Abstract
BACKGROUND AND OBJECTIVE:
Developmental language disorder (DLD) is a highly prevalent neurodevelopmental disorder associated with negative outcomes in different domains; the etiology of DLD is unknown. To investigate the genetic underpinnings of DLD, we performed genome-wide association and whole exome sequencing studies in a geographically isolated population with a substantially elevated prevalence of the disorder (ie, the AZ sample).
METHODS:
DNA samples were collected from 359 individuals for the genome-wide association study and from 12 severely affected individuals for whole exome sequencing. Multifaceted phenotypes, representing major domains of expressive language functioning, were derived from collected speech samples.
RESULTS:
Gene-based analyses revealed a significant association between SETBP1 and complexity of linguistic output (P = 5.47 × 10−7). The analysis of exome variants revealed coding sequence variants in 14 genes, most of which play a role in neural development. Targeted enrichment analysis implicated myocyte enhancer factor–2 (MEF2)-regulated genes in DLD in the AZ population. The main findings were successfully replicated in an independent cohort of children at risk for related disorders (n = 372).
CONCLUSIONS:
MEF2-regulated pathways were identified as potential candidate pathways in the etiology of DLD. Several genes (including the candidate SETBP1 and other MEF2-related genes) seem to jointly influence certain, but not all, facets of the DLD phenotype. Even when genetic and environmental diversity is reduced, DLD is best conceptualized as etiologically complex. Future research should establish whether the signals detected in the AZ population can be replicated in other samples and languages and provide further characterization of the identified pathway.
What’s Known on This Subject:
Genetic underpinnings of common forms of pediatric disorders of language are heavily understudied. Recent association studies identified several tentative candidate genes. However, thus far, none of these candidates has received strong support in replication or confirmation analyses.
What This Study Adds:
We established a statistically significant association between SETBP1 and language disorders in a geographically isolated population. Whole exome sequencing convergently implicated the myocyte enhancer factor–2–regulated pathways (of which SETBP1 is part) in language disorders in this special population.
Developmental language disorder (DLD) is a prevalent neurodevelopmental disorder, with 7-10% of children1,2 exhibiting atypical patterns of language development despite not having apparent sensorimotor/cognitive impairments or other medical conditions.3 DLD is lifelong,4 comorbid with other neurodevelopmental5 and psychiatric6 disorders, and associated with adverse academic7 and socio-emotional8 outcomes. It is phenotypically complex and genetically heterogeneous; although highly heritable,9 the etiology and pathogenesis of DLD are poorly understood.
A rare Mendelian type of DLD has been attributed to deleterious variants in the FOXP2 gene10–12 (7q31); however, it is not associated with the disorder’s common forms.13 For the latter, linkage studies have identified 3 susceptibility regions: 16q24, 19q13,14 and 13q2.15 Targeted association studies implicated CNTNAP216 (7q35; downregulated by FOXP2) and CMIP and ATP2C217 (16q) genes in phonological memory deficits. Four genome-wide association studies (GWAS) divulged no genome-wide significant signals,18–21 with the exception of gene-based associations for CDC2L1, CDC2L2, LOC728661, and RCAN3.19 A whole exome sequencing (WES) study of DLD in an admixed Chilean founder population suggested the involvement of a nonsynonymous single nucleotide variant (SNV) in NFXL122; however, its location is not in the previously identified linkage regions in this population.23
This pattern of findings highlights the complexity of DLD’s etiology, driven by the exclusionary nature of the diagnosis, the multicomponential nature of the phenotype, and the heterogeneity of the samples studied. The main objective of the present study was to identify genetic bases of DLD in a unique population (small, geographically secluded, and with an elevated prevalence of DLD [hereafter, the AZ population]) in which genetic and environmental variability is constrained. Genetic profiles of isolated populations are characterized by restricted genetic and allelic heterogeneity, thus rendering them ideal for studying the genetic bases of complex disorders.24
The study population resides in a remote cluster of villages in Russia’s rural north; it was founded in the 15th century by several nuclear families. Currently, the AZ population comprises ∼860 individuals (∼120 children aged 3–18 years). Of these, 74.6% are represented by a set of multigenerational family structures (6391 individuals), of whom 82% are interconnected through a single 11-generational pedigree. The environmental conditions in the population are relatively uniform: all children go to the same kindergarten and school, and the socioeconomic indicators such as parental education and income show little variation. The AZ population is relatively geographically isolated and is characterized by an atypically high prevalence of DLD25 (ie, ∼30% compared with 9% in the control rural population). This finding suggests the presence of a shared genetic component, potentially attributable to the founder effect(s).
Methods
Population and Sample
Altogether, 474 AZ individuals donated DNA. When considered in combination with first-degree relatives, 405 of these donors represented 79 nuclear and extended pedigrees (N = 1152; range, 3–474; median, 6). Of these, 359 underwent phenotyping and constituted the GWAS sample: 124 children (62 male subjects; age, 5.33–17.92 years) and 235 adults (102 male subjects; age, 18.83–83.42 years). A total of 149 were classified as affected (DLD) and 210 as typically developing individuals (Supplemental Information).
Phenotyping
Phenotyping was performed by clinical linguists using elicited semi-structured speech samples. These samples were scored by using previously described phenotyping procedures25 to produce 5 quantitative phenotypes representing the major facets of DLD: phonetic/prosodic characteristics (eg, phonological omissions, misarticulations); well-formedness (rate of grammatical/lexical errors); complex structures (frequency of complex syntactic structures); mean length of utterance in words; and semantic/pragmatic errors (rate of errors in sentence meaning). Age-adjusted z scores were computed by using data from healthy control subjects from the comparison population to determine impairment status (ie, a z score below –1). Individuals were classified as overall DLD if they met the impairment criterion for ≥2 facets. Principal component analysis revealed that the 5 phenotypes formed 2 independent components: linguistic errors (phonetic/prosodic characteristics, well-formedness, and semantic/pragmatic errors) and syntactic complexity (complex structures and mean length of utterance in words).
Single Nucleotide Polymorphism Genotyping
The DNA extracted from peripheral blood (n = 384) or saliva/buccal swabs (n = 21) underwent quality control (QC) assessment for purity and degradation after standard collection, storage, and extraction procedures recommended by the manufacturers (Qiagen N.V. [Hilden, Germany] and DNA Genotek, Inc [Ottawa, ON, Canada]), and prepared at a concentration of 50 ng/µL.
Samples were genotyped at the Yale Center for Genome Analysis using HumanCNV 370k-Duo (n = 315) or 610k-Quad (n = 90) BeadChips (Illumina, Inc, San Diego, CA). Language status and gender distributions across the plates were not statistically different from random. Allele calling was performed by using the GenCall algorithm in GenomeStudio version 2011.1.
Samples and markers underwent QC review with GenomeStudio and SNP & Variation Suite (SVS) version 7.7.8 (GoldenHelix, Inc, Bozeman, MT). Samples with call rates >95% and verified gender were retained. A total of 223 580 autosomal single nucleotide polymorphisms (SNPs) common to 2 genotyping platforms were retained after QC so that the GenCall score was >0.30, the call rate was >95%, and minor allele frequency was >1%.
Whole Exome DNA Sequencing
Four subpedigrees were chosen for WES based on the results of complex segregation analysis26 that suggested possible Mendelian transmission. From these subpedigrees, 12 severely affected individuals were selected. Nine control non-AZ individuals without DLD from the same geographical region also underwent sequencing.
Exome capture was completed by using NimbleGen EZ Exome SeqCap v2 (Roche NimbleGen, Madison, WI). One microgram of fragmented genomic DNA was used to prepare the library using the manufacturer’s protocol (Supplemental Information). The bar-coded libraries were sequenced by using Illumina’s HiSeq 2500 platform, producing 75-bp paired-end reads that were aligned to the hg19 human genome build using NovoAlign (http://www.novocraft.com; Novocraft Technologies Sn Bhd, Selangor, Malaysia). Variant calling was performed jointly for all samples by using the HaplotypeCaller algorithm in GATK.
Genetic Association Analysis
All of the quantitative trait loci association analyses were performed within the AZ sample for a set of 5 quantitative phenotypes. SNP-based association analysis of age- and gender-adjusted quantile-normalized phenotypes was performed by using mixed linear modeling (MLM) as implemented in GEMMA27 version 0.94. MLM tests for genetic association of SNPs with quantitative traits were performed under the additive model while controlling for sample structure estimated directly from data as a genetic relatedness matrix. MLM can be considered an example of the de-correlation approach to family-based data, and we chose it as our analytical framework for several reasons. First, although a number of transmission-based approaches (eg, family-based association testing, FBAT) have been developed, their use in a large complex multigenerational pedigree is problematic and computationally intensive in the presence of missing genotypic or phenotypic data, requiring splitting the larger pedigree into smaller units; this approach, coupled with conditioning on the founders’ genotypes, can lead to a loss of power. Second and most importantly, comparative studies suggest that decorrelation approaches (and MLM among them) tend to have higher (or at least comparable) statistical power than transmission approaches even in large and complex pedigrees.28
All 5 phenotypes were first used in a multivariate MLM analysis. Two multivariate MLMs were then fitted: 1 that modeled the genetic effects on the indicators of linguistic errors and the second that used syntactic complexity. We performed gene-based association analyses as implemented in KGG3 software29 version 3.0 by using the hybrid set–based test.
Copy number variant (CNV) association analysis was performed in the FBAT30 framework. Samples underwent additional CNV-specific QC (Supplemental Information). CNVs were identified by using a univariate Copy Number Analysis Method algorithm as implemented in SVS with a minimum of 5 markers per segment. Permutation testing was used to identify cut-points, and average segment intensity was used in the analyses.
Homozygosity mapping was completed by using the runs of homozygosity (ROH) detection algorithm in SVS. The minimum size was set to 250 kb and 25 SNPs, allowing for up to 1 heterozygote and 5 missing genotypes. The maximum gap between SNPs was 100 kb, and the minimum density was 18 kb. The total length of ROHs in 5 different length brackets was log-transformed to ensure normality before analysis. ROH association and burden analyses were performed by using univariate linear and logistic regression in R (R Foundation for Statistical Computing, Vienna, Austria).
WES data were analyzed by using a set of annotation and filtering tools. This analysis assumed that the most severely affected individuals in the AZ population from familial substructures with suggestive evidence for Mendelian transmission could provide additional information about the genetic architecture of DLD in the sample by focusing on: (1) the coding variants in candidate genes highlighted in the larger GWAS sample; or (2) the disruptive coding variants in other genes that could be conferring additional DLD risk in a subsample of the AZ population. Thus, we focused on coding sequence variants that were frequent among severely affected AZ probands (present in at least 4 of the 12 affected AZ individuals) but were not present in the control sample of 9 exomes. We then excluded variants observed in >5% of the National Heart, Lung, and Blood Institute Go Exome Sequencing Project (https://esp.gs.washington.edu) and the 1000 Genomes project (http://www.1000genomes.org/) Phase 1 exomes. We then retained only those variants that were located within the genes associated with any of the phenotypes in the GWAS (P < .05 for gene-level tests), disruptive frameshift variants, and variants prioritized by eXtasy31 based on the fusion of the information about their pathogenicity, haploinsufficiency predictions, and similarity to other genes linked to related phenotypes (Supplemental Information).
P values were corrected by using either standard or adjusted Bonferroni procedures (Supplemental Information). The study protocol was approved by the Yale University (New Haven, CT) and Northern State Medical University (Arkhangelsk, Russia) internal review boards.
Results
Genome-wide SNP Associations
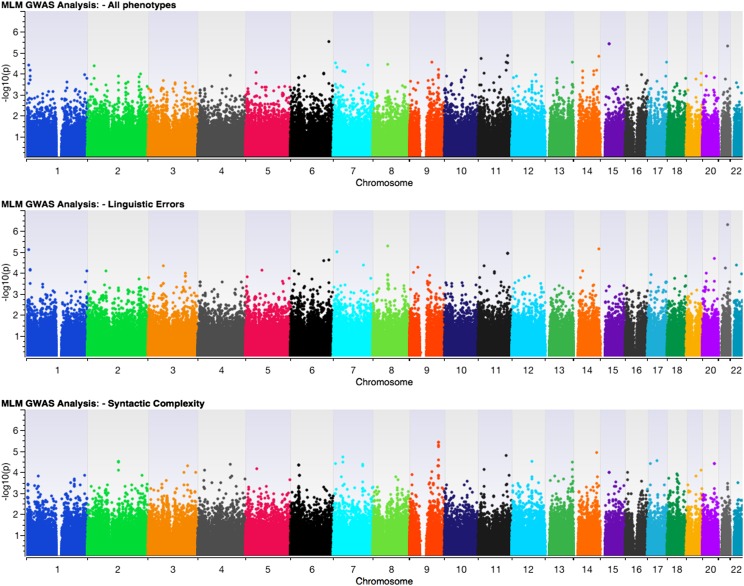
No single SNP reached genome-wide statistical significance (Fig 1). Table 1 lists the top 10 nominally significant SNPs for each analysis. For linguistic errors, the strongest association (P = 5.35 × 10−7) was for rs3787751 (21q22), located in the noncoding region of the HLCS (holocarboxylase synthetase) gene, involved in the biotinylation of apocarboxylases. Holocarboxylase synthetase deficiency syndrome (MIM#253270) is characterized by neurologic, developmental, and metabolic abnormalities in infancy.32 For syntactic complexity, the top 10 SNPs included 4 SNPs (rs378968, rs3789867, rs2480933, and rs2482078) located in intronic regions of the TNC gene on chromosome 9q33 (Fig 2). TNC codes for an extracellular matrix protein implicated in cochlear development33 and autosomal dominant deafness (MIM#615629). The univariate GWAS analyses produced similar results (Supplemental Information).
FIGURE 1.
Manhattan plots of P values for three multivariate GWAS analyses. Top row - MLM analysis of all five phenotypes; Middle row - MLM analysis of linguistic errors; Bottom row - MLM analysis of syntactic complexity.
TABLE 1.
Top 10 Nominally Significant SNP Associations for Each of the 3 Multivariate GWAS Analyses
| SNP | Phenotype | Chr | Min/Maj | MAF | PPC | WF | CS | MLU | SPE | P | Gene | Variant Type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs722208 | All | 6q25 | G/A | 0.35 | 0.38 | 0.18 | −0.17 | 0.08 | 0.22 | 3.09 × 10−6 | ESR1 | Intron |
| rs1559831 | All | 15q14 | T/C | 0.06 | −0.17 | −0.64 | 0.65 | 0.17 | −0.34 | 3.81 × 10−6 | Intergenic | |
| rs6496012 | All | 15q14 | G/A | 0.06 | −0.17 | −0.64 | 0.65 | 0.17 | −0.34 | 3.81 × 10−6 | Intergenic | |
| rs3787751 | All | 21q22 | C/T | 0.05 | −0.16 | −0.39 | 0.04 | 0.04 | −0.07 | 4.88 × 10−6 | HLCS | Intron |
| rs639373 | All | 11q23 | C/T | 0.38 | 0.30 | 0.08 | 0.19 | 0.05 | −0.10 | 1.37 × 10−5 | C2CD2L | Upstream |
| rs2180386 | All | 14q32 | A/C | 0.38 | −0.35 | −0.27 | 0.05 | −0.11 | −0.22 | 1.60 × 10−5 | MEG8 | Intron |
| rs780382 | All | 11p15 | A/G | 0.42 | −0.26 | −0.04 | 0.22 | 0.09 | −0.04 | 1.89 × 10−5 | SBF2 | Intron |
| rs585149 | All | 11q23 | T/C | 0.16 | 0.16 | 0.23 | −0.02 | 0.45 | 0.16 | 2.83 × 10−5 | SIK2, PPP2R1B | UTR′-3 |
| rs8066993 | All | 17q25 | T/C | 0.21 | −0.06 | −0.36 | 0.15 | −0.21 | 0.00 | 2.90 × 10−5 | RNF213 | Intron |
| rs2183850 | All | 13q34 | A/G | 0.48 | 0.03 | 0.04 | 0.28 | 0.21 | 0.16 | 2.98 × 10−5 | ||
| rs3789868 | Syn. comp. | 9q33 | G/A | 0.47 | — | — | 0.06 | −0.34 | — | 3.81 × 10−6 | TNC | Intron |
| rs3789867 | Syn. comp. | 9q33 | T/G | 0.47 | — | — | 0.05 | −0.35 | — | 4.02 × 10−6 | TNC | Intron |
| rs2480933 | Syn. comp. | 9q33 | A/G | 0.46 | — | — | 0.06 | −0.34 | — | 5.51 × 10−6 | TNC | Intron |
| rs2482078 | Syn. comp. | 9q33 | A/G | 0.42 | — | — | 0.05 | −0.33 | — | 6.57 × 10−6 | TNC | Intron |
| rs2011604 | Syn. comp. | 14q32 | T/C | 0.43 | — | — | 0.11 | −0.30 | — | 1.25 × 10−5 | Intergenic | |
| rs585149 | Syn. comp. | 11q23 | T/C | 0.16 | — | — | −0.01 | 0.45 | — | 1.70 × 10−5 | SIK2, PPP2R1B | |
| rs2893567 | Syn. comp. | 7p14-p13 | G/A | 0.27 | — | — | 0.23 | −0.28 | — | 2.04 × 10−5 | VPS41 | Intron |
| rs1537722 | Syn. comp. | 9q33 | T/C | 0.36 | — | — | −0.03 | 0.33 | — | 2.62 × 10−5 | ||
| rs4796604 | Syn. comp. | 17q21 | T/C | 0.44 | — | — | 0.36 | 0.04 | — | 2.96 × 10−5 | HAP1 | Missense |
| rs2699376 | Syn. comp. | 2q14 | T/C | 0.19 | — | — | 0.07 | −0.39 | — | 3.15 × 10−5 | CNTNAP5 | Intron |
| rs3787751 | Ling. err. | 21q22 | C/T | 0.45 | −0.16 | −0.39 | — | — | −0.07 | 5.35 × 10−7 | HLCS | Intron |
| rs10504229 | Ling. err. | 8q12 | A/G | 0.16 | 0.25 | 0.25 | — | — | 0.50 | 5.59 × 10−6 | LINC00588 | Intron |
| rs2180386 | Ling. err. | 14q32 | A/G | 0.38 | −0.33 | −0.27 | — | — | −0.21 | 7.37 × 10−6 | MEG8 | Intron |
| rs12121864 | Ling. err. | 1p36 | C/A | 0.09 | 0.64 | 0.14 | — | — | 0.28 | 8.00 × 10−6 | Intergenic | |
| rs10486031 | Ling. err. | 7p21 | G/A | 0.16 | 0.33 | −0.05 | — | — | −0.32 | 1.06 × 10−5 | DGKB | Intron |
| rs639373 | Ling. err. | 11q23 | C/T | 0.38 | 0.31 | 0.08 | — | — | −0.09 | 1.21 × 10−5 | C2CD2L | Upstream |
| rs643788 | Ling. err. | 11q23 | T/C | 0.41 | 0.32 | 0.11 | — | — | −0.06 | 1.22 × 10−5 | DPAGT1, H2AFX | Missense |
| rs17445063 | Ling. err. | 20q13 | G/T | 0.11 | 0.29 | 0.53 | — | — | 0.09 | 2.16 ×10−5 | Intergenic | |
| rs722208 | Ling. err. | 6q25 | G/A | 0.35 | 0.37 | 0.18 | — | — | 0.22 | 2.48 ×10−5 | ESR1 | Intron |
| rs4895970 | Ling. err. | 6q23 | T/C | 0.50 | 0.36 | 0.15 | — | — | 0.14 | 2.75 ×10−5 | Intergenic |
Coefficients for phonetic/prosodic characteristics (PPC), well-formedness (WF), complex structures (CS), mean length of utterance (in words) (MLU), and semantic/pragmatic errors (SPE) represent effect size estimates (regression coefficients in SD units) for the minor allele. Chr, chromosomal location (cytoband); Ling. err, linguistic errors; MAF, minor allele frequency; Min/Maj, minor/major allele; Syn. comp., syntactic complexity; — coefficient was not estimated in the analysis and is not available.
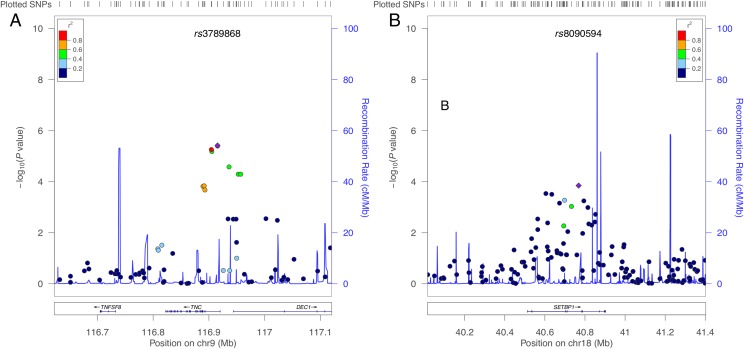
FIGURE 2.
Regional association plots for the TNC (left) and SETBP1 (right) genes and syntactic complexity phenotype. The purple diamond represents the SNP with the lowest P value in the plotted region.
Gene-based Associations
We found no genome-wide significant gene-based associations for the 5-phenotype multivariate analysis or linguistic errors. Importantly, such an association was established between SETBP1 (SET binding protein 1; 18q21) and the multivariate syntactic complexity phenotype (P = 5.47 × 10−7) (Fig 2). The nuclear protein encoded by SETBP1 binds the SET nuclear oncogene protein involved in DNA replication, apoptosis, transcription, and nucleosome assembly. Rare variants in SETBP1 are associated with Schinzel-Giedion syndrome (MIM#269150) characterized by severe developmental delays.
Table 2 presents the top 10 genes for the gene-based analyses. After SETBP1, the 2 next strongest associations with syntactic complexity were found for 2 genes on chromosome 11q23: PPP2R1B (P = 4.77 × 10−5), encoding a constant regulatory subunit of protein phosphatase 2A, and SIK2 (P = 5.00 × 10−5), a gene hypothesized to play a role in neuronal protection. These findings are likely driven by the top hit SNP rs585149 (P = 1.70 × 10−5), assigned to both genes and located in the 3′-UTR region of SIK2. We also found a nominally significant association of syntactic complexity with TNC (P = .0068).
TABLE 2.
Top 10 Gene-Based Associations for Each of the 3 Multivariate GWAS Analyses
| Phenotype | Gene | P | Chr | Length, bp |
|---|---|---|---|---|
| All | ESR1 | 4.76 × 10−5 | 6q25 | 297 602 |
| All | ABCG4 | 6.53 × 10−5 | 11q23 | 13 626 |
| All | SBF2 | 6.58 × 10−5 | 11p15 | 515 542 |
| All | PPP2R1B | 7.90 × 10−5 | 11q23 | 28 566 |
| All | SIK2 | 8.30 × 10−5 | 11q23 | 124 464 |
| All | HYOU1 | 8.44 × 10−5 | 11q23 | 13 022 |
| All | HINFP | 8.77 × 10−5 | 11q23 | 13 534 |
| All | SNORD113-9,-7,-8 | 9.05 × 10−5 | 14q32 | 72–74 |
| All | SNORD113-4,-5 | 9.06 × 10−5 | 14q32 | 75–78 |
| All | H2AFX | 9.10 × 10−5 | 11q23 | 1594 |
| Syn. comp. | SETBP1a | 5.47 × 10−7 | 18q21 | 388 337 |
| Syn. comp. | PPP2R1B | 4.77 × 10−5 | 11q23 | 28 566 |
| Syn. comp. | SIK2 | 5.00 × 10−5 | 11q23 | 124 464 |
| Syn. comp. | SERPINA1 | 7.83 × 10−5 | 14q32 | 13 947 |
| Syn. comp. | SPATA2 | 2.05 × 10−4 | 20q13 | 12 153 |
| Syn. comp. | EIF1 | 2.14 × 10−4 | 17q21 | 2773 |
| Syn. comp. | ST7-OT3 | 2.33 × 10−4 | 7q31 | 27 258 |
| Syn. comp. | RNF114 | 2.40 × 10−4 | 20q13 | 17 510 |
| Syn. comp. | HAP1 | 2.52 × 10−4 | 17q21 | 12 009 |
| Syn. comp. | GAST | 2.54 × 10−4 | 17q21 | 3645 |
| Ling. err. | LINC00588 | 3.44 × 10−5 | 8q12 | 5190 |
| Ling. err. | RP11-513O17.3 | 3.60 × 10−5 | 8q12 | 5387 |
| Ling. err. | SNORD113-9,-7,-8,-4,-5 | 4.17 × 10−5 | 14q32 | 72–78 |
| Ling. err. | HLCS | 4.40 × 10−5 | 21q22 | 239 358 |
| Ling. err. | SNORD113-3 | 4.76 × 10−5 | 14q32 | 72 |
| Ling. err. | SNORD113-6 | 4.90 × 10−5 | 14q32 | 75 |
| Ling. err. | SNORD113-2,-1 | 5.46 × 10−5 | 14q32 | 71–72 |
| Ling. err. | ABCG4 | 5.75 × 10−5 | 11q23 | 13 626 |
| Ling. err. | HYOU1 | 6.34 × 10−5 | 11q23 | 13 022 |
| Ling. err. | HINFP | 6.59 × 10−5 | 11q23 | 13 534 |
Chr, chromosomal location (cytoband); Ling. err., linguistic errors; Syn. comp., syntactic complexity.
Statistically significant after Bonferroni corrections for multiple testing.
Nominally significant associations were also established between linguistic errors and several genes (ABCG4, HYOU1, and HINFP) 7 Mb away from PPP2R1B (11q23); these genes and DPAGT1 and H2AFX were associated with the combined multivariate DLD phenotype, with the top hits being rs639373 (P = 1.21 × 10−5) and rs643788 (P = 1.22 × 10−5). The functional significance of the ABCG4 product is unknown; the DPAGT1 product is crucial for glycoprotein biosynthesis; and H2AFX encodes a histone involved in the maintenance of chromatin structure. The transcription factor encoded by HINFP plays an important role in DNA methylation. We found an association between the combined multivariate phenotype and estrogen-receptor 1 (ESR1; P = 4.76 × 10−5), with rs722208 (P = 3.09 × 10−6) as a top hit. There was a nominally significant association between linguistic errors and the HLCS gene (P = 4.40 × 10−5). Neither linguistic errors nor syntactic complexity was associated with previously identified candidate DLD genes.
CNV Analysis and Homozygosity Mapping
The multivariate FBAT CNV analysis revealed several nominally statistically significant and 1 highly statistically significant CNV. However, follow-up confirmation using real-time polymerase chain reaction (PCR) failed to substantiate the presence of these CNVs. An alternative pipeline that integrated 3 CNV detection algorithms yielded no genome-wide significant associations (Supplemental Information).
Overall, AZ-affected individuals, compared with unaffected individuals, had longer cumulative lengths of ROHs that were 250 to 500 kb long (P = .006) and 1000 to 1750 kb long (P = .004), corresponding to ∼10% and ∼1% increases in estimated autosomal homozygosity, respectively (Supplemental Fig 3). The association analysis did not reveal any ROHs that were genome-wide significantly enriched in affected individuals. None of the top 20 regions overlapped with the regions identified in the SNP analyses. Several potentially relevant identified regions are discussed in Supplemental Information.
Whole Exome DNA Sequencing
We identified 14 coding sequence variants, frequent in affected AZ individuals: 4 frameshift indels, 1 inframe insertion, 2 stop gain/loss, and 7 missense variants (Table 3). SNVs were predicted by polymorphism phenotyping (PolyPhen) to be possibly or probably damaging. Although any or all of these 14 variants could be implicated in the etiology of DLD in AZ, 2 sets of findings deserve special attention.
TABLE 3.
Prioritized Coding Variants Identified in WES Data
| ID | Type | Variant | N | Gene (GWAS Pa) | Brain Exp. | NHLBI Freq. | rsid | Protein Change | PolyPhen | GERP |
|---|---|---|---|---|---|---|---|---|---|---|
| AZW1 | Del (FS) | NC_000001.10:g.6727803_6727804delTC | 4/12 | DNAJC1 (P = .0509) | Low | 0.0000 | rs374290353 | NP_060668.2:p.Glu115fs | NA | NA |
| AZW2 | Ins (FS) | NC_000003.11:g.52559298_52559299insCAAA | 4/12 | NT5DC2 (P = .3050) | High (pn) | 0.0002 | rs745485897 | NP_001127703.1:p.Asp378fs | NA | 5.1 |
| AZW3 | Stop gain (SNV) | NC_000006.11:g.167591956C>T | 7/12 | TCP10L2 (P = .0198) | Medium | 0.0000 | rs2297462 | NP_001138593.1:p.Gln195Ter | NA | NA |
| AZW4 | Stop loss (SNV) | NC_000009.11:g.71152177A>G | 4/12 | TMEM252 (P = .0921) | Low/ medium | 0.0030 | rs147022534 | NP_694969.1:p.Ter171fs | NA | 0.4 |
| AZW5 | Ins | NC_000011.9:g.1651095insGGCTGTGGCTCCGGCTGTGGAGGCCTTGGCTCCGGCTGTGGG | 4/12 | KRTAP5-5 (P = .0163) | NA | 0.0000 | NA | NP_001001480.2:p.Gly9_Cys10insLeuTrpLeuArgLeuTrpArgProTrpLeuArgLeuTrpGly | NA | NA |
| AZW6 | Del (FS) | NC_000019.9:g.13255238delG | 4/12 | STX10 (P = .3610) | Medium (us) | 0.0018 | NA | NP_003756.1:p.Leu246fs | NA | 1.1 |
| AZW7 | Del (FS) | NC_000022.10:g.40161342_40161343delGA | 4/12 | ENTHD1 (P = .4250) | High (us) | 0.0075 | rs200712517 | NP_689725.2:p.Pro369fs | NA | 5.9 |
| AZW8 | Missense (SNV) | NC_000007.13:g.100465824G>A | 4/12 | TRIP6 (P = .4260) | Medium (us) | 0.0205 | rs2437100 | NP_003293.2:p.Arg111Gln | PrD | 4.5 |
| AZW9 | Missense (SNV) | NC_000008.10:g.91953077G>T | 4/12 | NECAB1 (P = .0246) | High (esp. temporal) | 0.0001 | rs115555424 | NP_071746.1:p.Ala271Thr | PrD | 4.4 |
| AZW10 | Missense (SNV) | NC_000009.11:g.124072992G>A | 4/12 | GSN (P = .8910) | Medium (us) | 0.0047 | rs41305623 | NP_000168.1:p.Val179Met | PrD | 5.3 |
| AZW11 | Missense (SNV) | NC_000011.9:g.3700876C>T | 4/12 | NUP98 (P = .9480) | Low/medium | 0.0164 | rs34446357 | NP_057404.2:p.Glu1661Lys | PrD | 5.6 |
| AZW12 | Missense (SNV) | NC_000011.9:g.6629343T>A | 4/12 | ILK (P = .1540) | Low | 0.0001 | rs200336608 | NP_001014794.1:p.Leu53Met | PrD | 3.9 |
| AZW13 | Missense (SNV) | NC_000017.10:g.56355397G>A | 4/12 | MPO (P = .1420) | No | 0.0135 | rs28730837 | NP_000241.1:p.Ala332Val | PoD | 4.3 |
| AZW14 | Missense (SNV) | NC_000018.9:g.25532304T>C | 6/12 | CDH2 (P = .0041) | High (pn) | 0.0266 | rs2289664 | NP_001783.2:p.Asn845Se | PoD | 5.5 |
Brain expression (Brain Exp.) information is based on the combination of the data from the BrainSpan Atlas of the Developing Human Brain (www.brainspan.org), the Human Protein Atlas (www.proteinatlas.org), and the Expression Atlas (www.ebi.ac.uk/gxa/). Del, deletion; FS, frameshift; GERP, Genomic Evolutionary Rate Profiling sequence conservation score; Ins, insertion; N, number of carriers (of 12 individuals); NA, not available; NHLBI Freq., variant frequency in National Heart, Lung, and Blood Institute European American exomes; pn, prenatally; PoD, possibly damaging; PrD, probably damaging; us, ubiquitous; rsid, Single Nucleotide Polymorphism Database submission number.
P value for the HYST gene-based analysis (multivariate, all phenotypes).
First, multiple individuals in the AZ population carried coding sequence variants in genes that regulate neural development or are highly expressed in the brain; that is, a frameshift insertion in NT5DC2 (3p21.1) and missense SNVs in NECAB1 (8q21.3) and ILK (11p15.4). NT5DC2 has been implicated in schizophrenia34 and borderline personality disorder.35 NECAB1 is a member of the neuronal calcium-binding family of proteins essential to Ca2+-mediated signaling and is highly expressed in the temporal lobe.36 The protein encoded by ILK is 1 of the key regulators of neural stem cell astrocytic differentiation37 and neurite outgrowth.38 We also found that 7 (58%) of 12 individuals in the AZ population carried a known missense variant in CDH2 (18q12) that was found only at a 2% frequency in the 1000 Genomes data set. CDH2 codes for a major cadherin that is widely expressed prenatally in neural stem cells and supports their differentiation and migration,39 regulating the laminar organization of the cortex.40 Moreover, 7 of 12 AZ individuals carried a stop-gain variant in TCP10L2 (6q27). It is unknown whether TCP10L2 codes for a functional protein; it is highly similar to TCP10L, a primate-specific transcription factor thought to evolve via segmental duplication41 from TCP10L2 or TCP10.
Second, a missense SNV in TRIP6 (7q22.1) and a frameshift deletion in ENTHD1 (22q13) indicate commonalities between the genetic pathways identified through GWAS and WES. TRIP6 is a transcription factor that has been identified as a regulator of postnatal neural stem cell maintenance in the subventricular zone.42 ENTHD1 codes for ENTH domain-containing protein 1. ENTH domain-containing proteins are involved in synaptic vesicle endocytosis at nerve terminals at the crucial stages that precede synapse formation.43 Importantly, TRIP6 interacts with and ENTHD1 is upregulated by the same family of genes, myocyte enhancer factor–2 (MEF2), labeled MEF2A-D. MEF2 are transcription factors implicated in muscle and central nervous system differentiation. In addition to ENTHD1, MEF2 targets in human neural stem cells include SETBP1, TNC, and DKGB (3 genes highlighted by our GWAS), as well as individual genes (BDNF, DMD, and NCAM2) and gene families (cadherins, contactins, semaphorins, and serpins) implicated in (a)typical central nervous system development. A targeted formal analysis of gene list enrichment using the Enrichr tool44 suggested that, combined, GWAS and WES hits in this population are indeed enriched for MEF2 targets (for MEF2A, P = 1.28 × 10−6) (Supplemental Information), providing support to this hypothesis.
Our WES analysis also revealed the presence of 2 heterozygous missense mutations in SETBP1, carried by 2 (rs3744825) and 1 (rs1064204) sequenced AZ individual, respectively. Both were common (for European ancestry, minor allele frequency >10% in National Heart, Lung, and Blood Institute exome database) known SNPs, projected to be tolerated according to 5 different functional prediction algorithms.
Replication
We interrogated the main loci highlighted in the GWAS or WES analyses of DLD in the AZ population in an independent sample (n = 372) of children at risk for developmental disorders of language (spoken and written) by using teachers’ ratings of student’s spoken and written language skills as the main phenotype (details are given in the Supplemental Information). Association analysis controlled for age and gender and was performed by using EMMAX,45 a MLM algorithm implemented in SVS.
Both main findings were replicated. First, a significant gene-based association was found between language scores and SETBP1 (P = .009360). The top signal originated at exm1383999/rs11082414 (P = .000359), a missense SNP located within exon 4 of SETBP1 that explained 3.41% of the variance in children’s language skills. Predicted to be tolerated according to sorting intolerant from tolerant/PolyPhen, this SNP may play a role in the regulation of expression of SETBP1. The analysis of the Braineac46 brain expression quantitative trait loci database suggested that it differentiates levels of SETBP1 expression in the brain, including the cerebellar cortex, hippocampus, and temporal cortex.
Second, genes nominally associated (at P < .05) with teacher ratings of students’ spoken and written language skills were enriched for MEF2A targets (P = .0007024), replicating the finding from the discovery cohort.
Discussion
We established a genome-wide association between syntactic complexity and the SETBP1 gene in the AZ sample and then replicated it in an independent sample. SETBP1 is relatively large (388 337 bp), has 2 isoforms, and is expressed widely. Although little is known about its function, it is implicated in several neurodevelopmental conditions: SETBP1 haploinsufficiency is documented in expressive DLD47–49 and intellectual disability.50 Moreover, several tentative SNP associations were found between syntactic complexity and TNC that encodes tenascin, an extracellular matrix glycoprotein involved in neural development; TNC-deficient mice exhibit structural and functional cortical abnormalities, including atypical neuronal density and abnormal dendrite morphology.51 However, the combined multivariate phenotype was also nominally associated with ESR1, a nuclear hormone receptor involved in regulation of gene expression, cell proliferation, and differentiation. Estrogen is involved in synaptogenesis, regulates neurotransmission, and modulates the activity of all types of neural cells.52 This finding is intriguing given the male bias in incidence of DLD and the recent report of associations between early postnatal gender hormone concentrations and later language development.53
Our WES highlighted 14 coding variants in a set of genes implicated in neural development and/or differentiation. Intriguingly, 2 of the WES-identified genes (ENTHD1 and TRIP6) and 3 of the GWAS-identified genes (SETBP1, TNC, and DKGB) interact with or are regulated by the MEF2 transcription factors. MEF2 isoforms are widely expressed in neural cells,54 and their activity is regulated by extracellular factors (eg, in neurons via neurotrophin stimulation or Ca2+ influx after the release of neurotransmitters). MEF2 targets show enriched expression in the central nervous system and implicate multiple signaling pathways, rendering MEF2 as a key regulator of activity-dependent synapse development.55 The complex transcriptional program of MEF2 results in the restriction of excitatory synaptic transmission via the reduction of the number of excitatory neurons, elimination of glutamatergic synapses,56 and postsynaptic differentiation of neurons (dendrite morphogenesis).57
The cascade of events regulated by the transcriptional activity of MEF2 is critical for learning and memory.58,59 A recent electrophysiological study partially attributed the DLD phenotype in the AZ population to atypicalities in the functioning of neural circuits that support attention and memory60 that were linked to syntactic complexity. It is plausible they at least partially stem from the dysregulation of common genetic pathways that orchestrate neural development.
This dysregulation can take multiple forms. Given the partial convergence of the results from the GWAS and WES, we hypothesized that the DLD phenotype in the AZ population emerged as the result of the interaction between common genetic variants that conferred background DLD susceptibility and rare variants that altered the development of language and memory circuits against that background. This extension of the threshold-dependent response model suggests that common variants in several genes (eg, SETBP1, TNC) formed the probabilistic landscape(s) of DLD vulnerability, and that coding variants in multiple different genes (eg, regulated by MEF2 such as such ENTHD1 and TRIP6 or other genes important for neural development such as CDH2 or NECAB1) conferred the critical amount of vulnerability and pushed this landscape into a critical state.
Finally, we established a higher rate of autosomal ROH burden among the affected AZ individuals compared with unaffected AZ individuals; this finding is not surprising given the isolated nature of the population and the role of ROHs in several developmental disorders.61 However, no single specific ROH was strongly associated with DLD. In addition, there was little overlap between the genetic loci identified in the GWAS analyses of the 2 multivariate phenotypes; this outcome raises an interesting hypothesis that the 2 global facets of DLD may be relatively independent at the level of their molecular neurobiology.
Our study has several limitations. First, it has a small sample size. Although it was modest for a GWAS study, however, the sample size was almost one-half of the total AZ population. Second, the unique nature of the population poses a complex issue for future research seeking to replicate these signals in other samples. Although we replicated the association finding for SETBP1 and the enrichment findings for GWAS-highlighted DLD genes for MEF2 targets in an independent sample of children at risk for a related disorder, further molecular and analytical studies in larger samples are necessary to better characterize the joint contribution of common and rare variants in the identified genes to DLD susceptibility and decipher the molecular pathways they affect.
Conclusions
This study presented a set of novel candidate genes and coding DNA sequence variants contributing to DLD phenotypes in the AZ population; the chief findings from this population have been replicated in an independent sample. Overall, the findings suggest that multiple genes (including a novel genome-wide significant candidate SETBP1) and genetic pathways (including the suggested MEF2-regulated pathway) are involved in DLD. This study underlines the complexity of the genetic architecture of DLDs and illustrates that even in populations with reduced genetic and environmental diversity, DLD is best conceptualized as a polygenic and etiologically complex disorder.
Acknowledgments
The authors thank the families who participated in the study for their cooperation and patience, and the local medical, kindergarten, and school officials of the AZ community for their help with data collection. They also thank Igor Pushkin, Anastasia Strelina, Liudmila Kniazeva, and various students, trainees, and employees from Northern State Medical University for their help with the logistics of the study; Dr Lesley Hart for her contributions to the early stages of the project; Drs Seongmin Han and Dean Palejev for their involvement at various stages of the project; Ms Mei Tan for her editorial assistance; and the late Dr Maria Babyonyshev for her contribution to the linguistic component of the study.
Glossary
- ACES
Academic Competence Evaluation Scales
- CNV
copy number variant
- DLD
developmental language disorder
- GWAS
genome-wide association study
- HWE
Hardy-Weinberg equilibrium
- MEF2
myocyte enhancer factor–2
- MLM
mixed linear modeling
- PCR
polymerase chain reaction
- PolyPhen
polymorphism phenotyping
- QC
quality control
- ROH
runs of homozygosity
- SNP
single nucleotide polymorphism
- SNV
single nucleotide variant
- SVS
SNP & Variation Suite
- WES
whole exome sequencing
Footnotes
Dr Kornilov collected the data, performed data analyses, interpreted the data, drafted the initial manuscript, and critically revised the manuscript; Dr Rakhlin collected the data, designed the scoring rubrics for phenotyping, scored the data, interpreted the data, and revised the manuscript; Dr Koposov supervised data collection on-site, collected the data, and revised the manuscript; Ms Lee and Dr Yrigollen managed DNA specimens, processed and interpreted the molecular genetic data, and drafted the initial manuscript; Drs Caglayan, Magnuson, and Mane contributed to data analyses, interpreted the data, and critically revised the manuscript; Dr Chang designed the study, developed the sampling strategy, performed data analyses, interpreted the data, drafted the initial manuscript, and critically revised the manuscript; Dr Grigorenko conceptualized and designed the study, supervised data collection, performed data analyses, interpreted the data, drafted the initial manuscript, and critically revised the manuscript; and all authors approved the final manuscript as submitted.
Grantees undertaking such projects are encouraged to express freely their professional judgment. This article, therefore, does not necessarily reflect the position or policies of the funding agencies, and no official endorsement should be inferred. The National Institutes of Health and the National Science Foundation were not involved in the study design, data collection and analysis, interpretation of findings, writing of this report, or the decision to submit the manuscript for publication.
FINANCIAL DISCLOSURE: The authors have indicated they have no financial relationships relevant to this article to disclose.
FUNDING: Supported by National Institute of Health grants R01 DC007665 (Dr Grigorenko, Principal Investigator) and P50 HD052120 (Richard Wagner, Principal Investigator), NIH Centers for Mendelian Genomics (5U54HG006504), National Science Foundation Integrative Graduate Education and Research Traineeship grant 114399 (Dr Magnuson, Principal Investigator), and grant 14.Z50.31.0027 from the Government of the Russian Federation (Dr Grigorenko, Principal Investigator). Funded by the National Institutes of Health (NIH).
POTENTIAL CONFLICT OF INTEREST: The authors have indicated they have no potential conflicts of interest to disclose.
References
- 1.Law J, Boyle J, Harris F, Harkness A, Nye C. Prevalence and natural history of primary speech and language delay: findings from a systematic review of the literature. Int J Lang Commun Disord. 2000;35(2):165–188 [DOI] [PubMed] [Google Scholar]
- 2.Tomblin JB, Records NL, Buckwalter P, Zhang X, Smith E, O’Brien M. Prevalence of specific language impairment in kindergarten children. J Speech Lang Hear Res. 1997;40(6):1245–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.American Psychiatric Association Diagnostic and Statistical Manual of Mental Disorders. 5th ed. Arlington, VA: American Psychiatric Publishing; 2013 [Google Scholar]
- 4.Clegg J, Hollis C, Mawhood L, Rutter M. Developmental language disorders—a follow-up in later adult life. Cognitive, language and psychosocial outcomes. J Child Psychol Psychiatry. 2005;46(2):128–149 [DOI] [PubMed] [Google Scholar]
- 5.McGrath LM, Hutaff-Lee C, Scott A, Boada R, Shriberg LD, Pennington BF Children with comorbid speech sound disorder and specific language impairment are at increased risk for attention-deficit/hyperactivity disorder. J Abnorm Child Psychol 2008;36(2):151–163 [DOI] [PubMed]
- 6.Im-Bolter N, Cohen NJ Language impairment and psychiatric comorbidities. Pediatr Clin North Am 2007;54(3):525–542 [DOI] [PubMed]
- 7.Snowling MJ, Adams JW, Bishop DV, Stothard SE. Educational attainments of school leavers with a preschool history of speech-language impairments. Int J Lang Commun Disord. 2001;36(2):173–183 [DOI] [PubMed] [Google Scholar]
- 8.Durkin K, Conti-Ramsden G. Young people with specific language impairment: a review of social and emotional functioning in adolescence. Child Lang Teach Ther. 2010;26(2):105–121 [Google Scholar]
- 9.Stromswold K. The heritability of language: a review and metaanalysis of twin, adoption, and linkage studies. Language. 2001;77(4):647–723 [Google Scholar]
- 10.Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413(6855):519–523 [DOI] [PubMed] [Google Scholar]
- 11.Palka C, Alfonsi M, Mohn A, et al. Mosaic 7q31 deletion involving FOXP2 gene associated with language impairment. Pediatrics 2012;129(1). Available at: www.pediatrics.org/cgi/content/full/129/1/e183 [DOI] [PubMed]
- 12.MacDermot KD, Bonora E, Sykes N, et al. Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. Am J Hum Genet. 2005;76(6):1074–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newbury DF, Bonora E, Lamb JA, et al. ; International Molecular Genetic Study of Autism Consortium . FOXP2 is not a major susceptibility gene for autism or specific language impairment. Am J Hum Genet. 2002;70(5):1318–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.SLI Consortium . A genomewide scan identifies two novel loci involved in specific language impairment. Am J Hum Genet. 2002;70(2):384–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartlett CW, Flax JF, Logue MW, et al. A major susceptibility locus for specific language impairment is located on 13q21. Am J Hum Genet. 2002;71(1):45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vernes SC, Newbury DF, Abrahams BS, et al. A functional genetic link between distinct developmental language disorders. N Engl J Med. 2008;359(22):2337–2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newbury DF, Winchester L, Addis L, et al. CMIP and ATP2C2 modulate phonological short-term memory in language impairment. Am J Hum Genet. 2009;85(2):264–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eicher JD, Powers NR, Miller LL, et al. ; Pediatric Imaging, Neurocognition, and Genetics Study . Genome-wide association study of shared components of reading disability and language impairment. Genes Brain Behav. 2013;12(8):792–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luciano M, Evans DM, Hansell NK, et al. A genome-wide association study for reading and language abilities in two population cohorts. Genes Brain Behav. 2013;12(6):645–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nudel R, Simpson NH, Baird G, et al. ; SLI Consortium . Genome-wide association analyses of child genotype effects and parent-of-origin effects in specific language impairment. Genes Brain Behav. 2014;13(4):418–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gialluisi A, Newbury DF, Wilcutt EG, et al. ; SLI Consortium . Genome-wide screening for DNA variants associated with reading and language traits. Genes Brain Behav. 2014;13(7):686–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villanueva P, Nudel R, Hoischen A, et al. ; SLI Consortium . Exome sequencing in an admixed isolated population indicates NFXL1 variants confer a risk for specific language impairment [published correction appears in PLoS Genet. 2015;11(6):e1005336]. PLoS Genet. 2015;11(3):e1004925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villanueva P, Newbury DF, Jara L, et al. Genome-wide analysis of genetic susceptibility to language impairment in an isolated Chilean population. Eur J Hum Genet 2011;19(6):687–695 [DOI] [PMC free article] [PubMed]
- 24.Shifman S, Darvasi A. The value of isolated populations. Nat Genet. 2001;28(4):309–310 [DOI] [PubMed] [Google Scholar]
- 25.Rakhlin N, Kornilov SA, Palejev D, Koposov RA, Chang JT, Grigorenko EL. The language phenotype of a small geographically isolated Russian-speaking population: implications for genetic and clinical studies of developmental language disorder. Appl Psycholinguist. 2013;34(5):971–1003 [Google Scholar]
- 26.S.A.G.E., Statistical Analysis for Genetic Epidemiology [computer program]. Version Release 6.3; 2012. Available at: http://darwin.cwru.edu. Accessed February 1, 2014
- 27.Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44(7):821–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li D, Zhou J, Thomas DC, Fardo DW Complex pedigrees in the sequencing era: to track transmission or decorrelate? Genet Epidemiol 2014;38(suppl 1):S29–S36 [DOI] [PMC free article] [PubMed]
- 29.Li MX, Kwan JS, Sham PC. HYST: a hybrid set-based test for genome-wide association studies, with application to protein-protein interaction-based association analysis. Am J Hum Genet. 2012;91(3):478–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ionita-Laza I, Perry GH, Raby BA, et al. On the analysis of copy-number variations in genome-wide association studies: a translation of the family-based association test. Genet Epidemiol. 2008;32(3):273–284 [DOI] [PubMed] [Google Scholar]
- 31.Sifrim A, Popovic D, Tranchevent LC, et al. eXtasy: variant prioritization by genomic data fusion. Nat Meth 2013;10(11):1083–1084 [DOI] [PubMed]
- 32.Suzuki Y, Yang X, Aoki Y, Kure S, Matsubara Y. Mutations in the holocarboxylase synthetase gene HLCS. Hum Mutat. 2005;26(4):285–290 [DOI] [PubMed] [Google Scholar]
- 33.Zhao Y, Zhao F, Zong L, et al. Exome sequencing and linkage analysis identified tenascin-C (TNC) as a novel causative gene in nonsyndromic hearing loss. PLoS One. 2013;8(7):e69549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ripke S, O'Dushlaine C, Chambert K, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet 2013;45(10):1150–1159 [DOI] [PMC free article] [PubMed]
- 35.Prados J, Stenz L, Courtet P, et al. Borderline personality disorder and childhood maltreatment: a genome-wide methylation analysis. Genes Brain Behav 2015;14(2):177–188 [DOI] [PubMed] [Google Scholar]
- 36.Wu H, Li D, Shan Y, et al. EFCBP1/NECAB1, a brain-specifically expressed gene with highest abundance in temporal lobe, encodes a protein containing EF-hand and antibiotic biosynthesis monooxygenase domains. DNA Seq. 2007;18(1):73–79 [DOI] [PubMed] [Google Scholar]
- 37.Pan L, North HA, Sahni V, et al. β1-Integrin and integrin linked kinase regulate astrocytic differentiation of neural stem cells. PLoS One. 2014;9(8):e104335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mills J, Digicaylioglu M, Legg AT, et al. Role of integrin-linked kinase in nerve growth factor-stimulated neurite outgrowth. J Neurosci 2003;23(5):1638–1648 [DOI] [PMC free article] [PubMed]
- 39.Zhang J, Woodhead GJ, Swaminathan SK, et al. Cortical neural precursors inhibit their own differentiation via N-cadherin maintenance of β-catenin signaling. Dev Cell 2010;18(3):472–479 [DOI] [PMC free article] [PubMed]
- 40.Kadowaki M, Nakamura S, Machon O, Krauss S, Radice GL, Takeichi M N-cadherin mediates cortical organization in the mouse brain. Dev Biol 2007;304(1):22–33 [DOI] [PubMed]
- 41.Zhong Z, Qiu J, Chen X, et al. Identification of TCP10L as primate-specific gene derived via segmental duplication and homodimerization of TCP10L through the leucine zipper motif. Mol Biol Rep 2008;35(2):171–178 [DOI] [PubMed]
- 42.Lai YJ, Li MY, Yang CY, Huang KH, Tsai JC, Wang TW. TRIP6 regulates neural stem cell maintenance in the postnatal mammalian subventricular zone. Dev Dyn. 2014;243(9):1130–1142 [DOI] [PubMed] [Google Scholar]
- 43.Ryan TA. A pre-synaptic to-do list for coupling exocytosis to endocytosis. Curr Opin Cell Biol 2006;18(4):416–421 [DOI] [PubMed]
- 44.Chen EY, Tan CM, Kou Y, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang HM, Sul JH, Service SK, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42(4):348–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramasamy A, Trabzuni D, Guelfi S, et al. ; UK Brain Expression Consortium; North American Brain Expression Consortium . Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17(10):1418–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Filges I, Shimojima K, Okamoto N, et al. Reduced expression by SETBP1 haploinsufficiency causes developmental and expressive language delay indicating a phenotype distinct from Schinzel-Giedion syndrome. J Med Genet. 2011;48(2):117–122 [DOI] [PubMed] [Google Scholar]
- 48.Marseglia G, Scordo MR, Pescucci C, et al. 372 Kb microdeletion in 18q12.3 causing SETBP1 haploinsufficiency associated with mild mental retardation and expressive speech impairment. Eur J Med Genet 2012;55(3):216–221 [DOI] [PubMed]
- 49.Bouquillon S, Andrieux J, Landais E, et al. A 5.3 Mb deletion in chromosome 18q12.3 as the smallest region of overlap in two patients with expressive speech delay. Eur J Med Genet 2011;54(2):194–197 [DOI] [PubMed]
- 50.Coe BP, Witherspoon K, Rosenfeld JA, et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet. 2014;46(10):1063–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Irintchev A, Rollenhagen A, Troncoso E, Kiss JZ, Schachner M. Structural and functional aberrations in the cerebral cortex of tenascin-C deficient mice. Cereb Cortex. 2005;15(7):950–962 [DOI] [PubMed] [Google Scholar]
- 52.Maggi A, Ciana P, Belcredito S, Vegeto E. Estrogens in the nervous system: mechanisms and nonreproductive functions. Annu Rev Physiol. 2004;66:291–313 [DOI] [PubMed] [Google Scholar]
- 53.Schaadt G, Hesse V, Friederici AD. Sex hormones in early infancy seem to predict aspects of later language development. Brain Lang. 2015;141:70–76 [DOI] [PubMed] [Google Scholar]
- 54.Lyons GE, Micales BK, Schwarz J, Martin JF, Olson EN. Expression of mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. J Neurosci. 1995;15(8):5727–5738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flavell SW, Kim TK, Gray JM, et al. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60(6):1022–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Flavell SW, Cowan CW, Kim TK, et al. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science 2006;311(5763):1008-1012 [DOI] [PubMed]
- 57.Shalizi A, Gaudillière B, Yuan Z, et al. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science 2006;311(5763):1012–1017 [DOI] [PubMed]
- 58.Barbosa AC, Kim MS, Ertunc M, et al. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proc Natl Acad Sci U S A. 2008;105(27):9391–9396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cole CJ, Mercaldo V, Restivo L, et al. MEF2 negatively regulates learning-induced structural plasticity and memory formation. Nature Neurosci 2012;15(9):1255–1264 [DOI] [PubMed]
- 60.Kornilov SA, Landi N, Rakhlin N, Fang SY, Grigorenko EL, Magnuson JS. Attentional but not pre-attentive neural measures of auditory discrimination are atypical in children with developmental language disorder. Dev Neuropsychol. 2014;39(7):543–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gamsiz E D, Viscidi E W, Frederick A M, et al. Intellectual disability is associated with increased runs of homozygosity in simplex autism. Am J Hum Genet 2013;93(1):103–109 [DOI] [PMC free article] [PubMed]