Abstract
Acute and chronic tissue injury results in the generation of a myriad of environmental cues that macrophages respond to by changing their phenotype and function. This phenotypic regulation is critical for controlling tissue inflammation and resolution. Here, we have identified the adaptor protein disabled homolog 2 (DAB2) as a regulator of phenotypic switching in macrophages. Dab2 expression was upregulated in M2 macrophages and suppressed in M1 macrophages isolated from both mice and humans, and genetic deletion of Dab2 predisposed macrophages to adopt a proinflammatory M1 phenotype. In mice with myeloid cell–specific deletion of Dab2 (Dab2fl/fl Lysm-Cre), treatment with sublethal doses of LPS resulted in increased proinflammatory gene expression and macrophage activation. Moreover, chronic high-fat feeding exacerbated adipose tissue inflammation, M1 polarization of adipose tissue macrophages, and the development of insulin resistance in DAB2-deficient animals compared with controls. Mutational analyses revealed that DAB2 interacts with TNF receptor–associated factor 6 (TRAF6) and attenuates IκB kinase β–dependent (IKKβ-dependent) phosphorylation of Ser536 in the transactivation domain of NF-κB p65. Together, these findings reveal that DAB2 is critical for controlling inflammatory signaling during phenotypic polarization of macrophages and suggest that manipulation of DAB2 expression and function may hold therapeutic potential for the treatment of acute and chronic inflammatory disorders.
Introduction
Functional heterogeneity is a hallmark of macrophages, and phenotypic polarization of macrophages is critically involved in the control of tissue inflammation and its resolution (1–3). The importance of macrophage phenotypes has been highlighted in many pathologies (3, 4) including inflammatory cancers (5), infectious and autoimmune diseases (6), and metabolic chronic diseases such as atherosclerosis (7–9) and diabetes (10, 11). Importantly, pharmacologic modulation of phenotypic polarization (“re-educating macrophages”) has been shown to affect disease progression and outcome in experimental models (12). Extracellular cues that induce macrophage phenotypic polarization are well characterized; however, much less is known regarding intracellular regulators of signaling mechanisms that control macrophage phenotypic polarization. Macrophages that are classically activated by IFN-γ and TLR ligands polarize to an M1 phenotype that induces a Th1 response and thus is important for effective host defense. Various forms of alternatively activated or M2 macrophage phenotypes are induced by IL-4, IL-10, IL-13, glucocorticoids, or immune complexes and are important immune regulators associated with tissue healing and the resolution of inflammation (13). M2 macrophages express Arg1, Mgl1, CD206, and CD163, while M1 macrophages are characterized by the expression of primarily NF-κB–dependent genes including Il6, Tnfa, Nos2, and Il1b (14). Furthermore, CD11c has been reported to be a marker for M1 macrophage activation in adipose tissue(10).
NF-κB–dependent gene expression plays a pivotal role in the classical (M1) activation of macrophages and was also shown to facilitate the resolution of inflammation (15, 16). Stimulation of cells with TLR ligands such as LPS or lipoteichoic acid (LTA) induces a common pathway of MyD88-dependent signaling. This pathway promotes the assembly of a complex containing TNF receptor–associated factor 6 (TRAF6) and TGF-activated kinase 1 (TAK1) (17), resulting in the activation of either IκB kinases (IKKs) or MKK6, which in turn leads to NF-κB– or p38 MAPK–dependent gene expression (18). NF-κB is a heterodimeric transcription factor typically composed of p65 (also known as RelA) and p50. In resting cells, NF-κB resides in the cytosol, bound to IκB. TLR ligands stimulate the proteasomal degradation of IκB through phosphorylation by IKKα and subsequent ubiquitination. These events promote the nuclear translocation of NF-κB and the transcriptional activation of its target genes. In this context, TAK1-dependent activation of IKKβ was shown to directly phosphorylate p65 in its transactivation domain (Ser536) (19), thereby enhancing its transcriptional activity (20). Although the activation mechanisms of NF-κB have been studied in considerable detail, much less is known about the mechanisms that restrain NF-κB activation at different stages of the inflammatory response.
To discover novel regulators of macrophage phenotypic polarization, we compared gene expression patterns of various macrophage phenotypes that had been polarized in vitro. We identified disabled homolog 2 (Dab2) (21) as a gene that is differentially regulated in M1 and M2 macrophages. DAB2 is a mitogen-responsive phosphoprotein with homology to Drosophila-disabled protein and was first identified in a murine macrophage cell line (22). Dab2 is evolutionarily conserved with high homology between species, especially in the N-terminal phosphotyrosine-binding (PTB) domain (23). Human DAB2 is highly similar to murine DAB2; both full-length proteins are composed of 15 exons with approximately 83% identity (24), and global Dab2 deficiency in mice results in lethality due to developmental defects (25). Because of its ability to bind to FXNPXY motifs, DAB2 was shown to promote LDL receptor endocytosis (26, 27) and to regulate endosomal recycling of the TGF-β receptor (28), the VEGF receptor (29), and CFTR (30, 31). In mice, the 2 major splice variants of Dab2 are p67 and p96 (22). The p67 splice variant of DAB2 lacks a region that includes 2 DPF amino acid motifs, NPF motifs, and a clathrin box, which participate in the interaction of DAB2 with AP-2 and its localization to clathrin-coated pits (32). Moreover, DAB2 was shown to regulate the Wnt signaling pathway (33) and the differentiation of Tregs (34). Because Dab2 is actively downregulated in various tumor cell lines, it has been proposed to be a tumor suppressor (35–39). IFN-γ, which promotes M1 phenotypic polarization in macrophages, induces the expression of IFN consensus sequence–binding protein (ICSBP, also known as IRF8), which in turn transcriptionally represses Dab2 (40). Upregulation of DAB2 was observed in spinal cord lesions in murine experimental autoimmune encephalomyelitis (41).
Here, we identify an unexpected role for DAB2 as an inhibitor of the NF-κB–signaling pathway and a regulator of macrophage phenotypic polarization. We show that DAB2 inhibits NF-κB–dependent transcription by binding to TRAF6 and repressing Ser536 phosphorylation in the p65 transactivation domain of NF-κB. As a result, Dab2 deficiency compromises the negative regulation of NF-κB and predisposes macrophages toward proinflammatory M1 phenotypic polarization. Through the analysis of multiple mouse models, we demonstrate that DAB2 controls the progression and severity of acute and chronic inflammation by regulating NF-κB–dependent macrophage phenotypic polarization.
Results
Dab2 expression is differentially regulated in M1 and M2 macrophages.
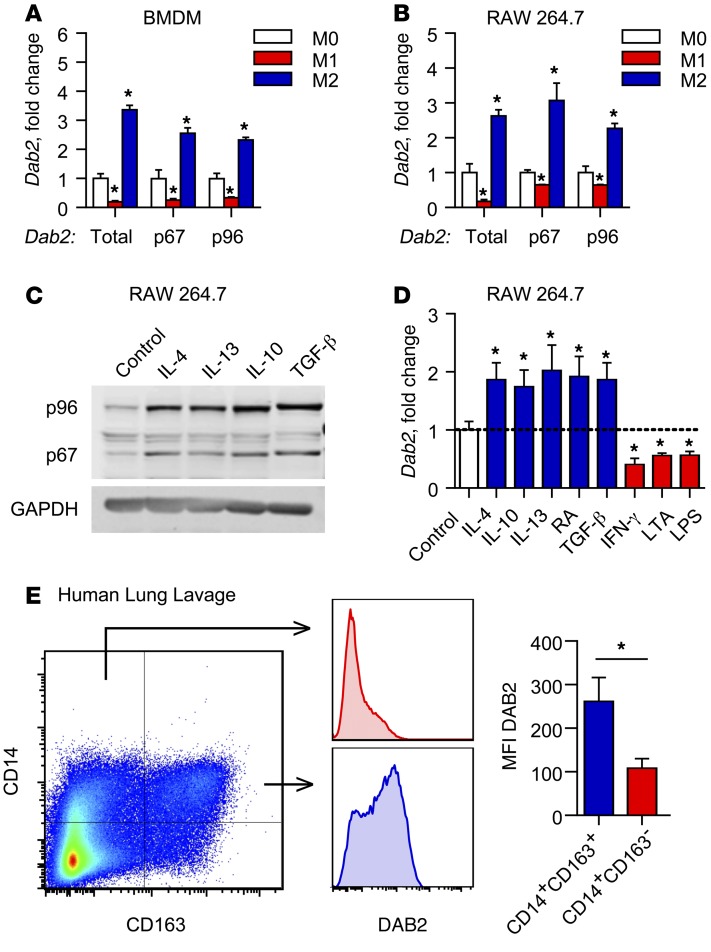
To identify novel regulators of macrophage phenotypic polarization, we compared gene expression patterns in M1 and M2 macrophages using Affymetrix gene arrays (9). We treated bone marrow–derived macrophages (BMDMs) isolated from C57Bl/6 mice with IFN-γ and LPS to produce classically activated M1 macrophages, or with IL-4 to produce alternatively activated M2 macrophages, as described previously (9). A screen for genes that are differentially regulated in M1 and M2 macrophages identified Dab2 among genes that are induced in M2 macrophages but downregulated in M1 macrophages (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI79590DS1). Using primers specifically designed to detect total Dab2 as well as its splice variants Dab2-p67 and Dab2-p96, we found that, compared with untreated macrophages (M0), expression of total Dab2 and both individual splice variants was downregulated in M1 BMDMs, but upregulated in M2 BMDMs (Figure 1A) as well as in phenotypically polarized RAW 264.7 macrophages (Figure 1B). Successful polarization to M1 or M2 macrophages was confirmed by induction of Il6 (M1) or Arg1 (M2) expression, respectively (Supplemental Figure 1B).
Figure 1. Dab2 expression is differentially regulated in M1 and M2 macrophages.
mRNA expression of total Dab2 and p67 and p96 Dab2 splice variants was determined by qPCR of BMDMs (A) and RAW 264.7 macrophages (B). For A and B, cells were treated with control media (M0), 1 μg/ml LPS and 10 U/ml IFN-γ (M1), or 10 ng/ml IL-4 (M2) for 3 hours. Results were normalized to B2m (encoding β-2-microglobulin) mRNA and are presented as fold change relative to M0. (A and B) Data are representative of 3 independent experiments. *P < 0.0001, by 2-way ANOVA with Dunnett’s multiple comparisons test. (C) Western blot analysis of p96-DAB2 and p67-DAB2 protein in RAW 264.7 macrophages treated for 24 hours with control media, 10 ng/ml IL-4, 10 ng/ml IL-13, 10 ng/ml IL-10, or 5 ng/ml TGF-β. GAPDH was used as a loading control. (D) qPCR analysis of total Dab2 mRNA expression in RAW 264.7 macrophages treated for 3 hours with control media (M0), 10 ng/ml IL-4, 10 ng/ml IL-10, 10 ng/ml IL-13, 10 μM 9-cis-retinoic acid (RA), 5 ng/ml TGF-β, 10 U/ml IFN-γ, 1 μg/ml LPS, or 1 μg/ml LTA. Results are normalized to B2m mRNA and are presented relative to M0. Data were compiled from 2 independent experiments and are presented as the mean ± SEM of quadruplicate treatments. *P < 0.05, by Student’s t test or Mann-Whitney U test. (E) DAB2 was preferentially expressed in M2-like human macrophages that expressed CD163. BAL fluid obtained from critically ill patients was analyzed by flow cytometry for CD14 and the macrophage M2 marker CD163. DAB2 MFI was determined for CD14+CD163– and CD14+CD163+ cells (bar graph shows the mean ± SEM, n = 6). *P < 0.02, by 2-tailed, paired Student’s t test.
Next, we tested a panel of pro- and antiinflammatory activators for their ability to regulate Dab2 mRNA expression. DAB2 protein and mRNA expression was induced in RAW 264.7 macrophages by various M2 polarization stimuli including IL-4, IL-13, and IL-10, as well as known inducers of Dab2 expression, retinoic acid and TGF-β (ref. 42 and Figure 1, C and D). Conversely, Dab2 mRNA expression was suppressed by IFN-γ (a known repressor of Dab2 expression, ref. 40), but also by LTA (a TLR2 ligand) and LPS (a TLR4 ligand) (Figure 1D and Supplemental Figure 1C). Consistent with DAB2 downregulation by IFN-γ, the expression of Irf8 — a known IFN-γ–dependent negative regulator of Dab2 transcription (40) — was induced in M1, but not M2, macrophages (Supplemental Figure 1D). Moreover, in the presence of cycloheximide, treatment with LPS significantly reduced the half-life of DAB2 protein in BMDMs (Supplemental Figure 1E).
To examine whether DAB2 is also preferentially expressed in human M2 macrophages, we analyzed macrophages in bronchoalveolar lavage (BAL) fluid from critically ill patients who were experiencing respiratory inflammation. DAB2 protein was predominantly expressed in CD14+CD163+ cells (Figure 1E), indicative of alternatively polarized M2 macrophages. Together, these data demonstrate that the expression level of DAB2 in macrophages is controlled in a phenotype-specific manner, implying that DAB2 may play a role in the regulation of inflammatory gene expression.
DAB2 expression levels control inflammatory gene expression.
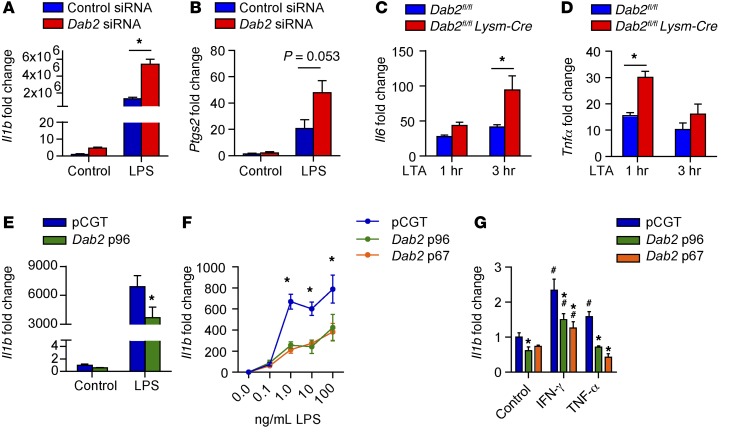
To examine whether DAB2 controls the expression of inflammatory genes, we performed loss-of-function and gain-of-function experiments using RAW 264.7 macrophages. First, we attenuated Dab2 expression in RAW 264.7 macrophages using siRNA (Supplemental Figure 2A), which resulted in significantly enhanced LPS-induced Il1b and Ptgs2 (cyclooxygenase 2) mRNA expression (Figure 2, A and B). Furthermore, LTA treatment of Dab2-deficient BMDMs (isolated from Dab2fl/fl Lysm-Cre mice) resulted in significantly increased expression of Il6 and Tnfa transcripts (Figure 2, C and D).
Figure 2. Dab2 expression controls inflammatory gene expression.
qPCR analysis of Il1b (A) or Ptgs2 (B), which encodes cyclooxygenase 2, in RAW 264.7 macrophages treated with control media only (control) or 100 ng/ml LPS for 3 hours. Before treatment, RAW 264.7 macrophages were transfected with control or Dab2 siRNA for 48 hours. Results were normalized to B2m mRNA and are presented relative to control. (A and B) *P < 0.0001, by 2-way ANOVA with Tukey’s multiple comparisons test. Data represent the mean ± SEM of of 4 technical replicates and are representative of 2 independent experiments. (C and D) BMDMs from Dab2fl/fl and Dab2fl/fl Lysm-Cre mice were cultured in the presence of GM-CSF (5 ng/ml) for 2 days before treatment with LTA for 1 or 3 hours. Il6 (C) and Tnfa (D) mRNA levels were then measured by qPCR. (C and D) *P < 0.001, by 2-way ANOVA with Sidak’s multiple comparisons test. (E) qPCR analysis of Il1b in RAW 264.7 macrophages transfected with pCGT or pCGT-DAB2 and treated with 100 ng/ml LPS for 3 hours. *P < 0.05, by 2-tailed, unpaired Student’s t test. (F) Il1b mRNA expression was measured in RAW 264.7 macrophages transfected with pCGT, pCGT-p96-Dab2, or pCGT-p67-Dab2 and treated with increasing concentrations of LPS. *P < 0.0001, by 2-way ANOVA with Tukey’s multiple comparisons test. (G) Il1b mRNA expression was measured in RAW 264.7 macrophages transfected with pCGT, pCGT-p96-Dab2, or pCGT-p67-Dab2 and treated with IFN-γ or TNF-α. *P < 0.0001 compared with pCGT and #P < 0.0001 compared with control, by 2-way ANOVA with Tukey’s multiple comparisons test. Data represent the mean ± SEM of quadruplicate treatments.
Conversely, overexpression of DAB2 by transient transfection of RAW 264.7 macrophages (Supplemental Figure 2B) significantly attenuated LPS-induced Il1b mRNA expression compared with cells transfected with the empty vector control plasmid (pCGT) (Figure 2E). Overexpression of individual splice variants Dab2-p67 and Dab2-p96 (Supplemental Figure 2C) inhibited LPS-induced Il1b mRNA expression to a similar extent (Figure 2F). The negative regulation of inflammatory gene expression by DAB2 was not restricted to TLR4/2 activation by LPS or LTA, since similar results were obtained when cells were treated with IFN-γ or TNF-α (Figure 2G) after overexpression of Dab2-p67 or Dab2-p96 (Supplemental Figure 2D).
Myeloid DAB2 controls the inflammatory response to endotoxemia.
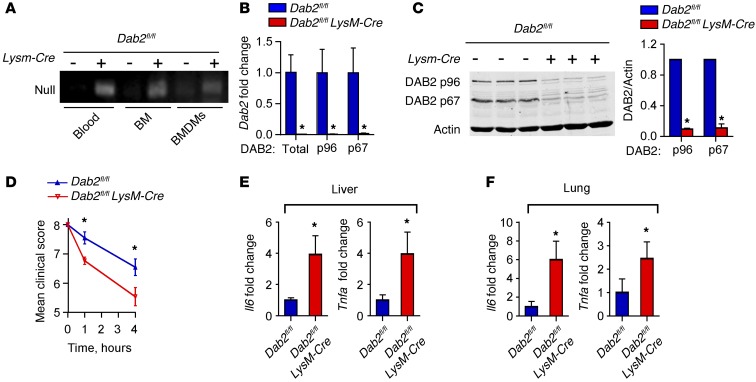
To study the role of DAB2 in macrophage polarization in vivo, we generated myeloid-specific Dab2-deficient mice by crossing Dab2fl/fl mice with Lysm-Cre mice (Dab2fl/fl Lysm-Cre). Genomic DNA analysis of blood, bone marrow (BM), BMDMs, and peritoneal lavage cells showed Cre recombinase–mediated deletion of the second coding exon of Dab2 in the Dab2fl/fl Lysm-Cre animals (Figure 3A). Deletion of myeloid Dab2 was further confirmed by quantitative PCR (qPCR) (Figure 3B) and Western blotting (Figure 3C), which demonstrated the absence of mRNA and protein for total Dab2 and both major splice variants Dab2-p67 and Dab2-p96 in Dab2fl/fl Lysm-Cre BMDMs.
Figure 3. Myeloid DAB2 regulates the inflammatory response to endotoxemia.
(A) PCR analysis of genomic DNA isolated from blood, BM, or BMDMs of Dab2fl/fl and Dab2fl/fl Lysm-Cre littermates. The presence of a null allele (213 bp) verified Cre recombinase activity (deletion of the second coding exon of Dab2). (B) qPCR analysis of total Dab2 or p96 and p67 splice variants of Dab2 mRNA expression in BMDMs from Dab2fl/fl and Dab2fl/fl Lysm-Cre littermates. Results were normalized to B2m mRNA and are presented relative to Dab2fl/fl mRNA levels. (C) Representative immunoblot analysis of DAB2 protein expression in BMDMs from 3 Dab2fl/fl and Dab2fl/fl Lysm-Cre littermates. Densitometric analysis is from 3 independent experiments with 3 littermate pairs, presented as the ratio of DAB2 to actin. Data represent the mean ± SD. *P < 0.05, by 2-tailed, unpaired Student’s t test. (D–F) Dab2fl/fl and Dab2fl/fl Lysm-Cre mice were injected i.p. with 2 mg/kg LPS (E. coli 0111:B4). (D) The endotoxemia score was analyzed on a scale of 8 (healthy) to 0 (poor) at 0, 1, and 4 hours after injection. Data represent the mean ± SEM; n = 9 (Dab2fl/fl) and n = 11 (Dab2fl/fl Lysm-Cre) mice. *P < 0.001, by 2-way ANOVA with Sidak’s multiple comparisons test. (E and F) Il6 and Tnfa mRNA expression levels in liver (E) and lung (F) homogenates were analyzed by qPCR. Data were normalized to B2m and are expressed relative to Dab2fl/fl levels as the mean ± SEM; n = 8 (Dab2fl/fl) and n = 10 (Dab2fl/fl Lysm-Cre) mice. *P < 0.05, by 2-tailed, unpaired Student’s t test.
Dab2fl/flLysm-Cre mice reproduced and developed normally, and neither showed differences in blood cell counts or characteristics (Supplemental Figure 3, A and B), nor in the abundance of monocyte subsets, as analyzed by Ly6Chi-, Ly6Cmid-, and Ly6Clo-expressing CD11b+ cells (Supplemental Figure 3C). We observed no overt phenotype, and there were no differences in body weight or food intake between Dab2fl/fl Lysm-Cre mice and their WT littermates (data not shown). Moreover, BM-derived cells from Dab2fl/fl Lysm-Cre mice readily differentiated into macrophages when cultured in macrophage-CSF–rich (M-CSF–rich) medium (data not shown).
To examine whether myeloid DAB2 would control inflammatory gene expression in vivo, Dab2fl/fl Lysm-Cre mice or their littermate controls were challenged with sublethal i.p. injection of LPS. As early as 1 and 4 hours after LPS challenge, Dab2fl/fl Lysm-Cre mice demonstrated a significantly poorer clinical score (ref. 43 and Figure 3D). Four hours after injection, mRNA expression of Il6 and Tnfa was significantly increased in the livers (Figure 3E) and lungs (Figure 3F) of Dab2fl/fl Lysm-Cre mice. After LPS injection into the peritoneum of Dab2fl/fl Lysm-Cre mice, total cell numbers in the peritoneum were not different from those in control mice injected with LPS, suggesting that the migratory capacity of macrophages was not impaired by DAB2 deficiency (Supplemental Figure 3D). Together, these data demonstrate that DAB2 expression levels in macrophages regulate inflammatory gene expression in vitro and in vivo, suggestive of a role for DAB2 in controlling macrophage phenotypic polarization.
Dab2 deficiency promotes M1 macrophage phenotypic polarization and regulates phenotypic switching.
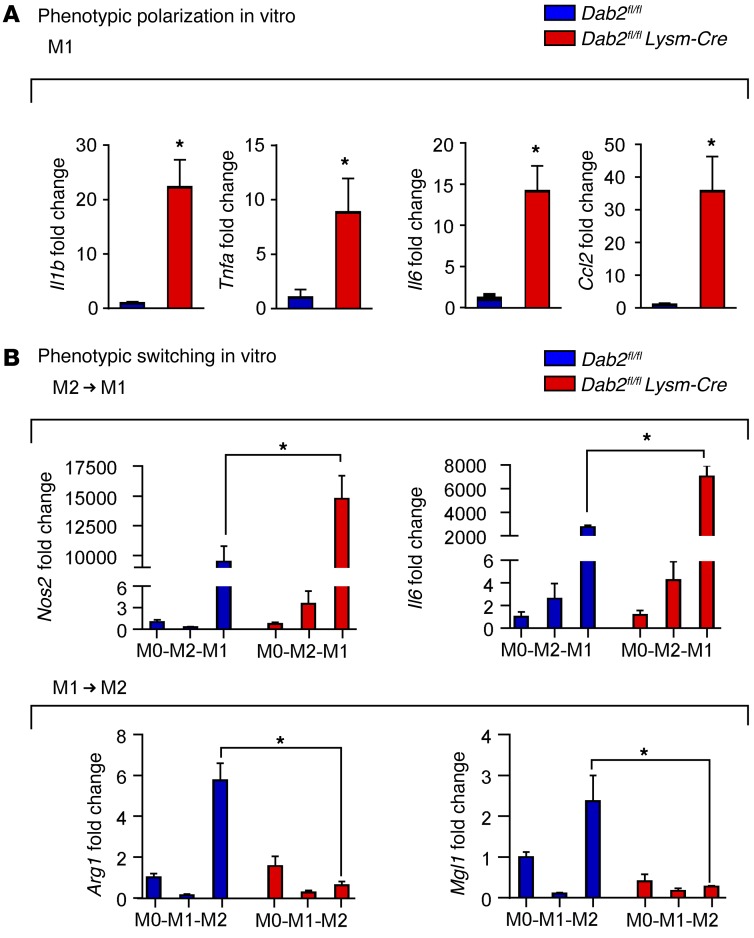
To address the hypothesis that DAB2 regulates macrophage phenotypic polarization, we treated BMDMs isolated from Dab2fl/fl or Dab2fl/fl Lysm-Cre mice with LPS and IFN-γ to induce M1 polarization. A panel of specific M1 marker genes was quantified by qPCR. We found that in the absence of DAB2, macrophages polarized with LPS and IFN-γ expressed significantly higher levels of the M1-associated genes Il1b, Tnfa, Il6, and Ccl2 (Figure 4A).
Figure 4. Dab2 deficiency promotes M1 macrophage phenotypic polarization and regulates phenotypic switching.
(A) qPCR analysis of Il1b, Tnfa, Il6, and Ccl2 mRNA expression in Dab2fl/fl and Dab2fl/fl Lysm-Cre BMDMs polarized to M1 for 3 hours. Results were normalized to B2m mRNA and are presented as fold change relative to Dab2fl/fl. Data represent the mean ± SEM of 4 technical replicates and are representative of 2 independent experiments. *P < 0.05, by 2-tailed, unpaired Student’s t test or Mann-Whitney U test. (B) BMDMs from Dab2fl/fl and Dab2fl/fl Lysm-Cre mice were serum starved for 3 hours (M0) and then polarized to M2 (IL-4) or M1 (LPS and IFN-γ) for 8 hours. Media were removed, and then M2 macrophages were treated with M1 stimuli (LPS and IFN-γ), and M1 macrophages were treated with M2 stimuli (IL-4) for an additional 8 hours. qPCR was performed to measure Nos2 and Il6 gene expression in M2 macrophages polarized to M1. Arg1 and Mgl1 expression was measured in M1 macrophages polarized to M2. Data represent the mean ± SEM of quadruplicate experiments. *P < 0.05, by 2-tailed, unpaired Student’s t test or Mann-Whitney U test.
Phenotypically polarized macrophages have the potential to switch their phenotype as tissue inflammation progresses and enters the resolution phase. This type of phenotypic switch can also be induced in vitro (9). To examine whether DAB2 plays a role in the phenotypic switching of macrophages, we polarized BMDMs isolated from Dab2fl/fl or Dab2fl/fl Lysm-Cre mice into M1 or M2 macrophages, before switching them back into M2 or M1 macrophages, respectively. Our results show that DAB2 deficiency significantly enhances the propensity of M2 macrophages to switch into an M1 phenotype, but inhibits the switch from M1 into M2 macrophages (Figure 4B). In the absence of DAB2, M2 macrophages that had been switched to M1 showed enhanced expression of Nos2 and Il6, while DAB2-deficient M1 macrophages that had been switched to M2 showed significantly lower expression levels of the M2-related genes Arg1 and Mgl1 (Figure 4B). Together, these data show that DAB2 deficiency promotes phenotypic polarization toward M1 and controls phenotypic switching of macrophages.
DAB2 in myeloid cells regulates high-fat-diet–induced adipose tissue inflammation and insulin resistance.
In obesity, low-grade inflammation of the adipose tissue is characterized by the infiltration and phenotypic polarization of macrophages in adipose tissue (10). Exacerbation of the inflammatory response by M1 macrophages may contribute to the development of insulin resistance (11). To examine whether DAB2 also regulates the response to inflammatory mediators that have been shown to be relevant in obesity-related insulin resistance, we treated BMDMs isolated from Dab2fl/fl and Dab2fl/fl Lysm-Cre mice with TNF-α or palmitate. Expression of Il1b, Il6, or Tnfa was significantly higher in DAB2-deficient cells when treated with palmitate (Supplemental Figure 4A) or TNF-α (Supplemental Figure 4B).
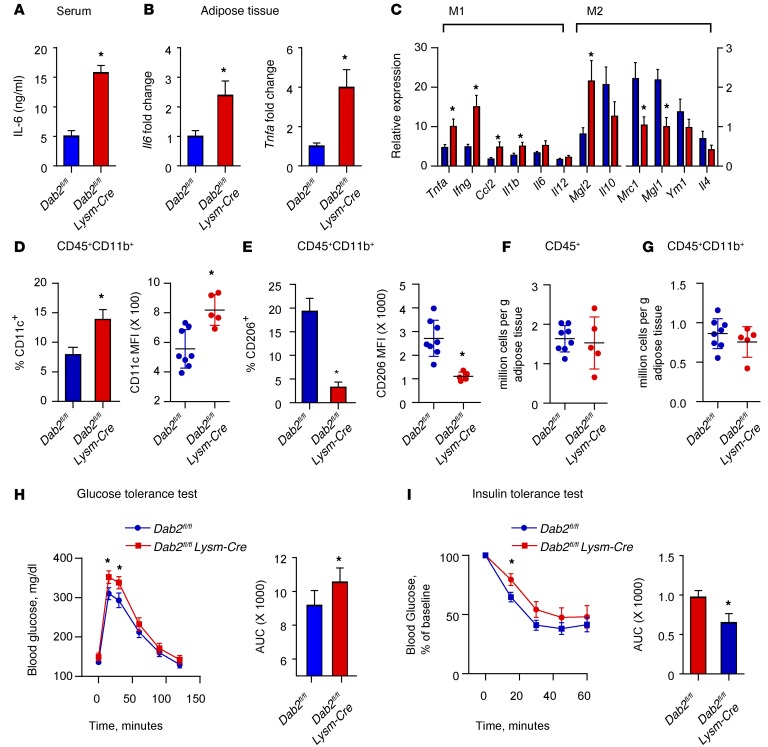
To examine the role of myeloid DAB2 in diet-induced inflammation, we fed Dab2fl/fl Lysm-Cre mice and their littermate Dab2fl/fl controls either a regular chow or a high-fat diet (HFD) for 12 weeks (conditions that induce obesity, adipose tissue inflammation, and insulin resistance). Of note, after HFD feeding, there was no difference between Dab2fl/fl and Dab2fl/fl Lysm-Cre animals in blood cell counts, body weight gain, serum insulin, food consumption, or serum lipid levels (Supplemental Figure 5, A–C, and data not shown). However, Dab2fl/fl Lysm-Cre mice on an HFD showed significantly increased serum IL-6 levels, indicative of an increased systemic inflammatory response to HFD feeding (Figure 5A). Moreover, in Dab2fl/fl Lysm-Cre mice on an HFD, Il6 and Tnfa mRNA levels were significantly increased in the adipose tissue (Figure 5B). Next, we analyzed gene expression in the adipose tissue stromal vascular fraction (SVF), which contains macrophages. Expression of a panel of proinflammatory M1–type genes was significantly increased in the SVF isolated from the adipose tissue of Dab2fl/fl Lysm-Cre mice on an HFD, while expression of antiinflammatory M2–like genes was either unchanged or decreased (Figure 5C). Analyses by flow cytometry (see Supplemental Figure 5D for gating strategy) of the CD11b+ cell population in the SVF, which contains mainly macrophages (44), demonstrated a significant increase in CD11c+ cells and mean fluorescence intensity (MFI) (Figure 5D and Supplemental Figure 5E) and a significant decrease in CD206+ cells and MFI (Figure 5E and Supplemental Figure 5F) in Dab2fl/fl Lysm-Cre mice on an HFD, consistent with a strong shift toward M1 polarization. Total numbers of CD45+ or CD45+CD11b+ cells were not different in the SVF of Dab2fl/fl Lysm-Cre mice on an HFD (Figure 5, F and G), indicating that DAB2 controls phenotypic polarization of macrophages, without affecting migration or proliferation.
Figure 5. DAB2 in myeloid cells regulates HFD-induced adipose tissue inflammation and insulin resistance.
Dab2fl/fl and Dab2fl/fl Lysm-Cre female mice were fed an HFD for 12 weeks. (A) Serum IL-6 levels after an HFD for 12 weeks as analyzed by ELISA. (B) Il6 and Tnfa mRNA levels in adipose tissue were analyzed by qPCR. (C) pPCR phenotypic characterization of cells isolated from the SVF in adipose tissue. (D–G) Cells isolated from the SVF in gonadal adipose tissue of HFD-fed Dab2fl/fl and Dab2fl/fl Lysm-Cre mice were stained with CD45-APC-Cy7, CD11b-FITC, CD206-PE, and CD11c-AF647 Abs and subjected to FACS analysis. CD45+CD11b+ cells were analyzed for the percentage of cells that were CD11c+ and for the MFI of CD11c (D), and CD45+CD11b+ cells were analyzed for the percentage of cells that were CD206+ and for the MFI of CD206 (E). Each point represents 1 mouse in the MFI graphs in D and E. *P = 0.003, by 2-tailed, unpaired Student’s t test (D) and *P = 0.004, by 2-tailed, unpaired Student’s t test with Welch’s correction (E). Absolute numbers of CD45+ (F) and CD45+CD11b+ (G) cells per gram of gonadal adipose tissue were quantified and showed no difference in cell infiltration levels between HFD-fed Dab2fl/fl and Dab2fl/fl Lysm-Cre mice. For glucose tolerance tests (H) and insulin tolerance tests (I), mice were fasted for 6 hours and injected i.p. with a 1 g/kg glucose bolus or 0.75 U/kg insulin, respectively, and blood glucose levels were measured in tail blood by glucometer. Data were analyzed by calculating the AUC and represent the mean ± SEM. n = 10–12. *P < 0.05, by 2-tailed, unpaired Student’s t test.
Increased macrophage-dependent inflammation due to phenotypic polarization in the adipose tissue has been shown to be associated with the development of insulin resistance (45). Indeed, glucose tolerance and insulin tolerance tests demonstrated increased insulin resistance in Dab2fl/fl Lysm-Cre mice on an HFD (Figure 5, H and I). There was no difference in fasting glucose levels between chow-fed Dab2fl/fl Lysm-Cre and Dab2fl/fl mice over the feeding period; however, HFD feeding increased fasting glucose levels in both groups (Supplemental Figure 5G). There was no difference in insulin or glucose tolerance between chow-fed Dab2fl/fl Lysm-Cre and Dab2fl/fl mice (Supplemental Figure 5, H and I), indicating that the observed differences in insulin tolerance in HFD-fed Dab2fl/fl Lysm-Cre mice resulted from increased inflammation. These data demonstrate that, in a model of chronic high-fat feeding, DAB2 in myeloid cells modulates the inflammatory status and phenotypic polarization of adipose tissue macrophages, which impacts the development of insulin resistance.
DAB2 regulates NF-κB–dependent gene expression via interaction with TRAF6.
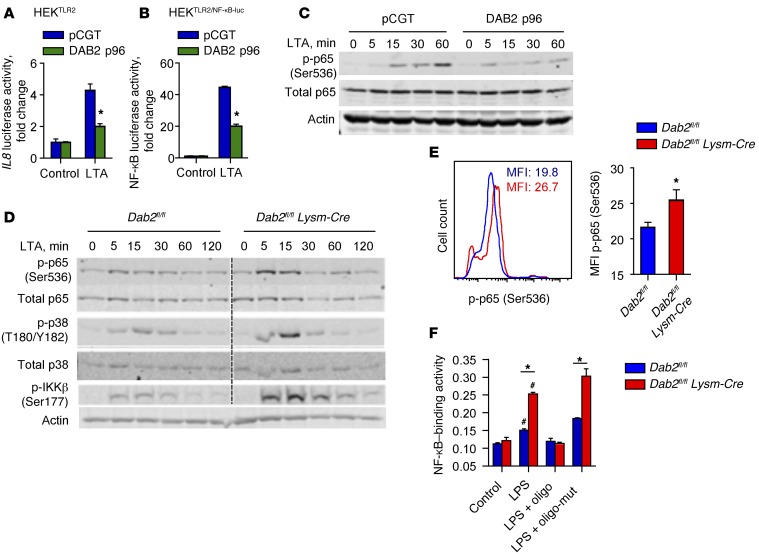
Expression of inflammatory genes in classically activated M1 macrophages is largely regulated by the transcription factor NF-κB. To test the hypothesis that DAB2 controls NF-κB–dependent gene expression, we used HEK293 cells, because they express very low levels of endogenous DAB2. In HEK293 cells stably expressing TLR2 (HEKTLR2), treatment with LTA induced the expression of a human IL8 promoter–luciferase reporter construct. Cotransfection of DAB2 significantly attenuated reporter gene expression (Figure 6A). Next, we produced HEK293 cells that stably coexpressed a 5× NF-κB promoter–luciferase reporter and TLR2 (HEKTLR2/NF-κB–luc). Ectopic expression of DAB2 in HEKTLR2/NF-κB–luc cells inhibited LTA-induced NF-κB–dependent luciferase expression (Figure 6B), as well as LTA-induced phosphorylation of Ser536 in the transactivation domain of NF-κB p65 (Figure 6C). Conversely, phosphorylation of NF-κB p65 at Ser536 was enhanced in DAB2-deficient BMDMs (Figure 6D) and in peritoneal macrophages isolated from LPS-challenged Dab2fl/fl Lysm-Cre mice (Figure 6E). In addition to enhanced phosphorylation of NF-κB p65, DAB2 deficiency also increased p38 MAPK and IKKβ phosphorylation in LTA-treated BMDMs (Figure 6D). Finally, in LPS-treated DAB2-deficient BMDMs, the binding of p65 to its consensus DNA sequence was significantly higher (Figure 6F). While LPS treatment induced NF-κB translocation to the nucleus, DAB2 remained in the cytosol (Supplemental Figure 6), further indicating that the inhibitory activity of DAB2 on NF-κB signaling involves interaction with signaling molecules outside of the nucleus.
Figure 6. DAB2 regulates NF-κB activity by suppressing phosphorylation of p65 (Ser536).
(A) HEKTLR2 cells were cotransfected with an IL8 promoter–luciferase reporter construct and either pCGT (control) or pCGT–DAB2 p96 and then treated overnight with control media only (control) or LTA (1 μg/ml). (B) HEKTLR2/NF-κB–luc cells were transfected with either pCGT or pCGT-DAB2 p96 and then stimulated with LTA (1 μg/ml) overnight. Luciferase activity was measured by luminometry (BioTek Synergy HT). (A and B) *P < 0.001, by 2-way ANOVA with Sidak’s multiple comparisons test. (C) Immunoblot analysis of total p65 and p-p65 (Ser536) in pCGT and pCGT-DAB2–transfected HEKTLR2 cells that were treated with 1 μg/ml LTA for 0, 5, 15, 30, and 60 minutes. Actin served as a loading control. (D) BMDMs from Dab2fl/fl and Dab2fl/fl Lysm-Cre mice were treated with LTA for the indicated times, and protein lysates were immunoblotted using Abs to detect p-p38 (T180/Y182), total p38, p-p65 (Ser536), total p65, p-IKKβ (Ser177), and β-actin. (E) Dab2fl/fl and Dab2fl/fl Lysm-Cre mice were injected i.p. with 2 mg/kg LPS (E. coli 0111:B4). After 4 hours, peritoneal cells were isolated and stained with anti–F4/80 Alexa Fluor 647, anti–CD11b Alexa Fluor 488, and anti–p-p65 (Ser536) Abs. F4/80hiCD11bhi cells were gated, and the MFI of Ser536 p-p65–specific fluorescence was analyzed. Data represent the mean ± SEM; n = 7. *P < 0.05, by 2-tailed, unpaired Student’s t test. (F) NF-κB DNA–binding activity in nuclear extracts from Dab2fl/fl and Dab2fl/fl Lysm-Cre BMDMs treated with 100 ng/ml LPS for 1 hour was analyzed by TransAM EMSA (Active Motif). For competitive binding studies, functional (oligo) or nonfunctional (oligo-mut) oligonucleotides were used according to the manufacturer’s instructions. Data are expressed as NF-κB–binding activity at A450 to A650 nm (mean ± SD) of triplicate measurements. A, absorbance. *P < 0.0001 and #P < 0.02 compared with control and compared with LPS plus oligonucleotide, by 2-tailed, unpaired Student’s t test.
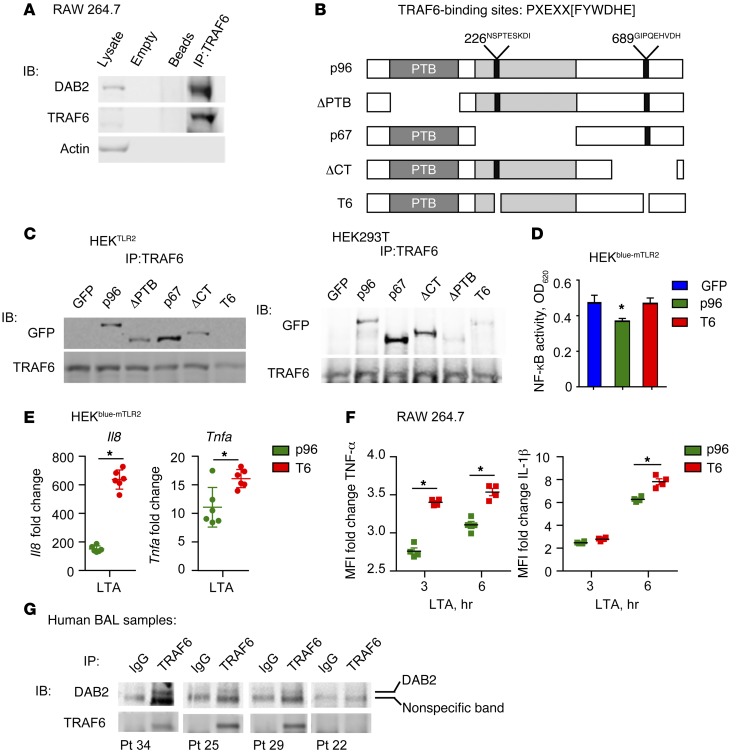
Together, these data demonstrate that DAB2 regulates NF-κB–dependent signaling by controlling phosphorylation of Ser536 in the p65 transactivation domain, most likely through IKKβ activation. The signaling complex directly upstream of both IKK and MKK6 that leads to activation of NF-κB and p38, respectively, is initiated by TRAF6 and TAK1. Indeed, co-IP from lysates derived from RAW 264.7 macrophages revealed an interaction between endogenous DAB2 and TRAF6 (Figure 7A). Through a screen for sequence domains, we identified 2 putative TRAF6-binding domains in DAB2 (Figure 7B) located at amino acid positions 226 and 689, which are conserved in human DAB2 and in NUMBL, another DAB2-interacting protein (refs. 46, 47, and Supplemental Table 1). Next, we tested a series of deletion mutants of DAB2 for their capacity to interact with TRAF6; these included full-length p96 DAB2; ΔPTB mutant DAB2, which lacks the PTB domain but contains both putative TRAF6-binding regions; p67 DAB2, which is a naturally occurring splice variant of DAB2 that lacks the endocytic domain containing one of the putative TRAF6-binding regions; C-terminal–deficient–mutant (ΔCT-mutant) DAB2, which lacks the carboxy terminal domain containing one of the putative TRAF6-binding regions; and T6-mutant DAB2, which lacks both of the putative TRAF6-binding regions (Figure 7B). Expression of these mutant DAB2 proteins in HEK293mTLR2 or HEK293T cells, followed by IP with a TRAF6 Ab, revealed that interaction with TRAF6 requires the presence of both putative TRAF6-binding regions in DAB2. Deletion of the PTB domain or the CT domain weakened the interaction of DAB2 with TRAF6, however, the interaction between DAB2 and TRAF6 was drastically reduced when both TRAF6-binding domains (T6) were deleted (Figure 7C).
Figure 7. DAB2 binding to TRAF6 controls NF-κB–dependent gene expression.
(A) Co-IP of endogenous DAB2 and TRAF6 from protein lysates of RAW 264.7 cells. Data are representative of at least 3 independent experiments. (B) The TRAF6-binding site consensus sequence is shown with 2 putative TRAF6-binding sites in DAB2 p96 at amino acid positions 226 and 689. Schematic of N-terminal GFP-tagged DAB2 deletion mutants. (C) TRAF6 IP with DAB2 deletion mutants in HEKTLR2 cells (left) and HEK293T cells (right). (D) HEKblue-mTLR2 cells were transfected with control GFP plasmid, DAB2 p96, or T6 mutant and treated with LTA (1 μg/ml) for 14 hours. Absorbance at 620 nm corresponds to NF-κB activity. The experiment was performed with 5 replicates. Data represent at least 2 independent experiments (C and D) and are expressed as the mean ± SEM. *P = 0.0276, by 1-way ANOVA with Fisher’s least significant difference test (LSD). (E) mRNA expression of Il8 and Tnfa in HEKblue-mTLR2 cells transfected with DAB2 p96 or T6 mutant and treated with LTA (1 μg/ml) for 6 hours. The experiment was performed with 6 replicates. Data represent the mean ± SEM. *P ≤ 0.0092, by 2-tailed, unpaired Student’s t test. Data were normalized to B2m and are expressed as the fold increase compared with untreated cells. (F) RAW 264.7 cells were transfected with either DAB2 p96 or T6 mutant before stimulation with LTA for 3 and 6 hours. Expression of IL-1β or TNF-α was analyzed by flow cytometry in GFP+ cells. Data represent at least 2 independent experiments and are expressed as the fold change in cytokine MFI compared with nontreated cells. *P < 0.001, by 2-way repeated-measures ANOVA with Sidak’s multiple comparisons test (mean ± SD). (G) Cells obtained from BAL fluid from critically ill patients were subjected to co-IP of TRAF6 and DAB2. Anti-TRAF6 Ab or control IgG was used in each sample for IP, followed by immunoblotting (IB). The specific DAB2 band is indicated.
Next, we tested whether the interaction with TRAF6 was required for DAB2 to inhibit NF-κB–dependent gene expression. Expression of DAB2 p96 inhibited LTA-induced NF-κB transcriptional activity in HEKblue-mTLR2 cells, while the TRAF6-binding–deficient mutant protein T6 had no effect (Figure 7D). Furthermore, the fold increase of LTA-induced Il8 or Tnfa mRNA expression was significantly higher when the T6 mutant was expressed, compared with expression of the DAB2 p96 WT protein (Figure 7E). Finally, the transfection of RAW 264.7 cells with the TRAF6-binding–deficient DAB2 mutant (T6) resulted in significantly higher LTA-induced TNF-α and IL-1β cytokine production when compared with cells transfected with WT DAB2 p96 (Figure 7F). These data demonstrate that the interaction of DAB2 with TRAF6 is required to inhibit TLR-induced inflammatory signaling and gene expression.
To test whether DAB2 also interacts with TRAF6 in humans, we performed co-IP in cells isolated from BAL fluid from critically ill patients who were experiencing respiratory inflammation (Figure 7G). DAB2 was pulled down together with TRAF6, indicating that DAB2-TRAF6 interactions play a role in controlling inflammatory signaling in human disease as well.
Discussion
Controlled changes in the phenotype and function of macrophages are essential for the progression of tissue inflammation and its successful resolution. However, the mechanisms that govern phenotypic polarization of macrophages in acute or chronic inflammatory conditions are not well understood. Here, we demonstrate an unexpected role for the adaptor protein and putative tumor suppressor DAB2 in controlling macrophage phenotypic polarization. Importantly, we found that DAB2 not only regulates the polarization of macrophages to the M1 or M2 phenotype, but is also critically involved in switching between these polarization phenotypes. We observed that Dab2 expression is increased in M2 macrophages and that its transcription is actively repressed in M1 macrophages. Accordingly, DAB2 is predominantly expressed in M2-type, CD163+ macrophages isolated from the lungs of critically ill patients. High DAB2 expression levels prevent classical (M1) activation of macrophages by inhibiting NF-κB–dependent gene expression, while low DAB2 levels facilitate M1 polarization. We show that mice with myeloid cell–specific deletion of Dab2 are significantly more prone to acute endotoxin–induced inflammation and to chronic HFD-induced inflammation, which exacerbates the development of insulin resistance.
Our findings have important implications for the numerous pathological processes in which macrophage phenotypic polarization plays a crucial role. Generally, the controlled balance between M1 and M2 macrophages is critical for the progression and resolution of inflammation (3). Thus, repression of Dab2 expression may be critical for the initiation and continuation of an inflammatory response. Since unrestrained inflammation invariably results in sustained tissue damage, increased expression of Dab2 during the resolution phase may be necessary for proper M2 polarization to prevent a switch back to the M1 phenotype. Indeed, our studies demonstrate that after challenge with LPS (endotoxin), Dab2fl/fl Lysm-Cre mice demonstrated increased inflammatory gene expression in the lungs and liver. In this model, leukocyte infiltration was not affected by the absence of DAB2, indicating that phenotypic and functional changes in myeloid cells accounted for the increased inflammation. Differences in monocyte subsets characterized by Ly6Chi- and Ly6Clo-expressing CD11b+ cells in the peripheral blood were shown to determine polarization of tissue macrophages to the M1 or M2 phenotype, respectively. However, Ly6C-expressing monocyte subsets in blood were not affected by the absence of DAB2, suggesting that DAB2 controls the phenotypic polarization of tissue macrophages during inflammatory cascades. Together, these findings indicate that DAB2 expression serves as a rheostat to dampen the inflammation mediated by activated myeloid cells and thus provides a crucial feedback mechanism to prevent the massive tissue damage that would result from an unrestrained immune response to microbial infection.
In addition to its function in acute inflammation, our results show that DAB2 plays an important role in the control of chronic inflammation. Deletion of Dab2 in myeloid cells exacerbated the inflammation seen in the adipose tissue of mice maintained on an HFD and promoted the development of insulin resistance. Macrophages isolated from the adipose tissue of DAB2-deficient mice on an HFD showed a strong preference for the M1 polarization phenotype, which previous studies have shown to promote insulin resistance (11).
DAB2 has been extensively demonstrated to function as an endocytic adaptor, regulating membrane traffic of a number of receptors. Part of this process requires association with clathrin/AP-2 through an endocytic region that is absent in p67, which is incapable of regulating traffic. Our studies reveal that both p96 and p67 can individually regulate LPS-induced cytokine gene expression, suggesting that regulation of membrane traffic is dispensable for this DAB2 function. Consistent with this, we observed no changes in steady-state cell-surface distribution or secretion of TLR2 to the plasma membrane when Dab2 was deleted (data not shown).
The NF-κB pathway is the prototypic signaling cascade that drives the classical (M1) activation of macrophages. We show that DAB2 negatively regulates NF-κB–dependent proinflammatory gene expression by repressing Ser536 phosphorylation of p65 (p-p65). In macrophages isolated from Dab2fl/fl Lysm-Cre mice treated with LTA, the level of p-p65, p-p38, and p-IKKβ were increased compared with those in WT macrophages at the early time points including 5 and 15 minutes; however, no significant difference was observed at later time points. It is possible that DAB2 plays a role in modulating the early, rather than later, phase of NF-κB activation. We exclude the possibility that DAB2 acts as a transcriptional regulator (48), since DAB2 did not translocate to the nucleus upon activation of the NF-κB pathway (Supplemental Figure 6). In an attempt to identify binding partners within the NF-κB pathway, we discovered two TRAF6-binding motifs [(P)X(E)XX(R)] in DAB2 that seem to be conserved in several proteins that have been described to play regulatory roles in controlling the NF-κB–signaling pathway (Supplemental Table 1). Recently, NUMBL, a PTB domain–containing adaptor protein related to DAB2, was shown to regulate NF-κB signaling by binding to TRAF6 via 2 conserved binding regions (46, 47). Although distinct motifs for NUMBL-TRAF6 interaction were not identified in this study, we found that both DAB2 and NUMBL share 2 conserved TRAF6-interacting domains in similar locations (Supplemental Table 1). Mutational deletion of the homologous TRAF6-binding motifs showed that DAB2 regulates NF-κB activation in the cytosol by binding to TRAF6. We were able to recapitulate the enhanced LTA-induced expression of Il1b or Tnfa observed in DAB2-deficient cells by generating a TRAF6-binding–deficient mutant of DAB2. This observation further highlights the importance of the interaction of DAB2 with TRAF6. These findings demonstrate that the interaction of DAB2 with TRAF6 is required for its inhibitory activity on NF-κB signaling and inflammatory gene expression. Furthermore, we observed that TRAF6 interaction is necessary for DAB2 association with TAK1 (data not shown), demonstrating that DAB2 regulates a signaling-capable complex in macrophages. Other signaling pathways that may potentially be affected by the DAB2-TRAF6-TAK1 complex include the TGF-β–, IL-1β–, and TCR-induced pathways (49), expanding the possible roles for DAB2 in immunity. Further studies will be required to elucidate the effect of DAB2 on these pathways. Importantly, DAB2 (also known as downregulated in ovarian cancer [DOC]) was proposed to be a tumor suppressor (38), and since NF-κB in tumor-associated macrophages (TAMs) plays an important role in tumor progression (12), control of NF-κB signaling by DAB2 may contribute to its tumor-suppressive properties.
In summary, our study identifies DAB2 as a novel regulator of macrophage phenotypic polarization that is independent of its function as a membrane traffic adaptor and reveals a function for DAB2 as a negative regulator of the NF-κB–signaling pathway via interaction with TRAF6. These findings have important implications for developing strategies to trigger resolution of inflammation by inducing phenotypic switching or “re-educating” of macrophages (12), which may be an effective treatment strategy for chronic inflammatory conditions or inflammatory cancers.
Methods
Cell culture and transfection.
RAW 264.7 and HEK293 cells (originally from ATCC but maintained by our laboratory) were maintained in DMEM supplemented with 10% FCS and antibiotics (DMEM 10%) at 37°C, 95% relative humidity, and 5% CO2. BMDMs were cultured for 7 days in RPMI media supplemented with 1% antibiotics, 10% FBS, and M-CSF–rich supernatant from L929 cells. For phenotypic polarization, BMDMs were starved for 3 hours in RPMI containing 2% FCS. For polarization toward the M1 or M2 phenotype, cells were treated with RPMI supplemented with 2% FCS and either 1 μg/ml LPS and 10 U/ml IFN-γ, or 10 ng/ml IL-4, respectively. BMDMs treated with LTA for cytokine analysis were treated with 5 ng/ml granulocyte macrophage–CSF (GM-CSF) in RPMI for 2 days prior to LTA treatment (1 μg/ml). HEK293 cells were transfected using FuGENE 6 (Promega) for 48 hours, PolyJet reagent (SL100688; SignaGen Laboratories) for 24 hours, or Lipofectamine 2000 reagent (Thermo Fisher Scientific) for 24 hours. RAW264.7 macrophages were transfected using the Amaxa Cell Line Nucleofector Kit V (VCA 1003; Lonza). For siRNA transfection, 2 × 106 cells were resuspended in 100 μl solution V containing DAB2 or control siRNA (Dharmacon, GE Healthcare). For plasmid transfection, 2 × 106 cells were resuspended in 100 μl solution V and pCGT or pCGT-DAB2 DNA. In all cases, cells were incubated in DMEM 10% for 48 hours before confirmation of knockdown or overexpression by qPCR and Western blot analysis. For HEK293 cells stably expressing TLR2 and NF-κB luciferase, cells were transfected with pUNO-hTLR2 (Invivogen) or cotransfected with pUNO-hTLR2 and pUBT–NF-κB–luc and then selected in DMEM 10% with 6 μg/ml blasticidin. Colonies were then amplified in DMEM 10% with 3 μg/ml blasticidin. Positive clones (HEK293TLR2 or HEK293TLR2/NF-κB–luc) were identified by immunoblotting for TLR2 and by screening for TLR2-induced luciferase activity using a Luciferase Reporter Assay Kit (Roche). HEKblue-mTLR2 cells were purchased from InvivoGen and were stably transfected with the Tlr2 gene and the mouse CD14 coreceptor. In addition, these cells stably express an optimized secreted alkaline phosphatase (SEAP) reporter gene under the control of a promoter inducible by NF-κB and activator protein 1 (AP-1) transcription factors. The level of SEAP protein released into the culture media was used to quantify the extent of NF-κB activation upon TLR2 stimulation by measuring absorbance at 620 nm with a BioTek Synergy HT microplate reader.
Mice.
Dab2fl/flLysm-Cre mice were generated through appropriate breeding of Lysm-Cre mice with Dab2fl/fl mice (25). Littermate controls were used for all experiments. Genomic DNA was isolated from tail clips by Proteinase K (Bioline) digestion in DirectPCR reagent (Viagen Biotech). Genomic DNA was subjected to PCR using Apex Taq Master Mix (Genesee Scientific) and specific primers. PCR products were resolved by agarose gel electrophoresis. Bands were recognized as WT 362 bp, floxed 432 bp, and null allele at 213 bp. A Cre-specific band was detected at 350 bp using Cre-specific primers. Randomization into groups was not formalized; however, mice were selected for use in experiments on the basis of being aged-matched littermates.
Endotoxemia model.
Endotoxemia was induced by i.p. injection into 8- to 12-week-old male mice of 2 mg/kg LPS (E. coli 0111:B4) for 4 hours. The clinical score was obtained by 2 independent, blinded observers and was analyzed as described previously (43).
HFD feeding, glucose, and insulin tolerance tests.
Female mice were fed an HFD containing 60% calories from fat and 0.2% from cholesterol (Bio-Serv) or normal chow (Teklad; 4% calories from fat) beginning at 6 weeks of age. For glucose tolerance tests, mice were fasted for 6 hours and then injected i.p. with 1 g/kg glucose, and blood glucose levels were measured from tail blood by glucometer (OneTouch Ultra; LifeScan Inc.) over a 2-hour period. Development of insulin resistance on the diet was evidenced by increased glucose excursion in the glucose tolerance test. For insulin tolerance tests, mice were fasted for 6 hours and then injected i.p. with 0.75 U/kg insulin, and blood glucose levels were measured from tail blood by glucometer (OneTouch Ultra) over a 1-hour period.
FACS analysis.
For analysis of adipose tissue macrophages, gonadal fat pads were minced and digested in 1 mg/ml collagenase type I (Worthington) in KRBH buffer (145 mM NaCl, 5.4 mM KCl, 1.4 mM CaCl2, 1.4 mM MgSO4, 0.2 mM NaPO4, 5 mM glucose, 10 mM HEPES, and 0.1% insulin-free BSA) for 1 hour with gentle agitation. Cells were passed over nylon mesh, and adipocytes were floated for separation. The stromal vascular cells present in the infranatant were pelleted and washed 3 times with KRBH buffer. SVF-pelleted cells were resuspended in 0.83% ammonium chloride to lyse rbc. After washing the cells in PBS, the cells were resuspended in FACS buffer and blocked with anti-mouse CD16/CD32, followed by incubation with specific mAbs for surface staining for 20 minutes at 4°C. Intracellular staining was performed by fixing and permeabilizing cells at 4°C using BD Cytofix/Cytoperm Plus (554715; BD Biosciences). Finally, cells were washed and resuspended in FACS buffer, and flow cytometry was performed on a BD FACScan (BD Biosciences). Data were analyzed using FlowJo software. The following Abs were used for staining: CD45-APC-Cy7, CD11b-FITC, CD206-PE, and CD11c-AF647. For analysis of peritoneal lavage cells, the peritoneal cavities of mice were washed with 10 ml PBS to isolate the peritoneal lavage. The lavage was pooled and centrifuged, and the pelleted cells were resuspended in 0.83% ammonium chloride to lyse rbc. After washing the cells in PBS, they were resuspended in FACS buffer and counted using a hemocytometer. Cells were blocked and stained as described above. The following Abs were used for surface staining: F4/80 (Alexa Fluor 647) and CD11c (APC–Alexa Fluor 780). The following Abs were used for intracellular staining: p65 (Cell Signaling Technology) and Alexa Fluor 610–RPE secondary Ab.
Statistics.
Statistics were performed using Prism software, version 4 (GraphPad Software). A 2-tailed, unpaired Student’s t test, Mann-Whitney U test, or ANOVA was used as appropriate. An F test was performed in Prism to determine whether variances were similar among groups. A P value of less than 0.05 was considered statistically significant.
Study approval.
All animal studies were approved by the Animal Care and Use Committee of the University of Virginia. BAL samples were obtained from critical care patients at the University of Virginia Hospital in accordance with the hospital’s IRB guidelines.
Additional Methods can be found in the Supplemental material.
Author contributions
SEA, RG, RM, WC, NL, and SS designed the experiments. SEA, RG, RM, GM, WC, JH, PRS, SS, GRM, and SAG performed the experiments. SEA, RG, RM, and SS analyzed the data. AK and KE provided the BAL samples. JAC, TJB, and TEH provided reagents, expertise, and advice. SEA, RG, and NL wrote the manuscript.
Supplementary Material
Acknowledgments
R. Griffiths and S.E. Adamson were supported by NIH Cardiovascular Training grant 5T32 HL007284, and R. Moravec and S.A. Gorski were supported by NIH Immunology Training grant 5T32AI007496. This study was supported by NIH grants R01 DK096076 and R01 HL084422; a Grant in Aid from the American Heart Association (to N. Leitinger); and by NIH grants R01 AI-15608 and Hl-33391 (to T.J. Braciale).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(4):1311–1322. doi:10.1172/JCI79590.
Contributor Information
Rachael Griffiths, Email: griffithsrachael@gmail.com.
Radim Moravec, Email: rm3r@virginia.edu.
Subramanian Senthivinayagam, Email: ss5zf@eservices.virginia.edu.
Garren Montgomery, Email: gsm4za@virginia.edu.
Wenshu Chen, Email: cwenshu@gmail.com.
Jenny Han, Email: jh4ad@virginia.edu.
Poonam R. Sharma, Email: prs6c@virginia.edu.
Alexandra Kadl, Email: AK5SC@hscmail.mcc.virginia.edu.
Kyle Enfield, Email: KE4Z@hscmail.mcc.virginia.edu.
Norbert Leitinger, Email: nl2q@virginia.edu.
References
- 1.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5(12):953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 2.Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol. 2011;11(11):750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 3.Mantovani A, Biswas SK, Galdiero MR, Sica A, Locati M. Macrophage plasticity and polarization in tissue repair and remodelling. J Pathol. 2012;229(2):176–185. doi: 10.1002/path.4133. [DOI] [PubMed] [Google Scholar]
- 4.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11(11):723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11(10):889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 6.Fairweather D, Cihakova D. Alternatively activated macrophages in infection and autoimmunity. J Autoimmun. 2009;33(3–4):222–230. doi: 10.1016/j.jaut.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mantovani A, Garlanda C, Locati M. Macrophage diversity and polarization in atherosclerosis: a question of balance. Arterioscler Thromb Vasc Biol. 2009;29(10):1419–1423. doi: 10.1161/ATVBAHA.108.180497. [DOI] [PubMed] [Google Scholar]
- 8.Adamson S, Leitinger N. Phenotypic modulation of macrophages in response to plaque lipids. Curr Opin Lipidol. 2011;22(5):335–342. doi: 10.1097/MOL.0b013e32834a97e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kadl A, et al. Identification of a novel macrophage phenotype that develops in response to atherogenic phospholipids via Nrf2. Circ Res. 2010;107(6):737–746. doi: 10.1161/CIRCRESAHA.109.215715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest. 2007;117(1):175–184. doi: 10.1172/JCI29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chawla A, Nguyen KD, Goh YPS. Macrophage-mediated inflammation in metabolic disease. Nat Rev Immunol. 2011;11(11):738–749. doi: 10.1038/nri3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagemann T, et al. “Re-educating” tumor-associated macrophages by targeting NF-κB. J Exp Med. 2008;205(6):1261–1268. doi: 10.1084/jem.20080108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25(12):677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 14.Martinez FO. Macrophage activation and polarization. Front Biosci. 2008;13(13): doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 15.Lawrence T, Bebien M, Liu GY, Nizet V, Karin M. IKKα limits macrophage NF-κB activation and contributes to the resolution of inflammation. Nature. 2005;434(7037):1138–1143. doi: 10.1038/nature03491. [DOI] [PubMed] [Google Scholar]
- 16.Fong CH, et al. An antiinflammatory role for IKKβ through the inhibition of “classical” macrophage activation. J Exp Med. 2008;205(6):1269–1276. doi: 10.1084/jem.20080124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adhikari A, Xu M, Chen ZJ. Ubiquitin-mediated activation of TAK1 and IKK. Oncogene. 2007;26(22):3214–3226. doi: 10.1038/sj.onc.1210413. [DOI] [PubMed] [Google Scholar]
- 18.Lee J, Mira-Arbibe L, Ulevitch RJ. TAK1 regulates multiple protein kinase cascades activated by bacterial lipopolysaccharide. J Leukoc Biol. 2000;68(6):909–915. [PubMed] [Google Scholar]
- 19.Jeong SJ, Pise-Masison CA, Radonovich MF, Park HU, Brady JN. A novel NF-κB pathway involving IKKβ and p65/RelA Ser-536 phosphorylation results in p53 Inhibition in the absence of NF-κB transcriptional activity. J Biol Chem. 2005;280(11):10326–10332. doi: 10.1074/jbc.M412643200. [DOI] [PubMed] [Google Scholar]
- 20.Sakurai H, Chiba H, Miyoshi H, Sugita T, Toriumi W. IκB kinases phosphorylate NF-κB p65 subunit on serine 536 in the transactivation domain. J Biol Chem. 1999;274(43):30353–30356. doi: 10.1074/jbc.274.43.30353. [DOI] [PubMed] [Google Scholar]
- 21.Finkielstein CV, Capelluto DGS. Disabled-2: a modular scaffold protein with multifaceted functions in signaling. Inside the Cell. 2016;1(1):48–58. doi: 10.1002/icl3.1021. [DOI] [PubMed] [Google Scholar]
- 22.Xu XX, Yang W, Jackowski S, Rock CO. Cloning of a novel phosphoprotein regulated by colony-stimulating factor 1 shares a domain with the Drosophila disabled gene product. J Biol Chem. 1995;270(23):14184–14191. doi: 10.1074/jbc.270.23.14184. [DOI] [PubMed] [Google Scholar]
- 23.Yun M, et al. Crystal structures of the Dab homology domains of mouse disabled 1 and 2. J Biol Chem. 2003;278(38):36572–36581. doi: 10.1074/jbc.M304384200. [DOI] [PubMed] [Google Scholar]
- 24.Albertsen HM, et al. Sequence, genomic structure, and chromosomal assignment of human DOC-2. Genomics. 1996;33(2):207–213. doi: 10.1006/geno.1996.0185. [DOI] [PubMed] [Google Scholar]
- 25.Morris SM. Dual roles for the Dab2 adaptor protein in embryonic development and kidney transport. EMBO J. 2002;21(7):1555–1564. doi: 10.1093/emboj/21.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maurer ME, Cooper JA. The adaptor protein Dab2 sorts LDL receptors into coated pits independently of AP-2 and ARH. J Cell Sci. 2006;119(20):4235–4246. doi: 10.1242/jcs.03217. [DOI] [PubMed] [Google Scholar]
- 27.Oleinikov AV, Zhao J, Makker SP. Cytosolic adaptor protein Dab2 is an intracellular ligand of endocytic receptor gp600/megalin. Biochem J. 2000;347(3): [PMC free article] [PubMed] [Google Scholar]
- 28.Penheiter SG, et al. Type II transforming growth factor-beta receptor recycling is dependent upon the clathrin adaptor protein Dab2. Mol Biol Cell. 2010;21(22):4009–4019. doi: 10.1091/mbc.E09-12-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakayama M, et al. Spatial regulation of VEGF receptor endocytosis in angiogenesis. Nat Cell Biol. 2013;15(3):249–260. doi: 10.1038/ncb2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madden DR, Swiatecka-Urban A. Tissue-specific control of CFTR endocytosis by Dab2. Commun Integr Biol. 2012;5(5):473–476. doi: 10.4161/cib.21375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu L, Rab A, Tang L, Rowe S, Bebok Z, Collawn J. Dab2 is a key regulator of endocytosis and post-endocytic trafficking of the cystic fibrosis transmembrane conductance regulator. Biochem J. 2011;441(2):633–643. doi: 10.1042/BJ20111566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris SM, Cooper JA. Disabled-2 colocalizes with the LDLR in clathrin-coated pits and interacts with AP-2. Traffic. 2001;2(2):111–123. doi: 10.1034/j.1600-0854.2001.020206.x. [DOI] [PubMed] [Google Scholar]
- 33.Hocevar BA. Regulation of the Wnt signaling pathway by disabled-2 (Dab2) EMBO J. 2003;22(12):3084–3094. doi: 10.1093/emboj/cdg286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jain N, et al. Cutting edge: Dab2 is a FOXP3 target gene required for regulatory T cell function. J Immunol. 2009;183(7):4192–4196. doi: 10.4049/jimmunol.0902041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karam JA, et al. Decreased DOC-2/DAB2 expression in urothelial carcinoma of the bladder. Clin Cancer Res. 2007;13(15):4400–4406. doi: 10.1158/1078-0432.CCR-07-0287. [DOI] [PubMed] [Google Scholar]
- 36.Bagadi SAR, Prasad CP, Srivastava A, Prashad R, Gupta SD, Ralhan R. Frequent loss of Dab2 protein and infrequent promoter hypermethylation in breast cancer. Breast Cancer Res Treat. 2006;104(3):277–286. doi: 10.1007/s10549-006-9422-6. [DOI] [PubMed] [Google Scholar]
- 37.Sheng Z, et al. Structure, sequence, and promoter analysis of human disabled-2 gene (DAB2) Genomics. 2000;70(3):381–386. doi: 10.1006/geno.2000.6383. [DOI] [PubMed] [Google Scholar]
- 38.Hannigan A, et al. Epigenetic downregulation of human disabled homolog 2 switches TGF-β from a tumor suppressor to a tumor promoter. J Clin Invest. 2010;120(8):2842–2857. doi: 10.1172/JCI36125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tong JH, et al. Putative tumour-suppressor gene DAB2 is frequently down regulated by promoter hypermethylation in nasopharyngeal carcinoma. BMC Cancer. 2010;10(1): doi: 10.1186/1471-2407-10-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenbauer F, et al. Disabled-2 is transcriptionally regulated by ICSBP and augments macrophage spreading and adhesion. EMBO J. 2002;21(3):211–220. doi: 10.1093/emboj/21.3.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jokubaitis VG, et al. Endogenously regulated Dab2 worsens inflammatory injury in experimental autoimmune encephalomyelitis. Acta Neuropathol Commun. 2013;1(1): doi: 10.1186/2051-5960-1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prunier C, Howe PH. Disabled-2 (Dab2) is required for transforming growth factor β-induced epithelial to mesenchymal transition (EMT) J Biol Chem. 2005;280(17):17540–17548. doi: 10.1074/jbc.M500974200. [DOI] [PubMed] [Google Scholar]
- 43.Kadl A, Pontiller J, Exner M, Leitinger N. Single bolus injection of bilirubin improves the clinical outcome in a mouse model of endotoxemia. Shock. 2007;28(5):582–588. doi: 10.1097/shk.0b013e31804d41dd. [DOI] [PubMed] [Google Scholar]
- 44.Talukdar S, et al. Neutrophils mediate insulin resistance in mice fed a high-fat diet through secreted elastase. Nat Med. 2012;18(9):1407–1412. doi: 10.1038/nm.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weisberg SP, et al. CCR2 modulates inflammatory and metabolic effects of high-fat feeding. J Clin Invest. 2006;116(1):115–124. doi: 10.1172/JCI24335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma Q, Zhou L, Shi H, Huo K. NUMBL interacts with TAB2 and inhibits TNFα and IL-1β-induced NF-κB activation. Cell Signal. 2008;20(6):1044–1051. doi: 10.1016/j.cellsig.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 47.Zhou L, Ma Q, Shi H, Huo K. NUMBL interacts with TRAF6 and promotes the degradation of TRAF6. Biochem Biophys Res Commun. 2010;392(3):409–414. doi: 10.1016/j.bbrc.2010.01.037. [DOI] [PubMed] [Google Scholar]
- 48.Cho SY, Jeon JW, Lee SH, Park SS. p67 isoform of mouse disabled 2 protein acts as a transcriptional activator during the differentiation of F9 cells. Biochem J. 2000;352(3): [PMC free article] [PubMed] [Google Scholar]
- 49.Landström M. The TAK1-TRAF6 signalling pathway. Int J Biochem Cell Biol. 2010;42(5):585–589. doi: 10.1016/j.biocel.2009.12.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.