Abstract
Precise epigenetic modifications in stem cells control developmental programs and cell fate decisions. In particular, the addition or removal of trimethylation of histone 3 lysine 27 (H3K27me3) at lineage-specific genes has been linked to the repression of gene expression, and a precise balance of methyltransferases and demethylases within cells determines H3K27me3 levels. The demethylase UTX is essential for development and tissue homeostasis; however, a role for UTX in stem cell–mediated tissue regeneration is unknown. In this issue of the JCI, Dilworth and colleagues reveal that UTX and its demethylase activity are required in the muscle stem cell lineage for muscle regeneration in response to injury. Specifically, UTX mediates the removal of H3K27me3 in the promoter of the transcription factor myogenin, which regulates myogenic differentiation. The results of this study provide important insight into the contribution of epigenetic regulation in stem cell–mediated regeneration of adult tissues.
H3K27me3 in development and tissue homeostasis
Modifications of histone proteins are among the most well studied epigenetic features of stem cells. One of these histone modifications, trimethylation of histone 3 lysine 27 (H3K27me3), is a mark of transcriptionally silenced chromatin and has been extensively studied in the context of developmental programs (1). It is generally believed that, during development, H3K27me3 is introduced at lineage-specific genes by the polycomb repressor complex 2 (PRC2) and is interpreted by PRC1, which recruits transcriptional repressors to the site of the H3K27me3 mark (2). The balance between methyltransferases (EZH1 and EZH2) and jumonji domain–containing demethylases (JMJD3 and UTX) maintains H3K27me3 levels in cells (3).
UTX (also known as KDM6A) has previously been shown to play important roles in both development and normal tissue homeostasis (4, 5), and germline KO of the UTX-encoding gene is embryonic lethal (5). Further evaluation of UTX-null embryos revealed a decrease in somite counts, defects in neural tube closure, and heart malformation between E9.5 and E13.5. Interestingly, female embryos exhibit a more severe phenotype than male embryos, which hemizygotically express the UTX paralog UTY — a paralog that does not possess H3K27 demethylase activity. Conditional deletion of Utx in adult animals compromises hematopoiesis due to defects in stem and progenitor cell migration (4). Many tissues, including muscle, rely on reserves of stem cells to facilitate repair and regeneration. However, the role of UTX in the context of tissue regeneration following injury in adult animals remains largely unexplored.
UTX in adult muscle regeneration
In this issue, Dilworth and colleagues provide important insight into the role of UTX in adult muscle regeneration (6). The authors generated a conditional KO mouse in which the expression of UTX can be specifically ablated in adult skeletal muscle stem cells (satellite cells) and used this mouse to study the effect of UTX deficiency on muscle regeneration following acute muscle injury. Indeed, loss of UTX in satellite cells impaired muscle regeneration in both male and female mice. Moreover, muscle regeneration was impaired in knock-in mice expressing a mutant UTX protein without demethylase activity, suggesting that the H3K27 demethylase activity in satellite cells is indispensable for adult muscle regeneration.
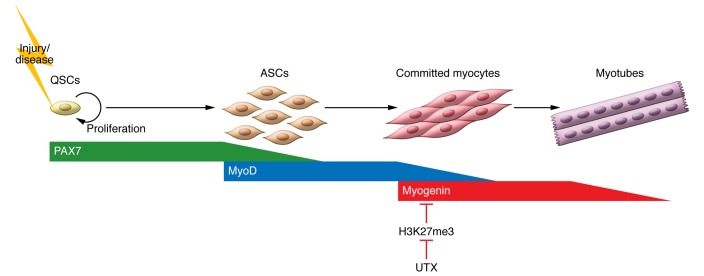
In order to explore the changes in cellular functions that are responsible for impaired muscle regeneration in Utx KO mice, Faralli et al. examined the activation and lineage progression of satellite cells (6). Satellite cells primarily reside in a quiescent state in adult animals due to the low turnover rate of mature muscle fibers. In response to injury or diseases that damage myofibers, satellite cells are activated and reenter the cell cycle. Progeny of satellite cells then proliferate and differentiate to form new myofibers. This process is tightly regulated by the temporal expression of key myogenic transcription factors, including PAX7, MyoD, and myogenin (Figure 1). Satellite cells from animals with conditionally deleted Utx or from animals expressing the enzymatically dead UTX properly entered the cell cycle and proliferated in response to muscle injury (6). However, there was a marked reduction in the number of differentiating satellite cell progeny, which are identified by expression of myogenin, a transcription factor that is critical for myogenic differentiation. The reduction in myogenin expression correlated with the accumulation of H3K27me3 at the myogenin promoter region. This observation is consistent with a previous report from the Dilworth group that demonstrated UTX is required for the induction of myogenin in myogenic cell lines in differentiation conditions in vitro (7).
Figure 1. Activation and differentiation of satellite cells require precise temporal expression of linage-specific transcription factors.
Quiescent satellite cells (QSCs), marked by high levels of the transcription factor PAX7, begin to proliferate in response to muscle damage as the result of injury or disease. The myogenic regulatory factor MyoD drives differentiation of activated satellite cells (ASCs) toward the myogenic lineage. In these committed myocytes, myogenin upregulation promotes differentiation and formation of new muscle fibers. In this issue, Faralli et al. reveal that the demethylase UTX is required for the differentiation of satellite cell progeny and mediates removal of the repressive H3K27me3 mark at the myogenin promoter (6).
Deciphering the balance between methyltransferases and demethylases is essential to understanding the regulation of H3K27me3 and its role in cellular function. Faralli et al. have shown that the absence of UTX leads to the accumulation of the repressive chromatin mark H3K27me3 at the promoter and/or enhancer regions of myogenin and a number of genes that are specifically expressed in terminally differentiated myofibers (6). Moreover, the increase of H3K27me3 at these promoters coincides with the decrease in their expression levels. It will be interesting to identify additional UTX target sites across the entire genome and to determine whether H3K27me3 at these loci dynamically changes upon satellite cell activation, proliferation, and differentiation. The pattern of H3K27me3 has been determined in quiescent satellite cells and their activated, proliferating progeny (8). The level of this repressive chromatin mark is low in quiescent satellite cells and rapidly increases when the cells are activated. This process is accompanied by an increase in the expression of EZH2. Consistent with this, loss of EZH2 expression impairs the activation of satellite cells and the expansion of their progeny (8, 9). EZH2 is also required for activation and proliferation in the skin stem cell compartment, as a subset of epidermal lineage–specific genes is regulated by H3K27me3 (10). It is interesting that these two enzymes, EZH2 and UTX, which have opposing enzymatic activity, function as different epigenetic switches for stem cell–fate determination. Whereas EZH2 appears to be the switch for the activation and proliferation of stem cells, UTX appears to be the switch for the differentiation of stem cell progeny. The expression of UTX remains constant throughout this process; therefore, it will be informative to determine whether the changes in H3K27me3 are due to changes in recruitment of UTX to its target sites across the genome or due to changes in its enzymatic activity.
The study by Faralli et al. also contributes to our understanding of the nonredundant roles of H3K27me3 demethylases in cellular functions (6). As discussed above, the global level of H3K27me3 is low in quiescent satellite cells, suggesting that H3K27 demethylase activity may be required to maintain this quiescent state. Therefore, it is surprising that the demethylase-dead UTX knock-in mouse develops normally and exhibits no defects in the homeostasis of satellite cells. The two known H3K27me3 demethylases, UTX and JMJD3, appear to have contrasting roles in certain disease conditions, such as T cell acute lymphoblastic leukemia (T-ALL). While JMJD3 is essential to initiate and maintain T-ALL, UTX functions as a tumor suppressor and is often genetically inactivated in this disease (11, 12). In satellite cells, JMJD3 is highly expressed in quiescent stem cells but downregulated when the cells are activated to enter the cell cycle (8). This decrease in JMJD3 expression and the concurrent increase in EZH2 expression lead to a rapid accumulation of H3K27me3 in activated satellite cells. Given these findings, it is possible that JMJD3 and UTX may regulate stem cell function under different contexts. It is intriguing to consider a model of epigenetic regulation of stem cell function in which JMJD3 is essential to maintain the plasticity and potency of quiescent stem cells, whereas UTX functions downstream to restrict cell fate decision.
Conclusions and future directions
From a signaling perspective, stem cell–mediated adult tissue regeneration has been regarded in many ways as a reactivation of the developmental program of the same tissue (13, 14). In addition to delineating an epigenetic switch that is specific to the lineage determination and differentiation process of satellite cells as they mediate muscle regeneration, the study by Faralli et al. also provides evidence that regenerative myogenesis in adult animals has distinct levels of regulation at the epigenetic level compared with developmental myogenesis (6). The regulation of other epigenetic marks during adult muscle regeneration remains largely unknown. Much work is also needed to understand the epigenetic regulation of the regeneration of other adult tissues with resident stem cells. These studies could be the foundation for developing epigenetic drugs to enhance tissue regeneration and to treat degenerative diseases.
Acknowledgments
This work was supported by funding from the Glenn Foundation for Medical Research, the NIH (P01 AG036695, R37 AG023806, R01 AR062185, and R01 AG047820), and the Department of Veterans Affairs (Merit Review) to T.A. Rando.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(4):1233–1235. doi:10.1172/JCI86798.
See the related article beginning on page 1555.
References
- 1.Steffen PA, Ringrose L. What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat Rev Mol Cell Biol. 2014;15(5):340–356. doi: 10.1038/nrm3789. [DOI] [PubMed] [Google Scholar]
- 2.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128(4):735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Agger K, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449(7163):731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 4.Thieme S, et al. The histone demethylase UTX regulates stem cell migration and hematopoiesis. Blood. 2013;121(13):2462–2473. doi: 10.1182/blood-2012-08-452003. [DOI] [PubMed] [Google Scholar]
- 5.Welstead GG, et al. X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. Proc Natl Acad Sci U S A. 2012;109(32):13004–13009. doi: 10.1073/pnas.1210787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faralli H, et al. UTX demethylase activity is required for satellite cell–mediated muscle regeneration. J Clin Invest. 2016;126(4):1555–1565. doi: 10.1172/JCI83239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seenundun S, et al. UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J. 2010;29(8):1401–1411. doi: 10.1038/emboj.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu L, et al. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 2013;4(1):189–204. doi: 10.1016/j.celrep.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juan AH, et al. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev. 2011;25(8):789–794. doi: 10.1101/gad.2027911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ezhkova E, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136(6):1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ntziachristos P, et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature. 2014;514(7523):513–517. doi: 10.1038/nature13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Haaften G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41(5):521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfefferli C, Jaźwińska A. The art of fin regeneration in zebrafish. Regeneration. 2015;2(2):72–83. doi: 10.1002/reg2.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaloulis K, Chera S, Hassel M, Gauchat D, Galliot B. Reactivation of developmental programs: the cAMP-response element-binding protein pathway is involved in hydra head regeneration. Proc Natl Acad Sci U S A. 2004;101(8):2363–2368. doi: 10.1073/pnas.0306512101. [DOI] [PMC free article] [PubMed] [Google Scholar]