Abstract
Label-free peptide quantification in LC-MS proteomics analyses is complicated by the presence of isobaric co-eluting peptides, as they generate the same extracted ion chromatogram corresponding to the sum of their intensities. Histone proteins are especially prone to this, as they are heavily modified by post-translational modifications (PTMs). Their proteolytic digestion leads to a large number of peptides sharing the same mass, while carrying PTMs on different amino acid residues. We present an application of MS data-independent acquisition (DIA) to confidently determine and quantify modified histone peptides. By introducing the use of low resolution MS/MS DIA, we demonstrate that the signals of 111 histone peptides could easily be extracted from LC-MS runs due to the relatively low sample complexity. By exploiting an LTQ-Orbitrap mass spectrometer, we parallelized MS and MS/MS scan events using the Orbitrap and the linear ion trap respectively, decreasing the total scan time. This, in combination with large windows for MS/MS fragmentation (50 m/z) and multiple full scan events within a DIA duty cycle, led to a MS scan cycle speed of ~45 full MS per minute, improving the definition of extracted LC-MS chromatogram profiles. By using such acquisition method, we achieved highly comparable results to our optimized acquisition method for histone peptide analysis (R2 correlation >0.98), which combines data-dependent acquisition (DDA) and targeted MS/MS scans, the latter targeting isobaric peptides. By using DIA, we could also re-mine our dataset and quantify 16 additional isobaric peptides commonly not targeted during DDA experiments. Finally, we demonstrated that by performing the full MS scan in the linear ion trap, we achieve highly comparable results as when adopting high resolution MS scans (R2 correlation 0.97). Taken together, results confirmed that histone peptide analysis can be performed using DIA and low resolution MS with high accuracy and precision of peptide quantification. Moreover, DIA intrinsically enables data re-mining to later identify and quantify isobaric peptides unknown at the time of the LC-MS experiment. These methods will open up epigenetics analyses to the proteomics community who do not have routine access to the newer generation high-resolution MS/MS generating instruments.
Keywords: data independent acquisition, histones, mass spectrometry, post-translational modifications, quantification
Graphical Abstract
Introduction
In eukaryotes, DNA is organized into protein-DNA complexes called nucleosomes. Nucleosomes are composed of 147 base pairs of DNA wrapped around a histone octamer containing two copies of each core histone protein-H2A, H2B, H3, and H4.1 Due to their intimate association with DNA, histone proteins play critical roles in many nuclear processes, including transcription, DNA damage repair, and heterochromatin formation. Each histone within the nucleosome has a globular core domain and a flexible N-terminal tail domain that protrudes from the nucleosomal surface.1 Histone proteins are extensively and dynamically post-translationally modified by nuclear proteins, mostly on the tail domains. These post-translational modifications (PTMs) are thought to comprise a “histone code” where each specific combinatorial PTM profile of a histone dictates its specific function, such as activating transcription.2 Aberrant regulations of PTM profiles have been linked to a wide array of diseases, including cancer, diabetes, and aging. Deciphering the histone code is therefore critical to understanding fundamental nuclear processes and how these are misregulated in disease.
Traditional methods to study histone PTMs rely on antibody-based methods such as western blots and immunoprecipitations. There are several drawbacks associated with the use of antibodies, including epitope occlusion from nearby PTMs, cross-reactivity, and inability to detect novel PTMs and PTM combinations.3 Mass spectrometry (MS)-based methods have therefore become the primary method to characterize histone PTM profiles as they allow for robust and unbiased quantitative analysis. Most MS methods employ data dependent acquisition (DDA), in which a defined number of precursor ions from the full scan are selected for fragmentation based on predetermined settings like intensity and charge state. Quantification is normally performed at the full MS level by integrating the area under the chromatographic peak in the extracted ion chromatogram.4 One drawback of DDA methodology is that the acquisition is somewhat stochastic in nature because precursor selection is dependent on elution time, presence of other abundant ions, and dynamic exclusion lists. Therefore, there can be a large degree of variability between runs as different precursors are selected for fragmentation, resulting in poor reproducibility compared to other MS methods.5 Moreover, this approach cannot accurately quantify co-eluting isobaric peptides because this quantification must be done based on their unique fragmentation ions at the MS/MS level.
To overcome the issue of quantifying isobaric peptides, targeted proteomic strategies are often employed. The most common method is including targeted MS/MS scans, where a given precursor or list of precursors are targeted for fragmentation followed by detection of all product ions. This targeting occurs every duty cycle to ensure accurate quantification across the chromatographic range. However, targeted scans can only be applied to known PTMs and the targeted nature of method precludes future data mining.6 Currently, the most common approach in case of histone analysis is to combine DDA and targeted scans into a single method7, where isobaric and co-eluting peptides are analyzed by targeted scans but other signals are subjected to DDA methodology. However, this combined approach requires previous knowledge about chromatography elution profiles, as targeting a long list of masses all over the MS run might lead to slow duty cycle.7 Furthermore, novel isobaric peptides might be discovered after the analysis, making it necessary to repeat the acquisition with a new mass inclusion list.
Data independent acquisition (DIA) methodology is an alternative option for proteomics analysis. In DIA, a full MS spectrum is acquired followed by a series of sequential MS/MS scans covering a relatively large m/z window (usually 5–25 m/z) that step across the desired m/z range. The idea behind generating m/z windows for MS/MS fragmentation is related to the fact that, on the contrary to fragmenting the entire range, this approach reduces MS/MS complexity and increases sensitivity.8 The Yates group was the first to highlight the strength of this strategy, using this methodology to measure differences in protein expression in two different stages of development in Caenorhabditis elegans.9 After a DIA run, the extracted ion chromatogram is then generated in silico using software tools, some of them freely available like Skyline10, OpenSWATH11 and EpiProfile12. Each of these tools relies on a spectral library of identified peptides that can be downloaded, manually programmed, or built from previous DDA experiments. Over the past decade, DIA methodology to study protein expression and modification has been increasing in popularity.13–17
Sciex was the first to automate this type of MS data acquisition by offering a DIA method called SWATH-MS® for their TripleTOF® high-resolution mass spectrometer. We previously demonstrated the power of SWATH-MS to quantify and identify histone samples, which contain a large number of co-eluting isobaric and highly modified peptides.17 We were able to identify 41 PTM profiles on H3, including discrimination of isobaric peptides. Quantification, performed at the MS/MS level, proved to be highly reproducible. Afterwards, Krautkramer et al. performed a DIA method on a Q-Exactive (Thermo) to quantify changes histone PTM profiles in breast cancer cells upon treatment with the deacetylase inhibitor SAHA.14
In this study we demonstrate that histone samples can be analyzed using DIA with low resolution MS/MS or both low resolution MS and MS/MS, using an LTQ-Orbitrap mass spectrometer (Thermo). For this purpose, we used histones extracted from mouse embryonic stem cells, and prepared the histones using propionic anhydride derivatization combined with trypsin digestion.7 The quantification results were highly comparable with our state-of-the-art acquisition method for histone peptide analysis, which combines DDA and targeted MS/MS scans. Moreover, we show that the new DIA method is less dependent on reproducible chromatography compared to our hybrid DDA/targeting mode because DIA does not require targeting of specific precursors at the time of their elution. Finally, DIA can more easily identify isobaric peptides containing different modification profiles, exemplified by re-mining data to quantify new co-eluting histone PTM isomers. Collectively, we show that DIA methods to analyze histone peptides do not require high resolution, and can be performed on previous generation Orbitraps. We also demonstrate that DIA is a much more convenient approach than the most widely adopted methods that combine DDA and targeted scans.
Materials and methods
Histone extraction and digestion
Mouse embryonic stem cells were grown using standard media and harvested as previously described.18 Nuclei were isolated and histone proteins were extracted as described in the protocol of Lin and Garcia with minor adjustments.7 Briefly, histones were acid extracted from nuclei with 0.2 M H2SO4 for 2 hours and precipitated with 33% trichloroacetic acid (TCA) overnight. Protein concentration was calculated using the Bradford assay. The derivatization and digestion were performed as previously described.7 Briefly, histones were dissolved in 30 μL of 50 mM NH4HCO3, pH 8.0. Derivatization reagent was prepared by mixing propionic anhydride with acetonitrile in a ratio of 1:3 (v/v), and such reagent was mixed with the histone sample in the ratio of 1:4 (v/v) for 20 minutes at room temperature. This reaction was performed twice. Histones were then digested with trypsin (enzyme:sample ratio 1:20, 6 hours, room temperature) in 50 mM NH4HCO3. After digestion, the derivatization reaction was performed again twice to derivatize peptide N-termini. Samples were desalted prior LC-MS analysis by using C18 Stage-tips.19
NanoLC–MS/MS
Samples were analyzed by using a nanoLC-MS/MS setup. NanoLC was configured with a 75 µm ID × 17 cm Reprosil-Pur C18-AQ (3 µm; Dr. Maisch GmbH, Germany) nano-column using an EASY-nLC nanoHPLC (Thermo Scientific, Odense, Denmark), packed in-house. The HPLC gradient was as follows: 2% to 28% solvent B (A = 0.1% formic acid; B = 95% MeCN, 0.1% formic acid) over 45 minutes, from 28% to 80% solvent B in 5 minutes, 80% B for 10 minutes at a flow-rate of 300 nL/min. nLC was coupled to an LTQ-Orbitrap Elite mass spectrometer (Thermo Scientific, Bremen, Germany). For DIA, a full scan MS spectrum (m/z 300–1100) was acquired in the Orbitrap with a resolution of 120,000 (at 200 m/z) and an AGC target of 5×10e5, or in the ion trap with an AGC target of 3×10e4. MS/MS was performed with an AGC target of 3×10e4 using an injection time limit of 30 or 60 msec. For DDA, dependent and targeted scans were performed. MS/MS was acquired using collision induced dissociation (CID) with normalized collision energy of 35. For DDA, MS acquisition was divided into three segments, each beginning with a full MS scan: (i) MS/MS of the top seven most abundant ions (14 min), (ii) targeted CID fragmentation of common isobaric species (H3 peptide aa 9–17 with 1 acetyl, H3 peptide aa 18–26 with 1 acetyl and histone H4 peptide aa 4–17 with 1/2/3 acetyl groups) followed by CID fragmentation of the top five most abundant ions (27 min), (iii) CID fragmentation of the top ten most abundant ions (19 min). MS/MS data were collected in centroid mode. All raw files are available on https://chorusproject.org at the project no. 923.
Data Analysis
For DIA analysis, the spectral library was manually generated using Skyline10, considering the peptides commonly detected during a traditional histone peptide analysis (as described in 12). Skyline peak extraction was optimized manually based on previous knowledge about peptide retention time. The template for peak extraction is available upon request. The peptide relative ratio was calculated using the total area under the extracted ion chromatograms of all peptides with the same aa sequence (including all of its modified forms) as 100%. For isobaric peptides, the relative ratio of two isobaric forms was estimated by averaging the ratio for each fragment ion with different mass between the two species. Statistical reproducibility was assessed by estimating the coefficient of variation and by linear regression. EpiProfile12 was used to retrieve the relative ratio of the isobaric forms for the two following peptides: aa 4–17 of histone H4 with 1 acetyl group and the same peptide with 3 acetyl groups. This was performed because EpiProfile is equipped with the script to estimate the relative ratio of peptides with more than two isobaric forms. The relative ratio value was used uniquely to divide the precursor area, extracted using Skyline, between the different isobaric forms.
Results and discussion
Histones extracted from mouse embryonic stem cells were used as a standard to compare sensitivity and precision of DDA and DIA acquisition methods in the analysis of isobaric and non-isobaric peptides. In order to minimize the variability between experiments, all runs were performed within two days with the same nLC-MS setup. The same sample was continuously injected from the same vial to avoid variability induced by sample preparation or pipetting. The DDA run was performed in triplicate, and these runs were used as a reference for peptide quantification in the DIA runs. The method was divided into three segments: (i) the first contained only DDA scans, spanning the first 27 minutes of the run; (ii) the second segment (20 minutes long) contained DDA scans and targeted scans for isobaric histone H3 and H4 peptides: H3 peptide aa 9–17 with 1 acetyl, H3 peptide aa 18–26 with 1 acetyl and histone H4 peptide aa 4–17 with 1/2/3 acetyl groups; (iii) the third segment contained only DDA scans. This method has been applied for several publications (e.g. 20–22), as it was the best option to perform the largest number of DDA events while still targeting co-eluting isobaric peptides to determine their relative abundances.
Duty cycle of data-independent acquisition methods
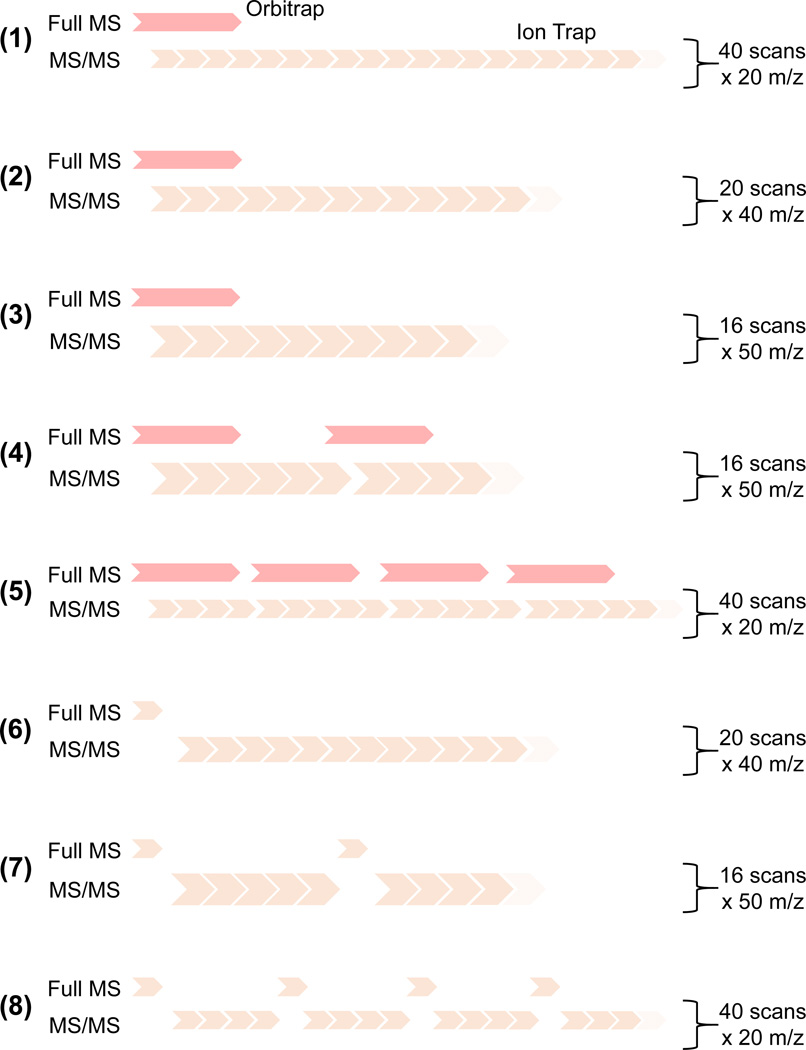
We tested eight different DIA methods (Fig. 1) to determine the optimal strategy for peptide quantification. All MS/MS scans were completed at low resolution in the ion trap. Each method was tested by performing three technical replicates of the same sample. We tested three different window sizes for MS/MS fragmentation: 20, 40, and 50 m/z, resulting in 40, 20, and 16 MS/MS scans per duty cycle, respectively. One potential drawback in DIA methodology is that collecting a large number of MS/MS acquisitions greatly increases the duty cycle compared to DDA, reducing the number of full MS scans collected. Collecting fewer full scans results in a less defined chromatographic peak, which can then reduce the accuracy of quantification when performed at the full MS level. To alleviate this potential problem, we designed four of the eight DIA methods to perform multiple full MS scans within a single duty cycle to improve the definition of the extracted ion chromatograms.
Figure 1. Data-independent acquisition (DIA) method experimental layouts.
We evaluated eight different DIA methods in which we varied the MS/MS fragmentation window size (from 20 to 50 m/z), the number of full MS scan events per cycle (from 1 to 4), and the choice of the mass analyzer for the full MS scan (Orbitrap or the ion trap). The scan event in the Orbitrap mass analyzer is represented as red arrows, while the brown boxes represent the ion trap scans. The alignment between full scan and MS/MS events indicates whether the two scans are performed in parallel. The size of the boxes is roughly proportional to the scan time required for the scan.
We also wanted to test whether DIA methodology performed using exclusively low-resolution mass analyzers guarantees results that are comparable to DIA experiments performed on high-resolution instruments. Low resolution instruments tend to be more ubiquitous, cost effective, and easier to maintain and can therefore be more convenient for use. To test this, we designed three methods that collect the full MS scans in the ion trap rather than in the Orbitrap (Fig. 1, methods 6–8). Moreover, we aimed to compare how changing the number and type of full MS scan events affects the length of the duty cycle and shape of the extracted ion chromatograms for precursor ions because these factors are critical for robust quantification. Positive results would confirm previous data obtained with DDA experiments22 and demonstrate that histone peptide analysis can be performed with cost-effective MS instrumentation.
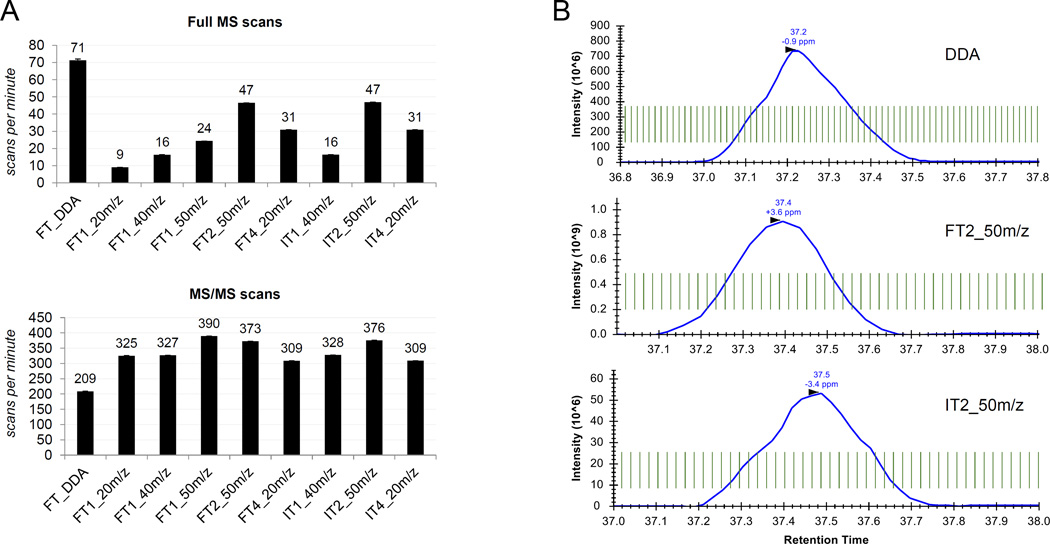
By analyzing the number of full MS and MS/MS scan events using RawMeat (Thermo), we observed that DDA methods have a much faster duty cycle than any of the DIA methods we tested (Fig. 2A). Specifically, the frequency of full MS scans was more than one per second, while the fastest DIA method tested collected a full MS every 1.28 seconds. However, all of the DIA methods obtained a much larger number of MS/MS events, up to almost double that of DDA. These results were expected because DIA methods perform a fixed number of MS/MS per cycle, which is always larger than the number of data-dependent scans. By introducing multiple full MS scans per cycle, we increased the frequency of full MS events. For instance, by using two full MS scans instead of one in the method with 50 m/z MS/MS windows, we increased the full MS frequency from 24 to 47 scans per minute. Furthermore, by introducing four full MS scans instead of one in the method with 20 m/z windows, we increased the full MS scans collected per minute from 9 to 31. In both cases, the number of MS/MS events changed minimally, indicating that the definition of the MS/MS chromatogram would remain mostly unaltered. Based on these results, we selected FT2_50m/z as the optimal method, which collects two full MS scans and uses 50 m/z windows, as it provided the highest frequency of full MS events without dramatically reducing the MS/MS frequency.
Figure 2. Scan frequency of the tested acquisition methods.
(A) Number of full MS scans (top) and MS/MS scans (bottom) per minute of the nine tested acquisition methods. The error bars represent standard deviation between three technical replicates. (B) Representation of the average full MS scan frequency in three selected acquisition methods. The green parallel lines represent the full MS scan event timing, while the blue line is the extracted ion chromatogram of the histone H3 peptide KQLATKAAR (aa 18–26) with 1 acetyl group.
Last, we compared the duty cycle time when performing the full MS scan in the Orbitrap or the ion trap. The ion trap is a much faster mass analyzer than the Orbitrap, especially when higher resolution scans are selected in the Orbitrap. On the other hand, scans collected in the Orbitrap can be parallelized to the MS/MS events in the ion trap, allowing MS/MS scans to be obtained simultaneously. Thus, the long scan time has a minimal effect on the instrument duty cycle, and the ion injection time is the only factor lengthening the duty cycle. We set a relatively high resolution for the full MS scan in the Orbitrap (120,000 at 200 m/z), leading to a scan time of about 0.54 sec. The same full MS scan in the ion trap elapsed 0.09 seconds. However, the frequency of MS and MS/MS scans was highly comparable between methods using the Orbitrap or the ion trap (Fig. 2A and 2B), demonstrating that the full MS acquisition had a similar effect on the duty cycle speed. Collectively, FT2_50m/z and IT2_50m/z methods achieved the best definition of MS and MS/MS chromatograms overall, both of which use two full MS scans and 50 m/z windows.
Characterization of isobaric peptides
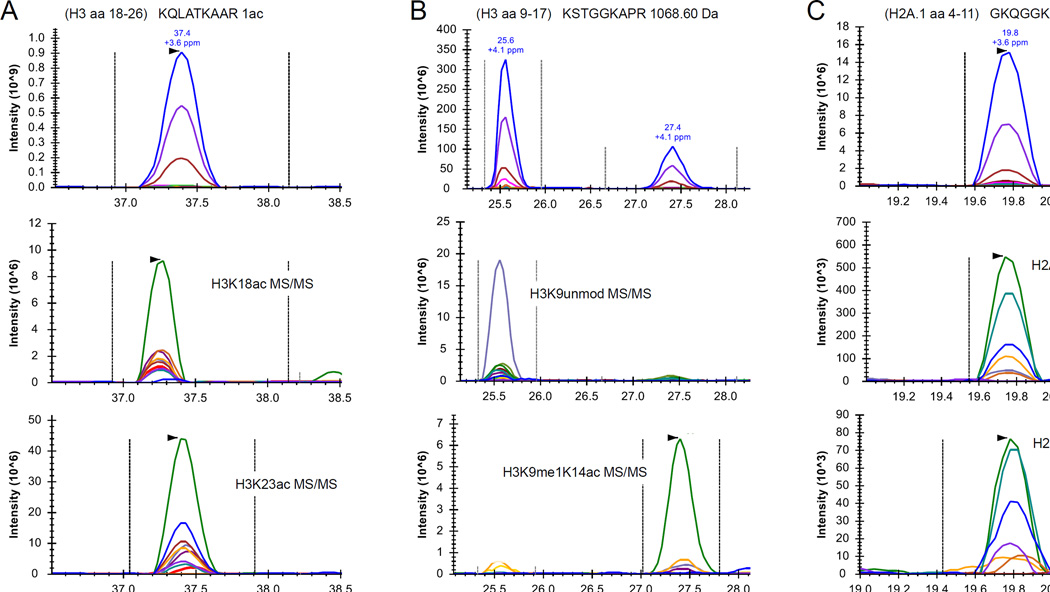
We evaluated the performances of the selected acquisition methods by testing the accurate LC-MS peak integration and quantification of 111 peptides (Table S1). Of these, 44 had isobaric forms, which we discriminated using the relative intensity of their fragment ions. For instance, the H3 peptide KQLATKAAR (aa 18–26) with 1 acetyl group can be present in two isobaric proteoforms, H3K18ac and H3K23ac (Fig. 3A). H3K18ac elutes slightly earlier than H3K23ac, but with traditional C18 chromatography (18 cm column, 3 µm particles), the two species cannot be baseline separated. The fragment ions spanning from b1 to b5 and from y4 to y8 have different masses between the two species. Thus, by integrating these MS/MS chromatograms, we could differentially quantify the two isobaric peptides.
Figure 3. Extracted ion chromatograms of isobaric peptides.
(A) Extracted ion chromatogram of the histone H3 peptide aa 18–26 with 1 acetyl group. The top panel displays the chromatogram of the monoisotopic precursor mass (in blue) and its respective first and second isotope (violet and brown, respectively). The panels below display the extracted ion chromatogram of the unique fragment ions for the peptide H3K18ac (middle) and H3K23ac (bottom). The figure demonstrates that the two peptides co-elute, but they can be distinguished based on their MS/MS chromatogram. (B) Extracted LC-MS chromatogram of the unmodified histone H3 peptide aa 9–17, which has the same mass as K9me1K14ac. The two signals present at the full MS scan level (top) can be discriminated by extracting their unique fragment ions (middle and bottom). (C) Ion chromatogram of the histone H2A peptide aa 4–11 with 1 acetyl group, which includes the two modified forms K5ac (middle) and K9ac (bottom).
Peptides with different types of PTMs may also have the same intact mass. For instance, the two H3 peptides KSTGGKAPR (aa 9–17) unmodified and K9me1K14ac have the same intact mass (1068.60 Da). This is because unmodified and monomethylated lysine residues are propionylated (+56.027 and +70.038 Da, respectively) with the presented histone derivatization protocol. The two peptides have the same mass because a propionyl group has the same mass of an acetylation (42.011 Da) plus a methylation (14.016 Da). However, the two peptides have different retention time due to their different modification states (Fig. 3B). However, without MS/MS identification, it is virtually impossible to define which LC-MS signal corresponds to which peptide. DIA allows for such discrimination without performing MS/MS database searching, as the unique fragment ions of the two species generate two different MS/MS chromatograms.
Finally, DIA methods, contrary to targeted methods, allow for data re-mining after the LC-MS run. As example, we re-mined our dataset searching for histone H2A variant peptides, which are commonly not considered in conventional acquisition methods that include DDA and targeted scans for peptides of histones H3 and H4. We characterized 16 more peptides from histone H2A variants with isobaric forms that could each be differentially quantified using DIA. These peptides include aa 4–11 of canonical histone H2A (Fig. 3C), H2A.J and H2A.X, and aa 12–17 of canonical histone H2A in different modified forms. We searched the ion chromatogram for other acetylated and methylated peptides from histone H2B and H1, but we could not find clear LC-MS chromatograms for those species (not shown). Taken together, DIA proved to be an effective acquisition method to confidently map peptides in the LC-MS chromatogram and discriminate isobaric species without generating any targeted mass list.
Relative quantification of histone peptides using different acquisition methods
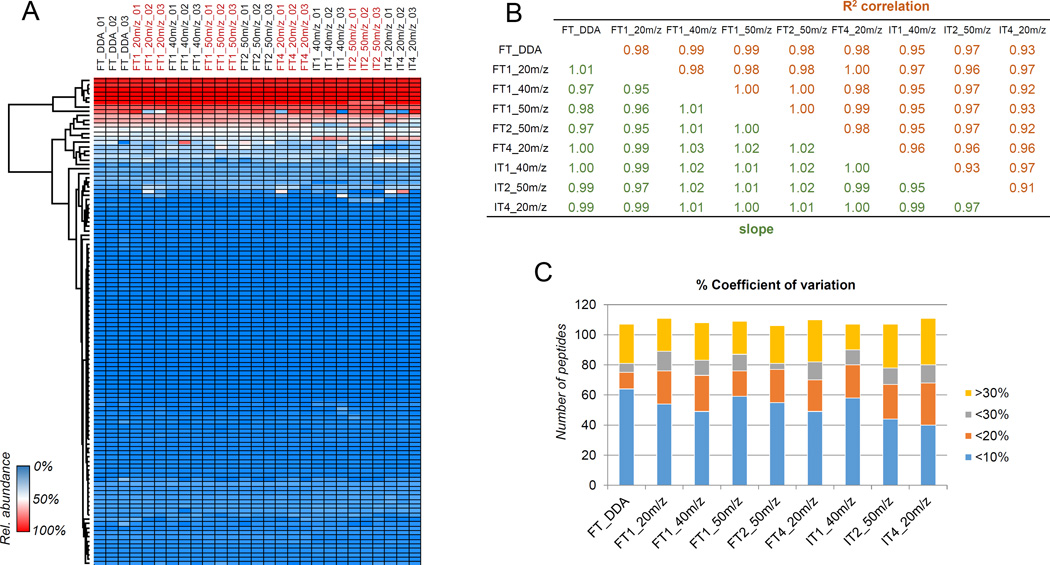
The final goal of this project was to ensure that DIA methods achieve the same results as our state-of-the-art DDA methodology.7 With the ability to re-mine data and eliminating the need for targeting, DIA is a more convenient method than DDA. To determine the variability between methods in terms of quantification, we compared the 27 LC-MS runs performed using DDA and the eight different DIA methods we tested (24 runs total) (Fig. 1). By comparing the relative abundances of the 111 quantified peptides, we observed high similarity between all the performed runs (Fig. 4A). Interestingly, variability was more pronounced between replicates rather than between methods. To determine if the methods obtained similar peptide quantifications, we averaged the three technical replicates for each acquisition method and determined the degree of correlation between each analysis (Fig. 4B). The results show that each comparison achieves a near perfect R2 linear correlation and slope between all methods, indicating that each method provided nearly identical results. Overall, the method that showed the most variability compared to the others was the IT4_20m/z, with R2 correlation values down to 0.91 with the other runs. This was probably due to the fact that this acquisition method has the lowest frequency of MS/MS scan events (Fig. 2A), leading to less defined LC-MS/MS chromatograms. As well, the method IT4_20m/z had the highest variability between runs (Fig. 4C), confirming that this DIA method is less precise than the other methods tested here.
Figure 4. Relative quantification of histone peptides and correlation between experimental methods.
(A) Heatmap representing 111 histone peptides in all LC-MS runs. Color scale represents the relative abundance estimated for each given peptide by each individual experiment. (B) R2 correlation (top) and slope (bottom) from the linear regression between all experiments, after averaging the quantifications from the technical replicates. (C) Histogram of the coefficient of variation estimated for all peptides in all performed runs. The y-axis represents the number of peptides within the specified range.
As previously discussed, the DIA method FT2_50m/z allowed for the highest scan frequency between full MS and MS/MS events (Fig. 2). By comparing the peptide quantification achieved using this method with the DDA approach, we observed high R2 correlation (0.98) and slope (0.97), demonstrating that results are highly comparable to our state-of-the-art acquisition method. Moreover, FT2_50m/z was among the methods with the lowest coefficient of variation between replicates (Fig. 4C), demonstrating high reproducibility. Similar results were obtained with the same method performed in the ion trap (IT2_50m/z). This method achieved an R2 correlation of 0.97 with both DDA and FT2_50m/z, with a nearly perfect slope of 0.99 and 1.02 with DDA and FT2_50m/z, respectively. These results demonstrate that DIA methodology, in both low- and high-resolution, is highly comparable to standard DDA methods. Collectively, we propose that a large m/z window for MS/MS fragmentation combined with two full MS scans within the duty cycle is a highly suitable DIA method to replace DDA in histone peptide analysis.
Conclusions
We demonstrated that low-resolution DIA methods can be used to replace DDA for the quantification of histone PTMs. In current literature, DIA is employed nearly exclusively in high-resolution mass spectrometers, as high resolution can increase confidence of fragment ion identifications in complex MS/MS scans. In addition, high speed instrumentation is required, as there are often a much larger number of MS/MS scans in the duty cycle for DIA methods than for DDA methods. It is important to specify that for this experiment we used an Orbitrap Elite mass spectrometer (Thermo Scientific). This instrument offers a good compromise between MS resolution and speed (60,000 FWHM at 4 Hz). Other types of mass spectrometers might perform optimally with different MS/MS scan numbers and m/z window sizes. The aim of our work was to determine the optimal DIA methodology for ion trap or hybrid instruments, with a particular focus on histone peptides that necessitate MS/MS-based quantification due to their isobaric species. This methodology could be applied to analysis of other highly modified proteins. The Skyline template files, including peptide specifics and obtained results, are available upon request. It is important for users aiming to adopt these templates to perform the experiment using the same chromatographic setup, in order to obtain the most reproducible retention time with our dataset, which facilitates peak alignment using Skyline. Moreover, we show that MS/MS-based extracted ion chromatography enabled more confident mapping of the correct ion signal in the LC-MS chromatogram. Finally, the intrinsic advantage of using DIA is that, contrary to standard DDA methods, data can be re-mined after the nLC-MS experiment. The datasets used in this study, including our raw files, are available in the Chorus repository (project no. 923) and can be fully re-mined for further characterization of other analyte species.
Supplementary Material
Acknowledgments
We gratefully acknowledge funding from NIH grants DP2OD007447, R01GM110174 and R01AI118891, and a National Science Foundation Early Faculty Career award.
Footnotes
Supporting information
Additional information includes Table S1. This material is available free of charge via the Internet at http://pubs.acs.org.
Caption Table S1: estimated relative abundance of modified peptides in all LC-MS runs.
References
- 1.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Britton L-MP, Gonzales-Cope M, Zee BM, Garcia BA. Expert Rev. Proteomics. 2011;8(5):631–643. doi: 10.1586/epr.11.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aebersold R, Mann M. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 5.Bern M, Finney G, Hoopmann MR, Merrihew G, Toth MJ, MacCoss MJ. Anal. Chem. 2010;82(3):833–841. doi: 10.1021/ac901801b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Picotti P, Aebersold R. Nat. Methods. 2012;9(6):555–566. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- 7.Lin S, Garcia BA. Methods in Enzymology. In: Wu Carl, Allis C David., editors. Nucleosomes, Histones & Chromatin Part A. Vol. 512. Academic Press; 2012. pp. 3–28. [DOI] [PubMed] [Google Scholar]
- 8.Gillet LC, Navarro P, Tate S, Röst H, Selevsek N, Reiter L, Bonner R, Aebersold R. Mol. Cell. Proteomics. 2012;11(6):O111.016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venable JD, Dong M-Q, Wohlschlegel J, Dillin A, Yates JR. Nat. Methods. 2004;1(1):39–45. doi: 10.1038/nmeth705. [DOI] [PubMed] [Google Scholar]
- 10.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Bioinformatics. 2010;26(7):966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Röst HL, Rosenberger G, Navarro P, Gillet L, Miladinović SM, Schubert OT, Wolski W, Collins BC, Malmström J, Malmström L, Aebersold R. Nat. Biotechnol. 2014;32(3):219–223. doi: 10.1038/nbt.2841. [DOI] [PubMed] [Google Scholar]
- 12.Yuan Z-F, Lin S, Molden RC, Cao X-J, Bhanu NV, Wang X, Sidoli S, Liu S, Garcia BA. Mol. Cell. Proteomics MCP. 2015;14(6):1696–1707. doi: 10.1074/mcp.M114.046011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porter CJ, Bereman MS. Anal. Bioanal. Chem. 2015:1–9. doi: 10.1007/s00216-015-8819-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krautkramer KA, Reiter L, Denu JM, Dowell JA. J. Proteome Res. 2015 doi: 10.1021/acs.jproteome.5b00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egertson JD, MacLean B, Johnson R, Xuan Y, MacCoss MJ. Nat. Protoc. 2015;10(6):887–903. doi: 10.1038/nprot.2015.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Croft NP, Verteuil DA, de Smith SA, Wong YC, Schittenhelm RB, Tscharke DC, Purcell AW. Mol. Cell. Proteomics. 2015;14(5):1361–1372. doi: 10.1074/mcp.M114.047373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sidoli S, Lin S, Xiong L, Bhanu NV, Karch KR, Johansen E, Hunter C, Mollah S, Garcia BA. Mol. Cell. Proteomics. 2015 doi: 10.1074/mcp.O114.046102. mcp.O114.046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas CE, Kelleher NL, Mizzen CA. J. Proteome Res. 2006;5(2):240–247. doi: 10.1021/pr050266a. [DOI] [PubMed] [Google Scholar]
- 19.Rappsilber J, Ishihama Y, Mann M. Anal. Chem. 2003;75(3):663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 20.Lin S, Wein S, Gonzales-Cope M, Otte GL, Yuan Z-F, Afjehi-sadat L, Maile T, Berger SL, Rush J, Lill JR, Arnott D, Garcia BA. Mol. Cell. Proteomics. 2014 doi: 10.1074/mcp.O113.036459. mcp.O113.036459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sidoli S, Yuan Z-F, Lin S, Karch K, Wang X, Bhanu N, Arnaudo AM, Britton L-M, Cao X-J, Gonzales-Cope M, Han Y, Liu S, Molden RC, Wein S, Afjehi-Sadat L, Garcia BA. Proteomics. 2015 doi: 10.1002/pmic.201400483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karch KR, Zee BM, Garcia BA. J. Proteome Res. 2014 doi: 10.1021/pr500902f. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.