Abstract
BACKGROUND:
The tumor suppressor p53 responds to a variety of environmental stressors by regulating cell cycle arrest, apoptosis, senescence, DNA repair, bioenergetics and mitochondrial DNA (mtDNA) copy number maintenance. Developmental abnormalities have been reported in p53-deficient mice, and altered p53 and p53-associated pathways in autism (AU). Furthermore, via the Pten-p53 crosstalk, Pten haploinsufficient-mice have autisticlike behavior accompanied by brain mitochondrial dysfunction with accumulation of mtDNA deletions.
METHODS:
mtDNA copy number and deletions, and p53 gene copy ratios were evaluated in peripheral blood monocytic cells from children aged 2–5 years with AU (n = 66), race-, gender-, and age-matched typically neurodeveloping children (n = 46), and both parents from each diagnostic group, recruited by the Childhood Autism Risk from Genes and Environment study at the University of California, Davis.
RESULTS:
mtDNA deletions and higher p53 gene copy ratios were more common in children with AU and their fathers. The incidence of mtDNA deletions in fathers of children with AU was increased 1.9-fold over fathers of typically neurodeveloping children, suggesting a role for deficient DNA repair capacity not driven by paternal age. Deletions in mtDNA and altered p53 gene copy ratios seem to result from genetics (children with severity scores ≥8) and/or act in concert with environmental factors (children with 6–7 severity scores).
CONCLUSIONS:
Given pro- and antioxidant activities of p53, and associations of genomic instability with disorders other than AU, our study suggests a link between DNA repair capacity, genomic instability in the 17p13.1 region influenced by environmental triggers, and AU diagnosis.
What’s Known on This Subject:
Developmental abnormalities have been observed in p53-deficient mice. Altered p53 and p53-dependent pathways have been reported in autism, and via the PTEN-p53 interplay, Pten haploinsufficient mice present autistic-like behavior accompanied by brain mitochondrial dysfunction with accumulation of mitochondrial DNA deletions.
What This Study Adds:
Altered p53 and mitochondrial DNA gene copy ratios segregate with autism. These outcomes seem to originate from deficient paternal DNA repair capacity (not age-related) at severity scores ≥8, whereas higher influence of gene x environment is observed at lower scores.
The p53 gene is responsive to a large number of environmental stressors by regulating maintenance of genomic stability,1 changes in oxidative stress,2–4 mitochondrial DNA (mtDNA) copy number (CN),5,6 and mitochondrial respiration1,6 as well as the neurotoxic response to flame retardants7,8 and pesticide exposures.9–12 Studies have linked p53 to developmental abnormalities13 (see also Supplemental Table 5), and regions adjacent to 17p13.1 encoding for proteins with important roles in brain function and neurodevelopment (eg, axonal dynein heavy chain 2 and Na+/K+ ATPase subunit β-2) and others have been found to be associated with autism (AU) (eg, ephrin-B314). Increased p53/Bcl2 protein ratios had been found in brain regions of a small set of autistic subjects15–17; however, no characterization of posttranslational modifications or functional status of p53 was undertaken. Neuronal Pten haploinsufficiency in mice results in a sustained Akt activation and decreased p53 protein expression and function (evidenced by downregulation of p53 targets), mitochondrial dysfunction, accumulation of mtDNA deletions,6 and the occurrence of aberrant social and repetitive behavior associated with AU.6,18
The mitochondrial electron transport chain is the major intracellular source of reactive oxygen species, and as such, mtDNA becomes oxidatively modified as it is evidenced by its relatively high mutation rate19 and accumulation of deletions with age.20,21 Mitochondria can compensate for these damages by responding with increased mtDNA replication without increases in oxidative phosphorylation22–27; however, increases in CN also have been associated with defective transcription, respiratory chain deficiency, and age-related accumulation of mtDNA deletions.28 Due to the involvement of p53 in genomic stability, mitochondrial function, and protection of mtDNA from oxidative stress,2 we hypothesized that a particular genetic background could ensue in p53 and mtDNA damage. On exposure to environmental stressors, this damage could be compounded (second-hit hypothesis). This is demonstrated by the enhanced neurotoxic effect of excitotoxic amino acids when oxidative phosphorylation is inhibited29–32 or the exacerbated neuronal mitochondrial toxicity to polybrominated diphenyl ethers in the presence of an AU-like background (PTEN deficiency).33 This hypothesis provides a framework for the observed mitochondrial dysfunction reported in peripheral blood mononuclear cells (PBMCs) from children with AU,34 deficits accompanied by increased oxidative stress, evidenced by higher rates of hydrogen peroxide production34 and increased mtDNA deletions.35
The major goal of the current study was to identify changes in mtDNA CN, mtDNA deletions, and p53 gene copy ratios in children diagnosed with AU and their biological parents. To this end, these outcomes were tested in PBMCs from children aged 2 to 5 years diagnosed with AU and race-, gender-, and age-matched, typically neurodeveloping (TD) children and their parents recruited by the Childhood Autism Risk from Genes and Environment study (CHARGE) at the University of California Davis.36
Methods
Clinical Selection of Individuals and Diagnosis
Specimens and data of children aged 2 to 5 years with clinically confirmed AU (n = 66; severity scores [SSs] ≥6), and race-, gender-, and age-matched, clinically confirmed TD children (n = 46) and both parents from each diagnostic group (when available) were obtained from the CHARGE study35,36 (Table 1). The Autism Diagnostic Observation Schedule (ADOS)-2 comparison scores were used to calculate the SSs.37,38 This metric measures severity on a 10-point scale and equates ADOS modules across the language level and age of children. The level of autism spectrum disorder (ASD)-related symptoms based on this scale is interpreted as follows: 1 to 3 = no ASD; 4 to 5 = ASD, low level; 6 to 10 = AU, with 6 to 7 = moderate level and 8 to 10 = high level37,39 (see also Supplemental Material).
TABLE 1.
Demographics of Groups Evaluated in This Study
| Outcome | TD | AU | P |
|---|---|---|---|
| Children, n | 46 | 66 | |
| Race, % | |||
| Asian | 7.7 | 7.7 | .719 |
| Hispanic | 35.9 | 42.3 | .628 |
| White | 48.7 | 50.0 | .955 |
| Multiracial | 7.7 | 3.8 | .636 |
| Age, y | 3.5 ± 0.1 | 3.9 ± 0.1 | .174 |
| Gender, % | |||
| Girls | 12.9 | 15.4 | |
| Boys | 87.1 | 84.6 | .638 |
| Test scores | |||
| Vineland | 107 ± 2 | 64 ± 2 | <.0001 |
| Mullen | 106 ± 3 | 61 ± 3 | <.0001 |
| Severity | n.a. | 7.7 ± 0.2 | |
| Parents (couples; n) | 37 | 48 | |
| Age, y | |||
| Father | 34.3 ± 0.9 | 34 ± 1 | .829 |
| Mother | 31.8 ± 0.8 | 35 ± 3 | .819 |
Age of parents is shown at birth of index child. Data (age, Vineland, Mullen, and SSs) were expressed as mean ± SEM. SSs for typical children are between 0 and 3; all TD children in CHARGE were administered a screening instrument to rule out ASD symptoms and scored very low; hence n.a., not applicable.
Evaluation of mtDNA CN, Deletions in mtDNA, and p53 Gene Copy Ratios
Genomic DNA was extracted from PBMCs and changes in mtDNA CN were evaluated by quantitative polymerase chain reaction (qPCR) by using dual-labeled probes.34,35 The gene CN of cytochrome b (CYTB), and NADH dehydrogenase subunits 1 (ND1) were normalized by a single-copy nuclear gene pyruvate kinase (PK).34 For mtDNA CN, the ratio of ND1/PK was used, for most of the deletions are present in the major arc.34 Deletions in mtDNA were estimated by evaluating the mitochondrial gene copy ratios of CYTB, located between origins of heavy-strand and light-strand mtDNA synthesis (associated with replication pausing and breakage40,41), normalized by ND1.34,35
Gene copy ratios of p53 were evaluated by qPCR (normalized to PK) in a segment with low incidence of mutations (<1.5%42,43). All individuals (regardless of diagnostic group, gender, or age) exhibited a mean p53 haploid CN >0.8, indicative of a normal CN status (more details in Supplemental Material). All individual data are included in Supplemental Table 6. Heritability of outcomes was estimated as h2 from coefficients for child outcomes regressed on midparent outcomes. Heritability is a ratio of variances: it is the proportion of total variance in a population for a particular measurement (ie, mtDNA CN), taken at a particular time, attributable to variation in additive genetic or total genetic values,44 reflecting all possible genetic contributions to populations’ phenotypic variances, including epistatic effects (multigene interactions) as well as maternal and paternal effects where individuals are directly affected by their parents’ phenotype.45 To estimate h2 in this study, the average values from spouses (midparent) versus offspring and single parent (values from either mothers or fathers) versus offspring models were used and calculated as described by Visscher et al.44
Statistical Analysis
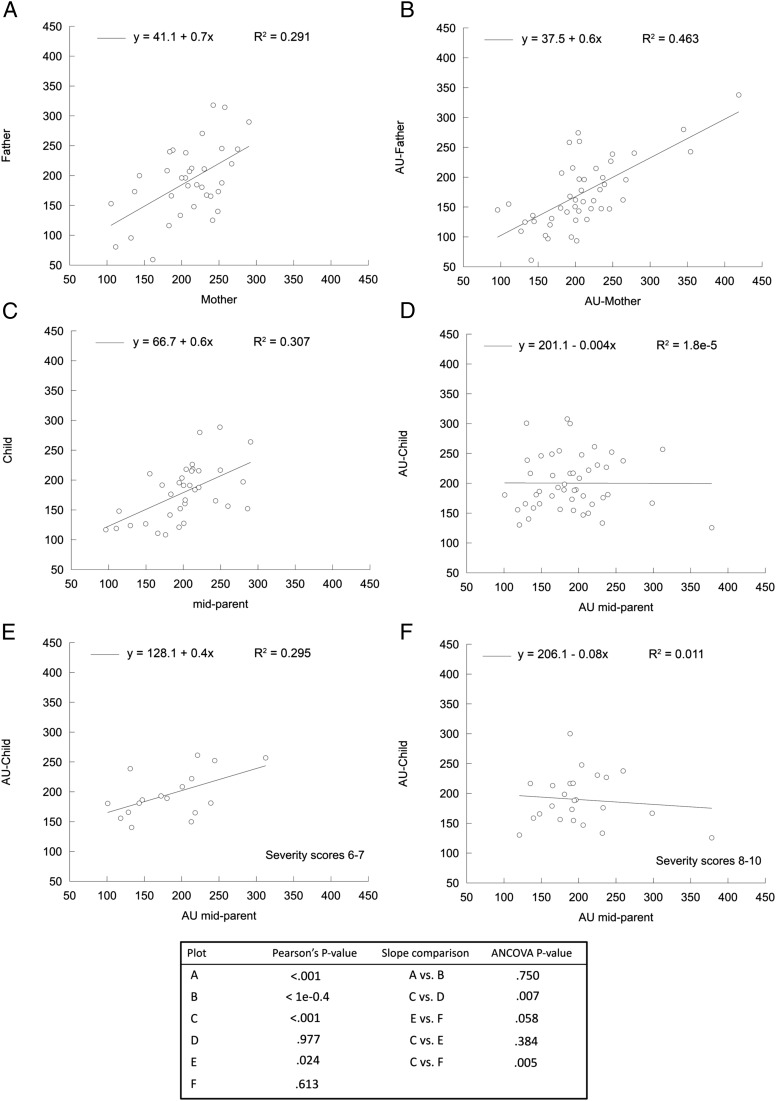
Experiments were run in triplicates and repeated 3 times on different days unless noted otherwise. Normal probability plots were generated to evaluate whether the variable observations followed a normal distribution. Significant differences between means of 2 groups were evaluated with the unequal variance, 2-tailed Student’s t test (GraphPad Prism 5 software, GraphPad software, La Jolla, CA). For frequencies, the χ2 test was used without Yates correction. All data collected in this study, except outliers identified by the Tukey test, are shown in the Supplemental Information. Analysis of covariance (ANCOVA) was carried out to evaluate the statistical significance for the correlations reported in Fig 1.
FIGURE 1.
mtDNA CN in PBMC from TD children and children with AU and their parents. The mtDNA CN per cell was determined as explained in the Methods section and evaluated in PBMC from children, mothers, and fathers. Statistical analyses were obtained by Pearson correlation and ANCOVA. A, Correlation between TD-mothers and TD-fathers. B, Correlation between AU-mothers and AU-fathers. C, Correlation of TD-midparents and TD children. D, Correlation between AU-midparents and AU-children. E, Correlation between AU-midparents and AU-children with SSs of 6 to 7. F, Correlation between AU-midparents and AU-children with SSs of 8 to 10. See other details in the text.
Results
Increased mtDNA CN/PBMC in Children With AU and Higher Incidence of mtDNA Deletions
This study was conducted on a sample of the population-based case-control CHARGE study, in which the major discriminators are diagnosis (AU versus TD, by design), and their accompanying adaptive behavior (Vineland scores) and cognitive function (Mullen Scales of Early Learning scores; Table 1). The diagnostic groups were matched for age, gender, and race. The mean mtDNA CN per lymphocyte, evaluated by qPCR, in children with AU was 212 ± 8 (mean ± SEM; n = 66), significantly higher than that of TD children (184 ± 7; n = 46; P = .012; Table 2). The incidence of children with AU with high mtDNA CN (where high was considered with a z score ≥2) was 1.4-fold that of TD children (P = .256; Table 3). We tested whether the higher average mtDNA CN was accompanied by increased mtDNA deletions. The mean mitochondrial gene ratio for TD children was 1.00 ± 0.02, not different from that of children with AU (0.98 ± 0.02; Table 2); however, the incidence of children with AU with mtDNA deletions was 2.6-fold of TD children (P = .026; Table 3). The mtDNA deletions detected in this study should not be considered pathogenic per se, for a mitochondrial defect is evidenced when a gene is deleted by >60%.46 Taken together, these results indicated that both the mtDNA CN (∼1.4-fold) and the incidence of mtDNA deletions were higher (∼3.0-fold) in children with AU than TD children. Increases in mtDNA CN and accumulation of deletions, without concomitant increases in mitochondrial mass and function, are consistent with increased oxidative stress triggered by environmental factor(s) and/or to an idiopathic form of chronic oxidative stress, confirming and extending our previous studies.35 Increases in mtDNA CN also have been associated with mitochondrial dysfunction and defects in the nucleoid structure.28
TABLE 2.
mtDNA CN/Cell, mtDNA Deletions, and p53 Gene Copy Ratios in TD and AU Trios
| Outcome | Children | Mothers of | Fathers of | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CN | TD | AU | P | TD | AU | P | TD | AU | P |
| mtDNA (±SEM) | 184 (±7) | 212 (±8) | .012 | 205 (±9) | 207 (±9) | .875 | 195 (±9) | 180 (±8) | .237 |
| n | 46 | 66 | 36 | 48 | 36 | 47 | |||
| Deletions in mtDNA (±SEM) | 1.00 (±0.02) | 0.98 (±0.02) | .458 | 0.83 (±0.01) | 0.87 (±0.01) | .206 | 0.86 (±0.02) | 0.88 (±0.02) | .520 |
| n | 46 | 66 | 35 | 46 | 37 | 48 | |||
| p53 gene copy ratio (±SEM) | 1.00 (±0.02) | 1.05 (±0.01) | .055 | 1.09 (±0.04) | 0.93 (±0.04) | .009 | 1.07 (±0.02) | 0.98 (±0.02) | .055 |
| n | 44 | 60 | 36 | 47 | 37 | 46 | |||
mtDNA deletions and p53 gene copy ratio values were normalized to TD children. Statistical analysis was performed with the 2-tailed Student’s t test.
TABLE 3.
Incidence of Changes in mtDNA CN, mtDNA Deletions, and p53 Gene Copy Ratios in TD and AU Trios
| Outcome | Children, % | P | Mothers of, % | P | Fathers of, % | P | |||
|---|---|---|---|---|---|---|---|---|---|
| TD | AU | TD | AU | TD | AU | ||||
| mtDNA CN | 10.9a | 15.2a | .256 | 8.1 | 6.3 | .357 | 10.8 | 12.5 | .409 |
| n | 46 | 66 | 36 | 48 | 36 | 47 | |||
| mtDNA deletions | 8.7 | 22.7b | .026 | 8.6 | 2.2 | .087 | 5.4 | 10.4b | .202 |
| n | 46 | 66 | 35 | 48 | 37 | 48 | |||
| p53 gene copy ratio | 9.3a | 25.0a | .017 | 13.9 | 21.3 | .193 | 8.1 | 19.6 | .070 |
| n | 44 | 60 | 36 | 47 | 37 | 46 | |||
The incidence of mtDNA deletions represents the number of individuals with deletions at the segment encoding for CYTB (z score ≤ –2 for adults and ≤ –1 for children). Incidence of p53 gene copy ratio was calculated as the percentage of individuals with high copy ratio (z score ≥ 2) or low gene copy ratio (z ≤–2). mtDNA deletions and p53 gene copy ratio values were normalized to TD children’s values and the z scores were also calculated by using the mean and SD from TD children. All TD versus AU comparisons were performed within each group (children, mothers, and fathers) by using 1-tailed χ2 test.
High mtDNA CN was considered as values with a z score ≥2.
Similar trend in outcomes between child and parents.
Higher Incidence of mtDNA Deletions in Fathers of Children With AU, Not Related to Age, Opposite to Mothers’ Trend
Fathers of TD children exhibited a mean mtDNA CN of 195 ± 9 (n = 36), not different from that of fathers with AU (180 ± 8; n = 47; Table 2). No difference in the average mtDNA CN between TD children and their fathers was observed; within the AU group, however, the fathers had significantly lower mean mtDNA CN than their affected children (P = .007). The possibility that few outliers in the AU father group had influenced the mean mtDNA CN was excluded because only 2 fathers, 1 from each diagnostic group, were considered to have mtDNA depletion (with mtDNA CN <60%).
The mtDNA deletions in TD and AU parents were within the same range (14%–17%), whereas in children, regardless of diagnosis, were <2% (P = .012 all parents versus all children; for all 6 groups: parents versus children of any diagnostic group P < .0001; Table 2). These results were consistent with an age-dependent imbalance between mtDNA repair and oxidative stress, not necessarily related to the diagnosis of the child.
As reported before, both children with AU and fathers with AU exhibited a higher incidence of mtDNA deletions compared with their respective TD group (2.6-fold children with AU versus TD children, P = .026; 1.9-fold fathers with AU versus TD fathers, P = .202; Table 3 and Napoli et al35). When stratified into decades of paternal age, the ratio of fathers with AU with mtDNA deletions versus TD fathers was 1.0 during the third, increasing to 1.4 at the fourth (P < .05) and 1.5 at the fifth (P < .05). Given that fathers’ mean age within each decade of life was statistically not different between AU and TD, a residual effect of age as a confounding factor is precluded. These results instead support age as an effect modifier of the relationship between paternal mtDNA deletions and risk for ASD. Thus, combined with the existing literature on paternal age and ASD risk,47–49 our findings are consistent with the concepts that (1) in fathers older than 30, defective repair systems governed by p53 confer an increased risk of the child developing ASD, independently of age; and (2) deficits in mtDNA repair systems may serve as one of the various processes (along with de novo mutations in nuclear DNA, epigenetic alterations, and so forth) or pathways by which advancing fathers’ age influences ASD risk.47–49
No differences in the mean mtDNA CN were observed between AU and TD mothers (207 ± 9; n = 48 and 205 ± 9; n = 36, respectively), consistent with the observations in fathers. No differences were observed in the mtDNA CN between children and mothers, regardless of their diagnostic group. The mean mitochondrial gene ratio in TD mothers was 0.83 ± 0.01 (n = 35), similar to that of mothers with AU (0.87 ± 0.01; n = 46) but both, as observed with fathers, lower than those from children (P < .0001), consistent with an age-dependent accumulation of deletions. Despite having similar mtDNA deletions (or similar mean mitochondrial gene ratios), the incidence of mothers with AU with damaged mtDNA was lower (26% of TD mothers; P = .087; Table 3).
Taken together, these results indicated that (1) the incidence of mtDNA deletions was higher in both children with AU and their fathers than their respective age-matched TD groups (approximately threefold and twofold, respectively), similar to the main paternal origin of de novo genomic point mutations49; and (2) the incidence of mothers with AU with mtDNA deletions was lower (one-third) than that expected for their age- and gender-matched group.
Contribution of Gene × Environment Interactions on the mtDNA CN in Children With AU
Considering that mtDNA CN is determined by genetic50 and environmental factors,51,52 the mtDNA CN was analyzed in AU and TD trios to elucidate the contribution of genetics and epigenetic-environmental factors (Fig 1). The correlation for the nongenetic, shared-environment or assortative-mating relationship between mtDNA CN of fathers and mothers was 0.72 for TD and 0.65 for AU (Fig 1 and Supplemental Table 8). Based on the lack of significant differences between the linear fits of AU and TD couples (as judged by ANCOVA analysis; Fig 1), the concept of assortative mating in either group is precluded. The significant and similar concordance between couples’ mtDNA CN for both diagnostic groups (and considering that they are not genetically related) supports the notion that environmental factors (eg, couples sharing at least some dietary habits, features of lifestyle, and household environment) have a strong influence on the mtDNA CN maintenance in PBMCs, as it has also been reported before.24,52
Heritability estimate (h2 or narrow sense heritability) for mtDNA CN was significant in TD child-midparent (0.56) but not for AU child-midparent (–0.004). Correlations for mtDNA CN between TD father-child (0.50) and TD mother-child (0.34) were significant, whereas those for AU were not, unless the SSs were taken into account (Supplemental Table 8). Further analysis based on the level of ASD-related symptoms (quantified as AU SSs) showed a clear pattern of h2 for mtDNA CN in children with AU: the h2 of mtDNA CN from children with AU with SSs of 6 to 7 was significant with midparent (0.37) and their mothers (0.36), reaching almost significance with their fathers as well (0.27), whereas the only correlation significant at high SSs was that of AU child-father (0.34) (Supplemental Table 8).
Taken together, these results suggest that (1) all mtDNA CN correlations involving children (0.3–0.4) were lower than the range of nongenetic, shared-environment observed between spouses (0.6–0.7), suggesting a larger variation contributed by the epigenetic/environmental influence at defining the mtDNA CN in children versus parents, possibly based on the habit differences between these 2 age groups; (2) a combination of parental and environmental influence was observed for the mtDNA CN in children with AU with moderate levels of severity (0.27–0.37), similar to that observed for TD children (0.34–0.56); and (3) the mtDNA CN in children with AU with higher SSs has a large paternal influence.
With regard to mtDNA deletions, the correlations were significant only between spouses for both diagnostic groups (0.36 and 0.39 for TD and AU, respectively), albeit at a lower level than the correlations for mtDNA CN, suggesting similar environmental influences (eg, shared habits or similar age) contributing to both mtDNA CN and deletions (Supplemental Table 9).
Higher Incidence of Altered p53 Gene Copy Ratio in Children With AU and Their Fathers
Several p53-associated proteins have been found mutated in ASD,49 lower p53 expression, and increased mtDNA CN and deletions have been observed in Pten haploinsufficient mice with ASD-like behavior.6 Further, the p53 gene is responsive to a large number of environmental stressors1–4 implicated in the maintenance of mtDNA CN5,6,28,53 and mitochondrial respiration.1,6 To this end, p53 gene CNs were evaluated by qPCR in the trios.
The mean p53 gene copy ratios of children with AU were on average 5% higher than TD (Table 2), and the incidence of high p53 gene copy ratio was higher in children with AU than TD children (2.7-fold; Table 3). In parents of children with AU, the mean p53 gene copy ratio was lower (P = .009 for mothers with AU, and P = .055 for fathers with AU) than TD parents (Table 2). A higher incidence of low p53 gene copy ratio also was observed in parents with AU (approximately twofold on average; Table 3).
Taken together, the incidence of high p53 gene copy ratio was higher in children with AU versus TD children and lower in parents with AU than TD parents.
AU Risk and Outcomes
The contribution of the previously described outcomes at influencing AU incidence was estimated with the odds ratio (OR). The risk of AU diagnosis in a child was threefold when bearing mtDNA deletions or high p53 gene copy ratio in PBMCs (Table 4). The risk of having a child with AU for fathers with mtDNA deletions or low p53 gene copy ratio was also two- to threefold.
TABLE 4.
Contribution of p53 and mtDNA Outcomes to AU Diagnosis in Children and Their Parents
| OR of AU Diagnosis Versus TD | OR | 95% Confidence Interval |
|---|---|---|
| High p53 gene copy ratio in child | 3.4 | 1.04–11.13 |
| Deletions in child mtDNA | 3.1 | 1.0–10.0 |
| Low paternal p53 gene copy ratio | 2.8 | 0.7–11.0 |
| Deletions in paternal mtDNA | 2.0 | 0.4–11.0 |
Outcomes shown have a P < .10. Cutoff values were indicated in Table 3.
Discussion
For the first time, p53 gene copy ratio and mtDNA, as molecular stress markers of nuclear and mitochondrial damage, respectively, have been studied in PBMC from a well-defined population of children with AU and their families. Although abnormalities in PBMCs may not reflect those in the brain, the impaired immune response observed in children with AU,54 the effect of neonatal immunity on neurodevelopment,55,56 and the central role of mitochondria in immunity57–59 highlight the relevance of using these cells in the context of AU. The higher incidence of an altered p53 gene copy ratio (Table 3) and increased mtDNA deletions in children with AU (Table 3) and their fathers (see text for age-stratified comparisons made previously and in Napoli et al35), compared with age- and gender-matched TD, is suggestive of a gender-dependent gene × environment interaction, exacerbated in children with AU, leading to the disruption of the link between p53 and mtDNA maintenance, possibly via increased oxidative stress and defective repair/antioxidant defenses. Contrary to other reports, our study supports the notion that increased paternal cumulative deletions (compared with age-matched group), rather than older age, confers the increased risk to AU diagnosis in the offspring.47
Increased mitochondrial reactive oxygen species production accompanied by accumulation of defective mitochondria is observed when p53 function is affected, either directly by allele loss (murine brain) or indirectly via Pten (murine brain and striatal neurons),6 resulting in lower transcription of genes encoding for antioxidant defenses.60 This is relevant in the central nervous system, given that the oxidative stress threshold required for the p53-induced pro-oxidant effects in neurons is lower than those reported in any other cell type.61 Taken together, instability in the p53 genomic region (haploinsufficiency, or altered CN) might set a background of heightened susceptibility for other detrimental triggers (environmental exposures,33,62 use of maternal medications and/or metabolic status,63,64 other diseases65–68) influencing the progeny outcome, especially during highly susceptible periods of development, such as perinatal stages.
Although >30 types of deletions are reported for the C-terminus of p53 (http://p53.iarc.fr), and CN variations in chromosome 17p13.1 (p53) and adjacent regions have been reported in cases of AU, mental retardation, and developmental delay (Supplemental Table 5), their impact on AU has not yet been fully elucidated with the exception of 1 report on ring chromosome 17.69 It is difficult to ascertain cause-effect in a case-control study; however, altered p53 gene copy ratios might underlie some of the mitochondrial defects observed before in children with AU with high SSs,34 as well as the reported genomic instability.70–72 The stronger paternal influence on mtDNA and p53 in children with AU at higher SSs is consistent with the concept that most of the de novo variants have a paternal origin.73,74 The environmental influence in children with lower SSs is consistent with the higher incidence in AU of damaging mutations in genes linked to epigenetic modification of histones,73 chromatin remodeling, and cell lineage determination.75,76,–
Given the antioxidant and pro-oxidant activities of p53,77,78 and that genomic instability also has been linked to disorders other than AU,79 future studies would need to address the role of p53 in nonimmune cells, and more importantly to define the mechanisms by which environmental factors shape this genetic susceptibility resulting in AU.
Acknowledgments
We thank the families that participated in this study, Ms Alicja Omanska-Klusek for her technical assistance, and Dr Ariel Singerman for providing statistical support.
Glossary
- ADOS
Autism Diagnostic Observation Schedule
- ANCOVA
analysis of covariance
- ASD
autism spectrum disorder
- AU
autism
- CHARGE
Childhood Autism Risk from Genes and Environment study
- CN
copy number
- CYTB
cytochrome b
- mtDNA
mitochondrial DNA
- ND1
NADH dehydrogenase subunits 1
- OR
odds ratio
- PBMC
peripheral blood mononuclear cell
- PK
pyruvate kinase
- qPCR
quantitative polymerase chain reaction
- SS
severity score
- TD
typically neurodeveloping
Footnotes
Ms. Wong performed all the experiments on mtDNA and p53, contributed to the writing of the manuscript, and performed statistical analyses; Dr. Napoli performed some of the statistical analysis and helped in drafting and revising the manuscript; Dr. Krakowiak calculated the severity scores and provided text associated with this analysis; Dr. Tassone provided genomic DNA from individuals; Dr. Hertz-Picciotto obtained funding for the Childhood Autism Risk from Genes and Environment study and oversaw demographic data and sample collection, provided intellectual input, and contributed to the editing of the manuscript; Dr. Giulivi obtained funding for this study, conceptualized and designed the study, wrote the manuscript, and performed some of the statistical analyses; and all authors approved the final manuscript as submitted.
FINANCIAL DISCLOSURE: Dr Hertz-Picciotto served on the Science Advisory Committee and received travel funds from Autism Speaks; the other authors have indicated they have no financial relationships relevant to this article to disclose.
FUNDING: This work was supported by a grant from the Simons Foundation (SFARI 271406) and National Institute of Environmental Health Sciences R01-ES011269, R01-ES015359, and R01-ES020392. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Funded by the National Institutes of Health (NIH).
POTENTIAL CONFLICT OF INTEREST: The authors have indicated they have no potential conflicts of interest to disclose.
COMPANION PAPER: A companion to this article can be found online at www.pediatrics.org/cgi/doi/10.1542/peds.2016-0049.
References
- 1.Lago CU, Sung HJ, Ma W, Wang PY, Hwang PM. p53, aerobic metabolism, and cancer. Antioxid Redox Signal. 2011;15(6):1739–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakthavatchalu V, Dey S, Xu Y, et al. Manganese superoxide dismutase is a mitochondrial fidelity protein that protects Polγ against UV-induced inactivation. Oncogene. 2012;31(17):2129–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pani G, Koch OR, Galeotti T. The p53-p66shc-Manganese Superoxide Dismutase (MnSOD) network: a mitochondrial intrigue to generate reactive oxygen species. Int J Biochem Cell Biol. 2009;41(5):1002–1005 [DOI] [PubMed] [Google Scholar]
- 4.Barone E, Cenini G, Sultana R, et al. Lack of p53 decreases basal oxidative stress levels in the brain through upregulation of thioredoxin-1, biliverdin reductase-A, manganese superoxide dismutase, and nuclear factor kappa-B. Antioxid Redox Signal. 2012;16(12):1407–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lebedeva MA, Eaton JS, Shadel GS. Loss of p53 causes mitochondrial DNA depletion and altered mitochondrial reactive oxygen species homeostasis. Biochim Biophys Acta. 2009;1787(5):328–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Napoli E, Ross-Inta C, Wong S, et al. Mitochondrial dysfunction in Pten haplo-insufficient mice with social deficits and repetitive behavior: interplay between Pten and p53. PLoS One. 2012;7(8):e42504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawashiro Y, Fukata H, Sato K, Aburatani H, Takigami H, Mori C. Polybrominated diphenyl ethers cause oxidative stress in human umbilical vein endothelial cells. Hum Exp Toxicol. 2009;28(11):703–713 [DOI] [PubMed] [Google Scholar]
- 8.Madia F, Giordano G, Fattori V, et al. Differential in vitro neurotoxicity of the flame retardant PBDE-99 and of the PCB Aroclor 1254 in human astrocytoma cells. Toxicol Lett. 2004;154(1–2):11–21 [DOI] [PubMed] [Google Scholar]
- 9.Nilufer Yonguc G, Dodurga Y, Kurtulus A, Boz B, Acar K. Caspase 1, caspase 3, TNF-alpha, p53, and Hif1-alpha gene expression status of the brain tissues and hippocampal neuron loss in short-term dichlorvos exposed rats. Mol Biol Rep. 2012;39(12):10355–10360 [DOI] [PubMed] [Google Scholar]
- 10.Loyant V, Jaffré A, Breton J, et al. Screening of TP53 mutations by DHPLC and sequencing in brain tumours from patients with an occupational exposure to pesticides or organic solvents. Mutagenesis. 2005;20(5):365–373 [DOI] [PubMed] [Google Scholar]
- 11.Dam K, Seidler FJ, Slotkin TA. Transcriptional biomarkers distinguish between vulnerable periods for developmental neurotoxicity of chlorpyrifos: implications for toxicogenomics. Brain Res Bull. 2003;59(4):261–265 [DOI] [PubMed] [Google Scholar]
- 12.Bagchi D, Balmoori J, Bagchi M, Ye X, Williams CB, Stohs SJ. Role of p53 tumor suppressor gene in the toxicity of TCDD, endrin, naphthalene, and chromium (VI) in liver and brain tissues of mice. Free Radic Biol Med. 2000;28(6):895–903 [DOI] [PubMed] [Google Scholar]
- 13.Armstrong JF, Kaufman MH, Harrison DJ, Clarke AR. High-frequency developmental abnormalities in p53-deficient mice. Curr Biol. 1995;5(8):931–936 [DOI] [PubMed] [Google Scholar]
- 14.Suda S, Iwata K, Shimmura C, et al. Decreased expression of axon-guidance receptors in the anterior cingulate cortex in autism. Mol Autism. 2011;2(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Araghi-Niknam M, Fatemi SH. Levels of Bcl-2 and P53 are altered in superior frontal and cerebellar cortices of autistic subjects. Cell Mol Neurobiol. 2003;23(6):945–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fatemi SH, Halt AR. Altered levels of Bcl2 and p53 proteins in parietal cortex reflect deranged apoptotic regulation in autism. Synapse. 2001;42(4):281–284 [DOI] [PubMed] [Google Scholar]
- 17.Sheikh AM, Malik M, Wen G, et al. BDNF-Akt-Bcl2 antiapoptotic signaling pathway is compromised in the brain of autistic subjects. J Neurosci Res. 2010;88(12):2641–2647 [DOI] [PubMed] [Google Scholar]
- 18.Page DT, Kuti OJ, Prestia C, Sur M. Haploinsufficiency for Pten and Serotonin transporter cooperatively influences brain size and social behavior. Proc Natl Acad Sci U S A. 2009;106(6):1989–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linnane AW, Marzuki S, Ozawa T, Tanaka M. Mitochondrial DNA mutations as an important contributor to ageing and degenerative diseases. Lancet. 1989;1(8639):642–645 [DOI] [PubMed] [Google Scholar]
- 20.Lee HC, Pang CY, Hsu HS, Wei YH. Differential accumulations of 4,977 bp deletion in mitochondrial DNA of various tissues in human ageing. Biochim Biophys Acta. 1994;1226(1):37–43 [DOI] [PubMed] [Google Scholar]
- 21.Fukui H, Moraes CT. Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons. Hum Mol Genet. 2009;18(6):1028–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee HC, Yin PH, Lu CY, Chi CW, Wei YH. Increase of mitochondria and mitochondrial DNA in response to oxidative stress in human cells. Biochem J. 2000;348(pt 2):425–432 [PMC free article] [PubMed] [Google Scholar]
- 23.Liu CS, Tsai CS, Kuo CL, et al. Oxidative stress-related alteration of the copy number of mitochondrial DNA in human leukocytes. Free Radic Res. 2003;37(12):1307–1317 [DOI] [PubMed] [Google Scholar]
- 24.Chou YF, Huang RF. Mitochondrial DNA deletions of blood lymphocytes as genetic markers of low folate-related mitochondrial genotoxicity in peripheral tissues. Eur J Nutr. 2009;48(7):429–436 [DOI] [PubMed] [Google Scholar]
- 25.Yeargin-Allsopp M, Rice C, Karapurkar T, Doernberg N, Boyle C, Murphy C. Prevalence of autism in a US metropolitan area. JAMA. 2003;289(1):49–55 [DOI] [PubMed] [Google Scholar]
- 26.Lee HC, Lu CY, Fahn HJ, Wei YH. Aging- and smoking-associated alteration in the relative content of mitochondrial DNA in human lung. FEBS Lett. 1998;441(2):292–296 [DOI] [PubMed] [Google Scholar]
- 27.Moreno-Loshuertos R, Acín-Pérez R, Fernández-Silva P, et al. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat Genet. 2006;38(11):1261–1268 [DOI] [PubMed] [Google Scholar]
- 28.Ylikallio E, Tyynismaa H, Tsutsui H, Ide T, Suomalainen A. High mitochondrial DNA copy number has detrimental effects in mice. Hum Mol Genet. 2010;19(13):2695–2705 [DOI] [PubMed] [Google Scholar]
- 29.Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, Beal MF, Wallace DC. Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat Genet. 1992;2(4):324–329 [DOI] [PubMed] [Google Scholar]
- 30.Yadava N, Nicholls DG. Spare respiratory capacity rather than oxidative stress regulates glutamate excitotoxicity after partial respiratory inhibition of mitochondrial complex I with rotenone. J Neurosci. 2007;27(27):7310–7317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noch E, Khalili K. Molecular mechanisms of necrosis in glioblastoma: the role of glutamate excitotoxicity. Cancer Biol Ther. 2009;8(19):1791–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kraft AD, Harry GJ. Features of microglia and neuroinflammation relevant to environmental exposure and neurotoxicity. Int J Environ Res Public Health. 2011;8(7):2980–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Napoli E, Hung C, Wong S, Giulivi C. Toxicity of the flame-retardant BDE-49 on brain mitochondria and neuronal progenitor striatal cells enhanced by a PTEN-deficient background. Toxicol Sci. 2013;132(1):196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giulivi C, Zhang YF, Omanska-Klusek A, et al. Mitochondrial dysfunction in autism. JAMA. 2010;304(21):2389–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Napoli E, Wong S, Giulivi C. Evidence of reactive oxygen species-mediated damage to mitochondrial DNA in children with typical autism. Mol Autism. 2013;4(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hertz-Picciotto I, Croen LA, Hansen R, Jones CR, van de Water J, Pessah IN. The CHARGE study: an epidemiologic investigation of genetic and environmental factors contributing to autism. Environ Health Perspect. 2006;114(7):1119–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gotham K, Pickles A, Lord C. Standardizing ADOS scores for a measure of severity in autism spectrum disorders. J Autism Dev Disord. 2009;39(5):693–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hus V, Lord C. The autism diagnostic observation schedule, module 4: revised algorithm and standardized severity scores. J Autism Dev Disord. 2014;44(8):1996–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lord CRM, Rutter M, DiLavore M, Risi S, Gotham K, Bishop S. Autism Diagnostic Observation Schedule, Second Edition (ADOS-2) Manual. Torrance, CA: Western Psychological Services; 2012 [Google Scholar]
- 40.Lenglez S, Hermand D, Decottignies A. Genome-wide mapping of nuclear mitochondrial DNA sequences links DNA replication origins to chromosomal double-strand break formation in Schizosaccharomyces pombe. Genome Res. 2010;20(9):1250–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bailey LJ, Cluett TJ, Reyes A, et al. Mice expressing an error-prone DNA polymerase in mitochondria display elevated replication pausing and chromosomal breakage at fragile sites of mitochondrial DNA. Nucleic Acids Res. 2009;37(7):2327–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petitjean A, Mathe E, Kato S, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28(6):622–629 [DOI] [PubMed] [Google Scholar]
- 43.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253(5015):49–53 [DOI] [PubMed] [Google Scholar]
- 44.Visscher PM, Hill WG, Wray NR. Heritability in the genomics era—concepts and misconceptions. Nat Rev Genet. 2008;9(4):255–266 [DOI] [PubMed] [Google Scholar]
- 45.Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. 4th ed. Essex, England: Longman; 1996 [Google Scholar]
- 46.Holt IJ, Harding AE, Cooper JM, et al. Mitochondrial myopathies: clinical and biochemical features of 30 patients with major deletions of muscle mitochondrial DNA. Ann Neurol. 1989;26(6):699–708 [DOI] [PubMed] [Google Scholar]
- 47.Kong A, Frigge ML, Masson G, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488(7412):471–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shelton JF, Tancredi DJ, Hertz-Picciotto I. Independent and dependent contributions of advanced maternal and paternal ages to autism risk. Autism Res. 2010;3(1):30–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Roak BJ, Vives L, Girirajan S, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485(7397):246–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niu R, Yoshida M, Ling F. Increases in mitochondrial DNA content and 4977-bp deletion upon ATM/Chk2 checkpoint activation in HeLa cells. PLoS One. 2012;7(7):e40572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clay Montier LL, Deng JJ, Bai Y. Number matters: control of mammalian mitochondrial DNA copy number. J Genet Genomics. 2009;36(3):125–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andreu AL, Martinez R, Marti R, García-Arumí E. Quantification of mitochondrial DNA copy number: pre-analytical factors. Mitochondrion. 2009;9(4):242–246 [DOI] [PubMed] [Google Scholar]
- 53.Noack H, Bednarek T, Heidler J, Ladig R, Holtz J, Szibor M. TFAM-dependent and independent dynamics of mtDNA levels in C2C12 myoblasts caused by redox stress. Biochim Biophys Acta. 2006;1760(2):141–150 [DOI] [PubMed] [Google Scholar]
- 54.Onore C, Careaga M, Ashwood P. The role of immune dysfunction in the pathophysiology of autism. Brain Behav Immun. 2012;26(3):383–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shanks N, Windle RJ, Perks PA, et al. Early-life exposure to endotoxin alters hypothalamic-pituitary-adrenal function and predisposition to inflammation. Proc Natl Acad Sci U S A. 2000;97(10):5645–5650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ellis S, Mouihate A, Pittman QJ. Early life immune challenge alters innate immune responses to lipopolysaccharide: implications for host defense as adults. FASEB J. 2005;19(11):1519–1521 [DOI] [PubMed] [Google Scholar]
- 57.West AP, Shadel GS, Ghosh S. Mitochondria in innate immune responses. Nat Rev Immunol. 2011;11(6):389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arnoult D, Soares F, Tattoli I, Girardin SE. Mitochondria in innate immunity. EMBO Rep. 2011;12(9):901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cloonan SM, Choi AM. Mitochondria: commanders of innate immunity and disease? Curr Opin Immunol. 2012;24(1):32–40 [DOI] [PubMed] [Google Scholar]
- 60.Li W, Kong AN. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol Carcinog. 2009;48(2):91–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chatoo W, Abdouh M, Bernier G. p53 pro-oxidant activity in the central nervous system: implication in aging and neurodegenerative diseases. Antioxid Redox Signal. 2011;15(6):1729–1737 [DOI] [PubMed] [Google Scholar]
- 62.Shelton JF, Hertz-Picciotto I, Pessah IN. Tipping the balance of autism risk: potential mechanisms linking pesticides and autism. Environ Health Perspect. 2012;120(7):944–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Croen LA, Grether JK, Yoshida CK, Odouli R, Hendrick V. Antidepressant use during pregnancy and childhood autism spectrum disorders. Arch Gen Psychiatry. 2011;68(11):1104–1112 [DOI] [PubMed] [Google Scholar]
- 64.Lyall K, Schmidt RJ, Hertz-Picciotto I. Maternal lifestyle and environmental risk factors for autism spectrum disorders. Int J Epidemiol. 2014;43(2):443–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Napoli E, Ross-Inta C, Wong S, et al. Altered zinc transport disrupts mitochondrial protein processing/import in fragile X-associated tremor/ataxia syndrome. Hum Mol Genet. 2011;20(15):3079–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krakowiak P, Walker CK, Bremer AA, et al. Maternal metabolic conditions and risk for autism and other neurodevelopmental disorders. Pediatrics. 2012;129(5). Available at: www.pediatrics.org/cgi/content/full/129/5/e1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Atladóttir HO, Henriksen TB, Schendel DE, Parner ET. Autism after infection, febrile episodes, and antibiotic use during pregnancy: an exploratory study. Pediatrics. 2012;130(6). Available at: www.pediatrics.org/cgi/content/full/130/6/e1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zerbo O, Iosif AM, Walker C, Ozonoff S, Hansen RL, Hertz-Picciotto I. Is maternal influenza or fever during pregnancy associated with autism or developmental delays? Results from the CHARGE (CHildhood Autism Risks from Genetics and Environment) study. J Autism Dev Disord. 2013;43(1):25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vazna A, Havlovicova M, Sedlacek Z. Molecular cloning and analysis of breakpoints on ring chromosome 17 in a patient with autism. Gene. 2008;407(1–2):186–192 [DOI] [PubMed] [Google Scholar]
- 70.Pinto D, Pagnamenta AT, Klei L, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466(7304):368–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sbacchi S, Acquadro F, Calò I, Calì F, Romano V. Functional annotation of genes overlapping copy number variants in autistic patients: focus on axon pathfinding. Curr Genomics. 2010;11(2):136–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bremer A, Giacobini M, Eriksson M, et al. Copy number variation characteristics in subpopulations of patients with autism spectrum disorders. Am J Med Genet B Neuropsychiatr Genet. 2011;156(2):115–124 [DOI] [PubMed] [Google Scholar]
- 73.Iossifov I, Ronemus M, Levy D, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74(2):285–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dong S, Walker MF, Carriero NJ, et al. De novo insertions and deletions of predominantly paternal origin are associated with autism spectrum disorder. Cell Reports. 2014;9(1):16–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gregory GD, Vakoc CR, Rozovskaia T, et al. Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes. Mol Cell Biol. 2007;27(24):8466–8479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Swigut T, Wysocka J. H3K27 demethylases, at long last. Cell. 2007;131(1):29–32 [DOI] [PubMed] [Google Scholar]
- 77.Liu B, Chen Y, St Clair DK. ROS and p53: a versatile partnership. Free Radic Biol Med. 2008;44(8):1529–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lozano G. Mouse models of p53 functions. Cold Spring Harb Perspect Biol. 2010;2(4):a001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carvalho CM, Zhang F, Lupski JR. Evolution in health and medicine Sackler colloquium: genomic disorders: a window into human gene and genome evolution. Proc Natl Acad Sci U S A. 2010;107(suppl 1):1765–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]