Abstract
The X chromosome–encoded histone demethylase UTX (also known as KDM6A) mediates removal of repressive trimethylation of histone H3 lysine 27 (H3K27me3) to establish transcriptionally permissive chromatin. Loss of UTX in female mice is embryonic lethal. Unexpectedly, male UTX-null mice escape embryonic lethality due to expression of UTY, a paralog that lacks H3K27 demethylase activity, suggesting an enzyme-independent role for UTX in development and thereby challenging the need for active H3K27 demethylation in vivo. However, the requirement for active H3K27 demethylation in stem cell–mediated tissue regeneration remains untested. Here, we employed an inducible mouse KO that specifically ablates Utx in satellite cells (SCs) and demonstrated that active H3K27 demethylation is necessary for muscle regeneration. Loss of UTX in SCs blocked myofiber regeneration in both male and female mice. Furthermore, we demonstrated that UTX mediates muscle regeneration through its H3K27 demethylase activity, as loss of demethylase activity either by chemical inhibition or knock-in of demethylase-dead UTX resulted in defective muscle repair. Mechanistically, dissection of the muscle regenerative process revealed that the demethylase activity of UTX is required for expression of the transcription factor myogenin, which in turn drives differentiation of muscle progenitors. Thus, we have identified a critical role for the enzymatic activity of UTX in activating muscle-specific gene expression during myofiber regeneration and have revealed a physiological role for active H3K27 demethylation in vivo.
Introduction
The development of complex organisms from a single genome requires that cell-specific gene expression programs be tightly controlled both spatially and temporally. This transcriptional control is partially mediated through epigenetic mechanisms where reversible modifications of chromatin alter the ability of specific genes to be transcribed. Experiments in Drosophila identified the polycomb (PcG) and trithorax (TrxG) group of proteins as key mediators of temporal and spatial regulation of transcription since mutations within these antagonizing genes lead to homeotic transformations (1, 2). As the grouping of PcG and TrxG proteins are based on a phenotype rather than conserved functional domains, these proteins play diverse roles in modulating transcriptional competency. The mechanisms through which many PcG and TrxG proteins contribute to transcriptional regulation remain to be elucidated. However, genome-wide studies have clearly established that genes repressed by PcG are marked by trimethylation of histone H3 lysine 27 (H3K27me3) (3, 4). This repressive H3K27me3 mark is established by the PcG protein EZH2 that functions as an H3K27 methyltransferase (5–7). The EZH2-mediated H3K27me3 mark contributes directly to gene repression, since mutant flies that cannot be methylated at the H3K27 position (H3K27R) fail to ensure PcG-mediated repression during development (8). Importantly, the H3K27me3 repressive mark can be epigenetically transmitted to daughter cells (9) and across generations (10). Thus, the removal of H3K27me3 marks is required to establish a transcriptionally permissive state at PcG target genes.
Active removal of H3K27me3 marks is mediated by the KDM6 family of histone demethylases. The KDM6 family includes the UTX (also known as KDM6A) protein that is coded by a gene on the X chromosome and the JMJD3 (also known as KDM6B) protein that is coded by an autosomal gene (11–15). The UTX gene is known to escape X chromosome inactivation (16), and therefore, females express both alleles of the gene. A paralog of UTX that shares a similar domain structure is found on the Y chromosome — it is termed UTY (16). The expression of UTY allows males to compensate for the presence of a single UTX allele. However, amino acid substitutions in the catalytic domain of UTY are known to prevent the UTX paralog from efficiently removing the H3K27me3 mark (15, 17).
Using cultured cell systems, the KDM6 family of demethylase enzymes have been shown to play an important role in a multitude of cellular processes, including differentiation (18, 19), senescence (12), somatic and germ cell reprogramming (20), inflammatory response (14), and cancer (21). These findings led to the postulation that H3K27 demethylase activity would be essential for altering gene expression programs during cell fate changes throughout development. Thus, it came as a surprise when studies in Drosophila and C. elegans demonstrated that mutants lacking H3K27 demethylase activity are morphologically indistinguishable from their WT counterparts in development (22, 23). Similarly, recent studies in mice have demonstrated that male mice that lack both UTX and JMJD3 are viable until E18.5, though their female littermates display embryonic lethality between E10.5 and E14.5 (24). This sex-specific difference in murine development is thought to be due to a functional rescue in males by the UTY protein. A demethylase-independent role for both UTX and JMJD3 has previously been reported where the KDM6 family members facilitate recruitment of the remodelling complex SWI/SNF to specific gene promoters (25). Consistent with a nonenzymatic role for UTX/UTY in mammalian development, the defect in mesodermal differentiation of Utx–/– embryonic stem cells can be rescued by the expression of a demethylase dead UTX (UTXKI) mutant (26). Similarly, cardiac and neural tube morphogenesis defects observed in the female Utx–/– mutant mice are not observed in the male Utx–/Y mice (17, 26, 27). Thus, in vivo evidence for a role of the H3K27 demethylase activity of either JMJD3 or UTX in any normal biological process is currently lacking.
Skeletal muscle regeneration is a well-characterized biological process in which myofiber injury induces a multistep process whereby muscle stem cells have the capacity to fully repair the tissue (28). Satellite cells (SCs), which reside along muscle fibers, are the primary muscle stem cells contributing to this tissue regeneration (29). SC-mediated muscle regeneration can be readily modeled using acute muscle injury in mice where the tibialis anterior (TA) muscle is injected with cardiotoxin (CTX) to induce muscle degeneration. Within the degenerating muscle, myofiber-associated SCs (identified by the presence of the transcription factor PAX7) become activated and transition to the cell fate of proliferating muscle progenitor cells (identified by the presence of the muscle-specific transcription factors MyoD and MYF5). After expansion of the MyoD+/MYF5+ progenitor cell population, induction of MYOG expression commits progenitor cells to exit the cell cycle and undergo terminal differentiation where they fuse into the regenerating myofibers (30, 31). We have previously shown an essential role for UTX in activating MYOG expression to mediate terminal muscle differentiation in cultured myoblasts (19). However, the lack of overt muscle defects in male UTX-null (Utx–/Uty) mice (17, 24, 26) suggests that UTX acts in a demethylase-independent manner to mediate myofiber formation during development. Here, we have used the CTX model of adult muscle regeneration to examine the requirement for UTX demethylase activity for SC-mediated myofiber repair in vivo.
Results
The UTX gene is thought to be expressed ubiquitously in mice and humans (16). To confirm that UTX is expressed in regenerating mouse muscle, we performed immunofluorescence (IF) analysis of TA muscle cross sections in healthy or CTX-injured muscle (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI83239DS1). As expected, we observe that UTX colocalizes with PAX7 in quiescent SCs of healthy adult muscle (Supplemental Figure 1B). Similarly, UTX is found to colocalize with MyoD in proliferating progenitor cells in CTX-treated muscle (Supplemental Figure 1C). Having established that UTX is expressed in SCs and their derived progenitors, we next wanted to determine the contribution of UTX to the process of muscle regeneration. For this, mice were bred to generate a tamoxifen-inducible KO of Utx specifically in SCs (Supplemental Figure 2) using Cre-ER recombinase expressed from the Pax7 locus (32). Treatment of mice with tamoxifen resulted in an excision rate of approximately 85% of the Utx alleles in SCs (Supplemental Figure 2B). The tamoxifen-treated mice that have undergone a SC-specific ablation of UTX are hereafter referred to as UTXmKO mice.
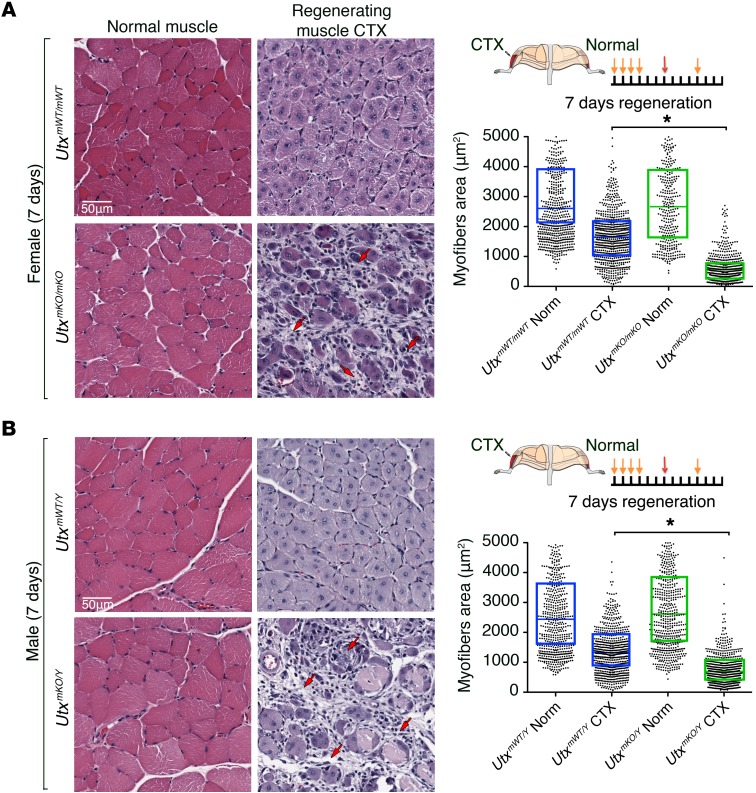
We next sought to determine the role of UTX in SC-mediated muscle regeneration. To eliminate the potential for confounding effects of UTY function in these experiments, we used female UTXmKO mice to analyze SC-mediated muscle regeneration after CTX injury. In the UTXWT mice, histological analysis showed efficient regeneration of healthy myofibers (as shown by characteristic centrally located nuclei) in the CTX-treated muscle 7 days after injury (Figure 1A, top right panel). In contrast, muscle cross sections from the CTX-injured UTXmKO muscle presented a decreased myofiber density with a marked increase in necrotic tissue and inflammatory cell infiltration compared with the UTXWT muscle (Figure 1A, lower right panel). Characterization of the centrally nucleated myofibers that were regenerated in UTXmKO mice showed they were significantly smaller in caliber (Figure 1A). This impaired regeneration observed in the UTXmKO mice persisted at least 21 days after CTX injury (Supplemental Figure 3). As a small number of newly regenerated myofibers were observed in the injured UTXmKO mice (Figure 1A), we examined whether they may originate from SCs that escape Cre-mediated excision. Using a TdTomato-based (TdT-based) lineage tracing methodology (UTXmKO Rosa26CAG(TdT)), we found that <1% of isolated myofibers possessing centrally localized nuclei were marked by the fluorescent marker 7 days after injury (Supplemental Figure 4). Thus, cells that undergo Cre-mediated excision of UTX are not contributing to newly formed myofibers. These findings clearly establish that UTX is required for SC-mediate muscle regeneration in vivo.
Figure 1. SC-mediated muscle regeneration is impaired in both male and female UTXmKO mice.
(A and B) Cross sections of TA muscles from female (A) or male mice (B) were visualized after H&E staining to evaluate muscle integrity in noninjured (normal muscle) and CTX-injected (regenerating muscle) muscle at 7 days after tissue injury. Necrotic tissues and the infiltrating interstitial cells are indicated by red arrows. Each image panel is visualized under ×20 magnification and represents a total area of 90,000 μm2. A schematic representation of the experimental procedure is shown, with tamoxifen injections indicated by orange arrows and CTX injection by red arrows. Myofiber calibers within the TA muscle of a representative mouse were calculated and plotted for each condition (n > 500 total fibers per condition). Each dot represents the area of a single myofiber; the horizontal bar corresponds to the mean within the 95% confidence interval (box). Statistical significance was determined using an ANOVA test where *P < 0.05. Measurements have been performed on a minimum of 9 mice taken from 3 independent experiments.
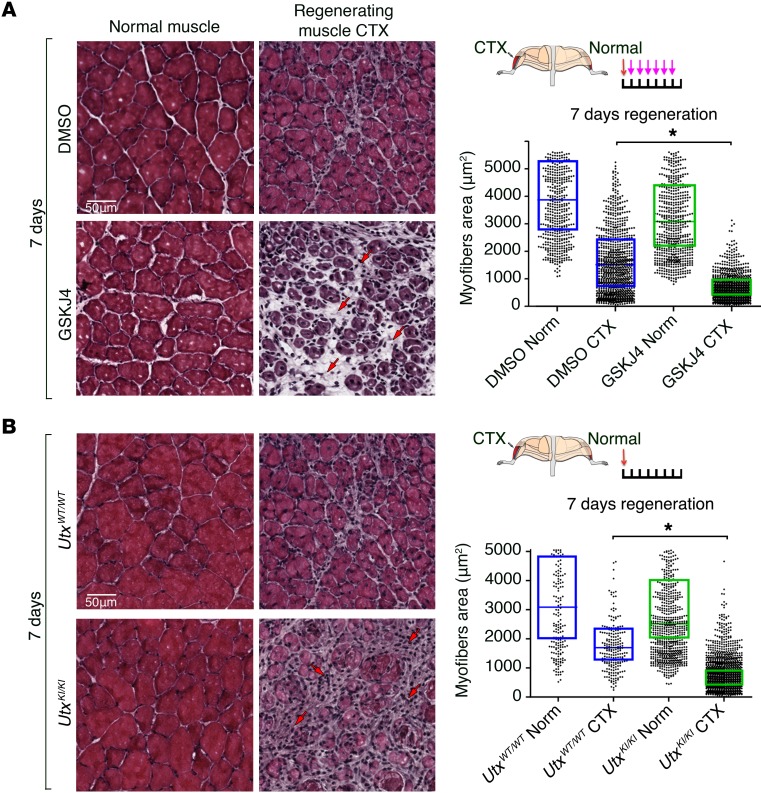
While UTX has been shown to play a critical role in embryonic development, the survival of male Utx-null embryos suggest that UTY could compensate for some functions of UTX in conditions when its H3K27me3 demethylase activity is not required (26, 33). Muscle progenitor cells from male mice express an amount of Utx/Uty similar to the total Utx amount expressed in females when compared at the RNA level (Supplemental Figure 1D). To determine whether UTY could compensate for UTX activity, we examined muscle regeneration in male mice that lack UTX (UtxmKO/Y). Similar to results observed in female UTXmKO mice, male UtxmKO/Y mice showed a profoundly impaired regenerative capacity with increased infiltration of inflammatory cells and reduced caliber for the limited number of fibers that did regenerate (Figure 1B and Supplemental Figure 3C). The regeneration defect in male UtxmKO/Y mice could reflect a haploinsufficiency phenotype where expression of the single allele of UTY is not sufficient to replace expression from the deleted Utx allele (see Supplemental Figure 1D). However, this does not appear to be the case, since heterozygous female mice (UtxmKO/WT) showed normal muscle regeneration (Supplemental Figure 5). The lack of compensation for loss of UTX in male mice suggests that the H3K27 demethylase activity of UTX may be required for adult muscle regeneration. To test this possibility directly, we first examined the ability of CTX-injured muscle to regenerate in WT mice treated with a small molecule inhibitor (GSK-J4) that specifically inactivates the H3K27 demethylase activity of KDM6 (both UTX and JMJD3) family members (34). Mice receiving a daily i.p. injection of GSK-J4 showed extensive fibrosis and poor myofiber repair compared with control-treated mice (Figure 2A), confirming a requirement for H3K27 demethylase activity to mediate muscle regeneration. Since GSK-J4 cannot discriminate between UTX and JMJD3, we have developed a knock-in mouse where the endogenous Utx gene is replaced by UTXKI (see Supplemental Figure 2). While UTX demethylase activity is inactivated in all cell types (26), UTXKI mice are viable and fertile (Supplemental Figure 2C). Western blot analysis shows that the UTXKI protein is stably expressed in SCs (Supplemental Figure 6A) and is efficiently incorporated into the MLL4 complex (Supplemental Figure 6B) where it continues to interact with core subunits PTIP, RbBP5, and MLL4 (13, 15, 35). Furthermore, muscle from adult UTXKI mice displayed normal myofiber integrity (Figure 2B and Supplemental Figure 4A; compare uninjured normal muscle between UTXWT and UTXKI mice). These findings suggest that developmental myogenesis can occur in the absence the H3K27 demethylase activity of UTX. To determine whether the enzymatic activity of UTX is necessary for SC-mediated muscle regeneration, we turned again to the model of CTX-induced muscle degeneration. Strikingly, we found that under these conditions where the muscle SCs are forced to mediate a rapid and coordinated response to repair the damaged myofibers, UTXKI mice show a clear defect in muscle repair at 7 days of regeneration (Figure 2B). Taken together, these results establish that the H3K27me3 demethylase activity of UTX is critically required for SC-mediated muscle regeneration in response to acute injury.
Figure 2. The demethylase activity of UTX plays a critical role in SC-mediated muscle repair.
(A and B) Analysis of muscle regeneration 7 days after CTX treatment in mice treated with daily i.p. injections of GSK-J4 (A) — an inhibitor of the demethylase activity of UTX and JMJD3 — and UTXKI mice (B). The experimental approach is schematically represented where CTX injections are indicated by red arrows, while GSK-J4 injections are shown by pink arrows. Cross sections of TA muscles were visualized after H&E staining to evaluate muscle integrity in noninjured (normal) and CTX-injected (regenerating) muscle at 7 days after tissue injury. Necrotic tissue and infiltrating immune cells are indicated by red arrows. Each image panel is visualized under ×20 magnification and represents a total area of 90,000 μm2. Myofiber calibers within the TA muscle were calculated and plotted for each condition (n > 500 total fibers per condition). Each dot represents the area of a single myofiber; the horizontal bar correspond to the mean within the 95% confidence interval (box). Statistical significance was determined using an ANOVA test where *P < 0.05. Measurements have been performed on a minimum of 9 mice taken from 3 independent experiments.
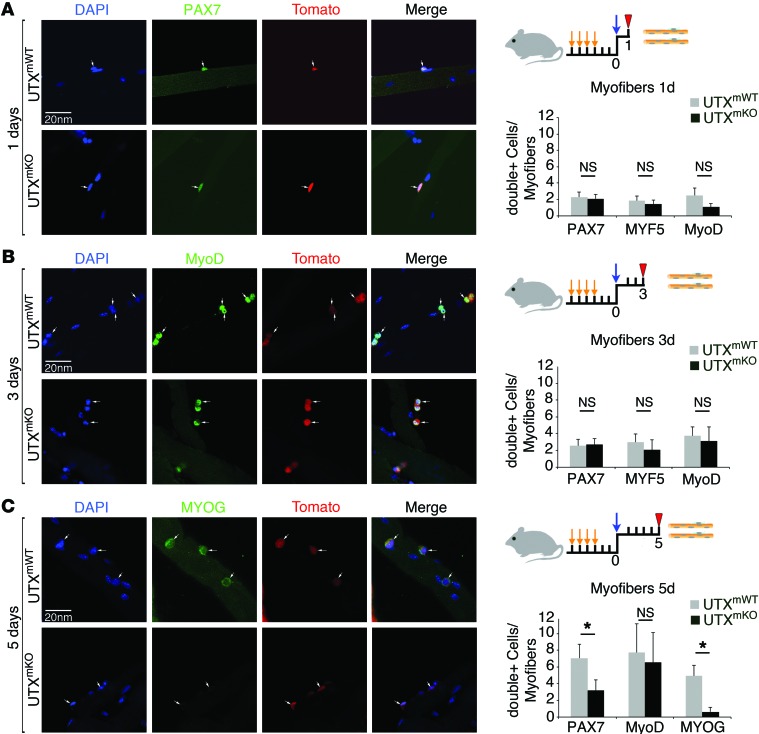
Having established a role for the enzymatic activity of UTX in the muscle regeneration process, we next wanted to identify the specific stages of regeneration where the H3K27 demethylase function is required. For this purpose, myofiber explants were prepared from mouse extensor digitorum longus (EDL) muscles (36). IF analysis in UTXmWT mice confirmed that UTX is expressed in SC-derived cells at all stages of muscle regeneration, including SC (PAX7+), muscle progenitor cell (MYF5+/MyoD+), and differentiating myoblast (MYOG+) populations (Supplemental Figure 7A). In order to facilitate identification of SCs on the myofibers that had undergone Cre-mediated excision, we crossed the UTXmKO mice with mice that express the lineage-tracing TdT marker from the Rosa26 locus when a floxed stop codon is excised; these mice are hereafter termed UtxmKO/TdT. Using myofiber explants from UtxmWT/TdT or UtxmKO/TdT mice, the different stages of regeneration were evaluated by IF looking at SC (Pax7+), muscle progenitor cell (Myf5+/MyoD+), and differentiating myoblast (Myog+) populations (36). Examination of SCs prior to their first cell division (day 1 after explant), we observe that the number of cells expressing Pax7, MyoD, or Myf5 is not affected by the loss of UTX in SCs (Figure 3A). This suggests that neither the short-term maintenance of the SC identity (Pax7+) nor the activation of the myogenic program (MyoD+/Myf5+) requires UTX activity. Next, we examined the role for UTX in facilitating the expansion of the muscle progenitor cell population. By approximately 40 hours, the majority of SCs will have undergone their first cell division and will have begun expanding the muscle progenitor cell population (37). Examination of the proliferating progenitor population in our explant cultures at day 3 again shows that the number of cells expressing PAX7, MyoD, and MYF5 is unaltered by the absence of UTX activity (Figure 3B). Furthermore, we observe that the loss of UTX does not affect progenitor cell proliferation (as measured by 5-ethynyl-2’-deoxyuridine [EdU] incorporation) or lead to increased apoptosis (as measured by active caspase activity) on fiber explants at any time point tested (Supplemental Figure 7, B and C). Finally, we examined the capacity of the progenitor cell population to undergo terminal differentiation. The expression of MYOG constitutes a point of no return, where progenitors commit to cell cycle exit and differentiation (30, 31). Examination of explanted fibers at day 5 showed that, in the absence of UTX, the proliferating progenitor cells were impaired in their ability to induce expression of MYOG to initiate differentiation (Figure 3C). A similar inability to activate MYOG expression was observed using myofiber explants from UTXKI mice (Supplemental Figure 8). These findings demonstrate an essential role for the demethylase activity of UTX in facilitating the transition of muscle progenitors to the terminally differentiated cell fate during muscle regeneration. Finally, we examined whether UTXmKO mice display a similar defect in transitioning from proliferating progenitor cells (MyoD+) to the terminally differentiating (MYOG+) state in vivo. IF analysis of TA muscle cross sections examined 7 days after CTX treatment show that the number of MyoD+ cells present within the regenerating muscle was not affected by the absence of UTX (Supplemental Figure 9). However, the MYOG+ cell population was markedly reduced within the regenerating muscle of both male and female mice lacking UTX or mice lacking UTX demethylase activity (Supplemental Figure 9). Consistent with a requirement for UTX-mediated demethylation during terminal differentiation, we observe that H3K27me3 levels are strikingly lower in MYOG+ cells compared with undifferentiated (Pax7+ or MyoD+) myoblasts in vitro (Supplemental Figure 8) and in vivo (Supplemental Figure 10). We note that some MYOG+ cells were observed in regenerating muscle of the UTXKI mice (Supplemental Figure 10), suggesting that a compensatory transcriptional network may be activated in SCs when the H3K27 demethylase activity of UTX is absent in development, permitting an inefficient terminal differentiation in response to acute injury. Taken together, these results establish that the demethylase activity of UTX facilitates the efficient transition between the proliferating and differentiating state of muscle progenitors during regeneration.
Figure 3. Terminal differentiation of SC-derived myoblasts is impaired in UTXmKO mice.
(A–C) Freshly isolated EDL myofibers from female UtxmWT/TdT or UtxmKO/TdT mice (blue arrow) were cultured in growth medium for 1 (A), 3 (B), or 5 days (C); fixed (red arrowhead); and then subjected to IF analysis for myogenic markers as indicated. Cells that are Tomato+ (indicated by white arrows) act as surrogate markers for cells that are expected to have undergone Cre-mediated excision of UTX (Tamoxifen injection, orange arrows). A representative myofiber visualized by confocal microscopy using ×60 magnification is shown on the left, while quantitation of all scored myofibers (n > 100) is shown on the right. (A) SC identity: After 1 day of culture, myofibers were stained and analyzed for total number of DAPI+ (blue), PAX7+ (green), and Tomato+ (red) cells present on each fiber. (B) Proliferating myoblasts: After 3 days of culture, myofibers were stained and analyzed for total number of DAPI+ (blue), MyoD+ (green), and Tomato+ (red) cells present on each fiber. (C) Terminal differentiation: After 5 days of culture, myofibers were stained and analyzed for total number of DAPI+ (blue), MYOG/SC+ (green), and Tomato+ (red) cells present on each fiber. The number of cells per myofiber that stain positive for both the myogenic marker of interest and Tomato are expressed as an average ± SD. Statistical significance was determined using an unpaired t test where *P < 0.05 (n ≥ 100).
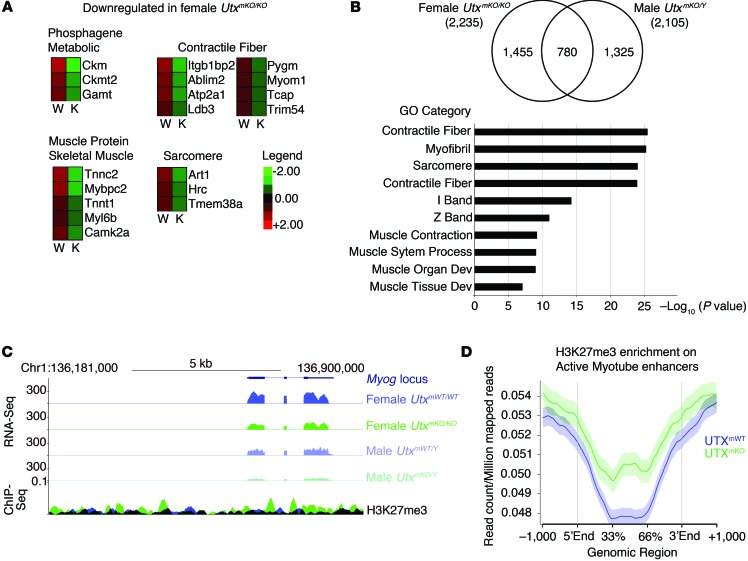
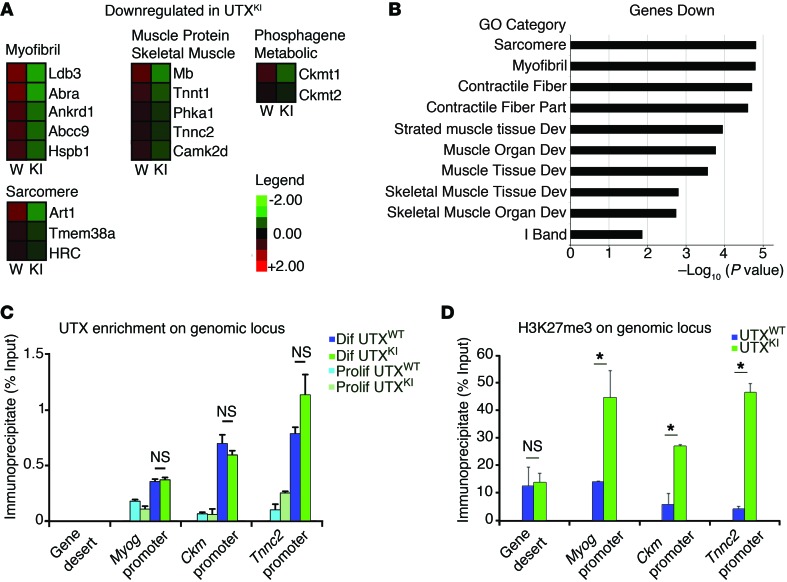
A role for UTX in mediating the transition between proliferating progenitors and differentiating myotubes is particularly interesting in light of previous studies showing that loss of the antagonizing H3K27me3 methyltransferase enzyme EZH2 leads to precocious differentiation of proliferating muscle progenitor cells (38, 39). This precocious differentiation in the absence of EZH2 is a result of transcriptional upregulation of the Myog and p16Ink4a genes (38, 39). To try to explain the impaired differentiation of progenitor cells in the absence of UTX, we used quantitative PCR (qPCR) to examine the expression of Myog and p16Ink4a in differentiating progenitors lacking UTX (Supplemental Figure 11). Though we saw no significant change in p16Ink4a expression, we did observe a 90% reduction in Myog expression in cells induced to differentiate in the absence of UTX (Supplemental Figure 11). To identify genes regulated by UTX during terminal differentiation, high-throughput sequencing of RNA (RNA-Seq) analysis was performed on UtxmWT/TdT- or UtxmKO/TdT-derived muscle progenitor cells (Figure 4). Comparative gene expression analysis identified a list of 780 genes that are downregulated by at least 2-fold in both male and female UtxmKO/TdT mice (Figure 4B). Consistent with our phenotypic results, gene ontology (GO) analysis of these 780 genes defined a role for UTX in regulating muscle development and function (Figure 4B and Supplemental Table 1A). Similarly, RNA-Seq analysis of differentiating myoblasts from UTXKI mice identified decreased expression of genes involved in muscle development and function (Figure 5, Supplemental Figure 11, and Supplemental Table 1B). Altered expression of a subset of these UTX-regulated genes during differentiation was confirmed by qPCR (Supplemental Figure 11). Among the muscle functional genes regulated by UTX, we show that expression of muscle regulatory gene Myog is significantly decreased upon differentiation of male and female UtxmKO/TdT progenitor cells (Figure 4C). Coincident with this decreased gene expression, ablation of UTX leads to an increased accumulation of H3K27me3 marks across the Myog locus (Figure 4C). Furthermore, we observe that loss of UTX causes H3K27me3 marks to begin accumulating (Figure 4D) at regions that had previously been described by the Dynlacht group as myotube-specific enhancers (40). These results strongly support a mechanism through which UTX regulates the expression of genes involved in terminal differentiation of muscle through a mechanism that entails removal of the transcriptionally repressive H3K27me3 modification. To confirm that UTX is acting to reduce H3K27me3 levels at muscle genes during differentiation of proliferating myoblasts, we performed ChIP-qPCR analysis for UTX. Indeed, upon differentiation, we observe an enrichment of UTX at the promoter of the Myog, Ckm, and Tnnc2 promoters (Figure 5C). Finally ChIP analysis for H3K27me3 was performed at the promoter of key target genes. Using muscle progenitor cells isolated from the UTXKI or UTXmWT mice, we observe that loss of UTX demethylase activity results in an accumulation of H3K27me3 marks at the promoter of Myog, CKm, and Tnnc2 in conditions that normally induce terminal differentiation (Figure 5D). Taken together, these findings establish that UTX mediates terminal differentiation of muscle progenitor cells through the removal of repressive H3K27me3 marks at key genes involved in the formation of functional myotubes.
Figure 4. UTX regulates the muscle gene expression program through demethylation of H3K27me3 marks.
WT primary myoblasts were isolated from female or male UtxmKO/TdT or UtxmWT/TdT mice. After expansion in culture for 5 days, cells were treated with 4-OH-tamoxifen for 24 hours. Cells that underwent recombination were isolated by FACS, plated, and allowed to adhere to the plate overnight. Myogenic differentiation was then induced for 24 hours prior to RNA isolation. (A and B) RNA-Seq analysis was performed to identify genes whose expression is modified in differentiation myoblasts from UtxmKO/TdT compared with UtxmWT/TdT mice. (A) A heatmap is shown for selected genes of different ontologies that are differentially expressed (P < 0.05). (B) The overlap of 780 genes that are downregulated in both male and female UtxmKO/TdT mice is represented by a Venn diagram. GO analysis of the 780 genes downregulated in myoblasts from both males and female UtxmKO/TdT shows highly significant enrichment of genes involved in muscle development and function. (C and D) H3K27me3 ChIP-Seq analysis was performed to examine H3K27me3 enrichment in differentiating myoblasts isolated from UtxmKO/TdT (green) or UtxmWT/TdT (blue) mice. (C) RNA-Seq and H3K27me3 ChIP-Seq tracks mapping to the Myog locus are presented in UtxmKO/TdT or UtxmWT/TdT conditions. (D) Analysis of H3K27me3 levels at enhancers that have previously been established as myotube-specific enhancers (39).
Figure 5. UTX demethylase activity is required for H3K27me3 removal on muscle differentiation gene.
Primary myoblasts from female UtxKI/KI or UtxWT/WT mice were isolated, sorted by FACS, and expanded in vitro. Myogenic differentiation was induced for 24 hours. (A and B) RNA-Seq analysis was then performed to identify genes whose expression is modified in differentiating myoblasts from UtxKI/KI mice compared with UtxWT/WT mice. (A) A heatmap is shown for selected genes of different ontologies that are differentially expressed in the UtxKI/KI myoblasts (P < 0.05). (B) GO analysis of the genes downregulated in myoblasts from the UtxKI/KI mice shows highly significant enrichment of genes involved in muscle development and function. (C) ChIP analysis was performed to analyze the enrichment of UTX at the Myog, CKm, and Tnnc2 promoters. (D) Native-ChIP analysis was performed to determine H3K27me3 enrichment at the Myog, CKm, and Tnnc2 promoters. Values are presented as the average enrichment as a percentage of input ± SD. Statistical analysis was performed using an unpaired t test where *P < 0.05, n = 3.
Discussion
Characterization of the Utx and Jmjd3 KO mice has yet to reveal an essential role for the enzymatic activity of KDM6 proteins in mammalian development (17, 24, 26, 33). Male mice lacking both UTX and JMJD3 are able to develop to E18.5, showing normal somite numbers at E10.5 (24). The nonessential nature of the UTX H3K27 demethylase activity in murine development is further supported by our finding that both male and female UTXKI mice are viable and fertile. Thus, contrary to expectations based on our previous studies of myoblast differentiation in cultured cell systems (19), the formation of muscle in vivo during murine development does not appear to require the enzymatic activity of UTX. However, our studies here reveal for the first time to our knowledge an essential role for the enzymatic activity of UTX in mediating efficient muscle regeneration in response to acute injury. Using an inducible KO in SCs, we have shown that muscle regeneration is severely compromised in the absence of H3K27 demethylase activity of UTX. Furthermore, we have been able to delineate this defect in regeneration to the cell fate transition between proliferating myoblasts and terminal differentiating myotubes. Therefore, our work firmly establishes a role for UTX-mediated demethylation of H3K27me3 in establishing the muscle-specific gene expression program during SC-mediated muscle regeneration.
Our finding that UTX plays a critical role in terminal muscle differentiation in vivo confirms findings previously demonstrated using in vitro cultured myoblasts (19). Dissection of the in vitro differentiation process revealed a direct role for UTX in mediating the activation of Myog gene expression. These studies revealed that UTX is targeted to Myog promoters by the transcription factor SIX4 where it mediates removal of the repressive H3K27me3 mark within the promoter region (19). The targeting of UTX to Myog promoters by SIX4 has recently been confirmed, and this recruitment mechanism has been shown to occur at a large number of MyoD target genes in differentiating muscle (41). Once associated with the muscle genes, UTX then becomes associated with the elongating RNA polymerase II, proceeding along the gene to remove H3K27me3 marks within the coding region of the gene to permit high levels of expression (19, 42, 43). Validating these previous findings in cultured myoblasts, our in vivo and ex vivo studies of injured muscle from UTXmKO and UTXKI mice show that MYOG activation crucially requires the enzymatic activity of UTX. While UTX plays a role in activating many genes involved in formation of functional myofibers, the inability to activate Myog expression likely represents one of the key target genes preventing UTXmKO muscle progenitor cells from undergoing terminal differentiation. Indeed, MYOG expression has been shown to play a key role in mediating cell cycle exit and commitment of the muscle progenitors to differentiation (30, 31). Interestingly, it has been shown that the SC-specific ablation of EZH2 (the methyltransferase that antagonizes UTX by establishing H3K27me3 marks) leads to defective muscle regeneration in vivo where a premature activation of MYOG expression induces muscle progenitor cells to undergo terminal differentiation before they have an opportunity to expand their population numbers (38, 39). This suggests that EZH2-UTX antagonism plays a key role in modulating the cell fate transition leading to terminal differentiation of proliferating myoblasts. Nevertheless, it is surprising that UTX activity is not required for the cell fate transition between the quiescent SC state and the proliferating progenitor cell. This could be explained by the previous finding that quiescent SCs of younger mice have relatively low levels of H3K27me3 marks when examined at the genomic level (44). It remains to be determined which epigenetic enzymes might contribute to the cell fate transition between the quiescent SC and the proliferating progenitor states.
The differential requirement for the demethylase activity of UTX in developmental versus regenerative myogenesis was unanticipated. Histological sections of undamaged TA muscles in the UTXKI mice revealed a normal muscle architecture with bundles of healthy-looking myofibers. Nevertheless, the myofiber-associated SCs within the muscle of UTXKI mice showed an inability to efficiently respond to acute muscle injury. The inability of SCs from UtxKI/KI to undergo terminal differentiation ex vivo suggests that the requirement for the demethylase activity is intrinsic to the SCs. However, we cannot rule out that the loss of UTX demethylase activity in additional cell types that contribute to efficient muscle regeneration (45, 46) may also be contributing to the impaired myofiber repair observed in the UTXKI mice in vivo. The absence of a muscle phenotype during development for the UTXKI mice could point to distinct mechanisms used by muscle progenitor cells to modulate their transition toward terminal differentiation in development compared with regeneration. A more likely explanation is that a compensating mechanism of H3K27 demethylation can be used to transition between the proliferating and differentiating progenitor states when the demethylase activity of UTX is blocked in development. This compensation could occur through a passive mechanism where H3K27me3 is removed from target genes by histone turnover during the S-phase of the cell cycle (24) or an active mechanism where an altered targeting of JMJD3/KDM6B to muscle gene promoters allows for efficient removal of H3K27me3 to permit gene activation. Given the fact that Utx–/yJmjd3–/– mice lacking H3K27 demethylase activity can survive to term and have normal embryonic morphology (24), a passive mechanism of H3K27 demethylation through histone turnover seems more likely. However, distinguishing between these various possibilities will require future studies using muscle progenitor cells isolated from the developing embryo.
The development of our UTXKI mice also revealed the unexpected finding that UTX demethylase activity of UTX is not required for fertility. Previous studies have suggested a requirement for UTX demethylase activity in germ cell development based on the observation that UTY could not compensate for the loss of UTX in ex vivo experiments looking at the reversion of primordial germ cells to the embryonic germ cells state (20). The viability and fertility of our UTXKI mice would suggest that the demethylase activity of UTX is not critically required for germ cell development. In fact, neither male (UtxKI/Y) or female (UtxKI/KI) UTXKI mice show overt phenotypes in development or reproduction, suggesting that the H3K27 demethylase activity of UTX can be compensated by alternate mechanisms in the absence of a stress to the organism. We highlight that the phenotypic data with our UTXKI mice are consistent with the observations in Drosophila and C. elegans that demethylase-dead UTX mutants are morphologically indistinguishable from their WT counterparts in development (22, 23). Thus, it remains to be determined how the mice compensate for the loss of UTX demethylase activity in the in vivo developmental setting.
In conclusion, our studies describe for the first time to our knowledge an essential role for the H3K27 demethylase enzyme UTX in muscle regeneration. This represents a physiological requirement for active removal of repressive H3K27me3 marks in vivo. Importantly, these findings demonstrate that the mechanism of myofiber repair after muscle injury displays some distinctions from that of myofiber formation during development. Since the demethylase activity of UTX is not required for the development of any tissues in mice, it will be important to examine whether the requirement for H3K27 demethylase activity of UTX in muscle regeneration is shared by other adult stem cell populations to ensure the efficient regenerative capacity of their nascent tissues.
Methods
Animals.
The Utxfl/fl (26) and Pax7-CreER (32) mouse strains have previously been described. UTXKI mice were derived from UtxKI male ES cells (UtxKI/Y), as previously described (26). The KI allele carries H1146A and H1148A mutations (26). The B6/129 (Cg)-Gt(ROSA)26sortm4 TdT mouse strain was purchased from the Jackson Laboratory. Generation of UTXmKO, UtxmKO/TdT, and UtxWT/TdT mice (see Supplemental Figure 2) has been achieved by successive breeding combinations. Primers used for genotyping are listed in Supplemental Table 2. The different strains used in this study were maintained on a mixed B6/129 background and have been inbred for at least 10 generations. All animals were studied at an age between 6–12 weeks old.
Ablation of the Utx allele in vivo using the CreER-recombinase was achieved by daily i.p. injection of tamoxifen (MP Biomedicals) dissolved in corn oil (150 μl at 10 μg/μl to achieve a dose of 75 mg/kg) over 4 consecutive days. For pharmacological inhibition of KDM6 demethylase activity in vivo, GSK-J4 (Cayman Chemical) dissolved in DMSO/Captisol (150 μl at 10 μg/μl to achieve a dose of 75 mg/kg) was administered by daily i.p. injection over the course of 6 days. Muscle damage was induced in mice by a single i.m. injection of CTX (Sigma-Aldrich) in saline 0.9% NaCl solution (50 μl at 10 μM to achieve a dose of 1.7 μg/muscle) into TA muscles.
Myofiber and SC isolation and culture.
The EDL muscles from UTXKI, UtxmKO/TdT, UtxWT/TdT mice were isolated and digested with 0.1% collagenase from clostridium histolyticum (Sigma-Aldrich) in F10 medium (Wisent Bio Products) according to established methods (47). The isolated myofibers were either fixed directly with 1% paraformaldehyde (PFA) or cultured in F10 medium with 20% FBS and bFGF (2.5 ng/ml) for 1, 3, or 5 days prior to fixation with 1% PFA.
SCs were isolated from hindlimb and forelimb of UTXKI, UtxmKO/TdT, UtxWT/TdT mice and according to established procedures (47). Briefly, muscles were minced and digested with a solution containing clostridium histolyticum collagenase (0.1%, Sigma-Aldrich) and Dispase II (2.4 U/ml final, Roche Diagnostics) in PBS. Dissociated cells were filtered consecutively through 100-μm and 40-μm cell strainers (BD Biosciences). Recovered mononuclear cells were preplated for a minimum of 1 hour on a plastic plate before either (i) transferring unbound cells to a fresh matrigel coated plates or (ii) isolating TdT+ SCs by FACS sorting (StemCore Facility, OHRI). The in vitro ablation of UTX in cultured SCs was achieved by treatment with 4-OH-tamoxifen (2.5 μM final concentration, Sigma-Aldrich) for 48 hours.
Proliferation assays were performed by adding 10 μM EdU to myofiber cultures for 2 hours prior to fixation. Incorporation of EdU into genomic DNA was visualized using the Click-iT EdU Imaging Kit (Invitrogen). Active caspase assays used to examine apoptosis were carried out with the FAM-FLICA in vitro Caspase Detection Kit (ImmunoChemistry Technologies).
Immunocytochemistry and IF staining.
For histological analysis, TA muscles were harvested and fixed in 4 % PFA overnight by rocking at 4°C. The muscle was then embedded in paraffin and cut at 5-μm cross sections (University of Ottawa Histology Services). Histological analysis was performed using H&E staining (Electron Microscopy Sciences). For IF analysis, TA muscles were harvested and directly embedded in Optimal Cutting Temperature solution (Tissue-Tek). Cryosections of 10 μM thickness were prepared on Superfrost Plus slides (Electron Microscopy Sciences). Muscle sections were then fixed in 4% PFA for 5 minutes and permeabilized using 1% Triton X-100 (Sigma-Aldrich) in PBS for 30 minutes. Samples were then blocked using a buffer of 2% donkey serum, 1% BSA, 0.1% gelatin from cold water fish skin, 0.1% Triton X-100, and 0.05% Tween20. For immunostaining using MyoD, MYOG, and PAX7 antibodies, antigen retrieval was performed by boiling in a buffer of 0.05% Tween20/10 mM citrate (pH 6) for 30 minutes prior to incubation in blocking buffer. Primary antibody incubation has been performed overnight at 4°C. After secondary antibody incubation (1 hour at 22°C), slides were quenched in a solution of 0.1% Black Suddan/70% ethanol (Sigma-Aldrich) for 10 minutes. Slides were then counterstained with DAPI (1 in 5,000) for nuclei visualization.
For IF staining on myofiber or primary SCs in culture, myofibers or cells were fixed in 1% PFA for 10 minutes, permeabilized in 0.5% Triton X-100 for 5 minutes, and incubated in the blocking buffer. Primary and secondary antibody incubations were performed at 22°C for 3 hours and 1 hour, respectively. After a wash with PBS, nuclei were visualized using DAPI (1 in 5,000).
IF images have been obtained using a LSM 510 Meta confocal microscope, or either an Axioplan2 or an Observer Z1 (all from Zeiss) fluorescent microscope. Histologic images were obtained using an Aperio CS2 Scan Scope (Leica).
Antibodies.
Primary antibodies used for immunostaining in this study include: rabbit anti-PAX7 (1 in 100, Abcam, catalog ab34360), rabbit anti-MyoD (1 in 100, Santa Cruz Biotechnology Inc., catalog sc304), rabbit anti-MYOG (1 in 100, Santa Cruz Biotechnology Inc., catalog sc576), rabbit anti-Myf5 (1 in 100, Santa Cruz Biotechnology Inc., catalog sc302), rabbit anti-H3K27me3 (1 in 100, Millipore, catalog 07449), homemade rabbit anti-UTX (19), mouse anti-PAX7 (1 in 5 of culture supernatant, Developmental Studies Hybridoma Bank [DSHB], catalog AB528428), mouse anti–MyHC/MF-20 (1 in 10 of culture supernatant, DSHB, catalog AB2147781), mouse anti-Myog/F5D (1 in 100 of ascites, DSHB, catalog AB2146602), rabbit anti-Myog (1 in 100, Santa Cruz Biotechnology Inc., catalog sc12732), and mouse anti-H3K27me2/3 (1 in 100, Active Motif, catalog 39535). Primary antibodies have been diluted in PBS containing 1% BSA and 0.1% gelatin from cold water fish skin. Species-specific secondary antibodies, donkey anti-rabbit conjugated to Alexa488 (Invitrogen, catalog A11034) or Cy3 (Jackson ImmunoResearch Laboratories Inc., catalog 111165006) and goat anti-mouse conjugated to Alexa 488 (Invitrogen, catalog A11001) were used at 1 in 1,000 dilution in PBS.
Histone chromatin IP.
Sorted α7-integrin+ SCs (AbLab, catalog 5300101) from Utxfl/TdT, or UtxWT/TdT or UtxKI/KI mice were expanded in culture prior to an in vitro ablation of UTX using 4-OH-tamoxifen. Alternatively, sorted α7-integrin+ SCs from UtxKI/KI mice were expanded in culture. Differentiation was induced 24 hours prior to chromatin isolation. ChIP assays on purified SCs were performed according to established protocols (48) where volumes were adjusted to accommodate a smaller number of cells. Briefly, 2.5 μg chromatin and 1 μg of rabbit anti-H3K27me3 (Millipore, catalog 07-449) were use for each experiment. Primers used for qPCR analysis of the ChIP are listed in Supplemental Table 2. For genome-wide analysis, IP DNA was amplified using the KAPA ChIP library kit (Kapa Biosystems), and paired-end sequencing was performed on a Illumina HiSeq 2000 at the McGill University and Génome Québec Innovation Centre (Montréal, Québec, Canada). Reads (50 bp) were aligned to the mm9 reference genome from UCSC genome browser with BWA version 0.7.10 (49) using default options. Reads mapping to multiple locations on the genome and repeated sequences were discarded for downstream analysis. Coverage plots were created using ngs.plot software (50).
UTX chromatin IP.
ChIP assays on purified SCs were performed according to established protocols (19) where volumes were adjusted to accommodate a smaller number of cells. Chromatin from purifed SCs (cross-linked using 1% formaldehyde) was sheared using a Bioruptor (Diagenode) to obtain a resolution of approximately 400 bp. UTX IP DNA was subjected to qPCR analysis using specific primers (see Supplemental Table 2). qPCR amplification of IP samples was normalized to genomic DNA for each primer set individually. Normalized values obtained from a mock IP (IgG) were subtracted from those observed in the UTX IP and then corrected for sample variation through division by normalized values observed from 1/50th of the input chromatin.
qPCR.
For qPCR assays, total RNA from cultured SCs was isolated using RNA-STAT60 reagent (Tel-Test Inc.) according to the manufacturer’s protocol. Purified RNA (1 μg) was subjected to reverse transcriptase reaction in the presence of 30 ng/ml random primers and 2.5 mM dNTP with 6 U/μl Moloney Murine Leukemia Virus Reverse Transcriptase (New England BioLabs Inc., catalog MO253L). Primers used for qPCR are listed in Supplemental Table 2.
RNA sequencing.
Sorted TdT+ SCs from UtxmKO/TdT or UtxWT/TdT from the hindlimbs of 10 mice were induced to differentiate for 24 hours. Total RNA was then isolated using RNesay kit (QIAGEN) using an on-column DNase digestion (RNase-Free DNase Set, QIAGEN) to avoid genomic DNA contamination. Library preparation and 150-bp paired-end RNA-Seq were performed using standard Illumina procedures for the NextSeq 500 platform. The RNA-Seq data were analyzed using the Bowtie, TopHat, CuffLinks, and CummeRbund software suite (51). Briefly, reads were mapped to the mm9 genome (University of California, Santa Cruz, California, USA) and known mm9 transcripts (RefSeq mm9 build37.2) using Bowtie version 2.2.4 (52) with default parameters. Splice junctions were determined from the mapped reads using TopHat version 2.0.12 (53) with default values. Transcript quantification was performed with HT-Seq version 0.6.1 (54). Identification of differentially expressed genes was performed with the edgeR Bioconductor package (55). RNA-Seq data were deposited in Gene Expression Omnibus (GEO GSE69968).
Statistics.
Statistical analysis was performed using one factorial ANOVA analysis when comparing 3 or more sample sets. For comparison between 2 sample sets, an unpaired, 2-tailed t test was performed. P ≤ 0.05 was considered statistically significant. Data were analyzed using Prism v4.0 (GraphPad Software).
Study approval.
All animal experiments were performed in accordance with the recommendations of the University of Ottawa Animal Care Facility, the guidelines published by the Canadian Council on the Animal Care (CCAC), and the Animal Care and Use Committee of National Institute of Diabetes and Digestive Kidney Diseases (NIDDK), NIH. CTX-induced muscle injury studies and myofiber isolation experiments were performed as described in protocol number ME-2371-A1 that had been previously approved by the University of Ottawa Animal Care Committee.
Author contributions
FJD, KG, and HF conceived and designed the experiments. HF was responsible for carrying out all the in vivo and ex vivo muscle regeneration studies and isolated tissues required for any biochemical analysis. CW and CL generated the Utxfl/fl and UTXKI mice, while CW characterized the integrity of the MLL4 complex in ES cells. KN performed the UTX-ChIP experiments and performed Western blots to confirm the stability of the UTXKI protein in muscle. AB performed the H3K27me3-ChIP experiments for ChIP-Seq analysis. SS performed the H3K27me3 ChIP-qPCR experiments. AC perform the bioinformatic analysis of RNA-Seq and ChIP-Seq studies. CGP, LZ, and BC provided unpublished reagents to the study. FJD, KG, MB, and HF interpreted the results, discussed the results’ implications, and contributed to the outline of the manuscript. FJD, HF, and KG wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Odile Neyret (Molecular Biology Platform at the Institut de Recherche Clinique de Montréal) for library preparation and high-throughput sequencing, and we thank University of Ottawa Histological Core Facility for preparation of histological slides. This work was funded by the Canadian Institutes of Health Research (MOP-136773 and FDN-143330 to F.J. Dilworth and MOP-89834 to M. Brand) and the NIH (Intramural Research Program of the NIDDK to K. Ge). H. Faralli was funded by a fellowship from the Association Française contre les Myopathies. F.J. Dilworth was supported by a Canada Research Chair in the Epigenetic Regulation of Transcription.
Footnotes
Soji Sebastian’s present address is: Canadian Nuclear Laboratories, Chalk River, Ontario, Canada.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(4):1555–1565. doi:10.1172/JCI83239.
See the related Commentary beginning on page 1.
Contributor Information
Hervé Faralli, Email: hfaralli@ohri.ca.
Chaochen Wang, Email: Chaochen.Wang@nih.gov.
Kiran Nakka, Email: knakka@ohri.ca.
Aissa Benyoucef, Email: abenyoucef@ohri.ca.
Soji Sebastian, Email: soji.sebastian@cnl.ca.
Lenan Zhuang, Email: lenan.zhuang@nih.gov.
Alphonse Chu, Email: alchu@ohri.ca.
Chengyu Liu, Email: liuch@nhlbi.nih.gov.
Brendan Camellato, Email: bcame020@uottawa.ca.
Marjorie Brand, Email: mbrand@ohri.ca.
Kai Ge, Email: kaig@niddk.nih.gov.
F. Jeffrey Dilworth, Email: jdilworth@ohri.ca.
References
- 1.Steffen PA, Ringrose L. What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat Rev Mol Cell Biol. 2014;15(5):340–356. doi: 10.1038/nrm3789. [DOI] [PubMed] [Google Scholar]
- 2.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the polycomb and trithorax group proteins. Annu Rev Genet. 2004;38(1):413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 3.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 5.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 6.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111(2):185–196. doi: 10.1016/S0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 7.Muller J, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111(2):197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 8.Pengelly AR, Copur O, Jackle H, Herzig A, Muller J. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science. 2013;339(6120):698–699. doi: 10.1126/science.1231382. [DOI] [PubMed] [Google Scholar]
- 9.Hansen KH, et al. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10(11):1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 10.Gaydos LJ, Wang W, Strome S. Gene repression. H3K27me and PRC2 transmit a memory of repression across generations and during development. Science. 2014;345(6203):1515–1518. doi: 10.1126/science.1255023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lan F, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449(7163):689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 12.Agger K, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449(7163):731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 13.Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci U S A. 2007;104(47):18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130(6):1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 15.Lee MG, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318(5849):447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 16.Greenfield A, et al. The UTX gene escapes X inactivation in mice and humans. Hum Mol Genet. 1998;7(4):737–742. doi: 10.1093/hmg/7.4.737. [DOI] [PubMed] [Google Scholar]
- 17.Shpargel KB, Sengoku T, Yokoyama S, Magnuson T. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet. 2012;8(9): doi: 10.1371/journal.pgen.1002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev. 2008;22(14):1865–1870. doi: 10.1101/gad.1673508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seenundun S, et al. UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J. 2010;29(8):1401–1411. doi: 10.1038/emboj.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mansour AA, et al. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature. 2012;488(7411):409–413. doi: 10.1038/nature11272. [DOI] [PubMed] [Google Scholar]
- 21.van Haaften G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41(5):521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Copur O, Müller J. The histone H3-K27 demethylase Utx regulates HOX gene expression in Drosophila in a temporally restricted manner. Development. 2013;140(16):3478–3485. doi: 10.1242/dev.097204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vandamme J, Lettier G, Sidoli S, Di Schiavi E, Norregaard Jensen O, Salcini AE. The C. elegans H3K27 demethylase UTX-1 is essential for normal development, independent of its enzymatic activity. PLoS Genet. 2012;8(5): doi: 10.1371/journal.pgen.1002647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shpargel KB, Starmer J, Yee D, Pohlers M, Magnuson T. KDM6 demethylase independent loss of histone H3 lysine 27 trimethylation during early embryonic development. PLoS Genet. 2014;10(8): doi: 10.1371/journal.pgen.1004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell. 2010;40(4):594–605. doi: 10.1016/j.molcel.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang C, et al. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc Natl Acad Sci U S A. 2012;109(38):15324–15329. doi: 10.1073/pnas.1204166109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thieme S, et al. The histone demethylase UTX regulates stem cell migration and hematopoiesis. Blood. 2013;121(13):2462–2473. doi: 10.1182/blood-2012-08-452003. [DOI] [PubMed] [Google Scholar]
- 28.Bentzinger CF, Wang YX, von Maltzahn J, Rudnicki MA. The emerging biology of muscle stem cells: implications for cell-based therapies. Bioessays. 2013;35(3):231–241. doi: 10.1002/bies.201200063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aziz A, Sebastian S, Dilworth FJ. The origin and fate of muscle satellite cells. Stem Cell Rev. 2012;8(2):609–622. doi: 10.1007/s12015-012-9352-0. [DOI] [PubMed] [Google Scholar]
- 30.Liu QC, et al. Comparative expression profiling identifies differential roles for Myogenin and p38alpha MAPK signaling in myogenesis. J Mol Cell Biol. 2012;4(6):386–397. doi: 10.1093/jmcb/mjs045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh K, Dilworth FJ. Differential modulation of cell cycle progression distinguishes members of the myogenic regulatory factor family of transcription factors. FEBS J. 2013;280(17):3991–4003. doi: 10.1111/febs.12188. [DOI] [PubMed] [Google Scholar]
- 32.Lepper C, Conway SJ, Fan CM. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460(7255):627–631. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Welstead GG, et al. X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. Proc Natl Acad Sci U S A. 2012;109(32):13004–13009. doi: 10.1073/pnas.1210787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kruidenier L, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488(7411):404–408. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho YW, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem. 2007;282(28):20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zammit PS, Golding JP, Nagata Y, Hudon V, Partridge TA, Beauchamp JR. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J Cell Biol. 2004;166(3):347–357. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siegel AL, Kuhlmann PK, Cornelison DD. Muscle satellite cell proliferation and association: new insights from myofiber time-lapse imaging. Skelet Muscle. 2011;1(1): doi: 10.1186/2044-5040-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Juan AH, et al. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev. 2011;25(8):789–794. doi: 10.1101/gad.2027911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woodhouse S, Pugazhendhi D, Brien P, Pell JM. Ezh2 maintains a key phase of muscle satellite cell expansion but does not regulate terminal differentiation. J Cell Sci. 2013;126(pt 2):565–579. doi: 10.1242/jcs.114843. [DOI] [PubMed] [Google Scholar]
- 40.Blum R, Vethantham V, Bowman C, Rudnicki M, Dynlacht BD. Genome-wide identification of enhancers in skeletal muscle: the role of MyoD1. Genes Dev. 2012;26(24):2763–2779. doi: 10.1101/gad.200113.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chakroun I, et al. Genome-wide association between Six4, MyoD, and the histone demethylase Utx during myogenesis. FASEB J. 2015;29(11):4738–4755. doi: 10.1096/fj.15-277053. [DOI] [PubMed] [Google Scholar]
- 42.Wang AH, et al. The histone chaperone Spt6 coordinates histone H3K27 demethylation and myogenesis. EMBO J. 2013;32(8):1075–1086. doi: 10.1038/emboj.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faralli H, Dilworth FJ. Chaperoning RNA Polymerase II through repressive chromatin. EMBO J. 2013;32(8):1067–1068. doi: 10.1038/emboj.2013.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu L, et al. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 2013;4(1):189–204. doi: 10.1016/j.celrep.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Judson RN, Zhang RH, Rossi FM. Tissue-resident mesenchymal stem/progenitor cells in skeletal muscle: collaborators or saboteurs? FEBS J. 2013;280(17):4100–4108. doi: 10.1111/febs.12370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saclier M, Cuvellier S, Magnan M, Mounier R, Chazaud B. Monocyte/macrophage interactions with myogenic precursor cells during skeletal muscle regeneration. FEBS J. 2013;280(17):4118–4130. doi: 10.1111/febs.12166. [DOI] [PubMed] [Google Scholar]
- 47.Shefer G, Van de Mark DP, Richardson JB, Yablonka-Reuveni Z. Satellite-cell pool size does matter: defining the myogenic potency of aging skeletal muscle. Dev Biol. 2006;294(1):50–66. doi: 10.1016/j.ydbio.2006.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brand M, Rampalli S, Chaturvedi CP, Dilworth FJ. Analysis of epigenetic modifications of chromatin at specific gene loci by native chromatin immunoprecipitation (N-ChIP) of nucleosomes isolated using hydroxyapatite chromatography. Nat Protoc. 2008;3(3):398–409. doi: 10.1038/nprot.2008.8. [DOI] [PubMed] [Google Scholar]
- 49.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen L, Shao N, Liu X, Nestler E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15: doi: 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anders S, Pyl PT, Huber W. HTSeq — a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.