Abstract
Although increased disease severity driven by intensive farming practices is problematic in food production, the role of evolutionary change in disease is not well understood in these environments. Experiments on parasite evolution are traditionally conducted using laboratory models, often unrelated to economically important systems. We compared how the virulence, growth and competitive ability of a globally important fish pathogen, Flavobacterium columnare, change under intensive aquaculture. We characterized bacterial isolates from disease outbreaks at fish farms during 2003–2010, and compared F. columnare populations in inlet water and outlet water of a fish farm during the 2010 outbreak. Our data suggest that the farming environment may select for bacterial strains that have high virulence at both long and short time scales, and it seems that these strains have also evolved increased ability for interference competition. Our results are consistent with the suggestion that selection pressures at fish farms can cause rapid changes in pathogen populations, which are likely to have long-lasting evolutionary effects on pathogen virulence. A better understanding of these evolutionary effects will be vital in prevention and control of disease outbreaks to secure food production.
Keywords: aquaculture, evolution, fish farming, Flavobacterium columnare, pathogen, virulence
1. Background
Food production creates environments where the ecology, epidemiology and evolution of disease differ from nature. Increased host density and decreased host diversity can reduce the costs of pathogen transmission, and, according to the predictions of virulence evolution theory, highly virulent strains may emergence as a consequence [1,2]. Indeed, intensive farming environments are considered evolutionary hotspots, where the enhanced transmission and frequency of infections could promote virulence evolution in pathogen populations [3–7]. This can be especially important for environmentally transmitted opportunistic pathogens, which often have the ability to survive and replicate outside hosts. These opportunistic pathogens do not suffer from the transmission–virulence trade-off that is characteristic of obligate pathogens, which are dependent on host survival during transmission, thus significantly altering pathogen virulence [8].
It is common that hosts in nature are co-infected by several pathogen strains. The within-host competitive interactions have been traditionally linked to virulence via faster growth of more virulent parasite strains, which is expected to influence dynamics of disease epidemics [1,2,9–14]. However, bacterial interactions towards each other are mainly negative [15]. Competition over limited resources can promote selective interference competition mechanisms targeted to exclude competitors through the release of toxins [16,17], and lead to decreased virulence during bacterial infection [18,19]. It is thus likely that the interactions between coexisting pathogen strains influence the development and persistence of disease epidemics, and the population dynamics of pathogens.
Although these abovementioned theories and laboratory studies have provided insight on pathogen evolution, very little is known about how anthropogenic changes, like intensive farming, influence virulence evolution [20–22]. Such empirical evidence is not only crucial for testing the theoretical predictions, but also for understanding disease ecology and evolution. More than anything, we need data outside of the laboratory from the economically relevant systems [21]. Information on the diseases threatening crop, livestock and aquaculture production is essential for securing world food production. Here, we study how selection in intensive aquaculture influences the virulence and the competitive ability, at both long and short time scales, in the globally significant fish pathogen Flavobacterium columnare (Bacteroidetes) [23,24]. First, we explore temporal changes in virulence and competitive ability using F. columnare strains isolated from disease outbreaks in 2003–2010. Second, we study locally occurring selection by comparing bacterial isolates originating from inlet water and outlet water of a fish farm during one outbreak season in summer 2010. We observe that intensive aquaculture seems to select for increased F. columnare virulence, as well as ability for interference competition, at both short and long time scales. This indicates that the more virulent strains have higher fitness under intensive farming conditions, and demonstrates that the intensive farming environments can be used as model systems to understand disease dynamics and evolution of virulence.
2. Material and methods
(a). Experimental set-up
In this study, we used two different sets of F. columnare strains to study evolution of bacterial virulence and competitive ability on both at temporal and spatial scale. In the first set, we used strains isolated during 2003–2010. In the second set, we compared the characteristics of bacteria isolated in 2010 from the inlet and the outlet water of a fish farm. In both study settings, we studied the bacterial growth rate, the bacterial virulence in zebra fish and the ability of the bacteria to inhibit the growth of other strains, and analysed if these traits changed in time and place. In addition, the data from 2003 to 2010 were used to analyse the bacterial competition in finite resources in liquid culture, and the growth of the inlet water and outlet water strains were compared in two different resource concentrations.
(b). Bacterial strains, their isolation and genetic characterization
The bacteria were isolated originally from three different fish farms in Central and Northern Finland during columnaris disease epidemics and from the environment in 2003–2010 (table 1) using standard culture methods on Shieh medium [26,27] or AO-agar [28]. Isolates for the dataset were randomly chosen from a large collection of uncharacterized bacterial strains without any a priori information on their competitive ability or virulence, except for strains H2, B067, B185, B245, B405 and B407, which have been used (separately) in our previous studies [29–31].
Table 1.
Flavobacterium columnare strains isolated in 2003–2010 and used in this study. Genetic group is based on MLSA analysis, which corresponds with the ARISA genotyping used with strains in table 2 [25].
| isolation source | bacterial strain | year of isolation | fish species/water sample | genetic group |
|---|---|---|---|---|
| fish farm A, Central Finland | B431 | 2003 | grayling Thymallus thymallus | A |
| B067 | 2007 | trout Salmo trutta | A | |
| B185 | 2008 | rearing tank water | G | |
| fish farm B, Central Finland | H2 | 2003 | rainbow trout Oncorhynchus mykiss | H |
| B429 | 2003 | pikeperch Zander lucioperca | H | |
| B430 | 2003 | pikeperch Z. lucioperca | E | |
| B425 | 2007 | rainbow trout O. mykiss | ||
| B245 | 2009 | rearing tank water | C | |
| B259 | 2009 | rearing tank water | C | |
| B402 | 2010 | whitefish Coregonus lavaretus | C | |
| B366 | 2010 | outlet water of a fish farm | C | |
| nature (lake) | B405 | 2010 | Lake Jyväsjärvi | C |
| nature (river) | B407 | 2010 | river upstream of fish farm B | G |
| fish farm C, Northern Finland | B428 | 2006 | Atlantic salmon Salmo salar | |
| B426 | 2006 | Atlantic salmon S. salar | C | |
| B420 | 2009 | Atlantic salmon S. salar | G | |
| B421 | 2009 | Atlantic salmon S. salar | C |
The bacterial strains from inlet water upstream of a fish farm [30] and from downstream and outlet water of the same farm were collected in summer 2010 (table 2). Both locations (inlet water and outlet water) were sampled on 8 and 21 June, on 5, 12, 19 and 26 July, and on 2, 16 and 30 August in 2010 (electronic supplementary material, table S1). Nine bacterial isolates were randomly selected from both locations for further analyses. After isolation, the bacteria were maintained frozen in −80°C with 10% glycerol and 10% fetal calf serum. For the analyses, the bacterial strains were grown in Shieh medium at room temperature (RT, approx. 24°C) under constant shaking (110 r.p.m.) for 24–48 h and enriched overnight.
Table 2.
Flavobacterium columnare strains used in this study. The strains were isolated from nature, from river upstream of a fish farm (inlet water) in Central Finland (farm B in table 1) and from the outlet water or from the river immediately downstream of the outlet water tube of the same farm in summer 2010. Genetic grouping is based on automated ribosomal intergenic spacer analysis (ARISA).
| isolate | inlet water | outlet water | genetic group | time of isolation (2010) | references |
|---|---|---|---|---|---|
| B395 | Xa | G | 21st June | [30] | |
| B396 | Xa | A | 21st June | [30] | |
| B397 | Xb | C | 21st June | [30] | |
| B398 | Xb | A | 21st June | [30] | |
| B400 | Xb | A | 19th July | [30] | |
| B404 | Xa | C | 2nd August | [30] | |
| B355 | X | A | 2nd August | this study | |
| B406 | Xb | C | 16th August | [30] | |
| B407 | Xb,c | G | 16th August | [30] | |
| B339 | Xd | C | 5th July | this study | |
| B340 | Xd | C | 5th July | this study | |
| B350 | Xa | E | 2nd August | this study | |
| B351 | Xa | E | 2nd August | this study | |
| B366 | Xd | C | 2nd August | this study | |
| B370 | Xb | E | 16th August | this study | |
| B374 | Xb | E | 16th August | this study | |
| B375 | Xb | E | 16th August | this study | |
| B379 | Xd | E | 30th August | this study |
aRiver, biofilm.
bRiver, free water.
c400 m upstream of the water intake.
dOutlet water of the farm.
The strains collected in 2003–2010 and the inlet water strains were genetically characterized in an earlier study using multilocus sequence analysis (MLSA) [25] and automated ribosomal intergenic spacer analysis (ARISA) [30]. The MLSA method produces genetic clustering comparable with the ARISA method [25], and the two methods can be used interchangeably. The outlet water strains and one inlet water strain (B355) were genotyped in this study using ARISA, as described earlier [30,32] (see the electronic supplementary material for more details). The possible presence of plasmids in the strains was studied from 3.8 ml of overnight-grown turbid cultures using a QIAprep Spin Miniprep kit (Qiagen) following the manufacturer's instructions. Flavobacterium sp. strain B330 harbouring a natural plasmid was used as a positive control. DNA was run in 0.8% agarose gel and visualized under UV light to detect the presence/absence of plasmids.
(c). Interference competition assays
The inhibitory activity of F. columnare strains was studied both in time (17 strains isolated in 2003–2010) and space (nine inlet water strains versus nine outlet water strains). Within each set of strains, inhibition was tested reciprocally using a double layer method and the assays were replicated three (strains from 2003 to 2010) or four (inlet versus outlet water strains from 2010) times. The optical density (OD, at 570 nm) of the bacterial cultures was measured with spectrophotometer and adjusted between 0.250 and 0.290. Three hundred microlitres of fresh overnight-grown ‘recipient’ bacterial culture was mixed with 3 ml of soft Shieh agar (0.7%) tempered to 47°C and poured on Shieh agar plates. Aliquot of the bacterial culture was centrifuged at 17 000g for 3 min in RT. Five microlitres of the supernatant of the ‘donor’ cultures were spotted on the surface of the top agar. Following an incubation of 48 h at RT, the plates were checked to see whether the ‘donor’ strain had caused a growth inhibition of the underlying ‘recipient’ bacterial lawn. The inhibition was ranked as 0, no inhibition; 1, inhibition.
The interaction between F. columnare strains B067 and B185 was further characterized with filtered supernatant (0.2 µm PES filter, VWR) and by cross-streaking on agar plates in 10 replicates. In the first experiment, turbid overnight-grown liquid culture and filtered supernatant of strain B185 were cross-streaked with B067 by using 1 µl loop. In the second experiment, 300 µl of turbid liquid culture of B067 was mixed with Shieh soft agar and plated, and 10 µl of B185 culture and filtered supernatant was applied on the soft agar. Growth inhibition was monitored after 48 h incubation.
(d). Bacterial growth measurements
A temperature-controlled spectrophotometer (Bioscreen C, Growth Curves Ltd, Helsinki, Finland) was used to monitor the growth of the bacterial strains. Before the growth measurements of the both study sets, the optical densities of the fresh overnight-grown bacterial cultures were adjusted to 0.10–0.20 in A570 to minimize the differences in the initial turbidity between strains. Forty microlitres of each bacterial strain was inoculated onto 400 µl of sterile Shieh culture media on a BioScreen Honeycomb plate (100-well plate, Oy Growth Curves Ab Ltd) in five replicates per strain, both in normal and diluted medium. The growth data were measured at 25°C for 96 h at 5 min intervals (absorbance at 420–580 nm, wide band option).
The growth parameters were calculated from the raw data by utilizing MATLAB script written by T.K. in which the maximal growth rate is found from log2-transformed data by fitting linear regressions on 25 time-point sliding windows. The highest linear (log transformation linearizes the exponential growth) slope found in sliding windows equals the maximal growth. The yield is found as a maximal average OD over 25 time-point sliding windows in the raw data. The area under curve (AUC) sums the OD data over the entire measurement period to indicate the cumulative amount of biomass attained during the time.
(e). Bacterial competition in liquid culture
We also studied competition in liquid culture (in finite resources) using the strains isolated in 2003–2010. Cultures and measurements were done similarly as above with the single strains, but with 1 : 1 mixture of bacterial strain pairs (compared with individual bacterial strain bacteria diluted 1 : 1 with dH2O) and OD at 600 nm was measured every 5 min for 95 h. The AUC, a measure that describes the cumulating amount of biomass a given strain and strain–strain combinations can produce within a given time, gave roughly the same results as the yield. From the obtained AUC, we calculated interaction indexes for the bacterial strains as
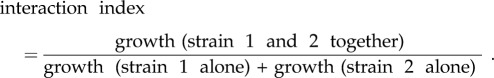 |
For the data analysis, the interaction index was arcsin-transformed (see data analysis below).
(f). Virulence in zebra fish
To measure the virulence of the studied bacterial strains, unsexed, adult, disease-free zebra fish were obtained from core facilities (COFA) and research services of Tampere (Tampere University, Finland). The zebra fish response to F. columnare infection is qualitatively similar to the common host of the pathogen in aquaculture, rainbow trout [33]. The OD of the overnight-grown bacterial culture (570 nm) was measured and the corresponding bacterial density in colony forming units (cfu) was calculated according to our previously fitted standard curve. The infection method and bacterial dose used were optimized in preliminary experiments.
The bacterial exposure with nine inlet water strains and nine outlet water strains was done using 14 replicate fish per strain. In addition, 14 control fish received sterile growth medium and served as a negative control group. In total, 266 individual zebra fish were used in the experiment. Five millilitres of sterile Shieh culture medium mixed with bacterial culture (pure culture medium in the negative control) was pipetted directly into each aquarium to reach the infective dose of 1 × 104 colony forming units (cfu) ml−1 in the water throughout the experiment, as a continuous exposure. The fish were then monitored for 11 d for disease symptoms and morbidity. For the first 3 days, during the most acute phase of the disease, the fish were monitored every hour and when the progression of the epidemic ceased the monitoring points were decreased accordingly, including at least two checks per day.
The virulence of strains isolated in 2003–2010 was tested similarly but with small modifications. Infection with each bacterial strain (11 strains of the dataset) was done to 10 replicate fish. In addition, 10 fish in the negative control group were exposed to sterile culture medium, thus the total number of individual fish in this experiment was 120. In this experiment, the bacteria were mixed in 550 µl of Shieh medium and pipetted into aquaria to reach a dose of 2.5 × 105 cfu ml−1. The fish were monitored every hour for 40 hours.
During the experiments the fish were held in individual 750 ml plastic aquaria with 500 ml of ground water (average t = 24.7°C). Morbid fish that had lost their natural swimming buoyancy and did not respond to external stimuli were considered dead and removed from the experiment. All the remaining healthy fish at the end of the experiment were euthanized by cutting the spinal cord under terminal aenaesthesia with MS-222 (Sigma). All fish were weighted and a bacterial culture sample was taken from the gills on Shieh agar supplemented with tobramycin [26] to ensure the cause of death to be columnaris disease.
(g). Data-analyses
The inhibition data of inlet water and outlet water strains were analysed using generalized linear mixed models implemented in function glm, in R. Inhibition data were modelled with quasi-Poisson distribution, explaining pooled replicates (within each reciprocal combination, considered as separate combination) with four-levels fixed factor representing all combinations of receiver and donor being an inlet or outlet strains (without correction for multiple comparisons). The inhibition data of strains isolated in 2003–2010 were analysed with a similar method to that from averaged data, but using year of isolation of donor and recipient strains as continuous covariates (both rank-transformed). The replicates were pooled due to problems in mixed model fitting of random effects of strain combination that prevented accurate estimates of 2003–2010 data. The results obtained from mixed models, in both datasets, give equivalent biological interpretation of the data that is presented here.
In competition in liquid culture there are no producer or receiver strains that could be distinguished, and we could only test the effects of average isolation year and difference of the isolation years on the interaction index, in addition to the strain combination identity. The growth traits of all bacterial isolates, and the competition indices for time series 2003–2010, were analysed with mixed models in SPSS.
Virulence of bacterial strains in both of the datasets (time series and the inlet versus outlet water strains) was first analysed by Kaplan–Meier survival analysis (log-rank Mantel cox). The fish alive at the end of experiments were treated as censored data. In addition to the log-rank analysis, we also used average longevity as a measure of virulence with strains isolated in 2003–2010. However, in the dataset of the inlet and outlet water strains only 10% of the fish infected with the inlet water strains (and 40% infected with the outlet water strains) entered a moribund state during the experiment, precluding the use of the fish longevity as a surrogate of virulence. Therefore, for the inlet and outlet water strains, we used the log-rank Mantel cox survival analysis, and also arcsin-square-transformed mortality percentage as a measure of virulence. The mortality percentages of fish were tested with t-tests allowing for the unequal variances [34].
3. Results
(a). Temporal evolution in Flavobacterium columnare strains isolated in 2003–2010
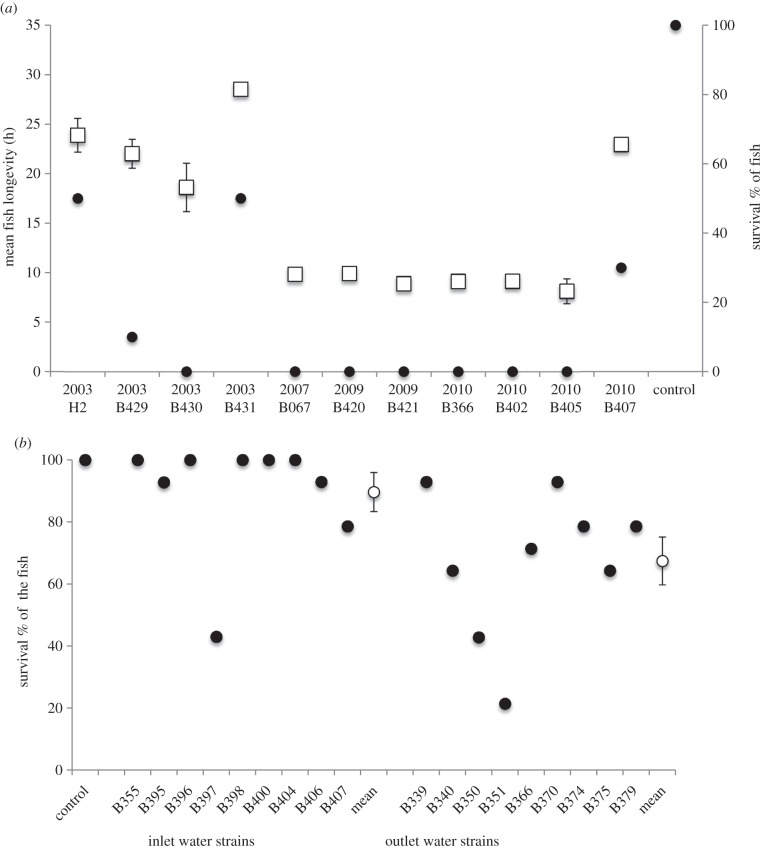
Inhibitory toxin production, competitive ability and virulence were significantly associated with the time of isolation in strains from different fish farms and environmental locations (2003–2010). The isolation year had a significant effect on bacterial virulence (log-rank Mantel cox, χ2 = 55.338, d.f. = 3, p < 0.001). The more recently isolated bacteria were significantly more virulent (average longevity of the infected fish) than the ones isolated earlier (b = −1.716, p = 0.014; figure 1a). In line with the virulence, the more recently isolated strains were more likely to inhibit the earlier isolates (b = 0.004, p = 0.0181). The sensitivity of the bacteria to the interference by the other strains was independent of the isolation time (year (rank-transformed) of isolation of the inhibited isolate b = 0.002, p = 0.2975; see electronic supplementary material, table S2a for original data). The year of isolation did not have an effect on the bacterial growth parameters (maximum growth rate, yield or AUC), whereas the strain identity did (electronic supplementary material, table S3).
Figure 1.
Virulence of Flavobacterium columnare in experimentally infected zebra fish (Danio rerio). (a) The mean longevity (±s.e., open squares, left axis) and survival percentage (filled circles, right axis) of the infected zebra fish (n = 10, in each bacterial exposure) after exposure to bacterial strains isolated in 2003–2010. (b) Survival percentage of zebra fish (n = 14 in each bacterial exposure) after exposure to bacterial strains isolated from inlet water (B355–B407) and outlet water (B399–B397) of a fish farm (filled circles for individual strains, open circles for mean survival ± s.e.).
To elaborate the benefits of interference in resource competition, we studied the intensity of competition between the isolates in co-culture, where the competition on finite resources is expected to lead to lower production of biomass if the competing strains use the same resource. The competition was more intense (indicated by lower interaction index) between strains that on average had been isolated recently than between the strains that were isolated earlier (table 3 and figure 2). The difference in the isolation times of the competing strains did not significantly affect the interaction index (table 3).
Table 3.
Effects of the average year of isolation and the difference of the isolation years on growth inhibition (i.e. interference, studied on agar plates) and competition (interaction studied in liquid medium) in pairs of Flavobacterium columnare isolates, collected during disease epidemics in 2003–2010.
| parameter | estimate | s.e. | d.f. | t | sig. |
|---|---|---|---|---|---|
| inhibition | |||||
| average year of isolation | 0.0034 | 0.002 | 2.078 | 0.039 | |
| difference in years of isolation | −0.0001 | 0.002 | 0.764 | 0.446 | |
| interaction indexa | |||||
| average year of isolation | −0.0255 | 0.009 | 104.707 | −2.759 | 0.007 |
| difference in years of isolation | 0.0024 | 0.007 | 103.641 | 0.355 | 0.724 |
aStrain combination: 0.0215, s.e.: 0.0036, Wald Z: 5.911, p < 0.001, residual: 0.008, s.e.: 0.001, Wald Z: 6.959, p < 0.001.
Figure 2.
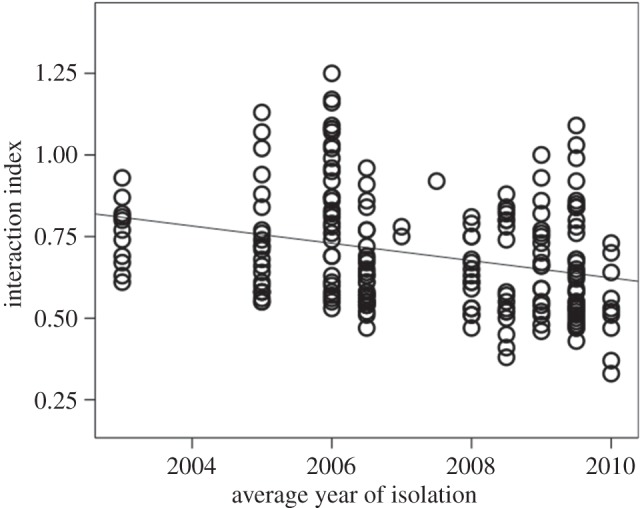
Changes in (arcsin-transformed) interaction index of the competing pairs of Flavobacterium columnare strains on limited resources in relation to the mean isolation time of the pair of isolates. Lower index indicates higher competition. Circle indicates the mean of the two replicates of the tested F. columnare strain pairs.
The cross-streaking experiments of the strains B185 and B067 indicated that toxin production in F. columnare could be contact-dependent. When cross-streaked on agar-plate, in nine out of ten replicates clear inhibitory zones (similar to Dienes lines) [35,36] were observed (electronic supplementary material, figure S1), but only when supernatant containing bacterial cells was used. Sterile filtered supernatant did not cause any inhibition in any of the experiments.
(b). Local, short-time-scale evolution
The farming environment was observed to have a significant impact on the population structure of the bacterial populations occurring in the inlet and outlet water. The bacterial population from the outlet water of the fish farm was genetically more homogeneous than the population from the inlet water upstream of the farm (table 2). Some of the genotypes in the inlet water were not detected in outlet water, indicating that the fish farming environment may select for specific genotypes.
Place of isolation (inlet or outlet water of a fish farm) had a significant effect on bacterial virulence (log-rank Mantel cox, χ2 = 33.471, d.f. = 2, p < 0.001). Mean percentage of mortality in fish infected with the outlet water isolates was 32.5%, and 10.3% with the inlet water strains (t = 3.155, p = 0.006, on arcsin-square transformed proportion of the dead fish; figure 1b). No background mortality in the fish exposed to sterile growth medium (negative control group) was observed.
When the outlet water isolates acted as donors in an inhibition test against the inlet water isolates, the risk of inhibition (0.225) seemed to be higher (z = −2.154, p = 0.033) than when the outlet water strains were let to inhibit other outlet water strains (0.167). Other types of inhibition pairs produced intermediate inhibition risks and did not statistically differ from the other pairs (table 4; electronic supplementary material, table S2b). Since level of significance (p = 0.033) of the only significant pairwise comparison is not dramatically different from 0.05, there is a possibility that significance is overstated without corrections for multiple testing, leading to acceptance error (type I error). However, several papers suggest that multiple corrections could very easily lead to rejection errors (type II), especially with small datasets, as here [37]. Hence, as we have not adopted corrections the obtained result should be considered tentative.
Table 4.
Pairwise comparisons of risk of inhibition (interference competition) by inlet water and outlet water strains in different combinations of donors and receivers (each reciprocal combination considered as separate combination). I, inlet water strains isolated from upstream of a fish farm; O, strains isolated from outlet water or downstream of the farm.
| donor O, receiver I | donor I, receiver O | donor O, receiver O | |
|---|---|---|---|
| donor I, receiver I | z = 1.084, p = 0.2790 | z = 0.098, p = 0.922 | z = −1.086, p = 0.278 |
| donor O, receiver I | z = −0.986, p = 0.325 | z = −2.154, p = 0.033 | |
| donor I, receiver O | z = 1.183, p = 0.238 |
To examine the effects of resource concentrations on bacterial growth we studied the growth of the inlet and the outlet water bacterial strains in two resource concentrations: in the standard growth medium and in the diluted (0.5×) medium. In yield and AUC, the outlet water strains (yield: 0.853, s.e.: 0.052; AUC: 647.6, s.e.: 40.98) excelled over the inlet water strains (yield: 0.668, s.e.: 0.052; AUC: 486.6, s.e.: 40.98), but the growth rates of the two groups were comparable (inlet water: 0.216, s.e.: 0.012; outlet water: 0.199, s.e.: 0.012; table 5; electronic supplementary material, figure S2). Interestingly, though, the growth of the outlet water strains was more sensitive to the differences in resource concentrations (low resource: 0.176, s.e.: 0.013; high resource: 0.222, s.e.: 0.013; table 5) than the growth of the inlet water strains (low resource: 0.208, s.e.: 0.013; high resource: 0.225, s.e.: 0.013).
Table 5.
Results of the mixed model analysis exploring the growth differences between the inlet water and the outlet water isolates (location) of F. columnare measured in high and low resource concentrations. The model also contains a random effect of the strain identity to control for the non-independency of observations arising from repeated growth measurements.
| F | d.f.1,d.f.2 | p-value | |
|---|---|---|---|
| maximal growth rate (change in OD h−1) | |||
| location | 0.968 | 116.054 | 0.34 |
| resource concentration | 27.113 | 1163.047 | <0.001 |
| location × resource concentration | 5.826 | 1163.047 | 0.017 |
| strain identity | Wald Z: | 2.477 | 0.013 |
| yield | |||
| location | 6.372 | 115.989 | 0.023 |
| resource concentration | 173.173 | 1163.002 | <0.001 |
| location × resource concentration | 0.004 | 1163.002 | 0.947 |
| strain identity | Wald Z: | 2.729 | 0.006 |
| AUC | |||
| location | 7.724 | 115.989 | 0.013 |
| resource concentration | 136.565 | 1163.002 | <0.001 |
| location × resource concentration | 1.302 | 1163.002 | 0.255 |
| strain identity | Wald Z: | 2.722 | 0.006 |
Fifteen bacterial strains (including strains from both datasets) were screened for the presence of plasmids, but none were observed (electronic supplementary material, figure S3).
4. Discussion
Since the mid-1990s F. columnare outbreak frequency, severity of symptoms and disease-related mortality have significantly increased in aquaculture, and the evolution of virulence has been suggested as an explanation [23]. Our study finds that the bacterial strains isolated most recently, during the period of more difficult disease outbreaks, are more virulent and have a higher competitive ability than the strains isolated earlier. In addition, our findings about the F. columnare populations from the inlet and outlet water of a fish farm are congruent with the suggestion of selection of more virulent strains in aquaculture. The strains isolated from outlet water were more virulent and able to produce higher maximum population sizes than the inlet water strains, indicating that the bacteria with the best ability to exploit fish populations benefit the most in the intensive aquaculture conditions.
Opportunistic lifestyle and ability to persist outside a host opens less stringent trajectories for virulence evolution, in contrast with the obligate pathogens restricted by the transmission–virulence trade-off [8], but factors that increase or limit virulence evolution in opportunistic pathogens are still poorly understood. F. columnare can be considered an opportunist, as it is known to survive long periods outside the host and transmit efficiently from dead hosts [38–40]. A weak trade-off between virulence and other fitness traits, like transmission, provided by the outside-host survival could thus promote the evolution of high virulence in F. columnare. Moreover, in aquaculture, new susceptible fish populations are introduced at the farm annually, creating conditions similar to serial passage, which further decreases the costs of virulence [9].
In addition to the factors related to host abundance, the bacterial communities in aquaculture are (in contrast to communities in nature) shaped by increased concentrations of nutrients, and chemical and antibiotic treatments within the rearing units. Eutrophication of the aquatic environment increases parasitic and bacterial diseases via direct and indirect effects in the food web [41,42]. While the role of eutrophication on F. columnare epidemics is still unclear, the increased nutrient concentrations may support the outside-host growth of this pathogen, giving the more virulent strains a greater advantage (electronic supplementary material, figure S2; see also [43]). On the other hand, the use of chemotherapy can relax the competitive interactions between bacteria by eliminating sensitive species. This may lead to more frequent or virulent outbreaks of opportunistic diseases if the use of antibiotics increases the intensity of the within-species competition [44] or selects for faster transmission rate [45]. The increasing amount of antibiotics used in food production [46] may thus affect microbial communities beyond the traditionally expected environmental effects [47–49]. Therefore, to secure the global food production it is vital to understand the factors that select for virulent pathogen strains.
On top of intensive farming practices per se, other ecological and evolutionary factors also underpin the evolution of more virulent and competitive bacteria at long time scales. A general increasing trend in disease diversity and outbreak frequency has been observed during recent decades, but the reasons for this are still largely unknown [50]. One contributing factor is the warming climate, which causes changes in disease ecology, outbreak dynamics and seasonality [51–53]. Owing to global warming, the longer outbreak period for columnaris disease [23] increases bacterium–host and bacterium–bacterium interactions, allowing greater opportunities for evolution of both virulence and competitive ability.
Strain–strain interactions can have a significant role in bacterial disease dynamics via competition in both within-host and outside-host environments [54–58], and the surrounding microbial community has been shown to have significant effects on the evolution of interference [59,60]. However, how interference competition is associated with intensive farming is not properly understood. Our data suggest that the most recently isolated bacteria seem to have the highest capacity for interference competition and, on average, the competition between the most recently isolated strains is the most intense. Although the strains isolated from the inlet water were able to inhibit each other and the outlet water strains, significant difference in inhibition was observed only when the outlet water isolates acted as inhibitors. The strains isolated from outlet water seemed to be able to inhibit the growth of the inlet water strains but tolerated well the toxins produced by other outlet water strains, probably due to more homogeneous population structure resulting in less competitive interactions [61]. While these results are in accordance with the general expectations of the evolution of interference competition in mixed populations [54,57], whether these interactions are relevant during disease outbreaks is unknown, and type I error in the interpretation of the results is possible. Nevertheless, different bacterial population structure was observed in these two locations even during the same sampling dates that are directly comparable (table 2). These results suggest that the competition pressures differ within and outside the farming environment, and that the interference, as well as toxin tolerance, could be beneficial for the virulent strains in the farming environment where invasions by multiple strains are frequent.
Although this study does not aim to characterize the mechanism of growth inhibition, bacteriocins have been reported previously in F. columnare [62]. Our data, however, indicate that the toxin production in F. columnare may require a direct contact between bacterial cells (see electronic supplementary material figure S1), but more studies are needed to identify the cell–cell interactions in this species in detail.
Previous experimental studies have often demonstrated a direct trade-off between toxin production and both growth rate and virulence. Virulence of co-infection with a toxin-producing and a toxin-sensitive bacterial strain leads to a decrease in the total virulence of infection [18,19,63]. In contrast with traditional assumptions, our data show no evidence of costs in toxin production for growth (growth rate, population size) or virulence in F. columnare. Although the competitive ability and virulence increased in time, the bacteria isolated in 2003–2010 did not differ in their growth features. Similarly, the growth rate of the virulent bacteria isolated from the outlet water did not exceed the growth rate of the less competitive and low-virulence inlet water strains. However, the outlet water population reached higher population sizes regardless of the nutrient conditions. It seems that in this study system the same factors selecting for increased virulence might simultaneously also select for increased competitive ability. It remains unknown whether virulence and competitive ability are genetically linked, and whether their benefits are traded off with other life-history traits.
To conclude, in accordance with the theoretical predictions [1,7–9,21], our data are consistent with the hypothesis that intensive farming conditions (high host densities, increased transmission opportunities, co-infections, possibility for serial passage, availability of nutrients, use of chemotherapy) can select for pathogen strains with the ability to produce large population sizes with high virulence that have high competitive ability under short time scales. This indicates a genetic difference in populations of high- and low-virulence bacterial strains resulting in selection for strains with an increased ability to exploit the fish host as a nutrient source. To reveal the possibility of horizontal gene transfer by conjugation as a mechanism for increased virulence, we assayed the presence of plasmids in the F. columnare bacterial strains, but, similarly to previous studies [29], none were found. Therefore, other possible genetic mechanisms causing the changes in virulence (transduction, chromosomal transformation, mutations) [64] remain to be solved, and will require whole-genome sequencing of several strains. Also, the recent achievements in genetic manipulation techniques [65,66] and genome sequencing [67] are likely to provide detailed insight into the mechanisms behind F. columnare pathogenicity.
Our results indicate selection for pathogen virulence and competitive ability in both long and short time scales. The bacterial strains isolated in 2003 were significantly less virulent than the strains isolated later, which correlates with the observed increase in the severity of columnaris outbreaks during the last decade [23]. Interestingly, a similar pattern was observed already during one outbreak season, as the strains isolated from the outlet water of a fish farm were more virulent than strains isolated from the inlet water. These results indicate that the selection pressures at fish farms can cause changes in pathogen populations, which may have long-lasting effects on pathogen virulence. The global changes in nutrients and climate can further select pathogen traits at a wider temporal scale. Indeed, we cannot rule out the possibility that the changes in bacterial characteristics observed in this paper are unrelated to aquaculture. Nevertheless, aquaculture has a major role in securing world protein production, but disease epidemics severely affect the profitability of the industry [68–70]. Understanding disease ecology and evolution in man-made environments is important in securing sustainable livestock and aquaculture production. In addition to the benefits in the applied field, the studies on pathogens in the intensive farming systems can provide much-needed empirical field data on the evolution of virulence.
Supplementary Material
Acknowledgements
We thank Dr Heidi Kunttu and Dr Päivi Rintamäki for donating the bacterial isolates used in this study, Dr Kunttu for help during the experiments, Dr Nina Pekkala, Mr Petri Papponen and Katja Neuvonen for technical assistance in the laboratory, and Dr Emily Burfield-Steel and Dr Andrés López-Sepulcre for helpful comments on the manuscript.
Ethics
The experiments were conducted according to the Finnish Act on the Use of Animals for Experimental Purposes, under permission ESAVI-2010–05569/Ym-23 granted for L-RS by the National Animal Experiment Board at the Regional State Administrative Agency for Southern Finland.
Data accessibility
All data used in this paper are publicly available in electronic supplementary material and in Dryad: http://dx.doi.org/10.5061/dryad.nk76k.
Authors' contributions
L.-R.S., T.K., J.K.H.B. and J.M. designed the study. L.-R.S., T.K., E.L., H.K. and R.P. collected and analysed the data. All authors participated in drafting and critically revising the manuscript. All authors approved the final version of the manuscript.
Competing interests
Authors do not have competing interests.
Funding
This work was supported by the Finnish Centre of Excellence Program of the Academy of Finland, CoE in Biological Interactions 2012–2017 (no. 252411, J.M. and J.K.H.B.), by Academy of Finland grants nos.272995 (L.-R.S.) and 278751 (T.K.) and by Maj and Tor Nessling Foundation (J.K.H.B.).
References
- 1.Ebert D, Herre E. 1996. The evolution of parasitic diseases. Parasitol. Today 12, 96–101. ( 10.1016/0169-4758(96)80668-5) [DOI] [PubMed] [Google Scholar]
- 2.Frank SA. 1996. Models of parasite virulence. Q. Rev. Biol. 71, 37–78. ( 10.1086/419267) [DOI] [PubMed] [Google Scholar]
- 3.Altizer S, Harvell D, Friedle E. 2003. Rapid evolutionary dynamics and disease threats to biodiversity. Trends Ecol. Evol. 18, 589–596. ( 10.1016/j.tree.2003.08.013) [DOI] [Google Scholar]
- 4.Galvani AP. 2003. Epidemiology meets evolutionary ecology. Trends Ecol. Evol. 18, 132–139. ( 10.1016/S0169-5347(02)00050-2) [DOI] [Google Scholar]
- 5.Schrag S, Wiener P. 2004. Emerging infectious disease: what are the relative roles of ecology and evolution? Trends Ecol. Evol. 10, 319–324. ( 10.1016/S0169-5347(00)89118-1) [DOI] [PubMed] [Google Scholar]
- 6.Peeler EJ, Feist SW. 2011. Human intervention in freshwater ecosystems drives disease emergence. Freshw. Biol. 56, 705–716. ( 10.1111/j.1365-2427.2011.02572.x) [DOI] [Google Scholar]
- 7.Kennedy DA, Kurath G, Brito IL, Purcell MK, Read AF, Winton JR, Wargo AR. 2015. Potential drivers of virulence evolution in aquaculture. Evol. Appl. 9, 344–354 ( 10.1111/eva.12342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown SP, Cornforth DM, Mideo N. 2012. Evolution of virulence in opportunistic pathogens: generalism, plasticity, and control. Trends Microbiol. 20, 336–342. ( 10.1016/j.tim.2012.04.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ebert D. 1998. Experimental evolution of parasites. Science 282, 1432–1436. ( 10.1126/science.282.5393.1432) [DOI] [PubMed] [Google Scholar]
- 10.Read AF, Taylor LH. 2001. The ecology of genetically diverse infections. Science 292, 1099–1102. ( 10.1126/science.1059410) [DOI] [PubMed] [Google Scholar]
- 11.Woolhouse MEJ, Webster JP, Domingo E, Charlesworth B, Levin BR. 2002. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat. Genet. 32, 569–577. ( 10.1038/ng1202-569) [DOI] [PubMed] [Google Scholar]
- 12.Karvonen A, Rellstab C, Louhi K-R, Jokela J. 2012. Synchronous attack is advantageous: mixed genotype infections lead to higher infection success in trematode parasites. Proc. R. Soc. B 279, 171–176. ( 10.1098/rspb.2011.0879) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leggett HC, Benmayor R, Hodgson DJ, Buckling A. 2013. Experimental evolution of adaptive phenotypic plasticity in a parasite. Curr. Biol. 23, 139–142. ( 10.1016/j.cub.2012.11.045) [DOI] [PubMed] [Google Scholar]
- 14.Susi H, Barrès B, Vale PF, Laine A-L. 2015. Co-infection alters population dynamics of infectious disease. Nat. Comm. 6, 5975 ( 10.1038/ncomms6975) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster KR, Bell T. 2012. Competition, not cooperation, dominates interactions among culturable microbial species. Curr. Biol. 22, 1845–1850. ( 10.1016/j.cub.2012.08.005) [DOI] [PubMed] [Google Scholar]
- 16.Brown SP, West SA, Diggle SP, Griffin AS. 2009. Social evolution in micro-organisms and a Trojan horse approach to medical intervention strategies. Phil. Trans. R. Soc. B 364, 3157–3168. ( 10.1098/rstb.2009.0055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. 2009. Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Micro. 8, 15–25. ( 10.1038/nrmicro2259) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Massey RC, Buckling A, Ffrench-Constant R. 2004. Interference competition and parasite virulence. Proc. R. Soc. B 271, 785–788. ( 10.1098/rspb.2004.2676) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inglis RF, Gardner A, Cornelis P, Buckling A. 2009. Spite and virulence in the bacterium Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA 106, 5703–5707. ( 10.1073/pnas.0810850106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nowak BF. 2007. Parasitic diseases in marine cage culture—an example of experimental evolution of parasites? Int. J. Parasitol. 37, 581–588. ( 10.1016/j.ijpara.2007.01.003) [DOI] [PubMed] [Google Scholar]
- 21.Mennerat A, Nilsen F, Ebert D, Skorping A. 2010. Intensive farming: evolutionary implications for parasites and pathogens. Evol. Biol. 37, 59–67. ( 10.1007/s11692-010-9089-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurath G, Winton J. 2011. Complex dynamics at the interface between wild and domestic viruses of finfish. Curr. Opin. Virol. 1, 73–80. ( 10.1016/j.coviro.2011.05.010) [DOI] [PubMed] [Google Scholar]
- 23.Pulkkinen K, Suomalainen LR, Read AF, Ebert D, Rintamaki P, Valtonen ET. 2010. Intensive fish farming and the evolution of pathogen virulence: the case of columnaris disease in Finland. Proc. R. Soc. B 277, 593–600. ( 10.1098/rspb.2009.1659) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Declercq AM, Haesebrouck F, Van den Broeck W, Bossier P, Decostere A. 2013. Columnaris disease in fish: a review with emphasis on bacterium-host interactions. Vet. Res. 44, 27 ( 10.1186/1297-9716-44-27) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashrafi R, Pulkkinen K, Sundberg L-R, Pekkala N, Ketola T. 2015. A multilocus sequence analysis scheme for characterization of Flavobacterium columnare isolates. BMC Microbiol. 15, 245 ( 10.1186/s12866-015-0576-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song YL, Fryer JL, Rohovec JS. 1988. Comparison of six media for the cultivation of Flexibacter columnaris. Fish Pathol. 23, 91–94. ( 10.3147/jsfp.23.91) [DOI] [Google Scholar]
- 27.Decostere A, Haesebrouck F, Devriese L. 1997. Shieh medium supplemented with tobramycin for selective isolation of Flavobacterium columnare (Flexibacter columnaris) from diseased fish. J. Clin. Microbiol. 35, 322–324. ( 10.1111/j.1365-2761.2006.00771.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anacker RL, Ordal EJ. 1959. Studies on the myxobacterium Chondrococcus columnaris: I. Serological typing. J. Bacteriol. 78, 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suomalainen LR, Tiirola M, Valtonen ET. 2006. Chondroitin AC lyase activity is related to virulence of fish pathogenic Flavobacterium columnare. J. Fish Dis. 9, 757–763. [DOI] [PubMed] [Google Scholar]
- 30.Kunttu HMT, Sundberg L-R, Pulkkinen K, Valtonen ET. 2012. Environment may be the source of Flavobacterium columnare outbreaks at fish farms. Environ. Microbiol. Rep. 4, 398–402. ( 10.1111/j.1758-2229.2012.00342.x) [DOI] [PubMed] [Google Scholar]
- 31.Laanto E, Bamford JKH, Laakso J, Sundberg L-R. 2012. Phage-driven loss of virulence in a fish pathogenic bacterium. PLoS ONE 7, e53157 ( 10.1371/journal.pone.0053157) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suomalainen LR, Kunttu H, Valtonen ET, Hirvela-Koski V, Tiirola M. 2006. Molecular diversity and growth features of Flavobacterium columnare strains isolated in Finland. Dis. Aquat. Organ. 70, 55–61. ( 10.3354/dao070055) [DOI] [PubMed] [Google Scholar]
- 33.Kinnula H, Mappes J, Valkonen J, Sundberg L-R. 2015. Influence of infection dose on the virulence of a generalist pathogen in a common and a novel host. PLoS ONE 10, e0139378 ( 10.1371/journal.pone.0139378.t003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruxton GD. 2006. The unequal variance t-test is an underused alternative to Student's t-test and the Mann–Whitney U test. Behav. Ecol. 17, 688–690. ( 10.1093/beheco/ark016) [DOI] [Google Scholar]
- 35.Gibbs KA, Urbanowski ML, Greenberg EP. 2008. Genetic determinants of self identity and social recognition in bacteria. Science 321, 256–259. ( 10.1126/science.1160033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Budding AE, Ingham CJ, Bitter W, Vandenbroucke-Grauls CM, Schneeberger PM. 2009. The Dienes phenomenon: competition and territoriality in swarming Proteus mirabilis. J. Bacteriol. 191, 3892–3900. ( 10.1128/JB.00975-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moran DM. 2003. Arguments for rejecting the sequential Bonferroni in ecological studies. Oikos 100, 403–405. ( 10.1034/j.1600-0706.2003.12010.x) [DOI] [Google Scholar]
- 38.Kunttu HMT, Valtonen ET, Jokinen EI, Suomalainen L-R. 2009. Saprophytism of a fish pathogen as a transmission strategy. Epidemics 1, 96–100. ( 10.1016/j.epidem.2009.04.003) [DOI] [PubMed] [Google Scholar]
- 39.Arias CR, Lafrentz S, Cai W, Olivares-Fuster O. 2012. Adaptive response to starvation in the fish pathogen Flavobacterium columnare: cell viability and ultrastructural changes. BMC Microbiol. 12, 266 ( 10.1186/1471-2180-12-266) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sundberg L-R, Kunttu HMT, Valtonen ET. 2014. Starvation can diversify the population structure and virulence strategies of an environmentally transmitting fish pathogen. BMC Microbiol. 14, 1–6. ( 10.1186/1471-2180-14-67) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKenzie VJ, Townsend AR. 2007. Parasitic and infectious disease responses to changing global nutrient cycles. EcoHealth 4, 384–396. ( 10.1007/s10393-007-0131-3) [DOI] [Google Scholar]
- 42.Johnson PTJ, Chase JM, Dosch KL, Hartson RB, Gross JA, Larson DJ, Sutherland DR, Carpenter SR. 2007. Aquatic eutrophication promotes pathogenic infection in amphibians. Proc. Natl Acad. Sci. USA 104, 15 781–15 786. ( 10.1073/pnas.0707763104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ketola T, Mikonranta L, Laakso J, Mappes J. 2016. Different food sources elicit fast changes to bacterial virulence. Biol. Lett. 12, 20150660 ( 10.1016/j.epidem.2009.04.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anttila J, Ruokolainen L, Kaitala V, Laakso J. 2013. Loss of competition in the outside host environment generates outbreaks of environmental opportunist pathogens. PLoS ONE 8, e71621 ( 10.1371/journal.pone.0071621.s004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mennerat A, Hamre L, Ebert D, Nilsen F, Dávidová M, Skorping A. 2012. Life history and virulence are linked in the ectoparasitic salmon louse Lepeophtheirus salmonis. J. Evol. Biol. 25, 856–861. ( 10.1111/j.1420-9101.2012.02474.x) [DOI] [PubMed] [Google Scholar]
- 46.Van Boeckel TP, Brower C, Gilbert M, Grenfell BT, Levin SA, Robinson TP, Teillant A, Laxminarayan R. 2015. Global trends in antimicrobial use in food animals. Proc. Natl Acad. Sci. USA 112, 5649–5654. ( 10.1073/pnas.1503141112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamminen M, Karkman A, Lõhmus A, Muziasari WI, Takasu H, Wada S, Suzuki S, Virta M. 2011. Tetracycline resistance genes persist at aquaculture farms in the absence of selection pressure. Environ. Sci. Technol. 45, 386–391. ( 10.1021/es102725n) [DOI] [PubMed] [Google Scholar]
- 48.Di Cesare A, Luna GM, Vignaroli C, Pasquaroli S, Tota S, Paroncini P, Biavasco F. 2013. Aquaculture can promote the presence and spread of antibiotic-resistant enterococci in marine sediments. PLoS ONE 8, e62838 ( 10.1371/journal.pone.0062838.t003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schubert AM, Sinani H, Schloss PD. 2015. Antibiotic-induced alterations of the murine gut microbiota and subsequent effects on colonization resistance against clostridium difficile. MBio 6, e00974–e01015. ( 10.1128/mBio.00974-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith KF, Goldberg M, Rosenthal S, Carlson L, Chen J, Chen C, Ramachandran S. 2014. Global rise in human infectious disease outbreaks. J. R. Soc. Interface 11, 20140950 ( 10.1098/rsif.2014.0950) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altizer S, dobson A, Hosseini P, Hudson P, Pascual M, Rohani P. 2006. Seasonality and the dynamics of infectious diseases. Ecol. Lett. 9, 467–484. ( 10.1111/j.1461-0248.2005.00879.x) [DOI] [PubMed] [Google Scholar]
- 52.Harvell CD, Mitchell CE, Ward JR, Altizer S, Dobson AP, Ostfeld RS, Samuel MD. 2002. Climate warming and disease risks for terrestrial and marine biota. Science 296, 2158–2162. ( 10.1126/science.1063699) [DOI] [PubMed] [Google Scholar]
- 53.Karvonen A, Rintamäki P, Jokela J, Valtonen ET. 2010. Increasing water temperature and disease risks in aquatic systems: climate change increases the risk of some, but not all, diseases. Int. J. Parasitol. 40, 1483–1488. ( 10.1016/j.ijpara.2010.04.015) [DOI] [PubMed] [Google Scholar]
- 54.Gardner A, West SA, Buckling A. 2004. Bacteriocins, spite and virulence. Proc. R. Soc. B 271, 1529–1535. ( 10.1098/rspb.2004.2756) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hawlena H, Bashey F, Mendes Soares H, Lively CM. 2010. Spiteful interactions in a natural population of the bacterium Xenorhabdus bovienii. Am. Nat. 175, 374–381. ( 10.1086/650375) [DOI] [PubMed] [Google Scholar]
- 56.Bashey F, Young SK, Hawlena H, Lively CM. 2012. Spiteful interactions between sympatric natural isolates of Xenorhabdus bovienii benefit kin and reduce virulence. J. Evol. Biol. 25, 431–437. ( 10.1111/j.1420-9101.2011.02441.x) [DOI] [PubMed] [Google Scholar]
- 57.Cordero OX, Wildschutte H, Kirkup B, Proehl S, Ngo L, Hussain F, Le Roux F, Mincer T, Polz MF. 2012. Ecological populations of bacteria act as socially cohesive units of antibiotic production and resistance. Science 337, 1228–1231. ( 10.1126/science.1219385) [DOI] [PubMed] [Google Scholar]
- 58.Pérez-Gutiérrez R-A, et al. 2012. Antagonism influences assembly of a Bacillus guild in a local community and is depicted as a food-chain network. ISME J. 7, 487–497. ( 10.1038/ismej.2012.119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Staves PA, Knell RJ. 2010. Virulence and competitiveness: testing the relationship during inter- and intraspecific mixed infections. Evolution 64, 2643–2652. ( 10.1111/j.1558-5646.2010.00999.x) [DOI] [PubMed] [Google Scholar]
- 60.Cornforth DM, Foster KR. 2013. Competition sensing: the social side of bacterial stress responses. Nat. Rev. Micro 11, 285–293. ( 10.1038/nrmicro2977) [DOI] [PubMed] [Google Scholar]
- 61.Buckling A, Brockhurst MA. 2008. Kin selection and the evolution of virulence. Heredity 100, 484–488. ( 10.1038/sj.hdy.6801093) [DOI] [PubMed] [Google Scholar]
- 62.Anacker RL, Ordal EJ. 1959. Studies on the myxobacterium Chondrococcus columnaris. II. Bacteriocins. J. Bacteriol. 78, 33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garbutt J, Bonsall MB, Wright DJ, Raymond B. 2011. Antagonistic competition moderates virulence in Bacillus thuringiensis. Ecol. Lett. 14, 765–772. ( 10.1111/j.1461-0248.2011.01638.x) [DOI] [PubMed] [Google Scholar]
- 64.Pallen MJ, Wren BW. 2007. Bacterial pathogenomics. Nature 449, 835–842. ( 10.1038/nature06248) [DOI] [PubMed] [Google Scholar]
- 65.Staroscik AM, Hunnicutt DW, Archibald KE, Nelson DR. 2008. Development of methods for the genetic manipulation of Flavobacterium columnare. BMC Microbiol. 8, 115 ( 10.1186/1471-2180-8-115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li N, Qin T, Zhang XL, Huang B, Liu ZX, Xie HX, Zhang J, McBride MJ, Nie P. 2015. Development and use of a gene deletion strategy to examine the two chondroitin lyases in virulence of Flavobacterium columnare. Appl. Environ. Microbiol. 81, 7394–7402. ( 10.1128/AEM.01586-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tekedar HC, et al. 2012. Genome sequence of the fish pathogen Flavobacterium columnare ATCC 49512. J. Bacteriol. 194, 2763–2764. ( 10.1128/JB.00281-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robertsen B. 2011. Can we get the upper hand on viral diseases in aquaculture of Atlantic salmon?. Aquacult. Res. 42, 125–131. ( 10.1111/j.1365-2109.2010.02671.x) [DOI] [Google Scholar]
- 69.Leung TLF, Bates AE. 2012. More rapid and severe disease outbreaks for aquaculture at the tropics: implications for food security. J. Appl. Ecol. 50, 215–222. ( 10.1111/1365-2644.12017) [DOI] [Google Scholar]
- 70.FAO. 2014. The state of world fisheries and aquaculture, opportunities and challenges. Rome, Italy: Food and Agriculture Organization of the United Nations. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this paper are publicly available in electronic supplementary material and in Dryad: http://dx.doi.org/10.5061/dryad.nk76k.