Abstract
Many intracellular pathogens cause disease by subverting macrophage innate immune defense mechanisms. Intracellular pathogens actively avoid delivery to, or directly target lysosomes, the major intracellular degradative organelle. Here, we demonstrate that AGS3 (Activator of G-protein Signaling 3), a lipopolysaccharide inducible protein in macrophages, affects both lysosomal biogenesis and activity. AGS3 binds the Gi family of G proteins via its G-protein regulatory (GPR/GoLoco) motif stabilizing the Gα subunit in its GDP-bound conformation. Elevated AGS3 levels in macrophages limited the activity of the mammalian target of rapamycin (mTOR) pathway, a sensor of cellular nutritional status. This triggered the nuclear translocation of transcription factor EB (TFEB), a known activator of lysosomal gene transcription. In contrast, AGS3 deficient macrophages had increased mTOR activity, reduced TFEB activity, and a lower lysosomal mass. High levels of AGS3 in macrophages enhanced their resistance to infection by Burkholderia cenocepacia J2315, Mycobacterium tuberculosis, and methicillin-resistant Staphylococcus aureus while AGS3 deficient macrophages were more susceptible. We conclude that LPS priming increases AGS3 levels, which enhances lysosomal function and increases the capacity of macrophages to eliminate intracellular pathogens.
Keywords: lysosomal biogenesis, intracellular pathogens, macrophages, AGS3
Pathogens especially those that survive and proliferate in intracellular niches have developed sophisticated strategies to subvert host defense systems (1). Subject to strong selective pressures, drug-resistant microorganisms have emerged that are unresponsive, or poorly responsive, to current antimicrobial agents (2). This provides an incentive to better understand host-pathogen interfaces; on the pathogen side, to unravel the mechanisms by which pathogens target and hijack host factors for their own benefit and, on the host side, to delineate how host cells marshal intracellular defense systems to combat infections. Such understanding may lead to alternative strategies to improve host defense and enhance cellular resistance (3). Lysosomes occupy a crucial position in host-pathogen interactions, both being targeted by pathogens and serving as a major mechanism for killing intracellular invaders.
Lysosomes maintain cellular homeostasis by acting as a degradative and recycling organelle in the cytosol. They recycle macromolecules reducing the energy intensive de novo synthesis of raw materials and dysfunction causes inefficient energy utilization diverting energy to synthesis pathways at the expense of energy storage depots. Lysosomes degrade substrates and metabolic waste products by fusing with endocytic, autophagic, and phagocytic vesicles (4). Both lysosomal mass and function are subject to regulation by the cellular physiologic state and in response to pathologic conditions including bacterial infections, neurodegenerative disorders, and lysosomal storage diseases (5). The coordinated upregulation of lysosomal volume and function is governed by the transcriptional factor EB (TFEB), which controls lysosomal biogenesis by enhancing the expression of the CLEAR (Coordinated Lysosomal Expression and Regulation) gene network. Among the induced genes are lysosomal hydrolases, lysosomal membrane proteins, and the components of vacuolar H+-ATPase (v-ATPase) that causes lysosomal acidification (6). Recently, TFEB was shown to be rapidly activated in murine macrophages upon S. aureus infection and required for the proper transcriptional induction of several proinflammatory cytokines and chemokines (7). Yet, the precise signaling pathways that govern TFEB activity and lysosome biogenesis in macrophages are largely unknown.
Here, we show the level of AGS3 (Activator of G protein signaling 3, also referred to as G-protein signaling modulator (Gpsm1)) in macrophages helps govern lysosomal biogenesis and function by affecting the translocation of TFEB into the nucleus through the regulation of the mTOR pathway. AGS3 was originally identified as a guanine nucleotide dissociation inhibitor (GDI) in a yeast-based functional screen of mammalian cDNA libraries for proteins that activate G-protein signaling independent of G-protein coupled receptors (8). As a GDI, AGS3 binds to the Gi/Go/transducin family of G proteins via its G-protein regulatory (GPR/GoLoco) motif and stabilizes the Gα subunit in its GDP-bound conformation. AGS3 has been implicated in a surprisingly broad array of intracellular functions including neuronal asymmetric cell divisions (9), adaptive responses to addiction and craving behavior (10–13), autophagy (14–18), membrane protein trafficking (19, 20), metabolism, cardiovascular function (21), polycystic kidney disease (22–24), and leukocyte migration (25–27). In macrophages, LPS stimulation raised AGS3 levels, which led to lysosomal enrichment and a decreased susceptibility to three different antibiotic resistant bacteria.
Materials and Methods
Mice
The animal experiments and protocols were performed according to the regulations of the National Institute of Allergy and Infectious Diseases Animal Care and Use Committee (ACUC) at the National Institutes of Health. All animals were bred and housed under pathogen-free conditions. Wild-type C57BL/6 mice were purchased from Jackson Laboratory. AGS3-deficient (Gpsm1−/−) mice have been described (21). All experiments were performed using sex and age matched animals, typically between 6 and 10 weeks old.
Antibodies and reagents
The AGS3 polyclonal antibody has been described previously and was kindly provided Dr. D. Ma (15). The commercial antibodies used as follows: LC3B antibody (L7543), β-actin-peroxidase antibody (A3854) [Sigma-Aldrich]; LAMP-1 (1D4B), LAMP-2 (H4B4), and CD-63/LAMP-3 (H5C6) antibodies [Developmental Studies Hybridoma Bank]; TGN-46 (ab16059) and LAMP-1 (ab25630) [Abcam]; cathepsin D (sc-6486) [Santa Cruz]; Akt (11E7), phospho-Akt (Ser473) (D9E), TSC2 (D93F12), phospho-TSC2 (Thr 1462) (5B12), 4E-BP1 (53H11), phospho-4E-BP1 (Thr37/46) (236B4), p70 S6 kinase (49D7), phospho-p70 S6 kinase (Thr389) (108D2) and GAPDH (14C10) [Cell Signaling]; and TFEB antibody (A303-672A) [Bethyl Laboratories]. The following reagents were used phorbol12-myristate 13-acetate (PMA) [Sigma-Aldrich]; bafilomycin A1 and glycogen synthase kinase 3 (GSK3) inhibitor VIII [Calbiochem]; G418 sulfate [Cellgro]; lipopolysaccharide (LPS) [Enzo Life Sciences]; Staphylococcus aureus (Wood strain without protein A) BioParticles, Escherichia coli (K-12 strain) BioParticles and DQ-Red BSA [Molecular Probes].
Cell isolation and culture
Bone marrow-derived macrophage (BMDM) cell cultures were prepared as described (18). Wild type (WT), MyD88-deficient and TRIF-deficient immortalized bone marrow macrophages (iBMDM) were provided by James Harris (Monash University, Melbourne, Australia) (28). The THP-1 cells (American Type Culture Collection) were maintained in RPMI-1640 medium containing 10% (vol/vol) fetal bovine serum along with 50 μM 2-mercaptoethanol. The THP-1 cells were treated with 50 nM PMA for at least 3 hours to promote their differentiation into macrophages. To generate AGS3 expressing THP-1 cell lines, a AGS3-GFP plasmid was nucleofected into THP-1 cells as previously described (29) using the human monocyte nucleofector kit (VPA-1007, Lonza). The cells were selected G418 (100 μg/ml) and later sorted on the basis of GFP expression into AGS3lo and AGS3hi cells using a FACSAria flow cytometer (Becton Dickinson). The AGS3-GFP stable HeLa cell lines were generated by transfection of the AGS3-GFP expression plasmid using Lipofectamine 2000. The cells were selected with 1000 μg/ml G418 and sorted based on GFP expression into HeLa AGS3lo and AGS3hi cell lines. The AGS3hi cells were periodically resorted to maintain high levels of GFP expression.
B. cenocepacia infection
B. cenocepacia J2315 bacteria were grown on blood agar plates at 37 °C for 3 days. Single colonies were picked and grown overnight at 37 °C in 10 ml Luria-Bertani (LB) broth while shaking. The OD of bacterial suspensions was measured at an absorbance of 600 nm and the CFU/ml calculated based on the previously established equation: CFU/ml/OD600 = 2.7X108. In addition, formalin-killed (FK) bacteria were prepared using 3.65% paraformaldehyde in PBS solution for 30 minutes and then washed with 150 mM NaCl. The infections were performed at MOI 1 into both THP-1 and BMDM cell cultures, which were seeded as 2.0×105 cells/ml density two days before. The plates were centrifuged at 1200 rpm for 5 minutes to synchronize the infection and this step corresponded to the zero time point. Next, the infected cells were incubated for an additional 1h in 5% CO2 at 37 °C. To remove the extracellular bacteria, the cells were further washed 3 times with culture medium and incubated 2h with an antibiotic cocktail (gentamicin 250 μg/ml; ceftazidime 500 μg/ml) to eradicate the remaining extracellular bacteria (30). At the end of 3- and 8-hours infections, the supernatant was aspirated and the macrophages were lysed by adding 200 μl of sterile water and shaking for 15–20 minutes at room temperature. For 24 hours infection, the cell supernatant and lysates were combined in 1:1 ratio to include the bacteria escaped from the cells at later stages of infection. The lysates were transferred to 96 well plates and serial dilutions were performed. The aliquots were plated on blood agar plates to determine the number of bacteria as CFU/ml unit.
M. tuberculosis infection
Mycobacterium tuberculosis H37Rv was grown in 7H9 medium supplemented with ADC (albumin, dextrose, catalase), glycerol and Tween-80 to mid-log phase (OD 0.6–0.8). Bacteria were washed and resuspended in PBS containing 0.05% Tween-80 prior to infection. THP-1 monocytes were maintained in RPMI 1640 supplemented with 10% heat inactivated fetal bovine serum. Cells were differentiated with 20ng/ml PMA for 20–24 hours, washed with PBS followed by addition of infection media containing 5% human serum. Bacteria were added to cells at MOI 0.5 for 4 hours at 37°C, following which extracellular bacteria were removed by PBS washes and chase medium without antibiotics was added. At indicated time points post infection, cells were lysed with 0.1% Triton X 100 in PBS and appropriate dilutions were plated on 7H11 medium in triplicate.
S. aureus infection
The clinical community-associated methicillin-resistant S. aureus isolates USA300 clone LAC and MRSA252 were grown in tryptic soy broth (TSB) at 37 °C with shaking at 180 rpm. For all experiments, 50-ml cultures were inoculated with 1/100 dilution of an overnight bacterial culture and grown for 2 hours in 125-ml baffled flasks. The bacteria were harvested and washed with sterile PBS and adjusted to the desired bacterial concentration in RPMI without phenol red and 20 mM HEPES (assay media). Bacteria were added to THP-1 cells or BMDMs, which had been seeded into 24 well plates (3.5 × 105 cells/ml) with assay media. Synchronized phagocytosis was initiated by centrifugation at 500 x g for 5 minutes. For BMDM infections, S. aureus USA300 clone LAC and MRSA252 were used at MOIs of 6:1 and 30:1, respectively. For THP-1 infections, S. aureus USA300 clone LAC and MRSA252 were used at MOIs of 2:1 and 4:1, respectively. Plates were incubated at 37 °C for 1 hour to allow phagocytosis of S. aureus. After the incubation period, THP-1 cells, but not BMDMs, were washed, to remove extracellular bacteria, and then replaced with RPMI without phenol red and 20 mM HEPES and incubated further at 37 °C. Supernatant was collected from assay wells every hour for lactate dehydrogenase (LDH) detection, which was subsequently detected using the Cytotoxicity Detection Kit (Roche). Cells, treated with 1% (v/v) Triton X-100 (Sigma), served as the 100% positive control. Images were taken using the EVOS FL microscope (Life Technologies).
siRNA knockdown
Control siRNA, AGS3 siRNA, and GFP-siRNA were purchased from Santa Cruz. HeLa cells were seeded at a density of 3.0×105 cells/well into 6-well plate one day prior to transfection. HeLa and HeLa AGS3lo/hi cells were transfected with 10–20 nM of scrambled, AGS3 and GFP siRNAs for 72 hours using Lipofectamine 2000 reagent according to manufacturer’s instructions. The knockdown results were confirmed by Western blot for each experiment.
Immunoblotting and nuclear fractionation
The immunoblot analysis of BMDM and THP-1 cell lysates were performed as described (18). HeLa cells were seeded in 6-well plate at 3.0×105 cells/well 36–40 hours prior to collection and preparation of cell lysates. Following the indicated treatments, HeLa cells were lysed with a buffer consisting of 25 mM HEPES, pH 7.4, 4% glycerol, 0.5 % (vol/vol) NP-40, 150 mM NaCl, 2mM CaCl2, protease inhibitor cocktail, and a PhosSTOP inhibitor cocktail (Roche Applied Sciences). Lysates were shaken at 4 °C for 30 minutes, transferred to Eppendorf tubes, and centrifuged at 10,000 rpm for 10 minutes at 4°C. The supernatant corresponds to cytosolic fraction. Using the same lysis buffer, the pellet was resuspended, washed and centrifuged twice at 5000 rpm for 5 minutes. After which, the pellet was resuspended with a lysis buffer including 0,5% SDS and incubated for 30 minutes at 4 °C, which corresponds to the nuclear fraction. The samples were subjected to 4–20% SDS-PAGE and analyzed by immunoblotting.
Immunocytochemistry
HeLa cells, BMDMs and PMA-differentiated THP-1 macrophages were cultured on 35 mm glass bottom dishes for 18–24 hours prior to treatments. The cells were washed with cold PBS, fixed with 4% paraformaldehyde for 20 minutes, permeabilized with 0.25% Triton-X for 5 minutes, and incubated with 4% normal goat serum for 30 minutes at room temperature. Subsequently, the fixed and blocked cells were incubated overnight at 4 °C with the indicated primary antibody, the following day washed three times, and incubated with Alexa Fluor-conjugated secondary antibodies for 1h at room temperature. The cells were imaged by confocal microscopy. For the B. cenocepacia infections, approximately 5.0×105 cells were grown on circular glass coverslips in 24-well plates. At the end of infections, the cells were fixed, permeabilized with ice-cold methanol for 5 minutes at −20 °C, and blocked with 3% BSA for 1 hour at room temperature. The same protocol was followed for primary and secondary antibody incubations as indicated above.
Intracellular calcium and flow cytometry
Cells were seeded at 5 × 104 cells per 100 μl loading medium (RPMI 1640, 10% FBS) into poly-D-lysine coated 96-well black wall, clear-bottom microtiter plates (Nalgene Nunc). An equal volume of assay loading buffer (FLIPR Calcium 4 assay kit, Molecular Devices) in Hanks’ balanced salt solution supplemented with 20 mM HEPES and 2 mM probenecid was added. Cells were incubated for 1 h at 37 °C before adding CXCL12 (100 ng/ml, R & D Systems) and then the calcium flux peak was measured using a FlexStation 3 (Molecular Devices). The data was analyzed with SOFT max Pro 5.2 (Molecular Devices). Data is shown as fluorescent counts and the y-axis labeled as Lm1. For flow cytometry the cells were immunostained with APC mouse Anti-human CD184 (clone 12G5, BD Biosciences). The background staining was performed with APC conjugated IgG2a,b (BD Biosciences). Data acquisition was done on FACSCanto II (BD) flow cytometer and the data analyzed with FlowJo software (Treestar).
Fluorescence confocal microscopy and image preparation
The images were captured using PerkinElmer Ultraview spinning-wheel confocal system mounted on a Zeiss Axiovert 200 equipped with an argon-krypton laser with 100Χ Plan-Aprochromax oil-immersion objective (numerical aperture 1.4). Volocity 4.1 (PerkinElmer) and Adobe Photoshop 7 (Adobe Systems) were used to process the images. For the co-localization experiments, approximately 100 cells were analyzed using Imaris (Bitplane Scientific Software). For each sample 8–15 1 μm Z sections were captured for analysis.
RNA isolation and quantitative RT-PCR
RNA was isolated from AGS3lo and AGS3hi THP-1 and HeLa cells using RNeasy Mini Kit (Qiagen) as per the manufacturer’s instructions. Complementary DNA (cDNA) was synthesized from 2 μg RNA using Omniscript RT Kit (Qiagen). Real-time PCR was performed in triplicate using StepOneTM Real Time PCR System (Applied Biosytems) according to Quantitect SYBR Green PCR kit (Qiagen) instructions. The following primer pairs were used:
| MAP1LC3B | F:5′-GAGAAGCAGCTTCCTGTTCTGG-3′ |
| R:5′-GTGTCCGTTCACCAACAGGAAG-3′ | |
| SQSTM1 | F:5′-GCACCCCAATGTGATCTGC-3′ |
| R:5′-CGCTACACAAGTCGTAGTCTGG-3′ | |
| ATP6V1H | F:5′-GGAAGTGTCAGATGATCCCCA-3′ |
| R:5′-CCGTTTGCCTCGTGGATAAT-3′ | |
| CTSD | F:5′-AACTGCTGGACATCGCTTGCT-3′ |
| R:5′-CATTCTTCACGTAGGTGCTGGA-3′ | |
| LAMP1 | F:5′-ACGTTACAGCGTCCAGCTCAT-3′ |
| R:5′-TCTTTGGAGCTCGCATTGG-3′ | |
| GAPDH | F:5′-TGCACCACCAACTGCTTAGC-3′ |
| R:5′-GGCATGGACTGTGGTCATGAG-3′ |
The results were normalized to the expression levels of the GAPDH mRNA and relative mRNA levels were calculated using the 2−ΔΔCt method.
Statistical analysis
Data are expressed as a mean value ± SEM. All results were confirmed in at least three independent experiments. Data were analyzed using Prism software (GraphPad) as Student’s t test or ANOVA and p< 0.05 was considered statistically significant (* p <0.05; ** p<0.005, *** p<0.001).
Results
Increased AGS3 expression reduces p62 levels without substantially affecting autophagic induction
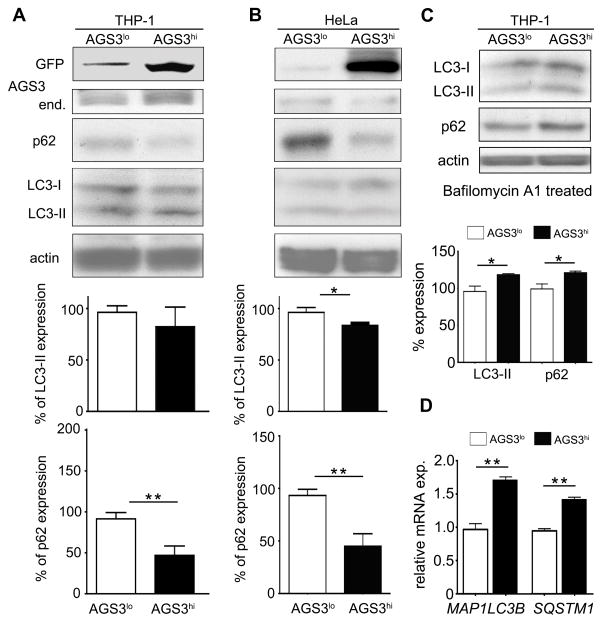
Considerable evidence implicates heterotrimeric G protein signaling and AGS3 in the regulation of autophagic processes (14, 15, 17, 31). However in primary macrophages, no essential role for AGS3 in starvation or nigericin induced autophagosome formation or autophagic flux was found (18). To reconcile this discrepancy, we generated 2 cell lines stably expressing AGS3 as a GFP fusion protein; THP-1 cells, a monocyte cell line that can be differentiated into macrophages, and HeLa cells. The cells lines were sorted into GFP high (AGS3hi) or GFP low (AGS3lo) cells. Initially, we assessed LC3-II protein levels, as a marker for autophagosome formation, and p62 levels, as a marker for long-lived protein degradation. We found similar LC3-II/actin ratios in the THP-1 AGS3lo and AGS3hi cells, and a slight reduction in the ratio in the HeLa AGS3hi compared to the AGS3lo cells (Fig. 1A and 1B). In contrast, the p62 protein levels were significantly depressed in both AGS3hi cell lines (Fig. 1A and 1B). To better interpret these results we treated the AGS3lo and AGS3hi THP-1 cell lines with Bafilomycin A1, an inhibitor of vacuolar-type H+ ATPases, which prevents the fusion between autophagosomes and lysosomes. Now when we immunoblotted for LC3 and p62, the THP-1 AGS3hi cells had elevated levels compared to AGS3lo cells (Fig. 1C). Consistent with this result, the untreated AGS3hi cells also had elevated SQSTM1 and MAP1LC3B mRNA transcripts, which encode p62 and LC3, respectively, compared to the AGS3lo cells (Fig. 1D). Together these findings suggested that the elevated AGS3 levels had predominately affected protein degradation, but not autophagy per se, perhaps by enhancing the lysosomal portion of the autophagy/lysosome degradation pathway.
Figure 1.
AGS3 stable cell lines exhibit increased proteolysis without an alteration in autophagic induction. (A,B,C) Immunoblot of cell lysates prepared from PMA-differentiated AGS3lo and AGS3hi THP-1 cells (A), or from AGS3lo and AGS3hi HeLa cells (B), or from Bafilomycin A1-treated AGS3lo and AGS3hi THP-1 cells (C) to examine the expression of the indicated proteins. The results obtained from three experiments are shown below and presented as % of the THP-1 AGS3lo (A) and HeLa AGS3lo (B). The LC3-II and p62 levels were normalized to the actin level in the corresponding lysates. (D) qRT-PCR of RNA prepared from AGS3lo and AGS3hi THP-1 cell lines for the indicated genes.
Elevated AGS3 expression enhances lysosomal mass
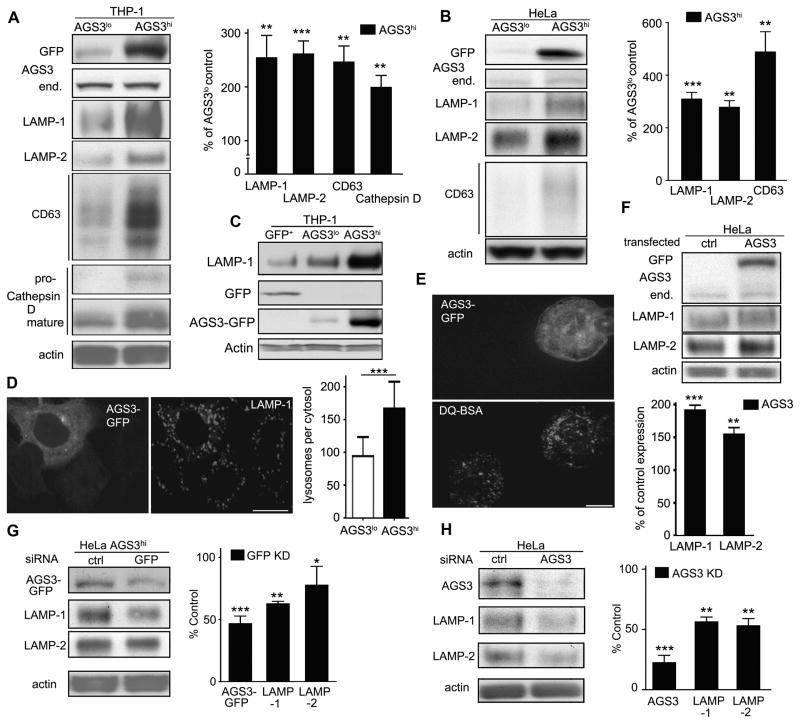
To examine the lysosome status in the stable cell lines, we assessed the cell lysates for the expression of LAMP-1, LAMP-2, CD63 (LAMP-3); integral membrane glycoproteins abundant in lysosomes, and for Cathepsin D, a lysosomal hydrolase (32). Both the THP-1 and HeLa AGS3hi cell lines possessed significantly elevated levels of these proteins compared to AGS3lo counterparts (Fig. 2A and B). To verify that the changes in LAMP-1 expression were not simply due to the overexpression of GFP we checked LAMP-1 expression in GFP-nucleofected stable THP-1 cell line. The THP-1 GFP cells had lower levels of LAMP-1 expression than did the AGS3lo cells arguing that GFP expression did not increase LAMP-1 expression (Fig. 2C). Confocal microscopy visualized immunostained LAMP-1 positive lysosomal vesicles in the HeLa (Fig. 2D) and THP-1 cell lines (data not shown). The lysosomal vesicles were larger and more extensively populated the cytosol in the AGS3hi cells. To assess lysosomal proteolytic activity, we used DQRedBSA, a substrate whose proteolysis leads to increased fluorescence. Based on this analysis the AGS3hi cells had increased lysosomal proteolytic activity (Fig. 2E). We validated the lysosomal enrichment in the permanent cell lines by transiently expressing AGS3-GFP in HeLa cells (Fig. 2F). Conversely, silencing AGS3 mRNA in the AGS3hi HeLa cells (Fig. 2G) or reducing endogenous AGS3 mRNA in HeLa cells (Fig. 2H) decreased LAMP1 expression. These data indicate that increased AGS3 expression enhances lysosomal biogenesis in both macrophages and HeLa cells.
Figure 2.
Lysosome levels are regulated by AGS3 expression. (A,B) Immunoblots of the indicated proteins in lysates from AGS3lo and AGS3hi stable THP-1 (A) and in AGS3lo and AGS3hi stable HeLa cells (B). Lysosome marker/actin band intensity ratios were normalized and the results shown as percentage of AGS3lo cells in the graphs to the right of the immunoblots. (C) Immunoblots of the indicated proteins in THP-1 stable cells expressing GFP, AGS3lo THP-1 cells, or AGS3hi THP-1 cells. (D) Confocal images of lysosomal vesicles in an AGS3lo HeLa cell (marked by asterisks) and AGS3hi (GFP positive) HeLa cell immunostained for LAMP-1 (right panel). The lysosomal mass was quantified by counting LAMP-1 positive vesicles in 50-70 cells of each cell type. The graph is shown to the right of the images. Scale bar: 10 μm. (E) Confocal images of an AGS3lo cell (** in the upper panel, visible in lower panel) and an AGS3hi THP-1 cell incubated with DQ-BSA overnight. Scale bar: 10 μm. (F) Immunoblot of indicated protein levels in cell lysates from HeLa cells transiently expressing AGS3 for 60 hours. Lysosome marker/actin band intensity ratios were normalized and shown as percentage of the control transfected cells. (G,H) Immunoblots of the indicated proteins in lysates from AGS3hi HeLa cells transfected with scrambled (ctrl) or GFP siRNAs (20nM), or HeLa cells transfected with scrambled or AGS3 siRNA (20 nM) for 72 hours. The results were normalized to actin levels and shown as a percentage of HeLa AGS3hi (G, right panel) or HeLa cells transfected with control siRNAs (H, right panel).
LPS stimulation upregulates macrophage AGS3 expression and increased AGS3 levels trigger the nuclear translocation of TFEB
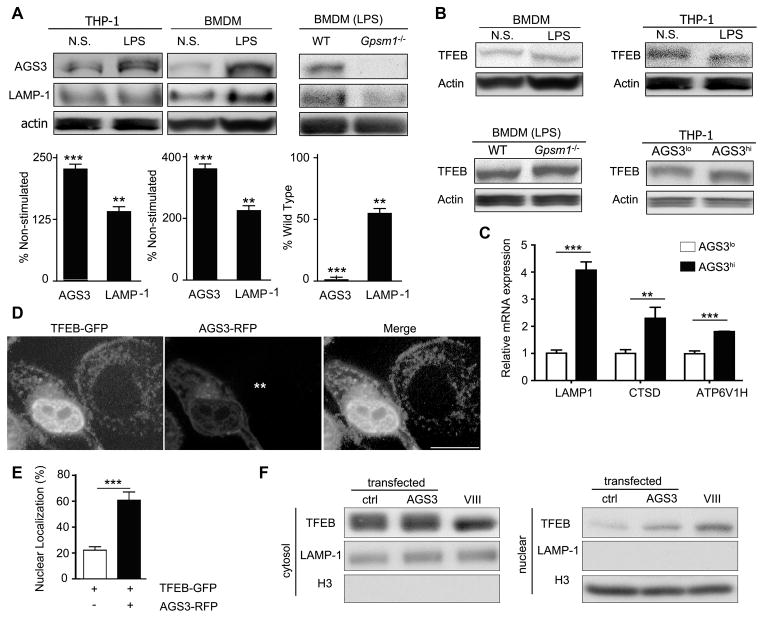
The levels of AGS3 are sensitive to a variety of stimuli in the cellular environment (25, 26). Here, we showed that lipopolysaccharide (LPS) stimulation raised AGS3 and LAMP-1 expression in both THP-1 cells and mouse bone marrow derived macrophages (BMDM, Fig. 3A). Consistent with a role for AGS3 in the induction of the LAMP-1 expression, LPS stimulated Gpsm1−/− BMDM had lower LAMP-1 levels than did WT BMDM (Fig. 3A). LPS stimulation of cardiomyocytes is known to increase LAMP1 and LAMP2 expression by triggering the nuclear translocation of TFEB (33). Increased TFEB transcriptional activity depends upon TFEB dephosphorylation, release from cytosolic 14-3-3 proteins, and nuclear translocation (34, 35). To test whether LPS affected TFEB phosphorylation, we stimulated BMDM and THP-1 cells with LPS and checked the mobility of TFEB via SDS-PAGE. LPS treatment altered TFEB mobility in a manner consistent with partial TFEB dephosphorylation (Fig. 3B, upper panels). Immunoblotting BMDM lysates from WT and AGS3 deficient mice revealed that under basal conditions TFEB migrated similarly, however following LPS treatment the mobility of TFEB shifted in WT cell lysates, but not in Gpsm1−/− cell lysates (Fig. 3B, lower left panel, and data not shown). Examination of cell lysates from the THP-1 stable cell lines revealed an increased TFEB mobility in the AGS3hi cells (Fig. 3B, lower right panel).
Figure 3.
AGS3 levels alter lysosomal biogenesis by regulating the nuclear translocation of TFEB. (A) Immunoblot of the indicated proteins in cell lysates from THP-1 cells, AGS3lo and AGS3hi stable THP-1 cells; and from BMDM derived from WT and Gpsm1−/− mice. In some instances the BMDM were stimulated with LPS overnight (100 ng/ml). Normalized AGS3 and LAMP-1 levels are shown as a percentage of non-stimulated (left and middle) or WT (right). (B) Immunoblot of TFEB and actin expression in cell lysates prepared from the indicated cells. The LPS treated cells were from overnight stimulation. (C) qRT-PCR of RNA prepared from AGS3lo and AGS3hi THP-1 cell lines for the indicated genes. (D) Confocal images of HeLa cells expressing TFEB-GFP and AGS3-RFP or only TFEB-GFP. The location of the AGS3lo cell in the middle panel is marked by asterisks. Scale bar: 10 μm. (E) Quantification of TFEB nuclear localization in HeLa cells as in part D. Localization assessed in 500 cells. (F) Immunoblots of the indicated proteins in cytosolic and nuclear fractions isolated from HeLa cells transfected with vector or AGS3 expression plasmids overnight or treated with a GSK3β inhibitor (VIII).
Assessment of steady state mRNA levels of several lysosomal genes revealed an increase in THP-1 AGS3hi cells (Fig. 3C). To demonstrate that increased AGS3 levels could shift TFEB into the nucleus, we co-expressed GFP-TFEB with AGS3-RFP, which resulted in a prominent nuclear localization of TFEB (Fig. 3D and E). These results were supported by cytosol/nuclear fractionation (Fig. 3F). The failure to observe an increase LAMP-1 in the co-transfection experiment is likely secondary to the collection of cell lysates after overnight transfection. These data show that increased levels of AGS3 in macrophages lead to the nuclear translocation of TFEB.
AGS3 basal expression and its upregulation by TLR4 engagement depends upon MyD88 and TRIF
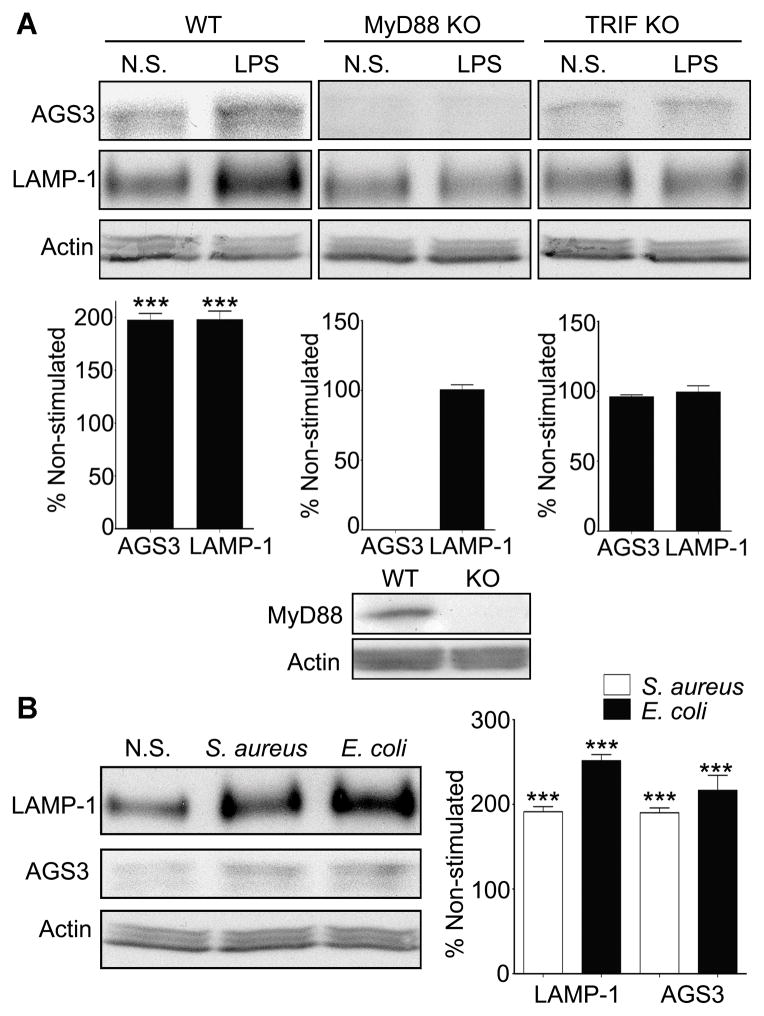
To examine the TLR4 signaling pathway that elevates AGS3 expression in macrophages we used immortalized BMDM from WT, MyD88−/− and Ticam1−/− (TRIF) deficient mice. We stimulated the cells, or not, with the TLR4 ligand LPS and immunoblotted for the expression of AGS3 and LAMP-1. The stimulated immortalized WT macrophages had higher AGS3 and LAMP-1 levels as we had previously noted with the stimulated THP-1 cells and BMDMs (Fig. 4A, left panel). Surprisingly, the MyD88 deficient cells had undetectable basal levels of AGS3, and no observable increased following LPS stimulation (Fig. 4A, middle panel). Furthermore, LPS failed to significantly upregulate LAMP-1 expression. LPS stimulation of the TRIF deficient cells also failed to raise AGS3 levels or increase LAMP-1 expression (Fig. 4A, right panel). These results suggest that MyD88 dependent signaling maintain the basal levels of AGS3 in macrophages and that TLR4 signaling through MyD88 and TRIF-dependent pathways raise AGS3 and LAMP-1 expression in macrophages.
Figure 4.
MyD88- and TRIF-dependent signaling pathways involved in AGS3 and LAMP-1 upregulation in macrophages. (A) Representative immunoblots of cell lysates prepared from WT, MyD88, and TRIF deficient immortalized bone marrow macrophages (iBMDMs) to examine AGS3 and LAMP-1 expression. The results of three separate experiments are shown below each of the immunoblots. The data is shown as a percentage increase versus the non-stimulated cells. The cells were exposed to LPS (100 ng/ml) overnight prior to cell lysis and immunoblotting. (B) Immunoblot of cell lysates from iBMDMs exposed overnight to S. aureus, E. coli bioparticles at MOI of 2.0, or not, to examine LAMP-1 and AGS3 expression. Quantification of three separate experiments is shown to the right.
We also checked whether exposure of the WT immortalized BMDMs to bacteria would raise AGS3 and LAMP-1 expression. We incubated the cells with either Staphylococcus aureus (S. aureus) or Escherichia coli (E. coli) bioparticles overnight, prepared lysates, and then immunoblotted for LAMP-1 and AGS3. The overnight exposures raised the expression of AGS3 and LAMP-1 approximately 2-fold compared to the non-exposed cells (Fig. 4B). These results suggest that macrophages may upregulate their expression of AGS3 and LAMP-1 in response to a variety of different pathogens.
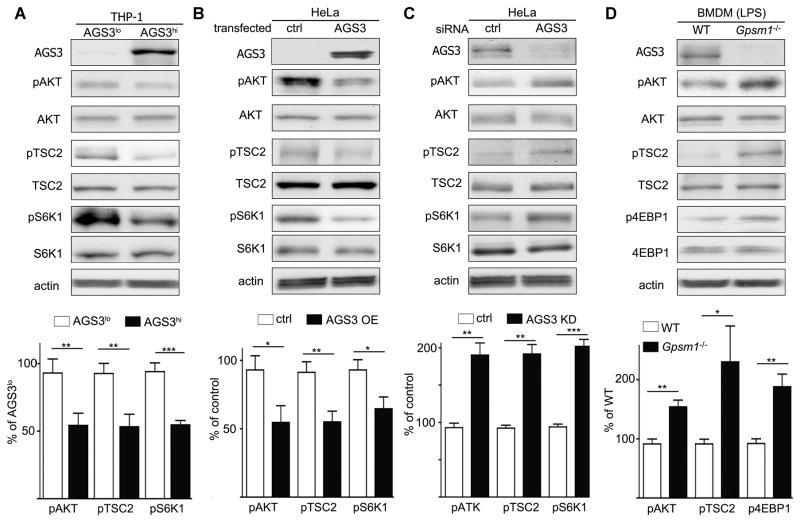
AGS3 regulates TFEB translocation through the AKT/TSC2/mTOR pathway
Previous reports have linked mTOR (mammalian target of rapamycin) activity to the nuclear translocation of TFEB (34, 35). Under basal conditions, activated mTOR on the lysosomal surface phosphorylates TFEB, preventing its nuclear translocation. Inhibition of mTOR activity reduces TFEB phosphorylation allowing it to translocate. To check mTOR activity in the cell lines, we assessed the phosphorylation status of a downstream target S6K1 (S6 kinase 1). We found a 50% reduction in pS6K1 in the AGS3hi cells (Fig. 5A). Rheb, an essential upstream protein in the mTOR pathway, is controlled by TSC1/2 (Tuberous Sclerosis Complex 1/2) (36, 37). TSC2 activity can be assessed by its phosphorylation status at threonine 1462, and we found pTSC2 levels lower in the AGS3hi cells. As the phosphatidylinositol 3-Kinase–AKT pathway is upstream of TSC2 (38), we also examined pAKT levels and found reduced pAKT levels in the AGS3hi THP-1 cells versus the AGS3lo cells (Fig. 5A). We recapitulated these results by transiently expressing AGS3 in HeLa cells (Fig. 5B). Moreover, reduced AGS3 expression in HeLa cells had the opposite phenotype (Fig. 5C). Finally, since LPS increases AGS3 expression in BMDM, we tested the AKT/TSC2/mTOR signaling axis in LPS-induced WT and Gpsm1−/− BMDM. Consistent with our previous results we noted higher levels of pAKT, pTSC2 and p4EBP1, another mTOR downstream target, in the LPS-induced Gpsm1−/− BMDM compared to WT cells (Fig. 5D). These results indicate that elevated AGS3 levels in macrophages decrease pAKT levels and dampen mTOR activity.
Figure 5.
AGS3 regulates mTOR complex via the AKT/TSC2/mTOR pathway. (A-D) Immunoblots of the indicated proteins in cell lysates from AGS3lo and AGS3hi THP-1 cells (A), control or HeLa cells transiently expressing AGS3 for 72 hours (B), HeLa cells treated with control or siRNA targeting AGS3 expression for 72 hours (C), or BMDM from WT or Gpsm1−/− mice stimulated with LPS overnight (100 ng/ml). Shown below each immunoblot is the summation of results from at least 3 experiments normalized to actin and presented as a percentage of AGS3lo, % of control, % of control, or % of WT as indicated.
Elevated level of AGS3 affects heterotrimeric G-protein signaling in macrophages
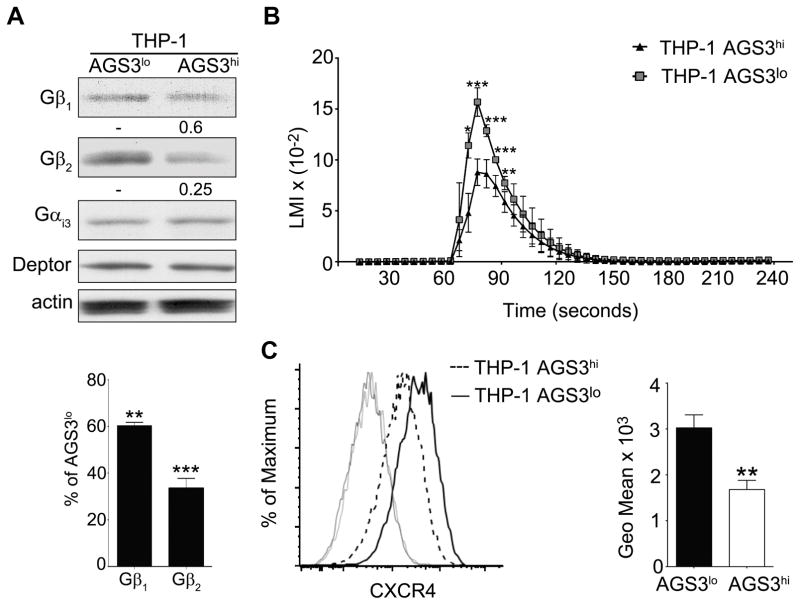
While a wide variety of upstream signals affect the PI3K–AKT pathway, a major upstream input is G-protein coupled receptor signaling. However, the role of AGS3 in regulating heterotrimeric G-protein signaling remains controversial. As AGS3 is a GDI, increased AGS3 levels compete with Gβγ for GDP-Gαi binding. When GDP-Gαi is sequestered by AGS3 docking, Gβγ remains free to signal or to be degraded. The increased expression of the AGS3 related protein LGN resulted in the ubiquitination and degradation Gβγ (39). Consistent with this scenario Gβ1 and Gβ2 levels were lower in the THP-1 AGS3hi cells (Fig. 6A). To determine whether elevated AGS3 level impacted Gαi signaling, we examined chemokine induced increases in intracellular calcium, which depend upon Gβγ release from Gαi. The THP-1 AGS3hi cells responded less well to the chemokine CXCL12 than did the AGS3lo cells (Fig. 6B). In addition, the THP-1 AGS3hi cells expressed less CXCR4, the cognate receptor for CXC12, possibility because of enhanced lysosomal degradation (Fig. 6C). In agreement with our results, overexpression of AGS3 in RBL-2H3-CXCR2 cells reduced chemokine induced increases in intracellular calcium and an AGS3 knockdown enhanced CXCR2 expression (27). Together, these data suggest that elevated levels of AGS3 in macrophages lowers pAKT levels, which leads to decreased mTOR activity, the translocation of TFEB into the nucleus, and enhanced lysosome biogenesis.
Figure 6.
High levels of AGS3 reduce Gβ expression and inhibit chemokine receptor signaling. (A) Immunoblot of cell lysates prepared from AGS3lo and AGS3hi THP-1 cells for the expression of Gαi and Gβ. The % reduction in Gβ1 and Gβ2 expression compared to the AGS3lo cells are shown below the immunoblot. (B) Trace of the intracellular calcium levels following exposure of AGS3hi and AGS3lo cells to different concentrations of CXCL12. (C) Flow cytometry to examine CXCR4 expression on AGS3lo and AGS3hi THP-1 cells. The geometric means are shown to the right of the flow cytometry plot.
AGS3-induced lysosomal enrichment mediates resistance against antibiotic resistant bacterial strains
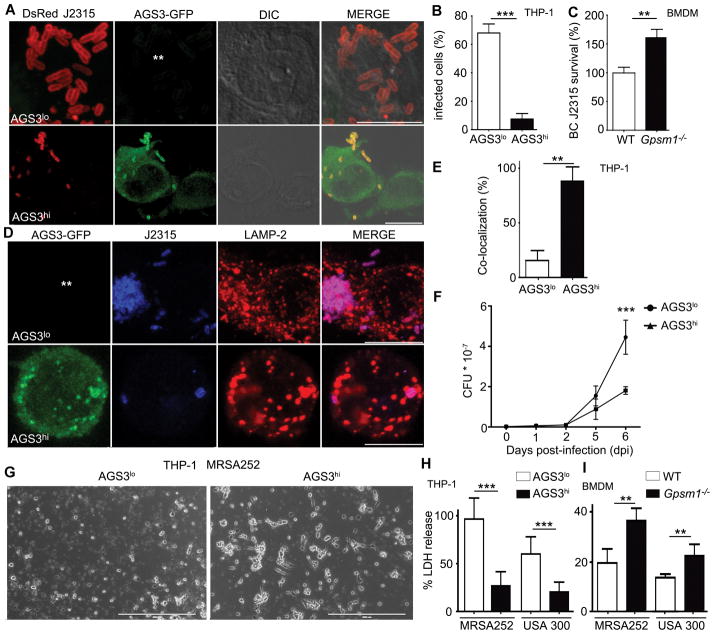
To assess whether AGS3 induced lysosomal enrichment offered a selective advantage against invasive pathogens, we challenged macrophages expressing different levels of AGS3 with three different bacteria. Burkholderia cenocepacia J2315 is a gram-negative bacterium that causes life threatening infections in cystic fibrosis patients (40). The intracellular replication kinetics in THP-1 cells is known (41). Analysis of the cells infected (MOI 1) with a DsRed- B. cenocepacia strain J2315 revealed that 24h post infection bacterial counts/cell exceeded 15 in 68% of AGS3lo cells, while only 11% of AGS3hi cells harbored similar numbers (Fig. 7A and B). The percentage of initially infected cells was similar (data not shown). Conversely, 24 hours after infection Gpsm1−/− BMDM had a 60% higher bacterial burden than did WT BMDM (Fig. 7C). Next, we infected the AGS3lo and AGS3hi cells and immunostained for the late endosomal/lysosomal protein LAMP-2. At 8 hours post infection only 15% of the bacteria co-localized with LAMP-2 in the AGS3lo cells while 88% co-localized in the AGS3hi cells (Fig. 7D and E). This argues that fewer bacteria had escaped the endo-lysosomal pathway in the AGS3hi cells. The second bacterium, we tested was Mycobacterium tuberculosis, which causes in excess 1.0 million deaths/year worldwide (42). We infected the THP-1 cell lines. While we noted little difference in initial CFU counts by 6 days post infection a marked decrease had occurred in the THP-1 AGS3hi cells indicating that the AGS3hi cells had better controlled the infection (Fig. 7F).
Figure 7.
AGS3-induced lysosomal enrichment mediates resistance against antibiotic resistant bacteria. (A) Confocal microscopy images of THP-1 AGS3lo and AGS3hi cells infected with DsRed-tagged B. cenocepacia J2315 strain at MOI of 1.0 for 24 hours. The location of the AGS3lo cell is indicated with asterisks in the GFP panel. Scale bars: 10 μm. (B) The percentage of THP-1 AGS3lo and AGS3hi cells harboring ≥15 bacteria per cytosol 24 hours post-infection. (C) Quantification of CFUs from WT and Gpsm1−/− BMDM infected with B. cenocepacia J2315 at an MOI 1.0 at 24 hours. Results were normalized to WT cells. The results obtained from three separate infection experiments are shown for each specific bacteria. (D) Confocal microscopy images of THP-1 AGS3lo and AGS3hi cells infected with B. cenocepacia J2315 at an MOI of 1.0 at 24 hours post-infection. The cells were immunostained with B. cenocepacia and LAMP-2 specific antibodies. The location of the AGS3lo cell is indicated with asterisks in the GFP panel. Scale bars: 10 μm. (E) Quantification of the co-localization rate between B. cenocepacia J2315 and LAMP-2 (F) Kinetic analysis of CFU counts upon Mycobacterium tuberculosis infection. THP-1 AGS3lo and AGS3hi cells were infected with M. tuberculosis at MOI 0.5 for 4 hours. The cells were lysed at various time points post infection and the lysates were plated on 7H11 media for CFU counts. The results are from three independent experiments performed in triplicate. (G) Brightfield microscopy images of THP-1 AGS3lo and AGS3hi cells infected with MRSA252 at MOI of 4:1 for 6 hours. Scale bar: 400 μm. (H,I) Quantification of LDH release from THP-1 AGS3lo and AGS3hi cells (H) and from WT and Gpsm1−/− BMDM (I) 6 hours post-infection with MRSA252 or USA300 clone LAC at MOIs of 6:1 and 30:1, respectively. Control cells were incubated with a 2% Triton-X solution to determine 100% LDH release. The amounts of LDH release following bacterial infections are shown as a percentage of the Triton-X triggered release. The results obtained from three separate infection experiments are shown for each specific bacteria.
Finally, we assessed whether AGS3 expression affected the course of a methicillin-resistant Staphylococcus aureus (MRSA) infection, a major cause of hospital-acquired infections (43). Representative strains of community-associated (CA)-MRSA (USA300 clone LAC) and hospital-acquired (HA)-MRSA 252 were used. While S. aureus produces an arsenal of virulence factors that assist in the escape from phagolysosomes, leading to host cell death (44), upon infection with MRSA252 the AGS3hi cells exhibited greater resistance displaying fewer signs of cell lysis and less cell death (Fig. 7G). In addition, USA300 clone LAC and MRSA252-infected AGS3hi cells released significantly lower levels of lactate dehydrogenase (LDH) than did the AGS3lo cells (Fig. 7H). Conversely, we found that Gpsm1−/− BMDM released more LDH than did WT BMDM incubated with both MRSA strains (Fig. 7I). Together these data indicate that elevated AGS3 levels help macrophages eliminate dangerous intracellular bacteria.
Discussion
Lysosomes degrade and recycle transported cellular components and internalized material by fusing with autophagosomes, phagosomes, and late endosomes. The resulting lysosomal breakdown products are used to generate new macromolecules and to provide energy in response to the nutritional needs of the cell. Recently, lysosomes functions have been expanded to include roles in nutrient sensing and energy metabolism (4). In this study, we report that the exposure of macrophages to LPS or heat killed bacteria raises AGS3 levels. The increases in AGS3 reduced signaling through the mTOR pathway. However, in a nutrient replete state this did not lead to a substantive increase in autophagy, but it did facilitate the nuclear translocation of TFEB, which transcriptionally activates many of the genes involved in lysosomal biogenesis and function. The subsequent increase in lysosomal biogenesis/function helped macrophages resist intracellular infections by several different strains of antibiotic resistant bacteria.
While AGS3 did not materially affect macrophage autophagosome formation (18), previous studies have documented increased intracellular proteolysis, vesiculation, and vesicle acidity following AGS3 overexpression in other cell types (14). While interpreted as increased autophagy, the accepted gold standard LC3-based assay was then unavailable. Also, the AGS3-induced increases in monodansylcadavarine staining noted previously (14) may have delineated lysosomes rather than autophagosomes (45). In this study when we overexpressed AGS3 as a GFP fusion protein, we found similar LC3-II levels in the AGS3hi and AGS3lo cell lines, irrespective of the cell line type, but decreased p62 protein in the AGS3hi cell lines. Conversely, we found a relative increase in p62 mRNA transcripts in the AGS3hi cells. When we blocked autophagic flux in the THP-1 cell lines using Bafilomycin A1, the LC3-II and p62 levels increased in the AGS3hi cells as compared to the AGS3lo cells. This indicates that the increased level of AGS3 had enhanced p62 degradation via the autophagosomal/lysosomal pathway without altering basal LC3-II levels in the cell. These observations suggested that the elevated AGS3 levels under nutrient replete conditions had not triggered autophagy, but rather had impacted lysosome function and/or biogenesis. By examining cells with elevated or reduced AGS3 levels we found a corresponding increase or decrease of LAMP-1 and other lysosomal markers.
Mouse BMDMs constitutively express AGS3 and following exposure to LPS further raise their expression level. Using immortalized BMDMs from WT, MyD88, and TRIF deficient mice stimulated, or not, with LPS; we showed that the basal level of AGS3 depended upon constitutive MyD88 signaling, and that both TRIF and MyD88 contributed to the increase in AGS3 that followed LPS stimulation. The loss of MyD88 or TRIF also impaired the induction of LAMP-1 expression following LPS stimulation. The combined role of MyD88 and TRIF in the early induction and sustained activity of NF-κB activity TLR4 signaling (46) suggests that AGS3 may be a NF-κB target gene in macrophages. An involvement of NF-κB in the induction of AGS3 is also consistent with the increase in AGS3 noted in lymphocytes following antigen receptor crosslinking (26).
Different physiological and pathological inputs affect the status of the mTOR complex 1, thereby affecting autophagy and controlling TFEB activity (4). Starvation induced autophagy depends upon ULK1 dephosphorylation, a consequence of mTORC1 inactivation and increased activity of the phosphatase PP2A (47). High AGS3 levels in a nutrient replete state reduced signaling through the mTOR pathway reducing TFEB phosphorylation and triggering its nuclear translocation, but evidently did not suppress the pathway sufficiently to trigger ULK1 dephosphorylation and visible increases in autophagosome formation. A well-defined input to the mTOR complex 1 is the phosphatidylinositol 3-Kinase (PI3K)–AKT pathway. In HeLa cells, THP-1 cells, and mouse BMDM we found that increased AGS3 expression led to decreased pAKT, pTSC2, and pS6K1 levels. While many upstream signals affect the PI3K–AKT pathway, a major input are signals generated by G-protein coupled receptors (GPCRs) (48). Ligand activated GPCRs triggers Gα nucleotide exchange and the functional dissociation of GTP-Gα and Gβγ, both of which activate downstream effectors that can lead to activation of the PI3K-AKT pathway. Following nucleotide exchange Gα remains only transiently GTP bound as its intrinsic GTPase activity returns it to its GDP bound state, which can recombine with Gβγ to terminate signaling. However, as AGS3 competes with Gβγ for binding to GDP-Gαi, AGS3 can potentially prolong Gβγ signaling, or conversely, trigger Gβγ degradation as free Gβγ in the absence of Gα is unstable. Consistent with the later scenario, we found a reduction in Gβ1 and Gβ2; decreased CXCR4 expression; and a reduced intracellular calcium response in the THP-1 AGS3hi cells as compared to the THP-1 AGS3lo cells. These results suggest that increasing levels of AGS3 in macrophages reduced the availability of Gi heterotrimers, which limits GPCR signaling through Gi-linked pathways, thereby decreasing an important input into the PI3K–AKT pathway. In contrast, low AGS3 levels likely boost the availability of Gi. Similar to our results, the overexpression of AGS3 in RBL-2H3-CXCR2 cells reduced chemokine induced increases in intracellular calcium (27).
Gpsm1−/− mice have increased energy expenditures, reduced body fat mass, and a lean body habitus (21). A similar deficiency in adipose deposition has been noted in mouse models of lysosomal storage diseases (LSDs) (49). A major function of lysosomes is to recycle macromolecules, reducing the energy intensive de novo synthesis of raw materials. Lysosomal dysfunction causes inefficient energy utilization diverting energy to synthesis pathways at the expense of energy storage depots. The phenotypic similarity of the Gpsm1−/− mice with the LSD model mice suggests a possible link between the metabolic changes and whole organism level lysosome activity in the Gpsm1−/− mice. This possibility warrants further investigation.
Pathogens often hijack the phagocytic and endocytic pathways to enter cells, but they must avoid being degraded in lysosomes (1). To this end, manipulation of lysosomes at the host-pathogen interface offers an alternative strategy to reduce or eliminate intracellular pathogens (3). To test this idea we infected the lysosome-enriched AGS3hi THP-1 macrophages with three different intracellular pathogens and in each instance the AGS3hi cells proved more resistant than the control cells. Most strikingly many of the AGS3hi THP-1 cells eliminated the highly antibiotic resistant B. cenocepecia J2315 likely due to a failure of the bacteria to subvert the endo-lysosomal pathway. The demonstration that the TFEB homolog, HLH-30 is a key transcription factor for host (C. elegans) defense (7) and that exposure of macrophages to S. aureus activates TFEB supports our findings that AGS3-mediated activation in TFEB leads to improved macrophage defense against intracellular bacteria.
While we demonstrated a role for mTOR inhibition in the enhanced lysosomal biogenesis, other AGS3 dependent mechanisms may contribute to this successful control. AGS3-induced changes in the trans-Golgi network could also promote endosome/lysosome enrichment. AGS3 can translocate to the trans-Golgi network (20), which leads to its fragmentation/vesiculation (19). Since the trans-Golgi network serves as a critical hub for the trafficking of resident lysosome proteins, high levels of AGS3 may enhance endosome and/or lysosome formation via this mechanism (32). AGS3 could also promote lysosomal fusion with phagosomes or autophagosomes bearing pathogens. While the factors that prepare autophagosomes or phagosomes for lysosomal fusion remain largely unknown, lysosomal LAMP1/2 expression are needed (50). The increased LAMP1/2 levels noted in the AGS3hi cells could enhance the fusion rate and explain the increased resistance to pathogens that actively try to avoid the fusion of bacterial laden phagosomes with lysosomes.
In conclusion, our study reveals AGS3-mediated lysosomal elevation through the inhibition of AKT/TSC2/mTOR pathway leads to TFEB nuclear translocation. This boosted intracellular resistance of macrophages against B. cenocepacia J2315, M. tuberculosis, and two clinically important strains of MRSA. Further studies with other intracellular pathogens may provide additional insights into the impact of raising AGS3 levels in macrophages. Finally, loss of AGS3 expression or function could contribute to pathological conditions associated with diminished lysosomal function.
Acknowledgments
The authors would like to thank Vee Yang Tang for his technical support and Dr. Anthony Fauci for his long standing support.
Footnotes
Author contributions
A.V. and J.H.K designed and analyzed the experiments; A.V. did the experimental work; S.K did BC J2315 infection experiments; G.C and A.Y did MRSA infection experiments; L.S did the M. tuberculosis infection experiments; T.J.M. did the autophagic proteolysis experiments; J.B., C.S.S, R.W, I.F., M.O, and V.B provided expert advice. A.V. and J.H.K. wrote and edited the manuscript.
Disclosures
The authors declare no competing financial interests.
The intramural program of the National Institute of Allergy and Infectious Diseases supported this research.
References
- 1.Baxt LA, Garza-Mayers AC, Goldberg MB. Bacterial subversion of host innate immune pathways. Science. 2013;340:697–701. doi: 10.1126/science.1235771. [DOI] [PubMed] [Google Scholar]
- 2.Schwegmann A, Brombacher F. Host-directed drug targeting of factors hijacked by pathogens. Science signaling. 2008;1:re8. doi: 10.1126/scisignal.129re8. [DOI] [PubMed] [Google Scholar]
- 3.Lebeis SL, Kalman D. Aligning antimicrobial drug discovery with complex and redundant host-pathogen interactions. Cell host & microbe. 2009;5:114–122. doi: 10.1016/j.chom.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 2013;14:283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneider L, Zhang J. Lysosomal function in macromolecular homeostasis and bioenergetics in Parkinson’s disease. Molecular neurodegeneration. 2010;5:14. doi: 10.1186/1750-1326-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, Banfi S, Parenti G, Cattaneo E, Ballabio A. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 7.Visvikis O, Ihuegbu N, Labed SA, Luhachack LG, Alves AM, Wollenberg AC, Stuart LM, Stormo GD, Irazoqui JE. Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity. 2014;40:896–909. doi: 10.1016/j.immuni.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blumer JB, Oner SS, Lanier SM. Group II activators of G-protein signalling and proteins containing a G-protein regulatory motif. Acta Physiol (Oxf) 2012;204:202–218. doi: 10.1111/j.1748-1716.2011.02327.x. [DOI] [PubMed] [Google Scholar]
- 9.Sanada K, Tsai LH. G protein betagamma subunits and AGS3 control spindle orientation and asymmetric cell fate of cerebral cortical progenitors. Cell. 2005;122:119–131. doi: 10.1016/j.cell.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Bowers MS, McFarland K, Lake RW, Peterson YK, Lapish CC, Gregory ML, Lanier SM, Kalivas PW. Activator of G protein signaling 3: a gatekeeper of cocaine sensitization and drug seeking. Neuron. 2004;42:269–281. doi: 10.1016/s0896-6273(04)00159-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao L, McFarland K, Fan P, Jiang Z, Inoue Y, Diamond I. Activator of G protein signaling 3 regulates opiate activation of protein kinase A signaling and relapse of heroin-seeking behavior. Proc Natl Acad Sci U S A. 2005;102:8746–8751. doi: 10.1073/pnas.0503419102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowers MS, Hopf FW, Chou JK, Guillory AM, Chang SJ, Janak PH, Bonci A, Diamond I. Nucleus accumbens AGS3 expression drives ethanol seeking through G betagamma. Proc Natl Acad Sci U S A. 2008;105:12533–12538. doi: 10.1073/pnas.0706999105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan P, Jiang Z, Diamond I, Yao L. Up-regulation of AGS3 during morphine withdrawal promotes cAMP superactivation via adenylyl cyclase 5 and 7 in rat nucleus accumbens/striatal neurons. Mol Pharmacol. 2009;76:526–533. doi: 10.1124/mol.109.057802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pattingre S, De Vries L, Bauvy C, Chantret I, Cluzeaud F, Ogier-Denis E, Vandewalle A, Codogno P. The G-protein regulator AGS3 controls an early event during macroautophagy in human intestinal HT-29 cells. J Biol Chem. 2003;278:20995–21002. doi: 10.1074/jbc.M300917200. [DOI] [PubMed] [Google Scholar]
- 15.Groves B, Abrahamsen H, Clingan H, Frantz M, Mavor L, Bailey J, Ma D. An inhibitory role of the G-protein regulator AGS3 in mTOR-dependent macroautophagy. PLoS One. 2010;5:e8877. doi: 10.1371/journal.pone.0008877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vural A, Oner S, An N, Simon V, Ma D, Blumer JB, Lanier SM. Distribution of activator of G-protein signaling 3 within the aggresomal pathway: role of specific residues in the tetratricopeptide repeat domain and differential regulation by the AGS3 binding partners Gi(alpha) and mammalian inscuteable. Mol Cell Biol. 2010;30:1528–1540. doi: 10.1128/MCB.01018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Marcos M, Ear J, Farquhar MG, Ghosh P. A GDI (AGS3) and a GEF (GIV) regulate autophagy by balancing G protein activity and growth factor signals. Mol Biol Cell. 2011;22:673–686. doi: 10.1091/mbc.E10-08-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vural A, McQuiston TJ, Blumer JB, Park C, Hwang IY, Williams-Bey Y, Shi CS, Ma DZ, Kehrl JH. Normal Autophagic Activity in Macrophages from Mice Lacking Galphai3, AGS3, or RGS19. PLoS One. 2013;8:e81886. doi: 10.1371/journal.pone.0081886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groves B, Gong Q, Xu Z, Huntsman C, Nguyen C, Li D, Ma D. A specific role of AGS3 in the surface expression of plasma membrane proteins. Proc Natl Acad Sci U S A. 2007;104:18103–18108. doi: 10.1073/pnas.0709282104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oner SS, Vural A, Lanier SM. Translocation of Activator of G-protein Signaling 3 to the Golgi Apparatus in Response to Receptor Activation and Its Effect on the trans-Golgi Network. J Biol Chem. 2013;288:24091–24103. doi: 10.1074/jbc.M112.444505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blumer JB, Lord K, Saunders TL, Pacchioni A, Black C, Lazartigues E, Varner KJ, Gettys TW, Lanier SM. Activator of G protein signaling 3 null mice: I. Unexpected alterations in metabolic and cardiovascular function. Endocrinology. 2008;149:3842–3849. doi: 10.1210/en.2008-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nadella R, Blumer JB, Jia G, Kwon M, Akbulut T, Qian F, Sedlic F, Wakatsuki T, Sweeney WE, Jr, Wilson PD, Lanier SM, Park F. Activator of G protein signaling 3 promotes epithelial cell proliferation in PKD. Journal of the American Society of Nephrology : JASN. 2010;21:1275–1280. doi: 10.1681/ASN.2009121224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Regner KR, Nozu K, Lanier SM, Blumer JB, Avner ED, Sweeney WE, Jr, Park F. Loss of activator of G-protein signaling 3 impairs renal tubular regeneration following acute kidney injury in rodents. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2011;25:1844–1855. doi: 10.1096/fj.10-169797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwon M, Pavlov TS, Nozu K, Rasmussen SA, Ilatovskaya DV, Lerch-Gaggl A, North LM, Kim H, Qian F, Sweeney WE, Jr, Avner ED, Blumer JB, Staruschenko A, Park F. G-protein signaling modulator 1 deficiency accelerates cystic disease in an orthologous mouse model of autosomal dominant polycystic kidney disease. Proc Natl Acad Sci U S A. 2012;109:21462–21467. doi: 10.1073/pnas.1216830110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blumer JB, Lanier SM. Activators of G protein signaling exhibit broad functionality and define a distinct core signaling triad. Mol Pharmacol. 2014;85:388–396. doi: 10.1124/mol.113.090068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Branham-O’Connor M, Robichaux WG, 3rd, Zhang XK, Cho H, Kehrl JH, Lanier SM, Blumer JB. Defective chemokine signal integration in leukocytes lacking activator of G protein signaling 3 (AGS3) J Biol Chem. 2014;289:10738–10747. doi: 10.1074/jbc.M113.515031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh V, Raghuwanshi SK, Smith N, Rivers EJ, Richardson RM. G Protein-coupled receptor kinase-6 interacts with activator of G protein signaling-3 to regulate CXCR2-mediated cellular functions. Journal of immunology. 2014;192:2186–2194. doi: 10.4049/jimmunol.1301875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris J, Hartman M, Roche C, Zeng SG, O’Shea A, Sharp FA, Lambe EM, Creagh EM, Golenbock DT, Tschopp J, Kornfeld H, Fitzgerald KA, Lavelle EC. Autophagy controls IL-1beta secretion by targeting pro-IL-1beta for degradation. J Biol Chem. 2011;286:9587–9597. doi: 10.1074/jbc.M110.202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schnoor M, Buers I, Sietmann A, Brodde MF, Hofnagel O, Robenek H, Lorkowski S. Efficient non-viral transfection of THP-1 cells. J Immunol Methods. 2009;344:109–115. doi: 10.1016/j.jim.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 30.Sajjan SU, Carmody LA, Gonzalez CF, LiPuma JJ. A type IV secretion system contributes to intracellular survival and replication of Burkholderia cenocepacia. Infection and immunity. 2008;76:5447–5455. doi: 10.1128/IAI.00451-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogier-Denis E, Houri JJ, Bauvy C, Codogno P. Guanine nucleotide exchange on heterotrimeric Gi3 protein controls autophagic sequestration in HT-29 cells. J Biol Chem. 1996;271:28593–28600. doi: 10.1074/jbc.271.45.28593. [DOI] [PubMed] [Google Scholar]
- 32.Saftig P, Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol. 2009;10:623–635. doi: 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- 33.Unuma K, Aki T, Funakoshi T, Yoshida K, Uemura K. Cobalt protoporphyrin accelerates TFEB activation and lysosome reformation during LPS-induced septic insults in the rat heart. PLoS One. 2013;8:e56526. doi: 10.1371/journal.pone.0056526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, Sabatini DM, Ballabio A. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. The EMBO journal. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, Manning BD. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell. 2014;156:771–785. doi: 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demetriades C, Doumpas N, Teleman AA. Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell. 2014;156:786–799. doi: 10.1016/j.cell.2014.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 39.Wan Y, Yang Z, Guo J, Zhang Q, Zeng L, Song W, Xiao Y, Zhu X. Misfolded Gbeta is recruited to cytoplasmic dynein by Nudel for efficient clearance. Cell research. 2012;22:1140–1154. doi: 10.1038/cr.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bazzini S, Udine C, Riccardi G. Molecular approaches to pathogenesis study of Burkholderia cenocepacia, an important cystic fibrosis opportunistic bacterium. Appl Microbiol Biotechnol. 2011;92:887–895. doi: 10.1007/s00253-011-3616-5. [DOI] [PubMed] [Google Scholar]
- 41.Al-Khodor S, Marshall-Batty K, Nair V, Ding L, Greenberg DE, Fraser ID. Burkholderia cenocepacia J2315 escapes to the cytosol and actively subverts autophagy in human macrophages. Cellular microbiology. 2014;16:378–395. doi: 10.1111/cmi.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zumla A, George A, Sharma V, Herbert N Baroness Masham of I. WHO’s 2013 global report on tuberculosis: successes, threats, and opportunities. Lancet. 2013;382:1765–1767. doi: 10.1016/S0140-6736(13)62078-4. [DOI] [PubMed] [Google Scholar]
- 43.Uhlemann AC, Otto M, Lowy FD, DeLeo FR. Evolution of community- and healthcare-associated methicillin-resistant Staphylococcus aureus. Infect Genet Evol. 2013;21:563–574. doi: 10.1016/j.meegid.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cole J, Aberdein J, Jubrail J, Dockrell DH. The role of macrophages in the innate immune response to Streptococcus pneumoniae and Staphylococcus aureus: mechanisms and contrasts. Adv Microb Physiol. 2014;65:125–202. doi: 10.1016/bs.ampbs.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 45.Bampton ET, Goemans CG, Niranjan D, Mizushima N, Tolkovsky AM. The dynamics of autophagy visualized in live cells: from autophagosome formation to fusion with endo/lysosomes. Autophagy. 2005;1:23–36. doi: 10.4161/auto.1.1.1495. [DOI] [PubMed] [Google Scholar]
- 46.Cheng Z, Taylor B, Ourthiague DR, Hoffmann A. Distinct single-cell signaling characteristics are conferred by the MyD88 and TRIF pathways during TLR4 activation. Science signaling. 2015;8:ra69. doi: 10.1126/scisignal.aaa5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong PM, Feng Y, Wang J, Shi R, Jiang X. Regulation of autophagy by coordinated action of mTORC1 and protein phosphatase 2A. Nature communications. 2015;6:8048. doi: 10.1038/ncomms9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vadas O, Dbouk HA, Shymanets A, Perisic O, Burke JE, Abi Saab WF, Khalil BD, Harteneck C, Bresnick AR, Nurnberg B, Backer JM, Williams RL. Molecular determinants of PI3Kgamma-mediated activation downstream of G-protein-coupled receptors (GPCRs) Proc Natl Acad Sci U S A. 2013;110:18862–18867. doi: 10.1073/pnas.1304801110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woloszynek JC, Coleman T, Semenkovich CF, Sands MS. Lysosomal dysfunction results in altered energy balance. J Biol Chem. 2007;282:35765–35771. doi: 10.1074/jbc.M705124200. [DOI] [PubMed] [Google Scholar]
- 50.Huynh KK, Eskelinen EL, Scott CC, Malevanets A, Saftig P, Grinstein S. LAMP proteins are required for fusion of lysosomes with phagosomes. The EMBO journal. 2007;26:313–324. doi: 10.1038/sj.emboj.7601511. [DOI] [PMC free article] [PubMed] [Google Scholar]