Abstract
Mutations to the RNA binding protein, fused in sarcoma (FUS) occur in ~5% of familial ALS and FUS-positive cytoplasmic inclusions are commonly observed in these patients. Altered RNA metabolism is increasingly implicated in ALS, yet it is not understood how the specificity with which FUS interacts with RNA in the cytoplasm can affect its aggregation in vivo. To further understand this, we expressed, in mice, a form of FUS (FUS ΔRRMcyt) that lacked the RNA recognition motif (RRM), thought to impart specificity to FUS-RNA interactions, and carried an ALS-associated point mutation, R522G, retaining the protein in the cytoplasm. Here we report the phenotype and results of histological assessment of the brain of transgenic mice expressing this isoform of FUS. Results demonstrated that neuronal expression of FUS ΔRRMcyt caused early lethality often preceded by severe tremor. Large FUS-positive cytoplasmic inclusions were found in many brain neurons; however, neither neuronal loss nor neuroinflammatory response was observed. In conclusion, the extensive FUS proteinopathy and severe phenotype of these mice suggests that affecting the interactions of FUS with RNA in vivo may augment its aggregation in the neuronal cytoplasm and the severity of disease processes.
Keywords: Amyotrophic lateral sclerosis, fused in sarcoma, RNA recognition motif, transgenic animals, RNA binding
Introduction
A growing body of evidence suggests that the aggregation of RNA binding proteins and disruptions to their normal functions in RNA metabolism play crucial roles in the pathways leading to the development and progression of motor neuron pathology in ALS (1). Mutations in the gene encoding the RNA binding protein, fused in sarcoma (FUS), cause ~5% of familial ALS and large cytoplasmic inclusions of FUS are frequently observed in these patients and occasionally in cases of sporadic ALS and other neurodegenerative diseases (2–8).
FUS comprises a low complexity prion-like N-terminal domain, a C-terminal nuclear localiztion signal (NLS) and several RNA binding domains including Arg-Gly-Gly repeat (RGG) domains, a zinc finger (ZF) and an RNA recognition motif (RRM) (9,10). This predominantly nuclear protein is involved in several aspects of global RNA metabolism, including roles in RNA polymerase II and III transcription (11–14), splicing (15–18) and transportation of mature transcripts (19–21). However, following the mislocalization of FUS to the cytoplasm, occurring as a direct result of ALS associated mutation in the NLS (22), post-transcriptional modification (23) or even DNA damage (24), aberrant interaction of FUS with cytoplasmic RNA may severely compromise homeostasis of vulnerable neurons.
In the cytoplasm of cultured cells, mutant FUS forms pathological ribonucleoprotein (RNP) granules, distinct from RNA transport granules, which coalesce and with further recruitment of RNA, form larger FUS aggregates (25). Interestingly, these large FUS aggregates can recruit components of stress granules (SGs) (25) – dynamic structures that function to attenuate non-essential translation as part of cytoprotective response (26). This sequestration may impair SG formation, having a detrimental effect on cell stress response. In a previous study we have shown that in transgenic mice, neuronal expression of a FUS isoform, which readily aggregates in the cytoplasm and is unable to be recruited into SGs in vitro (27), causes several signs of ALS-like pathology, including FUS-positive neuronal cytoplasmic inclusions, denervated neuromuscular junctions, degenerated motor neurons, progressive paralysis and early-onset death within two weeks after first clinical signs are noticed (28). Furthermore, as this form of FUS lacked major RNA binding domains, it provided the first in vivo evidence that aggregation alone may trigger toxicity, irrespective of the protein’s direct effect on RNA metabolism that inevitably occurs when expressing full-length forms of FUS. However, it is not yet understood how changes to the specificity with which FUS interacts with RNA can affect its aggregation and toxicity in an in vivo model system.
Although the presence of a consensus binding sequence for FUS is disputed (10,15,29), specificity of its RNA binding is thought to arise from the cooperation of the RRM domain with the less structurally complex RGG domains (10,19). Unlike C-terminus RGG and ZF domains, which were absent in our previous transgene, the RRM domain, although thought to impart specificity, is not a major domain required for FUS to bind RNA, which distinguishes it from canonical RRM domains (19,30–33).
Here we describe the production and characterization of transgenic mice expressing a cytoplasm-targeted form of human FUS lacking the RRM domain.
Materials and methods
Production of transgenic mice
A cDNA fragment encoding human FUS ΔRRMcyt mutant, which lacks amino acids 300–360 and harbours the R522G mutation, was inserted between XhoI sites of the Thy-1 promoter plasmid, 323-pTSC21k (34) as previously described (35), and used for pronuclear microinjection of B6CBAF1/J oocytes. PCR analysis with primers 5’-TCTTTGTGCAAGGCCTGGGT-3’and 5’-AGAAGCAAGACCTCTGCAGAG-3’was used for detection of the transgenic cassette in the mouse genome. Two female founders were produced; however, we were unable to establish transgenic lines due to the associated phenotype. As transgenic mice often died suddenly and unexpectedly, our analyses in many cases were limited to tissues that were of sufficient quality following post mortem collection and processing. Mice were housed in 12 h–12 h light-dark cycles with ad libitum access to food and water. All work carried out on animals was performed in accordance with the United Kingdom (Scientific Procedures) Act (1986) and European Directive 2010/63/EU.
Histology and immunohistochemistry
Tissues were fixed in paraformaldehyde, embedded in paraffin wax, and 8-μm-thick sections were cut and mounted onto poly-L-lysine coated slides (Thermo Fisher Scientific, Waltham, MA, USA) as described elsewhere (36). To assess motor neuron morphology in brainstem motor nuclei, cresyl violet staining was performed as previously described (28). Standard immunohistochemistry (IHC) was carried out as described in our previous publications (28,37).
Western blotting
Western blotting (WB) was performed as previously described (28).
Antibodies
The following commercial primary antibodies were used: FUS (mouse monoclonal, BD Biosciences, NJ, USA, #611385, IHC: 1:500; rabbit polyclonal, Proteintech, IL, USA, #11570-1-AP, IHC: 1:500; rabbit polyclonal, Bethyl Laboratories, TX, USA, #A300-302A, WB: 1:1000); Ubiquitin (mouse monoclonal, Santa Cruz Biotechnologies, #sc-8017, IHC: 1:100); NeuN (mouse monoclonal, Millipore, MA, USA, #MAB377, IHC: 1:1000); GFAP (rabbit polyclonal, Sigma-Aldrich, #G-9269, IHC: 1:500); TIAR (mouse monoclonal, BD laboratories, #610352, IHC: 1:1000); β-actin (mouse monoclonal, Sigma-Aldrich, #A5441, WB: 1:3000). Additionally, rabbit polyclonal anti-hFUS antibody (1480) and rabbit polyclonal anti-mFUS antibody (1482) (both a kind gift from D. Cleveland) specific to the N-terminus of human FUS and mouse FUS, respectively (28), were used at 1:1000 dilution for IHC and 1:4000 for WB.
Secondary anti-mouse or anti-rabbit biotinylated (Vector Laboratories, 1:1000) or Alexa Fluor-conjugated antibodies (Invitrogen, 1:1000) were used for detection.
RT-qPCR
Total RNA extraction and reverse transcription with random hexamers were carried out according to manufacturer’s (Qiagen, Hilden, Germany and Promega, WI, USA) instructions. cDNA was used for qPCR reaction (cycle parameters: 10 min at 95°C followed by 40 cycles of 15 s at 95°C and 60 s at 60°C) with GAPDH as a reference gene as previously described (35,38). Primer sequences: FUS (human & mouse), 5’-GGAACTCAGTCAACTCCCCA-3’& 5’-TACCGTAACTTCCCGAGGTG-3’; FUS (mouse only), 5’-AAGGCCTAGGCGAGAATGTTAC-3’& 5’-TGCCTCACCCTTCAACTTGC-3’; GAPDH, 5’-GTGGGTGCAGCGAACTTTAT-3’& 5’-CACTGAGCATCTCCCTCACA-3’.
Results
Expression of cytoplasm-targeted FUS ΔRRM causes a neurological phenotype and early lethality in mice
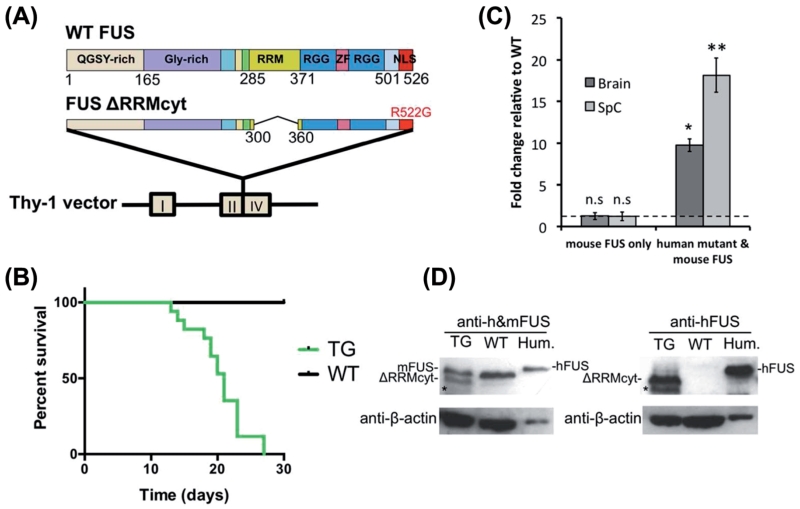
Two transgenic female founders were generated and mated with B6CBAF1/J males to produce offspring expressing a modified isoform of human FUS, ΔRRM-cyt, which lacked amino acids 300–360 and harboured the cytoplasm-targeting mutation, R522G, under the control of a Thy-1 neuronal promoter (Figure 1A). The first female founder produced five litters and the transgene displayed a Mendelian pattern of inheritance. All transgenic FUS ΔRRMcyt offspring (TG) were substantially smaller (~30% weight decrease) than wild-type (WT) littermates and exhibited very early lethality, with mice moribund at a mean age of 20 ± 1 days (n = 17) (Figure 1B). Approximately 25% died suddenly, prior to the observation of any additional phenotype(s). Interestingly, however, the remainder of these mice developed pronounced tremor on average two days prior to death, with the survival duration following tremor onset never exceeding more than five days. Tremor was constant and vigorous, affecting the whole body and was not confined to the limbs. Probably as a direct result of tremor, mice displayed a lack of balance; however, they were able to move around the cage freely and signs of limb paralysis were not observed (Supplementary Video 1, to be found online at http://informahealthcare.com/abs/doi/10.3109/21678421.2015.1040994). Because of the rapid nature of disease progression in these mice, we were unable to perform additional quantitative behavioural analyses. Although further breeding and production of TG lines was not possible as mice died prior to sexual maturation, we endeavored to characterize transgene expression and the associated pathology in this F1 generation.
Figure 1. Neuronal expression of cytoplasm-targeted FUS lacking RNA recognition motif causes early lethality in mice.
(A) Map of the DNA fragment used for pronuclear microinjection. Human FUS lacking RRM domain and targeted to the cytoplasm by the ALS associated R522G mutation was inserted between exons II and IV of the Thy-1 gene. (B) Survival plot of the F1 generation of mice used in this study which originated from the initial founder. TG mice were either found dead or were sacrificed at moribund stage (TG, n = 17; WT, n = 18). (C) The bar chart shows the mean ± S.E.M of FUS expression levels in the brain and spinal cord of moribund TG mice expressed as fold change from WT littermates (*p < 0.05, **p < 0.001, n = 3, Mann-Whitney test). The dashed line indicates the relative WT baseline of 1. No significant difference in endogenous mouse FUS expression was found between TG and WT littermates in either the brain or spinal cord (p > 0.05, n = 3, Mann-Whitney test). (D) Western blot analysis of FUS proteins expression in the total brain lysates of TG and WT mice, using antibodies against either only human FUS or both human and mouse FUS. Cell lysates from the human neuroblastoma cell line, SH-SY5Y, were included as a positive control (Hum.) for the full-length human FUS. Blots were reprobed for β-actin as a loading control. Asterisks indicate non-specific bands.
It is important to note that a second female founder gave four litters, which yielded two transgenic offspring. Again, both of these mice also died at the age of three weeks, with one developing a visually identical tremor to those from the initial founder, supporting the likelihood that the observed phenotype was a direct result of transgene expression.
RNA samples extracted from tissues of TG mice sacrificed at moribund stage in parallel with WT littermates were used to determine the level of transgene expression in the brain and spinal cord in these mice. RT-qPCR with a primer pair that detected both endogenous mouse FUS and the human ΔRRMcyt mutant FUS, showed that global FUS expression in the brain (9.8 ± 0.77-fold) and spinal cord (18.1 ± 2.07-fold) was significantly increased in TG mice compared to WT littermates (Figure 1C). As endogenous mouse FUS expression was not significantly altered from WT littermates in brain or spinal cord of TG mice, the increase in FUS RNA can be attributed directly to the expression of the transgene (Figure 1C). Furthermore, we analysed the expression of human ΔRRMcyt FUS protein in the brain of transgenic mice by Western blotting using an antibody specifically recognizing human FUS or an antibody recognizing both human and mouse proteins. The results obtained using the latter antibody demonstrated that the intensity of the band corresponding to FUS ΔRRMcyt protein is comparable to the intensity of the band for the endogenous mouse protein (Figures 1D, Supplementary Figure 1, to be found online at http://informahealthcare.com/abs/doi/10.3109/21678421.2015.1040994), suggesting that human FUS ΔRRMcyt protein is not massively overproduced in the brain of transgenic mice.
FUS ΔRRMcyt expression leads to cytoplasmic FUS-positive inclusion formation
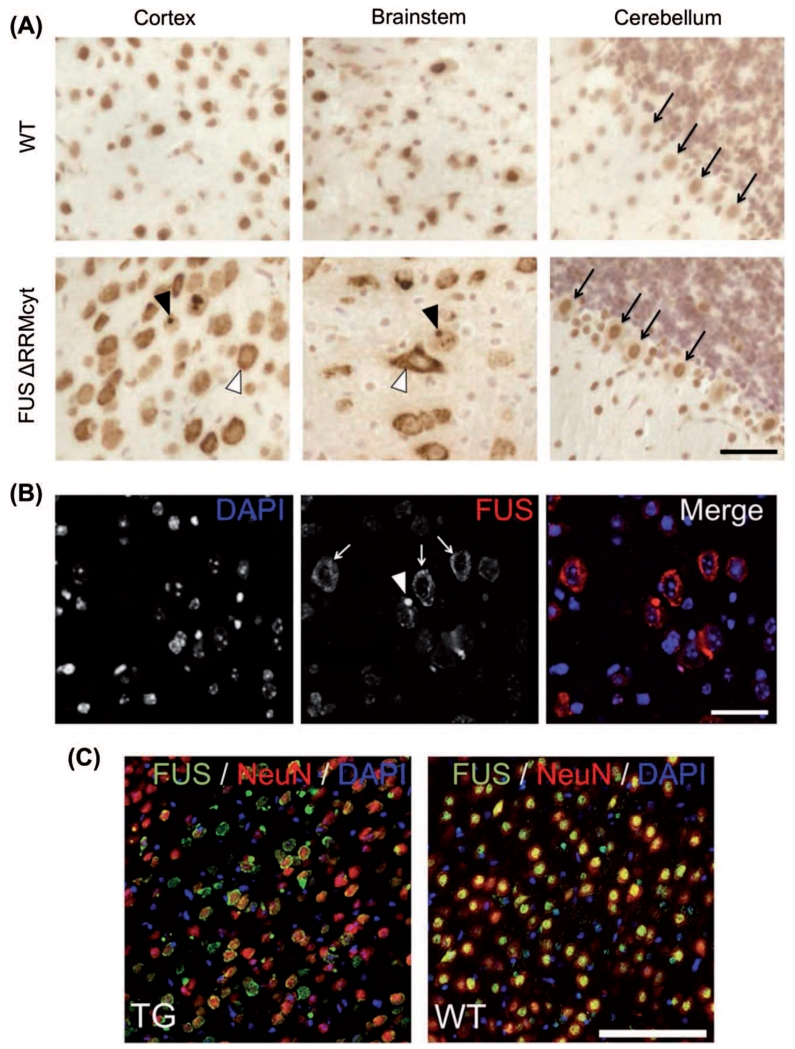
We have shown previously that FUS ΔRRMcyt, albeit N-terminally tagged with green fluorescent protein, forms pathological RNP granules and larger aggregates in the cytoplasm of transfected cultured cell lines and primary hippocampal neurons, dependent on its concentration (25). To determine whether this pattern of FUS localization was recapitulated in vivo, we performed IHC on brains taken post mortem from TG mice in conjunction with those from age-matched WT littermates. By using a monoclonal antibody that detects both endogenous mouse FUS and the human mutant FUS, prominent staining was observed in the cytoplasm of the cortex and brainstem cells of TG mice expressing the human FUS ΔRRMcyt but not in WT mice where endogenous FUS was predominantly localized to the nucleus (Figure 2A). In most instances FUS that was redistributed into the cytoplasm displayed a granule-like appearance, although frequently, round, dense inclusions, highly immunoreactive to FUS antibodies and resembling the pathological FUS inclusions seen in the neurons of ALS patients, were also observed, typically one per cell. This redistribution into the cytoplasm and pattern of FUS accumulation in TG mice was also confirmed using immunofluorescent staining of sections costained with the nuclear dye, DAPI (Figure 2B). The same pattern of FUS staining was also revealed by IHC with a different antibody, this time polyclonal, also recognizing mouse and human FUS (Figure 2C). Costaining with a neuronal marker, NeuN, demonstrated that mislocalization of FUS occurred in a large fraction of cortical neurons (Figure 2C). In cells with prominent cytoplasmic localization of FUS, the nucleus often appeared depleted of this protein (Figures 2, 3A). However, upon staining with an antibody specifically recognizing mouse FUS, we observed that the majority of cells maintained a nuclear localization of endogenous FUS, not dissimilar from WT litter mates (Supplementary Figure 2A to be found online at http://www.informahealthcare.com/doi/abs/10.3109/21678421.2015.1040994). Moreover, cytoplasmic inclusions positive for endogenous mouse FUS were rare (Supplementary Figure 2A, to be found online at http://www.informahealthcare.com/doi/abs/10.3109/21678421.2015.1040994). Taken together with the unaltered expression of endogenous FUS RNA, this suggests that endogenous FUS was affected to a minimal degree. Surprisingly, immunoreactivity of FUS in the cytoplasm of cerebellar Purkinje cells was comparatively weak (Figure 2A), although was detected more readily using an antibody highly specific to human FUS (Supplementary Figure 2B to be found online at http://www.informahealthcare.com/doi/abs/10.3109/21678421.2015.1040994). In these cells, FUS staining was predominantly nuclear and no FUS-positive inclusions or granule-like accumulations were observed (Figure 2A).
Figure 2. FUS mislocalization in cortical and brainstem neuronal populations in FUS ΔRRMcyt mice.
Mouse brain sections were immunostained with a mouse monoclonal antibody detecting both human mutant and mouse endogenous FUS proteins. (A) Cells of the cortex and brainstem in TG mice displayed prominent FUS immunoreactivity in the cytoplasm (white arrowheads). Large FUS-positive, round inclusions were also frequently observed in the cytoplasm of these cells (black arrowheads). Purkinje cells of the cerebellum (black arrows), however, displayed only very slight staining in the cytoplasm. In WT mice, the cellular localization of FUS is predominantly nuclear. (B) Immunofluorescent staining for FUS and DAPI counterstaining of cell nuclei revealed a granular pattern (white arrows) and the presence of FUS-positive inclusions (white arrowhead) in the cytoplasm of cortical cells. (C) Double immunofluorescent staining for FUS and the neuronal nuclear marker, NeuN, demonstrated that cytoplasmic mislocalization of FUS is widespread in cortical neurons of TG mice. Scale bars: A,B, 25 μm; C, 100 μm.
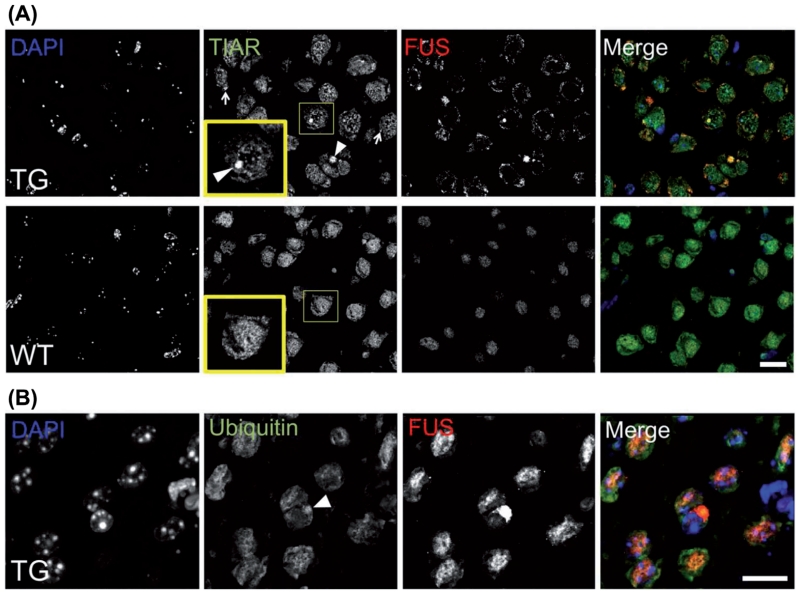
Figure 3. Characterization of FUS proteinopathy in FUS ΔRRMcyt mice.
Double immunofluorescent staining of mouse brain sections with a rabbit polyclonal antibody detecting both human mutant and mouse endogenous FUS proteins, and either mouse monoclonal anti-TIAR antibody or anti-ubiquitin antibody. (A) Large FUS-positive inclusions (arrowheads) and much smaller FUS-positive granular structures (arrows) colocalize with TIAR in cortical cells of TG mice. In contrast, in WT littermates, TIAR displays diffuse staining in the nucleus and cytoplasm. Insets show higher magnification of boxed areas. (B) Some FUS-positive inclusions (arrowhead) also displayed immunoreactivity to ubiquitin. Scale bars: 15 μm for all panels.
We have recently shown that pathological RNP granules containing FUS can recruit SG marker proteins and disrupt the cytoprotective formation of SGs (25). Therefore, we performed IHC to assess the presence or absence of SG marker proteins in the pathological FUS aggregates in these mice. The majority of large FUS-positive inclusions and smaller granule-like accumulations were positive for a SG marker, TIAR (Figure 3A). In some rare instances, large FUS-positive inclusions were also found to be ubiquitinated (Figure 3B). In addition to upper motor neurons in the cortex and brainstem motor nuclei, lower motor neurons in the spinal cord are also a key site of pathology in ALS, including FUS-ALS (39). However, all samples for histological analysis were collected post mortem and spinal cord tissues suffered from significant structural damage, preventing proper examination of FUS proteinopathy.
Signs of neurodegeneration are not apparent in brain and brainstem regions of TG mice
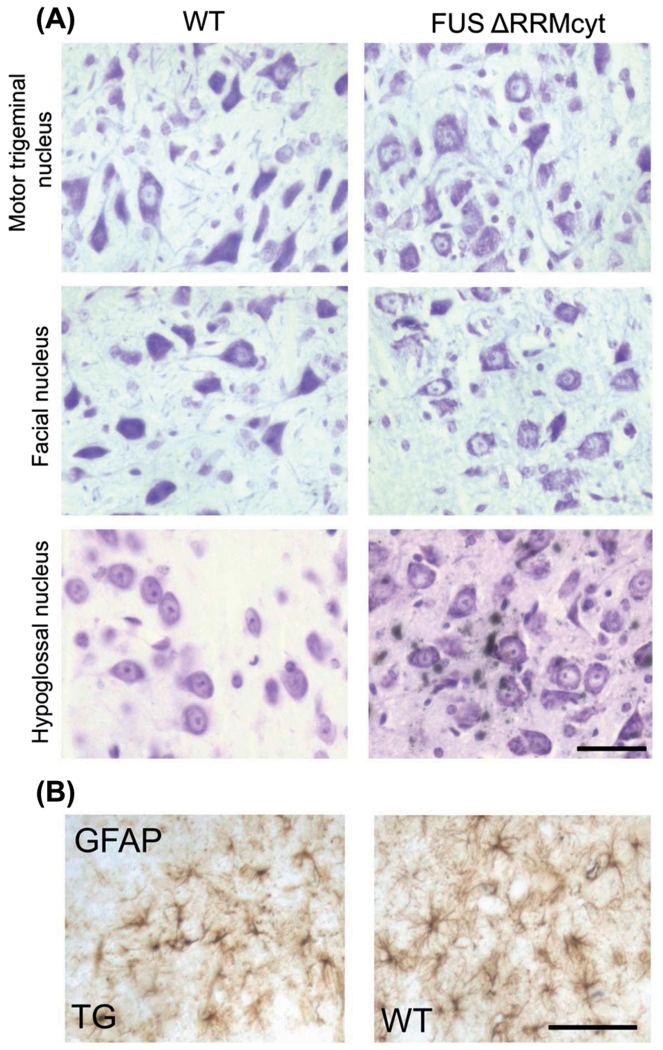
Because of an obvious tremor phenotype in all mice that survived beyond three weeks and the presence of neuronal FUS pathology in the brain of all analysed TG ΔRRMcyt animals, we sought to establish whether gross morphological changes to neuronal populations had occurred. However, in IHC experiments described above we observed no apparent loss or morphological differences of cortical neurons in TG mice compared to WT littermates (Figure 2A,C). Similarly, cerebellar Purkinje cells of TG mice were indistinguishable from those of WT littermates upon FUS IHC and cresyl violet staining (Figure 2A and data not shown). Furthermore, mice expressing FUS ΔRRMcyt did not display any obvious loss of motor neurons in motor trigeminal, facial or hypoglossal brainstem motor nuclei, all of which are commonly degenerated areas in ALS (40). Again, no changes to the morphology of motor neurons in these nuclei were apparent compared to WT controls (Figure 4A).
Figure 4. Brainstem motor nuclei of FUS ΔRRMcyt mice do not display obvious signs of neurodegeneration.
(A) Motor trigeminal, facial and hypoglossal nuclei in TG and WT mice were stained with cresyl violet to permit identification of motor neurons. No morphological differences were identified in TG mice compared with WT littermates. (B) Immunoreactivity to glial fibrillary acidic protein (GFAP) and morphology of astrocytes are indistinguishable between FUS ΔRRMcyt mice and WT littermates. Representative images shown. Scale bars: A, 25 μm; B, 50 μm.
A common feature of advanced neurodegenerative process is the presence of astro- and microgliosis in the immediate area. IHC with anti-GFAP antibody failed to reveal any indication of astrogliosis in the cortex or brainstem, namely differences in GFAP immunoreactivity or in the size or number of astrocytes were not observed between TG and WT mice (Figure 4B). Furthermore, using RCAI as a microglia marker, we were unable to detect microgliosis in these areas (data not shown).
Discussion
We have shown that the neuronal expression of FUS lacking its RRM domain and targeted to the cytoplasm causes abundant FUS pathology, a severe tremor phenotype and juvenile lethality in transgenic mice. In ALS caused by mutations of the FUS gene, localization of the predominantly nuclear protein dramatically shifts to the cytoplasm and large cytoplasmic FUS-positive inclusions are frequently observed in surviving motor neurons (3). However, FUS-positive inclusions in the cytoplasm were rarely observed following expression in transgenic animals of full-length forms of FUS, even when the protein becomes mislocalized due to the disruption of NLS function (41–43). In contrast, in transgenic FUS ΔRRMcyt mice we detected not only a dramatic mislocalization of FUS to the cytoplasm of neuronal cells in the cortex and in the brainstem but also the occurrence of large, FUS-positive cytoplasmic inclusions in many of these cells, reminiscent of those seen in patients’neurons. These results are consistent with our previously proposed model linking the absence of RNA binding of FUS to its increased aggregation propensity in the cell cytoplasm (for detail see (25)).
Cytoplasmically mislocalized FUS isoforms have been shown to recruit endogenous FUS in cultured cells and in a mouse model of FUSopathy (27,28). However, in FUS ΔRRMcyt animals, immunostaining with mouse specific antibodies showed that endogenous mouse FUS was not depleted from the nucleus and was very rarely included in human FUS-positive inclusions in the cytoplasm. It is possible that recruitment of endogenous FUS to cytoplasmic structures may represent a later stage of FUSopathy, which these mice did not reach.
Normally, FUS engages in nucleo-cytoplasmic shuttling (19) and has been identified as a constituent of RNA transport granules (21). Pertinent to this, FUS has been shown to interact directly with hundreds of mRNAs in the cytoplasm of motor neuron-like cells (44). In addition to the higher aggregation propensity of FUS ΔRRMcyt, it is feasible that the interactions of FUS with RNAs are altered in cells expressing this modified protein. As the RRM domain is proposed to impart specificity, one would expect an increase in non-specific RNA interactions for this isoform of FUS. Therefore, in addition to a deleterious effect on function or metabolism of normal RNA targets of FUS, this isoform might become engaged in interactions with, and compromise the normal functions of, novel RNA targets. Both of these mechanisms might contribute to the development of the severe early onset phenotype observed in these transgenic mice.
Despite a prominent proteinopathy, tremor and an early-onset lethality caused by expression of FUS ΔRRMcyt in the cytoplasm, we were not able to detect any signs of neurodegeneration or accompanying inflammatory glial response within either the cortex or brainstem motor nuclei that are commonly affected in ALS. In a recent murine model where wild-type human FUS was overexpressed, homozygous mice also developed a visually similar early-onset tremor, in this case leading to limb paralysis and lethality at a young, although significantly older, age than in FUS ΔRRMcyt mice. Interestingly, although FUS-positive inclusions were widespread, loss of neuronal populations and neuroinflammatory response were not found in the brain of these mice (45), which is consistent with our observations.
Although it was not possible to determine an exact mechanism by which severe pathology developed in this model, it is tempting to hypothesize that the very early death of FUS ΔRRMcyt mice before or soon after developing tremor but prior to the potential onset of any motor symptoms, reflects dramatic changes to intracellular regulatory/signalling processes that make neurons throughout the nervous system dysfunctional before any structural damage to these cells becomes evident. Such systemic dysregulation could have an insurmountable effect on bodily functions, for example by causing dysregulation of respiratory control, leading to death of the animal before changes more typical to ALS pathology occur in motor neurons.
Taken together with the previously reported evidence (28), our data further support the notion that expression of cytoplasmically mislocalized FUS with compromised RNA-binding capacity causes particularly prominent and harmful FUS pathology in the mouse nervous system. However, models allowing more precise spatial and temporal regulation of FUS isoforms with various tailored modifications are required to fully understand the relationship between pathological aggregation and compromised RNA binding of FUS in the development of neuronal dysfunction typical for FUS-ALS.
Supplementary Material
Acknowledgements
We are grateful to Don Cleveland for the kind gift of antibodies against both mouse FUS and human FUS. This work was supported by a Motor Neurone Disease Association Research Grant (Buchman/Apr13/6096) to VLB and Russian Scientific Fund No.14-14-01138 to NNN. Production of founders was partially supported by Transgenbank, IGB and the establishment of transgenic mouse colonies and phenotypical analysis–by Chernogolovka’s Resources Share Centre, State Contract 14.621.21.0008.
Footnotes
Declaration of interest: The authors declare no competing financial interests.
Supplementary material available online
Supplementary Figure 1 and 2, Video 1, to be found online at http://www.informahealthcare.com/doi/abs/10.3109/21678421.2015.1040994.
References
- 1.Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–38. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–8. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 3.Vance C, Rogelj B, Hortobagyi T, de Vos KJ, Nishimura AL, Sreedharan J, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–11. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hewitt C, Kirby J, Highley JR, Hartley JA, Hibberd R, Hollinger HC, et al. Novel FUS/TLS mutations and pathology in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol. 2010;67:455–61. doi: 10.1001/archneurol.2010.52. [DOI] [PubMed] [Google Scholar]
- 5.Munoz DG, Neumann M, Kusaka H, Yokota O, Ishihara K, Terada S, et al. FUS pathology in basophilic inclusion body disease. Acta Neuropathol. 2009;118:617–27. doi: 10.1007/s00401-009-0598-9. [DOI] [PubMed] [Google Scholar]
- 6.Neumann M, Roeber S, Kretzschmar HA, Rademakers R, Baker M, Mackenzie IR. Abundant FUS-immunoreactive pathology in neuronal intermediate filament inclusion disease. Acta Neuropathol. 2009;118:605–16. doi: 10.1007/s00401-009-0581-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doi H, Koyano S, Suzuki Y, Nukina N, Kuroiwa Y. The RNA binding protein FUS/TLS is a common aggregate-interacting protein in polyglutamine diseases. Neurosci Res. 2010;66:131–3. doi: 10.1016/j.neures.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Doi H, Okamura K, Bauer PO, Furukawa Y, Shimizu H, Kurosawa M, et al. RNA binding protein TLS is a major nuclear aggregate-interacting protein in Huntingtin exon 1 with expanded polyglutamine-expressing cells. J Biol Chem. 2008;283:6489–500. doi: 10.1074/jbc.M705306200. [DOI] [PubMed] [Google Scholar]
- 9.Iko Y, Kodama TS, Kasai N, Oyama T, Morita EH, Muto T, et al. Domain architectures and characterization of an RNA binding protein, TLS. J Biol Chem. 2004;279:44834–40. doi: 10.1074/jbc.M408552200. [DOI] [PubMed] [Google Scholar]
- 10.Lerga A, Hallier M, Delva L, Orvain C, Gallais I, Marie J, et al. Identification of an RNA binding specificity for the potential splicing factor TLS. J Biol Chem. 2001;276:6807–16. doi: 10.1074/jbc.M008304200. [DOI] [PubMed] [Google Scholar]
- 11.Tan AY, Manley JL. TLS inhibits RNA polymerase III transcription. Mol Cell Biol. 2010;30:186–96. doi: 10.1128/MCB.00884-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan AY, Riley TR, Coady T, Bussemaker HJ, Manley JL. TLS/FUS (translocated in liposarcoma/fused in sarcoma) regulates target gene transcription via single-stranded DNA response elements. Proc Natl Acad Sci U S A. 2012;109:6030–5. doi: 10.1073/pnas.1203028109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, et al. Induced ncRNAs allosterically modify RNA binding proteins in cis to inhibit transcription. Nature. 2008;454:126–30. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartz JC, Ebmeier CC, Podell ER, Heimiller J, Taatjes DJ, Cech TR. FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes Dev. 2012;26:2690–5. doi: 10.1101/gad.204602.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishigaki S, Masuda A, Fujioka Y, Iguchi Y, Katsuno M, Shibata A, et al. Position-dependent FUS-RNA interactions regulate alternative splicing events and transcriptions. Sci Rep. 2012;2:529. doi: 10.1038/srep00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakaya T, Alexiou P, Maragkakis M, Chang A, Mourelatos Z. FUS regulates genes coding for RNA binding proteins in neurons by binding to their highly conserved introns. RNA. 2013;19:498–509. doi: 10.1261/rna.037804.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orozco D, Tahirovic S, Rentzsch K, Schwenk BM, Haass C, Edbauer D. Loss of fused in sarcoma (FUS) promotes pathological tau splicing. EMBO Rep. 2012;13:759–64. doi: 10.1038/embor.2012.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Y, Liu S, Liu G, Ozturk A, Hicks GG. ALS associated FUS mutations result in compromised FUS alternative splicing and autoregulation. PLoS Genet. 2013;9:e1003895. doi: 10.1371/journal.pgen.1003895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zinszner H, Sok J, Immanuel D, Yin Y, Ron D. TLS (FUS) binds RNA in vivo and engages in nucleo-cytoplasmic shuttling. J Cell Sci. 1997;110:1741–50. doi: 10.1242/jcs.110.15.1741. [DOI] [PubMed] [Google Scholar]
- 20.Fujii R, Takumi T. TLS facilitates transport of mRNA encoding an actin-stabilizing protein to dendritic spines. J Cell Sci. 2005;118:5755–65. doi: 10.1242/jcs.02692. [DOI] [PubMed] [Google Scholar]
- 21.Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–25. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 22.Dormann D, Rodde R, Edbauer D, Bentmann E, Fischer I, Hruscha A, et al. ALS associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 2010;29:2841–57. doi: 10.1038/emboj.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dormann D, Madl T, Valori CF, Bentmann E, Tahirovic S, Abou-Ajram C, et al. Arginine methylation next to the PY-NLS modulates Transportin binding and nuclear import of FUS. EMBO J. 2012;31:4258–75. doi: 10.1038/emboj.2012.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng Q, Holler CJ, Taylor G, Hudson KF, Watkins W, Gearing M, et al. FUS is Phosphorylated by DNA-PK and Accumulates in the Cytoplasm after DNA Damage. J Neurosci. 2014;34:7802–13. doi: 10.1523/JNEUROSCI.0172-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shelkovnikova TA, Robinson HK, Southcombe JA, Ninkina N, Buchman VL. Multistep process of FUS aggregation in the cell cytoplasm involves RNA-dependent and RNA-independent mechanisms. Hum Mol Genet. 2014;23:5211–26. doi: 10.1093/hmg/ddu243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–50. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Shelkovnikova TA, Robinson H, Connor-Robson N, Buchman VL. Recruitment into stress granules prevents irreversible aggregation of FUS protein mislocalized to the cytoplasm. Cell Cycle. 2013;12:3194–202. doi: 10.4161/cc.26241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shelkovnikova TA, Peters OM, Deykin AV, Connor-Robson N, Robinson H, Ustyugov AA, et al. Fused in Sarcoma (FUS) Protein Lacking Nuclear Localization Signal (NLS) and Major RNA Binding Motifs Triggers Proteinopathy and Severe Motor Phenotype in Transgenic Mice. J Biol Chem. 2013;288:25266–74. doi: 10.1074/jbc.M113.492017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lagier-Tourenne C, Polymenidou M, Hutt KR, Vu AQ, Baughn M, Huelga SC, et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15:1488–97. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bentmann E, Neumann M, Tahirovic S, Rodde R, Dormann D, Haass C. Requirements for stress granule recruitment of fused in sarcoma (FUS) and TAR DNA-binding protein of 43 kDa (TDP-43) J Biol Chem. 2012;287:23079–92304. doi: 10.1074/jbc.M111.328757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X, Niu C, Ren J, Zhang J, Xie X, Zhu H, et al. The RRM domain of human fused in sarcoma protein reveals a non-canonical nucleic acid binding site. Biochim Biophys Acta. 2013;1832:375–85. doi: 10.1016/j.bbadis.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol. 2008;18:290–8. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 33.Liu X, Niu C, Ren J, Zhang J, Xie X, Zhu H, et al. The RRM domain of human fused in sarcoma protein reveals a non-canonical nucleic acid binding site. Biochim Biophys Acta. 2012;1832:375–85. doi: 10.1016/j.bbadis.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luthi A, van der Putten H, Botteri FM, Mansuy IM, Meins M, Frey U, et al. Endogenous serine protease inhibitor modulates epileptic activity and hippocampal long-term potentiation. J Neurosci. 1997;17:4688–99. doi: 10.1523/JNEUROSCI.17-12-04688.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ninkina N, Peters O, Millership S, Salem H, van der Putten H, Buchman VL. Gamma-synucleinopathy: neurodegeneration associated with overexpression of the mouse protein. Hum Mol Gen. 2009;18:1779–94. doi: 10.1093/hmg/ddp090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peters OM, Millership S, Shelkovnikova TA, Soto I, Keeling L, Hann A, et al. Selective pattern of motor system damage in gamma-synuclein transgenic mice mirrors the respective pathology in amyotrophic lateral sclerosis. Neurobiol Dis. 2012;48:124–31. doi: 10.1016/j.nbd.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shelkovnikova TA, Robinson HK, Troakes C, Ninkina N, Buchman VL. Compromised paraspeckle formation as a pathogenic factor in FUSopathies. Hum Mol Gen. 2014;23:2298–312. doi: 10.1093/hmg/ddt622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmittgen TD, Livak KJ. Analysing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 39.Blair IP, Williams KL, Warraich ST, Durnall JC, Thoeng AD, Manavis J, et al. FUS mutations in amyotrophic lateral sclerosis: clinical, pathological, neurophysiological and genetic analysis. J Neurol Neurosurg Psychiatry. 2010;81:639–45. doi: 10.1136/jnnp.2009.194399. [DOI] [PubMed] [Google Scholar]
- 40.DePaul R, Abbs JH, Caligiuri M, Gracco VL, Brooks BR. Hypoglossal, trigeminal, and facial motor neuron involvement in amyotrophic lateral sclerosis. Neurology. 1988;38:281–3. doi: 10.1212/wnl.38.2.281. [DOI] [PubMed] [Google Scholar]
- 41.Huang C, Zhou H, Tong J, Chen H, Liu YJ, Wang D, et al. FUS transgenic rats develop the phenotypes of amyotrophic lateral sclerosis and frontotemporal lobar degeneration. PLoS Genet. 2011;7:e1002011. doi: 10.1371/journal.pgen.1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu H, Lee S, Shang Y, Wang WY, Au KF, Kamiya S, et al. ALS associated mutation FUS-R521C causes DNA damage and RNA splicing defects. J Clin Invest. 2014;124:981–99. doi: 10.1172/JCI72723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shelkovnikova TA. Modelling FUSopathies: focus on protein aggregation. Biochem Soc Trans. 2013;41:1613–7. doi: 10.1042/BST20130212. [DOI] [PubMed] [Google Scholar]
- 44.Colombrita C, Onesto E, Megiorni F, Pizzuti A, Baralle FE, Buratti E, et al. TDP-43 and FUS RNA-binding proteins bind distinct sets of cytoplasmic messenger RNAs and differently regulate their post-transcriptional fate in motor neuron-like cells. J Biol Chem. 2012;287:15635–47. doi: 10.1074/jbc.M111.333450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitchell JC, McGoldrick P, Vance C, Hortobagyi T, Sreedharan J, Rogelj B, et al. Overexpression of human wild-type FUS causes progressive motor neuron degeneration in an age- and dose-dependent fashion. Acta Neuropathol. 2013;125:273–88. doi: 10.1007/s00401-012-1043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.