. 46, 343–368. ( 10.1146/annurev-ecolsys-112414-054441) [DOI] [Google Scholar]
- 9.D'Amen MD, Rahbek C, Zimmerman NE, Guisan A. 2015. Spatial predictions at the community level: from current approaches to future frameworks. Biol. Rev. ( 10.1111/brv.12222) [DOI] [PubMed] [Google Scholar]
- 10.Williams JW, Jackson ST. 2007. Novel climates, no-analog communities, and ecological surprises. Front. Ecol. Environ. 5, 475–482. ( 10.1890/070037) [DOI] [Google Scholar]
- 11.Veloz SD, Williams JW, Blois JL, He F, Otto-Bliesner B, Liu Z. 2012. No-analog climates and shifting realized niches during the Late Quaternary: implications for 21st-century predictions by species distribution models. Glob. Change Biol. 18, 1698–1713. ( 10.1111/j.1365-2486.2011.02635.x) [DOI] [Google Scholar]
- 12.Wisz MS, et al. 2013. The role of biotic interactions in shaping distributions and realised assemblages of species: implications for species distribution modelling. Biol. Rev. 88, 15–30. ( 10.1111/j.1469-185X.2012.00235.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis MB. 1981. Quaternary history and the stability of forest communities. In Forest succession (eds West DC, Shugart HH, Botkin DB), pp. 132–177. New York, NY: Spriner. [Google Scholar]
- 14.Williams JW, Shuman BN, Webb T III. 2001. Dissimilarity analyses of Late-Quaternary vegetation and climate in eastern North America. Ecology 82, 3346–3362. [Google Scholar]
- 15.Bonthoux S, Baselga A, Balent G. 2013. Assessing community-level and single-species models predictions of species distributions and assemblage composition after 25 years of land cover change. PLoS ONE 8, e54179 ( 10.1371/journal.pone.0054179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fitzpatrick MC, Sander NJ, Ferrier S, Longina JT, Weiser MD, Dunn R. 2011. Forecasting the future of biodiversity: a test of single- and multi-species models for ants in North America. Ecography 34, 836–847. ( 10.1111/j.1600-0587.2011.06653.x) [DOI] [Google Scholar]
- 17.Ovaskainen O, Soininen J. 2011. Making more out of sparse data: hierarchical modeling of species communities. Ecology 92, 289–295. ( 10.1890/10-1251.1) [DOI] [PubMed] [Google Scholar]
- 18.Hui F, Warton DI, Foster SD, Dunstan PK. 2013. To mix or not to mix: comparing the predictive performance of mixture models vs. separate species distribution models. Ecology 94, 1913–1919. ( 10.1890/12-1322.1) [DOI] [PubMed] [Google Scholar]
- 19.Madon B, Warton DI, Araújo MB. 2013. Community-level vs species-specific approaches to model selection. Ecography 36, 1291–1298. ( 10.1111/j.1600-0587.2013.00127.x) [DOI] [Google Scholar]
- 20.Elith J, et al. 2006. Novel methods improve prediction of species’ distributions from occurrence data. Ecography 29, 129–151. ( 10.1111/j.2006.0906-7590.04596.x) [DOI] [Google Scholar]
- 21.Olden JD, Joy MK, Death RG. 2006. Rediscovering the species in community-wide predictive modeling. Ecol. Appl. 16, 1449–1460. ( 10.1890/1051-0761(2006)016%5B1449:RTSICP%5D2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 22.Elith J, Leathwick J. 2007. Predicting species distributions from museum and herbarium records using multiresponse models fitted with multivariate adaptive regression splines. Divers. Distrib. 13, 265–275. ( 10.1111/j.1472-4642.2007.00340.x) [DOI] [Google Scholar]
- 23.Baselga A, Araújo MB. 2010. Do community-level models describe community variation effectively? J. Biogeogr. 37, 1842–1850. [Google Scholar]
- 24.Chapman DS, Purse BV. 2011. Community versus single-species distribution models for British plants. J. Biogeogr. 38, 1524–1535. ( 10.1111/j.1365-2699.2011.02517.x) [DOI] [Google Scholar]
- 25.North Greenland Ice Core Project Members. 2004. North Greenland ice core project oxygen isotope data. IGBP PAGES/World data Center for Paleoclimatology Data Contribution Series #2004-059. Boulder, CO: NOAA/NGDC Paleoclimatology Program. [Google Scholar]
- 26.NGRIP Dating Group. 2008. IGBP PAGES/World Data Center for Paleoclimatology Data Contribution Series #2008-034. Boulder, CO: NOAA/NCDC Paleoclimatology Program. [Google Scholar]
- 27.Williams JW, Blois JL, Gill JL, Gonzales LM, Grimm EC, Ordonez A, Shuman B, Veloz SD. 2013. Model systems for a no-analog future: species associations and climates during the last deglaciation. Ann. NY Acad. Sci. 1297, 29–43. ( 10.1111/nyas.12226) [DOI] [PubMed] [Google Scholar]
- 28.Maiorano L, et al. 2013. Building the niche through time: using 13 000 years of data to predict the effects of climate change on three tree species in Europe. Glob. Ecol. Biogeogr. 22, 302–317. ( 10.1111/j.1466-8238.2012.00767.x) [DOI] [Google Scholar]
- 29.Shuman B, Webb T III, Bartlein P, Williams JW. 2002. The anatomy of a climatic oscillation: vegetation change in eastern North America during the Younger Dryas chronozone. Q. Sci. Rev. 21, 1777–1791. ( 10.1016/S0277-3791(02)00030-6) [DOI] [Google Scholar]
- 30.Blois JL, Williams JW, Fitzpatrick MC, Ferrier S, Veloz SD, He F, Liu Z, Manion G, Otto-Bliesner B. 2013. Modeling the climatic drivers of spatial patterns in vegetation composition since the Last Glacial Maximum. Ecography 36, 460–473. ( 10.1111/j.1600-0587.2012.07852.x) [DOI] [Google Scholar]
- 31.Nieto-Lugilde D, Maguire KC, Blois JL, Williams JW, Fitzpatrick MC. 2015. Close agreement between pollen-based and forest inventory-based models of vegetation turnover. Glob. Ecol. Biogeogr. 24, 905–916. ( 10.1111/geb.12300) [DOI] [Google Scholar]
- 32.Prus-Głowacki W, Stephan BR, Bujas E, Alia R, Marciniak A. 2003. Genetic differentiation of autochthonous populations of Pinus sylvestris (Pinaceae) from the Iberian peninsula. Plant Syst. Evol. 239, 55–66. ( 10.1007/s00606-002-0256-3) [DOI] [Google Scholar]
- 33.Liu Z, et al. 2009. Transient simulation of last deglaciation with a new mechanism for Bølling-Allerød warming. Science 325, 310–314. ( 10.1126/science.1171041) [DOI] [PubMed] [Google Scholar]
- 34.Guisan A, Edwards TC Jr, Hastie T. 2002. Generalized linear and generalized additive models in studies of species distributions: setting the scene. Ecol. Modell. 157, 89–100. ( 10.1016/S0304-3800(02)00204-1) [DOI] [Google Scholar]
- 35.Yee TW. 2004. A new technique for maximum-likelihood canonical Gaussian ordination. Ecol. Monogr. 74, 685–701. ( 10.1890/03-0078) [DOI] [Google Scholar]
- 36.Yee TW. 2006. Constrained additive ordination. Ecology 87, 203–213. ( 10.1890/05-0283) [DOI] [PubMed] [Google Scholar]
- 37.Friedman JH. 1991. Multivariate adaptive regression splines. Ann. Stat. 19, 1–67. ( 10.1214/aos/1176347963) [DOI] [PubMed] [Google Scholar]
- 38.Leathwick JR, Elith J, Hastie T. 2006. Comparative performance of generalized additive models and multivariate adaptive regression splines for statistical modelling of species distributions. Ecol. Modell. 199, 188–196. ( 10.1016/j.ecolmodel.2006.05.022) [DOI] [Google Scholar]
- 39.Breiman L, Friedman JH, Olshen RA, Stone CJ. 1984. Classification and regression trees. Belmont, CA: Wadsworth International Group. [Google Scholar]
- 40.De'ath G, Fabricius KE. 2000. Classification and regression trees: a powerful yet simple technique for ecological data analysis. Ecology 81, 3178–3192. ( 10.1890/0012-9658(2000)081%5B3178:CARTAP%5D2.0.CO;2) [DOI] [Google Scholar]
- 41.Ferrier S, Drielsma M, Manion G, Watson G. 2002. Extended statistical approaches to modelling spatial pattern in biodiversity in northeast New South Wales. II. Community-level modelling. Biodivers. Conserv. 11, 2309–2338. ( 10.1023/A:1021374009951) [DOI] [Google Scholar]
- 42.Olden JD. 2003. A species-specific approach to modeling biological communities and its potential for conservation. Conserv. Biol. 17, 854–863. ( 10.1046/j.1523-1739.2003.01280.x) [DOI] [Google Scholar]
- 43.R Core Team. 2014. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; (http://www.R-project.org/) [Google Scholar]
- 44.Pearce J, Ferrier S. 2000. Evaluating the predictive performance of habitat models developed using logistic regression. Ecol. Modell. 133, 225–245. ( 10.1016/S0304-3800(00)00322-7) [DOI] [Google Scholar]
- 45.Brier GW. 1950. Verification of forecasts expressed in terms of probability. Mon. Weather Rev. 78, 1–3. () [DOI] [Google Scholar]
- 46.Tylianakis JM, Didham RK, Bascompte J, Wardle DA. 2008. Global change and species interactions in terrestrial ecosystems. Ecol. Lett. 11, 1351–1363. ( 10.1111/j.1461-0248.2008.01250.x) [DOI] [PubMed] [Google Scholar]
- 47.Colwell RK, Rangel TF. 2009. Hutchinson's duality: the once and future niche. Proc. Natl Acad. Sci. USA 106, 19 651–19 658. ( 10.1073/pnas.0901650106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiménez-Valverde A, Acevedo P, Barbosa AM, Lobo JM, Real R. 2013. Discrimination capacity in species distribution models depends on the representativeness of the environmental domain. Glob. Ecol. Biogeogr. 22, 508–516. ( 10.1111/geb.12007) [DOI] [Google Scholar]
- 49.Henderson EB, Ohmann JL, Gregory MJ, Roberts HM, Zald H. 2014. Species distribution modelling for plant communities: stacked single species or multivariate modelling approaches? Appl. Veg. Sci. 17, 516–527. ( 10.1111/avsc.12085) [DOI] [Google Scholar]
- 50.Ferrier S, Manion G, Elith J, Richardson K. 2007. Using generalized dissimilarity modelling to analyse and predict patterns of beta diversity in regional biodiversity assessment. Divers. Distrib. 13, 252–264. ( 10.1111/j.1472-4642.2007.00341.x) [DOI] [Google Scholar]
- 51.Ferrier S. 2002. Mapping spatial pattern in biodiversity for regional conservation planning: where to from here? Syst. Biol. 51, 331–363. ( 10.1080/10635150252899806) [DOI] [PubMed] [Google Scholar]
- 52.Ordonez A, Martinuzzi S, Radeloff VC, Williams JW. 2014. Combined speeds of climate and land-use change of the conterminous US until 2050. Nat. Clim. Change 4, 811–816. ( 10.1038/nclimate2337) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The presence/absence matrices of pollen taxa at each site per time slice and a list of final sites included in the analysis are available at Dryad data repository (http://dx.doi.org/10.5061/dryad.hk400).