Abstract
The alternatively spliced products of LMNA, lamin C and prelamin A (the precursor to lamin A), are produced in similar amounts in most tissues and have largely redundant functions. This redundancy suggests that diseases, such as Hutchinson-Gilford progeria syndrome (HGPS), that are caused by prelamin A–specific mutations could be treated by shifting the output of LMNA more toward lamin C. Here, we investigated mechanisms that regulate LMNA mRNA alternative splicing and assessed the feasibility of reducing prelamin A expression in vivo. We identified an exon 11 antisense oligonucleotide (ASO) that increased lamin C production at the expense of prelamin A when transfected into mouse and human fibroblasts. The same ASO also reduced the expression of progerin, the mutant prelamin A protein in HGPS, in fibroblasts derived from patients with HGPS. Mechanistic studies revealed that the exon 11 sequences contain binding sites for serine/arginine-rich splicing factor 2 (SRSF2), and SRSF2 knockdown lowered lamin A production in cells and in murine tissues. Moreover, administration of the exon 11 ASO reduced lamin A expression in wild-type mice and progerin expression in an HGPS mouse model. Together, these studies identify ASO-mediated reduction of prelamin A as a potential strategy to treat prelamin A–specific diseases.
Introduction
The nuclear lamina, an intermediate filament meshwork adjacent to the inner nuclear membrane, provides structural support for the cell nucleus, in addition to other important roles within the nucleus, including regulating chromatin structure and gene expression (1). The main protein components of the nuclear lamina are lamin A and lamin C (the A-type lamins) and lamin B1 and lamin B2 (the B-type lamins) (2, 3). All of the nuclear lamins have amino-terminal and carboxyl terminal globular domains flanked by a central coiled-coil rod domain (4, 5), and they self-associate to form higher-order filaments of the nuclear lamina (6).
The LMNA gene consists of 12 exons and yields transcripts for lamin C and prelamin A (the precursor to mature lamin A) by alternative splicing (7). In most cells, lamin C and prelamin A transcripts (and lamin C and lamin A proteins) are produced in similar amounts, but the mechanisms governing LMNA mRNA splicing have not yet been established. Lamin C and prelamin A are identical through their first 566 amino acids (encoded by exons 1–10), but their carboxyl terminal sequences diverge. Lamin C terminates with exon 10 sequences and contains 6 unique carboxyl terminal amino acids; prelamin A contains 98 unique amino acids at its carboxyl terminus (encoded by exons 11–12). The last 4 amino acids of prelamin A (CSIM) trigger protein farnesylation, protein methylation, and endoproteolytic processing steps that convert prelamin A to mature lamin A (8, 9). The processing of prelamin A to lamin A is very efficient; hence, prelamin A is almost undetectable in wild-type cells. The farnesylation of prelamin A and the subsequent processing steps are often assumed to assist in the targeting of lamin A to the nuclear rim (10). However, recent studies with genetically modified mice have shown that direct synthesis of mature lamin A (bypassing prelamin A synthesis and processing) results in no detectable pathology and has no obvious effects on the targeting of lamin A to the nuclear rim (11).
Studies with gene-targeted mice have indicated that lamin A and lamin C have largely redundant functions. A deficiency of both lamin A and lamin C causes death in mice from muscular dystrophy/cardiomyopathy (12), but the elimination of lamin C synthesis alone or lamin A synthesis alone has no obvious adverse effects (13–15). For example, “lamin C–only” knockin mice, in which all of the output of the Lmna gene is channeled into the production of lamin C, are free of disease phenotypes and have a normal life span (13).
LMNA mutations in humans cause a variety of diseases, including muscular dystrophy, cardiomyopathy, and progeriod disorders (1, 16). A subset of LMNA mutations, including those causing Hutchinson-Gilford progeria syndrome (HGPS), are located in carboxyl terminal sequences unique to prelamin A (exons 11 and 12) and therefore have no effect on lamin C. HGPS is caused by exon 11 point mutations that enhance usage of a suboptimal splice donor site, resulting in aberrant mRNA splicing and the production of progerin, a mutant prelamin A protein containing an internal deletion of 50 amino acids (17, 18). This deletion eliminates the site for ZMPSTE24-mediated endoproteolytic cleavage — the event that would normally convert farnesyl–prelamin A to mature lamin A. For this reason, progerin retains a farnesyl lipid anchor at its carboxyl terminus, which has been speculated to confer toxicity to progerin (19).
In cultured cells, progerin leads to an increased frequency of misshapen cell nuclei in a dose-dependent fashion (20), and the level of progerin expression in vivo dictates the severity of disease phenotypes, both in humans (21) and in mouse models (22–25). In recent years, several groups have tested the hypothesis that blocking the farnesylation of progerin with a protein farnesyltransferase inhibitor (FTI) would reduce disease phenotypes in HGPS. Indeed, FTI treatment did lead to statistically significant beneficial effects on disease phenotypes in mouse models of HGPS, but the FTI-treated mice still developed severe disease and died (22, 26). Similarly, open-label testing of an FTI in children with HGPS suggested modest benefits (27, 28). New treatment strategies are clearly needed.
The fact that lamin C–only mice are free of disease led us to consider a new potential strategy for treating HGPS. We reasoned that it might be possible to treat HGPS (as well as other diseases caused by prelamin A missense mutations) by changing the balance of LMNA mRNA splicing (i.e., channeling the output of LMNA more toward lamin C transcripts and away from prelamin A transcripts). In this study, we identified a sequence in LMNA that regulates lamin C/prelamin A splicing and were successful in identifying an antisense oligonucleotide (ASO) that reduced lamin A and progerin expression in cultured cells and mouse tissues.
Results
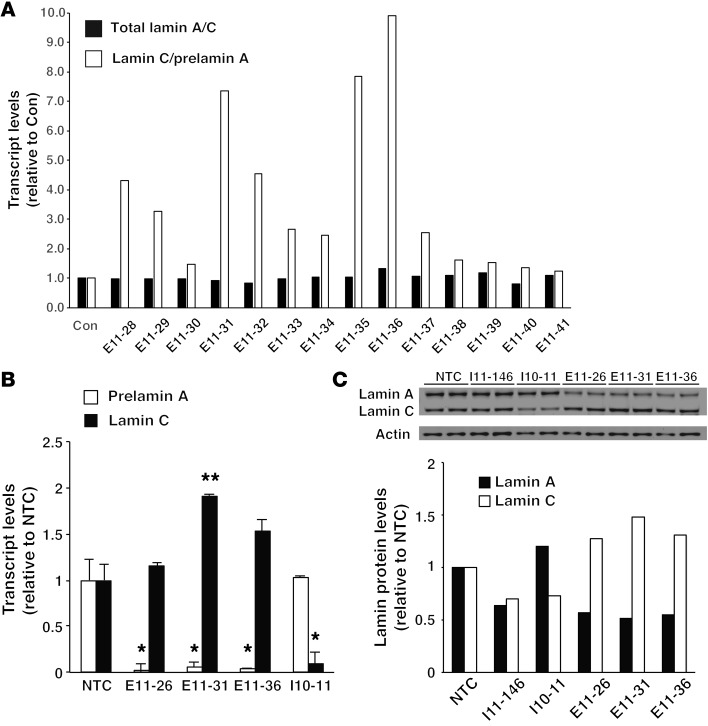
ASOs have been used to modulate splice-site utilization in precursor mRNAs (pre-mRNAs) (29). To identify sequences that regulate lamin C/prelamin A splicing, mouse fibroblasts were transfected with a series of 20-nt ASOs corresponding to exon 10, intron 10, and exon 11 sequences. All ASOs contained phosphorothioate linkages in the backbone and 2′-O-methoxyethyl substitutions of the sugar moiety; these ASOs have no capacity to promote RNase H–mediated removal of RNAs but are useful for modulating splicing (30). The ASOs were named based on their “binding site” (e.g., in the case of ASO E11-31, “E11” stands for exon 11 and “31” stands for nt 31 of exon 11). Our goal was to identify ASOs that shift the balance of lamin C/prelamin A splicing so as to increase lamin C production at the expense of lamin A. Most of the ASOs that we tested had no effect on lamin A/C production or only changed the expression of one protein isoform. For example, ASOs corresponding to sequences near lamin C’s 3′ UTR (in intron 10) tended to reduce lamin C expression, while ASOs that bound near the intron 10–exon 11 junction reduced lamin A expression (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI85908DS1). However, one ASO located in exon 11 (E11-31, corresponding to c.1735–1755) simultaneously reduced lamin A production and increased lamin C production. Next, we used quantitative real-time PCR (qRT-PCR) to test the effects of overlapping exon 11 ASOs (a series of ASOs that differed from E11-31 in 1-nt increments) on lamin C/prelamin A splicing. Several ASOs (with sequences –4 nt or +4 nt relative to E11-31) had effects similar to those of E11-31 (Figure 1A). This group of ASOs was called “lamin C–ASOs.” ASO E11-31 was selected for additional testing. Consistent with our screening studies (Figure 1A), ASO E11-31 increased lamin C transcript levels while lowering prelamin A transcript levels (Figure 1B; P < 0.05). ASO E11-31 also decreased lamin A protein levels and increased lamin C protein levels (relative to actin) (Figure 1C). The potency of ASOs was related to their length (20 nt > 18 nt > 16 nt) (Supplemental Figure 2A). As expected, the ASOs did not affect the expression of other nuclear lamins (e.g., lamin B1) produced from separate genes (Supplemental Figure 2A).
Figure 1. Identification of a region within exon 11 of Lmna that affects prelamin A and lamin C expression.
(A) qRT-PCR identifies several ASOs that increase the lamin C/prelamin A ratio. Wild-type MEFs were transfected with overlapping 18-nt ASOs corresponding to sequences in exon 11 of Lmna. After 2 days, mRNA levels for prelamin A, lamin C, and total lamin A/C were measured by qRT-PCR. Total lamin A/C levels (black bars) and the lamin C/prelamin A ratios (white bars) were calculated and expressed relative to control-treated cells (Con), set at a value of 1.0. (B) ASO E1-31 promotes lamin C splicing. Wild-type MEFs were transfected with ASOs E11-26, E11-31, E11-36, and I10-11 (I10-11 is located in lamin C’s 3′ UTR). After 2 days, mRNA levels for prelamin A and lamin C were measured by qRT-PCR. The expression was normalized to levels in cells that were treated with transfection reagent alone (nontemplate control [NTC]) (set at a value of 1.0). The mean ± SEM for 3 independent experiments is shown. Comparison with nontemplate control: *P < 0.05; **P < 0.01, t test. (C) Western blots showing that ASOs modulate lamin A and lamin C protein expression. MEFs were transfected in duplicate with ASOs I11-146, I10-11, E11-26, E11-31, and E11-36. After 2 days, cell extracts were prepared and analyzed by Western blotting with antibodies against lamin A/C and actin. The band intensities were measured by infrared laser scanning and normalized to actin. The results were expressed relative to the nontemplate control, set at a value of 1.0.
To determine whether the regulatory sequences that we uncovered in the mouse Lmna gene were relevant to lamin C/prelamin A splicing in humans, we generated overlapping ASOs corresponding to exon 11 sequences in human LMNA and then tested the ASOs in human skin fibroblasts. Human LMNA ASOs corresponding to the sequences identified in the mouse studies also shifted the balance of LMNA output more toward lamin C in human fibroblasts, as judged by both qRT-PCR (Figure 2A; P < 0.01) and Western blots (Supplemental Figure 2B). Of note, ASO E11-31 was also active in human hepatocyte, chondrocyte, and osteoblast cell lines (Figure 2, B and C).
Figure 2. ASOs modulate LMNA expression.
(A) ASOs modulate LMNA alternative splicing in human cells. Triplicate wells of wild-type human fibroblasts (clone CRL-2429) were transfected with ASO E11-26, E11-28, E11-31, E11-33, E11-36, E11-38, and E11-41. After 3 days, prelamin A and lamin C mRNA levels were measured by qRT-PCR. The data are expressed relative to cells treated with a scrambled ASO (Con) (set at a value of 1.0). The mean ± SD is shown. All yielded statistically significant differences compared with the control (P < 0.01, t test). (B) Modulating LMNA alternative splicing with ASO E11-31 in multiple cell types. Human skin fibroblasts (SKF; clone AG2492), HepG2 cells, HeLa cells, osteoblasts (OsteoB), chondrocytes (Chond), and smooth muscle cells (SMC) were transfected with ASO E11-31 or transfection reagent alone. After 2 days, prelamin A and lamin C transcript levels were measured by qRT-PCR and expressed relative to the nontemplate control (set at a value of 1.0). (C) Decreasing lamin A protein levels with ASO E11-31 in several human cell types. The same cell types described in B were transfected with ASO E11-31 or transfection reagent alone. After 3 days, the expression of lamin A and C was measured by Western blotting. Actin levels were measured as a loading control. Lamin protein expression was quantified and expressed relative to the nontemplate control (set at a value of 1.0). (D) ASO E11-31 decreases progerin levels in HGPS cells. Duplicate wells of HGPS cells were transfected once with ASO E11-31. Cells were harvested 1–5 days later, and the expression of lamin A, lamin C, and progerin was measured by Western blotting. Actin levels were measured as a loading control. The data are expressed relative to cells treated with a scrambled ASO (set at a value of 1.0).
Trace amounts of progerin transcripts (<1% of the amount of full-length prelamin A transcripts) can be detected in wild-type human cells (31, 32), a consequence of trace utilization of the same exon 11 splice donor site used in HGPS. Even though ASO E11-31 binds to sequences approximately 70 nt upstream of the HGPS mutation in codon 608 (c.1824C>T), it is possible that the lamin C–ASOs might increase utilization of the HGPS exon 11 splice donor site and result in larger amounts of progerin transcripts. To address this issue, we examined prelamin A transcripts by RT-PCR in cells that had been transfected with ASO E11-31 or a scrambled control ASO. Consistent with earlier studies (31, 32), trace amounts of progerin transcripts could be detected in human cells that had been transfected with a scrambled control ASO (Supplemental Figure 2C), and the low levels did not yield detectable progerin protein by Western blotting (see Figure 1C and Figure 2C). Trace amounts of progerin transcripts were also identified in ASO-E11-31–treated cells, but the levels of transcripts in those cells were similar to those found in cells transfected with the control ASO (or were perhaps even lower). Thus, lamin C–ASOs do not increase utilization of the HGPS splice donor site in normal cells. A second very faint RT-PCR product was detectable in the E11-31–transfected cells and in 2 of 3 cells transfected with the control ASO. The identity of the trace product was not identified by DNA sequencing, but its size (~77 bp) was consistent with a transcript lacking exon 11 sequences.
The low prelamin A transcript levels (and low lamin A protein levels) in wild-type mouse and human fibroblasts transfected with ASO E11-31 prompted us to test the efficacy of ASO E11-31 in human HGPS fibroblasts with the c.1824 (C>T) mutation. HGPS fibroblasts were transfected with ASO E11-31, and cell extracts were prepared 1 to 5 days later for Western blot with an antibody that binds to the amino terminus of lamin A (a region shared by lamin C, progerin, and lamin A). One day after the transfection, lamin A and progerin protein levels were lower in ASO E11-31–treated cells than in cells treated with the control ASO and lamin A, and progerin levels remained low through day 4 to 5 (Figure 2D). Lamin C levels did not appear to change on the first day after the ASO E11-31 transfection, but lamin C levels were higher on day 2 and peaked on day 3. The effects of ASO E11-31 on lamin A/C expression in the HGPS fibroblasts correlated with the dose of the ASOs (Supplemental Figure 2D) and were observed in each of 4 HGPS fibroblast cell lines tested (75-8, AG11513, AG1972, and AG11498; Supplemental Figure 2, E and F).
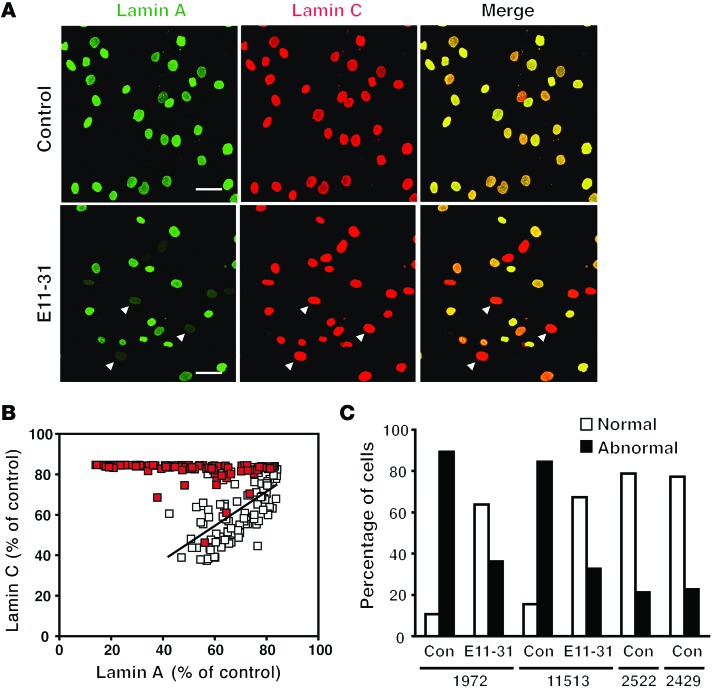
Reduced lamin A/progerin expression in ASO E11-31–treated HGPS fibroblasts was also apparent by immunocytochemistry. In these studies, ASO-transfected cells were stained with both a lamin A antibody (green) that recognizes lamin A and progerin and a lamin C antibody (red). In HGPS cells that had been transfected with the scrambled ASO, most cell nuclei were yellow or orange, reflecting the fact that lamin A/progerin and lamin C are expressed in roughly similar amounts (Figure 3A). In ASO E11-31–treated fibroblasts, the results were quite different. While lamin C was expressed in the nuclei of all cells, lamin A staining was minimal in some cells, resulting in many more red-colored nuclei in the merged image. Changes in lamin A and lamin C expression in individual cells were quantified by image processing and plotted. In control cells, the staining intensities for the lamin A and lamin C antibodies tended to be closely matched (i.e., cells with low lamin A expression typically had low lamin C levels, and cells with high lamin A levels typically had high lamin C levels; Figure 3B). However, after transfection with ASO E11-31, there was an easily detectable increase in the number of cells that expressed high levels of lamin C and low levels of lamin A/progerin (Figure 3, A and B).
Figure 3. ASOs lower lamin A levels and reduce the frequency of nuclear shape abnormalities in HGPS fibroblasts.
(A) Immunocytochemistry showing increased numbers of cells expressing more lamin C than lamin A after ASO E11-31 treatment. Human HGPS fibroblasts (clone 11513) were transfected once with ASO E11-31. After 5 days, the cells were processed for immunofluorescence microscopy with a lamin A antibody that binds lamin A and progerin (green) and a lamin C antibody (red). Cells transfected with a scrambled ASO served as a control. After transfection with ASO E11-31, lamin A expression was markedly reduced in many cells (arrowheads). Scale bar: 50 μm. (B) Quantitative measurements showing a higher number of cells expressing more lamin C than lamin A in ASO E11-31–treated cells. Control ASO–treated cells (white squares) expressed approximately equivalent amounts of lamin A and C. After ASO E11-31 treatment (red squares), there was an increase in the number of cells that expressed more lamin C than lamin A. Lamin C levels were scaled to the levels measured in control cells; thus, the highest levels for lamin C were limited to approximately 85 percent. (C) ASO E11-31 reduces the number of abnormally shaped nuclei in HGPS cells. Evaluation of nuclear shape after transfection with ASO E11-31. Two HGPS fibroblast cell lines (clones 1972 and 11513) were treated once with ASO E11-31 or a scrambled ASO. After 5 days, nuclear shape was assessed by two observers blinded to treatment group (n = 2 per group).
Misshapen cell nuclei are a hallmark of HGPS cells, and the frequency of misshapen nuclei depends on the amount of progerin expression (20). Since ASO E11-31 reduces progerin expression in HGPS fibroblasts, we suspected that the ASO would also reduce misshapen nuclei. Indeed, in two HGPS fibroblast cell lines examined (AG1972 and AG11513), the frequency of misshapen nuclei was lower in ASO E11-31–transfected cells (approaching the very low levels of misshapen nuclei in wild-type cells; Figure 3C; P < 0.05).
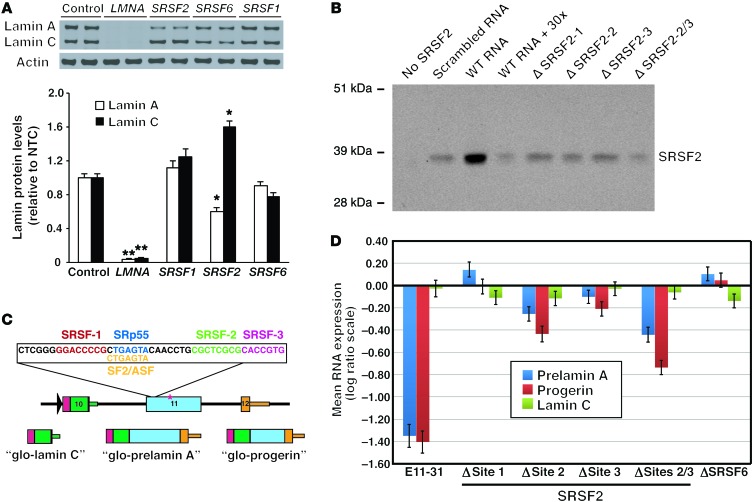
Because ASO E11-31 altered the pattern of lamin C/prelamin A splicing, we suspected that the exon 11 sequences corresponding to ASO E11-31 (and other “lamin C–ASOs”) might contain binding sites for exonic splice enhancer (ESE) proteins. A program to identify potential ESE-binding sites (ESEFinder; http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process=home) found 3 potential sites for serine/arginine-rich splicing factor 2 (SRSF2, also known as SC35), 2 for SRSF1 (also known as SF2 or ASF), and 1 for SRSF6 (also known as SRp55) (Supplemental Figure 3). To determine whether these ESEs regulate LMNA splicing, we began by performing siRNA knockdown studies. Wild-type human skin fibroblasts were transfected with siRNAs for SRSF1, SRSF2, and SRSF6; these siRNAs reduced SRSF1, SRSF2, and SRSF6 transcript levels by >80%. The effects of the siRNA knockdown on lamin A and lamin C were examined by Western blotting. As a positive control, we transfected cells with a LMNA siRNA and found, as expected, that the siRNA markedly reduced lamin A/C expression (Figure 4A). The knockdown of SRSF1 or SRSF6 had no effect on lamin A/C expression; however, the knockdown of SRSF2 shifted the output from the LMNA gene to increased amounts of lamin C and lower amounts of prelamin A (Figure 4A; P < 0.05).
Figure 4. SRSF2 modulates LMNA alternative splicing.
(A) siRNA knockdown of SRSF2 increases lamin C levels in human cells. Wild-type human fibroblasts were transfected with siRNAs against SRSF1, SRSF2, and SRSF6, and lamin A and C levels measured by Western blotting. Actin levels were measured as a loading control. Transcript levels for SRSF1, SRSF2, and SRSF6 were reduced by ≥80%. The mean ± SEM for 3 independent experiments are shown. *P < 0.05; **P < 0.01, t test. (B) SRSF2 binds to exon 11 RNA sequences. RNA molecules corresponding to nt 22–63 of exon 11 LMNA were biotinylated and bound to streptavidin beads. The binding of HA-tagged SRSF2 was assessed by Western blotting. Binding of SRSF2 to the wild-type RNA was compared with that to a scrambled RNA molecule, with the addition of a 30-fold excess of unlabeled RNA, and RNA molecules in which SRSF2 sites were mutated. (C) Schematic diagram of a LMNA reporter construct. A LMNA fragment (spanning exons 10–12) was cloned behind a glo fragment (colored red) driven by an RSV promoter (black arrowhead). The sequences of the 3 predicted SRSF2 sites and a single predicted SRSF6 (SRp55) site in exon 11 are highlighted. The red asterisk identifies the location of the HGPS mutation (c.1824; C>T). The reporter yields 3 transcripts (glo-prelamin A, glo-lamin C, and glo-progerin). (D) Reporter studies showing that SRSF2 sites 2 and 3 are important for LMNA prelamin A splicing. HeLa cells were cotransfected with the LMNA reporter and a “glo-only” plasmid. After 2 days, glo-prelamin A, glo-progerin, and glo-lamin C transcripts were quantified by qRT-PCR and normalized to levels of the glo-only transcript. Changes in expression, relative to the wild-type reporter (set at a value of 0), for 5 independent experiments (mean ± SEM) are shown.
The siRNA knockdown experiments supported a role for SRSF2 in LMNA splicing. To determine whether SRSF2 binds to the relevant LMNA sequences, we performed RNA pull-down studies. RNA molecules (41-nt long) spanning the entire 20-nt sequence in ASO E11-31 were biotinylated at the 3′-end and then bound to streptavidin beads. Nuclear extracts from HEK293 cells expressing a hemagglutinin-tagged (HA-tagged) SRSF2 were incubated with the beads; then, Western blotting was performed to define the binding of HA-tagged SRSF2 to the immobilized RNAs. Strong binding of SRSF2 was observed with the wild-type LMNA sequence (Figure 4B), but we observed only weak binding to the same wild-type RNA in the presence of an excess of unlabeled (nonbiotinylated) wild-type RNA. We also observed weak binding to a scrambled 41-nt RNA (Figure 4B). When the 41-nt LMNA RNA was mutated in order to eliminate the potential SRSF2-binding sites, the binding of SRSF2 to the RNAs was markedly reduced (Figure 4B).
To determine whether all 3 SRSF2-binding sites affect LMNA splicing, we generated a reporter construct to monitor LMNA splicing (Figure 4C). An exon-10 to exon-12 genomic fragment containing the HGPS point mutation (c.1824; C>T) was subcloned into the RHCglo reporter plasmid (33). The plasmid was transfected into HeLa cells, and transcripts derived from the reporter were examined by qRT-PCR. All 3 predicted transcripts (β-globin–prelamin A [glo–prelamin A], glo–lamin C, glo-progerin) were identified. To document that the system was capable of detecting changes in the expression of the different mRNA products, cells expressing the reporter were transfected with ASO E11-31. After 2 days, we observed a substantial decrease in prelamin A and progerin transcripts from the reporter construct (Figure 4D). We next asked whether any of the potential SRSF2-binding sites were relevant to the production of prelamin A transcripts by mutating each SRSF2 site in the reporter construct. When SRSF2 site 1 or the overlapping SRSF6/SRSF1 site was mutated, there was little change in the expression of prelamin A transcripts. However, when SRSF2 site 2 or site 3 was mutated, prelamin A and progerin transcript levels fell, and when both of those sites were mutated, the decrease in prelamin A transcripts was additive (Figure 4D; P < 0.05).
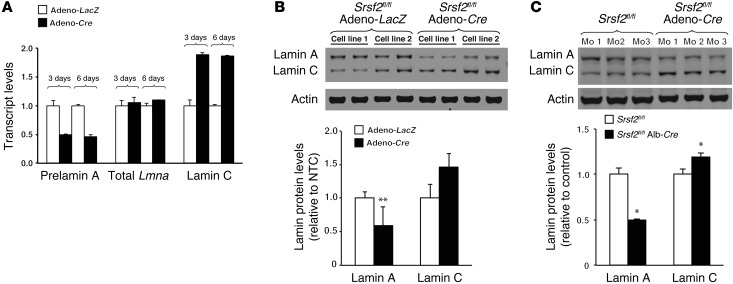
We next examined the impact of the absence of SRSF2 expression in Srsf2 knockout cells. To create Srsf2 knockout fibroblasts, Srsf2fl/fl mouse embryonic fibroblasts (MEFs) were infected with a Cre adenovirus (adeno-Cre), and prelamin A and lamin C transcript levels were measured 3 and 6 days later. At both time points, Srsf2 expression was reduced by >95% compared with that in cells transduced with a control virus (adeno-LacZ). In Srsf2-deficient cells, prelamin A mRNA levels were reduced by half, whereas lamin C mRNA levels almost doubled; total Lmna transcripts were unchanged (Figure 5A; P < 0.01). Similar changes were observed at the protein level. In adeno-Cre–transduced cells, lamin A levels were decreased by almost half (P < 0.01), whereas lamin C levels were increased by approximately 1.5-fold (Figure 5B).
Figure 5. Lamin C/prelamin A splicing in mouse cells and tissues depends on SRSF2 expression.
(A) Inactivating Srsf2 in mouse fibroblasts reduces prelamin A transcript levels and increases lamin C transcript levels without affecting total Lmna transcripts. A Srsf2fl/fl mouse fibroblast cell line was treated with adeno-Cre or adeno-LacZ. After 3 or 6 days, prelamin A and lamin C transcript levels were measured by qRT-PCR (mean ± SD). Srsf2 mRNA levels were reduced by >95% in the adeno-Cre–treated cells. (B) Western blots showing that Srsf2 inactivation in fibroblasts leads to reduced lamin A protein levels. Two Srsf2fl/fl fibroblast cell lines (cell lines 1 and 2) were treated with either adeno-Cre or adeno-LacZ. After 6 days, lamin A and lamin C levels were assessed by Western blotting. The bar graph shows changes in lamin expression (mean ± SEM) for 4 independent experiments. **P < 0.01, t test. (C) Western blots showing that an Srsf2 inactivation in the liver decreases lamin A levels and increases lamin C levels. Lamin A and lamin C protein levels were measured in isolated hepatocytes from Srsf2fl/fl mice (P23) expressing Cre in the liver (Srsf2fl/flAlb-Cre+). Actin levels were measured as a loading control. The bar graph shows the mean ± SEM for 3 mice per group. *P < 0.05.
To determine whether SRSF2 depletion affects Lmna alternative splicing in vivo, we measured steady-state levels of lamin A and lamin C in hepatocytes from liver-specific Srsf2 knockout mice (Srsf2fl/flAlb-Cre mice). Consistent with the cell culture studies, SRSF2 depletion changed the pattern of lamin A and lamin C expression in the liver. Lamin A protein levels in hepatocytes from Srsf2fl/fl mice tended to be higher than lamin C levels (or were similar), but lamin C levels in hepatocytes from Srsf2fl/flAlb-Cre+ mice were higher than lamin A levels (Figure 5C). Based on quantitative measurements, lamin A levels decreased by 50% and lamin C levels increased by approximately 20% (P < 0.05).
To test whether ASO E11-31 would be effective in manipulating Lmna splicing in vivo, we treated wild-type mice with ASO E11-31 for 4 weeks and then measured Lmna transcripts in the liver. Lamin C mRNA levels were 50% higher in ASO E11-31–treated mice than in mice given a scrambled ASO (Figure 6A; P < 0.01). Prelamin A levels trended lower (30% lower), but this change did not reach statistical significance. However, when lamin A protein levels in liver extracts from the same mice were examined, lamin A levels were reduced by >40% (Figure 6B; P < 0.02).
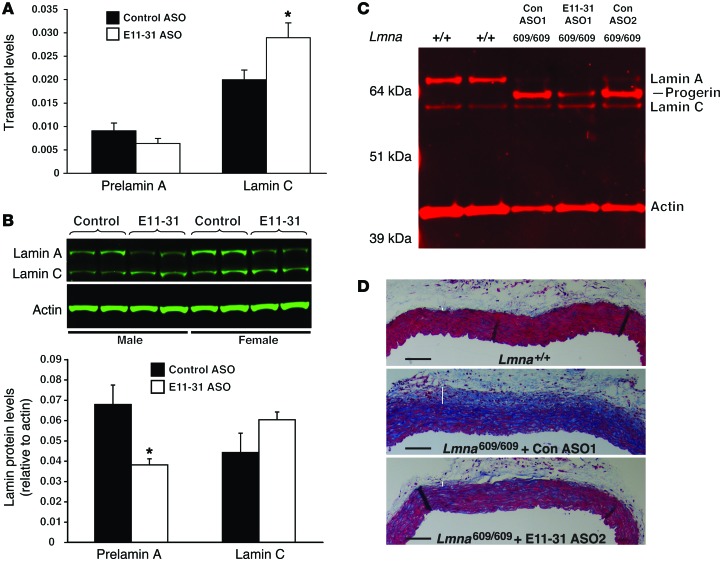
Figure 6. ASO E11-31 reduces the expression of lamin A in wild-type mice and progerin in LmnaG609G/G609G mice.
(A) ASO E11-31 increases lamin C transcript levels in vivo. Wild-type mice (n = 4 per group) were treated with ASO E11-31 or a scrambled oligonucleotide (Control ASO) for 4 weeks. Two days after the last injection, RNA was isolated from liver tissue. Prelamin A and lamin C transcript levels were measured by qRT-PCR (mean ± SEM). *P < 0.05, t test. (B) ASO E11-31 lowers lamin A protein levels in vivo. Lamin A and lamin C protein levels (relative to actin) in the livers of control- and E11-31–treated mice (described in A) were measured by Western blotting and graphed. *P < 0.05, t test. (C) ASO E11-31 decreases progerin levels in the aortas of LmnaG609G/G609G mice. One-month-old LmnaG609G/G609G mice were treated with ASO E11-31 or a scrambled oligonucleotide (n = 3) for 3 months. Two days after the last injection, the mouse aortas were dissected free of attached tissues. Lamin A, progerin, and lamin C protein levels in the aorta were measured by Western blotting with an antibody against the amino terminus of lamin A (a region common to all 3 isoforms). Actin levels were measured as a loading control. The results are shown for 2 wild-type mice (+/+), two LmnaG609G/G609G (609/609) mice treated with the control ASO (Con ASO1 and Con ASO2), and an LmnaG609G/G609G mouse treated with ASO E11-31 (E11-31 ASO1). (D) Histological images showing reduced disease in the ascending aortas of ASO E11-31–treated LmnaG609G/G609G mice. Images (original magnification, ×10) of Masson’s trichrome–stained cross sections through the ascending aortas from 1 wild-type mouse (Lmna+/+), 1 LmnaG609G/G609G mouse treated with a control ASO (Con ASO1), and 1 LmnaG609G/G609G mouse treated with ASO E11-31 (E11-31 ASO2). White bars identify the adventitia. Scale bar: 100 μm.
To investigate whether modulating Lmna alternative splicing might reduce progerin expression in vivo, we created a new HGPS mouse model by gene targeting. We introduced a C>T mutation into codon 609 of Lmna (corresponding to the most frequent mutation causing HGPS). As expected, the mutation increased utilization of the exon 11 HGPS splice donor site and led to abundant amounts of progerin expression (Figure 6C). The LmnaG609G mice exhibited disease phenotypes similar to those in previous HGPS mouse models (22, 25, 34), including a distinctive aortic pathology that was first reported in mice harboring a human LMNAG608G transgene (35) and later identified in an LmnaG609G knockin mouse (25). By 4 months of age, homozygous mice (LmnaG609G/G609G mice) invariably displayed a near-complete loss of smooth muscle cells in the media of the ascending aorta as well as advanced adventitial fibrosis. To explore the impact of ASO E11-31 on aortic pathology, we treated LmnaG609G/G609G mice with ASO E11-31 or a scrambled control ASO for 3 months, beginning at 1 month of age. In the aortas of LmnaG609G/G609G mice, we observed lamin C and large amounts of progerin expression but only small amounts of lamin A, indicating that the main product of the LmnaG609G allele is progerin (Figure 6C and Supplemental Figure 4A). The aortic pathology in LmnaG609G/G609G mice treated with the scrambled ASO was severe, identical to that in untreated mice, with a loss of medial smooth muscle cells and severe adventitial fibrosis, as judged by reduced cytoplasmic staining in the media and increased collagen staining in the adventitia. In contrast, in mice treated with ASO E11-31, the levels of progerin, normalized to actin, decreased by more than 50% as compared with that in mice treated with the scrambled ASO (Figure 6C and Supplemental Figure 4, A and B), and aortic pathology was markedly improved during examination of ascending aortas, which all had less adventitial fibrosis and more smooth muscle cells, as judged by Masson’s trichrome staining (Figure 6D and Supplemental Figure 4C).
Discussion
The expression of A-type lamins is essential for postnatal survival in both humans (36) and mice (12), but there is no requirement for synthesis of both lamin A and lamin C (13, 14). Lamin C–only mice (13) are free of disease and have a normal life span. The absence of detectable pathology in those mice, in which all of the output of the Lmna gene is channeled into lamin C, raised the possibility that HGPS and all other prelamin A diseases might be treated safely by tipping the balance of LMNA expression more toward lamin C production. To explore the feasibility of this approach, we investigated mechanisms that control LMNA splicing and were successful in identifying a 30-nt stretch in exon 11 of LMNA that regulates lamin C/prelamin A splicing. Targeting this region with a lamin C–ASO (e.g., E11-31) reduced prelamin A mRNA levels and lamin A protein levels in both mouse and human cell lines. Of note, treatment of human HGPS fibroblasts with a lamin C–ASO reduced progerin transcript and protein levels by >70%. The exon 11 sequence targeted by ASO E11-31 contains potential binding sites for 3 different ESEs, but only SRSF2 influenced lamin C/prelamin A splicing. Lowering SRSF2 levels in human cells by siRNA knockdown or inactivating mouse Srsf2 by Cre recombination shifted the output of the gene toward lamin C. RNA pull-down studies revealed that SRSF2 binds to exon 11 RNAs (spanning the sequence of ASO E11-31), and inactivating 2 SRSF2-binding sites in that sequence reduced prelamin A transcripts in reporter construct studies. In keeping with these in vitro findings, we observed a shift toward more lamin C synthesis in hepatocytes from hepatocyte-specific Srsf2 knockout mice. Finally, treating wild-type mice with ASO E11-31 shifted the output of Lmna in the liver toward lamin C, and treating LmnaG609G/G609G mice with ASO E11-31 reduced progerin levels in the aorta and reduced aortic pathology.
The exon 11 regulatory sequences of the human and mouse lamin A/C genes identified in our studies are nearly identical (97% sequence identity). The fact that these sequences are located in virtually the same position relative to the 3′ splice site supports the idea that these sequences play a role in splicing (37). According to a predicted secondary structure of exon 11 LMNA pre-mRNA sequences, proposed by Tazi and colleagues (38), the 30-nt regulatory region overlaps 2 stem-loop structures separated by a short stretch of unpaired nt. Since splicing regulatory proteins preferentially bind to single-stranded RNA (39), one would predict that an active ESE site would be located in the unpaired nt between the 2 stem-loop structures. Indeed, one of the two important SRSF2-binding sites identified in our reporter construct studies is located within that stretch of nt. We propose that the binding of SRSF2 to the regulatory sequence assists in defining exon 11 as an exon (40), facilitating the assembly of the spliceosome complex at the 3′ splice site and promoting exon 10 to exon 11 (prelamin A) splicing.
Our studies showed that ASO E11-31 reduces progerin levels in the aorta and suggested that ASO treatment reduced aortic pathology. In our studies, we initiated treatment at 1 month of age and euthanized the mice at 4 months of age, a time point when LmnaG609G/G609G mice invariably display very severe aortic pathology (a virtually complete loss of medial smooth muscle cells and extensive adventitial fibrosis). When we treated LmnaG609G/G609G mice with a scrambled control ASO, progerin levels in the aorta remained very high and all mice had severe pathology in the aorta. In contrast, ASO E11-31 lowered progerin levels in the aorta and clearly reduced aortic pathology. In one of the treated mice, aortic pathology was absent. We considered these results to be encouraging, particularly since the favorable results were obtained in homozygous knockin mice, which have extremely high levels of progerin expression. Also, the beneficial effects on aortic pathology were welcome, because disease in the large arteries almost always accounts for death in children with HGPS. In our studies, we terminated the ASO treatment studies when the mice were 4 months of age, a time point that is ideal for investigating arterial pathology. Longer-term studies in LmnaG609G/G609G and LmnaG609G/+ mice will be required to define the effects of the ASO treatment regimen on other disease phenotypes (e.g., longevity or skeletal abnormalities).
Our studies raised the possibility that ASO E11-31 could be used to shift the balance of LMNA splicing toward lamin C and that this strategy might be useful for treating HGPS (and other laminopathies caused by missense mutations in exon 11 of LMNA). In considering this strategy, the first question is whether this approach would be safe. In this regard, the fact that lamin C–only mice are free of disease is reassuring. Another reason for confidence comes from human genetics. Numerous nonsense and frameshift mutations have been identified within exons 1–10 of LMNA in patients with muscular dystrophy and cardiomyopathy (http://www.umd.be/), but human disease caused by a nonsense or frameshift mutation in exon 11 or exon 12 has not yet been described. This observation, albeit circumstantial, suggests that the loss of lamin A in the setting of preserved lamin C synthesis may not be harmful. The second question is whether an ASO that reduces progerin levels (but falls short of eliminating progerin synthesis) would be useful. Again, there are reasons for confidence. First, minor differences in progerin production associated with different HGPS mutations have a major influence on disease severity in humans (21). Second, studies with Zmpste24–/– mice indicate that complete elimination of farnesyl–prelamin A protein is not required to reduce the toxicity of the protein and obtain therapeutic benefit. When a single lamin C–only or Lmna knockout allele was introduced into Zmpste24–/– mice, the levels of the toxic farnesyl–prelamin A were reduced by only 50%, but the disease phenotypes of progeria were completely eliminated (13, 19). Finally, enhanced production of lamin C synthesis appears to act posttranscriptionally to alter the turnover of progerin and reduce progerin toxicity. In mice that carry a single “HGPS allele,” replacing the wild-type Lmna allele with a lamin C–only allele reduced steady-state levels of progerin in tissues and ameliorated disease phenotypes (23). The latter studies implied that a lamin C–ASO could have dual benefits for patients with HGPS — a direct benefit by lowering the production of progerin transcripts and a more indirect benefit from increasing lamin C production and thereby reducing the toxic effects of progerin.
In summary, we identified a 30-nt stretch in exon 11 of LMNA that is important for regulating lamin C/prelamin A mRNA splicing. We showed that ASOs that bind to this sequence tip the balance of LMNA production more toward lamin C and less toward lamin A. Mechanistic studies suggest that lamin C–ASOs reduce the binding of SRSF2 to exon 11 LMNA pre-mRNA and thereby reduce the production of prelamin A transcripts. The lamin C–ASO reduced lamin A expression in wild-type mice and also reduced progerin levels and aortic pathology disease in LmnaG609G/G609G mice. Our studies suggest that targeting LMNA mRNA alternative splicing could prove to be a safe and effective strategy for treating HGPS and all other diseases caused by prelamin A mutations.
Methods
Additional details are provided in the Supplemental Methods.
Cells.
Primary MEFs were prepared from E13.5 postcoitum wild-type embryos or Srsf2fl/fl embryos after intercrossing Srsf2fl/+ mice. Cells were cultured in DMEM (Gibco) containing 10% FBS (Hyclone) with 1 mM nonessential amino acids (Gibco), 1 mM sodium pyruvate (Gibco), 2 mM glutamine (Gibco), 10 units/ml penicillin, and 10 μg/ml streptomycin (Gibco). Human HGPS fibroblasts (AG11513, AG1972, AG11498), human wild-type fibroblasts (AG2522, AG2429), human chondrocytes (402-05a), human aortic smooth muscle cells (PCS-100-012), human osteoblasts (CRL-11372), HeLa cells, and HEK293 cells were obtained from ATCC or the Coriell Cell Repository and cultured according to instructions from the suppliers. hTERT-immortalized HGPS cell line 75-8 was provided by Junko Oshima (University of Washington, Seattle, Washington, USA) (41).
ASO transfections.
ASOs were synthesized by Ionis Pharmaceuticals and named based on their binding site within an exon (E) or intron (I). For example, E11-31 is an ASO that binds to exon 11, starting at nt 31 of exon 11. Cells were plated in 6- or 12-well plates and allowed to adhere overnight. The fibroblasts were washed with Opti-MEM I medium (Invitrogen), transfected with ASOs (31 nM) with 2 μl/ml Lipofectamine 2000 (Invitrogen), and then incubated at 37°C. After 6 hours, the transfection medium was replaced with fresh culture medium. In some cases, a second transfection was performed 2 days later, and the cells were cultured for 2 additional days before isolating RNA and protein extracts. All ASO sequences are listed in Supplemental Table 1.
siRNA transfection.
Wild-type and HGPS human fibroblasts were plated in 6- or 12-well plates and allowed to adhere overnight. The fibroblasts were washed with Opti-MEM I medium (Invitrogen) and transfected with siRNAs (25 nM) with 2 μl/ml Dharmafect I (Thermo Scientific) at 37°C for 6 hours. The transfection medium was removed and replaced with fresh culture medium. A second transfection was performed 2 days later, and the cells cultured for an additional 2 days before isolating RNA and protein extracts. Transfections were performed with the following siRNAs from Thermo Scientific: LMNA (L-004978-00), SRSF1 (L-018672-01), SRSF2 (L-019711-00), or SRSF6 (L-016067-01).
Adenovirus transduction.
Srsf2fl/fl MEFs were plated in 6-well plates and allowed to adhere overnight. Adeno-Cre or adeno-LacZ viruses (Gene Transfer Vector Core; University of Iowa) were added at 2,000 MOI and incubated at 37°C for 3 days before RNA and protein isolation.
Western blots.
Urea-soluble protein extracts from cells and tissues were prepared as described previously (19). Proteins were size fractionated on 4%–12% gradient polyacrylamide Bis-Tris gels (Invitrogen) and transferred to nitrocellulose membranes. The membranes were incubated with the following antibodies: a goat polyclonal antibody against lamin A/C (1:400; sc-6215, Santa Cruz Biotechnology); a goat polyclonal antibody against actin (1:2,000; sc-1616, Santa Cruz Biotechnology); infrared dye–labeled secondary antibodies (Rockland Immunochemicals). The infrared signals were quantified with an Odyssey infrared scanner (LI-COR Biosciences).
Immunofluorescence microscopy.
Cells on coverslips were fixed with ice-cold methanol, rinsed with acetone, and permeabilized with 0.2% Triton. The cells were processed for immunofluorescence microscopy as described previously (13). The following antibodies were used: a mouse monoclonal antibody against lamin A (Abcam); a rabbit polyclonal antibody against lamin C (Novus Bio); Alexa Fluor 488–labeled goat anti–mouse IgG (Invitrogen); and Alexa Fluor 568–labeled donkey anti–rabbit IgG (Invitrogen). Confocal fluorescence microscopy images were obtained with a Zeiss LSM700 laser-scanning microscopy with a Plan Apochromat ×20/0.80 NA objective (air), and images along the z axis were processed by Zen 2010 software (Zeiss) to generate maximum image projections.
Cell counting.
Maximum image projection cell images were imported into ImageJ and converted to gray scale, and the threshold was adjusted to identify nuclei. The intensity of the lamin A signal was recorded for each nucleus within an image using particle pixel sizes ranging from 200 to infinity. For each image, the mean intensity and standard deviation were calculated. Nuclei that were more than 1 standard deviation below the mean for lamin A intensity were evaluated for nuclear shape abnormalities by two observers blinded to treatment group.
qRT-PCR.
Total RNA was isolated and treated with DNase I (Ambion, Life Technologies). RNA was reverse-transcribed with random primers, oligo(dT), and SuperScript III (Invitrogen). qPCR reactions were performed on a 7900 Fast Real-Time PCR system (Applied Biosystems) with SYBR Green PCR Master Mix (Bioline). Transcript levels were determined by the comparative cycle threshold method and normalized to levels of cyclophilin A (for mouse samples) or GAPDH (for human samples). All primers are listed in Supplemental Table 1.
LMNA reporter assay.
HeLa cells were seeded in 6-well plates and allowed to adhere overnight. The cells were washed with Opti-MEM I medium (Invitrogen), transfected with ASOs (31 or 15.5 nM) using 4.5 μl/ml Lipofectamine 2000 (Invitrogen), and then placed at 37°C for 6 hours according to the manufacturer’s instructions. The transfection medium was then removed and replaced with fresh culture medium. On the following day, cells were cotransfected with the RHCglo-LMNA plasmid (1 ng/ml) and the RHCglo plasmid (0.5 ng/ml) using Fugene 6 (4 μl/ml) (Promega). The RHCglo plasmid was included to normalize for differences in transfection efficiency. Total RNA was isolated 2 days later, and gene expression was measured by qRT-PCR. Plasmid-specific prelamin A, lamin C, and progerin transcripts were amplified with a common forward primer and unique reverse primers. The common forward primer was 5′-CATTCACCACATTGGTGTGC-3′, and the unique reverse primers for prelamin A, lamin C, and progerin were 5′-AGGCAGAAGAGCCGAGGAGA-3′, 5′-AGCGGCGGCTACCACTCA-3′, and 5′-CATGATGCTGCAGTTCTGGGGGCTCTGGAC-3′, respectively. The RHCglo plasmid was amplified using the same common forward primer and reverse primer 5′-CGGTAAGAGCAAGCTGGTCA-3′.
RNA pull-down assay.
RNA molecules (41-nt long) were synthesized by Integrated DNA Technologies; 50 pmol of each RNA was biotinylated at the 3-end with desthiobiotinylated cytidine according to the manufacturer’s instructions (Life Technologies). The sequences for the wild-type, ΔSRSF-1, ΔSRSF-2, ΔSRSF-3, ΔSRSF-2/3, and control (scrambled) RNAs are described in Supplemental Table 1. The labeled RNA molecules were incubated with 50 μg of washed streptavidin magnetic beads (Dynabeads M-270, Life Technologies) in RNA capture buffer (20 mM Tris [pH 7.5], 1 M NaCl, 1 mM EDTA, and 0.1% Tween-20) at room temperature for 30 minutes. The beads were washed twice with wash buffer (20 mM Tris [pH 7.5] with 0.1% Tween-20) and once with protein-binding buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 20 mM MgCl2, and 0.5% Tween-20). The beads were incubated with 60 μg of nuclear protein extract in protein-binding buffer containing 100 units of SUPERASE-In RNase inhibitor (Life Technologies) at 4°C for 60 minutes. Nuclear extracts were prepared from HEK293 cells electroporated with an HA-SRSF2 expression vector (42) using the NE-PER extraction reagent (Life Technologies) containing protease (Life Technologies) and phosphatase (Biotool) inhibitors. The beads were washed 6 times with wash buffer (20 mM Tris [pH 7.5], 10 mM NaCl, 0.1% Tween-20), and bound proteins were eluted by incubating in protein sample buffer at 70°C for 10 minutes. The proteins were separated on 4% to 12% Bis-Tris SDS-polyacrylamide gels, and SRSF2 was detected by Western blotting with a mouse monoclonal antibody against the HA tag (ab18181, Abcam). The loading of the protein gel was adjusted to the amount of biotinylated RNA in the eluate, which was measured by incubating with Alexa Fluor 680–labeled streptavidin (S-21378, Life Technologies) and quantifying the signal with an Odyssey infrared scanner (LI-COR Biosciences).
Treatment of mice with ASOs.
Mice (wild-type or LmnaG609G/G609G) were injected intraperitoneally with E11-31 or a scrambled ASO (in normal saline) at a concentration of 150 mg/kg/wk (divided into 2 injections per week). The animals were sacrificed 2 days after the last injection, and the tissues were collected for analysis. The effects of E11-31 on transcript levels were quantified by qRT-PCR, and the effects on lamin protein levels were determined by quantitative Western blotting. For the LmnaG609G/G609G mice, the aortas were dissected free, and the aortic arch was processed for Western blotting, while a segment of the ascending aorta was collected for paraffin embedding and H&E staining.
Statistics.
Statistical analyses were performed with Microsoft Excel for Mac 2011. Differences in expression levels of lamin A, lamin C, and progerin in all experiments were analyzed by a 2-tailed Student’s t test. Differences in the numbers of abnormally shaped nuclei were analyzed by the χ2 test.
Study approval.
This study was carried out according to guidelines in the Guide for the Care and Use of Laboratory Animals from the National Institutes of Health (8th ed. National Academy Press. 2011). UCLA’s Animal Research Committee approved all procedures performed using animals.
Author contributions
JML, HJJ, SGY, and LGF designed research studies. JML, CN, YT, CC, SHY, and LGF conducted experiments. TAV, FR, and CFB provided reagents. JML, SGY, and LGF wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH (HL089781 and AG047192 to L.G. Fong and AG035626 to S.G. Young). We would like to thank Thomas A. Cooper (Baylor College of Medicine, Houston, Texas, USA) for providing the RHCglo reporter plasmid and Junko Oshima (University of Washington, Seattle, Washington, USA) for providing hTERT-HGPS cell lines 75-8 and AG03513D. We would also like to acknowledge the assistance of Tiffany Dang and Kara Hamamoto in nuclei counting.
Footnotes
John M. Lee’s present address is: CHI St. Luke’s Episcopal Hospital, Houston, Texas, USA.
Conflict of interest: S.G. Young has received income from and been an expert witness for Regeneron. T.A. Vickers, F. Rigo, and C.F. Bennett are paid employees of Ionis Pharmaceuticals.
Reference information:J Clin Invest. 2016;126(4):1592–1602. doi:10.1172/JCI85908.
See the related Commentary beginning on page 1.
Contributor Information
Chika Nobumori, Email: c.nobumori@gmail.com.
Yiping Tu, Email: ytu@mednet.ucla.edu.
Catherine Choi, Email: cchoi1229@gmail.com.
Hea-Jin Jung, Email: uranushj@g.ucla.edu.
Timothy A. Vickers, Email: TVickers@isisph.com.
Frank Rigo, Email: FRigo@isisph.com.
C. Frank Bennett, Email: fbennett@isisph.com.
Stephen G. Young, Email: sgyoung@mednet.ucla.edu.
References
- 1.Worman HJ, Bonne G. “Laminopathies”: a wide spectrum of human diseases. Exp Cell Res. 2007;313(10):2121–2133. doi: 10.1016/j.yexcr.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerace L, Blum A, Blobel G. Immunocytochemical localization of the major polypeptides of the nuclear pore complex-lamina fraction. Interphase and mitotic distribution. J Cell Biol. 1978;79(2 pt 1):546–566. doi: 10.1083/jcb.79.2.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehner CF, Kurer V, Eppenberger HM, Nigg EA. The nuclear lamin protein family in higher vertebrates. Identification of quantitatively minor lamin proteins by monoclonal antibodies. J Biol Chem. 1986;261(28):13293–13301. [PubMed] [Google Scholar]
- 4.Fisher DZ, Chaudhary N, Blobel G. cDNA sequencing of nuclear lamins A and C reveals primary and secondary structural homology to intermediate filament proteins. Proc Natl Acad Sci U S A. 1986;83(17):6450–6454. doi: 10.1073/pnas.83.17.6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKeon FD, Kirschner MW, Caput D. Homologies in both primary and secondary structure between nuclear envelope and intermediate filament proteins. Nature. 1986;319(6053):463–468. doi: 10.1038/319463a0. [DOI] [PubMed] [Google Scholar]
- 6.Kolb T, Maass K, Hergt M, Aebi U, Herrmann H. Lamin A and lamin C form homodimers and coexist in higher complex forms both in the nucleoplasmic fraction and in the lamina of cultured human cells. Nucleus. 2011;2(5):425–433. doi: 10.4161/nucl.2.5.17765. [DOI] [PubMed] [Google Scholar]
- 7.Lin F, Worman HJ. Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J Biol Chem. 1993;268(22):16321–16326. [PubMed] [Google Scholar]
- 8.Weber K, Plessmann U, Traub P. Maturation of nuclear lamin A involves a specific carboxy-terminal trimming, which removes the polyisoprenylation site from the precursor; implications for the structure of the nuclear lamina. FEBS Lett. 1989;257(2):411–414. doi: 10.1016/0014-5793(89)81584-4. [DOI] [PubMed] [Google Scholar]
- 9.Sinensky M, Fantle K, Trujillo M, McLain T, Kupfer A, Dalton M. The processing pathway of prelamin A. J Cell Sci. 1994;107(pt 1):61–67. doi: 10.1242/jcs.107.1.61. [DOI] [PubMed] [Google Scholar]
- 10.Lutz RJ, Trujillo MA, Denham KS, Wenger L, Sinensky M. Nucleoplasmic localization of prelamin A: Implications for prenylation-dependent lamin A assembly into the nuclear lamina. Proc Natl Acad Sci U S A. 1992;89(12):3000–3004. doi: 10.1073/pnas.89.7.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young SG, Fong LG, Michaelis S. Prelamin A, Zmpste24, misshapen cell nuclei, and progeria — new evidence suggesting that protein farnesylation could be important for disease pathogenesis. J Lipid Res. 2005;46(12):2531–2558. doi: 10.1194/jlr.R500011-JLR200. [DOI] [PubMed] [Google Scholar]
- 12.Sullivan T, et al. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147(5):913–919. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fong LG, et al. Prelamin A and lamin A appear to be dispensable in the nuclear lamina. J Clin Invest. 2006;116(3):743–752. doi: 10.1172/JCI27125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coffinier C, et al. Direct synthesis of lamin A, bypassing prelamin A processing, causes misshapen nuclei in fibroblasts but no detectable pathology in mice. J Biol Chem. 2010;285(27):20818–20826. doi: 10.1074/jbc.M110.128835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez-Mejia IC, et al. Antagonistic functions of LMNA isoforms in energy expenditure and lifespan. EMBO Rep. 2014;15(5):529–539. doi: 10.1002/embr.201338126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schreiber KH, Kennedy BK. When lamins go bad: nuclear structure and disease. Cell. 2013;152(6):1365–1375. doi: 10.1016/j.cell.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Sandre-Giovannoli A, et al. Lamin A truncation in Hutchinson–Gilford progeria. Science. 2003;300(5628): doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- 18.Eriksson M, et al. Recurrent de novo point mutations in lamin A cause Hutchinson–Gilford progeria syndrome. Nature. 2003;423(6937):293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fong LG, et al. Heterozygosity for Lmna deficiency eliminates the progeria-like phenotypes in Zmpste24-deficient mice. Proc Natl Acad Sci U S A. 2004;101(52):18111–18116. doi: 10.1073/pnas.0408558102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glynn MW, Glover TW. Incomplete processing of mutant lamin A in Hutchinson-Gilford progeria leads to nuclear abnormalities, which are reversed by farnesyltransferase inhibition. Hum Mol Genet. 2005;14(20):2959–2969. doi: 10.1093/hmg/ddi326. [DOI] [PubMed] [Google Scholar]
- 21.Moulson CL, et al. Increased progerin expression associated with unusual LMNA mutations causes severe progeroid syndromes. Hum Mutat. 2007;28(9):882–889. doi: 10.1002/humu.20536. [DOI] [PubMed] [Google Scholar]
- 22.Yang SH, et al. A farnesyltransferase inhibitor improves disease phenotypes in mice with a Hutchinson-Gilford progeria syndrome mutation. J Clin Invest. 2006;116(8):2115–2121. doi: 10.1172/JCI28968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang SH, Qiao X, Farber E, Chang SY, Fong LG, Young SG. Eliminating the synthesis of mature lamin A reduces disease phenotypes in mice carrying a Hutchinson-Gilford progeria syndrome allele. J Biol Chem. 2008;283(11):7094–7099. doi: 10.1074/jbc.M708138200. [DOI] [PubMed] [Google Scholar]
- 24.Varela I, et al. Combined treatment with statins and aminobisphosphonates extends longevity in a mouse model of human premature aging. Nat Med. 2008;14(7):767–772. doi: 10.1038/nm1786. [DOI] [PubMed] [Google Scholar]
- 25.Osorio FG, et al. Splicing-directed therapy in a new mouse model of human accelerated aging. Sci Transl Med. 2011;3(106): doi: 10.1126/scitranslmed.3002847. [DOI] [PubMed] [Google Scholar]
- 26.Yang SH, Qiao X, Fong LG, Young SG. Treatment with a farnesyltransferase inhibitor improves survival in mice with a Hutchinson-Gilford progeria syndrome mutation. Biochim Biophys Acta. 2008;1781(1–2):36–39. doi: 10.1016/j.bbalip.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gordon LB, et al. Clinical trial of a farnesyltransferase inhibitor in children with Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A. 2012;109(41):16666–16671. doi: 10.1073/pnas.1202529109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordon LB, Massaro J, D’Agostino RB., Sr Campbell SE, Brazier J, Brown WT, Kleinman ME, and Kieran MW. Impact of farnesylation inhibitors on survival in Hutchinson-Gilford progeria syndrome. Circulation. 2014;130(1):27–34. doi: 10.1161/CIRCULATIONAHA.113.008285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rigo F, Seth PP, Bennett CF. Antisense oligonucleotide-based therapies for diseases caused by pre-mRNA processing defects. Adv Exp Med Biol. 2014;825:303–352. doi: 10.1007/978-1-4939-1221-6_9. [DOI] [PubMed] [Google Scholar]
- 30.Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 31.Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312(5776):1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez S, Coppede F, Sagelius H, Eriksson M. Increased expression of the Hutchinson-Gilford progeria syndrome truncated lamin A transcript during cell aging. Eur J Hum Genet. 2009;17(7):928–937. doi: 10.1038/ejhg.2008.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh G, Cooper TA. Minigene reporter for identification and analysis of cis elements and trans factors affecting pre-mRNA splicing. Biotechniques. 2006;41(2):177–181. doi: 10.2144/000112208. [DOI] [PubMed] [Google Scholar]
- 34.Bergo MO, et al. Zmpste24 deficiency in mice causes spontaneous bone fractures, muscle weakness, and a prelamin A processing defect. Proc Natl Acad Sci U S A. 2002;99(20):13049–13054. doi: 10.1073/pnas.192460799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varga R, et al. Progressive vascular smooth muscle cell defects in a mouse model of Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A. 2006;103(9):3250–3255. doi: 10.1073/pnas.0600012103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Engelen BG, Muchir A, Hutchison CJ, van der Kooi AJ, Bonne G, Lammens M. The lethal phenotype of a homozygous nonsense mutation in the lamin A/C gene. Neurology. 2005;64(2):374–376. doi: 10.1212/01.WNL.0000149763.15180.00. [DOI] [PubMed] [Google Scholar]
- 37.Witten JT, Ule J. Understanding splicing regulation through RNA splicing maps. Trends Genet. 2011;27(3):89–97. doi: 10.1016/j.tig.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez-Meijia IC, et al. A conserved splicing mechanism of the LMNA gene controls premature aging. Hum Mol Genet. 2011;20(23):4540–4555. doi: 10.1093/hmg/ddr385. [DOI] [PubMed] [Google Scholar]
- 39.Hiller M, Zhang Z, Backofen R, Stamm S. Pre-mRNA secondary structures influence exon recognition. PLoS Genet. 2007;3(11): doi: 10.1371/journal.pgen.0030204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graveley BR, Maniatis T. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol Cell. 1998;1(5):765–771. doi: 10.1016/S1097-2765(00)80076-3. [DOI] [PubMed] [Google Scholar]
- 41.Huang S, et al. Correction of cellular phenotypes of Hutchinson-Gilford Progeria cells by RNA interference. Hum Genet. 2005;118(3–4):444–450. doi: 10.1007/s00439-005-0051-7. [DOI] [PubMed] [Google Scholar]
- 42.Xiao R, et al. Splicing regulator SC35 is essential for genomic stability and cell proliferation during mammalian organogenesis. Mol Cell Biol. 2007;27(15):5393–5402. doi: 10.1128/MCB.00288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.